Abstract
Bacterial cell-cell signaling, or quorum sensing, is characterized by the secretion and group-wide detection of small diffusible signal molecules called autoinducers. This mechanism allows cells to coordinate their behavior in a density-dependent manner. A quorum-sensing cell may directly respond to the autoinducers it produces in a cell-autonomous and quorum-independent manner, but the strength of such self-sensing effect and its impact on bacterial physiology are unclear. Here, we explored the existence and impact of self-sensing in the Bacillus subtilis ComQXP and Rap-Phr quorum-sensing systems. By comparing the quorum-sensing response of autoinducer-secreting and non-secreting cells in co-culture, we found that secreting cells consistently showed a stronger response than non-secreting cells. Combining genetic and quantitative analyses, we demonstrated this effect to be a direct result of self-sensing and ruled out an indirect regulatory effect of the autoinducer production genes on response sensitivity. In addition, self-sensing in the ComQXP system affected persistence to antibiotic treatment. Together, these findings indicate the existence of self-sensing in the two most common designs of quorum-sensing systems of Gram-positive bacteria.
Introduction
In bacterial quorum-sensing systems, a secreted, diffusible signal molecule called an autoinducer, activates a cognate receptor to control a wide array of quorum-sensing responses in a density-dependent manner [1]. The design of quorum-sensing systems may allow the receptor to interact cell autonomously with autoinducer produced by the same cell. The overall concentration sensed by the receptor will be the sum of the local effect and the average concentration in the environment (Supplementary Discussion) [2]. We term the autocrine component, self-sensing, to distinguish it from the non-autonomous, quorum-sensing sources [3]. Mathematical analysis suggests that the self-sensing may depend on the rate of autoinducer secretion, diffusion and degradation, and on possible compartmentalization of the autoinducer and its receptor (Supplementary Discussion).
Recently, strong self-sensing was observed in a synthetic yeast quorum-sensing system based on the alpha mating factor [4], but the impact of self-sensing in endogenous quorum-sensing systems has not been significantly explored. This work aimed to explore the existence and impact of self-sensing in the quorum-sensing, Gram-positive bacterium B. subtilis. This species codes for two types of quorum-sensing systems; ComQXP and Rap-Phr, which represent the two main types of quorum-sensing families found in Gram-positive bacteria – a membranal receptor sensing a long or modified peptide and a cytoplasmic receptor sensing a short unmodified peptide (Supplementary Fig. 1) [5, 6]. Both the ComQXP system and many Rap-Phr systems control the response regulator ComA, which controls the production of surfactin and the induction of the K-state through its regulation of the srfA operon [7]. In the K-state, bacteria become competent for DNA transformation and persist in the presence of antibiotics [8, 9].
The ComQXP system is encoded by the comQXP operon. In this system, the ComX autoinducer is encoded by the comX gene and post-translationally cleaved and prenylated by ComQ [10–12]. ComX binds to and activates ComP, a membranal histidine kinase receptor, which then phosphorylates the response regulator ComA [13]. The Rap-Phr systems on the other hand, code for cytoplasmic Rap receptors and short, unmodified Phr autoinducers. The Phr autoinducer is expressed as a pre-peptide, which is secreted through the general secretory pathway and undergoes further extracellular cleavage events to form the mature, unmodified autoinducer peptide. The Phr peptide is then imported through the oligopeptide permease system into the cytoplasm, where it prevents its cognate Rap receptor from repressing its target response regulators and is degraded by peptidases.
Here, we found that in both systems, the autoinducer-secreting cells had a stronger quorum-sensing response than non-secreting cells when co-cultured. A combination of genetic and quantitative analyses ascribed this difference to a self-sensing mechanism, as opposed to a regulatory one.
Results
Autoinducer-secreting cells have a stronger quorum-sensing response than non-secreting cells
To monitor for the effect of self-sensing, we adopted an approach previously used to analyze a synthetic yeast cell-cell signaling system [4], where autoinducer-secreting and non-secreting cells, coding for the same reception pathway, were co-cultured in well-mixed conditions (Fig. 1a). This process ensures that the two cell types are exposed to the same average concentration of autoinducer, and any difference in the response of the two strains can then be attributed to a cell autonomous effect.
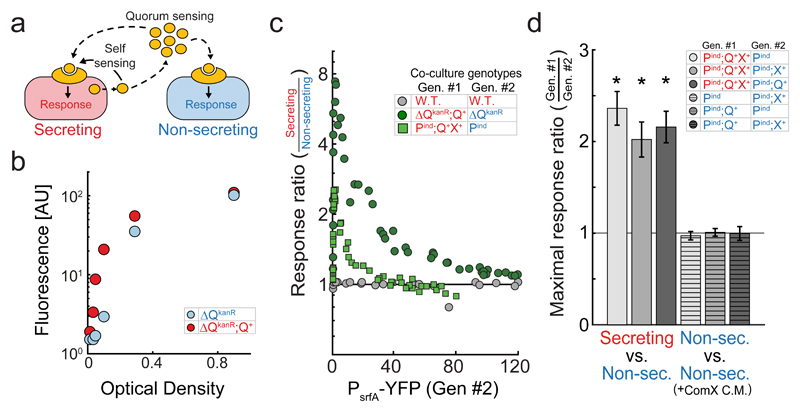
Figure 1. In co-culture, ComX-secreting cells have a stronger quorum-sensing response than non-secreting cells.
