Abstract
Type 2 diabetes mellitus (T2DM) is associated with oxidative stress and perturbed iron metabolism. Serotransferrin (Trf) and ceruloplasmin (Cp) are two key proteins involved in iron metabolism and anti-oxidant defense. Non-enzymatic glycation and oxidative modification of plasma proteins are known to occur under hyperglycemia and oxidative stress. In this study, shotgun proteomics and 2H2O-based metabolic labeling were used to characterize post-translational modifications and assess the kinetics of Trf and Cp in T2DM patients and matched controls in vivo. Six early lysine (Amadori) and one advanced arginine glycation were detected in Trf. No glycation, but five asparagine deamidations, were found in Cp. T2DM patients had increased fractional catabolic rates of both Trf and Cp that correlated with HbA1c (p < 0.05). The glycated Trf population was subject to an even faster degradation compared to the total Trf pool, suggesting that hyperglycemia contributed to an increased Trf degradation in T2DM patients. Enhanced production of Trf and Cp kept their levels stable. The changes in Trf and Cp turnover were associated with increased systemic oxidative stress without any alteration in iron status in T2DM. These findings can help better understand the potential role of altered Trf and Cp metabolism in the pathogenesis of T2DM and other diseases.
Keywords: Serotransferrin, Ceruloplasmin, Non-enzymatic glycation, Deamidation, Oxidative stress, Iron metabolism, Type 2 diabetes mellitus, Proteome dynamics, Heavy water metabolic labeling, High resolution mass spectrometry, LC-MS/MS
1. Introduction
As an essential micronutrient, iron plays a crucial role in cell survival and viability [1]. Its functional importance as a cofactor for a wide range of enzymatic reactions is mainly due to its ability to readily undergo redox cycling between its two major oxidation states, Fe3+ and Fe2+. While oxidized Fe3+ is benign, non-protein-bound reduced Fe2+ has the potential to cause oxidative damage by catalyzing the formation of hydroxyl radicals and other reactive species via Fenton reaction. Therefore, organisms have evolved elaborate mechanisms to regulate iron uptake, transport and export, and maintain its levels within safe limits [2]. In humans, dietary Fe3+ is first reduced to Fe2+, then absorbed by duodenal enterocytes and released into the blood stream through ferroportin channels. Additional influx of iron originates from the catabolism of hemoglobin-derived heme in the macrophages. The excess of iron is stored in the liver and may result in hepatic iron overload. The Fe2+ released from enterocytes, macrophages and hepatocytes is oxidized to Fe3+ by plasma ferroxidases, such as hephaestin and ceruloplasmin (Cp), for incorporation into serotransferrin (Trf), which delivers iron to all cells [3]. The specific interaction between Cp and Trf with iron depends on the structural integrity of these proteins that is critical to iron mobilization and transport [4,5].
As part of the “positive” and “negative” control mechanism of the acute phase response (APR) cascade, Cp and Trf levels are regulated in response to inflammation, infection, malnutrition and perturbations in copper and iron metabolism. These factors affect Trf and Cp expression] via a complex interplay of transcriptional and post-transcriptional factors [6–8]. While hypoxia and copper induce Cp synthesis [9], failure to incorporate copper during Cp synthesis leads to Cp degradation [10]. Inflammation, estrogen and insulin transcriptionally regulate Trf and Cp synthesis, whereas iron deficiency and hypoxia control their synthesis at a post-transcriptional level [11]. Diabetes is associated with altered iron metabolism, mild inflammation and oxidative stress [12]. While reducing circulating iron levels by iron chelation therapy is known to reverse diabetes, iron transfusion used to treat hereditary hemochromatosis, an iron overload-related disease, is associated with an increased risk of diabetes [12]. Mechanistically, iron overload is deemed to contribute to diabetes development via iron-induced beta cell dysfunction and altered glucose metabolism [13,14]. Cp and Trf serum levels are positively associated with hyperglycemia and insulin resistance whereas Trf saturation, i.e. the fraction of Trf that is saturated with iron, is negatively associated with impaired glucose metabolism, suggesting that changes in Cp and Trf metabolism could be used as surrogate markers of altered iron status in diabetes [15]. However, there is no consensus on the status of Cp and Trf metabolism in diabetes; elevated, normal, even decreased Cp and Trf levels have been reported in type 1 and type 2 diabetes mellitus (T2DM) [16–21]. It is also unknown whether hyperglycemia and hyperinsulinemia are involved in the altered Cp and Trf metabolism of diet-controlled T2DM patients.
In diabetes, hyperglycemia induces different oxidative modifications to the proteins leading to their altered biological functions. Non-enzymatic post-translational glycation and deamidation are the hallmarks of hyperglycemia- and oxidative stress-related disorders [22,23]. Non-enzymatic glycation involves covalent binding of an aldose – mostly glucose – to the terminal amino group of lysine or arginine residues to form Amadori glycation adducts (AGAs). AGAs can then give rise to a myriad of elimination, oxidation and cross-link/polymerization products, generally known as the “advanced glycation end-products” (AGEs) that promote immune response and cell damage through further protein modification and aggregation [24]. Asparagine and glutamine deamidation are other prevalent protein modifications which occur as a result of aging and/or oxidative stress [22]. Dysfunctional modified proteins are usually detected and salvaged through conjugation/secretion or proteolysis mechanisms depending on the nature of modification and pathological conditions [24].
Mass spectrometry (MS) is one of the most widely used tools to study proteins and their post-translational modifications (PTMs). Recent advancements in chromatography, MS instrumentation, data acquisition and processing have allowed for efficient detection and characterization of often low-abundance modified proteins in complex mixtures [25]. Another important application of MS analysis – isotopic labeling – has also become a widespread technique to assess protein quantities and dynamics. Heavy water (2H2O)-based metabolic labeling allows the study of different classes of biomolecules including lipids, peptides and proteins in vivo [26–30]. In this approach, the 2H from 2H2O gets incorporated into the structure of amino acids, and by that the proteins, at a rate that reflects the rate of protein synthesis. As a non-radioactive and low-cost tracer, 2H2O can be administered in drinking water and is thus widely used in human studies.
In this study, high-resolution high-accuracy MS-based proteomics was employed to detect hyperglycemia-induced PTM of two key components of iron metabolism – Trf and Cp – in T2DM patients. Heavy water labeling mass spectrometry was then used to assess in vivo dynamics of these proteins. Subsequent biochemical tests were conducted to probe the impact of the identified modifications on biological properties of Trf and Cp in T2DM patients.
2. Materials and methods
2.1. Chemicals and reagents
2H2O was purchased from Cambridge Isotopes (Tewksbury, MA). Sequencing-grade modified trypsin was from Promega (Madison, WI) and HPLC-grade water, methanol and formic acid were purchased from Fisher Scientific (Fair Lawn, NJ). The Pierce BCA protein assay kit was obtained from Thermo Fisher Scientific (Waltham, MA). All other reagents (analytical grade) were purchased from Sigma-Aldrich (St. Louis, MI).
