Graded induction of regulatory light chain (RLC) activators MLCK and ROCK were used to explore the relationship between RLC phosphorylation and actin-myosin stress fiber viscoelasticity. MLCK controls peripheral stress fiber mechanics by monophosphorylation of RLC, whereas ROCK acts on central stress fibers via diphosphorylation.
Abstract
The assembly and mechanics of actomyosin stress fibers (SFs) depend on myosin regulatory light chain (RLC) phosphorylation, which is driven by myosin light chain kinase (MLCK) and Rho-associated kinase (ROCK). Although previous work suggests that MLCK and ROCK control distinct pools of cellular SFs, it remains unclear how these kinases differ in their regulation of RLC phosphorylation or how phosphorylation influences individual SF mechanics. Here, we combine genetic approaches with biophysical tools to explore relationships between kinase activity, RLC phosphorylation, SF localization, and SF mechanics. We show that graded MLCK overexpression increases RLC monophosphorylation (p-RLC) in a graded manner and that this p-RLC localizes to peripheral SFs. Conversely, graded ROCK overexpression preferentially increases RLC diphosphorylation (pp-RLC), with pp-RLC localizing to central SFs. Interrogation of single SFs with subcellular laser ablation reveals that MLCK and ROCK quantitatively regulate the viscoelastic properties of peripheral and central SFs, respectively. The effects of MLCK and ROCK on single-SF mechanics may be correspondingly phenocopied by overexpression of mono- and diphosphomimetic RLC mutants. Our results point to a model in which MLCK and ROCK regulate peripheral and central SF viscoelastic properties through mono- and diphosphorylation of RLC, offering new quantitative connections between kinase activity, RLC phosphorylation, and SF viscoelasticity.
INTRODUCTION
A single mammalian cell can exert tensile forces on its surroundings, which can regulate cell shape, motility, and in the case of stem cells, differentiation (Prager-Khoutorsky et al., 2011; Downing et al., 2013; Burnette et al., 2014). At the multicellular level, such forces contribute significantly to collective cell migration, tissue morphogenesis during development, and wound healing (Tamada et al., 2007; Tambe et al., 2011; Heisenberg and Bellaïche, 2013). Actomyosin stress fibers (SFs) are partly responsible for generating and transmitting these forces to the extracellular matrix (ECM) through direct attachment to focal adhesions as well as through interactions with other cytoskeletal structures (Chang and Kumar, 2013; Kassianidou and Kumar, 2015; Kassianidou et al., 2017; Soiné et al., 2015; Lee and Kumar, 2016). SFs are composed of F-actin, cross-linking proteins such as α-actinin, and in some cases, the force-generating motor protein nonmuscle myosin II (NMMII).
NMMII is composed of two essential light chains (ELCs), two regulatory light chains (RLCs), and two heavy chains (Vicente-Manzanares et al., 2009; Beach et al., 2014). Each heavy chain contains a globular head domain, which can bind to F-actin and hydrolyze ATP. This ATP hydrolysis is needed to power the contractile sliding of myosin filaments against actin filaments, leading to a build-up of tension within the SF (Sekine and Yamaguchi, 1963). Myosin motor activity and filament formation are strongly regulated by phosphorylation of RLC at Ser19 and Thr18, which allows NMMII to assemble into linear thick filaments (Vicente-Manzanares et al., 2009). Monophosphorylation (p-RLC) at Ser19 alters the conformation of the NMMII head domains to permit ATPase activity (Wendt et al., 2001) and diphosphorylation (pp-RLC) of Thr18 and Ser19 further enhances ATPase activity (Umemoto et al., 1989; Kamisoyama et al., 1994; Mizutani et al., 2006). Each RLC phosphospecies appears to play different roles in governing SF assembly and tension generation, even though both can coexist within a single SF (Beach et al., 2014). For example, while p-RLC has been reported to contribute to SF assembly and to distribute along the entire SF length, pp-RLC preferentially localizes to the most contractile regions of the SF interior as observed during time-lapse imaging (Watanabe et al., 2007). While these and other observations hint that p-RLC and pp-RLC contribute differently to SF tensile functions, a causal relationship has not been established.
RLC phosphorylation is driven by two orthogonal kinases: the Ca2+/calmodulin-dependent myosin light chain kinase (MLCK) and the RhoA effector Rho-associated kinase (ROCK). MLCK is encoded by one gene and exists in two forms: long MLCK (∼211 kDa) and short MLCK (∼150 kDa) which lacks the N-terminal extension thought to be associated with actin (Blue et al., 2002). Both MLCK forms directly phosphorylate RLC. On the other hand, ROCK promotes RLC phosphorylation either by direct phosphorylation of RLC or by phosphorylating and inactivating RLC phosphatase. Precisely how these kinases differentially contribute to RLC phosphorylation remains unclear, with the few studies focused on this question producing differing results depending on the cellular system and method of perturbation. For example, pharmacologic ROCK inhibition has been reported to reduce pp-RLC but not p-RLC levels in epithelial cells and thrombin-activated porcine aortic endothelial cells, whereas pharmacologic MLCK inhibition (via ML-9 or Ca2+ depletion) has been observed to affect neither phosphorylation state significantly (Watanabe et al., 2007; Hirano and Hirano, 2016). In contrast, RLC monophosphorylation in human platelets is Ca2+ dependent, implying activation by MLCK (Getz et al., 2010). In reconstituted systems, however, MLCK has been reported to produce both p-RLC and pp-RLC (Ikebe and Hartshorne, 1985; Ikebe et al., 1986; Umemoto et al., 1989). Complicating matters further, there are two mammalian isoforms of ROCK (ROCK1 and ROCK2), and recent isoform-specific knockdown studies have shown that ROCK1 induces pp-RLC and regulates actin microfilament bundle formation in fibroblasts, whereas ROCK2 preferentially regulates RLC monophosphorylation, adhesion maturation and cortical contractility (Yoneda et al., 2005 , 2007; Newell-Litwa et al., 2015).
In addition to differences in phosphorylation state, MLCK and ROCK appear to act upon different subcellular pools of SFs, with MLCK preferentially contributing to the assembly of SFs at the periphery of the cell and ROCK preferentially contributing to the assembly of SFs at the cell center (Totsukawa et al., 2000; Katoh et al., 2001; Tanner et al., 2010). Subcellular laser ablation (SLA) measurements reveal corresponding differences in the SF viscoelastic properties of these peripheral and central SFs, with peripheral SFs releasing more elastic energy than central SFs when photo-severed (Tanner et al., 2010). The spatial distribution of p-RLC and pp-RLC appears to be much more nuanced, as both species are observed within both peripheral and central SFs. Perhaps for this reason, it has remained unclear how p-RLC and pp-RLC differentially contribute to the mechanical functions of each SF subpopulation (Sakurada et al., 1998; Saitoh et al., 2001; Vicente-Manzanares and Horwitz, 2010). Overall, these observations raise the question of whether ROCK and MLCK preferentially control central and peripheral SF formation and mechanical properties by preferential phosphorylation of RLC.
In this study, we investigate mechanistic connections between MLCK and ROCK activity, RLC phosphorylation states, and SF viscoelastic properties using a combination of cell biological and single-cell biophysical approaches. We find that both ROCK1 and ROCK2 regulate central SF retraction kinetics via increased diphosphorylation of RLC, whereas MLCK regulates peripheral SF retraction kinetics via increased monophosphorylation of RLC. An important innovation in our approach is the use of inducibly graded expression of ROCK and MLCK, which enables us to construct quantitative relationships between RLC phosphorylation and SF viscoelastic properties.
RESULTS
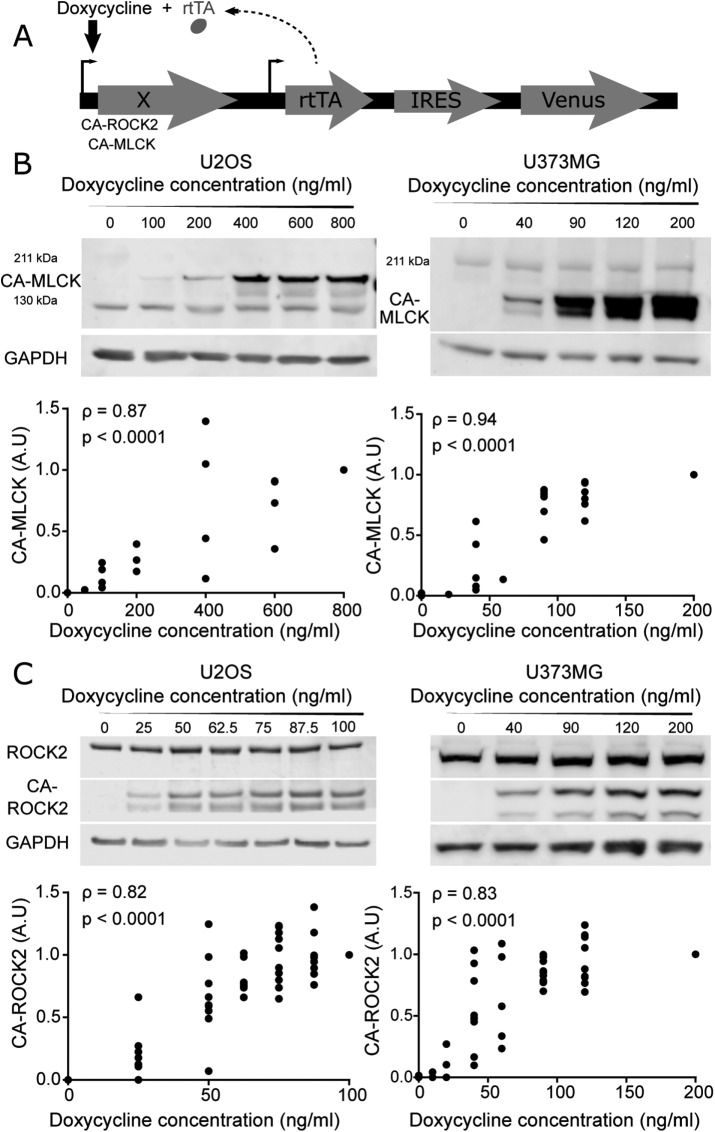
To investigate functional contributions of MLCK and ROCK to RLC phosphorylation and SF function, we stably overexpressed constitutively active (CA) mutants of ROCK and MLCK under a doxycycline-inducible promoter in two cell lines: U2OS human osteosarcoma and U373MG human glioblastoma cells (Figure 1A; MacKay and Kumar, 2014; Wong et al., 2015). The mutants p160ROCK Δ3 (human CA-ROCK1) and ROCK CAT (bovine CA-ROCK2) lack the RhoA binding domain, thereby unleashing kinase activity in the absence of RhoA-GTP binding, whereas rabbit smooth muscle short MLCK ED785-786KK (CA-MLCK) lacks a functional autoinhibition domain (Gallagher et al., 1993; Leung et al., 1995; Ishizaki et al., 1997). Importantly, doxycycline induction allows titration of gene expression over a continuous range, which in turn enables elucidation of quantitative relationships between expression and mechanobiological phenotype in a manner not possible with pharmacological inhibition or transient plasmid overexpression (MacKay et al., 2012 , 2014; MacKay and Kumar, 2014; Hughes and Kumar, 2016). Understanding this dose-response relationship is an important experimental design consideration given that the relationship between myosin activation and mechanobiological phenotype is often highly nonlinear (MacKay and Kumar, 2014; Rape et al., 2015).
