ABSTRACT
Healthy feto-maternal communication is critical during pregnancy and is orchestrated by the placenta. Dysfunction of the placenta leads to fetal growth complications; however, the underlying biological mechanisms have yet to be fully elucidated. Circulating extracellular microRNAs (exmiRNAs) in the blood have been implicated in cell-to-cell communication. Therefore, exmiRNAs may provide useful biological information about communication between the mother, the fetus, and the placenta during pregnancy. We used logistic regression to determine the association of exmiRNAs with abnormal fetal growth by comparing mothers of infants classified as small-for-gestational age (SGA) (n = 36) and large-for-gestational age (LGA) (n = 13) to appropriate-for-gestational age (AGA), matched by gestational age at delivery and infant sex. In addition, we used linear regression to determine associations between exmiRNAs and birth weight-for-gestational age (BWGA) z-score (n = 100), adjusting for maternal age, body mass index, and parity. We found that higher levels of miR-20b-5p, miR-942-5p, miR-324-3p, miR-223-5p, and miR-127-3p in maternal serum were associated with lower odds for having a SGA vs. AGA infant, and higher levels of miR-661, miR-212-3p, and miR-197-3p were associated with higher odds for having a LGA vs. AGA infant. We also found associations between miR-483-5p, miR-10a-5p, miR-204-5p, miR-202-3p, miR-345-5p, miR-885-5p, miR-127-3p, miR-148b-3p, miR-324-3p, miR-1290, miR-597-5p, miR-139-5p, miR-215-5p, and miR-99b-5p and BWGA z-score. We also found sex-specific associations with exmiRNAs and fetal growth. Our findings suggest that exmiRNAs circulating in maternal blood at second trimester are associated with fetal growth. Validation of our findings may lead to the development of minimally-invasive biomarkers of fetal growth during pregnancy.
KEYWORDS: Exosomes, extracellular vesicles, fetal growth, intrauterine growth restriction, large-for-gestational age, miR-127-3p, miR-20b-5p, miR-483-5p, miR-942-5p, small-for-gestational age
Abbreviations
- AGA
Appropriate-for-gestational age
- BMI
Body mass index
- BWGA
Birth weight-for-gestational age
- C14MC
Chromosome 14 microRNA cluster
- C19MC
Chromosome 19 microRNA cluster
- Cq
Quantification cycle
- EVs
Extracellular vesicles
- exmiRNAs
Extracellular microRNAs
- FC
Fold change
- LGA
Large-for-gestational age
- SGA
Small-for-gestational age
- SD
Standard deviation
Introduction
Extracellular RNAs in blood circulation are found either encased in extracellular vesicles (EVs) or bound to proteins such as Argonaute 2 and low/high-density-lipoproteins. Emerging evidence shows that extracellular RNAs, and in particular extracellular microRNAs (exmiRNAs), are involved in functional cell-to-cell communication.1 In fact, microRNAs that are small noncoding RNA molecules (∼22 nucleotides) can recognize and inhibit translation of multiple mRNAs in the recipient cells, either by initiating their degradation or by inhibiting the translational machinery.2 Therefore, exmiRNAs in maternal blood circulation provide a unique opportunity to non-invasively access biologic information that may be involved in feto-maternal communication.
During pregnancy, the number of EVs in the blood circulation increases significantly as compared with non-pregnant women.3 In fact, previous work demonstrated that EVs are an active communication mechanism between mother, fetus, and the placenta, and that fetal and placental RNAs are found in EVs in the maternal blood circulation.4,5 Several members of the placental-specific microRNA clusters on chromosomes 14 (C14MC) and 19 (C19MC) that are implicated in placental regulation have been found to be differentially expressed in normal and abnormal placentas, and were detected in maternal blood EVs as well.6 In addition, a few studies have identified microRNAs that were differentially expressed in placentas from pregnancies complicated by fetal growth restriction.7-9 However, no studies have assessed the presence of exmiRNAs in maternal blood circulation during pregnancy using a high-throughput method and determined their association with fetal growth.
Healthy placental communication between mother and fetus during pregnancy is critical, but our knowledge of the mechanisms involved in this communication and how they affect maternal, fetal, and placental health, is very limited. In this study, we screened for 754 exmiRNAs in serum samples prospectively collected at second trimester from pregnant women (n = 100) who participated in the Programming Research in Obesity, Growth Environment, and Social Stress (PROGRESS) birth cohort study, and determined exmiRNA association with fetal growth. We hypothesized that exmiRNAs circulating in maternal blood at second trimester are associated with fetal growth.
Results
Characteristics of study participants
Mothers of large-for-gestational age (LGA) infants were older than those of small-for-gestational age (SGA) infants (P = 0.03) and had higher body mass index (BMI) (P = 0.003) than mothers of either SGA or appropriate-for-gestational age (AGA) infants. As expected, there was a significant difference in birth weight between SGA, AGA, and LGA infants (P < 0.0001). No differences were observed in any of the matching variables (i.e., gestational age at delivery and infant sex), or any other characteristics between SGA, AGA, and LGA mother-infant pairs (Table 1).
