Abstract
Oxidative stress has been proposed to be one of the main causes of aging and has been implicated in the pathogenesis of many diseases. Sensitivity to oxidative stress can be measured by quantifying survival following exposure to a reactive oxygen species (ROS)-generating compound such as paraquat or juglone. Sensitivity to oxidative stress is a balance between basal levels of ROS, the ability to detoxify ROS, and the ability to repair ROS-mediated damage.
Keywords: Oxidative stress, C. elegans, Paraquat, Juglone, Stress resistance, Reactive oxygen species
Background
A number of approaches have been used to test sensitivity to oxidative stress in Caenorhabditis elegans including exposure to paraquat, juglone, t-BOOH, arsenite, H2O2, or hyperbaric oxygen( Keith et al., 2014 ). All of these assays serve to increase the levels of ROS in the worm to a point where survival is decreased. The assays differ in the primary type of ROS that the worm is exposed to (e.g., superoxide, hydrogen peroxide), the rate of exposure (acute versus chronic) and the subcellular compartment believed to be most affected (e.g., paraquat increases superoxide levels primarily in the mitochondria ( Castello et al., 2007 ). Based on these differences, it is possible that a particular strain of worm exhibits increased sensitivity or increased resistance to oxidative stress in one assay, but does not show a difference in another assay. It is also possible that a strain of worms is sensitive to oxidative stress at one age, but resistant to that same form of oxidative stress at a different age. Thus, to obtain a full understanding of sensitivity to oxidative stress in a particular strain it is necessary to use multiple assays at different time points.
Materials and Reagents
Petri dishes 60 x 15 mm 500/cs (Thermo Fisher Scientific, Fisher Scientific, catalog number: FB0875713A)
Petri dishes 35 x 10 mm 500/cs (Thermo Fisher Scientific, Fisher Scientific, catalog number: FB0875711YZ)
Autoclave tape
Aluminum foil
99.95% Platinum, 0.05% Iridium Wire (3 ft/pk) (Tritech Research, catalog number: PT-9901)
OP50 E. Coli bacteria (University of Minnesota, C. elegans Genetics Center, N/A)
Experimental and control C. elegans strains (University of Minnesota, C. elegans Genetics Center, N/A)
Eggs from experimental and control C. elegans strains
Methyl viologen dichloride hydrate (paraquat) (Sigma-Aldrich, catalog number: 856177)
FUdR (5-fluoro-2’-deoxyuridine) (Sigma-Aldrich, catalog number: F0503)
5-hydroxy-1,4-naphthoquinone (juglone) (Sigma-Aldrich, catalog number: H47003)
100% ethanol
Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: S7653)
Bacto peptone (BD, catalog number: 211677)
Agar (Sigma-Aldrich, catalog number: A1296)
Cholesterol (Sigma-Aldrich, catalog number: C8667)
Magnesium sulfate heptahydrate (MgSO4.7H2O) (Sigma-Aldrich, catalog number: M1880)
Calcium chloride dihydrate (CaCl2.2H2O) (Sigma-Aldrich, catalog number: C3881)
Potassium phosphate dibasic (K2HPO4) (Sigma-Aldrich, catalog number: P2222)
Potassium phosphate monobasic (KH2PO4) (Sigma-Aldrich, catalog number: P5655)
Tryptone (Sigma-Aldrich, catalog number: T7293)
Yeast extract (Sigma-Aldrich, catalog number: 70161)
0.5% cholesterol (see Recipes)
1 M MgSO4 (see Recipes)
1 M CaCl2 (see Recipes)
KPI (see Recipes)
Nematode growth medium (NGM) (see Recipes)
1 M paraquat (see Recipes)
3.33 M paraquat (see Recipes)
0.1 M FUdR stock solution (see Recipes)
Juglone stock solution (12 mM) (see Recipes)
2 YT medium (see Recipes)
Equipment
Erlenmeyer flask (Thermo Fisher Scientific, Fisher Scientific, catalog number: FB5006000)
M50 stereomicroscope (Leica, model: 10450154)
Pipetor (Gilson, catalog number: F167300)
Autoclave (Tuttnauer, model: 6690)
Stirring hotplate (Corning, catalog number: 6795-620)
Centrifuge (Eppendorf, model: 5430)
Refrigerated incubator (Thermo Fisher Scientific, Thermo scientificTM, model: 51028064; 37-20)
Bunsen burner (Humbolt, catalog number: H5870)
Software
GraphPad Prism software (we use Version 5.01)
Procedure
-
Paraquat development assay - sensitivity to chronic oxidative stress during development
Prepare NGM media (see Recipes) in Erlenmeyer flask (Note 1).