FIGURE 1:
Graded control over the expression of a constitutively active form of MLCK (CA-MLCK) and ROCK2 (CA-ROCK2). (A) Schematic of doxycycline-inducible lentiviral system, where X encodes MLCK, ROCK1, or ROCK2. (B) Representative Western blot showing expression levels of endogenous MLCK, CA-MLCK, and GAPDH in U2OS (left) and U373MG cells (right) as a function of doxycycline concentration. U2OS cells were probed with rabbit ant-MLCK (Abcam 76092) and U373MG cells were probed with mouse anti-MLCK (Sigma M7905). Expression levels of CA-MLCK were quantified, normalized to GAPDH and to the highest doxycycline concentration for each cell line, and plotted below the respective Western blots (n = 4 blots for U2OS and n = 6 blots for U373MG). (C) Representative Western blot showing expression levels of endogenous ROCK2, CA-ROCK2, and GAPDH in U2OS (left) and U373MG (right) cells in the presence of various amounts of doxycycline. Expression levels of CA-ROCK2 were quantified, normalized to GAPDH and to the highest doxycycline concentration, and plotted below the respective Western blots (n = 10 blots for U2OS and n = 10 blots for U373MG at the maximum doxycycline concentration). Statistical parameters shown represent the Spearman’s rank correlation coefficient (ρ) and p value.
We first confirmed that we can indeed express each kinase in a gradient by quantifying CA-MLCK (Figure 1B) and CA-ROCK2 (Figure 1C) levels as a function of doxycycline concentration for both cell lines. As expected, CA-MLCK and CA-ROCK2 were undetectable in the absence of doxycycline for both cell lines. The expression of each kinase increased in a statistically significant manner with increasing doxycycline concentration (quantification shown in Figure 1, B and C; Spearman correlation coefficients ρU2OS, CA-MLCK = 0.87, ρU373, CA-MLCK = 0.94, ρU2OS, CA-ROCK2 = 0.82, ρU373, CA-ROCK2 = 0.83). We also successfully produced similarly graded expression of CA-ROCK1 as shown by increasing intensity of the Myc tag with doxycycline (Supplemental Figure S1). Moreover, we compared the expression of the CA constructs relative to the levels of the endogenous kinases. We observed that the expression levels of CA-ROCK2 relative to endogenous were 0.85-fold for U2OS and 1.66-fold at the highest doxycycline concentration for U373MG cells, respectively (Supplemental Figure S2). The expression levels of CA-MLCK relative to endogenous MLCK were higher for both cell lines ranging around 1-fold for U2OS and 4.8-fold for U373MG cells at the highest doxycycline concentrations (Supplemental Figure S2; U2OS CA-MLCK was normalized to 130 kDa MLCK, whereas U373MG CA-MLCK was normalized to 211 kDa MLCK). Overall, we observe that our system allows us to produce graded but modest overexpression of ROCK2 and MLCK in both cell lines over a range that enables us to study the relationship between kinase expression and mechanobiological phenotypes.
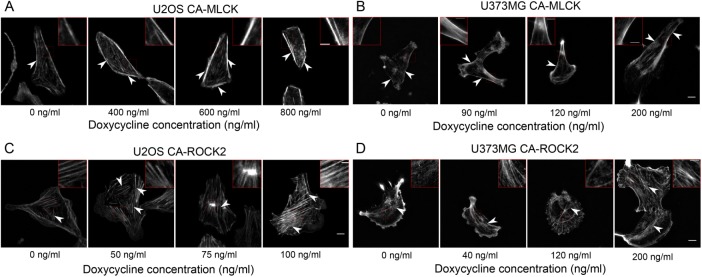
As described earlier, our and others’ pharmacological studies have demonstrated that ROCK and MLCK regulate the formation of different subsets of SFs, with ROCK inhibition disrupting central SFs and MLCK inhibition disrupting peripheral SFs (Katoh et al., 2001; Tanner et al., 2010). We therefore hypothesized that graded increases in the expression of each kinase would produce graded changes in each corresponding SF subpopulation, which may in turn drive alterations in cell morphology. The two cell lines chosen exhibit different central SF architectures: U2OS cells feature prominent ventral SFs (Figure 2, A and C, at 0 ng/ml doxycycline), which localize to the cell rear and terminate in FAs, whereas U373MG cells exhibit more transverse arcs, which lie parallel to the leading edge and anchor internally within the SF network (Figure 2, B and D, at 0 ng/ml doxycycline). With increasing doxycycline concentration, both U2OS CA-MLCK and U373MG CA-MLCK cells exhibited brighter and slightly thicker peripheral SFs (Figure 2, A and B; highlighted insets and arrowheads point to peripheral SFs). Additionally, expression of CA-MLCK in U373MG cells resulted in the formation of smaller actin fibers close to the peripheral SFs (Figure 2B; 200 ng/ml inset). In contrast, CA-ROCK2 expression increased the density of central SFs for both cell lines (Figure 2, C and D). Specifically, U2OS CA-ROCK2 cells exhibited thicker central SFs with increasing doxycycline concentrations compared with the 0 ng/ml doxycycline condition (Figure 2C; highlighted insets and arrowheads pointing to central ventral SFs). At higher concentrations, central SFs sometimes formed mesh-like structures with indistinguishable SFs. Similarly, expression of CA-ROCK2 in U373MG cells led to the formation of ventral SFs within the cell center as compared with cells cultured in the 0 ng/ml doxycycline condition, which exhibited a more poorly defined SF network (Figure 2D; highlighted inset, and arrowheads point to central ventral SFs). At higher doxycycline concentrations, U373MG CA-ROCK2 cells also exhibited brighter and thicker central SFs (Figure 2D; 120 ng/ml doxycycline inset). We also saw similar effects on central SF architecture with expression of CA-ROCK1 (Supplemental Figure S3). Thus, both cell lines exhibit similar ROCK-dependent enhancement of central SFs and MLCK-dependent enhancement of peripheral SFs.
FIGURE 2:
Graded expression of CA-MLCK alters peripheral SF architecture, whereas CA-ROCK2 expression alters central SF architecture. F-actin images of (A) U2OS CA-MLCK and (B) U373MG CA-MLCK cultured in various doxycycline concentrations. Arrowheads point to peripheral SFs and insets highlight peripheral SFs of interest. F-actin images of (C) U2OS CA-ROCK2 and (D) U373MG CA-ROCK2 cultured in various doxycycline concentrations. Arrowheads point to central SFs and insets highlight central SFs of interest. Fluorescence intensity was normalized to the 0 ng/ml doxycycline concentration for all panels with the exception of B where the 0 ng/ml doxycycline condition is set at a higher intensity than the others. Scale bars = 10 µm; inset scale bars = 2 µm.
To determine whether the CA constructs exhibit preferential localization, we performed immunostaining in both U2OS and U373MG CA-MLCK and CA-ROCK2 cells cultured in the presence and absence of doxycycline. Previous work has shown that MLCK has an actin-binding domain in its N-terminus (Smith and Stull, 2000). Consequently, long MLCK preferentially localizes to SFs, whereas short MLCK exhibits a more cytoplasmic localization (Blue et al., 2002). In both U2OS and U373MG cells, endogenous MLCK exhibits diffuse localization (Supplemental Figure S4). The addition of doxycycline increases the fluorescence intensity due to expression of short CA-MLCK but does not change the localization patterns. Endogenous ROCK2 and CA-ROCK2 also exhibit diffuse localization for both U2OS and U373MG cell lines. Similar localization patterns were also observed with overexpression of CA-ROCK1 in U2OS cells. Overall, the kinases do not seem to differ in their localization despite the distinct changes in SF architecture observed with expression of each kinase.