(a) Quorum- and self-sensing can be decoupled by co-culturing fluorescently distinguishable secreting and non-secreting strains, both encoding for a fluorescent reporter of quorum sensing response. Secreting and non-secreting strains are correspondingly denoted by a red and blue colors throughout the paper. (b) ComX-secreting (ΔQkanR;Q+) and non-secreting (ΔQkanR) PsrfA-YFP reporter expression, measured simultaneously at different optical densities during co-culture, by introducing distinguishing fluorescent markers into the strains (Supplementary Fig. 4). (c) Quorum-sensing response ratios of ComX- secreting and non-secreting variants in co-culture as a function of YFP fluorescence of the non-secreting strain. Shown are results for the overexpressed system (dark green) and physiologically expressed system (light green). Additionally, results are shown for a control co-culture comprised of a pair of wild-type strains (gray). (d) Maximal response ratio (mean±50th percentile expected variation) for six co-cultures of ComQXP variants, which differ in the presence or absence of the comQ and comX genes, as described in the table. Maximal values are calculated by interpolation from response profiles presented in Supplementary Fig. 6. Conditioned medium from a ComX-producing E. coli strain was added to co-cultures of two non-secreting strains. Asterisk mark results which are statistically different from a ratio of one (strict quorum-sensing null hypothesis). Results in (c) were taken from ≥4 biological repeats for each co-culture pair. In (c,d), each co-culture pair was measured over ≥5 time points at different optical densities (All data is given in Supplementary File 1).
A previous study on B. subtilis which used a similar approach, found that during co-culture, a wild-type autoinducer-secreting strain had a weaker quorum-sensing response (as measured by srfA promoter activity) than a non-secreting strain. This was interpreted to imply that the signaling genes repressed the reception of the signal [14]. By direct measurement of RNA levels and complementation analysis, we found that the non-secreting mutant used in that work (a disruption of the comQ gene with a kanamycin resistance cassette, designated as ΔQKanR below [14–16], Supplementary Fig. 2a) had a polar effect, leading to increased expression of the downstream comX and comP genes (Supplementary Fig. 2b,c). This polar effect is sufficient to explain the co-culture results of the previous work (Supplementary Fig. 2d).
To overcome the difference in ComP expression, we utilized the ΔQKanR allele and a comQ complementation strain (ΔQKanR;Q+) to study the interaction between strong ComX-secreting and non-secreting variants in a background where the quorum-sensing receptors are equally over-expressed (Supplementary Fig. 2b, Supplementary Fig. 3). To this end, the two strains were co-cultured, and PsrfA-YFP reporter activity was measured in both strains simultaneously using flow cytometry [17]. Additional fluorescent reporters marked each cell type with no significant effect on the results (Supplementary Fig. 4). Strikingly, we found that YFP expression of the non-secreting ΔQKanR strain was always lower than that of the ComX-secreting ΔQKanR;Q+ strain (Fig. 1b,c, Supplementary Fig. 4). The quorum-sensing response ratio, measured as the ratio of mean YFP expression between the co-cultured strains, rapidly increased to a maximal level of ˜8 at intermediate-low cell densities and then gradually decreased with increasing cell densities (Fig. 1c). The initial rise in response ratio stems from the contribution of auto-fluorescence and constitutive leakiness of the pathway at low density (See below and in Supplementary Discussion).
To monitor the cell-autonomous response at physiological expression levels, we constructed a non-secreting strain expressing comP under an IPTG-inducible promoter in a ΔcomQXP background (Pind allele, methods). A secreting strain was constructed by introducing the comQX bi-cistronic operon regulated by its native promoter into a different locus (Q+X+ allele, methods). The quorum-sensing response profile of the Pind;Q+X+ strain was similar to that of the wild-type strain, when the former was induced with 100µM of IPTG, suggesting that it reflects the physiological behavior of the system (Supplementary Fig. 5). The quorum-sensing response ratio between the ComX-secreting (Pind; Q+X+) and non-secreting (Pind) strains showed a similar, but weaker, dependence on quorum-sensing response levels compared to that measured for the ΔQKanR;Q+ and ΔQKanR co-culture. This is reflected by the lower maximal response ratio, which was ˜2.5 for this pair (Fig. 1c).
The response difference between ComX-secreting and non-secreting cells is due to self-sensing
The difference in quorum-sensing response observed between ComX-secreting and non-secreting strains can be ascribed to either self-sensing (Fig. 2a) or to a regulatory over-reception effect of the signaling genes (Fig. 2b). For example, ComQ may also modify the receptor and increase its sensitivity (Supplementary Fig. 1). To distinguish between these two options, a combined genetic and quantitative analysis was performed. We first examined whether both comQ and comX are needed for the differential cell autonomous response. If one of them is sufficient, the response difference must arise from regulatory over-reception, as both are required for autoinducer secretion (Supplementary Fig. 1). To this end, we measured the quorum-sensing response ratio in all six possible co-cultures of variants that encode comP (Pind) with or without either of the genes (Fig. 1d, methods). In three co-culture pairs, in which both strains did not secrete ComX, quorum-sensing was activated by addition of conditioned medium from a ComX-producing E. coli strain (methods) [12, 15]. A difference between the quorum-sensing responses of the co-cultured strains was only observed when a ComX-secreting cell was co-cultured with a non-secreting strain (Fig. 1d, Supplementary Fig. 6). Thus, the cell autonomous effect requires both the comQ and comX genes.
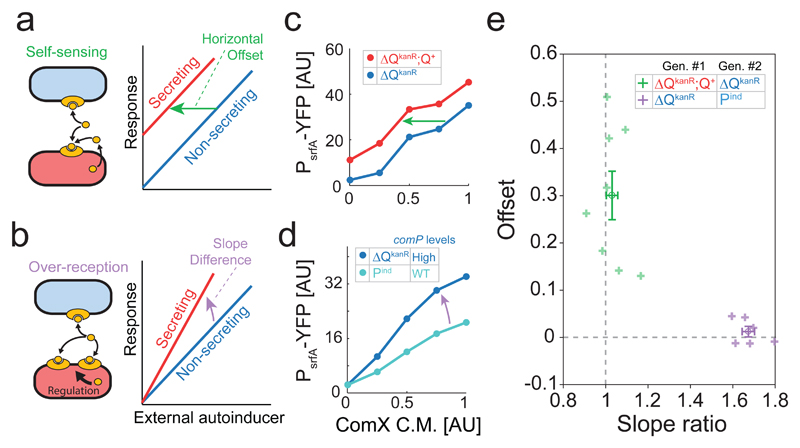
Figure 2. The cell autonomous effect of ComX-secretion fits a self-sensing model with no over-reception.