Table 1.
Summary of study participants' characteristics.
| Matched groups |
|||||
|---|---|---|---|---|---|
| Characteristica | All Participantsb | SGAb | AGAb | LGAb | |
| n | 100 | 36 | 51 | 13 | P valued |
| Maternal | |||||
| Age (years) | 27.8 ± 6.1 | 26.0 ± 6.1 | 28.4 ± 5.8 | 30.6 ± 6.4 | 0.03 |
| Height (cm) | 154.4 ± 5.1 | 153.4 ± 5.4 | 154.7 ± 4.8 | 156.0 ± 4.9 | 0.09 |
| Body weight (kg) | 62.7 ± 9.5 | 61.0 ± 10.8 | 61.8 ± 7.9 | 71.2 ± 7.6 | 0.001 |
| Body mass index (BMI) (kg/m2) | 26.3 ± 3.7 | 25.9 ± 4.1 | 25.8 ± 3.3 | 29.2 ± 2.6 | 0.003 |
| Hemoglobin levels (g/dL) | 12.8 ± 1.0 | 13.0 ± 1.1 | 12.7 ± 0.9 | 12.7 ± 1.4 | 0.44 |
| Multiparous; n (%) | 65 (65.0) | 20 (55.6) | 34 (66.7) | 11 (84.6) | 0.17 |
| Alcohol consumption; n (%) | 0.72 | ||||
| Yes | 6 (6.0) | 3 (8.3) | 3 (5.9) | 0 (0.0) | |
| No | 94 (94.0) | 33 (91.7) | 48 (94.1) | 13 (100.0) | |
| Socioeconomic status; n (%) | 0.93 | ||||
| Low | 49 (49.0) | 17 (47.2) | 25 (49.0) | 7 (53.8) | |
| Medium | 40 (40.0) | 15 (41.7) | 21 (41.2) | 4 (30.8) | |
| High | 11 (11.0) | 4 (11.1) | 5 (9.8) | 2 (15.4) | |
| Smoke exposure in the homee; n (%) | 0.49 | ||||
| Yes | 28 (28.6) | 9 (25.7) | 17 (33.3) | 2 (16.7) | |
| No | 70 (71.4) | 26 (74.3) | 34 (66.7) | 10 (83.3) | |
| Infant | |||||
| Birth weight (g) | 3014 ± 633 | 2396 ± 220 | 3190 ± 393 | 4038 ± 331 | <0.0001 |
| Gestational age (weeks)f | 38.3 ± 1.0 | 38.2 ± 1.0 | 38.3 ± 1.1 | 38.3 ± 1.0 | 0.89 |
| Birthweight-for-gestational ageg (z-score) | −0.6 ± 1.3 | −1.9 ± 0.4 | −0.2 ± 0.7 | 1.5 ± 0.3 | <0.0001 |
| Sexf; n (%) | 1.00 | ||||
| Males | 51 (51.0) | 18 (50.0) | 26 (51.0) | 7 (53.8) | |
| Females | 49 (49.0) | 18 (50.0) | 25 (49.0) | 6 (46.2) | |
All maternal information was collected during the second trimester visit (16-22 weeks).
All values are shown as mean ± SD, unless otherwise noted.
Participants in the SGA, AGA, and LGA groups were matched by gestational age at delivery and infant's sex.
P value corresponds to Chi squared (continuous) or Fisher's exact (categorical) test for the difference in study participants' characteristics between SGA, AGA, and LGA groups.
Data are missing for 2 participants.
Variables used to match controls (LGA) to cases (SGA or LGA).
Adjusted for gestational age and infant sex.
Second trimester exmiRNAs in maternal blood are associated with fetal growth
A total of 444 exmiRNAs were found in at least one of the serum samples analyzed (n = 100), but only 124 exmiRNAs were detected in ≥50% of the samples (Table S1).
When we compared the matched pairs (by gestational age and infant's sex), we found that differential expression of specific exmiRNAs was associated with fetal growth. In particular, we found that mothers had lower odds of having SGA vs. AGA infant with increased levels of miR-20b-5p [odds ratio (OR) = 0.49, P = 0.02], miR-942-5p (OR = 0.48, P = 0.02), miR-324-3p (OR = 0.44, P = 0.03), miR-223-5p (OR = 0.47, P = 0.05), and miR-127-3p (OR = 0.61, P = 0.06) (Fig. 1). We also found higher odds of having an LGA vs. AGA infant with increased levels of miR-661 (OR = 4.00, P = 0.07), miR-212-3p (OR = 3.20, P = 0.08), and miR-197-3p (OR = 4.73, P = 0.10) (Fig. 1). Although the associations were significant, when we adjusted for multiple comparisons no significance was sustained.
Figure 1.
