Abstract
In addition to occupational exposures to acrylamide (AA), concerns about AA health risks for the general population have been recently raised due to the finding of AA in food. In this study, we evaluated the genotoxicity of AA and its metabolite glycidamide (GA) in L5178Y/ Tk+/− mouse lymphoma cells. The cells were treated with 2–18 mM of AA or 0.125–4 mM of GA for 4 h without metabolic activation. The DNA adducts, mutant frequencies and the types of mutations for the treated cells were examined. Within the dose range tested, GA induced DNA adducts of adenine and guanine [N3-(2-carbamoyl-2-hydroxyethyl)-adenine and N7-(2-carbamoyl-2-hydroxyethyl)-guanine] in a linear dose-dependent manner. The levels of guanine adducts were consistently about 60-fold higher across the dose range than those of adenine. In contrast, no GA-derived DNA adducts were found in the cells treated with any concentrations of AA, consistent with a lack of metabolic conversion of AA to GA. However, the mutant frequency was significantly increased by AA at concentrations of 12 mM and higher. GA was mutagenic starting with the 2 mM dose, suggesting that GA is much more mutagenic than AA. The mutant frequencies were increased with increasing concentrations of AA and GA, mainly due to an increase of proportion of small colony mutants. To elucidate the underlying mutagenic mechanism, we examined the loss of heterozygosity (LOH) at four microsatellite loci spanning the entire chromosome 11 for mutants induced by AA or GA. Compared to GA induced mutations, AA induced more mutants whose LOH extended to D11Mit22 and D11Mit74, an alteration of DNA larger than half of the chromosome. Statistical analysis of the mutational spectra revealed a significant difference between the types of mutations induced by AA and GA treatments (P = 0.018). These results suggest that although both AA and GA generate mutations through a clastogenic mode of action in mouse lymphoma cells, GA induces mutations via a DNA adduct mechanism whereas AA induces mutations by a mechanism not involving the formation of GA adducts.
Keywords: Acrylamide, Glycidamide, Genotoxicity, Loss of heterozygosity, DNA adducts
1. Introduction
Acrylamide (AA) is an important industrial compound with many uses. Recently, new concerns about health risks for the general population have been raised by the finding that AA is formed in food during cooking (Mottram et al., 2002; Stadler et al., 2002; Tareke et al., 2000; Tornqvist, 2005). Although a large number of studies and the updated reviews on the genotoxicity of AA and its metabolite glycidamide (GA) have been reported (Besaratinia and Pfeifer, 2007; Dearfield et al., 1988; Dearfield et al., 1995; Rice, 2005), the mechanisms for the genotoxicity of AA are not completely clear.
The activation of AA by cytochrome P450 2E1-mediated biotransformation to GA has been clearly demonstrated both in humans and experimental animals, and GA can subsequently reacts with cellular DNA, leading to multiple forms of toxicity including mutation and tumor induction (Adler et al., 2000; Ghanayem et al., 2005; Rice, 2005; Sumner et al., 1999). Several GA–DNA adducts have been identified in mice and rats treated with AA and GA (Doerge et al., 2005a; Gamboa da Costa et al., 2003; Segerback et al., 1995). The predominant DNA adduct of N7-(2-carbamoyl-2-hydroxyethyl)-guanine (N7-GA-Gua) is formed in all tested organs at 100-fold higher levels than is N3-(2-carbamoyl-2-hydroxyethyl)-adenine (N3-GA-Ade) (Gamboa da Costa et al., 2003). Increased incidences in a number of benign and malignant tumors in a variety of organs have been observed in rodents exposed to AA (Bull et al., 1984; Friedman et al., 1995; Johnson et al., 1986; Rice, 2005).
The genotoxicity of AA has been studied extensively, but there are fewer experimental data available with GA as the test substance (Baum et al., 2005; Besaratinia and Pfeifer, 2004; Koyama et al., 2006; Martins et al., 2006; Paulsson et al., 2003). AA is not mutagenic in bacterial assays, and it is mutagenic in mammalian somatic and germinal cells in vitro and in vivo (Dearfield et al., 1995). AA in vitro induces chromosomal aberrations, micronuclei, sister chromatid exchanges, polyploidy, aneuploidy and other mitotic disturbances in the absence of metabolic activation. GA is positive for micronucleus induction in vivo in both mice and rats. The micronuclei result from chromosome breakage rather than aneuploidy (Dearfield et al., 1995; Martins et al., 2006). Recently, Manjanatha et al. have reported that AA and GA in drinking water increased micronucleated reticulocytes, lymphocyte Hprt mutant frequencies (MFs), and liver cII MFs in Big Blue mice (Manjanatha et al., 2006).
The L5178Y mouse lymphoma assay (MLA) is widely used as a short-term mutagenicity assay (Chen and Moore, 2004; Clements, 2000). The MLA is particularly useful for evaluating the ability of mutagens to induce a wide variety of mutational events, because it detects not only intragenic events (mainly point mutations) but also loss of heterozygosity (LOH) including Tk gene loss and karyotypically visible alterations of the Tk+-bearing chromosome 11b (Applegate et al., 1990; Chen et al., 2002; Hozier et al., 1982; Moore et al., 1985). AA has been found to be both mutagenic and clastogenic in mouse lymphoma cells in the absence of an exogenous activation system (Moore et al., 1987). In the present study, we investigated the ability of GA and AA to form DNA adducts and to induce mutations using L5178Y/Tk+/− mouse lymphoma cells. In addition, the underlying mechanisms for mutation induction by AA and GA were explored by LOH analysis of the induced mutants.