Cover the top of Erlenmeyer flask with aluminum foil and use autoclave tape to secure it in place.
Autoclave NGM media and MgSO4, CaCl2 and KPI solutions (see Note 2).
Cool media to ~55 °C (see Note 3) while stirring.
Add MgSO4, CaCl2 and KPI (according to Recipes) while stirring.
Pour control plates (no paraquat): 10 ml of media in 60 mm Petri dish (see Notes 4 and 5).
Add paraquat (methyl viologen) (according to Recipes) while stirring (see Note 6). Allow ~60 sec for paraquat to be mixed into the media.
Pour plates: 10 ml of media in 60 mm Petri dish.
Repeat steps A7 and A8 for each concentration of paraquat (see Note 7).
Allow plates to solidify and dry overnight at room temperature.
Seed plates with 100 µl of OP50 bacteria overnight culture.
Allow bacteria to dry and grow at room temperature for 2 days.
Transfer equal numbers of eggs from each strain being tested to control plates and plates containing paraquat (Note 8) ensuring that you don’t transfer any eggs that have hatched into the first larval stage of development (L1 worms).
Check to ensure that eggs have hatched the following day.
Monitor the developmental stage of the worms daily (see Note 9).
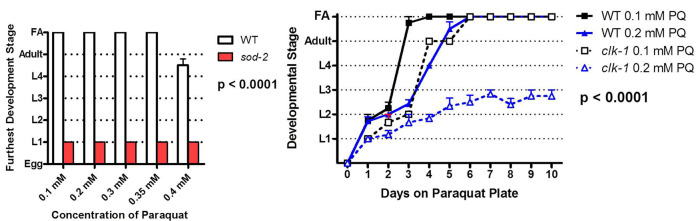
Expected results: Wild-type N2 worms should develop to fertile adults on plates up to about 0.35 mM paraquat.
Positive controls: clk-1, sod-2 both arrest at an early larval stage at concentrations of paraquat that permit WT worms to develop to fertile adults (e.g., 0.2 mM). References in which this protocol was used: Van Raamsdonk and Hekimi, 2012; Van Raamsdonk et al., 2010 .
Standard output format: see Figure 1.
-
Acute paraquat sensitivity assay
Prepare NGM media (see Recipes) in Erlenmeyer flask (Note 1).
Cover the top of Erlenmeyer flask with aluminum foil and use autoclave tape to secure it in place.
Autoclave NGM media and MgSO4, CaCl2 and KPI solutions (see Note 2).
Cool media to ~55 °C (see Note 3) while stirring.
Add MgSO4, CaCl2 and KPI (according to Recipes) while stirring.
Pour control plates (no paraquat): 4 ml of media in 35 mm Petri dish (see Notes 4 and 5).
Pour NGM media equivalent to the number of plates required plus one into a new Erlenmeyer flask or bottle. E.g., for 5 plates pour 24 ml of media into a new vessel (see Note 10).
Add paraquat (methyl viologen) (according to Recipes) while stirring (see Notes 6 and 11).
Pour plates: 4 ml of media in 35 mM Petri dish.
Repeat steps B7 and B8 for each concentration of paraquat (see Note 7).
Allow plates to solidify and dry overnight.