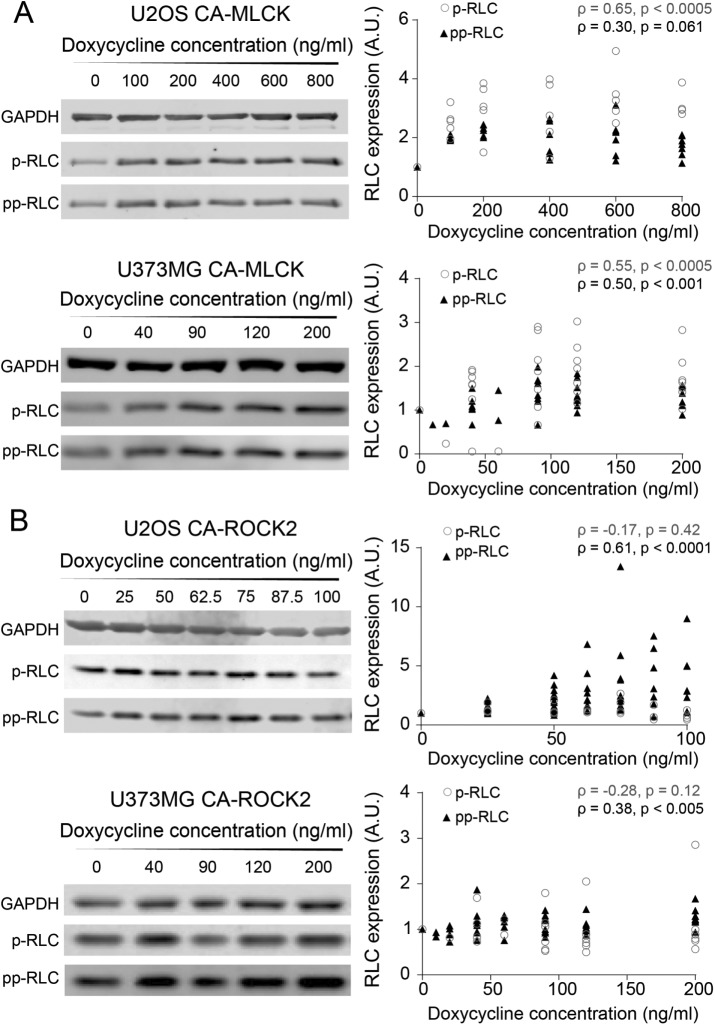
To quantify how changes in the expression of each kinase translate into RLC mono- and diphosphorylation levels, we performed Western blots using phospho-specific antibodies. First, we explored whether any changes in RLC phosphorylation were observed in the presence and absence of doxycycline induction of MLCK and ROCK (Supplemental Figure S5). Induction of CA-MLCK produced a statistically significant increase in p-RLC but not in pp-RLC for both cell lines (pU2OS, p-RLC (rabbit) = 0.035, pU2OS, pp-RLC (rabbit) = 0.75, pU373, p-RLC (mouse) = 0.0092, pU373, pp-RLC (rabbit) = 0.061; Kruskal–Wallis followed by Dunn’s nonparametric test for U2OS and Student’s t test for U373MG). In contrast, induction of CA-ROCK2 increased pp-RLC but not p-RLC in both cell lines (pU2OS, p-RLC (rabbit) = 0.10, pU2OS, pp-RLC (rabbit) = 0.0006, pU373, p-RLC (mouse) = 0.072, pU373, pp-RLC (rabbit) = 0.019; Student’s t test of U373MG and analysis of variance followed by Student’s t test for U2OS). We next asked how graded variations in the activity of each kinase altered phosphorylation states (Figure 3). In both cell lines, graded induction of CA-MLCK expression produced a monotonic increase in p-RLC (Figure 3A, empty gray circles; Spearman correlation coefficients ρU2OS, p-RLC (mouse) = 0.65, ρU373, p-RLC (mouse) = 0.55). Interestingly, CA-MLCK in both U2OS and U373MG cells slightly increased pp-RLC in a graded manner as well, consistent with a sequential phosphorylation mechanism (Figure 3A, solid triangles; Spearman correlation coefficients ρU2OS, pp-RLC (rabbit) = 0.30, ρU373, pp-RLC (rabbit) = 0.50). In both cell lines, increasing the expression of CA-ROCK2 increased pp-RLC (Figure 3B, solid triangles circles; Spearman correlation coefficients ρU373, pp-RLC (rabbit) = 0.38, ρU2OS, pp-RLC (rabbit) = 0.61), while no graded change was observed in p-RLC in both U2OS and U373MG cells (Figure 3B, empty gray circles; Spearman correlation coefficients ρU2OS, p-RLC (mouse) = −0.17, ρU373, p-RLC = −0.28). We also observed the same trends with CA-ROCK1 expression in U2OS cells (Supplemental Figure S5), suggesting that both ROCK1 and ROCK2 isoforms preferentially produce pp-RLC.
FIGURE 3:
Graded increases in CA-MLCK and CA-ROCK2 produce graded changes in p-RLC and pp-RLC. (A) Representative Western blots probed for p-RLC and pp-RLC in U2OS CA-MLCK (top) and U373MG CA-MLCK (bottom). Phosphorylation levels were quantified, normalized to GAPDH and CA-MLCK + 0 ng/ml doxycycline for each cell line, and plotted below the respective Western blots. p-RLC is shown by empty gray circles, whereas pp-RLC is shown by black triangles (U2OS: n = 6 blots for p-RLC [mouse] and 7 blots for pp-RLC [rabbit] blots; U373MG: n = 8 blots for p-RLC [mouse] and n = 9 blots for pp-RLC [rabbit]). (B) Representative Western blots probed for pp-RLC and p-MCL in U2OS CA-ROCK2 (top) and U373MG CA-ROCK2 (bottom). Phosphorylation levels were quantified, normalized to GAPDH and CA-ROCK2 + 0 ng/ml doxycycline for each cell line, and plotted below the respective Western blots. p-RLC is shown as empty, black circles, whereas pp-RLC is shown as solid, black triangles (U2OS: n = 4 blots for p-RLC [mouse] and n = 11 blots for pp-RLC [rabbit] expression; U373MG: n = 9 blots for pp-RLC [rabbit] and n = 6 blots for p-RLC [mouse]). Statistical parameters shown represent the Spearman’s rank correlation coefficient (ρ) and p value.
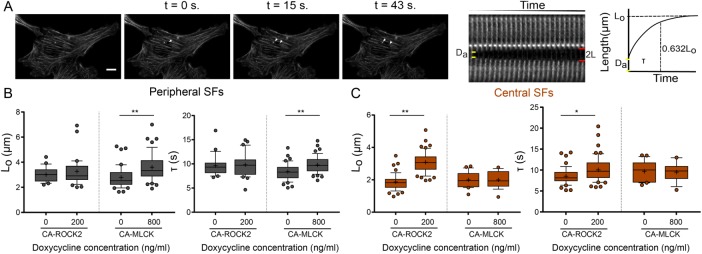
FIGURE 5:
CA-MLCK and CA-ROCK2 regulate the viscoelastic properties of distinct SF subpopulations. (A) SF retraction analysis. Da: SF material destroyed by ablation; 2L: distance between SF ends over time (L is the retraction distance of a severed SF fragment); t: time. L-t curves for each stress fiber are fitted to a Kelvin-Voigt model to determine Lo, whose magnitude correlates with the SF’s dissipated elastic energy, and τ, the viscoelastic time constant, which reflects the ratio of viscosity to elasticity. (B) Lo and τ values of peripheral SF ablation for U2OS CA-ROCK2 and CA-MLCK cells cultured in the presence and absence of doxycycline (n = 21, 32 for U2OS CA-ROCK2, n = 42, 47 for U2OS CA-MLCK). (C) Lo and τ values of central SF ablation for U2OS CA-ROCK2 and CA-MLCK cells cultured in the presence and absence of doxycycline (n = 49, 51 for U2OS CA-ROCK 2, n = 22, 19 for U2OS CA-MLCK). Boxes represent 25th and 75th percentiles; whiskers represent 10th and 90th percentiles. Cross represents the mean of the distribution. Statistical differences calculated using Mann-Whitney (*p < 0.005, **p < 0.0005). Scale bars = 10 µm.
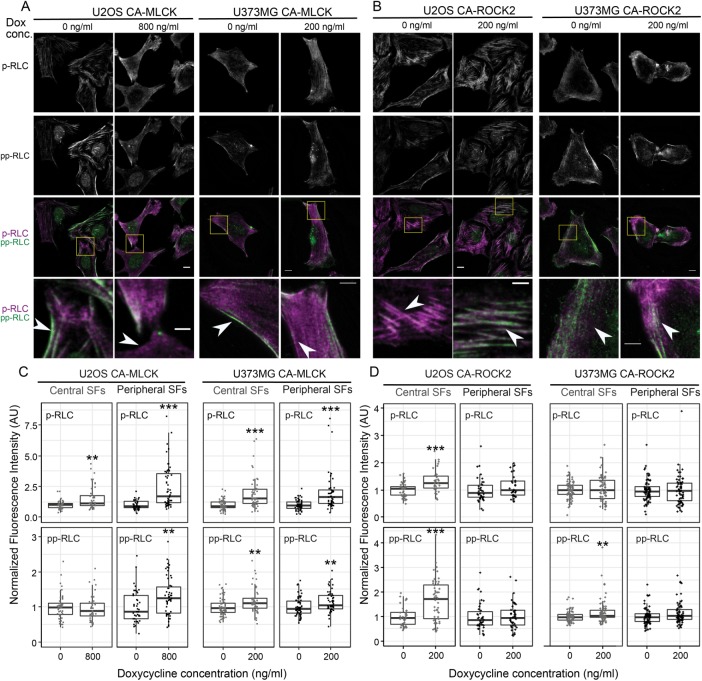
Given the strong influence of ROCK1/2 on central SFs and pp-RLC levels, and that of MLCK on peripheral SFs and p-RLC levels, we wondered whether the observed kinase-dependent RLC phospho-species localized to the respective kinase-dependent SF subpopulations. RLC-phospho-specific immunostaining of CA-MLCK (Figure 4A) and CA-ROCK2 cells (Figure 4B) indeed revealed an MLCK-dependent increase in p-RLC in the cell periphery (Figure 4A, highlighted inset) and a ROCK-dependent increase in pp-RLC in the cell center (Figure 4B, highlighted inset). Quantification of this localization revealed a statistical increase in p-MLC in both peripheral (black) and central SFs (gray) for both U2OS and U373MG CA-MLCK cells (Figure 4C, top row). The details of this relationship varied with cell line. U2OS cells exhibited a much greater enhancement of p-RLC in peripheral SFs than central SFs (U2OS: 2.6-fold for peripheral vs. 1.6-fold for central; U373MG: 2.0-fold for peripheral vs. 1.92-fold for central) as well as a slight increase in the amount of pp-RLC in peripheral SFs (U2OS: 1.3-fold increase; U373MG: 1.1-fold increase; Figure 4C, bottom row). Induction of CA-ROCK2 produced an increase in pp-RLC in central SFs for both cell lines (U2OS: 1.94-fold; U373MG: 1.2-fold; Figure 4D, bottom row). For U2OS cells, a small increase in the amount of p-RLC was also observed in central SFs (U2OS: 1.28-fold increase; Figure 4D, top row). Taken together with our earlier SF morphometric observations (Figure 2) and Western blots (Figure 3), these results indicate that ROCK promotes formation of central SFs and an associated central localization of pp-RLC, whereas MLCK promotes formation of peripheral SFs and an associated peripheral localization of p-RLC.