2. Materials and methods
2.1. Chemical agents
AA was purchased from the Sigma Chemical Co. (St. Louis, MO). GA was purchased from Toronto Research Chemicals (North York, Ontario). Compound purity was determined using 1H and 13C NMR, LC–UV (200 nm), GC-FID, and full scan LC–ES/MS. The purity for AA was >99.9%, and the purity for GA was >99.5% with <1% AA. Fischer’s medium was purchased from Quality Biological Inc. (Gaithersburg, MD), and all cell culture supplies were purchased from Invitrogen Life Technologies (Carlsbad, CA). PCR Master Mix was purchased from Promega Company (Madison, WI). The primers used for detection of LOH at the Tk locus and the D11Mit59, D11Mit22, and D11Mit74 loci were purchased from Invitrogen Life Technologies, and the primers for Cyp2e1 were purchased from Sigma–Genosys (Woodlands, TX). The PowerScript First Strand Synthesis System for RT-PCR was purchased from Clontech (Mountain View, CA) and SybrGreen Master Mix was purchased from Applied Biosystems (Foster City, CA).
2.2. Cells and culture conditions
The L5178Y/Tk+/− 3.7.2 C mouse lymphoma cell line was utilized for the mutation assay. Cells were grown according to the methods described by Chen and Moore (2004). Briefly, the basic medium was Fischer’s medium for leukemic cells of mice with L-glutamine supplemented with pluronic F68 (0.1%), sodium pyruvate (1 mM), penicillin (100 units/mL), and streptomycin (100 μg/mL). The treatment medium (F5p), growth medium (F10p), and cloning medium (F20p) were the basic medium supplemented with 5%, 10%, and 20% heat-inactivated horse serum, respectively. The cultures were maintained in a humidified incubator with 5% CO2 in air at 37 °C.
2.3. Cell treatment with AA and GA
The AA and GA working solutions (100×) were prepared just prior to use by dissolving the compounds in distilled water. The cells were suspended in 100-mm diameter tissue culture dishes at a concentration of 6 × 106 cells in 10 mL of treatment medium. Aliquots (100 μL) of the working solutions were added to give final concentrations of 2–18 mM for AA and 0.125–4 mM for GA. In all cases, including the untreated controls and positive controls [0.53 μM (0.1 μg/mL) 4-nitroquinoline-1-oxide (4-NQO)], the cells were incubated for 4 h at 37 °C. After treatment, the cells were centrifuged, washed once with fresh medium, and then resuspended in growth medium at a density of 3 × 105 cells/mL in 25 cm2 cell culture flasks to begin the 2-day phenotypic expression.
2.4. Tk microwell mutation assay
Mutant selection was performed as described previously (Chen and Moore, 2004). Briefly, the cells were counted and the densities were adjusted with fresh growth medium at approximately 1 and 2 days following exposure. For mutant enumeration, trifluorothymidine (TFT, 3 μg/ mL) was added to the cells in cloning medium. Cells were seeded into four 96-well flat-bottom microtiter plates, 200 μL/well for a final density of 2000 cells/well. For the determination of plating efficiency, approximately 1.6 cells were aliquoted in 200 μL/well into two 96-well flat-bottom microtiter plates. All plates were incubated at 37 °C in a humidified incubator with 5% CO2 in air. After 11 days of incubation, colonies were counted and mutant colonies were categorized as small or large. Small colonies are defined as those smaller than 25% of the well diameter. Mutant frequencies (MFs) were calculated by use of the Poisson distribution. Cytotoxicity was measured via relative total growth (RTG), which includes a measure of growth during treatment, expression, and cloning (Chen and Moore, 2004).
2.5. Tk mutant evaluation for LOH at the thymidine kinase (Tk1) and three other microsatellite loci spanning the entire chromosome 11
Mutant clones were directly taken from TFT selection plates. Forty-eight large and 48 small mutant colonies resulting from treatment with 16 mM AA or 4 mM GA were analyzed. The mutant cells were washed once with phosphate-buffered saline (PBS) by centrifugation, and cell pellets were quickly frozen and stored at −80 °C. Genomic DNA was extracted by digesting the cells in lysis buffer [10 mM Tris-HCl (pH 7.5), 5 mM MgCl2, 1% (v/v) Triton X-100, and 1% (v/v) Tween 20] with 200 μg/ mL of proteinase K at 60 °C for 90 min, followed by inactivation of proteinase K at 95 °C for 10 min. For polymerase chain reaction (PCR) analysis of LOH at Tk and other loci (D11Mit59, D11Mit22 and D11Mit74 loci), the amplification reactions were carried out in a total volume of 20 μL using of 2× PCR Master Mix and pairs of primers described previously (Singh et al., 2005). The thermal cycling conditions were as follows: initial incubation at 94 °C for 3 min; 40 cycles of 94 °C denaturation for 30 s, 55 °C annealing for 30 s, and 72 °C extension for 30 s; and a final extension at 72 °C for 7 min. The amplification products were scored for the presence of one band (indicating LOH) or two bands (retention of heterozygosity at the given locus) after running 2% agarose gel electrophoresis.
2.6. DNA adduct assay
After treatment, the cells were centrifuged, washed once with PBS, and then stored at −80 °C. DNA was purified and GA–DNA adducts (N7-GA-Gua and N3-GA-Ade) were quantified after neutral thermal hydrolysis using an isotope dilution LC–ES/MS/MS procedure detailed previously (Doerge et al., 2005a; Gamboa da Costa et al., 2003). The limit of quantification (LOQ) for N3-GA-Ade was approximately 1.5 adducts in 108 nucleotides and the limit of detection (LOD) was approximately 0.5 adducts in 108 nucleotides. For N7-GA-Gua, the LOQ was approximately 1 adduct in 108 nucleotides and the LOD was approximately 0.5 adducts in 108 nucleotides. This corresponds to approximately 500 and 250 fg on-column, respectively, from injections of approximately 50 μg DNA equivalents on-column. As previously reported for GA-modified DNA standards, the method precision for adduct determination was 1.5–5.6% relative standard deviation (Gamboa da Costa et al., 2003).