2.2. Study design
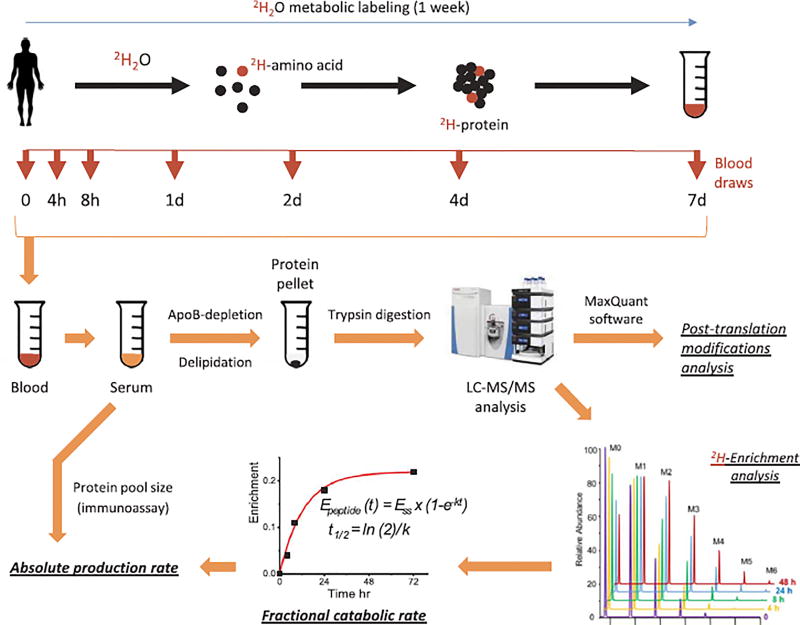
Nine newly diagnosed diet-controlled T2DM patients (4:5, male/female) and eight age- and body mass index (BMI)-matched non-diabetic control (4:4, male/female) subjects were enrolled to this study. The characteristics of study participants are shown in Table 1. The study protocol was approved by the Cleveland Clinic's Institutional Review Board. All volunteers gave their informed written consent to participate in the study after having the procedures and potential risks fully explained. T2DM patients were diagnosed based on the oral glucose tolerance test (≥ 200 mg/dl after 2 h of 75 g dextrose challenge), HbA1c (> 6.5%), or both. All patients were insulin-naive individuals with average diabetes duration of 1–3 months and were not taking any oral hypoglycemic drugs at the time of diagnosis. Since enrollment, patients were advised to adhere to lifestyle modification: hypocaloric carbohydrate-controlled diet and moderate physical activity. Follow-up counseling was provided by endocrinologist. For 3 days immediately prior to and during the one week of the kinetics study, all subjects were advised to avoid strenuous exercise and to consume an isocaloric diet to ensure weight stability and prevent diet- or exercise-induced change (stress) in protein metabolism. Individuals were excluded if they had undergone significant weight loss (> 2 kg), or had a history of alcohol and/or drug abuse, were smokers or had quit smoking within the past 3 months, or if they showed any evidence of cardiovascular, renal, hepatic, hypothyroid or hematological diseases. A 2H2O metabolic labeling approach was used to assess Trf and Cp turnover in human serum (Fig. 1). Briefly, study participants received 0.7% 2H2O in their drinking water (4.0 ml 2H2O/kg body mass) in 5 doses over a 4-h period (given at 0, 1, 2, 3 and 4 h) during the first day of the study, followed by an additional daily maintenance dose of 10% of the loading dose/day in drinking water for the next 6 days. Fasting blood samples were collected at different time-points (Fig. 1). Plasma serum was isolated and subjected to biochemical and proteomics analysis, and protein turnover measurement.
Table 1.
Demographic and biochemical characteristics of the studied subjects.
| Factor | Control (n = 8) |
T2DM (n = 9) |
p-value |
|---|---|---|---|
| Age (yr) | 48.9 ± 12.7 | 59.3 ± 9.1 | 0.07 |
| Body mass index (kg/m2) | 28.7 ± 3.3 | 31.2 ± 3.5 | 0.15 |
| Glucose (mg/dl) | 92.5 ± 11.9 | 113.3 ± 14.3 | 0.005 |
| Insulin (mIU/dl) | 8.5 ± 4.7 | 19.0 ± 7.9 | 0.005 |
| HOMA IR | 1.9 ± 1.04 | 5.3 ± 2.0 | < 0.001 |
| HbA1c(%) | 5.5 ± 0.24 | 6.3 ± 0.32 | < 0.001 |
| Triglycerides (mg/dl) | 96.1 ± 40.7 | 100.1 ± 37.3 | 0.84 |
| hsCRP (mg/l) | 1.5 ± 1.1 | 5.3 ± 4.6 | 0.15 |
| Cp (mg/dl) | 19.3 ± 2.8 | 21.1 ± 3.2 | 0.25 |
| Cp ferroxidase activity (µM/min) | 530.9 ± 64.7 | 538.8 ± 27.8 | 0.74 |
| Serum copper (µg/dl) | 72.3 ± 9.0 | 68.6 ± 13.7 | 0.57 |
| Serum iron (µg/dl) | 78.3 ± 32.6 | 78.4 ± 28.5 | 0.99 |
| Ferritin (ng/ml) | 76.3 ± 36.7 | 144.1 ± 8.5 | 0.10 |
| Trf (g/l) | 2.57 ± 0.35 | 2.52 ± 0.43 | 0.94 |
| Trf unsaturated iron-binding capacity (µg/dl) | 216.3 ± 66.1 | 215.0 ± 66.3 | 0.97 |
| Trf total iron-binding capacity (µg/dl) | 294.6 ± 39.8 | 293.4 ± 44.7 | 0.96 |
| Trf iron saturation | 0.28 ± 0.12 | 0.28 ± 0.13 | 0.94 |
| Cp/Trf ratio | 7.7 ± 1.2 | 8.6 ± 1.2 | 0.21 |
| TBARS (µM) | 0.48 ± 0.16 | 0.43 ± 0.08 | 0.43 |
| Anti-oxidant capacity of plasma (RFU/µl/min) | 37.2 ± 8.2 | 33.7 ± 7.1 | 0.45 |
Statistics presented as mean ± SD. p-Values are from Student's t-test.
Cp: ceruloplasmin, HbA1c: glycated hemoglobin, HOMA IR: homeostatic model assessment of insulin resistance, hsCRP: high-sensitivity C-reactive protein, RFU: relative fluorescence unit, TBARS: thiobarbituric acid reactive substances, Trf: serotransferrin.
Fig. 1.
Experimental design and the workflow of the study. Control (n = 8) and diabetic (n = 9) subjects received 4.0 ml/kg bolus dose of 2H2O during the first day of the study and 10% of the loading dose/day for the following 6 days. Blood samples were drawn at 0, 4, 8, 24, 48, 96 and 168 h. Serum proteins were isolated and analyzed by LC-MS/MS after depletion of apolipoprotein B-associated lipoproteins, delipidation and trypsin-digestion. High-resolution mass spectra were collected and processed to (i) identify proteins and their post-translational modifications, and (ii) assess protein fractional catabolic rates (k) based on the rate of 2H-incorporation. Production rate of serotransferrin and ceruloplasmin was calculated as the product of protein pool size and their respective fractional catabolic rate.
2.3. Analytical procedures
Fasting plasma glucose, triglycerides, total cholesterol, and high-density lipoprotein (HDL) cholesterol were measured by enzymatic analysis on an automated platform (Roche Modular Diagnostics, Indianapolis, IN). Plasma insulin concentration was determined by radioimmunoassay (Diagnostic Products, Los Angeles, CA). Human Trf and Cp were quantified by immunoassay methods on an Abbott Architect ci8200 Integrated Analyzer System (Abbott Labs, Abbott Park, IL). Serum ferritin levels were quantified using human ferritin enzyme-linked immunosorbent assay (ELISA) kit (Thermo Fisher Scientific) according to manufacturer's protocol. High-sensitivity C-reactive protein (hsCRP) serum levels were measured using a standard clinical assay. Serum iron and total iron-binding capacity (TIBC), i.e. the total amount of iron that can bind Trf, were measured photometrically using commercially available kits. Unsaturated iron-binding capacity (UIBC), i.e. the portion of Trf that is not saturated with iron, was calculated as UIBC = TIBC – iron. Trf saturation was calculated by the formula: serum iron/TIBC. Cp N-oxidase activity was measured by spectrophotometry at 540 nm with o-dianisidine as substrate, using the method from Wei et al. [31] modified based on Lehmann et al. [32]. Cp ferroxidase activity was measured at 530 nm with p-phenylenediamine as described [33].
2.3.1. Total body water enrichment measurement
2H-enrichment of total body water was measured using a modification of the acetone exchange method [34]. Briefly, serum (5 µl) was incubated with 10 M potassium hydroxide (5 µl) and pure acetone (5 µl) in a 2-ml glass screw-cap GC vial at room temperature for 4 h. One µl of acetone vapor from the headspace was directly injected for gas chromatography-mass spectrometry (GC-MS) analysis. Isotopic enrichment of acetone was determined using electron impact ionization and selected ion monitoring. Acetone isotopomers were monitored at m/z 58 (M0), 59 (M1) and 60 (M2). Biological samples were analyzed in parallel with a set of calibration curve samples containing 0–5% 2H2O. The regression equation of the calibration curve was used for 2H2O enrichment measurement in the biological sample.
2.3.2. Oxidative stress assessment
The effect of altered Cp and Trf metabolism on systemic oxidative stress was assessed based on two independent assays: lipid peroxidation assay and anti-oxidant capacity of plasma.
2.3.2.1. Lipid peroxidation assay
The extent of lipid peroxidation was determined by measuring thiobarbituric acid reactive substances (TBARS) including malondialdehyde in EDTA-treated plasma samples using a commercial kit provided by Cayman Chemical (Ann Arbor, MI). In brief, serum (30 µl) was mixed with 10% tricholoroacetic acid (30 µl) and a color reagent (1.33 ml) composed of 37 mM thiobarbituric acid, 1.75 M acetic acid and 0.35 M sodium hydroxide. The mixture was incubated at 100 °C for 1 h, the precipitate was removed, and TBARS were measured in the supernatants using fluorometry at 530:550 nm (excitation/emission).