(a,b) Schemes of the self-sensing (a) and over-reception (b) models and the expected difference between the responses of secreting and non-secreting strains to an external autoinducer under the two models. (c,d) YFP expression of each strain during co-culture, as a function of the relative volume of ComX-conditioned medium added (methods). (c) ComX-secreting (ΔQkanR;Q+, red) and Non-secreting (ΔQkanR, blue). (d) Non-secreting strains with High (ΔQkanR, dark blue) and low (Pind, light blue) ComP levels. (e) Optimal slope ratio and x-offset between co-cultured strains, under a model that allows the two parameters to vary independently. Shown is the fit for the ΔQkanR;Q+ (ComX-secreting) and ΔQkanR (non-secreting) pair (purple, n=8) and for the ΔQkanR (High ComP levels) and Pind (Low ComP levels) pair (green, n=6). Average ± St. Err. marks are shown for each pair of strains in a darker tone. Each point in (e) arises from a biological repeat of a series of measurements as shown in (c,d). Experiments were repeated over multiple days. All data points are given in Supplementary File 1.
Self-sensing should lead to a measureable response in an autoinducer-secreting strain even at very low densities, where the average autoinducer concentration is insufficient to elicit a quorum-sensing response. Theory, however, predicts that the level of self-sensing may depend on measurement sensitivity and the steepness of the autoinducer response curve (Supplementary Discussion). Due to these reasons, we have used the comQXP overexpression strains (ΔQKanR;Q+ and ΔQKanR) to study self-sensing at low cell densities. In agreement with a self-sensing model, we find that at very low cell density (an optical density of ˜0.001, methods), the strong quorum-sensing autoinducer secreting variant had a higher response than its co-cultured non-secreting variant (Supplementary Fig. 7, p<10-6, two sampled t-test, n=42). We find that quorum-sensing was negligible at the cell densities used, as the reporter expression level of each strain was the same in co-culture and pure culture (Supplementary Fig. 7, p>0.1, two-sampled t-test, n>30 for both comparisons). Notably, the non-secreting strain had a small, but measurable, expression level above auto-fluorescence, (p<10-5, two-sampled t-test, n=27).
To further study the relative impact of self-sensing and over-reception, we examined another expected difference between self-sensing and over-reception. A general model of quorum-sensing response (see model in Supplementary Discussion) predicts that the addition of external autoinducer would introduce a constant horizontal shift between the response curves of autoinducer-secreting and non-secreting strains if the difference between strains arise from self-sensing (Fig. 2a). In contrast, over-reception is expected to lead to a steeper response curve of the autoinducer-secreting strain than that of the non-secreting strain (Fig. 2b).
To quantify the self-sensing and over-reception components of the cell-autonomous response, the strong ComX-secreting (ΔQKanR;Q+) and non-secreting (ΔQKanR) variants were co-cultured at low density. Response curves of each strain to external ComX were measured by adding varying volumes of conditioned medium, collected from a ComX-producing E. coli strain (Fig. 2c, Methods). As a control, we similarly measured the response curves of two non-secreting strains known to differ in comP expression levels (Fig. 2d).
The response curves of each co-cultured pair were fitted to a model which allowed both for a left shift and a change of slope between the curves (Methods). Compared to the non-secreting strain, the response curve of the ComX-secreting strain was shifted to the left by a fraction of 30%±15% (Fig. 2e, average +/- st. dev, p<10-3, t-test, n=8) of the maximal ComX levels in the conditioned medium, but with no significant difference in the slope of the curves (p=0.3, t-test, n=8). In contrast, the ComP over-expressing strain (ΔQKanR) displayed a difference in the slope of the response curve compared to that of the physiologically expressing ComP strain (Pind, Fig. 2d), with an average slope ratio of 1.67±0.07 (Fig. 2e, average ± st. dev., p<10-7, t-test, n=6), but with no significant shift between the curves (p=0.32, t-test, n=6). Together, these results strongly suggest that the cell-autonomous response of a ComX-secreting strain is due to self-sensing.
Autoinducer self-sensing results in elevated antibiotic persistence during co-culture
As the impact of self-sensing on gene expression occurs mostly at low cell densities, it is unclear whether it would have an observable physiological effect. ComA-dependent induction of the slow growing K-state raises the possibility that self-sensing may lead to increased persistence to antibiotic treatment [8, 9]. To asses this effect, the persistence levels of the strong (ΔQkanR;Q+) and physiological (Pind;Q+X+) quorum-sensing strains and of the non-secreting (ΔQkanR) strain were first monitored in pure cultures by transient exposure to the cell-wall targeting antibiotic ampicillin (Methods). In all three strains, persistence increased with cell density and, at a given density, was always highest in the strong quorum-sensing strain and lowest in the ΔQKanR autoinducer-secreting mutant (Supplementary Fig. 8a). These findings ascribe a physiological role to the ComQXP system, at low cell densities where self-sensing is apparent.