Seed plates with 50 µl of 10x concentrated OP50 bacteria overnight culture (see Note 12).
Allow bacteria to and grow at room temperature for 2 days.
Transfer 25 worms from each experimental strain to paraquat plates.
Check the survival of the worms hourly until all worms have died (see Note 13).
Expected results: The majority of wild-type N2 worms will die by 15 h. For young adults, 50% survival occurs around 8 h.
Positive controls: clk-1 worms have decreased survival compared to WT, isp-1 worms have increased survival compared to WT. References in which this protocol was used: Schaar et al., 2015 .
Standard output format: see Figure 2.
-
Acute juglone sensitivity assay
Juglone plates lose their toxicity with time ( Cooper et al., 2016 ). They need to be made fresh on the day of the assay and it is best to have a constant amount of time between pouring the plates and putting the worms on. They cannot be used for a chronic assay.
Prepare NGM media (see Recipes) in Erlenmeyer flask (Note 1).
Cover the top of Erlenmeyer flask with aluminum foil and use autoclave tape to secure it in place.
Autoclave NGM media and MgSO4, CaCl2 and KPI solutions (see Note 2).
Make juglone solution (see Recipes), stir in the dark for at least an hour (see Note 14).
Cool media to ~55 °C (see Note 3) while stirring.
Add MgSO4, CaCl2 and KPI (according to Recipes) while stirring.
Pour control plates (no juglone): 10 ml of media in 60 mm Petri dishes (see Notes 4 and 5).
-
Add juglone to desired concentration:
300 μM = 2.5 ml juglone/100 ml NGM
240 μM = 2 ml juglone/100 ml NGM
180 μM = 1.5 ml juglone/100 ml NGM
120 μM = 1 ml juglone/100 ml NGM
60 μM = 0.5 ml juglone/100 ml NGM
You can make multiple concentrations by progressively adding more juglone.
Pour juglone plates: 10 ml of media in 60 mm Petri dishes. More concentrated plates will be a darker yellow color.
Leave plates uncovered for 30 min to solidify agar. Plates should be in a single layer on the bench top, ideally next to a flame for sterility.
Add 40 μl of 5x concentrated OP50 bacteria with a pipet and spread to a thin layer by tilting plate in a circle (so bacteria will dry quicker). Do not let bacteria reach the edge of the plate, to limit the number of worms that will crawl up the sides.
Once bacteria is dry (~30 min), add 25 young adult worms per strain to the juglone plates to begin assay.
Check worm survival periodically (e.g., 1, 2 and 4 h).
Expected results: At 240 μM, wild-type N2 worms will start to die at 2 h and most will be dead by 4 h. Increasing juglone concentration to 300 μM results in more rapid death, while decreasing juglone concentration results in increased survival.
Positive controls: sod-1 worms have decreased survival compared to WT. References in which this protocol was used: Van Raamsdonk and Hekimi, 2009; 2012; Van Raamsdonk et al., 2010 ; Cooper et al., 2015 ; Schaar et al., 2015 ; Dues et al., 2016 ; Machiela et al., 2016 .
Standard output format: see Figure 3.
-
Chronic paraquat assay during adulthood
Prepare NGM media (see Recipes) in Erlenmeyer flask (Note 1).
Cover the top of Erlenmeyer flask with aluminum foil and use autoclave tape to secure it in place.
Autoclave NGM media and MgSO4, CaCl2 and KPI solutions (see Note 2).
Cool media to ~55 °C (see Note 3) while stirring.
Add MgSO4, CaCl2 and KPI (according to Recipes) while stirring.
Add FUdR to a concentration of 100 µM (see Note 15).
Pour control plates (no paraquat): 10 ml of media in 60 mm Petri dishes (see Notes 4 and 5).
Add paraquat (methyl viologen) (according to Recipes) while stirring (see Note 6).
Pour paraquat plates: 10 ml of media in 60 mm Petri dishes.