2.7. Real-time PCR
The powerscript first strand synthesis system for RT-PCR (Clontech, Mountain View, CA) was used to synthesize cDNA in a 20 μL reaction containing 1.0 μg total RNA, 0.5 μg oligo(dT) 12–18, 4 μL 5× first strand buffer, 2 μL 10 mM dNTP, 2 μL 100 mM DTT, and 1 μL powerscript reverse transcriptase. RNA and oligo(dT) were mixed first, heated to 70 °C for 10 min, and cooled on ice until addition of the remaining reaction components. The reaction was incubated at 42 °C for 90 min, and terminated by heating at 70 °C for 15 min. The forward primer for Cyp2e1 was 5′-TTCACACTGCACCTGGGTCA-3′ and the reverse primer was 5′-CTTGTAGCCTGCAGGACCA-3′. As an internal control, housekeeping gene β-tubulin was co-amplified to normalize the expression profile. Real-time PCR was performed in a total volume of 25 μL on an ABI Prism 7000 using a 96-well format. Each PCR reaction contained 1.0 μL of cDNA, 12.5 μL of 2× SybrGreen Master Mix (Applied Biosystems, Foster City, CA), and 50 μM each of the forward and reverse primers. Each reaction was run in triplicate. The thermocycler parameters were as follows: 50 °C for 2 min; 95 °C for 10 min; 40 cycles of 95 °C for 15 s and 60 °C for 1 min.
2.8. Data analyses
The data evaluation criteria developed by the mouse lymphoma assay expert workgroup of the international workgroup for genotoxicity tests (IWGT) were used to determine whether or not a response was positive or negative (Moore et al., 2006). Positive responses are defined as those where the induced MF in one or more treated cultures exceeds the global evaluation factor (GEF) of 126 mutants per 106 cells and there is also a dose related increase with MF. LOH patterns of mutants were compared using the computer program (Cariello et al., 1994) for Monte Carlo analysis developed by Adams and Skopek (1987).
3. Results
3.1. Cytotoxicity and mutagenicity of AA and GA
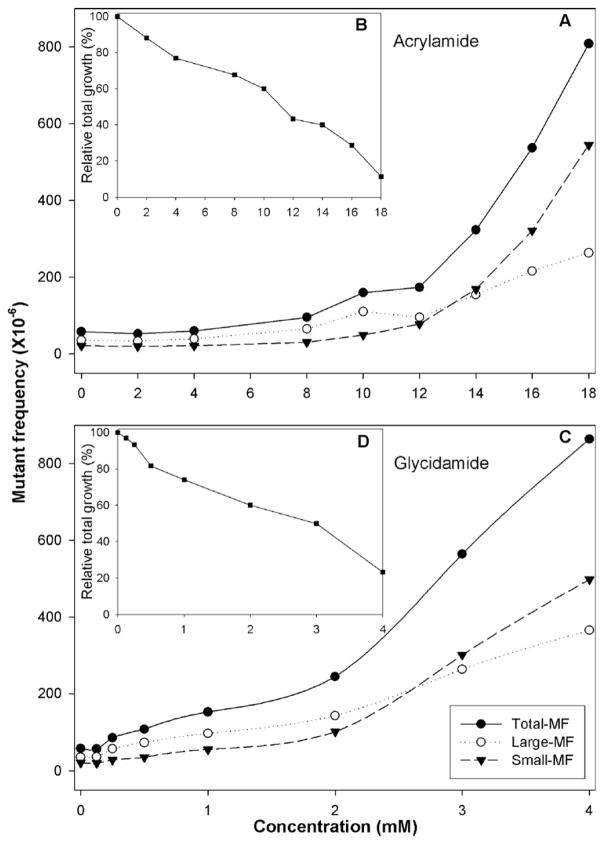
The relative total growth (RTG) values, Tk mutant frequency (MF), small colony MF, and large colony MF from one representative MLA experiment with treatments of AA and GA are presented in Table 1. These results were replicated in 2–3 additional experiments and the data are displayed as means in Fig. 1. Treatment of mouse lymphoma cells with various concentrations of AA or GA resulted in dose-dependent increases for both the cytotoxicity and mutagenicity (Fig. 1). Both small and large colony Tk mutants were induced by both chemicals. Concentrations of 14–18 mM AA and 2–4 mM GA induced a mutant frequency exceeding the global evaluation factor (GEF) of 126 × 10−6 (Moore et al., 2003), indicating positive mutagenic responses. The MFs for the negative control, 4 mM of GA, and 18 mM of AA were 55 ± 14 × 10−6, 864 ± 151 × 10−6, and 808 ± 109 × 10−6, respectively (Fig. 1).
Table 1.
Toxicity and mutagenicity of acrylamide and glycidamide from one representative experiment in L5178Y/Tk+/− mouse lymphoma cells
| Treatment | Concentration (mM) | Mutant frequency (MF, ×10−6) | Relative total growth (%) | MF of small colonies (×10−6) | MF of large colonies (×10−6) | (%) of small colonies |
|---|---|---|---|---|---|---|
| Control | 0 | 83 | 100 | 26 | 57 | 31 |
| Acrylamide | 8 | 137 | 72 | 36 | 101 | 26 |
| 10 | 159 | 60 | 49 | 110 | 31 | |
| 12 | 235 | 45 | 111 | 124 | 47 | |
| 14 | 323 | 36 | 169 | 154 | 52 | |
| 16 | 623 | 29 | 355 | 268 | 57 | |
| 18 | 956 | 15 | 622 | 334 | 65 | |
| Glycidamide | 0.25 | 94 | 91 | 28 | 66 | 30 |
| 0.5 | 121 | 93 | 31 | 90 | 26 | |
| 1 | 172 | 78 | 60 | 112 | 35 | |
| 2 | 272 | 62 | 98 | 174 | 36 | |
| 3 | 564 | 43 | 301 | 263 | 53 | |
| 4 | 1009 | 26 | 540 | 469 | 53 | |
| 4-NQOa | 5.3 ×10−4 | 598 | 51 | 256 | 342 | 43 |
4-NQO was used as a positive control.
Fig. 1.
Comparison of cytotoxicity (B and D) and mutagenicity (A and C) of acrylamide (A and B) and glycidamide (C and D) in mouse lymphoma cells treated with different concentrations. The data points represent the mean of three (acrylamide) or four (glycidamide) independent experiments. (●), total mutant frequency (MF) from large and small colonies; (○), MF from large colonies; (▼), MF from small colonies; (■), relative total growth.