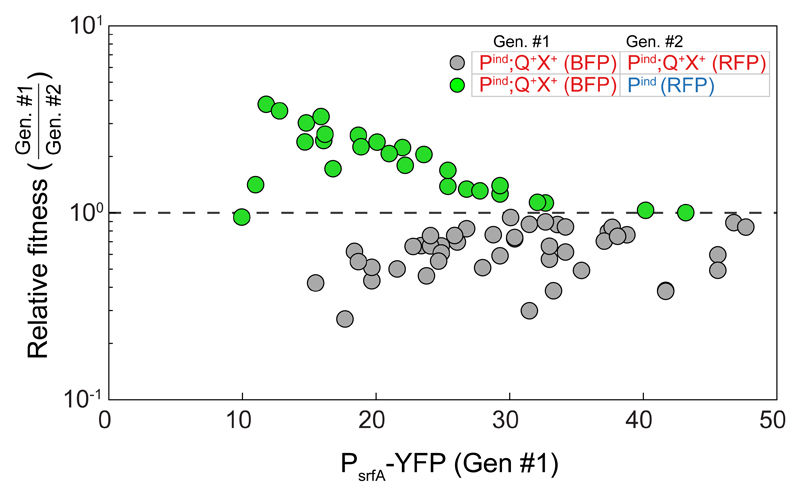
Next, we co-cultured the physiological ComX-secreting (Pind;Q+X+) and non-secreting (Pind or Pind;Q+) strains and measured their relative fitness following the transient admission of ampicillin. These measurements were correlated with measurements of the co-culture’s density or PsrfA-YFP expression of the secreting strain (Fig. 3, Supplementary Fig. 8b, Methods). Under these conditions, the secreting strain was more fit than its co-cultured non-secreting strain (Fig. 3, green, two-sample t-test, p<10-12, n=76), despite carrying a BFP marker, which has a fitness disadvantage compared to the RFP marker (Fig. 3, gray, Supplementary Fig. 8c, p<10-13, ANCOVA analysis, n=49). Despite the positive correlation between increased level of persistence and optical density, the relative fitness of the secreting strain was reduced with increasing cell density and response levels (ANCOVA, p=0.02 two-tailed test, n=28). This corresponds with the reduction in the quorum-sensing response ratio between the two strains. Finally, a similar but much stronger effect was observed when the quorum-sensing overexpressing ComX-secreting (ΔQkanR;Q+) and non-secreting (ΔQkanR) strains were co-cultured (Supplementary Fig. 8b). In summary, these results indicate that self-sensing results in enhanced persistence at low cell densities.
Figure 3. Self-sensing contributes to antibiotic persistence.
The relative fitness of co-cultured strains is plotted as a function of the PsrfA-YFP expression of genotype #1 prior to administration of antibiotics (see legend). Relative fitness was calculated as the ratio of relative frequency of the strains at the end of the experiment to that prior to the addition of antibiotics [19]. Shown are results for two differentially marked Pind;Q+X+-secreting strains (gray), and between the physiological ComX-secreting (Pind;Q+X+) and the corresponding non-secreting (Pind) strains (green). A value of one (dashed line) indicates no change in frequency. Each data point represents a separate measurement. Data was collected over ≥3 different optical densities in each series of experiments. Experiments were repeated multiple times over ≥3 days for each co-culture type. All data points are given in Supplementary File 1.
Self-sensing is a general feature of B. subtilis quorum-sensing systems
To assess whether self-sensing is a general feature of B. subtilis quorum-sensing systems, the ComA-regulating RapP-phrP was used as a model for the second major family of quorum-sensing systems found in its genome [17–19]. A rapP+ strain was constructed by using an active rapP allele [17], whose native, ComA-regulated promoter was replaced by the constitutive comQXP promoter, to prevent a ComA-dependent negative feedback on reception [17]. This allele strongly represses the expression of the PsrfA-YFP reporter (Supplementary Fig. 9a); Introduction of the phrP gene, under the control of an inducible promoter (phrP+ allele), into the genome of this strain restored YFP expression to near wild-type levels (Supplementary Fig. 9a).
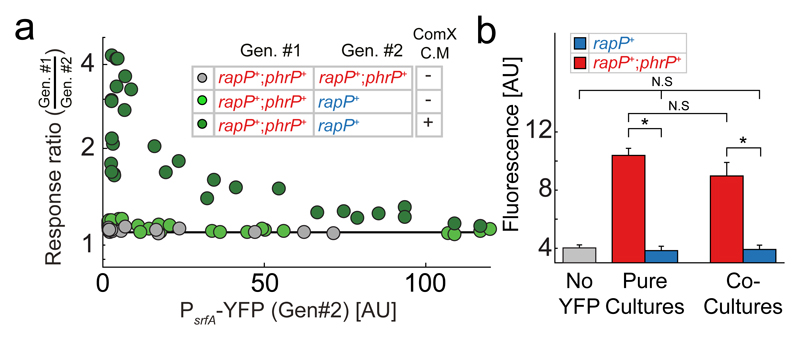
To monitor self-sensing, the PhrP-secreting (rapP+;phrP+) and non-secreting (rapP+) strains were co-cultured and quorum-sensing response ratios at different optical densities, were measured. Obtained response ratios were very close to one, suggesting a negligible cell-autonomous effect (Fig. 4a, Supplementary Fig. 9b,c). Since Rap repression of ComA activity is dependent on prior phosphorylation of ComA by the ComQXP quorum-sensing system, the lack of a substantial cell-autonomous effect may stem from the low levels of phosphorylated ComA at low densities (Supplementary Fig. 1) [20]. Response ratios were therefore re-measured in the presence of ComX-conditioned medium. Under these conditions, the PhrP-secreting strain had a significantly higher quorum-sensing response than the non-secreting strain at low densities (Fig. 4a). The maximal response-ratio was dependent on the level of ComX in the medium and reached a maximal factor of four at high concentrations of exogenously added ComX (Fig. 4a, Supplementary Figs. 9b,c).
Figure 4. Self-sensing in also apparent in the Rap-Phr system.