Repeat steps D7 and D8 for each concentration of paraquat (see Note 16).
Allow plates to solidify and dry overnight.
Seed plates with 150 µl of 10x concentrated OP50 bacteria overnight culture (see Note 12).
Allow bacteria to and grow at room temperature for 2 days.
Transfer 25 worms from each experimental strain to control and paraquat plates.
Check the survival of the worms daily until all worms have died.
Transfer worms to new plates every seven days.
Expected results: Wild-type N2 worms should have an average survival of about 10 days.
-
Positive controls: sod-1 and sod-2 worms have decreased survival compared to WT (Figure 4).
References in which this protocol was used: Van Raamsdonk and Hekimi, 2009; 2012; Dues et al., 2016 ; Schaar et al., 2015 .
Standard output format: see Figure 4.
Figure 1. Paraquat development assay.
Results are displayed as either a graph of the furthest developmental stage reached by worms of each genotype (Left) or the average stage of development each day after transferring to paraquat plates (Right). Typically, wild-type worms can develop to fertile adults (FA) at paraquat concentrations up to 0.35 mM. Increasing concentrations of paraquat result in slower development times.
Figure 2. Acute paraquat sensitivity assay.
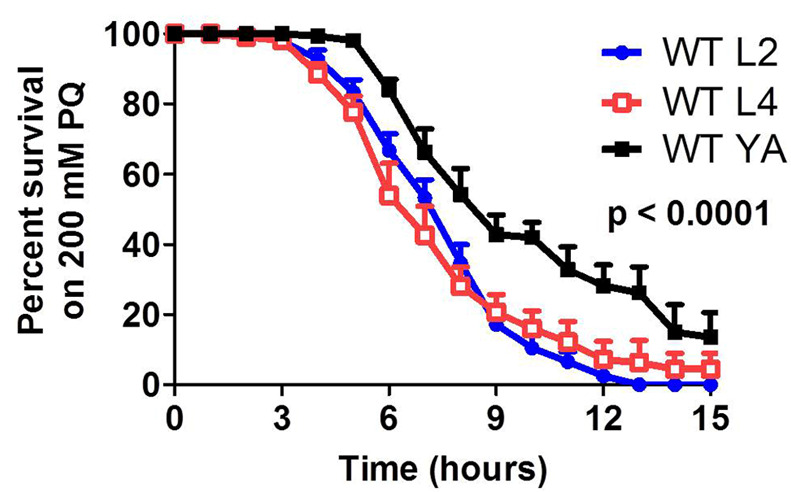
Results are displayed as the percentage of worms that are still alive at each time point. The majority of wild-type worms do not survive past 15 h. Developing L2 and L4 worms have decreased survival compared to young adults (YA). Results represent the average of three biological replicates of 25 worms per replicate.
Figure 3. Acute juglone sensitivity assay.
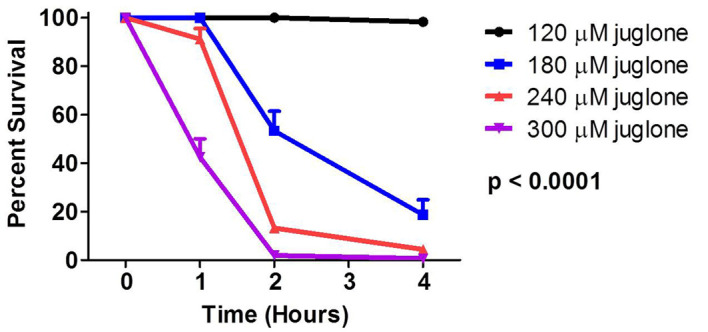
Results are displayed as the percentage of worms that are still alive at each time point. There is a dose-dependent decrease in survival with increasing juglone concentrations. Results represent the average of three biological replicates of 25 worms per replicate.
Figure 4. Chronic paraquat assay.
