Abstract
Drug-metabolizing enzymes are an important battery of proteins that are involved in drug metabolism, xenobiotic detoxification, and drug-induced toxicity. Systematic, efficient, and simultaneous evaluation of drug-metabolizing gene expression in response to chemicals has a wide variety of implications in drug development, disease prevention, and personalized medicine and nutrition. In the current study, the authors have systematically and simultaneously evaluated the hepatic expression profile of drug-metabolizing enzymes in cultured human hepatocytes exposed to the xenobiotics rifampicin, omeprazole, and 3-methylcholanthrene (3-MC) using the Drug Metabolism RT2Profiler™ PCR Arrays. This new high-throughput tool allowed the authors to evaluate the expression of genes coding for 84 drug-metabolizing enzymes (including phase 1 and phase 2 drug-metabolizing enzymes and transporters) simultaneously, in a 96-well format using a small amount of experimental materials. To validate the quality of the Drug Metabolism RT2Profiler™ PCR Arrays, the PCR Array was compared with the well-documented platform TaqMan assay, and a high concordance was shown between these 2 methods, indicating the high reliability of the Drug Metabolism RT2Profiler™ PCR Arrays. In addition, increasing or decreasing the expression of drug-metabolizing enzymes by these 3 compounds was observed, and underlying mechanisms are discussed.
Keywords: real time-PCR, gene expression, drug-metabolizing enzyme, drug-metabolizing gene
INTRODUCTION
Administration of some pharmacologically active substances in humans can result in induction or inhibition of drug-metabolizing enzymes (DMEs). Induction, a phenomenon in which a substance leads to an increase in enzyme activity, usually results from overexpression of the respective gene; one of the major mechanisms for enzyme inhibition is due to a downregulation of gene expression in response to xenobiotics.1 Systematic examination of DME expression profiles provides important information for clinical implications as well as the potential toxicity of a given drug.
Primary human hepatocytes remain differentiated and sustain the major DME activities for a relatively long period of time in culture; therefore, they represent a unique in vitro system and serve as a gold standard for induction and inhibition studies of DMEs.2 Numerous studies with cultured primary human hepatocytes have demonstrated that this system is a valuable model for evaluation of DMEs in response to inductive/inhibitory substances. For example, the antibiotic rifampicin has exhibited an inductive ability in the expression of cytochrome (CYP)2C8 and CYP2C9 in human hepatocytes,3,4 the proton-pump inhibitor omeprazole has been reported to increase expression levels of CYP1A1 and CYP1A2,5 and the carcinogen 3-methylcholanthrene (3-MC) is believed to be an inducer of CYP1A genes.6,7 However, few studies have systematically evaluated the effects of these chemicals on the expression of human drug-metabolizing genes (DMGs) in primary human hepatocytes. In addition, inhibitory effects on human DMEs by these xenobiotics have been underinvestigated.
Traditional enzymatic activity assays have been the primary tools for assessing drug metabolisms. However, these assays are time-consuming and require a large amount of experimental materials. High-throughput, simultaneous, and efficient measurements are currently lacking. Enzymatic induction is primarily mediated by increasing the transcription levels of DMGs, whereas downregulation of these genes is one of the mechanisms for enzymatic inhibition. Thus, assessing gene expression at the mRNA level is an important approach in identifying drug-induced effects on DMGs. Real-time reverse transcriptase polymerase chain reaction (RT-PCR) analysis is one of the most sensitive, reliable, and quantitative methods and is widely used to evaluate and measure gene expression at mRNA level.
In this study, we employed a new high-throughput method, the Human Drug Metabolism RT2 Profiler™ PCR Array, to profile 84 DMGs in primary human hepatocytes systematically and simultaneously, using 3 well-known DME inducers (rifampicin, omeprazole, and 3-MC) as treatment models. This PCR Array combines the quantitative performance of SYBR Green–based real-time PCR with the multiple gene-profiling capabilities of a microarray. We also assessed the technical performance and reproducibility of the PCR Array by comparing our data to the TaqMan data generated from the MicroArray Quality Control study (MAQC).8,9
MATERIALS AND METHODS
Primary human hepatocytes and cell treatments
Primary human hepatocytes from 3 anonymous male donors were obtained from Dr. Steve Strom at the University of Pittsburgh as part of the Liver Tissue Cell Distribution System. Primary hepatocytes were plated on collagen in T-25 flasks containing approximately 2 × 106 cells. Upon arrival, the shipping media was removed and replaced with serum-free hepatocyte maintenance media (HMM) supplemented with insulin and GA-1000 using HMM SingleQuots (BioWhittaker, Rockland, ME). The cultured hepatocytes were incubated at 37 °C in a humidified atmosphere of 5% CO2 for 4 h and then dosed with 20 µM rifampicin, 50 µM omeprazole, or 8 µM 3-MC, respectively, for 24 h. The doses for rifampicin, omeprazole, and 3-MC were based on literature describing the induction of DMEs in primary human hepatocytes.4,5,7 Rifampicin, omeprazole, and 3-MC were purchased from Sigma-Aldrich (St. Louis, MO). All chemicals were prepared as a 1000-times stock solution in DMSO and added to the cultured human primary hepatocytes in the indicated concentrations. The same amount of DMSO (0.1% v/v) was added as the vehicle to the nontreatment control group. This project was approved by the Research Involving Human Subjects Committee of the Food and Drug Administration.
