Abstract
Nepetalactones are iridoid monoterpenes with a broad range of biological activities produced by plants in the Nepeta genus. However, none of the genes for nepetalactone biosynthesis have been discovered. Here we report the transcriptomes of two Nepeta species, each with distinctive profiles of nepetalactone stereoisomers. As a starting point for investigation of nepetalactone biosynthesis in Nepeta, these transcriptomes were used to identify candidate genes for iridoid synthase homologs, an enzyme that has been shown to form the core iridoid skeleton in several iridoid producing plant species. Iridoid synthase homologs identified from the transcriptomes were cloned, heterologously expressed, and then assayed with the 8-oxogeranial substrate. These experiments revealed that catalytically active iridoid synthase enzymes are present in Nepeta, though there are unusual mutations in key active site residues. Nevertheless, these enzymes exhibit similar catalytic activity and product profile compared to previously reported iridoid synthases from other plants. Notably, four nepetalactone stereoisomers with differing stereochemistry at the 4α and 7α positions – which are generated during the iridoid synthase reaction – are observed at different ratios in various Nepeta species. This work strongly suggests that the variable stereochemistry at these 4α and 7α positions of nepetalactone diastereomers is established further downstream in the iridoid pathway in Nepeta. Overall, this work provides a gateway into the biosynthesis of nepetalactones in Nepeta.
Keywords: Terpenoid, Iridoid, Short chain alcohol dehydrogenase, Natural product biosynthesis
Graphical abstract
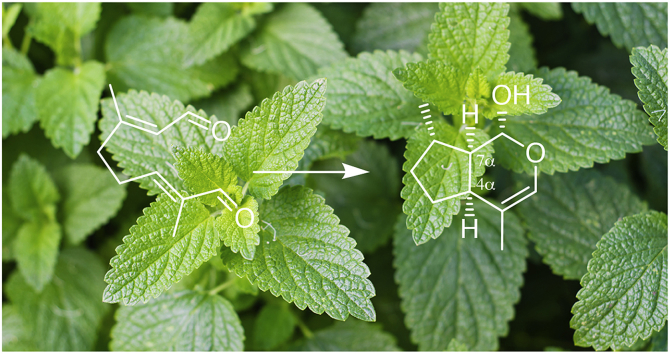
Iridoid synthase from Nepeta cateria (catnip) and Nepeta mussinii, have been cloned and characterized.
Highlights
-
•
Species within the Nepeta genus (such as catnip) produce nepetalactone iridoids.
-
•
The enzymes that produce the iridoid scaffold of nepetalactone were identified from two species of Nepeta.
-
•
The iridoid synthase enzymes are not responsible for the stereochemical variation in these iridoids.
1. Introduction
Nepeta is a large genus of approximately 250 species within the Lamiaceae (Nepetoideae) family (Formisano et al., 2011, Kaufmann and Wink, 1994). Nepeta species are well-known for production of nepetalactone (McElvain et al., 1941), the molecule responsible for cat euphoria as exemplified in Nepeta cataria (catnip) and other Nepeta species (Sakurai et al., 1988, Waller et al., 1969). Notably, nepetalactones are also produced by a number of insects, most notably aphids where they act as sex pheromones (Dawson et al., 1987, Fernández-Grandon et al., 2013, Fernández-Grandon and Poppy, 2015, Goldansaz et al., 2004, Hardie et al., 1997, Smith et al., 1979, Stewart-Jones et al., 2007, Symmes et al., 2012). Not surprisingly, nepetalactone biosynthesis in plants influences plant-insect interactions, raising the possibility that nepetalactones could be utilized as crop protection agents (Aldrich et al., 2016, Birkett and Pickett, 2003, Birkett et al., 2011, Eisner, 1964, Khan et al., 2012, Koczor et al., 2015, van Tol et al., 2009, Zhu et al., 2012).