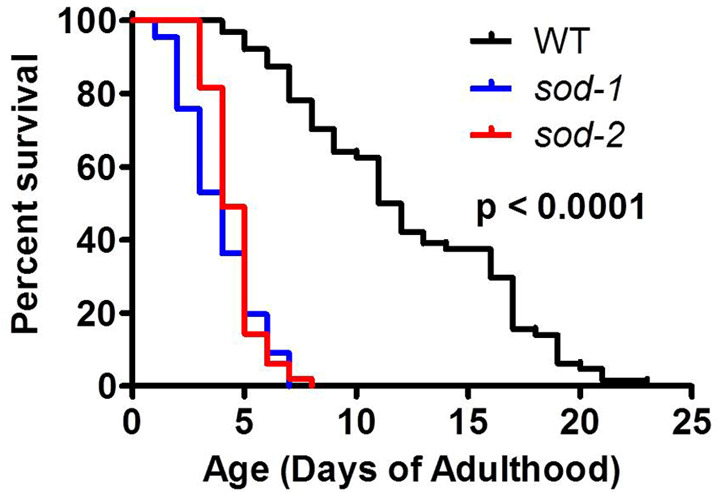
Results are typically shown as a Kaplan-Meier survival plot. Results represent the mean survival rate from a minimum of three biological replicates of 25 worms per replicate.
Data analysis
In the paraquat development assay, acute paraquat sensitivity assay and acute juglone sensitivity assay, we normally assess statistical significance using a two-way ANOVA with Bonferroni post-test. For the chronic paraquat assay, we assess significance using the Log-rank test. GraphPad Prism software is used to prepare all graphs and perform data analysis. Assays are performed such that the experimenter is blinded to the genotypes of the strains being tested. We perform a minimum of three independent biological replicates of at least 25 worms per strain. Worms that die due to internal hatching of progeny, externalization of internal organs, or crawling up the side of plates are censored.
Notes
Be sure not to overfill flask. If liquid is too near to the top it can spill over during autoclaving leading to lost volume. With small volumes, glass bottles can be used instead of Erlenmeyer flasks.
We typically use a 45 min sterilization cycle. Total time in autoclave is approximately 1 h and 15 min.
For 2 L, we normally cool for 45 min. Smaller volumes will cool more rapidly, so it is necessary to adjust cooling times.
We pour the control plates and oxidative stress plates from the same batch of media. As a result the only difference in the plates is the compound added. If we are making multiple concentrations, we make all of the concentrations from the same batch of media by adding additional amounts of the compound (e.g., paraquat or juglone) to prepare the higher concentrations.
To keep the volumes precise, we prefer to use a pipetman to ‘hand pour’ the plates. We add 10 ml to each 60 mm plate.
We make up a 1 M paraquat solution for the paraquat development assay and the chronic paraquat sensitivity assay. This solution can be stored at 4 °C for many months.
In the paraquat development assay, we normally test multiple concentrations of paraquat: 0.1 mM, 0.2 mM, 0.3 mM and 0.4 mM. Wild-type worms should be able to develop to fertile adults at a concentration of 0.35 mM but not at higher concentrations.
In the paraquat development assay, we typically transfer at least 50 eggs. For strains with markedly different embryonic development time, this assay can be adapted to look at development to adulthood beginning with L1 worms. In that case, transfer 200-300 eggs from each strain to an NGM plate. 3 h later pick L1 worms to plates containing paraquat.
For the paraquat development assay we typically use the furthest developmental stage attained (L1, L2, L3, L4, adult, fertile adult) as the outcome measure. A figure showing the different developmental stages of C. elegans can be found in the wormatlas (see Figure 6 at http://www.wormatlas.org/ver1/handbook/anatomyintro/anatomyintro.htm). The furthest developmental stage attained can be measured as the average for the population of worms, the maximum or the minimum, since there can be variability within a strain, especially close to its threshold. For a more quantitative approach, determine the percentage of worms from each strain that reach adulthood. It is also possible to record the average developmental stage of the population of worms each day to compare rate of development.