RNA isolation and quality control
Total RNA from primary human hepatocytes was isolated using an RNeasy system (Qiagen, Hilden, Germany). The yield of the extracted RNA was determined spectrophotometrically by measuring the optical density at 260 nm. The purity and quality of extracted RNA were evaluated using the RNA 6000 LabChip and Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). Only high-quality RNA with RNA integrity numbers greater than 9.0 were used for PCR analysis.
Reference RNA sample
Universal Human Reference RNA (UHRR) was purchased from Stratagene (La Jolla, CA), and Human Brain Reference RNA (HBRR) was purchased from Ambion (Austin, TX). The same batches were used in this study as were used by the MAQC project.
Human Drug Metabolism RT2Profiler™ PCR Array
First Strand cDNA Synthesis Kit and Human Drug Metabolism RT2Profiler™ PCR Arrays were from SuperArray Bioscience (Frederick, MD).
Real-time RT-PCR
First-strand cDNA synthesis. One microgram of total RNA was reverse transcribed in a final volume of 20 µl with random primers at 37 °C for 60 min according to the manufacturer’s instructions (SuperArray Bioscience). Briefly, reverse transcriptase was inactivated by heating at 95 °C for 5 min, and the cDNA was diluted to 100 µl by adding RNase-free water and stored at −20 °C. The PCR was carried out using an ABI 7000 instrument (Applied Biosystems, Foster City, CA). For one 96-well plate of the PCR Array, a 2450 µl PCR master mix containing 1× PCR master mix and 98 µl of diluted cDNA was prepared, and an aliquot of 25 µl was added to each well. For quality control, no reverse transcription control and no template control were performed. A series of 10-fold cDNA dilutions for β-actin amplification was performed to validate the linearity of PCR Array assays. Universal cycling conditions (10 min at 95 °C, 15 s at 95 °C, 1 min at 60 °C for 40 cycles) were used. Five replicates were run for each reference RNA sample; 3 technical replicates were run for each treated sample and controls. Table 1 lists the genes measured in this assay.
Table 1.
Functional Gene Grouping of Human Drug Metabolism PCR Array
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | ABCB1 | ABCC1 | ABP1 | ADH1B | ADH1C | ADH4 | ADH5 | ADH6 | AHR | ALAD | ALDH1A1 | ALOX12 |
| B | ALOX15 | ALOX5 | APOE | ARNT | ASNA1 | BLVRA | BLVRB | CES2 | CES4 | CHST1 | COMT | CYP11B2 |
| C | CYP17A1 | CYP19A1 | CYP1A1 | CYP2B6 | CYP2C19 | CYP2C8 | CYP2C9 | CYP2D6 | CYP2E1 | CYP2F1 | CYP2J2 | CYP3A5 |
| D | CYP5R3 | EPHX1 | FAAH | FBP1 | GAD1 | GCKR | GGT1 | GPI | GPX1 | GPX2 | GPX3 | GPX4 |
| E | GPX5 | GSR | GSTA3 | GSTA4 | GSTM2 | GSTM3 | GSTM5 | GSTP1 | GSTT1 | GSTZ1 | HK2 | HSD17B1 |
| F | HSD17B2 | HSD17B3 | LPO | MARCKS | MGST1 | MGST2 | MGST3 | MPO | MT2A | MT3 | MTHFR | NAT1 |
| G | NAT2 | NOS3 | NQO1 | PKLR | PKM2 | PON1 | PON2 | PON3 | SMARCAL1 | SNN | SRD5A1 | SRD5A2 |
| H | 18SrRNA | HPRT1 | RPL13A | GAPDH | ACTB | ACTB | ACTB | ACTB | ACTB | ACTB | ACTB | ACTB |
Fill color: red = phase 1 drug-metabolizing genes (DMGs); white = phase 2 DMGs; green = transporters; blue = other related genes; pink = housekeeping genes.
Data normalization and analysis
Four endogenous control genes—hypoxanthine phosphoribosyltransferase (HPRT1), ribosomal protein L13a (RPL13A), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and β-actin (ACTB)—present on the PCR Array were used for normalization. Each replicate cycle threshold (CT)was normalized to the average CT of 4 endogenous controls on a per plate basis. The comparative CT method was used to calculate the relative quantification of gene expression.10 The following formula was used to calculate the relative amount of the transcripts in the chemical-treated samples (treat) and the vehicle-treated samples (control), both of which were normalized to the endogenous controls. ΔΔCT = ΔCT (treat) −ΔCT (control) for biological RNA samples or ΔΔCT = ΔCT (HBRR) − ΔCT (UHRR) for reference RNA samples. ΔCT is the log2 difference in CT between the target gene and endogenous controls by subtracting the average CT of controls from each replicate. The fold change for each treated sample (or HBRR) relative to the control sample (or UHRR) = 2−ΔΔCT.