FIGURE 4:
CA-MLCK expression regulates RLC phosphorylation in peripheral SFs, whereas CA-ROCK2 expression regulates RLC phosphorylation in central SFs. Representative fluorescence images of (A) U2OS CA-MLCK (left) and U373MG CA-MLCK (right) cells and (B) U2OS CA-ROCK2 and U373MG CA-ROCK2 cells cultured in the presence and absence of doxycycline stained for p-RLC (top row, third and fourth rows in magenta) and pp-RLC (second row, third and fourth rows in green). Scale bars = 10 µm; inset scale bars = 5 µm. Fluorescence intensity of all images is normalized to that of 0 ng/ml doxycycline for each condition. (C) Quantification of immunofluorescence intensity of p-RLC (top row) and pp-RLC (bottom row) within central (gray) and peripheral (black) SFs for U2OS and U373MG CA-MLCK cells cultured in the presence and absence of doxycycline (n = 46, 46, 41, 55, 62, 53, 60, and 51 cells collected from three independent experiments for each condition, respectively, from left to right). Intensities were normalized to the average intensity value of either U2OS or U373MG CA-MLCK cells cultured in the absence of doxycycline for each experiment. (D) Quantification of immunofluorescence intensity of p-RLC (top row) and pp-RLC (bottom row) within central (gray) and peripheral (black) SFs for U2OS and U373MG CA-ROCK2 cells cultured in the presence and absence of doxycycline (n = 41, 32, 39, 31, 75, 72, 75, and 71 cells collected from three independent experiments analyzed per condition, respectively, from left to right). Intensities were normalized to the average intensity value of either U2OS or U373MG CA-ROCK2 cells cultured in the absence of doxycycline per each experiment (**p < 0.01; ***p < 0.001; Mann-Whitney test).
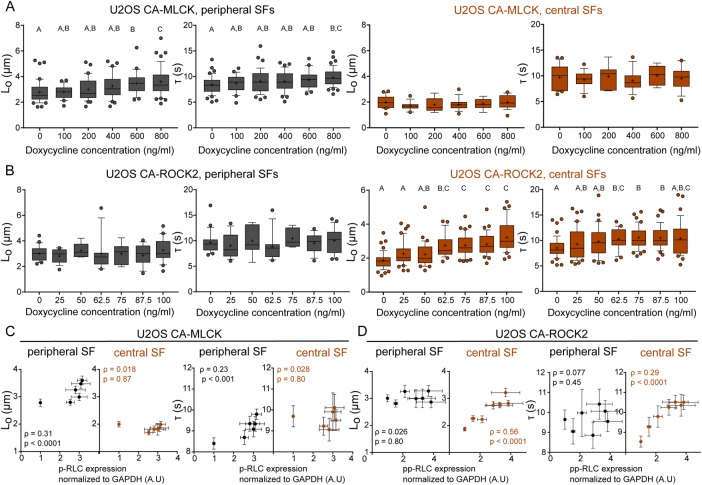
To determine whether the localized changes in RLC phosphorylation caused by each kinase produced changes in the mechanical properties of the associated SFs, we performed SLA to sever individual central and peripheral SFs of U2OS CA-ROCK2 and U2OS CA-MLCK cells cultured in the presence and absence of doxycycline. As in our previous studies, we photo-severed single SFs and fitted the time-dependent retraction of the two SF ends to the Kelvin-Voigt model of viscoelasticity. This model is described by two parameters: a viscoelastic time constant (τ), reflecting the SF’s effective viscosity to elasticity ratio, and a plateau retraction distance (Lo), reflecting the elastic energy dissipated by half of the severed SF (Figure 5A; Kumar et al., 2006; Chang and Kumar, 2015; Kassianidou et al., 2017). Whereas Lo and τ for peripheral SFs were insensitive to CA-ROCK2 induction (Figure 5B), both parameters statistically increased with CA-MLCK induction (Figure 5B). For central SFs, both Lo and τ were influenced by CA-ROCK2 induction but not by CA-MLCK induction (Figure 5C). Notably, both CA-ROCK1 and CA-ROCK2 influenced SF mechanics in a similar manner (Supplemental Figure S6 and Figure 5, B and C). Thus, ROCK and MLCK preferentially regulate the viscoelastic properties of central and peripheral SFs, respectively.
FIGURE 6:
SF mechanical properties increase in a graded manner following graded expression of RLC phosphorylation. (A) Lo and τ values of peripheral (left, dark gray) and central (right, orange) SF ablation of CA-MLCK cells cultured in various concentrations of doxycycline (n = 42, 24, 39, 37, 25, and 47 cells for peripheral SF ablation of CA-MLCK, n = 22, 14, 7, 14, 9, and 19 cells for central SF ablation). (B) Lo and τ values of peripheral (left, dark gray) and central (right, orange) SF ablation of CA-ROCK2 cells cultured in various concentrations of doxycycline (n = 21, 15, 6, 16, 6, 14, and 21 cells for peripheral SF ablation; n = 49, 41, 31, 25, 40, and 38 cells for central SF ablation). A, B, and C statistical families show statistical differences (p < 0.05) determined using Dunn’s test for multiple comparisons of nonnormally distributed data. Boxes represent 25th and 75th percentiles; whiskers represent 10th and 90th percentiles. Cross represents the distribution mean. (C) Viscoelastic retraction parameters Lo and τ of peripheral (black) and central SFs (orange) of U2OS CA-MLCK cells plotted vs. the observed increase in p-RLC (replotted from Figure 3A). (D) Viscoelastic retraction parameters Lo and τ of peripheral (black circles) and central SFs (solid orange circles) and peripheral (black circles) of U2OS CA-ROCK2 cells plotted vs. the observed increase in pp-RLC (replotted from Figure 3B). Orange values correspond to central SF ablation Spearman’s rank correlation coefficient analysis, whereas black values correspond to Spearman’s rank correlation coefficient analysis for peripheral SF ablation. Error bars of x-axis values were determined based on Western blot quantifications shown in Figure 3. All error bars represent SEM.
As noted earlier, an important advantage of graded, inducible expression systems is the ability to construct quantitative relationships between effector level and phenotype. Given our ability to associate RLC phosphorylation levels at a specific doxycycline concentration (Figure 3) and our ability to elucidate SF viscoelastic properties at these same doxycycline concentrations, we were uniquely well positioned to explore correlations between phospho-RLC levels and SF mechanics. To answer this question, we performed SLA on U2OS cells expressing either CA-MLCK (Figure 6A) or CA-ROCK2 (Figure 6B) cultured in various doxycycline concentrations. We observed statistical differences in the dissipated elastic energy (Lo) and viscoelastic time constant (τ) for peripheral SFs of CA-MLCK (Figure 6A, gray) and in both Lo and τ for central SFs of CA-ROCK2 cells (Figure 6B, orange; Kruskal–Wallis test followed by Dunn’s nonparametric test). We also observed that graded expression of CA-MLCK, which increases p-RLC (x-axis is from Figure 3A), preferentially increased both the elastic energy dissipated by the fiber (Lo) and the viscoelastic time constant (τ) of peripheral SFs (dark-gray circles) in a graded manner but did not alter the properties of central SFs (Figure 6C, orange circles; Spearman correlation for Lo: ρperipheral = 0.31, ρcentral = 0.018; Spearman correlation for τ: ρperipheral = 0.23, ρcentral = 0.028). The increase in Lo and τ of peripheral SFs with increased p-RLC follows a nonlinear relationship with the properties not changing significantly until p-RLC reaches at least a 2.5-fold increase (Figure 6C). Graded expression of CA-ROCK2, which led to an increase in the amount of pp-RLC (x-axis is from Figure 3B), preferentially increased the elastic energy dissipated by the fiber after SLA (Lo) and the viscoelastic time constant (τ) of central SFs (Figure 6D, orange circles; Spearman correlation for Lo: ρperipheral = 0.026, ρcentral = 0.56; Spearman correlation for τ: ρperipheral = 0.077, ρcentral = 0.29). Lo appeared to increase linearly with pp-RLC, and at approximately a 3.5-fold increase in pp-RLC above basal levels, the viscoelastic character of central SFs matched that of peripheral SFs. The increase in τ followed a decaying nonlinear curve, eventually reaching a plateau at approximately a 3.5-fold increase in pp-RLC (Figure 6D). These results suggest that SF mechanical properties are indeed tunable based on the type and amount of phosphorylated RLC present.
Our results indicate that ROCK and MLCK phosphorylate RLC in a preferential manner; MLCK primarily monophosphorylates RLC (Ser19), whereas ROCK 1 and ROCK 2 preferentially diphosphorylate it (Thr18 and Ser19; Figure 3 and Supplemental Figure S2). We also show that these changes in phosphorylation have specific localizations: MLCK-induced p-RLC localizes primarily in the periphery of the cell, whereas ROCK-induced pp-RLC localizes in the center (Figure 4). Finally, we show that SF mechanical properties are also regulated preferentially via the kinases: ROCK controls central SF retraction kinetics, whereas MLCK controls peripheral retraction kinetics (Figures 5 and 6). However, these results leave open the question of whether ROCK and MLCK-induced changes in RLC phosphorylation and SF properties are causally linked as opposed to unrelated epiphenomena. To provide a direct link between the kinases, MLC phosphorylation states, and SF mechanics, we transduced U2OS RFP-LifeAct cells with monophosphomimetic RLC, where Ser19 was mutated to Asp (RLC-AD), or with diphosphomimetic RLC, where both Thr18 and Ser19 were mutated to Asp (RLC-DD; Vicente-Manzanares and Horwitz, 2010). We first noted that U2OS phosphomimetic-expressing cells phenocopy the SF architecture of U2OS CA-MLCK and CA-ROCK2 cells. Specifically, U2OS GFP RLC-AD cells show an elongated phenotype with bright peripheral SFs, whereas U2OS GFP RLC-DD cells exhibit bright central SFs (Figure 7, A and B). We then wished to determine whether these phosphomimetic species localize in similar patterns as those observed earlier (Figure 3). GFP-RLC AD localizes strongly on peripheral SFs (Figure 7, A and B, white arrows), whereas GFP-RLC DD localizes primarily on central SFs (yellow arrows). We quantified the GFP signal of RLC-AD and RLC-DD on both peripheral and central SFs and calculated a localization ratio in which >1 indicates preferential localization to peripheral SFs, whereas <1 indicates preferential localization to central SFs. Analogous to the immunostaining studies (Figure 3), we observed that GFP RLC-AD localizes preferentially to peripheral SFs, whereas GFP RLC-DD localizes to central SFs (Figure 7C). Finally, we performed SLA on peripheral (Figure 7D) and central SFs (Figure 7E) of RLC-AD and RLC-DD cells. Overexpression of RLC-AD affected only the elastic energy (Lo) dissipated by peripheral SFs (p = 0.029) and not central SFs (p = 0.63), phenocopying the results seen with CA-MLCK expression (Figure 7D). In turn, overexpression of RLC-DD affected only the elastic energy (Lo) dissipated by central SFs (p < 0.0001) and not peripheral SFs (p = 0.72), phenocopying the results obtained via CA-ROCK2 expression (Figure 7E). These results suggest that the changes in the viscoelastic retraction parameters observed from the increased expression of kinases are directly due to the changes in RLC phosphorylation.