Because of the high concentration of paraquat used in the acute paraquat sensitivity assay we use small 35 mm plates to minimize the amount of paraquat required. In addition, we only use a small excess of media again to minimize the amount of paraquat required. Due to the small volumes of media used, it can cool and solidify quickly. As a result it is important to pour these plates as quickly as possible. Alternatively, one could keep the media in a 55 °C water bath or on a hotplate to prevent solidification.
For the acute paraquat sensitivity assay, we make a concentrated paraquat stock solution of 3.33 M to minimize changes in volume when added to the NGM media. We add 1,167 μl of water to 1 g of paraquat powder.
To concentrate bacteria, we centrifuge at 2,935 × g (5,000 RPM) for 10 min in a 15 ml or 50 ml conical flask. The supernatant is removed and then the bacteria are resuspended by vortexing.
In the acute paraquat sensitivity assay, for some developmental stages and certain strains, we have observed worms surviving past 15 h. In this case, we stop the assay at 15 h as this is typically sufficient to observe a difference, if there is one. However, it is possible that when comparing two very resistant strains that the assay may need to be extended past 15 h.
The ease with which juglone goes into solution varies with the batch. Typically, juglone takes a long time to dissolve. We stir the juglone in ethanol for at least 1 h (while the media is autoclaving and cooling). We will normally weigh out ~0.05 g juglone and mix with 100% ethanol (23.926 ml ethanol/0.05 g juglone). Cover with foil while stirring since juglone is light sensitive.
Paraquat causes an increased rate of internal hatching of progeny. To prevent this, we add FUdR (5-fluorodeoxyuridine), which inhibits the development of progeny. It should be noted that this compound has been shown to affect the lifespan of specific genetic mutants (Van Raamsdonk and Hekimi, 2011).
In the chronic paraquat sensitivity assay, we typically test worms at 4 mM paraquat. We have also tested worms at lower concentrations such as 2 mM but this increases the duration of the assay.
-
Additional general notes
Always include all strains that you want to compare in each assay. There will be some variability in the results depending on the batch of paraquat/juglone and how fresh the plates are. It is not appropriate to compare experimental strains tested in one assay with control samples tested in a different assay.
All assays are performed at 20 °C. Worms are kept in 20 °C incubators during the assay and only taken out of the incubator for scoring.
To determine whether a worm is alive or dead, we start by simply observing the worm. If the worm is not moving, we tap the tail of the worm. If the worm doesn’t respond to the tail tap, we gently lift the head of the worm and let it fall to the agar. If the worm still doesn’t move, it is recorded as dead and removed from the plate.
Worms that crawl up the side of the plate, exhibit internal hatching of progeny or externalization of internal organs are censored.