Sensitivity detection and identification of differentially expressed genes
PCR Array quantification was based on the CT number. A gene was considered not detectable when CT >32. CT was defined as 35 for the ΔCT calculation when the signal was under detectable limits. A list of differentially expressed genes was identified using a 2-tailed t-test. The criteria were a p value less than 0.05 and a mean difference equal to or greater than 2-fold. The statistical calculation was based on ΔCT values.
RESULTS
Gene expression patterns associated with exposure to DME inducers
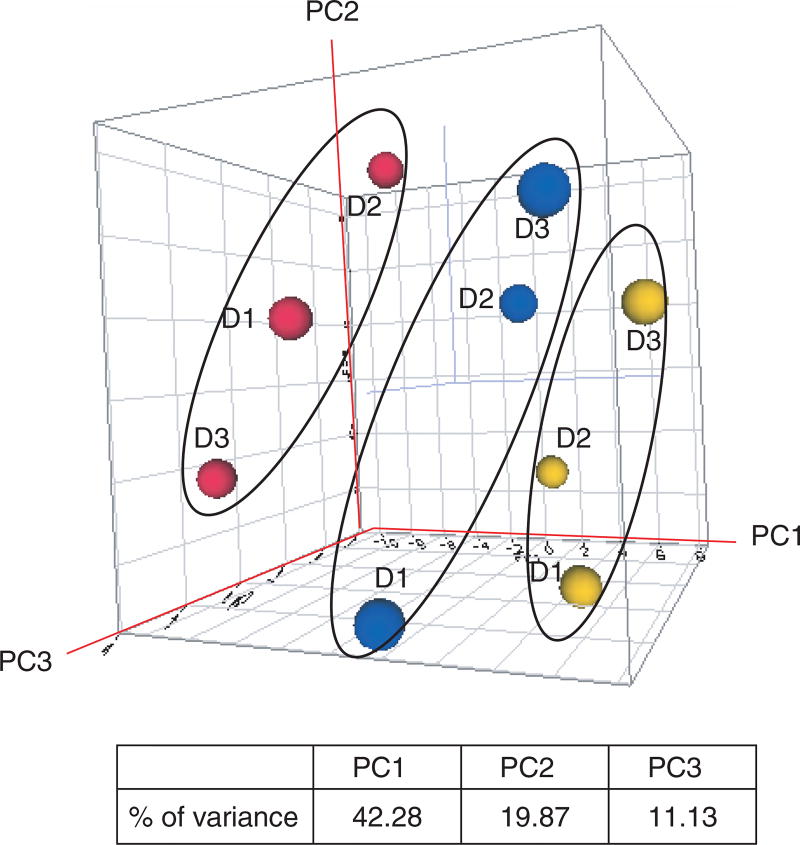
Primary human hepatocytes isolated from 3 donors were exposed to 3 classic DME inducers: 20 µM rifampicin, 50 µM omeprazole, and 8 µM 3-MC, respectively. Following a 24-h treatment, RNA was isolated, and the Human Drug Metabolism PCR Array analysis was performed. A comparative CT method was used to calculate the relative quantification of gene expression, designated as fold change. Log2 fold changes (ΔΔCT) of 84 DMGs for each treatment were calculated against the control group and used for clustering analysis. Figure 1 displays a principal component analysis 3-dimensional view using the first 3 principal components for gene expression profiles from the samples. This analysis revealed 3 groups, indicating 3 distinct patterns of gene expression associated with the treatments. The clustering analysis is an unbiased data analysis procedure as it does not consider the class assignment of compounds. The results indicate that a treatment effect was detected based on the compound rather than individual subjects.
FIG. 1.
Principal component analysis for gene expression profiles from 3 chemical treatments and distribution of variance in the first 3 components. The log2 fold change of 84 drug-metabolizing genes present on the PCR array was used. The red, blue, and orange dots indicate rifampicin, 3-methylcholanthrene, and omeprazole samples, respectively. D1 = donor 1; D2 = donor 2; D3 = donor 3.
Induction of gene expression by DME inducers
Based on gene selection criteria (p <0.05 and fold change ≥ 2), there were 11, 13, and 13 genes that showed an altered expression with rifampicin treatment; 12, 15, and 16 genes that were altered by omeprazole; and 11, 15, and 13 genes that were altered by 3-MC for donor 1, 2, and 3, respectively.