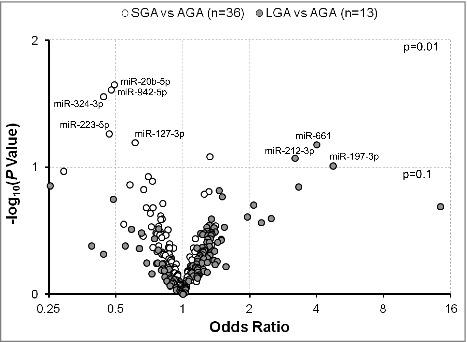
Odds ratios for having an SGA or LGA vs. AGA infant in association with levels of extracellular microRNAs (exmiRNAs) in maternal serum. Odds ratios are adjusted for gestational age and infant's sex; -log10 (P Value) indicates transformed P values of the association between exmiRNAs and SGA or LGA. Odds ratios between SGA and AGA are shown in white and between LGA and AGA in gray.
We also found that lower levels of miR-483-5p (β = −0.19, P = 0.002), miR-10a-5p (β = −0.33, P = 0.002), miR-204-5p (β = −0.29, P = 0.01), miR-202-3p (β = −0.28, P = 0.01), miR-345-5p (β = −0.18, P = 0.01), miR-885-5p (β = −0.23, P = 0.01), miR-127-3p (β = −0.24, P = 0.02), miR-148b-3p (β = −0.28, P = 0.02), miR-324-3p (β = −0.24, P = 0.02), miR-1290 (β = −0.19, P = 0.02), miR-597-5p (β = −0.30, P = 0.02), miR-139-5p (β = −0.18, P = 0.03), miR-215-5p (β = −0.25, P = 0.05), and miR-99b-5p (β = −0.15, P = 0.05) in second trimester maternal serum were associated with lower birth weight-for-gestational age (BWGA) z-score (Fig. 2; Table S2). Again, when we adjusted for multiple comparisons, no significance was observed for any of the abovementioned exmiRNAs.
Figure 2.
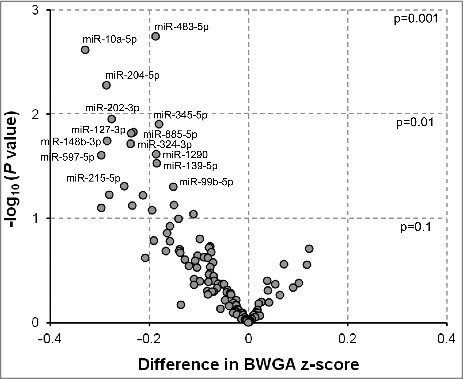
Associations between extracellular microRNAs (exmiRNAs) in maternal serum and birth weight-for-gestational age z-score (n = 100). Volcano plot showing the association between serum exmiRNAs at second trimester and birth weight-for-gestational age z-score; estimates are adjusted for maternal age, gestational age, body mass index, parity, and infant's sex; -log10 (P value) indicates transformed P values of the association between exmiRNAs and birth weight-for-gestational age z-score.
In silico pathway analysis showed that 7 of the most associated exmiRNAs with fetal growth (miR-10a-5p, miR-127-3p, miR-197-3p, miR-202-3p, miR-204-4p, miR-215-5p, and miR-483-5p) could interact with 43 experimentally validated mRNAs that regulate several biological pathways downstream. These pathways were enriched for functions and diseases related to embryo and fetal development, including the size of placenta and embryo, development of body axis and body trunk, size and formation of bones, angiogenesis and others (Fig. 3). The full list of functions and diseases regulated by these exmiRNAs, together with the molecules involved and P values are provided in Table S3.
Figure 3.
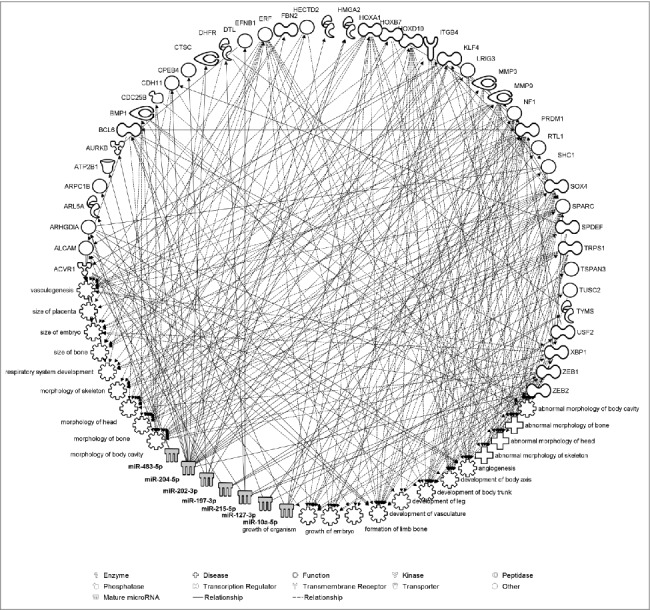
Pathway analysis illustrating experimentally validated mRNA targets of the most associated exmiRNAs with fetal growth. Biological relevance with functions and diseases related to fetal growth is also identified by the Ingenuity Pathway Analysis®. Direct relationships are represented with solid lines and indirect relationships with dotted lines.