FIGURE 7:
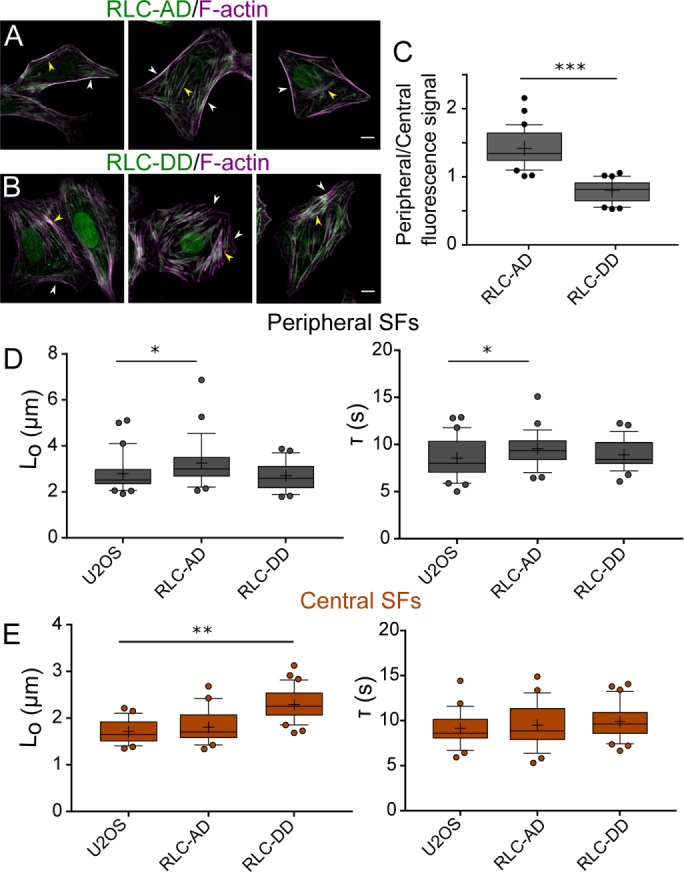
Expression of phosphosmimetic p-RLC and phosphomimetic pp-RLC phenocopy the changes in SF viscoelasticity induced by CA-MLCK and CA-ROCK. Representative images of (A) U2OS RFP-LifeAct GFP-RLC AD and (B) U2OS RFP-LifeAct GFP-RLC DD. Images are taken using the GFP channel for the phosphomimetic constructs and phalloidin for SFs. White arrows point to peripheral SFs, whereas yellow arrows point to central SFs. (C) Quantification of GFP signal localization as a ratio of localization on peripheral over central SFs (n = 36 for GFP-RLC AD and 30 for GFP-RLC DD). (D) Lo and τ values of peripheral SF ablation for U2OS RFP-LifeAct, U2OS RFP-LifeAct GFP RLC-AD, and U2OS RFP-LifeAct GFP RLC-DD cells (n = 21, 28, and 28 cells, respectively). (E) Lo and τ values of central SF ablation for U2OS RFP-LifeAct, U2OS RFP-LifeAct GFP RLC-AD, and U2OS RFP-LifeAct RFP RLC-DD cells (n = 22, 23, and 31 cells, respectively). Boxes represent 25th and 75th percentiles; whiskers represent 10th and 90th percentiles. Cross represents the distribution mean. Statistical differences calculated using Mann-Whitney tests (*< 0.05, **p < 0.001, ***p < 0.0001). Scale bars = 10 µm.
DISCUSSION
ROCK and MLCK are broadly understood to govern RLC phosphorylation, thereby regulating the assembly and contraction of SFs. While there is much evidence that ROCK and MLCK respectively regulate the assembly and contractility of central and peripheral SFs (Totsukawa et al., 2000; Katoh et al., 2001; Tanner et al., 2010), it has remained unclear how each kinase controls RLC phosphorylation states or how these states influence SF viscoelastic properties. By combining cell and molecular biological approaches with mechanical measurements of single SFs in living cells, we have provided support for a model in which MLCK and ROCK distinctly regulate peripheral and central SF mechanics via differential phosphorylation of RLC. Specifically, MLCK stimulates production of p-RLC, which localizes to and controls peripheral SF viscoelasticity. In contrast, both ROCK isoforms stimulate production of pp-RLC, which localizes to and controls central SF viscoelasticity (Figure 8). The mechanical effects of each kinase can be recapitulated with overexpression of a corresponding mono- or diphosphomimetic RLC, strongly supporting a causal link between kinase activity, RLC phosphorylation states, and SF viscoelastic properties.
FIGURE 8:
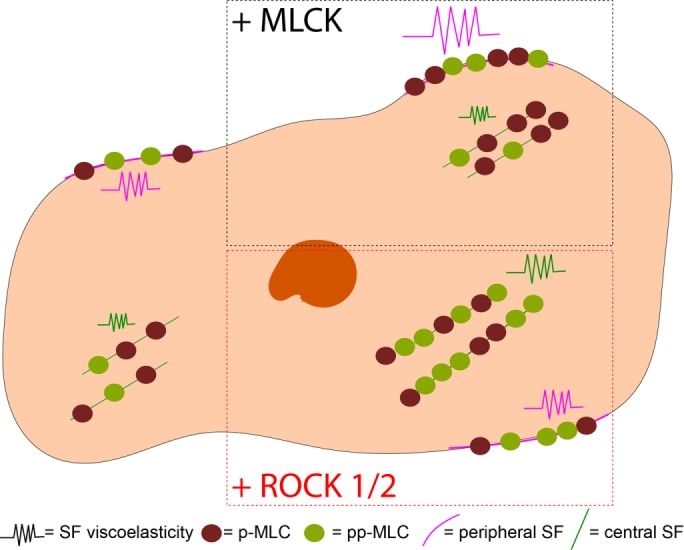
Model of subcellular regulation of RLC phosphorylation and SF viscoelastic properties. MLCK-induced p-RLC localizes and regulates the viscoelastic properties of peripheral SFs, whereas ROCK1 and 2–induced pp-RLC localizes and regulates viscoelastic properties of central SFs.
As noted earlier, previous efforts to dissect contributions of ROCK and MLCK to RLC phosphorylation have produced results that vary with the method used to study and perturb each kinase. In reconstituted preparations, MLCK has been observed to produce both p-RLC and pp-RLC; however, pp-RLC requires comparatively high MLCK concentrations (0.1–1 µM), leaving open the question of which phosphospecies is favored under more physiological conditions (Ikebe and Hartshorne, 1985; Ikebe et al., 1986; Itoh et al., 1989). While pharmacological inhibition in cell culture of either ROCK or MLCK has been observed to reduce diphosphorylation of RLC (Watanabe et al., 2007), the interpretation of these results is complicated by the fact that MLCK and ROCK inhibitors can produce SF and FA disassembly at sufficiently high dose, create off-target effects, and lack isoform selectivity. Moreover, studies of ROCK/MLCK effects on RLC phosphorylation have not been systematically coupled to measurements of contractile function. Our study begins to close this loop by combining controlled expression of each kinase with measurements of viscoelastic properties of individual SFs. We show that overexpression of CA-ROCK1 and CA-ROCK2 preferentially influences the viscoelastic parameters of central SFs (Figures 5 and 6 and Supplemental Figure S3). Our results also reveal that overexpression of a short CA-MLCK that does not localize to SFs preferentially increases the stored elastic energy (as reflected by Lo) and viscoelastic time constant (τ) of peripheral SFs. It should be informative to apply long MLCK mutants of varying actin-binding abilities and determine how the viscoelastic properties of peripheral SFs are altered by direct MLCK binding.
Although SF tension generation has been shown to depend on RLC phosphorylation, it has remained unclear whether graded changes in myosin activation produce graded changes in SF tension generation, or whether there are instead activation thresholds at which SF tension changes in a concerted manner (Kaneko-Kawano et al., 2012). We find that graded increases in the expression of either CA-MLCK or CA-ROCK2 produce monotonic increases in both RLC phosphorylation and SF elastic energy. To our knowledge, this represents the first indication that SF viscoelastic properties can be tuned over a continuous range based on kinase activity. Furthermore, we were able to quantitatively map the relationship between whole-cell RLC phosphorylation levels and individual SF properties. Surprisingly, the viscoelastic properties of peripheral SFs depend much more nonlinearly on p-RLC levels than central SF mechanics depend on pp-RLC levels, indicating that central SFs may be more sensitive to small perturbations above basal RLC phosphorylation levels than peripheral SFs. Moreover, these correlations juxtapose properties of single SFs against whole-cell measurements of p-RLC and pp-RLC, which is a consequence of our inability to perform SLA in live cells while simultaneously performing antibody-based detection of phospho-RLC levels. It should be noted that our measured fold changes in RLC phosphorylation cannot be used to infer stochiometric ratios of p-RLC and pp-RLC within a given cell or SF. It would be valuable to revisit these MLCK and ROCK manipulations, measure the effects on p-RLC/pp-RLC ratios (e.g., with mass spectrometry or urea/glycerol gel electrophoresis), and ask if these ratios are predictive of SF viscoelastic properties. Additionally, fiber-by-fiber correlations of phospho-RLC states and viscoelastic properties may be facilitated in the future by geometric standardization of SFs (Kassianidou et al., 2017) or through the use of live-cell kinase probes. Finally, further work is required to discern the relationship of each kinase to NMMIIA and IIB heavy isoform and their respective localization to central and peripheral SFs (Beach et al., 2014, 2017; Chang and Kumar, 2015).