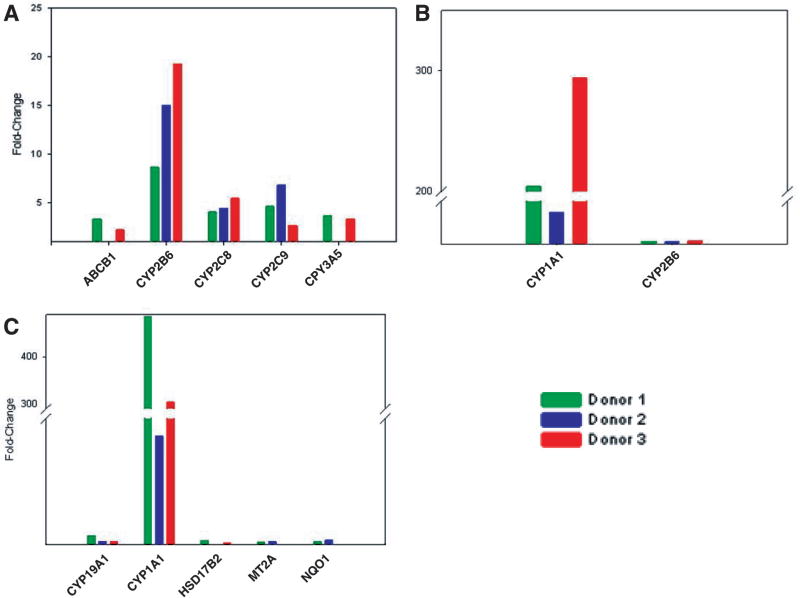
Rifampicin is a well-known inducer for many DMEs including the CYP2 family, CYP3A,11–13 MDR1,MDR2, and UGT1A1.14 In comparison to vehicle (DMSO) treatment, our data showed that the 20-µM rifampicin treatment elevated the gene expression levels of CYP2B6, CYP2C8, and CYP2C9 in hepatocytes isolated from all 3 donors (Fig. 2A). Of these, CYP2B6 had the highest expression level (8- to 20-fold), followed by CYP2C8 (4- to 5.5-fold) and CYP2C9 (2.5- to 7-fold). In addition, ABCB1 and CYP3A5 showed a moderate increase in expression, although this was observed only in hepatocytes from 2 of 3 donors. In agreement with another report,5 Figure 2B demonstrates that the well-known inducer of CYP1A1 and CYP1A2, omeprazole, had the greatest induction ability on CYP1A1 expression, with an overexpression level more than 40-fold higher. Notably, omeprazole increased CYP2B6 expression 3- to 5-fold higher in hepatocytes from all 3 donors, whereas it elevated LPO and MT2A expression in hepatocytes from only 1 donor (data not shown). The effects of 3-MC on human DMGs are shown in Figure 2C, in which the hepatic CYP1A1 mRNA expression levels in all 3 donors were elevated 60- to 480-fold by 3-MC treatment. The CYP19A1 expression levels were also increased 2.7- to 5–9 fold. In addition, HSD17B2 expression was increased in donor 1 and 3, whereas MT2A and NQ01 expression levels were moderately increased 2.5- to 3.4-fold in 2 donors (donor 1 and 2).
FIG. 2.
Induction of gene expression by treatment with (A) 20 µM rifampicin, (B) 50 µM omeprazole, and (C) 8 µM 3-methylcholanthrene. Green = donor 1; blue = donor 2; red = donor 3.
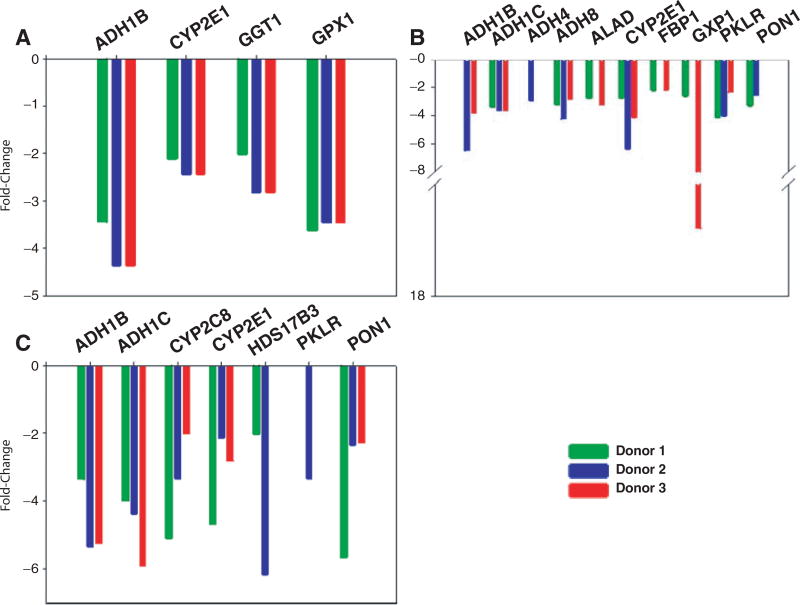
Interestingly, the downregulation of DMEs was also observed in our study. As shown in Figure 3A, rifampicin exhibits 3- to 4-fold suppression on the expression of ADH1B and GPX1. It has been reported that activation of PXR by rifampicin promotes PXR interaction with hepatic nuclear factor 4 alpha (HNF4 alpha), which blocks PGC-1 alpha (peroxisome proliferator-activated receptor gamma coactivator) and results in inhibition of CYP7A1 gene transcription.15 On the other hand, rifampicin could induce hepatic injury through an oxidative stress pathway,16 thus decreasing the expression of oxidative stress-related genes. Inhibition of GPX1 expression by rifampicin may contribute to this process. Omeprazole has shown inhibitory effects on the expression of ADH1B, ADH1C, ADH8, CYP2E1, GPX1, and PKLR by 2- to 17-fold (Fig. 3B). Likewise, 3-MC had very similar inhibitory effects on the expression of some of these genes (Fig. 3C). However, the underlying mechanisms for such inhibition are not clear. The aromatic hydrocarbon receptor (AHR) is a ligand-activated transcription factor that is involved in the regulation of the expression of many DMGs. Accumulative evidence indicates that many AHR ligands participate in signal transduction pathways that deal with drug metabolism and toxicant detoxification.17 Several studies have shown that the AHR cross-talks, directly or indirectly, with other transcription factors, coactivators, and corepressors to modulate gene transcriptional programs.18 Therefore, more intensive and well-designed studies are warranted for elucidating the underlying mechanisms.