Sex-specific associations between exmiRNAs and fetal growth
To further examine any differences between male and female infants in the expression of serum exmiRNAs, we stratified our analysis in mothers of SGA/LGA vs. AGA infants. In mothers of SGA vs. AGA infants, we found that most of the significantly associated exmiRNAs were concordantly regulated in both male and female infants, such as miR-127-3p, miR-20b-5p, miR-324-3p, and miR-942-5p (Fig. 4). All together, we found that 50 exmiRNAs were concordantly downregulated and 22 concordantly upregulated in mothers of both male (n = 18) and female (n = 18) SGA vs. AGA infants. We also found that 47 exmiRNAs were downregulated in male, but upregulated in female infants, and that 7 exmiRNAs were upregulated in male, but downregulated in female infants (Table S4). Similarly, when we analyzed the expression levels of exmiRNAs in mothers of LGA vs. AGA infants by gender, we found that the most exmiRNAs were downregulated in males (n = 7) and upregulated in females (n = 6). Interestingly, 3 of the top hits, miR-139-5p, miR-215-5p and miR-30d-5p, were downregulated in males, but upregulated in females (Fig. 5). Overall, there were 8 exmiRNAs concordantly downregulated and 29 concordantly upregulated in mothers of both male and female LGA vs. AGA infants. On the contrary, there were 84 exmiRNAs that were downregulated in male, but upregulated in female infants, and 5 exmiRNAs that were upregulated in male, but downregulated in female infants (Table S5).
Figure 4.
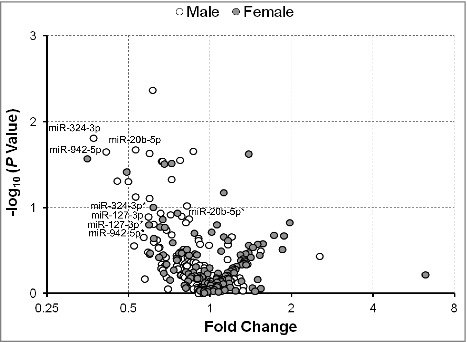
Fold change of serum extracellular microRNAs (exmiRNAs) between mothers of small- and appropriate-for-gestational age infants by gender. Volcano plot showing the fold change of all tested serum exmiRNAs at second trimester from mothers of male (white) and female (gray) SGA vs. AGA infants. *Denotes exmiRNAs in female mother-infant pairs.
Figure 5.
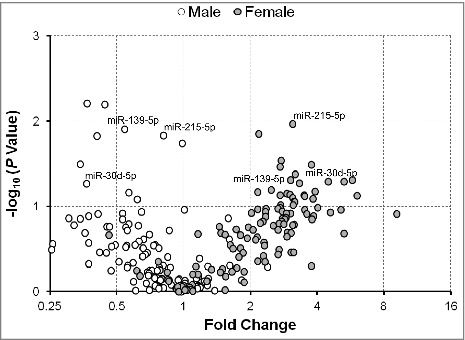
Fold change of serum extracellular microRNAs (exmiRNAs) between mothers of large- and appropriate-for-gestational age infants by gender. Volcano plot showing the fold change of all tested serum exmiRNAs at second trimester from mothers of male (white) and female (gray) LGA vs. AGA infants.
Discussion
In this exploratory study, we conducted a high-throughput screening of extracellular microRNAs from 100 serum samples collected from pregnant women at second trimester. Overall, we observed lower exmiRNAs levels in serum of mothers of SGA infants, and higher exmiRNAs levels in mothers of LGA infants, when compared with mothers of AGA infants. To the best of our knowledge, several of these exmiRNAs have not been previously associated with fetal growth.
Previous studies have identified differentially expressed microRNAs in placentas from complicated pregnancies, suggesting their role in placental regulation and fetal development. Emerging evidence shows that several of these microRNAs can also be detected in the maternal blood circulation during pregnancy. Several of these studies focused on microRNAs of the placenta-specific C14MC (e.g., miR-376c-3p) and C19MC (e.g., miR-517a-3p, miR-520h, and miR-525-3p) clusters, which were associated with pregnancy-related complications, including impaired fetal growth.10,11 In our study, we found no significant associations with any of the detected C19MC microRNA members and fetal growth. Consistent with our findings, Hromadnikova et al. investigated C19MC placenta-specific microRNAs (i.e., miR-516-5p, miR-517-5p, miR-518b, miR-520a-5p, miR-520h, miR-525-3p, and miR-526a-3p) in the plasma of normal and fetal growth restricted pregnancies, and found no significant differences.8 Similarly, Mouillet et al. compared the levels of 12 placenta-specific microRNAs, including C19MC cluster members (i.e., miR-517-5p, miR-518b, miR-518e, and miR-524), in plasma samples of gestational age-matched normal and fetal growth restricted pregnancies; the investigators reported no significant differences in any of the screened C19MC microRNAs.9
We found the most robust association between abnormal fetal growth and miR-127-3p—a member of the C14MC cluster—which had not been evaluated in relation to fetal growth by previous studies. In particular, we found that mothers that had higher levels of extracellular miR-127-3p in their blood had lower odds of having SGA infant. We also compared the expression levels of miR-127-3p in mothers of SGA, AGA, and LGA and found that mothers of SGA had the lowest levels, followed by mothers of AGA and LGA; miR-127-3p levels in serum of mothers of SGA infants were significantly lower (P < 0.05) than those of LGA infants, although these pairs were unmatched (Fig. S1). When we further tested for linear association of miR-127-3p and BWGA z-score using data from all 100 analyzed samples, we found that lower levels of miR-127-3p were associated with lower BWGA z-score. Together, these findings suggest a dose-response relationship between miR-127-3p and fetal growth. Labialle et al. showed that while knockout of the C14MC cluster in mice was not associated with specific placental adverse phenotypes, these mice exhibited partially penetrant neonatal lethality.12 A more recent study by Ito et al. showed that when miR-127-3p was deleted in mice, the placenta developed insufficiencies in the labyrinthine zone, which is analogous to the human placenta villous area that is directly involved in the transfer of gases and nutrients between the mother and the fetus.13 Although in mice, these findings are consistent with ours, suggesting that miR-127-3p may be associated with placental development and fetal growth.