3.2. LOH analysis of mutants of AA and GA
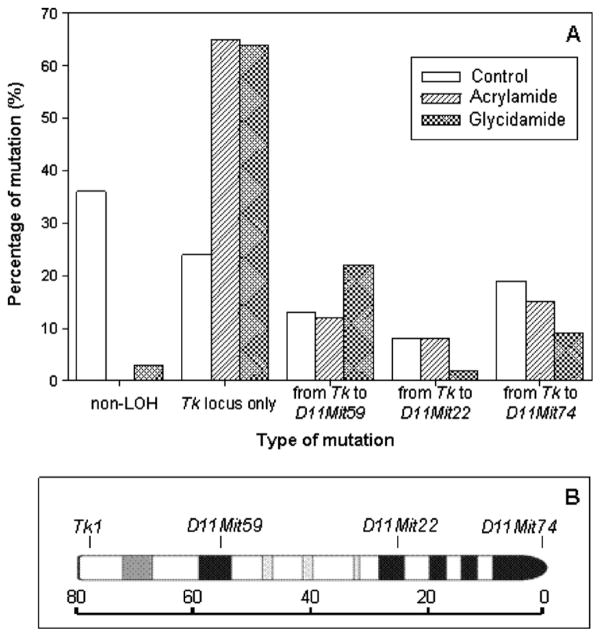
LOH analysis of the mutants was conducted using four microsatellite loci spanning the entire chromosome 11 (Fig. 2B) to determine the types of the mutations. DNA samples were isolated from 48 large and 48 small mutant colonies for 4 mM GA and 16 mM AA, which had similar cytotoxicity. More than 94% of the large and 100% of the small colony mutants from the two treatment groups of AA or GA lost heterozygosity at the Tk1 locus (Table 2). Fig. 2A shows the percentages of different types of mutations in all (large and small) mutant colonies. The most common type of mutation for AA and GA treatments (more than 60%) was LOH with a small deletion limited to the Tk locus, while the major type of mutation in untreated controls has been previously shown to be non-LOH (Wang et al., 2007). Compared to the GA induced mutations, AA induced more mutants whose LOH extended to D11Mit22 and D11Mit74, indicating an alteration of DNA larger than half of the chromosome. Statistical analysis of the different types of mutations revealed that the mutational types induced by AA and GA were significantly different from the published data for untreated controls (P < 0.001) (Wang et al., 2007). In addition, there was a significant difference between the types of mutations induced by AA and GA (P = 0.018).
Fig. 2.
(A) Comparison of the percentage of mutational types for all (large and small) colonies produced in mouse lymphoma cells untreated or treated with acrylamide (16 mM) or glycidamide (4 mM). The data are the weighted sum of mutant frequencies from large and small colonies (the proportion of small colony mutants was 64% for acrylamide and 57% for glycidamide). The data for the untreated control are from the literature (36). For the mutational spectra, the treatment groups were significantly different from the untreated control (P < 0.0001), and there was also a significant difference between the two treatment groups (P < 0.05). (B) The loci that were analyzed for LOH (Tk1, D11Mit59, D11Mit22, and D11Mit74) are marked. The ruler in centimorgans indicates the distance from the top of the chromosome.
Table 2.
LOH at different loci along chromosome 11 in 48 large- and 48 small-colony Tk mutants from cells treated with acrylamide or glycidamide
| Locus | Position (cM)b | Acrylamide | Glycidamide | Controla | |||
|---|---|---|---|---|---|---|---|
|
|
|
|
|||||
| No. in large colonies (%) | No. in small colonies (%) | No. in large colonies (%) | No. in small colonies (%) | No. in large colonies (%) | No. in small colonies (%) | ||
| D11Mit74 | 0 | 9 (19) | 6 (13) | 5 (10) | 4 (8) | 5 (14) | 9 (27) |
| D11Mit22 | 25 | 15 (31) | 9 (19) | 7 (15) | 4 (8) | 7 (19) | 13 (39) |
| D11Mit59 | 56 | 27 (56) | 11 (23) | 29 (60) | 6 (13) | 11 (31) | 18 (55) |
| Tk | 78 | 48 (100) | 48 (100) | 45 (94) | 48 (100) | 15 (42) | 32 (97) |
| Non-LOHc | 0 (0) | 0 (0) | 3 (6) | 0 (0) | 21 (58) | 1 (3) | |
The negative control data are from the literature (Wang et al., 2007).
Locus position in centimorgan (cM) is the distance to the top of chromosome 11.
Retain heterozygosity.
3.3. DNA adduct formation for AA and GA
To investigate whether or not the mutagenic effects of AA and GA are associated with the formation of DNA adducts, we analyzed the DNA adducts formed in mouse lymphoma cells by LC–ES/MS/MS. Within the dose range evaluated (0.5–4 mM) for GA-treated cells, GA-derived DNA adducts of adenine and guanine (N3-GA-Ade and N7-GA-Gua) were formed in a linear dose response (Fig. 3). However, no N3-GA-Ade was detected at a concentration of 0.5 mM. At the highest dose of 4 mM used in this study, GA induced 137.4 ± 22.4 N7-GA-Gua and 2.3 ± 0.4 N3-GA-Ade per 106 nucleotides. The levels of N7-GA-Gua adduct were consistently about 60-fold higher than those for N3-GA-Ade. In contrast, there were no GA-derived DNA adducts in the cells treated with 8–20 mM AA for 4 h.
Fig. 3.
Levels of N3-GA-Ade (A) and N7-GA-Gua (B) DNA adducts in mouse lymphoma cells treated with glycidamide (GA) for 4 h at different concentrations. The data is expressed as the number of adducts in 106 nucleotides and represents the means ± 1 S.D. (n = 4 or 8).
3.4. Real-time PCR analysis of the gene expression of Cyp2e1
To confirm a lack of bioactivation potential for AA to GA in L5178Y mouse lymphoma cells, we determined the gene expression of Cyp2e1 using real-time RT-PCR. No PCR product of Cyp2e1 RNA was detected in the mouse lymphoma cells compared to that in mouse liver tissue used as a positive control (data not shown).