2.3.2.2. Anti-oxidant capacity of plasma
This assay measures the anti-oxidant capacity of plasma proteins against Cu2+-induced oxidative stress. Briefly, oxidation was initiated in apolipoprotein B (ApoB)-depleted plasma by adding Cu2+ and the rate of total oxidation was quantified by 2′,7′-dichlorodihydrofluorescein (DCFH) in a microtiter plate at 37 °C. Fluorescent emission was measured at 530 nm after serial excitation at 485 nm.
2.4. LC-MS/MS analysis
Since Trf and Cp are associated with HDL [35], less-dense lipid-rich ApoB-associated particles that can interfere with tryptic digestion were removed prior to the LC-MS/MS analysis [36]. Serum (20 µl) was diluted with phosphate buffer saline pH 7.4 (1:1 v/v), ApoB-depleted using an Mg2+/dextran sulfate reagent (4 µl) from Stanbio (Boerne, TX), and delipidated with ice-cold acetone (1:10, v/v). The pellet was resolubilized in 100 mM ammonium bicarbonate pH 8.0 (200 µl). An aliquot equivalent to 10 µg protein (BCA assay) was reduced (15 mM dithiothreitol, 30 min, 60 °C), alkylated (45 mM 2-iodoacetamide, 30 min, 25 °C, in the dark) and digested by trypsin at a 1:40 w/w enzyme-to-protein ratio at 37 °C overnight. Digestion mixture was acidified (2% formic acid, 5 µl) and cleaned up on a HyperSEP Retain PEP (Thermo Fisher Scientific) solid-phase extraction cartridge using water and methanol. Sample was evaporated to dryness under vacuum (2 h, 45 °C, Savant Universal, Thermo Fisher Scientific) and reconstituted in 0.1% formic acid. A volume approximately equivalent to 0.1 µg protein was injected into a Q Exactive Plus hybrid quadrupole-Orbitrap mass spectrometer (Thermo Fisher Scientific) using an UltiMate 3000 RSLC nano-HPLC system (Dionex, Sunnyvale, CA). Liquid chromatography was conducted at 0.3 µl/min (40 °C) on an Acclaim PepMap RSLC (C18) 0.075 × 150 mm column (Dionex) with a gradient elution of 5–35% B in 90 min then up to 66.7% in 1 min followed by another increase to 89% B in 1 min held for 5 min whereas solvents A and B were water and acetonitrile (0.1% formic acid), respectively. Mass spectrometry analysis was performed at m/z 380–1300 (MS) with 70,000 resolution (200 m/z). MS/MS spectra were collected in data-dependent acquisition mode for the 12 most abundant product ions within m/z 110–2000 with an isolation window of 2.0 ± 0.4 m/z and 17,500 resolution (200 m/z). MS and MS/MS spectra were acquired for 100 ms with the automatic gain control (AGC) target set at 1.0 × 106 and 2.0 × 104 ions for MS and MS/MS scans, respectively. Higher-energy collisional dissociation (HCD) was performed at normalized collision energy of 25%. Dynamic exclusion was enabled for a duration of 17 s. Samples were analyzed in triplicate. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium [37] via the PRIDE partner repository [38] with the dataset identifier PXD005755.
2.4.1. Detection of post-translational modifications
Andromeda search engine was used within the MaxQuant software environment (version 1.5) [39] for PTM identification at 1% false discovery rate. To maximize detection efficiency, similar PTMs were grouped and searched together. The search was performed against TRFE_HUMAN.FASTA (P02787) and CERU_HUMAN.FASTA (P00450) entries from the UniProt Consortium website (http://www.uniprot.org). Trypsin was chosen as the protease allowing up to two missed cleavages per peptide. Cysteine carbamidomethylation was added as a fixed modification. Methionine oxidation, lysine early glycation and the most common AGEs including carboxymethyl lysine, carboxyethyl lysine, lysine pyrraline, arginine pyrimidine and arginine hydroimidazolone derivatives (G-H1, MG-H1, 3DG-H1) were also set as variable modifications. Since AGAs are known to be susceptible to HCD-induced elimination of up to three water molecules per glycation sites [40], peptide identification method was modified to include a –H6O3 neutral loss for lysine glycation. Other searched PTMs were 4-hydroxynonenal and/or 4-oxononenal addition to arginine and lysine residues, lysine acetylation and acetoacetylation, tyrosine and tryptophan nitration, tyrosine chlorination, and asparagine/glutamine deamidation. A matching score threshold of 40 was set for all Andromeda searches. To account for LC retention shifts, the “match between runs” option was enabled with a match time window of 0.7 min and an alignment time window of 20 min.
2.4.2. Protein turnover measurement
LC-MS/MS data files were searched using Mascot software (Matrix Science, London, UK) version 2.3 against the human subset of UniProt protein database released on June 29th, 2016 (containing 149,730 entries) with cysteine carbamidomethylation and methionine oxidation as fixed and variable modifications and trypsin digestion with a maximum of two missed cleavages per peptide. A custom-built software application (http://ispace.utmb.edu/users/rgsadygo/proteomics/heavywater) was employed to calculate isotopic enrichment (E) of the 2H atoms incorporated in the proteins of interest at each time-point. To correct for body water enrichment variations, E values were normalized to the subject's total body water 2H-enrichment.
The rate constant (k) representing the protein's fractional catabolic rate (FCR) was determined using a one-compartmental model based on the exponential growth curve fitting of net normalized 2H- enrichment values of tryptic peptides against time (t) (Eq. (1)) after the removal of statistical outliers within the Prism (GraphPad, La Jolla, CA) software (version 5).
| (1) |
Here, Ess is the plateau enrichment of a tryptic peptide. Protein half-life (t½) was later obtained from Eq. (2)
| (2) |
Finally, production rate (PR) was calculated from the protein's FCR and its total pool size (P) using Eq. (3).
| (3) |
2.4.3. Quantification of Trf glycation
Trf glycation was quantified through the calculation of the averaged intensity ratios from the extracted ion chromatograms corresponding to both native and glycated peptides. A Visual Basic application was developed for accurate quantification of the modified peptides. Quantitation was based on the following restriction criteria. First, to reduce the interference of background signals, only the MS scans with monoisotopic (M0) peak intensity > 104 were quantified. Second, outlier scans in terms of their mass isotopomer intensity ratios were excluded. The scans generating a coefficient of variation > 50% relative to the averaged scans were also considered outliers.
The effect of glycation on Trf stability was assessed based on the half-lives of one glycated peptide and the total Trf pool consisting of mainly non-modified Trf peptides. 2H-enrichment of both peptides were extracted from their high resolution MS spectra whereas molar percent enrichments were calculated based on isotopomer distribution analysis, as described in the previous section.
2.5. Data presentation and statistical analysis
Continuous variables were evaluated for normality using the Shapiro-Wilk test. Only glycation measures showed strong departures from normality, and were compared between groups defined by diabetic status using nonparametric Wilcoxon rank sum tests and were summarized using medians and quartiles. Normally-distributed continuous measures were summarized using means and standard deviations and were compared between groups using two-sample t-tests. To evaluate the impact of demographic factors on associations observed, linear regression models, with and without adjustment factors, one at a time, were performed. If necessary, log transformations (with offsets) were performed to achieve more normally distributed measures. Mean differences between groups with 95%-confidence intervals were presented. Categorical factors were compared using Fisher exact tests. To assess correlations between continuous factors, Pearson and Spearman correlations were used for normally and non-normally distributed factors, respectively. Analysis was performed using SAS (Cary, NC) software (version 9.4).
3. Results
3.1. Clinical characteristics
As expected, compared to healthy controls, T2DM patients had significantly higher levels of blood glucose, glycated hemoglobin (HbA1c), insulin, and insulin resistance assessed based on homeostatic model assessment (HOMA) (Table 1). All other biochemical characteristics, including body mass index (BMI) and lipid profile were similar between the two groups as well as their age. Although ferritin and hsCRP levels were higher in diabetes compared to the controls, they did not reach significance due to large variabilities among the subjects.