FIG. 3.
Inhibition of gene expression by treatment with (A) 20 µM rifampicin, (B) 50 µM omeprazole, and (C) 8 µM 3-methylcholanthrene. Green = donor 1; blue = donor 2; red = donor 3.
Concordance between Human Drug Metabolism PCR Array and TaqMan assays
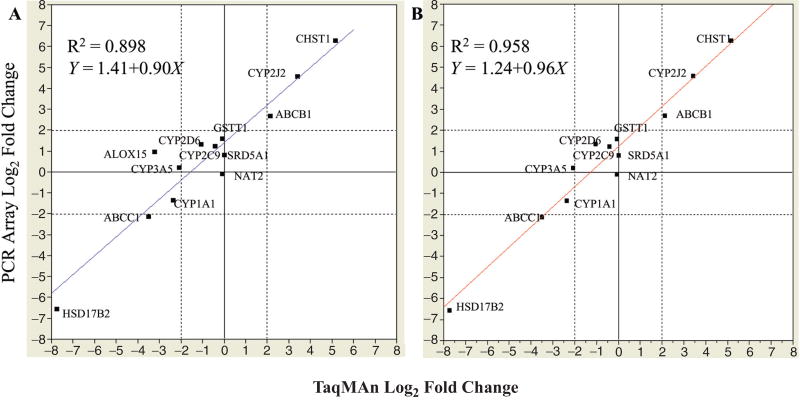
To address the technical performance and reproducibility of the PCR Array, we compared the PCR Array data with those generated from TaqMan RT-PCR using 2 RNA reference samples from the MAQC project (http://edkb.fda.gov/MAQC/MainStudy/upload). The 2 RNA reference samples are UHRR, which is composed of RNA from 10 different human cell lines, and HBRR. By matching GenBank accession numbers, 13 genes were found present in both the TaqMan assays and the PCR Arrays. Using these 13 identified genes in a pairwise manner, a regression analysis revealed that the R2 and slope for the PCR Array versus TaqMan was 0.898 and 0.90, respectively (Fig. 4A). Notably, ALOX15 demonstrated discordant expressions between the TaqMan and PCR Array, with opposite directions in fold-change between the 2 platforms. With careful examination, we found that the average CT numbers were relatively high, 31.8 (sample UHRR) and 39.2 (sample HBRR) in the TaqMan data, whereas the CT numbers remained in the detectable range (<32) with the PCR Array. The discrepancies could be caused by a coamplification of genomic DNA in the PCR Array due to the upstream and downstream primers being designed to target the same exon of the gene. When this gene was removed from analysis, the R2 and slope increased to 0.958 and 0.96 (Fig. 4B), respectively demonstrating a high degree of similarity in detecting differential gene expression between the TaqMan assay and the PCR Array.
FIG. 4.
The scatterplots showing the correlation of the log2 fold change values (sample Human Brain Reference RNA over Universal Human Reference RNA) of common genes between the TaqMan and PCR Array. The log2 fold change of each gene was subjected to bivariate analysis. (A) Based on 13 common genes. (B) Based on 12 common genes. The line shown is a linear regression fit. Linear fit: Y = 1.41 + 0.90X, R2 = 0.898 (A), Y = 1.24 + 0.96X, R2 = 0.958 (B).
DISCUSSION
In pharmacological or toxicological studies, the modifications (induction or inhibition) of DMEs in liver samples are traditionally evaluated by performing enzymatic activity assays; however, gene expression profiling is an efficient and relatively reliable approach. It has been demonstrated that there is a high degree of correlation for phase 1 enzymes (e.g., CYP1A2 and CYP3A4) and phase 2 enzymes (e.g., GST, UDP glycosyltransferase, sultotransferase, N-acetyl transferase, thiopurine methyl transferase, and COMT) between the fold inductions of the enzymatic activity and mRNA expression in human liver samples.19,20 Indeed, real-time PCR is one of the most sensitive, reliable methods for gene expression quantification,21 evidently proved by many studies.7,22,23 However, many real-time PCR assays are not able to be performed simultaneously.