(a) Co-culture response ratio of PhrP secreting (rapP+;phrP+) and non-secreting (rapP+) strains as a function of YFP response of the latter strain. Shown are results with (dark green) or without (light green) the addition of conditioned medium collected from a ComX-producing E. coli. Response ratio of a control co-culture of two phrP+ secreting strains is also shown (gray). Each data point represents a separate measurement. Series of experiments over ≥3 varying optical densities were repeated ≥3 times, on different days. Line at response ratio of 1 represents the null hypothesis of no cell-autonomous response. (b) Fluorescence levels (mean ± st. err.) of a PsrfA-YFP reporter integrated into PhrP secreting (red) and non-secreting (blue) strains. Response was measured at very low densities with ComX conditioned medium added to the culture (methods). The response is measured for pure cultures of the two strains and in co-culture. Also shown is the auto-flurescence of a similarly measured wild-type with no YFP reporter (gray). Note that auto-fluorescence here is larger than that measured in Supplementary Fig. 7, due to the addition of conditioned medium. Asterisks and N.S. mark statistically significant and non-significant differences accordingly. All data points are given in Supplementary File 1.
To verify that the cell-autonomous effect depends on self-sensing, we measured PhrP-secreting and non-secreting strains expression at very low densities (optical density ˜0.001, methods), where quorum-sensing is insignificant (Fig. 4b). As for the ComQXP system, this was verified by the lack of significant difference between expression of the strains in pure and co-cultures (p>0.15, two-sample t-test, n=24 for both comparisons). We found that the PhrP-secreting strain had an expression level significantly higher than auto-fluorescence (p=3×10-4, two-sample t-test, n=20). In contrast, no significant difference was found between the expression of the non-secreting strain and background auto-fluorescence (p>0.75, two-sampled t-test, n=16), indicating that expression of the non-secreting strain is below the measurement errors. This leads to a background-subtracted response-ratio which is larger than 10, strongly suggesting that self-sensing underlies the cell autonomous effect of PhrP.
The extracellular maturation of the Phr peptide supposedly protects it from direct interaction with its intracellular receptor [5]. Self-sensing in this system may result from secretion failure which lead to direct intracellular interaction between the Phr peptide (or pre-peptide) and the corresponding Rap receptor [21] (Supplementary Fig. 1). To test the feasibility of this scenario, we constructed an inducible PhrPint allele, which lacks the N-terminal secretion signal sequence of the pre-PhrP peptide and codes only for the last 15 amino-acids of pre-PhrP (Methods). Expression of the PhrPint allele together with RapP, fully restored PsrfA-YFP reporter expression, but did not result in any activation of quorum-response of a co-cultured rapP+ strain (Supplementary Fig. 9a, Supplementary Fig. 10), supporting notion that secretion failure leads to self-sensing in this system.
Discussion
Formation of concentration gradients around autoinducer-secreting cells is a natural consequence of the diffusion process, even in a well-mixed environment. However, our theoretical estimation suggests that a self-sensing mechanism which is exclusively diffusion-based cannot explain the level of self-sensing observed in our analysis of both the ComQXP and Rap-Phr systems (Supplementary Discussion). Enhanced self-sensing may occur if the autoinducer and the receptor interact within a subcellular compartment (Supplementary Discussion), but the design of both B. subtilis quorum-sensing systems seems to prevent compartmentalization of signal and receptor within the cytoplasm. For the Rap-Phr system, the presented data suggest that self-sensing may arise from pre-peptide secretion failures, which will lead to cytoplasmic co-occurrence of Rap and the cell-autonomously produced mature Phr peptide (Supplementary Fig. 9, Supplementary Fig. 10). The impact of self-sensing in the Rap-Phr system is mitigated in the wild-type, by co-regulation of ComA by the ComQXP system (Supplementary Fig. 9b,c) and by the transcriptional regulation of many Rap-Phr systems by ComA [17, 22]. The underlying mechanism for self-sensing in the ComQXP system is unknown. However, the membranal localization of the ComP receptor and the hydrophobicity of the ComX prenyl chain suggest that self-sensing in the ComQXP system may occur through membranal compartmentalization of the receptor with the autoinducer prior to its secretion (Supplementary Discussion) [11–13].
The specific mechanisms underlying quorum sensing can have a significant impact on the quantitative aspects of quorum-sensing response and on its evolutionary fate [23, 24], raising the question whether self-sensing is an adaptive feature of these systems, and results from a direct selective pressure. Quorum sensing is known to control various types of activities with a different impact on individual and group fitness [25]. Self-sensing is disadvantageous when controlling a public benefit, but provides an advantage when controlling private traits, such as antibiotic persistence (Fig. 3). This explanation for the existence of self-sensing is problematic, as a similar private benefit would arise from constitutive activation of the quorum-sensing regulated factors [26]. Finally, some activities are intermediate between individual and public, leading to a selective advantage for the trait in aggregated, but not in planktonic form [27–29]. A hydrophobic autoinducer, such as ComX, may better inform cells on their aggregate status than a hydrophilic one [30]. In this case, self-sensing would be a tolerable side effect of aggregation-sensing.
Altogether, our results demonstrate that self-sensing is observed in the two most common designs of Gram-positive quorum-sensing systems – a membranal extracellular receptor, with a modified or long peptide autoinducer (ComQXP) and a cytoplasmic receptor, with an unmodified peptide autoinducer (Rap-Phr). Theoretically, these designs better compartmentalize signal production and sensing than the design of Acyl homoserine lactone based systems, where both signal production and reception are intracellular, yet they still show a self-sensing behavior. Further work will be required to identify the mechanisms underlying self-sensing, its impact on the design and evolution of quorum-sensing systems and its prevalence in other types of quorum-sensing systems.