3.2. Proteomics analysis of Trf and Cp
3.2.1. Post-translational modification of serotransferrin and ceruloplasmin
To evaluate the role of diabetes on Trf and Cp structural integrity and stability, a proteomics approach combining high-resolution data-dependent mass spectrometry with nano-flow liquid chromatography was optimized to enhance peptide identification efficiency and maximize protein sequence coverage in human serum samples. Applying this method to T2DM and non-diabetic samples, Trf and Cp were identified with average sequence coverage of 89% and 84%, respectively.
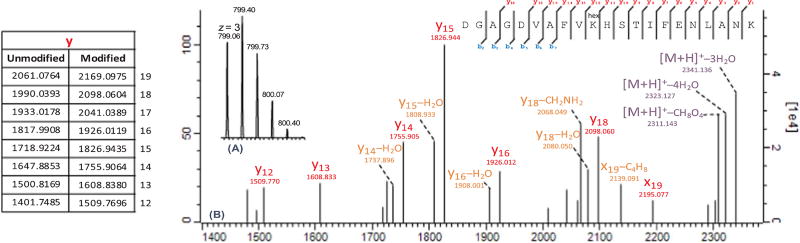
A recently developed approach was used for efficient detection and characterization of glycated peptides by HCD fragmentation in Orbitrap mass spectrometers [40]. Six lysine residues including Lys-103, Lys-206, Lys-276, Lys-296, Lys-534 and Lys-640 were found in Trf to be the targets of early Amadori glycation in both T2DM and control subjects with high score (MaxQuant score > 40) and mass accuracy (< 1 ppm) (Table 2). However, the PTM search for Trf AGEs led to the detection of merely one arginine pyrimidine (Argpyr) modification at Arg-678 in a limited number of diabetic subjects. Argpyr is one of the cyclic AGEs known to form through modification of arginine residues with methylglyoxal, a secondary reactive oxygen species [41]. Peptide structures were confirmed using the “expert system” integrated into the MaxQuant software for accurate annotation of collision induced dissociation (CID)–generated MS/MS spectra [42]. As an example, Fig. 2 depicts MS and MS/MS spectra (m/z > 1400) of the Trf peptide containing the Lys-206 glycation site. The neutral losses of H6O3 (3H2O) and CH8O4 (3H2O + HCOH), observed for glycated peptides, are associated with post-activation fragmentation of the AGA (M + 162) into the relatively more stable pyrilium (M + 108) and furylium/immonium (M + 78) ions, respectively. Annotated MS/MS spectra of other glycated peptides can be found under Section A of the Supplemental Figures.
Table 2.
List of peptides containing serotransferrin early (bold) and advanced (underlined) glycation sites. Location of the modified residue in the protein as well as MaxQuant score, theoretical m/z value, mass accuracy (in ppm), charge state (z) and retention time of the corresponding peptide are included.
| Modified peptide | Site | MaxQuant score | m/z (ppm) | z | Retention time (min) |
|---|---|---|---|---|---|
| KDSGFQMNQLR | K-103 | 48 | 495.9049 (0.01) | 3 | 31.6 |
| DGAGDVAFVKHSTIFENLANK | K-206 | 138 | 799.0640 (0.03) | 3 | 73.2 |
| SMGGKEDLIWELLNQAQEHFGKDK | K-276 | 53 | 734.6090 (0.02) | 4 | 107.0 |
| DLLFKDSAHGFLK | K-296 | 66 | 551.6225 (0.03) | 3 | 54.1 |
| GDVAFVKHQTVPQNTGGK | K-534 | 49 | 682.3481 (0.01) | 3 | 40.5 |
| DDTVCLAKLHDR | K-640 | 101 | 535.5910 (0.02) | 3 | 29.8 |
| CSTSSLLEACTFRR | R-678 | 52 | 884.4113 (0.01) | 2 | 63.9 |
Fig. 2.
MS (A) and MS/MS (B) spectra of the peptide containing glycated Lys-206 residue of human serotransferrin. Product ions detected at m/z 2341.1 and 2311.1 represent the pyrilium (M + 108) and furylium/immonium (M + 78) ions formed by post-activation fragmentation of the relatively less stable Amadori compound (M + 162). Here, the MS/MS spectrum has been zoomed in to include y-ions containing the glycated residue. The detection of pyrilium y-ions (y + C6H4O2) together with multiple losses of H2O are particular to glycated peptides. Theoretical values for selected y-ions are listed in the table on the left.
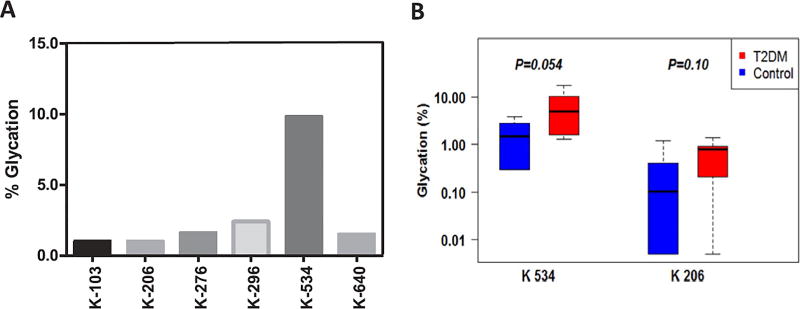
Accurate peak integration of glycated peptides in the T2DM sample with the highest extent of glycation revealed that Lys-534 is the primary site of modification accounting for nearly 60% of total protein glycation and 13% more than other residues altogether (Fig. 3A and Supplementary Table S1). Taking this lysine residue as a surrogate estimate of protein glycation, Trf was found to be more extensively glycated in T2DM patients than in control samples (5.6 ± 1.6% vs. 1.8 ± 0.5%, p = 0.054) (Fig. 3B, left panel). Although a notably higher Lys-206 glycation was observed in T2DM patients, the differences didn't reach significance due to large variabilities between the subjects (Fig. 3B, right panel).
Fig. 3.
Enhanced serotransferrin glycation in T2DM patients. Extent of modification (%) of serotransferrin glycation sites in a selected T2DM patient highlighting Lys-534 as the major site of glycation (A). Lys-206 and Lys-534 are positioned on the two GDVAFVKH glycation motifs located at the entrance of the iron-binding pockets. Glycation at these sites can sterically hinder iron (III) coordination impairing the protein's iron-transporting function. Serotransferrin was glycated at a higher extent at Lys-206 and Lys-534 in T2DM patients than in controls (B). Percent glycation was calculated by dividing peak area of the glycated peptide over the sum of all peptide species (of different length and charge state) containing the site of modification.
In contrast to Trf, no non-enzymatic glycation was detected in Cp. However, five asparagine residues were found to be deamidated in Cp in T2DM and control subjects (Supplementary Table S2). Peptides containing these residues were identified under stringent parameters (MaxQuant score > 50, mass accuracy < 1 ppm) and were confirmed based on their parent (MS) and product (MS/MS) spectra (Supplemental Figures, Section B). Asparagine deamidation takes place during protein aging and/or oxidative stress and may affect proteins’ structure and function via altering their total charge states. Particularly, oxidative stress-induced deamidation of Cp at Asn-943 can potentially affect its ferroxidase activity [4]. Due to large variabilities between the samples, the deamidated peptides could not be quantified accurately.
3.3. Serotransferrin and ceruloplasmin kinetics in T2DM
To evaluate the effect of early diabetes on Trf and Cp metabolism a 2H2O metabolic labeling approach was applied (Fig. 1). The FCRs were measured from the isotopic enrichment of unique proteolytic peptides corresponding to each protein. The results show that both Trf and Cp had significantly increased FCR in T2DM patients compared to the healthy controls (Trf: 0.013 ± 0.003 vs. 0.006 ± 0.001 pool/h, p < 0.001 and Cp: 0.027 ± 0.004 vs. 0.018 ± 0.004 pool/h, p = 0.002). Enhanced FCR of Trf and Cp resulted in their shorter half-lives in T2DM patients compared to the controls (Trf: 55.9 ± 11.8 vs. 120.5 ± 20.3 h and Cp: 34.8 ± 6.8 vs. 42.4 ± 11.3 h, both p < 0.005). However, changes in Trf and Cp FCR did not have any effect on their pool sizes. Both Trf and Cp had higher PRs in T2DM patients than in healthy controls indicating that the fluxes of Trf and Cp have increased in T2DM to keep their levels stable (Table 3).
Table 3.