We also found an association between abnormal fetal growth and miR-20b-5p, a member of the miR-17–92 cluster, which is expressed in a wide spectrum of tissues14 and has been suggested to play an important role in embryonic development.15,16 Members of this cluster were previously found to be associated with pregnancy complications, suggesting their importance in placental and fetal development.17 We found that miR-20b-5p levels were significantly lower in serum of mothers of SGA, as compared with mothers of AGA infants. Wang et al. found that expression of miR-20b-5p was altered in placentas collected from women with pre-eclampsia, suggesting its regulating potential in placental angiogenesis and trophoblast development,18 processes that are important for normal fetal development.
One of the exmiRNAs most significantly associated with SGA in our study was miR-942-5p, which is implicated in angiogenesis, an important process in normal placental development. To the best of our knowledge, no studies investigated the association of miR-942-5p and fetal growth; however, 2 studies that examined its association with preeclampsia found no significant results.19,20 Other exmiRNAs (i.e., miR-10a-5p, miR-197-3p, miR-204-5p, and miR-483-5p) that we found to be associated with abnormal fetal growth and were enriched in related biologic pathways in silico, were also found to be dysregulated under pregnancy-related adverse outcomes, such as preterm delivery, pregnancy-induced hypertension, and pre-eclampsia.21-23
Previous studies have shown that regulation of several microRNAs under certain conditions is sexually dimorphic.24 In our study, we observed notable sex-specific differences in the levels of exmiRNAs when comparing mothers of LGA vs. AGA, but not in SGA vs. AGA infants. In a recent study, Muralimanoharan et al. found that the expression of miR-210 in the placenta was regulated by fetal sex via the NFkB1 pathway and estrogen levels in obese pregnant women, a condition known to be a risk factor for LGA pregnancies.25 Sexual dimorphism in fetal cardiac miRNAs under intrauterine growth restriction conditions has also been linked to the NFkB1 pathway.26 Lastly, Bouhaddioui et al. showed that specific miRNAs involved in mouse fetal lung development are androgen-depended.27 Whether these sex-specific differences under certain conditions during pregnancy can be attributed to fetal sex or other biological mechanisms that are associated with, it remains to be fully elucidated.
Our study has certain limitations. Despite the large number of known microRNAs screened (n = 754), we acknowledge the possibility that other exmiRNAs, not screened in our analysis, may be present in the maternal blood circulation and associated with fetal growth. In addition, due to the relatively small sample size of this exploratory study, no significant associations were sustained after correction for multiple comparisons. Nonetheless, in silico analysis of most associated exmiRNAs revealed their significant enrichment in biologically relevant pathways for placental and fetal development. Therefore, we encourage external validation of our findings in larger and more diverse epidemiological studies. Strengths of our study include the nested case-control design within a prospective cohort study, with careful matching by gestational age at delivery and infant's sex. In addition, we very carefully evaluated several potential confounders in the association between exmiRNAs levels and fetal growth, and adjusted for maternal age, BMI, and parity. Last, the homogeneity of the study population helped reduce any unmeasured confounding that may be attributed to differences at the population level.
This exploratory study suggests that circulating exmiRNAs early in pregnancy—some of which placenta-specific—are associated with fetal growth. Our findings highlight a unique opportunity to easily collect biological information implicated in feto-maternal communication, which may help us develop minimally-invasive biomarkers of fetal growth monitoring during pregnancy.
Materials and methods
Study design and sample selection
Study participants were enrolled during their first prenatal care visit (<20 weeks of gestation) at the Mexican Social Security Institute clinics in Mexico City, as part of the prospective birth cohort study PROGRESS. The profile of the cohort is described elsewhere.28 Our base population was composed of pregnant women who agreed to participate (n = 948) and had a live birth. The study was approved by the Institutional Review Boards of Brigham and Women's Hospital, Harvard T.H. Chan School of Public Health, Icahn School of Medicine at Mount Sinai, and the Mexican National Institute of Public Health (#14265-101; #14706-101; #2006-P-001416; #2006-P-001792; #12-00751; #560). All study participants provided written informed consent in accordance with the Declaration of Helsinki ethical principles for medical research.