In the current study, we used the Drug Metabolism RT2Profiler™ PCR Arrays to systematically and simultaneously evaluate the hepatic expression profiles of DMEs in cultured primary human hepatocytes exposed to the xenobiotics rifampicin, omeprazole, and 3-MC. This PCR Array combines the quantitative performance of SYBR Green–based real-time PCR with the multiple gene-profiling capabilities of a microarray. Moreover, as little as 25 ng of RNA was needed for measuring 84 genes simultaneously, much less than is required for conventional RT-PCR.
Using this high-throughput method, all 84 DMGs were examined simultaneously. As shown in Table 1, the Drug Metabolism RT2Profiler™ PCR Array contains 84 important DMGs, including 20 phase 1 genes, 51 phase 2 genes, and 5 transporter genes. Although the array is relatively comprehensive and informative, some important DMEs such as CYP3A4 and CYP1A2 are not included in the array. Subsequently, the array has been reformatted to include 42 major human isoforms of CYPs and other important DMEs. More practically, custom RT2Profiler™ PCR arrays can be made to order by the users (http://www.superarray.com/rt_pcr_product/HTML/PAHS-068A.html).
To validate the reliability of the data generated by the PCR Arrays, we compared this method with the well-documented platform TaqMan assays. By using 2 RNA reference samples from the MAQC project, we obtained a high concordance between the 2 methods, with an R2 and slope of 0.958 and 0.96, respectively (Fig. 4B). The consistency of the PCR Array was evaluated by intralaboratory experiments (technical replicates) as well as interlaboratory reproducibility (data collected between 2 laboratories). High concordance within technical replicates was observed, with correlation coefficients of >0.99. Good concordances were also seen between the 2 laboratories (>0.97), indicating the high reproducibility of the PCR Array.
The notable increases in gene expression levels following the 3 treatments (e.g., upregulation of CYP2B6, CYP2C8, CYP2C9, and CYP3A5 by rifampicin; upregulation of CYP1A1 by omeprazole; upregulation of CYP1A1 by 3-MC) are in agreement with previous findings.3–7,12,24 CYP2B6 is an inducible enzyme in humans and is present in almost all samples of human liver microsomes.25,26 Treatment of primary human hepatocytes with rifampicin resulting in a marked increase in CYP2B6 production has been reported by many studies.27,28 It has been shown that rifampicin binds to the nuclear pregnane X receptor (PXR), which activates the phenobarbital-responsive enhancer module region of the CYP2B6 gene.28 Likewise, CYP2C8, CYP2C9, and CYP3A5 are upregulated transcriptionally by rifampicin through binding the PXR, which cross-talks with other nuclear receptors, such as constitutive androstane receptor (CAR), glucocorticoid receptor, and HNF4 alpha.3,29 The fold induction of CYP2B6, CYP2C8, and CYP2C9 by rifampicin was highly variable in our experiment, suggesting that either an optimal induction of these enzymes was not achieved or that environmental exposure conditions for these liver donors were complicated.
Omeprazole is thought to be an AHR ligand that has a role in controlling the expression of human CYP1A1 and CYP1A2 genes.30 Omeprazole has been shown to bind the AHR and translocate into the nucleus. In the nucleus, the activated AHR-AHR dimer interacts with the xenobiotic responsive element that is located upstream of the CYP1A genes and enhances CYP1A gene transcription.30,31 Despite the finding that many chemicals can enhance CYP2B6 induction, very little information is available regarding the chemical diversity, efficacy, and potency of human CYP2B6 inducers. Several studies have reported that CYP2B6 is inducible by binding human PXR and CAR, and studies have shown a significant correlation between CYP2B6 and CAR/PXR mRNA levels in human liver samples.32 However, there is no evidence in the literature that CYP2B6 may be regulated by omeprazole. Therefore, it is worthwhile to investigate whether omeprazole binding to AHR upregulates CYP2B6 expression.
The carcinogen 3-MC, a polycyclic aromatic hydrocarbon, is a prototypical inducer of several CYPs, especially CYP1A1 and CYP1A2. Experiments have revealed that induction is basically due to an increase in the rate of transcription of these genes, although posttranscriptional effects on CYP1A2/1B1 have been reported. Biochemical and genetic evidence suggests that the induction of CYP1A1 transcription is mediated by AHR.33
In summary, we have used the PCR array to profile the gene expression in primary human hepatocytes exposed to 3 well-known DME inducers and also evaluated its technical performance by comparing the results to those generated by TaqMan assay. Our results demonstrated that this high-throughput, efficient method can be applied to determine human DMG expression after exposure to drugs or toxins.
Footnotes
DISCLAIMER
This document has been reviewed in accordance with United States Food and Drug Administration (FDA) policy and approved for publication. Approval does not signify that the contents necessarily reflect the position or opinions of the FDA nor does mention of trade names or commercial products constitute endorsement or recommendation for use. The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the FDA.