Serotransferrin and ceruloplasmin kinetics in T2DM patients (n = 9) and matched controls (n = 8).
| Trf | Cp | |||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| Parameter | Control | T2DM | p-value | Control | T2DM | p-value |
| Pool size (g) | 9.0 ± 1.1 | 10.5 ± 2.1 | 0.13 | 0.7 ± 0.1 | 0.8 ± 0.2 | 0.08 |
| FCR (pool/h) | 0.006 ± 0.001 | 0.013 ± 0.003 | < 0.001 | 0.018 ± 0.004 | 0.027 ± 0.004 | 0.002 |
| Half-life (h) | 120.5 ± 20.3 | 55.9 ± 11.8 | < 0.001 | 42.4 ± 11.3 | 34.8 ± 6.8 | 0.003 |
| PR (mg/kg.h) | 0.68 ± 0.12 | 1.44 ± 0.47 | < 0.001 | 0.15 ± 0.04 | 0.25 ± 0.07 | 0.002 |
Statistics presented as mean ± SD. p-Values are from Student's t-test.
FCR: fractional catabolic rate, PR: production rate.
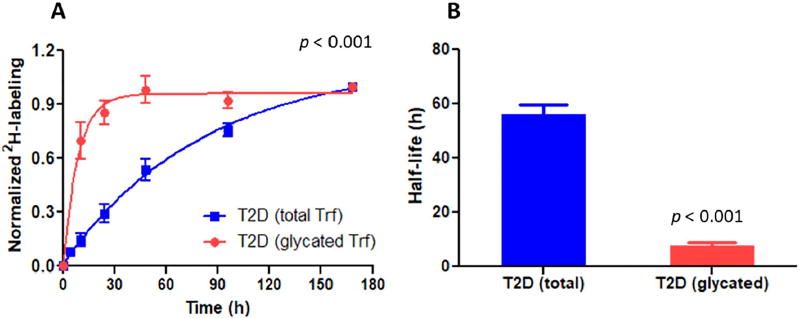
To find out if glycation has contributed to increased Trf degradation, the kinetics of the Trf peptide GDVAFVKHQTVPQNTGGK modified at the Lys-534 site was quantified. The results showed that, in T2DM patients, glycated Trf had a significantly higher FCR (0.105 ± 0.048 pool/ h, p < 0.001) than total Trf, based on both the modified and unmodified peptides (Fig. 4A). PTM-induced increased degradation of Trf resulted in a shorter half-life of the modified protein variant (7.7 ± 2.9 h, p < 0.001) compared to the total pool of Trf consisting the modified and unmodified species together (Fig. 4B).
Fig. 4.
Effect of glycation on serotransferrin kinetics in T2DM patients. The glycated Trf peptide reaches maximum 2H enrichment levels at a higher rate than the total Trf pool (consisting modified and unmodified Trf variants) which is an indicator of its faster catabolism (A) and thus shorter half-life (B).
3.4. Effect of structural modifications on Trf and Cp function
Since Trf glycation and Cp deamidation may affect their biological functions [4,5], we measured serum iron and copper levels, Cp enzymatic activity, Trf iron saturation, TIBC, UIBC, and the extent of systemic oxidative stress as surrogates to Trf and Cp function. Our results did not show any loss in Trf iron-binding capacity and iron saturation in T2DM patients. Similarly, no difference was observed in serum copper levels and Cp ferroxidase and N-oxidase (not shown) activities between healthy controls and patients. Likewise, no significant difference was observed in systemic lipid peroxidation measured as TBARS in EDTA-treated plasma (Table 1). To further determine the functional consequence of T2DM-induced change in the dynamics of Trf and Cp, we also measured the anti-oxidant capacity of plasma. This test determines the capacity of plasma to either inhibit or aggravate Cu2+-induced total oxidation. Consistent with other functional results, we could not detect any differences in anti-oxidant capacity of plasma between healthy controls and T2DM patients, indicating that the alterations in Trf and Cp metabolism were not associated with systemic oxidative stress.
3.5. Correlations
To assess the contribution of different biochemical variables to altered Trf and Cp metabolism in diabetes, we performed a linear regression analysis between selected variables and measured parameters. We also evaluated whether the alterations in Trf and Cp kinetics were associated with serum iron levels and oxidative stress-related parameters. To identify independent predictors of the observed differences between the controls and T2DM subjects, we performed adjustments for multiple clinical variables, including age, gender, BMI, glucose, insulin, insulin resistance (HOMA), total cholesterol, triglycerides and HbA1c. The regression analysis showed that Cp FCR was directly associated with HbA1c in all the studied subjects (Table 4). However, a direct association between Trf FCR and HbA1c was only observed for the T2DM patients, suggesting that hyperglycemia could be involved in increased degradation of Trf and Cp in diabetes. There was a trend towards a positive correlation between Trf glycation and Trf PR (p = 0.07), suggesting that increased Trf production may have compensated for its glycation-induced degradation. Furthermore, a strong direct correlation was observed between Trf and Cp kinetics (Table 4), indicating coordinated upregulation of Trf and Cp flux in T2DM to keep their levels stable. Despite no significant change in iron metabolism on the onset of T2DM, TIBC of Trf had marginal inverse association with Cp FCR (p = 0.05), implying that increased clearance of Cp may contribute to altered iron metabolism in severe diabetes. The adjustment for age, glucose, insulin and HOMA substantially weakened the differences between the groups in Cp FCR, suggesting that aging and insulin resistance may contribute to increased Cp degradation. Interestingly, the differences in Cp FCR were completely abolished only after the adjustment for HbA1c (Supplementary Table S3). Similarly, the differences in Trf glycation were eliminated after the adjustment for diabetes-related parameters (glucose, insulin, HOMA and HbA1c) (Supplementary Table S4).
Table 4.
Correlations between kinetic parameters, HbA1c, and selected oxidative stress-related variables.
| Parameter | Variable | Groupa | Rhoa | 95% CI | p-valuea |
|---|---|---|---|---|---|
| FCR Trf (pool/h) | HbA1c | Ct+T2DM | 0.24 | (−0.30, 0.77) | 0.360 |
| FCR Trf (pool/h) | HbA1c | T2DM | 0.77 | (−1.00, −0.20) | 0.015 |
| FCR Cp (pool/h) | HbA1c | Ct+T2DM | 0.65 | (0.23, 1.00) | 0.005 |
| PR Trf (mg/kg h) | FCR Cp (pool/h) | Ct+T2DM | 0.43 | (−0.09, 0.95) | 0.096 |
| PR Trf (mg/kg h) | PR Cp (mg/kg h) | Ct+T2DM | 0.670 | (0.24, 1.00) | 0.005 |
| PR Trf (mg/kg h) | Trf % glycationb | Ct+T2DM | 0.470 | (−0.04, 0.97) | 0.070 |
| Iron (µg/dl) | hsCRP | Ct+T2DM | −0.47 | (−0.97, 0.04) | 0.069 |
| TIBC (µg/dl) | FCR Cp (pool/h) | Ct+T2DM | −0.49 | (0.99, 0.01) | 0.054 |
Cp: ceruloplasmin, HbA1c: glycated hemoglobin, hsCRP: high-sensitivity C-reactive protein, FCR: fractional catabolic rate, PR: production rate, TIBC: Trf total iron-binding capacity, Trf: serotransferrin.
Ct: control. Rho and p-values are from Spearman's method.
Protein glycation at the Lys-534 residue.
4. Discussions
In this study, we utilized 2H2O-metabolic labeling and shotgun proteomics to investigate the effect of diet-controlled T2DM on the metabolism of Trf and Cp, two key proteins involved in iron metabolism. Our results demonstrated that even at this early stage of the disease, T2DM has a profound effect on Trf and Cp metabolism: both proteins were degraded 1.5–2-fold faster in the patients than in healthy controls. Increased clearance of Trf and Cp was associated with hyperglycemia, and proteomics analysis revealed that post-translational glycation contributed to reduced stability of Trf in T2DM. However, coordinated increase in Trf and Cp production keeps their levels and the iron status stable.