Methods
Growth media
Routine B. subtilis growth was performed in Luria–Bertani (LB) broth: 1% tryptone (Difco), 0.5% yeast extract (Difco), 0.5% NaCl. Experiments were performed using Spizizen minimal medium (SMM; 2 g L−1 (NH4)2SO4, 14 g L−1K2HPO4, 6 g L−1KH2PO4, 1 g L−1disodium citrate, 0.2 g L−1MgSO4⋅7H2O), supplemented with trace elements (125 mg L−1MgCl2⋅6H2O, 5.5 mg L−1CaCl2, 13.5 mg L−1FeCl2⋅6H2O, 1 mg L−1MnCl2⋅4H2O, 1.7 mg L−1ZnCl2, 0.43 mg L−1CuCl2⋅4H2O,0.6 mg L−1CoCl2⋅6H2O, 0.6 mg L−1Na2MoO4⋅2H2O). 0.5% glucose served as a carbon source. E. coli cultures were grown for their conditioned medium in M9 Minimal medium (12.8 g L-1 Na2HPO4*7H2O, 3 g L-1 KH2PO4, 0.5 g L-1 NaCl, 1 g L-1 NH4Cl, 1 mM MgSO4 and 0.1 mM CaCl2), supplemented with 0.4% glucose (Merck) as a carbon and energy source. When preparing plates, medium was solidified by addition of 1% agar. Antibiotics were added (when necessary) at the following concentrations: spectinomycin: 100 µg ml-1, tetracycline: 10 µg ml-1, chloramphenicol: 5 µg ml-1, kanamycin: 10 µg ml-1, erythromycin: 3 µg ml-1, phleomycin: 2.7 µg ml-1, MLS: 3 µg ml-1 erythromycin + 25 µg ml-1 lincomycin, ampicillin for E. coli: 100 µg ml-1. Isopropyl-β-D-thiogalactopyranoside (IPTG- Sigma) was added to the liquid medium when appropriate, at the concentrations indicated in the text.
Strain constructions
All bacterial strains used are listed in Table 1. Table 2 lists all the primers used in this study. In order to construct new B. subtilis strains, standard transformation and Spp1 transduction protocols were used for genomic integration and plasmid transformation [31]. To generate the amyE::Phs-comP construct, the open reading frame (ORF), with its native ribosome binding site (RBS), was amplified from the PY79 strain using the ComP-NheI-R & ComP-native-RBS-F primer pair. After the amplification, the DNA fragment and the pDR111 vector were digested with NheI-HF and SalI-HF restriction enzymes (NewEngland BioLabs), followed by ligation. The final construct has the insert downstream of a hyperspank inducible promoter found in the pDR111 vector. To generate sacA::PcomQXP-comQX and sacA::PcomQXP-comQ constructs, the native promoter of the comQXP operon with the ORF of comQX or comQ only, were amplified from the PY79 strain, using the forward primer ComQ-into-ECE174-F-[BamHI], either with ComQX-into-ECE174-R-[EcoRI] or with ComQ-into-ECE174-R-[EcoRI], respectively, and cloned into plasmid ECE174 using the designated restriction enzymes. The sacA::PcomQXP-comX construct was generated by amplifying the whole ECE174::PcomQXP-ComQX (AEC840) plasmid without the comQ ORF, using the dcomQ-R and dcomQ-F primer pair. The dcomQ-R primer exists at the end of comQ in the forward direction, and dcomQ-F primer exists at the beginning of comQ in the reverse direction. The amplified fragment was treated with DpnI and then T4 Polynucleotide Kinase (NewEngland BioLabs), followed by self-ligation. To generate the pDL30::PcomQXP-rapPT236N construct, the ORF, with its native RBS, was amplified from the ECE174::PrapP-rapPT236N (AEC735) plasmid using the RapP-SphI-R & hsRapP-F primer pair. In addition, the DNA fragment pDL30::PcomQXP was amplified from the pDL30::PcomQXP-3xYFP (AEC962) plasmid using the PQXP-NheI-R & pDL30-SphI-F primer pair. After the amplification, the DNA fragments were digested with NheI-HF and SphI-HF restriction enzymes, followed by ligation. To generate the pDR111::Phs-phrP construct, the ORF, with its native RBS, was amplified from the NCIB3610 strain using the PhrP-SalI-F & PhrP-NheI-R primer pair. After the amplification, the DNA fragment and the pDR111 vector were digested with NheI-HF and SalI-HF restriction enzymes, followed by ligation. To generate pDR111::Phs-phrPint, the entire pDR111::Phs-phrP (AEC1272) plasmid was amplified using PhrP-NO-signal-seq-R & PhrP-NO-signal-seq-F primer pair. The purified DNA was treated with DpnI and then T4 Polynucleotide Kinase, followed by self-ligation
Flow cytometry analysis
Flow cytometry was performed to quantify gene expression at the single-cell level, using a Beckman-Coulter Gallios flow-cytometer equipped with four lasers (405 nm, 488 nm co-linear with 561 nm, 638 nm). The emission filters used were: BFP – 450/50, YFP – 525/40, mCherry – 620/30.
Two methods were used to distinguish between co-cultured cells. In Figs. 1B,C and Supplementary Fig. 2, cells were distinguished by the expression of a constitutive ppsB::(PtrpE-mCherry Ph) construct [24]. Alternatively, plasmids carrying either an mCherry or mTag2-BFP genes under a constitutive promoter were introduced into each of the co-cultured genotypes. This method was used to gather the rest of the data. We note that in both cases, all co-cultures of different genotypes were performed with the two swapped options of distinguishing reporter with the same results (See Supplementary Fig. 4).
YFP levels were measured relative to a set voltage which was approximately set such that a value of 1 will be given to auto-fluorescence of strain PY79 in SMM medium. Detailed analysis of auto-fluorescence was done when needed (Supplementary Fig. 7 and Fig. 4b).