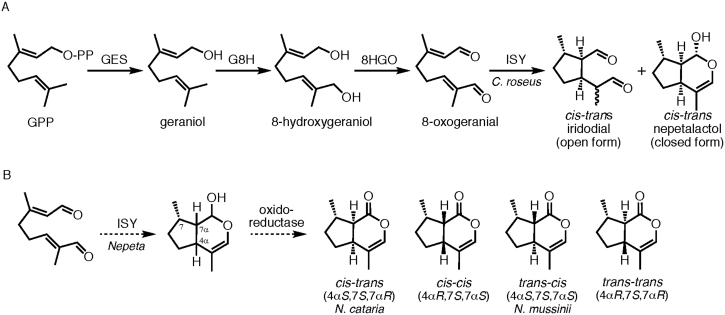
Early biosynthetic work suggested that nepetalactones are a specific type of monoterpene known as iridoids (Bellesia et al., 1984, Dawson et al., 1996, Hallahan et al., 1992, Meinwald et al., 1966, Regnier et al., 1968). In iridoid biosynthesis in the medicinal plant Catharanthus roseus, geranyl-pyrophosphate (GPP) is converted to geraniol by geraniol synthase (GES) (Iijima et al., 2004), hydroxylated to form 8-hydroxygeraniol by geraniol 8-hydroxylase (G8H) (Hallahan et al., 1992, Collu et al., 2001) and then oxidized to 8-oxogeranial by the 8-hydroxygeraniol oxidoreductase (8HGO) (Miettinen et al., 2014) (Fig. 1A). A non-canonical monoterpene synthase, iridoid synthase (ISY) (Geu-Flores et al., 2012), then converts 8-oxogeranial to nepetalactol, along with the open form of this lactol, iridodial. In Nepeta, to complete the nepetalactone biosynthetic pathway, an oxidoreductase is believed to convert the nepetalactol to the lactone (Hallahan et al., 1998) (Fig. 1B). The genes that encode GES, G8H, 8HGO, and ISY have been cloned from several plant species, though none have been cloned from a Nepeta species, and the gene for the final oxidoreductase to form the lactone has not been identified to date.
Fig. 1.
Iridoid biosynthesis. A. Formation of the open (iridodial) and closed (nepetalactol) forms of the iridoid skeleton from geranyl pyrophosphate (GPP). GES, geraniol synthase; G8H, geraniol-8-hydroxylase; 8HGO, 8-hydroxygeraniol oxidoreductase; ISY, iridoid synthase. B. Nepetalactone biosynthesis entails the conversion of 8-oxogeranial to nepetalactol by ISY as shown in panel A, and an uncharacterized oxidoreductase oxidizes nepetalactol to nepetalactone. Four nepetalactone diastereomers are found in Nepeta, with differing stereochemistry at the 4α and 7α carbons. This study used a cultivar of N. cataria that produced primarily cis-trans-nepetalactone, and a cultivar of N. mussinii that produced primarily trans-cis-nepetalactone.
The stereochemical variation of the nepetalactones is an intriguing aspect of this biosynthetic pathway. Four nepetalactone stereoisomers with differing stereochemistry at the 4α and 7α positions are observed at different ratios in various Nepeta species (referred to as cis-cis; cis-trans; trans-cis; and trans-trans; Fig. 1B) (Bates et al., 1958, Clark et al., 1997, Eisenbraun et al., 1980). The ratio of the stereoisomers appears to play a role in the plants’ ability to deter insects (Birkett et al., 2011, Glinwood et al., 1999). However, it is not known how the stereochemistry of these centers is set during the biosynthesis of these compounds. Iridoid synthase (ISY), which converts 8-oxogeranial to nepetalactone, could establish the stereochemistry of the 4α and 7α carbons during the cyclization of 8-oxogeranial. Alternatively, downstream isomerase enzymes could be responsible for this stereochemical variation. As a first step to understanding the nepetalactone pathway, we set out to discover and characterize the ISYs from Nepeta (Fig. 1B).
In this study, we report the identification of two Nepeta species, N. cataria and N. mussinii, that produce different profiles of nepetalactone diastereomers in leaf tissue: N. cataria accumulates primarily the cis-trans isomer, while N. mussinii accumulates primarily the trans-cis isomer (Fig. 1B). Leaf tissue from both species was subjected to a transcriptomic analysis, to provide a resource for understanding the molecular basis for the biosynthesis of these compounds. Candidate ISY enzymes were identified by searching for C. roseus ISY homologs in the transcriptomes. These genes were then cloned, heterologously expressed and functionally characterized by an in vitro assay. Assays of these Nepeta ISY (NISY) enzymes with 8-oxogeranial revealed that both Nepeta species each harbored two ISY homologs, one of which was substantially more active than the other. These enzymes produced product profiles that were identical to the product profile of the previously reported ISY from Catharanthus roseus, an enzyme known to produce the cis-trans stereoisomer. These results strongly suggest that the variable stereochemistry of the nepetalactone diastereomers is not established during the cyclization reaction catalyzed by ISY, but that a downstream epimerization step must occur. Notably, the Nepeta ISY homologs show surprisingly low sequence identity, as well as changes in the active site residues, to previously characterized ISY enzymes. This discovery highlights the plasticity of genes and encoded enzymes responsible for the synthesis of the iridoid scaffold.