Blood samples were collected in EDTA tubes (Becton Dickinson, Cat. No. 366643) at the second trimester visit (16-22 weeks of gestation), and serum fraction was separated within 2 h by centrifugation at 1,500 × g for 15 min, following standard operating procedures. Serum was aliquoted in cryovial tubes, stored at −80°C, and shipped to the Harvard T.H. Chan School of Public Health, Laboratory of Environmental Epigenetics for further analysis. Infants were considered as SGA if they were in the <10th percentile, AGA if they were between the 10th - 90th percentile, and LGA if they were in the >90th percentile of the birth weight-for-gestational age (BWGA) distribution according to the international infant growth charts.29
For exmiRNAs screening, all samples were selected from pregnant women who delivered between 37 to 40 weeks of gestation (n = 786). More specifically, we randomly selected 36 serum samples from women with SGA infants and 13 samples from women with LGA infants. We also selected 51 samples from women with AGA infants that were used as controls for SGA and/or LGA samples, for a total of 100 serum samples analyzed in our study (Fig. S2). Each SGA and/or LGA sample was matched to an AGA control sample (1:1 ratio) by gestational age at delivery and infant's sex. Due to the lack of ultrasound data, gestational age at the time of enrollment was calculated using the date of last menstrual period.
RNA extraction and microRNA profiling
Serum samples (0.7 mL) were thawed on ice and filtered using a 0.8-μm membrane unit (Millipore Corp., Bedford, MA) to remove any cell debris and large aggregates. Following this step, we used the exoRNeasy Serum/Plasma Maxi kit (Qiagen, Cat. No. 77064), to extract total extracellular RNA.30 Total RNA was eluted in ∼12–μL volumes and subsequently concentrated to 6-μL volumes using the RNA Clean & Concentrator-5 (Zymo Research, Cat. No. R1015), before microRNA expression analysis. We used the TaqMan OpenArray technology to measure the expression levels of 754 known microRNAs (miRBase v14).
All RNA samples were reverse transcribed and pre-amplified (16 cycles) using Megaplex Reverse Transcription Primers, Human Pool A v2.1 (Life Technologies, Cat. No. 4399966) and Human Pool B v3.0 (Life Technologies, Cat. No. 4444281), and Megaplex PreAmp Primers, Human Pool A v2.1 (Life Technologies, Cat. No. 4399233) and Human Pool B v3.0 (Life Technologies, Cat. No. 4444748), following the manufacturer's protocol. The quantitative polymerase chain reaction (qPCR) step was performed on the QuantStudio 12K Flex Real-Time PCR System (Life Technologies) and relative quantification cycle (Cq) values were calculated. Cq values were defined as the PCR cycle where the amplification curve exceeded background fluorescence threshold.
Data analysis
Descriptive characteristics of the study participants are reported as mean ± standard deviation (SD), unless otherwise specified. We performed quality control on raw qPCR data according to the following criteria, suggested by the manufacturer: (a) Cq values <35, (b) amplification scores ≥1.1, and (c) Cq confidence ≥0.8. Data that did not meet these criteria were treated as missing values in the analysis. Following this step, exmiRNAs were ranked based on their frequency of detection across all analyzed samples, and for reasons of statistical power, we only considered exmiRNAs detected in ≥50% of the analyzed samples (n = 124) in the analysis. Due to the lack of reference genes for exmiRNAs analysis, we used the global mean method to normalize our data and calculated Delta Cq (ΔCq) values according to the ΔCq_microRNAi = (Cq_microRNAi - Cq_microRNAi_global_mean) formula.31
We used logistic regression (adjusted for gestational age and infant's sex) to determine the association [odds ratio (OR)] between serum exmiRNAs at second trimester and SGA or LGA among matched pairs. We also used linear regression to determine the association between serum exmiRNAs levels at second trimester and BWGA z-score (continuous). All models were adjusted for maternal age (continuous), body mass index (continuous), gestational age (continuous), parity (multiparous or nulliparous), and infant's sex. Other covariates such as maternal hemoglobin levels, socioeconomic status, smoking exposure in the home, and alcohol consumption were also evaluated as confounders in the analysis, but not included in our final models as they did not confound our estimates. To balance for the potential bias due to oversampling of SGA and LGA cases in our analysis, we used inverse probability weights. We allocated weights to each group by using the probability that each sample could have been selected from our base population. We then used these weights to estimate weighted effect estimates (β coefficients), which can be interpreted as the weighted change in BWGA z-score for every 1 unit (ΔCq) increase in the expression of a specific exmiRNA.
Last, to examine any differences by gender in the expression levels of exmiRNAs between pregnant women who delivered male vs. female SGA, AGA, and LGA infants, we used the fold change method according to the 2−ΔΔq formula. To test statistical significance after correcting for multiple comparisons we used the Benjamini-Hochberg method. All statistical analyses were performed in SAS version 9.4 (SAS Institute Inc., Cary, NC). Ingenuity Pathway Analysis (IPA) software (Ingenuity Systems®, Redwood City, CA) was used to identify experimentally validated mRNA targets of the most associated exmiRNAs with fetal growth, and further explore relevant downstream biological pathway and functions.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank the faculty and staff of the American British Cowdray (ABC) Hospital in Mexico City who provided space and valuable assistance in the data collection.