Recipes
-
0.5% cholesterol
Dissolve 1 g of cholesterol in 200 ml of 100% ethanol
Do not autoclave
Store at room temperature
-
1 M MgSO4
Dissolve 246.47 g MgSO4 in 1 L of dH2O
Autoclave
Store at room temperature
-
1 M CaCl2
Dissolve 147.01 g CaCl2 in 1 L of dH2O
Autoclave
Store at room temperature
-
KPI
Dissolve 35.6 g K2HPO4 and 108.3 g KH2PO4 to 1 L of dH2O
Adjust pH to 6.0
Autoclave
Store at room temperature
-
Nematode growth medium (NGM)
Volume (Liters) 0.5 1 1.5 2 2.5 3 NaCl 1.5 3 4.5 6 7.5 9 g Peptone 1 2 3 4 5 6 g Agar 8.5 17 25.5 34 42.5 51 g 0.5% Cholesterol 0.5 1 1.5 2 2.5 3 ml To be added after autoclaving and cooling, before pouring plates: 1 M MgSO4 0.5 1 1.5 2 2.5 3 ml 1 M CaCl2 0.5 1 1.5 2 2.5 3 ml 1 M KPI 12.5 25 37.5 50 62.5 75 ml -
1 M paraquat
Add 3.89 ml of dH2O to 1 g of paraquat
Do not autoclave
Store at 4 °C
-
3.33 M paraquat
Add 1,167 μl of dH2O to 1 g of paraquat
Do not autoclave
Store at 4 °C
-
0.1 M FUdR stock solution
Dissolve 1 g FUdR in 40.62 ml dH2O
Do not autoclave
Store in -80 °C freezer in aliquots
-
Juglone stock solution (12 mM)
Dissolve 0.05 g of juglone in 23.93 ml of 100% ethanol
Stir in dark for at least 1 h
Do not autoclave
Use immediately as toxicity is lost with time
-
2 YT medium
Dissolve 16 g tryptone, 10 g yeast extract and 5 g NaCl in 1 L of dH2O
Adjust pH to 7.0
Autoclave
Store at room temperature
Acknowledgments
This work was supported by the Van Andel Research Institute. Many other researchers have utilized similar protocols to test sensitivity to oxidative stress. These protocols are the way that we measure sensitivity to oxidative stress in our lab.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1.Castello P. R., Drechsel D. A. and Patel M.(2007). Mitochondria are a major source of paraquat-induced reactive oxygen species production in the brain. J Biol Chem 282(19): 14186-14193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dues D. J., Andrews E. K., Schaar C. E., Bergsma A. L., Senchuk M. M. and Van Raamsdonk J. M.(2016). Aging causes decreased resistance to multiple stresses and a failure to activate specific stress response pathways. Aging(Albany NY) 8(4): 777-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cooper J. F., Dues D. J., Spielbauer K. K., Machiela E., Senchuk M. M. and Van Raamsdonk J. M.(2015). Delaying aging is neuroprotective in Parkinson’s disease: a genetic analysis in C. elegans models . npj Parkinson's Disease 1: 15022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cooper J.F., Dues D.J., and Van Raamsdonk J.M.(2016). Measuring sensitivity to oxidative stress: the superoxide-generator juglone rapidly loses potency with time. Worm Breeder's Gazette. [Google Scholar]
- 5.Keith S. A., Amrit F. R., Ratnappan R. and Ghazi A.(2014). The C. elegans healthspan and stress-resistance assay toolkit . Methods 68(3): 476-486. [DOI] [PubMed] [Google Scholar]
- 6.Machiela E., Dues D. J., Senchuk M. M. and Van Raamsdonk J. M.(2016). Oxidative stress is increased in C. elegans models of Huntington's disease but does not contribute to polyglutamine toxicity phenotypes . Neurobiol Dis 96: 1-11. [DOI] [PubMed] [Google Scholar]
- 7.Schaar C. E., Dues D. J., Spielbauer K. K., Machiela E., Cooper J. F., Senchuk M., Hekimi S. and Van Raamsdonk J. M.(2015). Mitochondrial and cytoplasmic ROS have opposing effects on lifespan. PLoS Genet 11(2): e1004972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Raamsdonk J. M. and Hekimi S.(2009). Deletion of the mitochondrial superoxide dismutase sod-2 extends lifespan in Caenorhabditis elegans . PLoS Genet 5(2): e1000361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Raamsdonk J. M. and Hekimi S.(2011). FUdR causes a twofold increase in the lifespan of the mitochondrial mutant gas-1. Mech Ageing Dev 132(10): 519-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Raamsdonk J. M. and Hekimi S.(2012). Superoxide dismutase is dispensable for normal animal lifespan. Proc Natl Acad Sci U S A 109(15): 5785-5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Raamsdonk J. M., Meng Y., Camp D., Yang W., Jia X., Benard C. and Hekimi S.(2010). Decreased energy metabolism extends life span in Caenorhabditis elegans without reducing oxidative damage . Genetics 185(2): 559-571. [DOI] [PMC free article] [PubMed] [Google Scholar]