Growth protocols
Cells were grown to OD600 <0.1, in SMM medium containing trace elements and glucose, then diluted by a factor of 106 or 107 into fresh SMM medium, and grown for about 16 hours in exponential phase. In co-culture gene expression experiments, each strain was grown from a single colony in SMM to OD600 <0.1, or diluted to 0.1 prior to strain mixing. The two strains were mixed at equal volumes and then diluted by a factor of 106 or 107 in fresh SMM medium. Samples were taken from cultures at several time points. At each time point OD600nm was measured using a spectrophotometer and fluorescence was measured using a flow cytometer.
For measurements at very low densities (Fig. 4b, Supplementary Fig. 7), cells were grown over night for only 13 hours. They were then further diluted by a factor of 10 and grown for an additional hour, prior to first measurements (Supplementary Fig. 7) or to the addition of conditioned medium (Fig. 4b). Optical density at time of measurement was estimated by measuring optical density of the culture at later times and back extrapolation.
Conditioned medium assays
An MG1655 E. coli strain containing the ECE174::PcomQXP-comQX plasmid was grown in M9 minimal medium with ampicillin to an OD600 of >1. The cells were centrifuged and the supernatant was filtered through an 0.45 µm filter. To generate dilutions of the ComX-containing conditioned medium, the MG1655 WT strain was grown at the same time under identical conditions. Different concentrations [0%, 25%, 50%, 75% and 100%] of the ComX-containing conditioned medium were prepared by mixing volume fractions of the ComX-containing supernatant with the WT supernatant. The sterile, conditioned medium was added to the B. subtilis growth medium ˜16 hours after the B. subtilis strains were mixed and diluted, and YFP expression levels of both strains were measured ˜1hr later or more. When conditioned medium was added to B. subtilis cultures, the cell density was still low.
Persistence assay
B. subtilis strains were diluted by a factor of 106 or 107 in fresh SMM, supplemented with glucose and trace elements. The optical density was measured 16 hours thereafter and serial dilutions of the cells were plated on LB agar to determine colony forming units (CFU)/ml before the antibiotic treatment. Ampicillin (1mg/ml) was then added to the growth medium. The cells were incubated for one hour and then washed twice in SMM by centrifuging and re-suspending. Serial dilutions of the treated and washed cells were plated on LB agar. The percentage of the persistent cells was calculated by dividing the CFU/ml after the antibiotic treatment, by the CFU/ml determined before antibiotic treatment.
In co-culture assays, each strain was diluted to an OD of 0.1 before co-culturing. The co-cultured strains were grown and treated with antibiotics, as described above. YFP expression levels and frequency of each strain were measured before the antibiotic was added. In addition, frequency of each strain was measured after re-growth overnight in fresh SMM. Relative fitness was calculated as the ratio of relative frequency of the strains at the end of the experiment to that prior to the addition of antibiotics [19].
Real Time qPCR Measurements
Total RNA was extracted from B. subtilis PY79 cells, using a High Pure RNA Isolation kit (Roche). To this end, cells were grown in SMM, supplemented with glucose and trace elements, to an OD of 0.5-0.8. One microgram of RNA was reverse-transcribed to cDNA using a qScript™ cDNA Synthesis Kit (Quanta BioSciences). Real-time qPCR was performed on a Step One Plus Real Time PCR system (Applied Biosystems), using SYBR Green (Roche). The transcript level of comP was normalized to levels of the reference genes: rpoB, bglA. Results were analyzed using the Step One™ V2.3 software. Each strain was measured across three biological repeats, where each biological repeat included three technical repeats.
Data analysis
Relative left-shift and slope ratio (Fig. 2e) were calculated by fitting each response curve to a line and then calculating the shift and change in slope between the two lines. Notably, in most cases, response curves were nearly linear, justifying this linear analysis (Supplementary File 1, Supplementary Discussion).
Supplementary Material
Acknowledgments
This work was supported by European Research Council grants 281301 and 724805. We thank Ron D. Oshri and Niv Antonovsky for comments and Nadejda Sigal for help with qPCR.
Footnotes
Data availability
All data generated or analyzed during this study are included in this published article (and its supplementary information files).
Authors contribution
T.B.,S.P. and A.E. designed the experiments, T.B. performed the experiments, T.B., S.P. and A.E. analyzed the data and wrote the manuscript.