4. Discussion
There are at least two theories on the mutagenic action of AA (Dearfield et al., 1995). The first one is that AA is oxidized to GA, an epoxide metabolite that is very reactive with nucleophilic sites on DNA bases. Cytochrome P450 2E1 (CYP2E1) is involved in this metabolic conversion (Adler et al., 2000; Ghanayem et al., 2005; Sumner et al., 1999). It has been suggested that this is the primary pathway responsible for the genotoxicity of AA in animal experiments (Gamboa da Costa et al., 2003) and in mammalian cells (Besaratinia and Pfeifer, 2004). Another possible mechanism is a direct Michael-type reactivity of AA with nucleophiles in DNA and proteins, in particular with soft nucleophiles such as thiols. For example, AA can react directly with glutathione (GSH), a molecule protecting the cell against endogenous and exogenous oxidants and electrophiles (Dearfield et al., 1995). In this study, we explored the mechanisms of mutagenicity of AA by investigating DNA adducts, mutations and types of mutations induced by AA and GA.
When mouse lymphoma cells were exposed to AA or GA for 4 h, both compounds were cytotoxic and mutagenic. AA significantly increased the mutant frequency at concentrations of 12 mM and higher, and the cytotoxicity of 16 mM AA was similar to that of 4 mM GA (Fig. 1). Koyama et al. also reported that AA was weak genotoxic and 14 mM AA was positive by TK assay and MN test in human lymphoblastoid TK6 cells which are also widely used for the Tk gene mutation assay. Compared to AA, GA (2–4 mM) was much more mutagenic, about 5–10-fold more potent. This is consistent with several previously reported genotoxicity studies conducted using mammalian cells (Baum et al., 2005; Besaratinia and Pfeifer, 2003; Koyama et al., 2006; Martins et al., 2006). Since the genotoxic responses at higher concentration are reproducible, LOH analysis of the mutants induced by 4 mM GA and 16 mM AA treatments was conducted to determine the types of the mutations. Their mutagenic effects were mainly due to LOH involving chromosome 11 (Table 2 and Fig. 2). Although both AA and GA induced LOH type of mutations, GA induced DNA adducts of N7-GA-Gua and N3-GA-Ade (Fig. 3) while AA did not induce these two adducts, probably due to the absence of Cyp2e1 activity in mouse lymphoma cells.
We analyzed the Tk mutants for LOH using loci distributed along chromosome 11. LOH is the loss of the remaining normal allele of a heterozygous locus, resulting in either hemizygous or homozygous status for the mutant allele. LOH can be caused by a deletion or mitotic recombination. In the mouse lymphoma assay, compounds that induce exclusively or almost exclusively point mutations result in a high proportion of large colony Tk mutants and little LOH at the Tk locus. Chemicals that act primarily as clastogens result in a high proportion of small colony mutants and LOH at the Tk locus in both large and small colony mutants (Applegate et al., 1990; Harrington-Brock et al., 2003). In our study, more than 94% of the mutants induced by AA and GA lost heterozygosity at the Tk locus (Table 2), indicating that the mutations mainly resulted from extensive damage to the chromosome; and that mutagenicity of AA or GA was generated by a clastogenic mode of action rather than a point mutation mode-of-action in mouse lymphoma cells. We also observed that there was a significant difference (P = 0.018) between the mutational spectra of AA and GA (Fig. 2). The difference suggests that AA and GA may produce chromosomal mutations via different mechanisms.
AA is slow to react with DNA and only forms adducts under forced chemical conditions and after extended reaction time (up to 40 days) (Solomon et al., 1985). AA is not mutagenic in bacterial cell systems with or without metabolic activation (Knaap et al., 1988; Tsuda et al., 1993; Zeiger et al., 1987). AA is an unsaturated amide and can react with cellular nucleophiles, such as GSH (detoxification pathway). These activities of AA may be responsible for its mutagenic effects. In previously published studies, using V79 cells that do not express CYP2E1, Puppel et al. (2005) demonstrated significant inductions of DNA strand breaks with AA treatment. Thus, the enhanced effects should not result from the generation of GA and respective DNA adduct formation, and may result from the interaction between AA and DNA. They suggested that the DNA breakage caused by AA was associated with the enhanced oxidative stress because of the depletion of GSH, an oxidative defense system (Puppel et al., 2005). Others have also reported that AA generates reactive oxygen species (ROS) that can attack all cellular constituents, including protein, nucleic acids, and lipid, and induces oxidative DNA damage, which is reduced by the spin trap of α-phenyl-N-tert-butyl nitrone (PBN) and antioxidants of vitamins C and E (Blasiak et al., 2004).
Oxidative stress can result in DNA damage, mainly DNA breakage and deletions. The DNA strand breaks and deletions can be repaired by recombination or other mechanisms in cells, which will result in LOH. These chromosomal types of mutation cannot be detected by bacterial test systems. In a recent study investigating the mutagenicity of the lipid peroxidation product 4-hydroxy-2-nonenal (4-HNE) using mouse lymphoma cells (Singh et al., 2005), we found that 4-HNE is mutagenic and the major type of mutation is LOH involving the Tk locus, similar to what we observed in this study with AA. Therefore, mutagenicity of AA might result from increasing reactive oxygen species and/or decreasing oxidative defense system like GSH.
GA, the metabolite of AA, is a direct mutagen via DNA adduct formation. GA can form DNA adducts in cells and can induce point mutations in bacterial test systems (Hashimoto and Tanii, 1985), in mammalian cell test systems (Barfknecht et al., 1988; Besaratinia and Pfeifer, 2004) and in transgenic mutation test systems (Manjanatha et al., 2006). GA DNA adduct-specific mutations (i.e., G→ T transversions) have also been detected in mammalian cells (Besaratinia and Pfeifer, 2004) and animals (Manjanatha et al., 2006). Our data also indicate that GA induces DNA adducts in vitro in mouse lymphoma cells; the levels of N7-GA-Gua adduct were consistently much higher than those for N3-GA-Ade as previously reported (Doerge et al., 2005b,c; Gamboa da Costa et al., 2003), and that GA is a potent mutagen.