While very little is known about in vivo regulation of Cp, Trf metabolism is well studied in human metabolic diseases and stress conditions such as inflammation and iron imbalance [8,43–45]. In particular, Trf flux is known to be increased in the patients with alcoholic fatty liver disease [45] and nephrosis [8] without any significant correlation with serum Trf and iron levels. In the children with severe protein-energy malnutrition, lower Trf plasma levels were found to be related to its decreased synthesis and increased degradation [8]. These studies did however not explain why Trf would degrade faster in the diseased state. It is also not yet clear how Trf metabolism is regulated in T2DM. In this study, the FCRs of both Trf and Cp increased in T2DM and this was associated with their PTM. Our results suggest that neither iron deficiency nor inflammation is the main drive of increased Trf and Cp turnover in T2DM patients, as no correlation was found between Trf or Cp fluxes and serum iron or hsCRP, which is a marker of systemic inflammation. In contrast, increased FCR of Trf and Cp in T2DM patients directly correlated with HbA1c, a marker of long-term hyperglycemia. While adjustment for glucose weakened the differences in Cp FCR, adjustment for HbA1c completely eliminated the latter, enforcing that chronic hyperglycemia contributed to increased Cp degradation. Although our data show that hyperglycemia-induced glycation enhanced Trf degradation, the cause of increased Cp degradation in T2DM is not clear. It is possible that oxidative stress-induced deamidation in hyperglycemic environment may have contributed to the enhanced Cp degradation in T2DM. Further studies are warranted to assess the role of deamidation and other oxidative modifications on Cp structural alteration and stability.
In T2DM and other conditions associated with insulin resistance, in addition to hyperglycemia, hyperinsulinemia also might be involved in altered Trf and Cp metabolism. Although the effect of hyperinsulinemia on Trf and Cp turnover in T2DM is not known, it has been shown that physiological increment of plasma insulin concentration affects the synthesis of other secreted liver proteins in healthy human subjects [46]. In vitro studies in HepG2 cells have shown that insulin plays a dual role in the transcriptional regulation of Cp: it increases Cp transcription under normal physiological conditions, while inhibiting Cp gene expression in the presence of interleukin 6, a pro-inflammatory cytokine. Interestingly, both effects of insulin on Cp metabolism are mediated via hypoxia inducible factor 1 (HIF-1), reflecting the important role of insulin in inflammation and iron homeostasis [47]. In our study, we found that the adjustments for insulin and HOMA markedly reduced the differences in Cp FCR between the groups (Supplementary Table S3), suggesting that hyperinsulinemia could contribute to altered Cp metabolism in T2DM. As a modulator of HIF-1, insulin also increases Trf and Trf receptor-1 (Trfr-1) transcription and attenuates inflammation-induced suppression of Trf in human hepatocytes [48–50]. Although we did not observe any correlation between Trf kinetics and insulin, hyperinsulinemia-induced increase in Trfr-1 and Trf transcription could potentially facilitate iron uptake through increased Trf flux in T2DM. Our results on altered Trf and Cp turnover in T2DM complement the existing knowledge on Cp regulation and Trf metabolism. The unique combination of shotgun proteomics with 2H metabolic labeling in our study suggests that hyperglycemia-induced modification is an important determinant of Trf, and possibly Cp, turnover. Thus, while transcriptional and post-transcriptional factors are the key regulators of Trf and Cp synthesis, post-translational modifications may play role in their degradation.
Our findings may have clinical implications pertaining to the oxidative stress and iron metabolism in T2DM. While in vitro experiments suggest that Trf glycation impairs its Fe3+–binding function, the mechanisms of altered iron metabolism in diabetes are not defined. An in vitro study by Silva et al. demonstrated that Lys-206 and Lys-534 sites of Trf are the most vulnerable residues to glycation due to their low side-chain pKa values [51]. The X-ray structure analysis of Trf shows that Lys-206 and Lys-534 are positioned on the two GDVAFVKH “glycation motifs” located at the entrance of Trf iron-binding pockets of the N- and C-terminal lobes of the protein [52]. The side-chain amino group of these lysine residues participates in hydrogen bonding crucial for Fe3+ coordination [53,54]. Our study showed that Lys-206 and Lys-534 were both glycated in T2DM patients (Fig. 3B) and Lys-534 is the primary site of Trf glycation in vivo (Fig. 3A and Supplementary Table S1). It is plausible that post-translational glycation at Lys-534 site (and potentially Lys-206, to a lesser extent) can sterically hinder iron-binding capacity of Trf, rendering it dysfunctional. In support of this notion, an in vitro study has shown that non-enzymatic glycation of Trf reduces its iron-binding capacity contributing to the pro-oxidant effects of unbound iron [5].
Similarly, it has been shown that oxidative stress-induced deamidation of Cp reduces its ferroxidase activity which may further exacerbate Fe2+-induced oxidative stress [55]. In particular, oxidative deamidation at Asn-943 of Cp produces an isoDGR motif resulting in the loss of Cp ferroxidase activity and acquisition of integrin-binding properties [4,56]. Our study revealed that five Cp Asn residues, including Asn-943, were deamidated in T2DM patients (Supplementary Table S2). It is conceivable that Cp modification, particularly at the Asn-943 site, could compromise its antioxidant function fueling oxidative stress in poorly controlled diabetic patients. A correlation of Cp FCR with Trf TIBC in our study suggests that increased Cp degradation may contribute to altered iron metabolism in T2DM. However, in contrast to observed alterations in the Trf and Cp kinetics, no difference was seen between the patients’ and the controls’ iron status, including Trf, Cp, ferritin and iron levels, Trf/Cp ratio, Cp ferroxidase activity, Trf UIBC, TIBC and iron saturation (Table 1). This is in agreement with the previous reports demonstrating that Trf glycation is only minimally responsible for the iron-binding antioxidant capacity in diabetic patients [21] and that Trf glycation does not correlate with serum iron or TIBC [57]. Consistent with the unaltered iron metabolism, the present study failed to detect any measurable oxidative stress in T2DM patients. Although more targeted studies are required to delineate the role of hyperinsulinemia and hyperglycemia in Trf and Cp metabolism in T2DM, we speculate that while at the early stages of the disease, hyperinsulinemia-associated increase in Trf and Cp production compensates for their hyperglycemia-induced degradation, with the progression of the disease, uncompensated degradation may result in altered iron metabolism and oxidative stress.
In summary, our study demonstrated that Trf and Cp turnovers were increased in T2DM patients and that was associated with hyperglycemia-induced non-enzymatic Trf glycation, and potentially Cp deamidation. Kinetics analysis revealed that Trf glycation was associated with reduced Trf stability in vivo; suggesting that hyperglycemia-induced degradation of Trf and Cp may impair Fe3+/Fe2+ homeostasis at the outbreak of oxidative stress in the patients with severe T2DM. This proteomics and stable-isotope labeling approach for accurate measurement of the dynamics of native and modified proteins could be applied to assess potential mechanisms involved in the pathogenesis of hyperglycemia-induced complications of diabetes, and other diseases. However, additional studies are required to examine the links between these protein modifications and their impact on the biology of iron metabolism and T2DM development.
Supplementary Material
Abbreviations
- AGA
Amadori glycation adduct
- AGE
advanced glycation end-product
- ApoB
apolipoprotein B
- APR
acute phase response
- Argpyr
arginine pyrimidine
- BMI
body mass index
- Cp
ceruloplasmin
- FCR
fractional catabolic rate
- GC-MS
gas chromatography mass spectrometry
- HbA1c
glycated hemoglobin
- HCD
higher-energy collisional dissociation
- HDL
high-density lipoprotein
- hsCRP
high-sensitivity C-reactive protein
- HOMA IR
homeostatic model assessment of insulin resistance
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- PR
production rate
- PTM
post-translational modification
- T2DM
type 2 diabetes mellitus
- TBARS
thiobarbituric acid reactive substances
- TIBC
total iron-binding capacity
- Trf
serotransferrin
- UIBC
unsaturated iron-binding capacity
Footnotes
Appendix A. Supporting information
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.freeradbiomed.2017.10.373.