References
- 1.Barry M, Feely J. Enzyme induction, inhibition. Pharmacol Ther. 1990;48:71–94. doi: 10.1016/0163-7258(90)90019-x. [DOI] [PubMed] [Google Scholar]
- 2.LeCluyse EL. Human hepatocyte culture systems for the in vitro evaluation of cytochrome P450 expression, regulation. Eur J Pharm Sci. 2001;13:343–368. doi: 10.1016/s0928-0987(01)00135-x. [DOI] [PubMed] [Google Scholar]
- 3.Gerbal-Chaloin S, Daujat M, Pascussi JM, Pichard-Garcia L, Vilarem MJ, Maurel P. Transcriptional regulation of CYP2C9 gene: role of glucocorticoid receptor, constitutive, rostane receptor. J Biol Chem. 2002;277:209–217. doi: 10.1074/jbc.M107228200. [DOI] [PubMed] [Google Scholar]
- 4.Raucy JL, Mueller L, Duan K, Allen SW, Strom S, Lasker JM. Expression, induction of CYP2C P450 enzymes in primary cultures of human hepatocytes. J Pharmacol Exp Ther. 2002;302:475–482. doi: 10.1124/jpet.102.033837. [DOI] [PubMed] [Google Scholar]
- 5.Diaz D, Fabre I, Daujat M, Saint Aubert B, Bories P, Michel H, et al. Omeprazole is an aryl hydrocarbon-like inducer of human hepatic cytochrome P450. Gastroenterology. 1990;99:737–747. doi: 10.1016/0016-5085(90)90963-2. [DOI] [PubMed] [Google Scholar]
- 6.Nebert DW, Dalton TP, Okey AB, Gonzalez FJ. Role of aryl hydrocarbon receptor-mediated induction of the CYP1 enzymes in environmental toxicity, cancer. J Biol Chem. 2004;279:23847–23850. doi: 10.1074/jbc.R400004200. [DOI] [PubMed] [Google Scholar]
- 7.Girault I, Rougier N, Chesne C, Lidereau R, Beaune P, Bieche I, et al. Simultaneous measurement of 23 isoforms from the human cytochrome P450 families 1 to 3 by quantitative reverse transcriptase-polymerase chain reaction. Drug Metab Dispos. 2005;33:1803–1810. doi: 10.1124/dmd.105.005173. [DOI] [PubMed] [Google Scholar]
- 8.Shi L, Reid LH, Jones WD, Shippy R, Warrington JA, Baker SC, et al. The MicroArray Quality Control (MAQC) project shows inter-, intraplatform reproducibility of gene expression measurements. Nat Biotechnol. 2006;24:1151–1161. doi: 10.1038/nbt1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Canales RD, Luo Y, Willey JC, Austermiller B, Barbacioru CC, Boysen C, et al. Evaluation of DNA microarray results with quantitative gene expression platforms. Nat Biotechnol. 2006;24:1115–1122. doi: 10.1038/nbt1236. [DOI] [PubMed] [Google Scholar]
- 10.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR, the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 11.Chang TK, Yu L, Maurel P, Waxman DJ. Enhanced cyclophosphamide, ifosfamide activation in primary human hepatocyte cultures: response to cytochrome P-450 inducers, autoinduction by oxazaphosphorines. Cancer Res. 1997;57:1946–1954. [PubMed] [Google Scholar]
- 12.Gerbal-Chaloin S, Pascussi JM, Pichard-Garcia L, Daujat M, Waechter F, Fabre JM, et al. Induction of CYP2C genes in human hepatocytes in primary culture. Drug Metab Dispos. 2001;29:242–251. [PubMed] [Google Scholar]
- 13.Rae JM, Johnson MD, Lippman ME, Flockhart DA. Rifampin is a selective, pleiotropic inducer of drug metabolism genes in human hepatocytes: studies with cDNA, oligonucleotide expression arrays. J Pharmacol Exp Ther. 2001;299:849–857. [PubMed] [Google Scholar]
- 14.Kuypers DR, Verleden G, Naesens M, Vanrenterghem Y. Drug interaction between mycophenolate mofetil, rifampin: possible induction of uridine diphosphate-glucuronosyltransferase. Clin Pharmacol Ther. 2005;78:81–88. doi: 10.1016/j.clpt.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 15.Li T, Chiang JY. Mechanism of rifampicin, pregnane X receptor inhibition of human cholesterol 7 alpha-hydroxylase gene transcription. Am J Physiol Gastrointest Liver Physiol. 2005;288:G74–G84. doi: 10.1152/ajpgi.00258.2004. [DOI] [PubMed] [Google Scholar]
- 16.Sodhi CP, Rana SV, Mehta SK, Vaiphei K, Attari S, Mehta S. Study of oxidative-stress in isoniazid-rifampicin induced hepatic injury in young rats. Drug Chem Toxicol. 1997;20:255–269. doi: 10.3109/01480549709003881. [DOI] [PubMed] [Google Scholar]