Within the region of ROCK isoform activities probed, we did not observe strong differences between ROCK1 and ROCK2 in regulation of RLC phosphorylation, SF formation, or SF viscoelastic properties. Recent ROCK isoform-specific knockdown studies have revealed that ROCK1 preferentially regulates pp-RLC, whereas ROCK2 preferentially regulates p-RLC in CHO.K1 and REF52 cells, and that the two isoforms play differential roles in migration (Mertsch and Thanos, 2014; Newell-Litwa et al., 2015). Moreover, ROCK1 knockdown in MCF-7 cells specifically reduced p-RLC localization to adherens junctions suggesting that ROCK1 preferentially regulates contractility in adherens junctions (Priya et al., 2017). On the other hand, ROCK1 and ROCK2 have also been observed to serve redundant functions in other settings; ROCK1- and ROCK2-null fibroblasts showed similarly small reductions in RLC diphosphorylation and cell contractility relative to wild-type controls with statistically significant effects seen only when both ROCK isoforms were removed (Kümper et al., 2016). Additional differences may arise from the method of ROCK isoform perturbation (i.e., overexpression vs. knockdown). These distinct regimes of kinase activity may produce divergent effects on RLC phosphorylation states due to the complex and potentially nonlinear relationships between these RLC activators, ROCK-inhibited RLC phosphatase and RLC phosphorylation (Wang et al., 2009; Kaneko-Kawano et al., 2012). It is important to acknowledge that previous work has suggested that phosphomimetic RLCs (TE or EE) may not mimic endogenous phosphorylated RLC in in vitro assays and in Drosophila. We observed that phosphomimetic RLCs used in this study (AD and DD) emulated the localization patterns of p-RLC and pp-RLC, phenocopied the SF architecture changes triggered by MLCK-induced p-RLC and ROCK-induced pp-RLC, and congruently altered SF viscoelastic changes (Kamisoyama et al., 1994; Sweeney et al., 1994; Vasquez et al., 2016). Nonetheless, additional characterization of these and related RLC mutants would help assess the degree to which they capture the effects of phosphorylation.
Overall, our work highlights a potential mechanism for cells to spatially and temporally regulate the distribution of contractile forces via preferential phosphorylation from different kinases. The distinct contributions of each phosphospecies to SF viscoelasticity may involve multiple protein–protein interactions, given that highly reductionist myosin sliding assays do not detect differences in velocities between p-RLC and pp-RLC (Umemoto et al., 1989). Nevertheless, with the many recent efforts to develop multiscale mechanochemical models of SF function (Besser, 2007; Stachowiak and O’Shaughnessy, 2008; Colombelli et al., 2009; Russell et al., 2009; Stachowiak et al., 2014), data such as ours may offer valuable new inputs for these models and facilitate incorporation of distinct subcellular pools of SFs.
MATERIALS AND METHODS
Cell lines and reagents
Myc-tagged human p160ROCK Δ3 (kindly provided by S. Narumiya, Kyoto University, Japan), Flag-tagged rabbit smooth muscle MLCK ED785-786KK (kindly provided by P. J. Gallagher, Indiana University), and bovine ROCK CAT (kindly provided by K. Kaibuchi, Nagoya University, Japan) were subcloned into the lentiviral vector pSLIK (Addgene #84647 for CA-MLCK and #84649 for CA-ROCK2; MacKay and Kumar, 2014; Wong et al., 2015). This vector contains a tet response element (TRE) doxycycline-inducible promoter, along with constitutive expression of a reverse tetracycline-controlled transactivator (rtTA) and Venus selection marker separated by an internal ribosomal entry site (IRES) (Figure 1A). p160ROCK Δ3 is a CA mutant of the ROCK1 isoform, whereas ROCK CAT is a CA mutant of the ROCK2 isoform. Our sequencing reveals that this CA-MLCK bears 98% identity to rabbit smooth muscle MLCK (∼150 kDa) but harbors a mutation within the autoinhibitory site (ED773-774KK of construct, which aligns with ED785-786KK in wild-type MLCK; GenBank accession number: MG189932). This mutation has previously been shown to confer CA function in bovine smooth muscle MLCK through disruption of autoinhibition (Gallagher et al., 1991, 1993). Empty pSLIK vectors were also used to establish control cell lines. Viral particles for each pSLIK plasmid and for the pFUG-RFP-LifeAct vector were packaged in 293T cells. U2OS osteosarcoma cells (ATCC HBT-96) were transfected with pFUG-RFP-LifeAct and sorted on a DakoCytomation MoFlo High Speed Sorter based on red fluorescent protein (RFP) fluorescence (Lee et al., 2016). U2OS RFP-LifeAct cells were further stably transduced with the pSLIK vectors at a multiplicity of infection (MOI) of 0.5 IU/cell. Cells were further sorted based on RFP and Venus fluorescence. U373MG glioblastoma cells (ATCC HTB-17, also known as U-373 MG) were transfected with the pSLIK plasmids at an MOI of 0.5 IU/cell, and cells receiving the construct were selected based on Venus fluorescence. U373MG cells containing the pSLIK plasmid were then transfected with pFUG-RFP-LifeAct at an MOI of 1.5 IU/cell and cells receiving the LifeAct vector were selected using 0.6 μg/ml puromycin. ATCC U373MG cells have been established to be derived from a common progenitor with U251 cells and SNB 19 cells, although the lines have diverged and exhibit some phenotypic and karyotypic differences (Stepanenko and Kavsan, 2014). Both cell lines were confirmed by short tandem repeat profiling, and mycoplasma testing was carried out every 4 mo.
Plasmids containing phosphomimetic myosin light chains (pEGFP RLC-DD, pEGFP RLC-AD) were kindly provided by A. R. Horwitz (University of Virginia; Vicente-Manzanares and Horwitz, 2010). The RLC-GFP constructs were digested from the plasmid backbone using EcoRI and XhoI and ligated into the lentiviral vector pLVX-AcGFP-N1 (Clonetech). Successful ligation was verified via sequencing. Viral particles for each plasmid were packaged in 293T cells. U2OS RFP-LifeAct cells were stably transduced with the viral particles at an MOI of 0.5 IU/cell. Cells were then sorted based on RFP and green fluorescent protein (GFP) fluorescence.
U2OS cells were cultured in DMEM supplemented with 10% fetal bovine serum (JR Scientific), 1% penicillin/streptomycin (Thermo Fisher Scientific), and 1% nonessential amino acids (Life Technologies). U373MG cells were cultured in DMEM supplemented with 10% calf serum (JR Scientific), 1% penicillin/streptomycin, 1% nonessential amino acids, and 1% sodium pyruvate (Thermo Fisher Scientific). Doxycycline (Fisher Bioreagents) was added at the required concentration 2 d before all experiments to activate the CA constructs.
Western blots
As described previously, cells were lysed in radioimmunoprecipitation assay (RIPA) buffer with phosphatase and protease inhibitors (EMD Millipore, Billerica, MA; Wong et al., 2015; Kassianidou et al., 2017). Protein content was measured by bicinchoninic acid (BCA) assay and used to normalize samples to the lowest concentration. Lysates were boiled, run on 4–12% Bis-Tris gels, and transferred onto a polyvinylidene difluoride (PVDF) membrane. The following primary antibodies were used: anti-phosphorylated myosin light chain 2 (Thr18/Ser19; Cell Signaling Technology), anti-phosphorylated myosin light chain 2 (Ser19) produced in rabbit or in mouse (both obtained from Cell Signaling Technology), anti-GAPDH (Sigma-Aldrich, St. Louis, MO), mouse anti-MLCK (Sigma-Aldrich, St. Louis, MO) and rabbit anti-MLCK (abcam), anti-ROCK 2 (Sigma-Aldrich, St. Louis, MO), anti-Myc tag (Cell Signaling Technology), and anti-ROCK1 (Cell Signaling Technology). The following secondaries were used: IRDye 800 Goat anti-mouse IgG, IRDye 700 Goat anti-rabbit IgG (Licor), and HRP-conjugated anti-mouse (Life Technologies). All bands except Myc tag for CA-ROCK1 visualization (Supplemental Figure S1) were visualized using an Odyssey system and were quantified with the built-in gel analyzer tool in ImageJ (National Institutes of Health [NIH], Bethesda, MD). Myc-tag bands were visualized using enhanced chemiluminiscence (ECL; Thermo Fisher) reagent.
Immunofluorescence staining
Cells were seeded on glass coverslips coated with 25 μg/ml fibronectin (EMD Millipore Corporation). After doxycycline incubation, cells were fixed with 4% paraformaldehyde for 10 min at room temperature. After phosphate-buffered saline (PBS) washes, cells were permeabilized in 0.5% Triton-X for 15 min, and blocked in 5% goat serum (GS; Thermo Fisher Scientific) for at least 1 h. U2OS cells were incubated in 1% GS and primary antibody for 2 h at room temperature or overnight at 4°C in a humidity chamber. U373MG cells were incubated in 1% GS and primary antibody overnight at 4°C. Following primary incubation, cells were washed in 1% GS (3 × 5 min) and then incubated in secondary antibody for 1 h at room temperature. We used the following antibodies: anti-phosphorylated myosin light chain 2 (Thre18/Ser19) produced in rabbit (Cell Signaling Technology), anti-phosphorylated myosin light chain 2 (Ser19) produced in mouse (Cell Signaling Technology), anti-MLCK (Sigma-Aldrich, St. Louis, MO), anti-ROCK2 (Sigma-Aldrich, St. Louis, MO), anti-ROCK1 (Cell Signaling Technologies), Alexa Fluor 647 anti-mouse, Alexa Fluor 488 anti-rabbit, and Alexa Fluor 633 anti-rabbit (Thermo Fisher Scientific). F-Actin was stained with 546-phalloidin. U373MG cells were mounted on glass slides using ProLong Gold Antifade Mountant (Thermo Fisher Scientific). Figure 4 immunofluorescence images were obtained using a swept-field upright confocal microscope equipped with a 60× water-immersion lens (Prairie Technologies) and a Nikon TE2000 microscope equipped with a 60× oil immersion lens. Supplemental Figure S4 images were obtained on a Zeiss LSM 510 Meta Confocal microscope equipped with a 63× oil immersion objective and a Nikon TE2000 microscope equipped with a 60× oil immersion lens. For presentation purposes, the contrast and brightness of fluorescence images were optimized using ImageJ (NIH).
Analysis of immunofluorescence images
All analyses were performed using ImageJ. To quantify the localization of p-RLC and pp-RLC, images were overlaid with phalloidin and background was subtracted. Alignment of images was verified using Template Matching plug-in (Tseng et al., 2012). A line of 0.6 µm thickness was manually drawn over peripheral or central SFs for multiple SFs per cell and a measurement of raw integrated intensity of p-RLC and pp-RLC was recorded across each traced line. The intensities were then normalized to the length of the drawn line and averages of normalized intensities were calculated per cell. To account for experiment-to-experiment variations in fluorescence intensity, values were normalized to the mean value of the appropriate control, that is, CA-ROCK2 or CA-MLCK cells cultured in 0 ng/ml doxycycline for each specific experiment.