References
- 1.Schneider BD, Leibold EA. Regulation of mammalian iron homeostasis. Curr. Opin. Clin. Nutr. Metab. Care. 2000;3(4):267–273. doi: 10.1097/00075197-200007000-00005. [DOI] [PubMed] [Google Scholar]
- 2.Wallace DF. The regulation of iron absorption and homeostasis. Clin. Biochem. Rev. 2016;37(2):51–62. [PMC free article] [PubMed] [Google Scholar]
- 3.Gkouvatsos K, Papanikolaou G, Pantopoulos K. Regulation of iron transport and the role of transferrin. Biochim. Biophys. Acta. 2012;1820(3):188–202. doi: 10.1016/j.bbagen.2011.10.013. [DOI] [PubMed] [Google Scholar]
- 4.Barbariga M, Curnis F, Spitaleri A, Andolfo A, Zucchelli C, Lazzaro M, Magnani G, Musco G, Corti A, Alessio M. Oxidation-induced structural changes of ceruloplasmin foster NGR motif deamidation that promotes integrin binding and signaling. J. Biol. Chem. 2014;289(6):3736–3748. doi: 10.1074/jbc.M113.520981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van Campenhout A, Van Campenhout C, Lagrou AR, Manuel YKB. Effects of in vitro glycation on Fe3+ binding and Fe3+ isoforms of transferrin. Clin. Chem. 2004;50(9):1640–1649. doi: 10.1373/clinchem.2004.033811. [DOI] [PubMed] [Google Scholar]
- 6.Baumann H, Gauldie J. The acute phase response. Immunol. Today. 1994;15(2):74–80. doi: 10.1016/0167-5699(94)90137-6. [DOI] [PubMed] [Google Scholar]
- 7.Fleck A. Clinical and nutritional aspects of changes in acute-phase proteins during inflammation. Proc. Nutr. Soc. 1989;48(3):347–354. doi: 10.1079/pns19890050. [DOI] [PubMed] [Google Scholar]
- 8.Morlese JF, Forrester T, Del Rosario M, Frazer M, Jahoor F. Transferrin kinetics are altered in children with severe protein-energy malnutrition. J. Nutr. 1997;127(8):1469–1474. doi: 10.1093/jn/127.8.1469. [DOI] [PubMed] [Google Scholar]
- 9.Martin F, Linden T, Katschinski DM, Oehme F, Flamme I, Mukhopadhyay CK, Eckhardt K, Troger J, Barth S, Camenisch G, Wenger RH. Copper-dependent activation of hypoxia-inducible factor (HIF)-1: implications for ceruloplasmin regulation. Blood. 2005;105(12):4613–4619. doi: 10.1182/blood-2004-10-3980. [DOI] [PubMed] [Google Scholar]
- 10.Gitlin JD, Schroeder JJ, Lee-Ambrose LM, Cousins RJ. Mechanisms of caeruloplasmin biosynthesis in normal and copper-deficient rats. Biochem. J. 1992;282(Pt 3):835–839. doi: 10.1042/bj2820835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Laurell CB, Kullander S, Thorell J. Effect of administration of a combined estrogen-progestin contraceptive on the level of individual plasma proteins, Scand. J. Clin. Lab Investig. 1968;21(4):337–343. doi: 10.3109/00365516809077003. [DOI] [PubMed] [Google Scholar]
- 12.Fernandez-Real JM, Lopez-Bermejo A, Ricart W. Cross-talk between iron metabolism and diabetes. Diabetes. 2002;51(8):2348–2354. doi: 10.2337/diabetes.51.8.2348. [DOI] [PubMed] [Google Scholar]
- 13.Simcox JA, McClain DA. Iron and diabetes risk. Cell Metab. 2013;17(3):329–341. doi: 10.1016/j.cmet.2013.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dongiovanni P, Valenti L, Ludovica Fracanzani A, Gatti S, Cairo G, Fargion S. Iron depletion by deferoxamine up-regulates glucose uptake and insulin signaling in hepatoma cells and in rat liver. Am. J. Pathol. 2008;172(3):738–747. doi: 10.2353/ajpath.2008.070097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Memisogullari R, Bakan E. Levels of ceruloplasmin, transferrin, and lipid peroxidation in the serum of patients with type 2 diabetes mellitus. J. Diabetes Complicat. 2004;18(4):193–197. doi: 10.1016/S1056-8727(03)00032-1. [DOI] [PubMed] [Google Scholar]
- 16.McMillan DE. Increased levels of acute-phase serum proteins in diabetes. Metabolism. 1989;38(11):1042–1046. doi: 10.1016/0026-0495(89)90038-3. [DOI] [PubMed] [Google Scholar]
- 17.Daimon M, Susa S, Yamatani K, Manaka H, Hama K, Kimura M, Ohnuma H, Kato T. Hyperglycemia is a factor for an increase in serum ceruloplasmin in type 2 diabetes. Diabetes Care. 1998;21(9):1525–1528. doi: 10.2337/diacare.21.9.1525. [DOI] [PubMed] [Google Scholar]
- 18.Collier A, Wilson R, Bradley H, Thomson JA, Small M. Free radical activity in type 2 diabetes. Diabet. Med. 1990;7(1):27–30. doi: 10.1111/j.1464-5491.1990.tb01302.x. [DOI] [PubMed] [Google Scholar]
- 19.Awadallah R, El-Dessoukey EA, Doss H, Khalifa K, el-Hawary Z. Blood-reduced glutathione, serum ceruloplasmin and mineral changes in juvenile diabetes. Z. Ernahrungswiss. 1978;17(2):79–83. doi: 10.1007/BF02021114. [DOI] [PubMed] [Google Scholar]
- 20.Asayama K, Uchida N, Nakane T, Hayashibe H, Dobashi K, Amemiya S, Kato K, Nakazawa S. Antioxidants in the serum of children with insulin-dependent diabetes mellitus. Free Radic. Biol. Med. 1993;15(6):597–602. doi: 10.1016/0891-5849(93)90162-n. [DOI] [PubMed] [Google Scholar]
- 21.Van Campenhout A, Van Campenhout C, Lagrou AR, Moorkens G, De Block C, Manuel-y-Keenoy B. Iron-binding antioxidant capacity is impaired in diabetes mellitus. Free Radic. Biol. Med. 2006;40(10):1749–1755. doi: 10.1016/j.freeradbiomed.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 22.Hao P, Adav SS, Gallart-Palau X, Sze SK. Recent advances in mass spectrometric analysis of protein deamidation. Mass Spectrom. Rev. 2016 doi: 10.1002/mas.21491. [DOI] [PubMed] [Google Scholar]
- 23.Simm A. Protein glycation during aging and in cardiovascular disease. J. Proteom. 2013;92:248–259. doi: 10.1016/j.jprot.2013.05.012. [DOI] [PubMed] [Google Scholar]
- 24.Chondrogianni N, Petropoulos I, Grimm S, Georgila K, Catalgol B, Friguet B, Grune T, Gonos ES. Protein damage, repair and proteolysis. Mol. Asp. Med. 2014;35:1–71. doi: 10.1016/j.mam.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 25.Mayne J, Ning Z, Zhang X, Starr AE, Chen R, Deeke S, Chiang CK, Xu B, Wen M, Cheng K, Seebun D, Star A, Moore JI, Figeys D. Bottom-up proteomics (2013–2015): keeping up in the era of systems biology. Anal. Chem. 2016;88(1):95–121. doi: 10.1021/acs.analchem.5b04230. [DOI] [PubMed] [Google Scholar]
- 26.Landau BR, Wahren J, Chandramouli V, Schumann WC, Ekberg K, Kalhan SC. Use of 2H2O for estimating rates of gluconeogenesis. Application to the fasted state. J. Clin. Investig. 1995;95(1):172–178. doi: 10.1172/JCI117635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kasumov T, Willard B, Li L, Li M, Conger H, Buffa JA, Previs S, McCullough A, Hazen SL, Smith JD. 2H2O-based high-density lipoprotein turnover method for the assessment of dynamic high-density lipoprotein function in mice. Arterioscler. Thromb. Vasc. Biol. 2013;33(8):1994–2003. doi: 10.1161/ATVBAHA.113.301700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li L, Willard B, Rachdaoui N, Kirwan JP, Sadygov RG, Stanley WC, Previs S, McCullough AJ, Kasumov T. Plasma proteome dynamics: analysis of lipoproteins and acute phase response proteins with 2H2O metabolic labeling. Mol. Cell Proteom. 2012;11(7) doi: 10.1074/mcp.M111.014209. (M111 014209) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang D, Liem DA, Lau E, Ng DC, Bleakley BJ, Cadeiras M, Deng MC, Lam MP, Ping P. Characterization of human plasma proteome dynamics using deuterium oxide. Proteom. Clin. Appl. 2014;8(7–8):610–619. doi: 10.1002/prca.201400038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Price JC, Khambatta CF, Li KW, Bruss MD, Shankaran M, Dalidd M, Floreani NA, Roberts LS, Turner SM, Holmes WE, Hellerstein MK. The effect of long term calorie restriction on in vivo hepatic proteostatis: a novel combination of dynamic and quantitative proteomics. Mol. Cell Proteom. 2012;11(12):1801–1814. doi: 10.1074/mcp.M112.021204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wei H FB, Beckman JS, Zhang WJ. Copper chelation by tetrathiomolybdate inhibits lipopolysaccharide-induced inflammatory responses in vivo. Am. J. Physiol. – Heart Circ. Physiol. 2011;301(3):H712–H720. doi: 10.1152/ajpheart.01299.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lehmann HP SK, Beeler MF. Standardization of serum ceruloplasmin concentrations in international enzyme units with o-dianisidine dihydrochloride as substrate. Clin. Chem. 1974;20(12):1564–1567. [PubMed] [Google Scholar]