2. Results
2.1. Selection of plant material
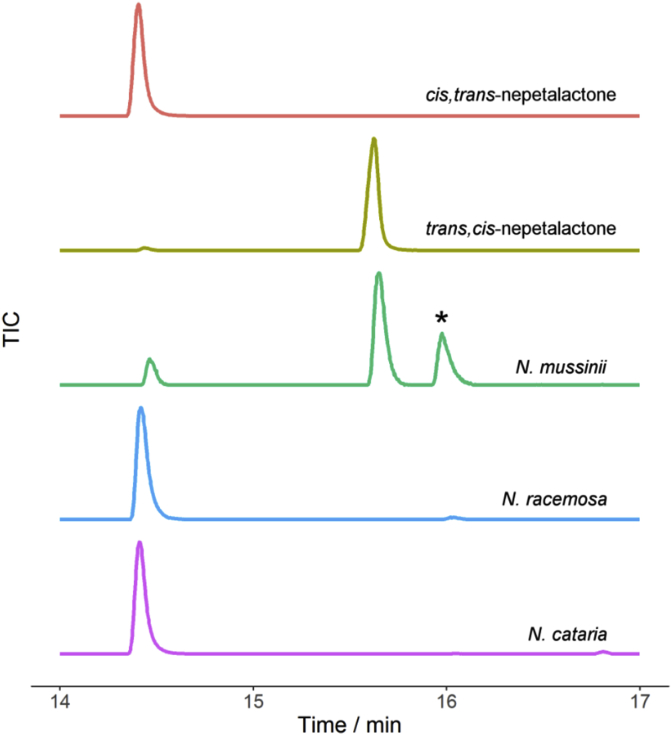
Different species and cultivars of Nepeta produce different profiles of nepetalactone diastereomers (Birkett et al., 2011, Clark et al., 1997). We obtained local N. cataria and N. mussinii plants and screened the leaves of these plants by GC-MS for production of nepetalactone diastereomers (Fig. 2). We found that N. mussinii plants produced primarily the trans-cis (4αS,7S,7αS) lactone, while N. cataria produced primarily the cis-trans (4αS,7S,7αR) isomer as evidenced by co-elution with authentic standards on a GC-MS (Fig. 2). Both N. cataria and N. mussinii have been reported to produce additional nepetalactone isomers, but the nepetalactone profile likely varies widely from cultivar to cultivar. A third Nepeta species, N. racemosa, was also screened (Fig. 2). This species showed a product profile identical to N. cataria and was not investigated further.
Fig. 2.
GC-MS chromatograms of leaf tissue extract of three Nepeta species.N. cataria produced primarily cis-trans-nepetalactone, while N. mussinii produced primarily trans-cis-nepetalactone. A third Nepeta species, N. racemosa, was also analysed but was found to have the same chemical composition as N. cataria, and was not analysed further. Authentic standards of cis-trans and trans-cis-nepetalactones are shown for comparison. One representative GC-MS chromatogram from each species is shown; three biological replicates showed consistent nepetalactone profiles. The peak highlighted with a star is an unknown isomer of nepetalactone.
2.2. Cloning, heterologous expression and biochemical assay of ISY from Nepeta
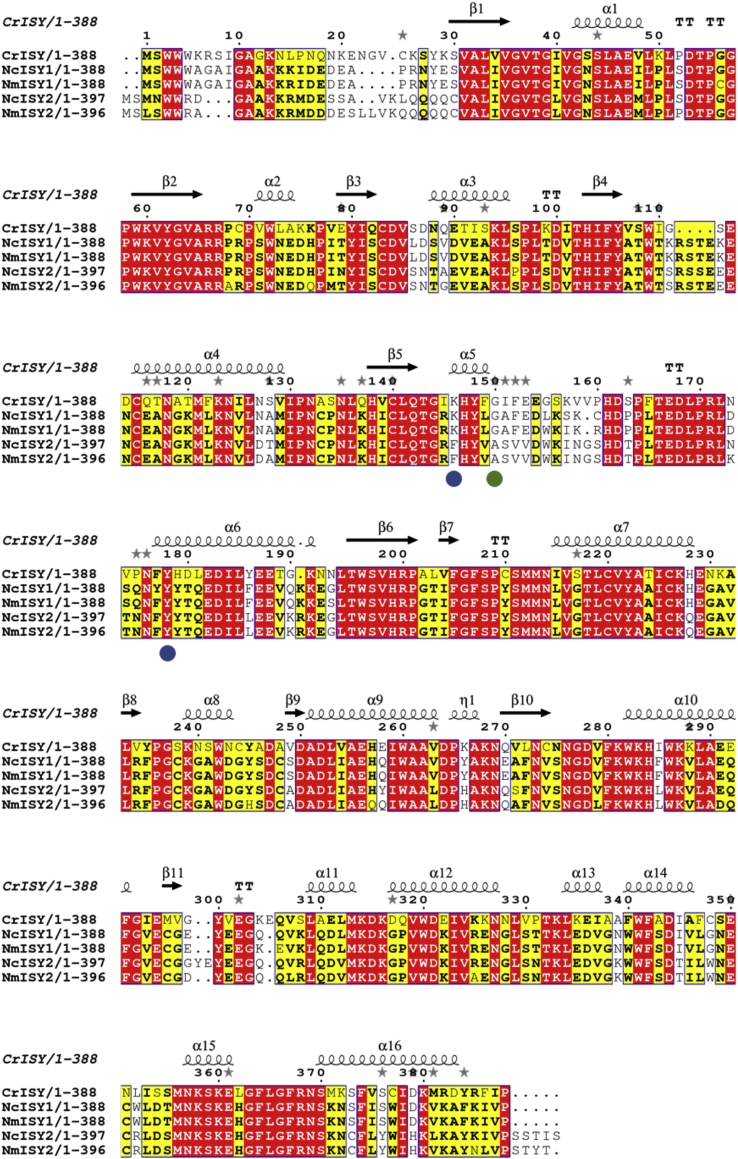
Transcriptomes were obtained for leaf tissue of N. mussinii and N. cataria. ISY homologs, which are short chain dehydrogenases, are typically annotated as progesterone 5-beta-reductase, the closest known homolog to this enzyme. Two homologs of ISY, from C. roseus (Geu-Flores et al., 2012) and Olea europaea (olive) (Alagna et al., 2016), that have been functionally characterized were used to search