Previous reports indicate that GA induces point mutations (Besaratinia and Pfeifer, 2004; Hashimoto and Tanii, 1985; Manjanatha et al., 2006), and these assays cannot detect the LOH mutations. We found that the major type of mutations induced by 4 mM GA were chromosomal mutations in mouse lymphoma cells. Using human lymphoblastoid TK6 cells, Koyama et al. found that GA of 2.2 mM induced primarily non-LOH type mutations. The differences between these two Tk gene mutation assays may due to the different sensitivity to GA treatment and the mutations analyzed from different concentrations. Although GA can react readily with glutathione and produce oxidative stress like that proposed for AA, GA induced chromosomal mutations from oxidative DNA damage are unlikely because GA is mutagenic at much lower concentrations than AA in mouse lymphoma cells. Those chromosomal mutations are most likely associated with GA-derived DNA adducts. A recent study shows that there is a strong correlation between the levels of N7-GA-Gua and the extent of sister-chromatid exchange induction in GA-treated V79 cells (Martins et al., 2006). It has been suggested that the clastogenicity results from formation of depurinating DNA lesions and the formation of DNA breaks when repairing the DNA adducts.
In summary, although both AA and GA were cytotoxic and mutagenic, GA treatment resulted in DNA adduct formation while AA treatment did not in the mouse lymphoma cells. Both AA, at concentrations above 12 mM, and GA, at concentrations above 1 mM, induced mutations, mainly chromosomal mutations in mouse lymphoma cells. Our data suggest that AA induces those mutations probably by an indirect effect through an increase of cellular oxidative stress while GA induces the mutations via a DNA adduct mechanism. Regardless of the mechanistic difference, AA and GA generate mutations through a clastogenic mode of action in mouse lymphoma cells.
Acknowledgments
This research was partially supported by appointment (J.H.) to the Postgraduate Research Program at the National Center for Toxicological Research, administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the US Department of Energy and the US Food and Drug Administration. The views presented in this article do not necessarily reflect those of the Food and Drug Administration.
References
- Adams WT, Skopek TR. Statistical test for the comparison of samples from mutational spectra. J Mol Biol. 1987;194:391–396. doi: 10.1016/0022-2836(87)90669-3. [DOI] [PubMed] [Google Scholar]
- Adler ID, Baumgartner A, Gonda H, Friedman MA, Skerhut M. 1-Aminobenzotriazole inhibits acrylamide-induced dominant lethal effects in spermatids of male mice. Mutagenesis. 2000;15:133–136. doi: 10.1093/mutage/15.2.133. [DOI] [PubMed] [Google Scholar]
- Applegate ML, Moore MM, Broder CB, Burrell A, Juhn G, Kasweck KL, Lin PF, Wadhams A, Hozier JC. Molecular dissection of mutations at the heterozygous thymidine kinase locus in mouse lymphoma cells. Proc Natl Acad Sci USA. 1990;87:51–55. doi: 10.1073/pnas.87.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barfknecht TR, Mecca DJ, Naismith RW. The genotoxic activity of acrylamide. Environ Mol Mutagen. 1988;11(Suppl):11. [Google Scholar]
- Baum M, Fauth E, Fritzen S, Herrmann A, Mertes P, Merz K, Rudolphi M, Zankl H, Eisenbrand G. Acrylamide and glycidamide: genotoxic effects in V79-cells and human blood. Mutat Res. 2005;580:61–69. doi: 10.1016/j.mrgentox.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Besaratinia A, Pfeifer GP. Weak yet distinct mutagenicity of acrylamide in mammalian cells. J Natl Cancer Inst. 2003;95:889–896. doi: 10.1093/jnci/95.12.889. [DOI] [PubMed] [Google Scholar]
- Besaratinia A, Pfeifer GP. Genotoxicity of acrylamide and glycidamide. J Natl Cancer Inst. 2004;96:1023–1029. doi: 10.1093/jnci/djh186. [DOI] [PubMed] [Google Scholar]
- Besaratinia A, Pfeifer GP. A review of mechanisms of acrylamide carcinogenicity. Carcinogenesis. 2007;28:519–528. doi: 10.1093/carcin/bgm006. [DOI] [PubMed] [Google Scholar]
- Blasiak J, Gloc E, Wozniak K, Czechowska A. Genotoxicity of acrylamide in human lymphocytes. Chem Biol Interact. 2004;149:137–149. doi: 10.1016/j.cbi.2004.08.002. [DOI] [PubMed] [Google Scholar]
- Bull RJ, Robinson M, Laurie RD, Stoner GD, Greisiger E, Meier JR, Stober J. Carcinogenic effects of acrylamide in Sencar and A/J mice. Cancer Res. 1984;44:107–111. [PubMed] [Google Scholar]
- Cariello NF, Piegorsch WW, Adams WT, Skopek TR. Computer program for the analysis of mutational spectra: application to p53 mutations. Carcinogenesis. 1994;15:2281–2285. doi: 10.1093/carcin/15.10.2281. [DOI] [PubMed] [Google Scholar]