- 17.Puga A, Tomlinson CR, Xia Y. Ah receptor signals cross-talk with multiple developmental pathways. Biochem Pharmacol. 2005;69:199–207. doi: 10.1016/j.bcp.2004.06.043. [DOI] [PubMed] [Google Scholar]
- 18.Marlowe JL, Puga A. Aryl hydrocarbon receptor, cell cycle regulation, toxicity, tumorigenesis. J Cell Biochem. 2005;96:1174–1184. doi: 10.1002/jcb.20656. [DOI] [PubMed] [Google Scholar]
- 19.Roymans D, Van Looveren C, Leone A, Parker JB, McMillian M, Johnson MD, et al. Determination of cytochrome P450 1A2, cytochrome P450 3A4 induction in cryopreserved human hepatocytes. Biochem Pharmacol. 2004;67:427–437. doi: 10.1016/j.bcp.2003.09.022. [DOI] [PubMed] [Google Scholar]
- 20.Iyer KR, Sinz MW. Characterization of phase I, phase II hepatic drug metabolism activities in a panel of human liver preparations. Chem Biol Interact. 1999;118:151–169. doi: 10.1016/s0009-2797(99)00007-1. [DOI] [PubMed] [Google Scholar]
- 21.Klein D. Quantification using real-time PCR technology: applications, limitations. Trends Mol Med. 2002;8:257–260. doi: 10.1016/s1471-4914(02)02355-9. [DOI] [PubMed] [Google Scholar]
- 22.Rodriguez-Antona C, Jover R, Gomez-Lechon MJ, Castell JV. Quantitative RT-PCR measurement of human cytochrome P-450s: application to drug induction studies. Arch Biochem Biophys. 2000;376:109–116. doi: 10.1006/abbi.2000.1697. [DOI] [PubMed] [Google Scholar]
- 23.Perez G, Tabares B, Jover R, Gomez-Lechon MJ, Castell JV. Semi-automatic quantitative RT-PCR to measure CYP induction by drugs in human hepatocytes. Toxicol In Vitro. 2003;17:643–649. doi: 10.1016/s0887-2333(03)00109-7. [DOI] [PubMed] [Google Scholar]
- 24.Whitlock JP., Jr Induction of cytochrome P4501A1. Annu Rev Pharmacol Toxicol. 1999;39:103–125. doi: 10.1146/annurev.pharmtox.39.1.103. [DOI] [PubMed] [Google Scholar]
- 25.Gervot L, Rochat B, Gautier JC, et al. Human CYP2B6: expression, inducibility, catalytic activities. Pharmacogenetics. 1999;9:295–306. [PubMed] [Google Scholar]
- 26.LeCluyse E, Madan A, Hamilton G, Carroll K, DeHaan R, Parkinson A. Expression, regulation of cytochrome P450 enzymes in primary cultures of human hepatocytes. J Biochem Mol Toxicol. 2000;14:177–188. doi: 10.1002/(sici)1099-0461(2000)14:4<177::aid-jbt1>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 27.Edwards RJ, Price RJ, Watts PS, Renwick AB, Tredger JM, Boobis AR, et al. Induction of cytochrome P450 enzymes in cultured precision-cut human liver slices. Drug Metab Dispos. 2003;31:282–288. doi: 10.1124/dmd.31.3.282. [DOI] [PubMed] [Google Scholar]
- 28.Goodwin B, Moore LB, Stoltz CM, McKee DD, Kliewer SA. Regulation of the human CYP2B6 gene by the nuclear pregnane X receptor. Mol Pharmacol. 2001;60:427–431. [PubMed] [Google Scholar]
- 29.Ferguson SS, Chen Y, LeCluyse EL, Negishi M, Goldstein JA. Human CYP2C8 is transcriptionally regulated by the nuclear receptors constitutive rostane receptor, pregnane X receptor, glucocorticoid receptor, hepatic nuclear factor 4alpha. Mol Pharmacol. 2005;68:747–757. doi: 10.1124/mol.105.013169. [DOI] [PubMed] [Google Scholar]
- 30.Quattrochi LC, Tukey RH. Nuclear uptake of the Ah (dioxin) receptor in response to omeprazole: transcriptional activation of the human CYP1A1 gene. Mol Pharmacol. 1993;43:504–508. [PubMed] [Google Scholar]
- 31.Daujat M, Charrasse S, Fabre I, Lesca P, Jounaidi Y, Larroque C, et al. Induction of CYP1A1 gene by benzimidazole derivatives during Caco-2 cell differentiation: evidence for an aryl-hydrocarbon receptor-mediated mechanism. Eur J Biochem. 1996;237:642–652. doi: 10.1111/j.1432-1033.1996.0642p.x. [DOI] [PubMed] [Google Scholar]
- 32.Urquhart BL, Tirona RG, Kim RB. Nuclear receptors, the regulation of drug-metabolizing enzymes, drug transporters: implications for interindividual variability in response to drugs. J Clin Pharmacol. 2007;47:566–578. doi: 10.1177/0091270007299930. [DOI] [PubMed] [Google Scholar]
- 33.Denison MS, Whitlock JP., Jr Xenobiotic-inducible transcription of cytochrome P450 genes. J Biol Chem. 1995;270:18175–18178. doi: 10.1074/jbc.270.31.18175. [DOI] [PubMed] [Google Scholar]