Funding
This work was supported by the Harvard T. H. Chan School of Public Health-NIEHS Center for Environmental Health (ES000002) and the National Institute of Environmental Health Science grants R01 ES013744, R01 ES014930, R01 ES021357, R01 ES020268, and P30 ES023515.
References
- 1.Yanez-Mo M, Siljander PR, Andreu Z, Zavec AB, Borras FE, Buzas EI, Buzas K, Casal E, Cappello F, Carvalho J, et al.. Biological properties of extracellular vesicles and their physiological functions. J Extracellular Vesicles. 2015;4:27066. doi: 10.3402/jev.v4.27066. PMID:25979354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215-33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Salomon C, Torres MJ, Kobayashi M, Scholz-Romero K, Sobrevia L, Dobierzewska A, Illanes SE, Mitchell MD, Rice GE. A gestational profile of placental exosomes in maternal plasma and their effects on endothelial cell migration. PLoS One. 2014;9:e98667. doi: 10.1371/journal.pone.0098667. PMID:24905832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ge Q, Zhu Y, Li H, Tian F, Xie X, Bai Y. Differential expression of circulating miRNAs in maternal plasma in pregnancies with fetal macrosomia. Int J Mol Med. 2015;35:81-91. doi: 10.3892/ijmm.2014.1989. PMID:25370776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Williams Z, Ben-Dov IZ, Elias R, Mihailovic A, Brown M, Rosenwaks Z, Tuschl T. Comprehensive profiling of circulating microRNA via small RNA sequencing of cDNA libraries reveals biomarker potential and limitations. Proc Natl Acad Sci U S A. 2013;110:4255-60. doi: 10.1073/pnas.1214046110. PMID:23440203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mouillet JF, Ouyang Y, Coyne CB, Sadovsky Y. MicroRNAs in placental health and disease. Am J Obstet Gynecol. 2015;213:S163-172. doi: 10.1016/j.ajog.2015.05.057. PMID:26428496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Higashijima A, Miura K, Mishima H, Kinoshita A, Jo O, Abe S, Hasegawa Y, Miura S, Yamasaki K, Yoshida A, et al.. Characterization of placenta-specific microRNAs in fetal growth restriction pregnancy. Prenat Diagn. 2013;33:214-22. doi: 10.1002/pd.4045. PMID:23354729 [DOI] [PubMed] [Google Scholar]
- 8.Hromadnikova I, Kotlabova K, Doucha J, Dlouha K, Krofta L. Absolute and relative quantification of placenta-specific micrornas in maternal circulation with placental insufficiency-related complications. J Mol Diagn. 2012;14:160-7. doi: 10.1016/j.jmoldx.2011.11.003. PMID:22251611 [DOI] [PubMed] [Google Scholar]
- 9.Mouillet JF, Chu T, Hubel CA, Nelson DM, Parks WT, Sadovsky Y. The levels of hypoxia-regulated microRNAs in plasma of pregnant women with fetal growth restriction. Placenta. 2010;31:781-4. doi: 10.1016/j.placenta.2010.07.001. PMID:20667590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Braun JM, Wright RJ, Just AC, Power MC, Tamayo YOM, Schnaas L, Hu H, Wright RO, Tellez-Rojo MM. Relationships between lead biomarkers and diurnal salivary cortisol indices in pregnant women from Mexico city: a cross-sectional study. Environ Health. 2014;13:50. doi: 10.1186/1476-069X-13-50. PMID:24916609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang H, Wen Y, Hu L, Miao T, Zhang M, Dong J. Serum MicroRNAs as diagnostic biomarkers for Macrosomia. Reprod Sci. 2015;22:664-71. doi: 10.1177/1933719114561557. PMID:25519717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Labialle S, Marty V, Bortolin-Cavaille ML, Hoareau-Osman M, Pradere JP, Valet P, Martin PG, Cavaille J. The miR-379/miR-410 cluster at the imprinted Dlk1-Dio3 domain controls neonatal metabolic adaptation. EMBO J. 2014;33:2216-30. doi: 10.15252/embj.201387038. PMID:25124681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.IIto M, Sferruzzi-Perri AN, Edwards CA, Adalsteinsson BT, Allen SE, Loo TH, Kitazawa M, Kaneko-Ishino T, Ishino F, Stewart CL, et al.. A trans-homologue interaction between reciprocally imprinted miR-127 and Rtl1 regulates placenta development. Development. 2015;142:2425-30. doi: 10.1242/dev.121996. PMID:26138477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mogilyansky E, Rigoutsos I. The miR-17/92 cluster: a comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ. 2013;20:1603-14. doi: 10.1038/cdd.2013.125. PMID:24212931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Houbaviy HB, Murray MF, Sharp PA. Embryonic stem cell-specific MicroRNAs. Dev Cell. 2003;5:351-8. PMID:12919684 [DOI] [PubMed] [Google Scholar]