- 33.Ravin HA. An improved colorimetric enzymatic assay of ceruloplasmin. J. Lab Clin. Med. 1961;58:161–168. [PubMed] [Google Scholar]
- 34.Shah V, Herath K, Previs SF, Hubbard BK, Roddy TP. Headspace analyses of acetone: a rapid method for measuring the 2H-labeling of water. Anal. Biochem. 2010;404(2):235–237. doi: 10.1016/j.ab.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 35.Toth PP, Barter PJ, Rosenson RS, Boden WE, Chapman MJ, Cuchel M, D'Agostino RB, Sr, Davidson MH, Davidson WS, Heinecke JW, Karas RH, Kontush A, Krauss RM, Miller M, Rader DJ. High-density lipoproteins: a consensus statement from the National Lipid Association. J. Clin. Lipidol. 2013;7(5):484–525. doi: 10.1016/j.jacl.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 36.Li L, Bebek G, Previs SF, Smith JD, Sadygov RG, McCullough AJ, Willard B, Kasumov T. Proteome dynamics reveals pro-inflammatory remodeling of plasma proteome in a mouse model of NAFLD. J. Proteome Res. 2016;15(9):3388–3404. doi: 10.1021/acs.jproteome.6b00601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vizcaino JA, Deutsch EW, Wang R, Csordas A, Reisinger F, Rios D, Dianes JA, Sun Z, Farrah T, Bandeira N, Binz PA, Xenarios I, Eisenacher M, Mayer G, Gatto L, Campos A, Chalkley RJ, Kraus HJ, Albar JP, Martinez-Bartolome S, Apweiler R, Omenn GS, Martens L, Jones AR, Hermjakob H. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 2014;32(3):223–226. doi: 10.1038/nbt.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vizcaino JA, Csordas A, del-Toro N, Dianes JA, Griss J, Lavidas I, Mayer G, Perez-Riverol Y, Reisinger F, Ternent T, Xu QW, Wang R, Hermjakob H. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016;44(D1):D447–D456. doi: 10.1093/nar/gkv1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26(12):1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 40.Keilhauer EC, Geyer PE, Mann M. HCD fragmentation of glycated peptides. J. Proteome Res. 2016;15(8):2881–2890. doi: 10.1021/acs.jproteome.6b00464. [DOI] [PubMed] [Google Scholar]
- 41.Shipanova IN, Glomb MA, Nagaraj RH. Protein modification by methylglyoxal: chemical nature and synthetic mechanism of a major fluorescent adduct. Arch. Biochem. Biophys. 1997;344(1):29–36. doi: 10.1006/abbi.1997.0195. [DOI] [PubMed] [Google Scholar]
- 42.Neuhauser N, Michalski A, Cox J, Mann M. Expert system for computer-assisted annotation of MS/MS spectra. Mol. Cell Proteom. 2012;11(11):1500–1509. doi: 10.1074/mcp.M112.020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prinsen BH, de Sain-vander Velden MG, Kaysen GA, Straver HW, van Rijn HJ, Stellaard F, Berger R, Rabelink TJ. Transferrin synthesis is increased in nephrotic patients insufficiently to replace urinary losses. J. Am. Soc. Nephrol. 2001;12(5):1017–1025. doi: 10.1681/ASN.V1251017. [DOI] [PubMed] [Google Scholar]
- 44.Afolabi PR, Jahoor F, Jackson AA, Stubbs J, Johnstone AM, Faber P, Lobley G, Gibney E, Elia M. The effect of total starvation and very low energy diet in lean men on kinetics of whole body protein and five hepatic secretory proteins. Am. J. Physiol. Endocrinol. Metab. 2007;293(6):E1580–E1589. doi: 10.1152/ajpendo.00169.2007. [DOI] [PubMed] [Google Scholar]
- 45.Potter BJ, Chapman RW, Nunes RM, Sorrentino D, Sherlock S. Transferrin metabolism in alcoholic liver disease. Hepatology. 1985;5(5):714–721. doi: 10.1002/hep.1840050503. [DOI] [PubMed] [Google Scholar]
- 46.De Feo P, Volpi E, Lucidi P, Cruciani G, Reboldi G, Siepi D, Mannarino E, Santeusanio F, Brunetti P, Bolli GB. Physiological increments in plasma insulin concentrations have selective and different effects on synthesis of hepatic proteins in normal humans. Diabetes. 1993;42(7):995–1002. doi: 10.2337/diab.42.7.995. [DOI] [PubMed] [Google Scholar]
- 47.Seshadri V, Fox PL, Mukhopadhyay CK. Dual role of insulin in transcriptional regulation of the acute phase reactant ceruloplasmin. J. Biol. Chem. 2002;277(31):27903–27911. doi: 10.1074/jbc.M203610200. [DOI] [PubMed] [Google Scholar]
- 48.O'Riordain MG, Ross JA, Fearon KC, Maingay J, Farouk M, Garden OJ, Carter DC. Insulin and counterregulatory hormones influence acute-phase protein production in human hepatocytes. Am. J. Physiol. 1995;269(2 Pt 1):E323–E330. doi: 10.1152/ajpendo.1995.269.2.E323. [DOI] [PubMed] [Google Scholar]
- 49.Thompson D, Harrison SP, Evans SW, Whicher JT. Insulin modulation of acute-phase protein production in a human hepatoma cell line. Cytokine. 1991;3(6):619–626. doi: 10.1016/1043-4666(91)90489-z. [DOI] [PubMed] [Google Scholar]
- 50.Wenger RH, Rolfs A, Marti HH, Bauer C, Gassmann M. Hypoxia, a novel inducer of acute phase gene expression in a human hepatoma cell line. J. Biol. Chem. 1995;270(46):27865–27870. doi: 10.1074/jbc.270.46.27865. [DOI] [PubMed] [Google Scholar]
- 51.Silva AM, Sousa PR, Coimbra JT, Bras NF, Vitorino R, Fernandes PA, Ramos MJ, Rangel M, Domingues P. The glycation site specificity of human serum transferrin is a determinant for transferrin's functional impairment under elevated glycaemic conditions. Biochem. J. 2014;461(1):33–42. doi: 10.1042/BJ20140133. [DOI] [PubMed] [Google Scholar]
- 52.MacGillivray RT, Moore SA, Chen J, Anderson BF, Baker H, Luo Y, Bewley M, Smith CA, Murphy ME, Wang Y, Mason AB, Woodworth RC, Brayer GD, Baker EN. Two high-resolution crystal structures of the recombinant N-lobe of human transferrin reveal a structural change implicated in iron release. Biochemistry. 1998;37(22):7919–7928. doi: 10.1021/bi980355j. [DOI] [PubMed] [Google Scholar]
- 53.Dewan JC, Mikami B, Hirose M, Sacchettini JC. Structural evidence for a pH-sensitive dilysine trigger in the hen ovotransferrin N-lobe: implications for transferrin iron release. Biochemistry. 1993;32(45):11963–11968. doi: 10.1021/bi00096a004. [DOI] [PubMed] [Google Scholar]
- 54.Luck AN, Mason AB. Transferrin-mediated cellular iron delivery. Curr. Top. Membr. 2012;69:3–35. doi: 10.1016/B978-0-12-394390-3.00001-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Olivieri S, Conti A, Iannaccone S, Cannistraci CV, Campanella A, Barbariga M, Codazzi F, Pelizzoni I, Magnani G, Pesca M, Franciotta D, Cappa SF, Alessio M. Ceruloplasmin oxidation, a feature of Parkinson's disease CSF, inhibits ferroxidase activity and promotes cellular iron retention. J. Neurosci. 2011;31(50):18568–18577. doi: 10.1523/JNEUROSCI.3768-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barbariga M, Curnis F, Andolfo A, Zanardi A, Lazzaro M, Conti A, Magnani G, Volonte MA, Ferrari L, Comi G, Corti A, Alessio M. Ceruloplasmin functional changes in Parkinson's disease-cerebrospinal fluid. Mol. Neurodegener. 2015;10:59. doi: 10.1186/s13024-015-0055-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Van Campenhout A, Van Campenhout C, Olyslager YS, Van Damme O, Lagrou AR, Manuel-y-Keenoy B. A novel method to quantify in vivo transferrin glycation: applications in diabetes mellitus. Clin. Chim. Acta. 2006;370(1–2):115–123. doi: 10.1016/j.cca.2006.01.028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.