To quantify the localization of GFP RLC-AD and GFP RLC-DD, cells were seeded on 25 µg/ml fibronectin-coated coverslips and fixed using 4% paraformaldehyde. Cells were permeabilized and incubated with Alexa 546-tagged phalloidin. Images of SFs and phosphomimetic species were obtained using a 63× oil immersion objective. A line of 0.6 µm was drawn over peripheral and central SFs and the intensity of the GFP signal was recorded. The intensities were then normalized to the length of the drawn line and averages of normalized intensities were calculated per cell. The ratio of peripheral to central SF localization was then determined.
SF photodisruption
SF SLA experiments were performed on a Zeiss LSM 510 Meta Confocal microscope equipped with a MaiTai Ti:sapphire femtosecond laser (Spectra Physics, Newport Beach, CA; Tanner et al., 2010; Chang and Kumar, 2013, 2015). Cells (10,000) were seeded on 35-mm glass bottom dishes (MatTek Corporation) coated with 25μg/ml fibronectin (EMD Millipore Corporation) and incubated in doxycycline for 2 d. Media was changed to Live Cell Imaging Solution (Invitrogen) before SLA. For SF photodisruption, the femtosecond laser was used at 770 nm resulting in an energy deposition of 1–2 nJ on a single SF (Tanner et al., 2010; Chang and Kumar, 2015; Kassianidou et al., 2017). All images were acquired with a 40× water-immersion objective (N.A. = 0.8).
Data analysis of SF retraction
SF retraction distance was recorded every 2 s for 49 s following SLA. The SF ends were manually traced using Image J to determine the retraction dynamics. Results were fitted to a Kelvin-Voigt model defined by the following equation:
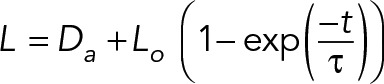 |
where L is defined as half the distance between the two severed SF ends, Da is the length of SF destroyed by the laser, Lo is the retraction plateau distance, and τ is the viscoelastic time constant. Curve fitting to extract parameters Lo and τ was performed using CurveFit (MATLAB; Tanner et al., 2010; Chang and Kumar, 2015; Kassianidou et al., 2017).
Statistical analysis
All statistical analyses and graph generation were performed using GraphPad Prism and R. Unless otherwise noted, samples were compared using nonparametric t tests such as Mann-Whitney. Normality was assessed based on the Shapiro-Wilk Normality test. Experiments that used cells seeded and assayed on different days were deemed independent, and at least three independent experiments were performed for each assay.
Supplementary Material
Acknowledgments
Subcellular laser ablation and confocal imaging were performed at the CRL Molecular Imaging Center supported by NIH 3R01EY015514-01S1. Confocal images were also obtained at the CIRM/QB3 Stem Cell Shared Facility. Fluorescence-activated cell sorting was performed with the help of H. Nolla in the Flow Cytometry Facility at the University of California, Berkeley. We gratefully acknowledge financial support from the following sources: Howard Hughes Medical Institute International Student Research Fellowship; Natural Sciences and Engineering Research Council (NSERC) of Canada (PGS D 555229) and Siebel Scholars Program (E.K. and J.H.H.); the National Institutes of Health (R21CA174573, R21EB016359, R01GM122375, and R01NS074831), the National Science Foundation (CMMI 1055965), and the W.M. Keck Foundation (S.K.).
Abbreviations used:
- MLCK
myosin light chain kinase
- NMMII
nonmuscle myosin II
- p-RLC
monophosphorylated myosin regulatory light chain
- pp-RLC
diphosphorylated regulatory light chain
- RLC
myosin regulatory chain
- ROCK
Rho-associated kinase
- SF
stress fiber
- SLA
subcellular laser ablation.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E17-06-0401) on October 18, 2017.
REFERENCES
- Beach JR, Bruun KS, Shao L, Li D, Swider Z, Remmert K, Zhang Y, Conti MA, Adelstein RS, Rusan NM, et al. Actin dynamics and competition for myosin monomer govern the sequential amplification of myosin filaments. Nat Cell Biol. 2017;19:85–93. doi: 10.1038/ncb3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beach JR, Shao L, Remmert K, Li D, Betzig E, Hammer JA. Nonmuscle myosin II isoforms coassemble in living cells. Curr Biol. 2014;24:1160–1166. doi: 10.1016/j.cub.2014.03.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besser A, Schwarz US. Coupling biochemistry and mechanics in cell adhesion: a model for inhomogeneous stress fiber contraction. New J Phys. 2007;9:425. [Google Scholar]
- Blue EK, Goeckeler ZM, Jin Y, Hou L, Dixon SA, Herring BP, Wysolmerski RB, Gallagher PJ. 220- and 130-kDa MLCKs have distinct tissue distributions and intracellular localization patterns. Am J Physiol Cell Physiol. 2002;282:C451–C460. doi: 10.1152/ajpcell.00333.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnette DT, Shao L, Ott C, Pasapera AM, Fischer RS, Baird MA, Der Loughian C, Delanoe-Ayari H, Paszek MJ, Davidson MW, et al. A contractile and counterbalancing adhesion system controls the 3D shape of crawling cells. J Cell Biol. 2014;205:83–96. doi: 10.1083/jcb.201311104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C-W, Kumar S. Vinculin tension distributions of individual stress fibers within cell-matrix adhesions. J Cell Sci. 2013;126((Pt 14)):3021–3030. doi: 10.1242/jcs.119032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C-W, Kumar S. Differential contributions of nonmuscle myosin II isoforms and functional domains to stress fiber mechanics. Sci Rep. 2015;5:13736. doi: 10.1038/srep13736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombelli J, Besser A, Kress H, Reynaud EG, Girard P, Caussinus E, Haselmann U, Small JV, Schwarz US, Stelzer EH. Mechanosensing in actin stress fibers revealed by a close correlation between force and protein localization. J Cell Sci. 2009;122:1665–1679. doi: 10.1242/jcs.042986. [DOI] [PubMed] [Google Scholar]
- Downing TL, Soto J, Morez C, Houssin T, Fritz A, Yuan F, Chu J, Patel S, Schaffer DV, Li S. Biophysical regulation of epigenetic state and cell reprogramming. Nat Mater. 2013;12:1154–1162. doi: 10.1038/nmat3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher PJ, Herring BP, Griffin SA, Stull JT. Molecular characterization of a mammalian smooth muscle myosin light chain kinase. J Biol Chem. 1991;266:23936–23944. [PMC free article] [PubMed] [Google Scholar]
- Gallagher PJ, Herring BP, Trafny A, Sowadski J, Stull JT. A molecular mechanism for autoinhibition of myosin light chain kinases. J Biol Chem. 1993;268:26578–26582. [PMC free article] [PubMed] [Google Scholar]
- Getz TM, Dangelmaier CA, Jin J, Daniel JL, Kunapuli SP. Differential phosphorylation of myosin light chain (Thr)18 and (Ser)19 and functional implications in platelets. J Thromb Haemost. 2010;8:2283–2293. doi: 10.1111/j.1538-7836.2010.04000.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisenberg C-P, Bellaïche Y. Forces in tissue morphogenesis and patterning. Cell. 2013;153:948–962. doi: 10.1016/j.cell.2013.05.008. [DOI] [PubMed] [Google Scholar]
- Hirano M, Hirano K. Myosin di-phosphorylation and peripheral actin bundle formation as initial events during endothelial barrier disruption. Sci Rep. 2016;6:20989. doi: 10.1038/srep20989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes JH, Kumar S. Synthetic mechanobiology: engineering cellular force generation and signaling. Curr Opin Biotechnol. 2016;40:82–89. doi: 10.1016/j.copbio.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikebe M, Hartshorne DJ. Phosphorylation of smooth muscle myosin at two distinct sites by myosin light chain kinase. J Biol Chem. 1985;260:10027–10031. [PubMed] [Google Scholar]
- Ikebe M, Hartshorne DJ, Elzinga M. Identification, phosphorylation, and dephosphorylation of a second site for myosin light chain kinase on the 20,000-dalton light chain of smooth muscle myosin. J Biol Chem. 1986;261:36–39. [PubMed] [Google Scholar]
- Ishizaki T, Naito M, Fujisawa K, Maekawa M, Watanabe N, Saito Y, Narumiya S. p160ROCK, a Rho-associated coiled-coil forming protein kinase, works downstream of Rho and induces focal adhesions. FEBS Lett. 1997;404:118–124. doi: 10.1016/s0014-5793(97)00107-5. [DOI] [PubMed] [Google Scholar]
- Itoh T, Ikebe M, Kargacin GJ, Hartshorne DJ, Kemp BE, Fay FS. Effects of modulators of myosin light-chain kinase activity in single smooth muscle cells. Nature. 1989;338:164–167. doi: 10.1038/338164a0. [DOI] [PubMed] [Google Scholar]
- Kamisoyama H, Araki Y, Ikebe M. Mutagenesis of the phosphorylation site (serine 19) of smooth muscle myosin regulatory light chain and its effects on the properties of myosin. Biochemistry. 1994;33:840–847. doi: 10.1021/bi00169a027. [DOI] [PubMed] [Google Scholar]
- Kaneko-Kawano T, Takasu F, Naoki H, Sakumura Y, Ishii S, Ueba T, Eiyama A, Okada A, Kawano Y, Suzuki K. Dynamic regulation of myosin light chain phosphorylation by Rho-kinase. PLoS One. 2012;7:e39269. doi: 10.1371/journal.pone.0039269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassianidou E, Brand CA, Schwarz US, Kumar S. Geometry and network connectivity govern the mechanics of stress fibers. Proc Natl Acad Sci USA. 2017;114:2622–2627. doi: 10.1073/pnas.1606649114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassianidou E, Kumar S. A biomechanical perspective on stress fiber structure and function. Biochim Biophys Acta. 2015;1853:3065–3074. doi: 10.1016/j.bbamcr.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Kano Y, Amano M, Kaibuchi K, Fujiwara K. Stress fiber organization regulated by MLCK and Rho-kinase in cultured human fibroblasts. Am J Physiol Cell Physiol. 2001;280:1669–1679. doi: 10.1152/ajpcell.2001.280.6.C1669. [DOI] [PubMed] [Google Scholar]