- 16.Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR, et al.. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875-886. doi: 10.1016/j.cell.2008.02.019. PMID:18329372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manokhina I, Wilson SL, Robinson WP. Noninvasive nucleic acid-based approaches to monitor placental health and predict pregnancy-related complications. Am J Obstet Gynecol. 2015;213:S197-206. doi: 10.1016/j.ajog.2015.07.010. PMID:26428499 [DOI] [PubMed] [Google Scholar]
- 18.Wang W, Feng L, Zhang H, Hachy S, Satohisa S, Laurent LC, Parast M, Zheng J, Chen DB. Preeclampsia up-regulates angiogenesis-associated microRNA (i.e., miR-17, −20a, and −20b) that target ephrin-B2 and EPHB4 in human placenta. J Clin Endocrinol Metab. 2012;97:E1051-1059. doi: 10.1210/jc.2011-3131. PMID:22438230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hromadnikova I, Kotlabova K, Hympanova L, Doucha J, Krofta L. First trimester screening of circulating C19MC microRNAs can predict subsequent onset of gestational hypertension. PLoS One. 2014;9:e113735. doi: 10.1371/journal.pone.0113735. PMID:25502889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luque A, Farwati A, Crovetto F, Crispi F, Figueras F, Gratacos E, Aran JM. Usefulness of circulating microRNAs for the prediction of early preeclampsia at first-trimester of pregnancy. Sci Rep. 2014;4:4882. doi: 10.1038/srep04882. PMID:24811064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mayor-Lynn K, Toloubeydokhti T, Cruz AC, Chegini N. Expression profile of microRNAs and mRNAs in human placentas from pregnancies complicated by preeclampsia and preterm labor. Reprod Sci. 2011;18:46-56. doi: 10.1177/1933719110374115. PMID:21079238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mei Z, Huang B, Mo Y, Fan J. An exploratory study into the role of miR-204-5p in pregnancy-induced hypertension. Exp Ther Med. 2017;13:1711-8. doi: 10.3892/etm.2017.4212. PMID:28565757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Truong G, Guanzon D, Kinhal V, Elfeky O, Lai A, Longo S, Nuzhat Z, Palma C, Scholz-Romero K, Menon R, et al.. Oxygen tension regulates the miRNA profile and bioactivity of exosomes released from extravillous trophoblast cells - Liquid biopsies for monitoring complications of pregnancy. PLoS One. 2017;12:e0174514. doi: 10.1371/journal.pone.0174514. PMID:28350871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dai R, Ahmed SA. Sexual dimorphism of miRNA expression: a new perspective in understanding the sex bias of autoimmune diseases. Ther Clin Risk Manag. 2014;10:151-63. doi: 10.2147/TCRM.S33517. PMID:24623979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muralimanoharan S, Guo C, Myatt L, Maloyan A. Sexual dimorphism in miR-210 expression and mitochondrial dysfunction in the placenta with maternal obesity. Int J Obes (Lond). 2015;39:1274-81. doi: 10.1038/ijo.2015.45. PMID:25833255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muralimanoharan S, Li C, Nakayasu ES, Casey CP, Metz TO, Nathanielsz PW, Maloyan A. Sexual dimorphism in the fetal cardiac response to maternal nutrient restriction. J Mol Cell Cardiol. 2017;108:181-93. doi: 10.1016/j.yjmcc.2017.06.006. PMID:28641979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bouhaddioui W, Provost PR, Tremblay Y. Expression profile of androgen-modulated microRNAs in the fetal murine lung. Biol Sex Differ. 2016;7:20. doi: 10.1186/s13293-016-0072-z. PMID:27042289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodosthenous RS, Burris HH, Svensson K, Amarasiriwardena CJ, Cantoral A, Schnaas L, Mercado-Garcia A, Coull BA, Wright RO, Tellez-Rojo MM, et al.. Prenatal lead exposure and fetal growth: Smaller infants have heightened susceptibility. Environ Int. 2016. doi: 10.1016/j.envint.2016.11.023. PMID:27923585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fenton TR, Nasser R, Eliasziw M, Kim JH, Bilan D, Sauve R. Validating the weight gain of preterm infants between the reference growth curve of the fetus and the term infant. BMC Pediatr. 2013;13:92. doi: 10.1186/1471-2431-13-92. PMID:23758808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Enderle D, Spiel A, Coticchia CM, Berghoff E, Mueller R, Schlumpberger M, Sprenger-Haussels M, Shaffer JM, Lader E, Skog J, et al.. Characterization of RNA from exosomes and other extracellular vesicles isolated by a novel spin column-based method. PLoS One. 2015;10:e0136133. doi: 10.1371/journal.pone.0136133. PMID:26317354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mestdagh P, Van Vlierberghe P, De Weer A, Muth D, Westermann F, Speleman F, Vandesompele J. A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol. 2009;10:R64. doi: 10.1186/gb-2009-10-6-r64. PMID:19531210 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