- Chen T, Harrington-Brock K, Moore MM. Mutant frequency and mutational spectra in the Tk and Hprt genes of N-ethyl-N-nitrosourea-treated mouse lymphoma cells dagger. Environ Mol Mutagen. 2002;39:296–305. doi: 10.1002/em.10075. [DOI] [PubMed] [Google Scholar]
- Chen T, Moore MM. Screening for chemical mutagens using the mouse lymphoma assay. In: Yan Z, Caldwell GW, editors. Optimization in Drug Discovery: In-vitro Methods. Humana Press; Totowa, New Jersey: 2004. pp. 337–352. [Google Scholar]
- Clements J. The mouse lymphoma assay. Mutat Res. 2000;455:97–110. doi: 10.1016/s0027-5107(00)00066-x. [DOI] [PubMed] [Google Scholar]
- Dearfield KL, Abernathy CO, Ottley MS, Brantner JH, Hayes PF. Acrylamide: its metabolism, developmental and reproductive effects, genotoxicity, and carcinogenicity. Mutat Res. 1988;195:45–77. doi: 10.1016/0165-1110(88)90015-2. [DOI] [PubMed] [Google Scholar]
- Dearfield KL, Douglas GR, Ehling UH, Moore MM, Sega GA, Brusick DJ. Acrylamide: a review of its genotoxicity and an assessment of heritable genetic risk. Mutat Res. 1995;330:71–99. doi: 10.1016/0027-5107(95)00037-j. [DOI] [PubMed] [Google Scholar]
- Doerge DR, da Costa GG, McDaniel LP, Churchwell MI, Twaddle NC, Beland FA. DNA adducts derived from administration of acrylamide and glycidamide to mice and rats. Mutat Res. 2005a;580:131–141. doi: 10.1016/j.mrgentox.2004.10.013. [DOI] [PubMed] [Google Scholar]
- Doerge DR, Young JF, McDaniel LP, Twaddle NC, Churchwell MI. Toxicokinetics of acrylamide and glycidamide in B6C3F1 mice. Toxicol Appl Pharmacol. 2005b;202:258–267. doi: 10.1016/j.taap.2004.07.001. [DOI] [PubMed] [Google Scholar]
- Doerge DR, Young JF, McDaniel LP, Twaddle NC, Churchwell MI. Toxicokinetics of acrylamide and glycidamide in Fischer 344 rats. Toxicol Appl Pharmacol. 2005c;208:199–209. doi: 10.1016/j.taap.2005.03.003. [DOI] [PubMed] [Google Scholar]
- Friedman MA, Dulak LH, Stedham MA. A lifetime oncogenicity study in rats with acrylamide. Fundam Appl Toxicol. 1995;27:95–105. doi: 10.1093/toxsci/27.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamboa da Costa G, Churchwell MI, Hamilton LP, Von Tungeln LS, Beland FA, Marques MM, Doerge DR. DNA adduct formation from acrylamide via conversion to glycidamide in adult and neonatal mice. Chem Res Toxicol. 2003;16:1328–1337. doi: 10.1021/tx034108e. [DOI] [PubMed] [Google Scholar]
- Ghanayem BI, McDaniel LP, Churchwell MI, Twaddle NC, Snyder R, Fennell TR, Doerge DR. Role of CYP2E1 in the epoxidation of acrylamide to glycidamide and formation of DNA and hemoglobin adducts. Toxicol Sci. 2005;88:311–318. doi: 10.1093/toxsci/kfi307. [DOI] [PubMed] [Google Scholar]
- Harrington-Brock K, Collard DD, Chen T. Bromate induces loss of heterozygosity in the thymidine kinase gene of L5178Y/Tk+/−-3.7.2C mouse lymphoma cells. Mutat Res. 2003;537:21–28. doi: 10.1016/s1383-5718(03)00044-5. [DOI] [PubMed] [Google Scholar]
- Hashimoto K, Tanii H. Mutagenicity of acrylamide and its analogues in Salmonella typhimurium. Mutat Res. 1985;158:129–133. doi: 10.1016/0165-1218(85)90075-8. [DOI] [PubMed] [Google Scholar]
- Hozier J, Sawyer J, Clive D, Moore M. Cytogenetic distinction between the TK+ and TK− chromosomes in the L5178Y Tk+/− 3.7.2C mouse-lymphoma cell line. Mutat Res. 1982;105:451–456. doi: 10.1016/0165-7992(82)90193-2. [DOI] [PubMed] [Google Scholar]
- Johnson KA, Gorzinski SJ, Bodner KM, Campbell RA, Wolf CH, Friedman MA, Mast RW. Chronic toxicity and oncogenicity study on acrylamide incorporated in the drinking water of Fischer 344 rats. Toxicol Appl Pharmacol. 1986;85:154–168. doi: 10.1016/0041-008x(86)90109-2. [DOI] [PubMed] [Google Scholar]
- Knaap AG, Kramers PG, Voogd CE, Bergkamp WG, Groot MG, Langebroek PG, Mout HC, van der Stel JJ, Verharen HW. Mutagenic activity of acrylamide in eukaryotic systems but not in bacteria. Mutagenesis. 1988;3:263–268. doi: 10.1093/mutage/3.3.263. [DOI] [PubMed] [Google Scholar]
- Koyama N, Sakamoto H, Sakuraba M, Koizumi T, Takashima Y, Hayashi M, Matsufuji H, Yamagata K, Masuda S, Kinae N, Honma M. Genotoxicity of acrylamide and glycidamide in human lymphoblastoid TK6 cells. Mutat Res. 2006;603:151–158. doi: 10.1016/j.mrgentox.2005.11.006. [DOI] [PubMed] [Google Scholar]
- Manjanatha MG, Aidoo A, Shelton SD, Bishop ME, McDaniel LP, Lyn-Cook LE, Doerge DR. Genotoxicity of acrylamide and its metabolite glycidamide administered in drinking water to male and female Big Blue mice. Environ Mol Mutagen. 2006;47:6–17. doi: 10.1002/em.20157. [DOI] [PubMed] [Google Scholar]
- Martins C, Oliveira NG, Pingarilho M, da Costa GG, Martins V, Marques MM, Beland FA, Churchwell MI, Doerge DR, Rueff J, Gaspar JF. Cytogenetic damage induced by acrylamide and glycidamide in mammalian cells: correlation with specific glycidamide-DNA adducts. Toxicol Sci. 2006;95:383–390. doi: 10.1093/toxsci/kfl155. [DOI] [PubMed] [Google Scholar]