- Kumar S, Maxwell IZ, Heisterkamp A, Polte TR, Lele TP, Salanga M, Mazur E, Ingber DE. Viscoelastic retraction of single living stress fibers and its impact on cell shape, cytoskeletal organization, and extracellular matrix mechanics. Biophys J. 2006;90:3762–3773. doi: 10.1529/biophysj.105.071506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kümper S, Mardakheh FK, McCarthy A, Yeo M, Stamp GW, Paul A, Worboys J, Sadok A, Jørgensen C, Guichard S, Marshall CJ. Rho-associated kinase (ROCK) function is essential for cell cycle progression, senescence and tumorigenesis. eLife. 2016;5:e12994. doi: 10.7554/eLife.12203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JP, Kassianidou E, MacDonald JI, Francis MB, Kumar S. N-terminal specific conjugation of extracellular matrix proteins to 2-pyridinecarboxaldehyde functionalized polyacrylamide hydrogels. Biomaterials. 2016;102:268–276. doi: 10.1016/j.biomaterials.2016.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Kumar S. Actomyosin stress fiber mechanosensing in 2D and 3D. F1000Research. 2016;5:F1000 Faculty Rev-2261. doi: 10.12688/f1000research.8800.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung T, Manser E, Tan L, Lim L. A novel serine/threonine kinase binding the Ras-related RhoA GTPase which translocates the kinase to peripheral membranes. J Biol Chem. 1995;270:29051–29054. doi: 10.1074/jbc.270.49.29051. [DOI] [PubMed] [Google Scholar]
- MacKay JL, Keung AJ, Kumar S. A genetic strategy for the dynamic and graded control of cell mechanics, motility, and matrix remodeling. Biophys J. 2012;102:434–442. doi: 10.1016/j.bpj.2011.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKay JL, Kumar S. Simultaneous and independent tuning of RhoA and Rac1 activity with orthogonally inducible promoters. Integr Biol. 2014;6:885–894. doi: 10.1039/c4ib00099d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKay JL, Sood A, Kumar S. Three-dimensional patterning of multiple cell populations through orthogonal genetic control of cell motility. Soft Matter. 2014;10:2372–2380. doi: 10.1039/c3sm52265b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertsch S, Thanos S. Opposing signaling of ROCK1 and ROCK2 determines the switching of substrate specificity and the mode of migration of glioblastoma cells. Mol Neurobiol. 2014;49:900–915. doi: 10.1007/s12035-013-8568-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani T, Haga H, Koyama Y, Takahashi M, Kawabata K. Diphosphorylation of the myosin regulatory light chain enhances the tension acting on stress fibers in fibroblasts. J Cell Physiol. 2006;209:726–731. doi: 10.1002/jcp.20773. [DOI] [PubMed] [Google Scholar]
- Newell-Litwa KA, Badoual M, Asmussen H, Patel H, Whitmore L, Horwitz AR. ROCK1 and 2 differentially regulate actomyosin organization to drive cell and synaptic polarity. J Cell Biol. 2015;210:225–242. doi: 10.1083/jcb.201504046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prager-Khoutorsky M, Lichtenstein A, Krishnan R, Rajendran K, Mayo A, Kam Z, Geiger B, Bershadsky AD. Fibroblast polarization is a matrix-rigidity-dependent process controlled by focal adhesion mechanosensing. Nat Cell Biol. 2011;13:1457–1465. doi: 10.1038/ncb2370. [DOI] [PubMed] [Google Scholar]
- Priya R, Liang X, Teo JL, Duszyc K, Yap AS, Gomez GA. ROCK1 but not ROCK2 contributes to RhoA signaling and NMIIA-mediated contractility at the epithelial zonula adherens. Mol Biol Cell. 2017;28:12–20. doi: 10.1091/mbc.E16-04-0262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rape AD, Zibinsky M, Murthy N, Kumar S. A synthetic hydrogel for the high-throughput study of cell–ECM interactions. Nat Commun. 2015;6:8129. doi: 10.1038/ncomms9129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell RJ, Xia S-L, Dickinson RB, Lele TP. Sarcomere mechanics in capillary endothelial cells. Biophys J. 2009;97:1578–1585. doi: 10.1016/j.bpj.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh T, Takemura S, Ueda K, Hosoya H, Nagayama M, Haga H, Kawabata K, Yamagishi A, Takahashi M. Differential localization of non-muscle myosin II isoforms and phosphorylated regulatory light chains in human MRC-5 fibroblasts. FEBS Lett. 2001;509:365–369. doi: 10.1016/s0014-5793(01)03186-6. [DOI] [PubMed] [Google Scholar]
- Sakurada K, Seto M, Sasaki Y. Dynamics of myosin light chain phosphorylation at Ser 19 and Thr 18/Ser 19 in smooth muscle cells in culture. Am J Physiol Cell Physiol. 1998;274:1563–1672. doi: 10.1152/ajpcell.1998.274.6.C1563. [DOI] [PubMed] [Google Scholar]
- Sekine T, Yamaguchi M. Effect of ATP on the binding of N-ethylmaleimide to SH groups in the active site of myosin ATPase. J Biochem. 1963;54:196–198. [PubMed] [Google Scholar]
- Smith L, Stull JT. Myosin light chain kinase binding to actin filaments. FEBS Lett. 2000;480:298–300. doi: 10.1016/s0014-5793(00)01931-1. [DOI] [PubMed] [Google Scholar]
- Soiné JRD, Brand CA, Stricker J, Oakes PW, Gardel ML, Schwarz US. Model-based traction force microscopy reveals differential tension in cellular actin bundles. PLoS Comput Biol. 2015;11:e1004076. doi: 10.1371/journal.pcbi.1004076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachowiak MR, O’Shaughnessy B. Kinetics of stress fibers. New J Phys. 2008;10:25002. [Google Scholar]
- Stachowiak MR, Smith MA, Blankman E, Chapin LM, Balcioglu HE, Wang S, Beckerle MC, O’Shaughnessy B. A mechanical-biochemical feedback loop regulates remodeling in the actin cytoskeleton. Proc Natl Acad Sci USA. 2014;111:17528–17533. doi: 10.1073/pnas.1417686111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanenko AA, Kavsan VM. Karyotypically distinct U251, U373, and SNB19 glioma cell lines are of the same origin but have different drug treatment sensitivities. Gene. 2014;540:263–265. doi: 10.1016/j.gene.2014.02.053. [DOI] [PubMed] [Google Scholar]
- Sweeney HL, Yang Z, Zhi G, Stull JT, Trybus KM. Charge replacement near the phosphorylatable serine of the myosin regulatory light chain mimics aspects of phosphorylation. Proc Natl Acad Sci USA. 1994;91:1490–1494. doi: 10.1073/pnas.91.4.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamada M, Perez TD, Nelson WJ, Sheetz MP. Two distinct modes of myosin assembly and dynamics during epithelial wound closure. J Cell Biol. 2007;176:27–33. doi: 10.1083/jcb.200609116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tambe DT, Hardin CC, Angelini TE, Rajendran K, Park CY, Serra-Picamal X, Zhou EH, Zaman MH, Butler JP, Weitz DA, et al. Collective cell guidance by cooperative intercellular forces. Nat Mater. 2011;10:469–475. doi: 10.1038/nmat3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner K, Boudreau A, Bissell MJ, Kumar S. Dissecting regional variations in stress fiber mechanics in living cells with laser nanosurgery. Biophys J. 2010;99:2775–2783. doi: 10.1016/j.bpj.2010.08.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totsukawa G, Yamakita Y, Yamashiro S, Hartshorne DJ, Sasaki Y, Matsumura F. Distinct roles of ROCK (Rho-kinase) and MLCK in spatial regulation of MLC phosphorylation for assembly of stress fibers and focal adhesions in 3T3 fibroblasts. J Cell Biol. 2000;150:797–806. doi: 10.1083/jcb.150.4.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng Q, Duchemin-Pelletier E, Deshiere A, Balland M, Guillou H, Filhol O, Théry M. Spatial organization of the extracellular matrix regulates cell-cell junction positioning. Proc Natl Acad Sci USA. 2012;109:1506–1511. doi: 10.1073/pnas.1106377109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umemoto S, Bengur AR, Sellers JR. Effect of multiple phosphorylations of smooth muscle and cytoplasmic myosins on movement in an in vitro motility assay. J Biol Chem. 1989;264:1431–1436. [PubMed] [Google Scholar]
- Vasquez CG, Heissler SM, Billington N, Sellers JR, Martin AC. Drosophila non-muscle myosin II motor activity determines the rate of tissue folding. eLife. 2016;5:e20828. doi: 10.7554/eLife.20828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Manzanares M, Horwitz AR. Myosin light chain mono- and di-phosphorylation differentially regulate adhesion and polarity in migrating cells. Biochem Biophys Res Commun. 2010;402:537–542. doi: 10.1016/j.bbrc.2010.10.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Manzanares M, Ma X, Adelstein RS, Horwitz AR. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat Rev Mol Cell Biol. 2009;10:778–790. doi: 10.1038/nrm2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zheng XR, Riddick N, Bryden M, Baur W, Zhang X, Surks HK. ROCK isoform regulation of myosin phosphatase and contractility in vascular smooth muscle cells. Circ Res. 2009;104:531–540. doi: 10.1161/CIRCRESAHA.108.188524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Hosoya H, Yonemura S. Regulation of myosin II dynamics by phosphorylation and dephosphorylation of its light chain in epithelial cells. Mol Biol Cell. 2007;18:605–616. doi: 10.1091/mbc.E06-07-0590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt T, Taylor D, Trybus KM, Taylor K. Three-dimensional image reconstruction of dephosphorylated smooth muscle heavy meromyosin reveals asymmetry in the interaction between myosin heads and placement of subfragment 2. Proc Natl Acad Sci USA. 2001;98:4361–4366. doi: 10.1073/pnas.071051098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong SY, Ulrich TA, Deleyrolle LP, MacKay JL, Lin J-MG, Martuscello RT, Jundi MA, Reynolds BA, Kumar S. Constitutive activation of myosin-dependent contractility sensitizes glioma tumor-initiating cells to mechanical inputs and reduces tissue invasion. Cancer Res. 2015;75:1113–1122. doi: 10.1158/0008-5472.CAN-13-3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneda A, Multhaupt HAB, Couchman JR. The Rho kinases I and II regulate different aspects of myosin II activity. J Cell Biol. 2005;170:443–453. doi: 10.1083/jcb.200412043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneda A, Ushakov D, Multhaupt HAB, Couchman JR. Fibronectin matrix assembly requires distinct contributions from Rho kinases I and -II. Mol Biol Cell. 2007;18:66–75. doi: 10.1091/mbc.E06-08-0684. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.