References
- 1.Waters CM, Bassler BL. Quorum Sensing: Cell-to-Cell Communication in Bacteria. Annual Review of Cell and Developmental Biology. 2005;21(1):319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 2.Berg HC. Random walks in biology. Princeton University Press; 1993. [Google Scholar]
- 3.Doğaner BA, Yan LK, Youk H. Autocrine signaling and quorum sensing: Extreme ends of a common spectrum. Trends in cell biology. 2016;26(4):262–271. doi: 10.1016/j.tcb.2015.11.002. [DOI] [PubMed] [Google Scholar]
- 4.Youk H, Lim WA. Secreting and sensing the same molecule allows cells to achieve versatile social behaviors. Science. 2014;343(6171):1242782. doi: 10.1126/science.1242782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pottathil M, Lazazzera BA. The extracellular Phr peptide-Rap phosphatase signaling circuit of Bacillus subtilis. Front Biosci. 2003;8:d32–45. doi: 10.2741/913. [DOI] [PubMed] [Google Scholar]
- 6.Grossman AD. Genetic Networks Controlling the Initiation of Sporulation and the Development of Genetic Competence in Bacillus Subtilis. Annual Review of Genetics. 1995;29(1):477–508. doi: 10.1146/annurev.ge.29.120195.002401. [DOI] [PubMed] [Google Scholar]
- 7.Comella N, Grossman AD. Conservation of genes and processes controlled by the quorum response in bacteria: characterization of genes controlled by the quorum-sensing transcription factor ComA in Bacillus subtilis. Molecular Microbiology. 2005;57(4):1159–1174. doi: 10.1111/j.1365-2958.2005.04749.x. [DOI] [PubMed] [Google Scholar]
- 8.Yüksel M, et al. Fitness Trade-Offs in Competence Differentiation of Bacillus subtilis. Frontiers in Microbiology. 2016;7 doi: 10.3389/fmicb.2016.00888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnsen PJ, Dubnau D, Levin BR. Episodic selection and the maintenance of competence and natural transformation in Bacillus subtilis. Genetics. 2009;181(4):1521–1533. doi: 10.1534/genetics.108.099523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Magnuson R, Solomon J, Grossman AD. Biochemical and genetic characterization of a competence pheromone from B. subtilis. Cell. 1994;77(2):207–216. doi: 10.1016/0092-8674(94)90313-1. [DOI] [PubMed] [Google Scholar]
- 11.B K, Palmer TM, Grossman AD. Characterization of comQ and comX, Two Genes Required for Production of ComX Pheromone in Bacillus subtilis. J Bacteriol. 2002;184(2):410–419. doi: 10.1128/JB.184.2.410-419.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ansaldi M, et al. Specific activation of the Bacillus quorum-sensing systems by isoprenylated pheromone variants. Molecular Microbiology. 2002;44(6):1561–1573. doi: 10.1046/j.1365-2958.2002.02977.x. [DOI] [PubMed] [Google Scholar]
- 13.Piazza F, Tortosa P, Dubnau D. Mutational analysis and membrane topology of ComP, a quorum-sensing histidine kinase of Bacillus subtilis controlling competence development. Journal of bacteriology. 1999;181(15):4540–4548. doi: 10.1128/jb.181.15.4540-4548.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oslizlo A, et al. Private link between signal and response in Bacillus subtilis quorum sensing. Proceedings of the National Academy of Sciences. 2014;111(4):1586–1591. doi: 10.1073/pnas.1316283111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tortosa P, et al. Specificity and Genetic Polymorphism of the Bacillus Competence Quorum-Sensing System. J Bacteriol. 2001;183(2):451–460. doi: 10.1128/JB.183.2.451-460.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Msadek T, et al. DegS-DegU and ComP-ComA modulator-effector pairs control expression of the Bacillus subtilis pleiotropic regulatory gene degQ. Journal of bacteriology. 1991;173(7):2366–2377. doi: 10.1128/jb.173.7.2366-2377.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bendori SO, et al. The RapP-PhrP Quorum-Sensing System of Bacillus subtilis Strain NCIB3610 Affects Biofilm Formation through Multiple Targets, Due to an Atypical Signal-Insensitive Allele of RapP. Journal of bacteriology. 2015;197(3):592–602. doi: 10.1128/JB.02382-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parashar V, et al. A plasmid-encoded phosphatase regulates Bacillus subtilis biofilm architecture, sporulation, and genetic competence. Journal of bacteriology. 2013;195(10):2437–2448. doi: 10.1128/JB.02030-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pollak S, Bendori SO, Eldar A. A complex path for domestication of B. subtilis sociality. Current genetics. 2015:1–4. doi: 10.1007/s00294-015-0479-9. [DOI] [PubMed] [Google Scholar]
- 20.Baker MD, Neiditch MB. Structural basis of response regulator inhibition by a bacterial anti-activator protein. PLoS-Biology. 2011;9(12):2624. doi: 10.1371/journal.pbio.1001226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lazazzera BA, Solomon JM, Grossman AD. An exported peptide functions intracellularly to contribute to cell density signaling in B. subtilis. Cell. 1997;89(6):917–25. doi: 10.1016/s0092-8674(00)80277-9. [DOI] [PubMed] [Google Scholar]
- 22.Lazazzera BA, et al. An autoregulatory circuit affecting peptide signaling in Bacillus subtilis. J Bacteriol. 1999;181(17):5193–200. doi: 10.1128/jb.181.17.5193-5200.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Drees B, et al. A modular view of the diversity of cell-density-encoding schemes in bacterial quorum-sensing systems. Biophysical journal. 2014;107(1):266–277. doi: 10.1016/j.bpj.2014.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Even-Tov E, et al. Social evolution selects for redundancy in bacterial quorum sensing. PLOS Biology. 2016;14(2):e1002386. doi: 10.1371/journal.pbio.1002386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schuster M, et al. Acyl-homoserine lactone quorum sensing: from evolution to application. Annual Review of Microbiology. 2013;67:43–63. doi: 10.1146/annurev-micro-092412-155635. [DOI] [PubMed] [Google Scholar]
- 26.Schuster M, Sexton DJ, Hense BA. Why Quorum Sensing Controls Private Goods. Frontiers in Microbiology. 2017;8 doi: 10.3389/fmicb.2017.00885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gore J, Youk H, van Oudenaarden A. Snowdrift game dynamics and facultative cheating in yeast. Nature. 2009;459(7244):253–256. doi: 10.1038/nature07921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koschwanez JH, Foster KR, Murray AW. Sucrose utilization in budding yeast as a model for the origin of undifferentiated multicellularity. PLoS biology. 2011;9(8):e1001122. doi: 10.1371/journal.pbio.1001122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ratzke C, Gore J. Self-organized patchiness facilitates survival in a cooperatively growing Bacillus subtilis population. Nature Microbiology. 2016:16022. doi: 10.1038/nmicrobiol.2016.22. [DOI] [PubMed] [Google Scholar]
- 30.Hense BA, et al. Does efficiency sensing unify diffusion and quorum sensing? Nat Rev Micro. 2007;5(3):230–239. doi: 10.1038/nrmicro1600. [DOI] [PubMed] [Google Scholar]
- 31.Harwood CR, Cutting SM. Molecular biological methods for Bacillus. Wiley; 1990. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.