- Moore MM, Amtower A, Doerr C, Brock KH, Dearfield KL. Mutagenicity and clastogenicity of acrylamide in L5178Y mouse lymphoma cells. Environ Mutagen. 1987;9:261–267. doi: 10.1002/em.2860090305. [DOI] [PubMed] [Google Scholar]
- Moore MM, Clive D, Howard BE, Batson AG, Turner NT. In situ analysis of trifluorothymidine-resistant (TFTr) mutants of L5178Y/Tk+/− mouse lymphoma cells. Mutat Res. 1985;151:147–159. doi: 10.1016/0027-5107(85)90193-9. [DOI] [PubMed] [Google Scholar]
- Moore MM, Honma M, Clements J, Bolcsfoldi G, Burlinson B, Cifone M, Clarke J, Delongchamp R, Durward R, Fellows M, Gollapudi B, Hou S, Jenkinson P, Lloyd M, Majeska J, Myhr B, O’Donovan M, Omori T, Riach C, San R, Stankowski LF, Jr, Thakur AK, Van Goethem F, Wakuri S, Yoshimura I. Mouse lymphoma thymidine kinase gene mutation assay: follow-up meeting of the International Workshop on Genotoxicity Testing— Aberdeen, Scotland, 2003—Assay acceptance criteria, positive controls, and data evaluation. Environ Mol Mutagen. 2006;47:1–5. doi: 10.1002/em.20159. [DOI] [PubMed] [Google Scholar]
- Moore MM, Honma M, Clements J, Bolcsfoldi G, Cifone M, Delongchamp R, Fellows M, Gollapudi B, Jenkinson P, Kirby P, Kirchner S, Muster W, Myhr B, O’Donovan M, Oliver J, Omori T, Ouldelhkim MC, Pant K, Preston R, Riach C, San R, Stankowski LF, Jr, Thakur A, Wakuri S, Yoshimura I. Mouse lymphoma thymidine kinase gene mutation assay: International Workshop on Genotoxicity Tests Workgroup report—Plymouth, UK 2002. Mutat Res. 2003;540:127–140. doi: 10.1016/j.mrgentox.2003.07.003. [DOI] [PubMed] [Google Scholar]
- Mottram DS, Wedzicha BL, Dodson AT. Acrylamide is formed in the Maillard reaction. Nature. 2002;419:448–449. doi: 10.1038/419448a. [DOI] [PubMed] [Google Scholar]
- Paulsson B, Kotova N, Grawe J, Henderson A, Granath F, Golding B, Tornqvist M. Induction of micronuclei in mouse and rat by glycidamide, genotoxic metabolite of acrylamide. Mutat Res. 2003;535:15–24. doi: 10.1016/s1383-5718(02)00281-4. [DOI] [PubMed] [Google Scholar]
- Puppel N, Tjaden Z, Fueller F, Marko D. DNA strand breaking capacity of acrylamide and glycidamide in mammalian cells. Mutat Res. 2005;580:71–80. doi: 10.1016/j.mrgentox.2004.11.009. [DOI] [PubMed] [Google Scholar]
- Rice JM. The carcinogenicity of acrylamide. Mutat Res. 2005;580:3–20. doi: 10.1016/j.mrgentox.2004.09.008. [DOI] [PubMed] [Google Scholar]
- Segerback D, Calleman CJ, Schroeder JL, Costa LG, Faustman EM. Formation of N-7-(2-carbamoyl-2-hydroxyethyl)guanine in DNA of the mouse and the rat following intraperitoneal administration of [14C]acrylamide. Carcinogenesis. 1995;16:1161–1165. doi: 10.1093/carcin/16.5.1161. [DOI] [PubMed] [Google Scholar]
- Singh SP, Chen T, Chen L, Mei N, McLain E, Samokyszyn V, Thaden JJ, Moore MM, Zimniak P. Mutagenic effects of 4-hydroxynonenal triacetate, a chemically protected form of the lipid peroxidation product 4-hydroxynonenal, as assayed in L5178Y/Tk+/− mouse lymphoma cells. J Pharmacol Exp Ther. 2005;313:855–861. doi: 10.1124/jpet.104.080754. [DOI] [PubMed] [Google Scholar]
- Solomon JJ, Fedyk J, Mukai F, Segal A. Direct alkylation of 2′-deoxynucleosides and DNA following in vitro reaction with acrylamide. Cancer Res. 1985;45:3465–3470. [PubMed] [Google Scholar]
- Stadler RH, Blank I, Varga N, Robert F, Hau J, Guy PA, Robert MC, Riediker S. Acrylamide from Maillard reaction products. Nature. 2002;419:449–450. doi: 10.1038/419449a. [DOI] [PubMed] [Google Scholar]
- Sumner SC, Fennell TR, Moore TA, Chanas B, Gonzalez F, Ghanayem BI. Role of cytochrome P450 2E1 in the metabolism of acrylamide and acrylonitrile in mice. Chem Res Toxicol. 1999;12:1110–1116. doi: 10.1021/tx990040k. [DOI] [PubMed] [Google Scholar]
- Tareke E, Rydberg P, Karlsson P, Eriksson S, Tornqvist M. Acrylamide: a cooking carcinogen? Chem Res Toxicol. 2000;13:517–522. doi: 10.1021/tx9901938. [DOI] [PubMed] [Google Scholar]
- Tornqvist M. Acrylamide in food: the discovery and its implications: a historical perspective. Adv Exp Med Biol. 2005;561:1–19. doi: 10.1007/0-387-24980-X_1. [DOI] [PubMed] [Google Scholar]
- Tsuda H, Shimizu CS, Taketomi MK, Hasegawa MM, Hamada A, Kawata KM, Inui N. Acrylamide; induction of DNA damage, chromosomal aberrations and cell transformation without gene mutations. Mutagenesis. 1993;8:23–29. doi: 10.1093/mutage/8.1.23. [DOI] [PubMed] [Google Scholar]
- Wang J, Chen T, Honma M, Chen L, Moore MM. 3′-azido-3′-deoxythymidine induces deletions in L5178Y mouse lymphoma cells. Environ Mol Mutagen. 2007;48:248–257. doi: 10.1002/em.20263. [DOI] [PubMed] [Google Scholar]
- Zeiger E, Anderson B, Haworth S, Lawlor T, Mortelmans K, Speck W. Salmonella mutagenicity tests: III. Results from the testing of 255 chemicals. Environ Mutagen. 1987;9(Suppl 9):1–109. [PubMed] [Google Scholar]