the Nepeta transcriptomes for ISY homologs using BLAST. Both Nepeta species each contained a total of two distinct homologs of ISY, which were named family 1 and family 2 (amino acid sequence identity between N. cataria family 1 (NcISY1) and N. cataria family 2 (NcISY2) was 80%; between N. mussinii family 1 (NmISY1) and family 2 (NmISY2) was 79%). (Identity between N. cataria and N. mussinii family 1 is 98%; between N. cataria and N. mussinii family 2 is 99%). Nepeta homologs exhibited amino acid sequence identities to the ISY from C. roseus ranging from 53 to 58% (Fig. 3). All of these proteins are members of the short chain dehydrogenase SDR75U family (http://sdr-enzymes.scilifelab.se/).
Fig. 3.
ISY homologs. Alignment of the ISY homologs that were cloned from Nepeta compared to ISY from C. roseus.
The Nepeta ISY enzymes were heterologously expressed in E. coli for biochemical characterization, though despite substantial optimization, these enzymes could not be purified to complete homogeneity (Fig. 4). The enzymes were incubated with the ISY substrate 8-oxogeranial and NADPH according to previously reported ISY assay conditions, and product formation was monitored by GC-MS (Geu-Flores et al, 2012, Kries et al., 2016). If the stereochemistry of the 4α and 7α carbons is set by the ISY catalyzed cyclization, then it would be expected that at least one of the N. mussinii ISY enzymes would produce large amounts of trans-cis product. Authentic standards of cis-trans-nepetalactol and iridodials as well as the trans-cis-iridodials (the trans-cis-nepatalactol is not stable (Liblikas et al., 2005) could be resolved on the GC-MS chromatogram (Fig. 5A).
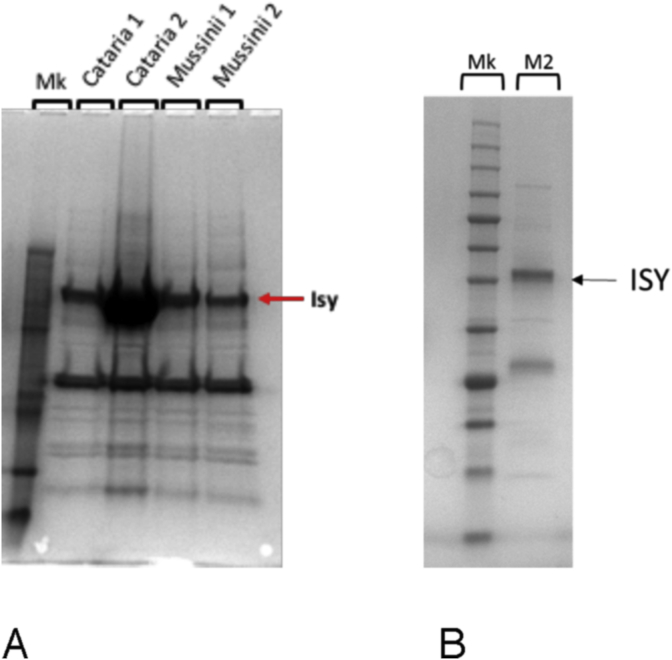
Fig. 4.
SDS-PAGE of ISY proteins. A. All four ISY proteins from Nepeta after expression optimization and purification by Ni-NTA chromatography and gel filtration chromatography. Despite extensive optimization of purification conditions (see Experimental), proteins were not entirely homogenous. B. SDS-PAGE of N. mussinii ISY 2 after Ni-NTA and gel filtration chromatography used for kinetic analysis.
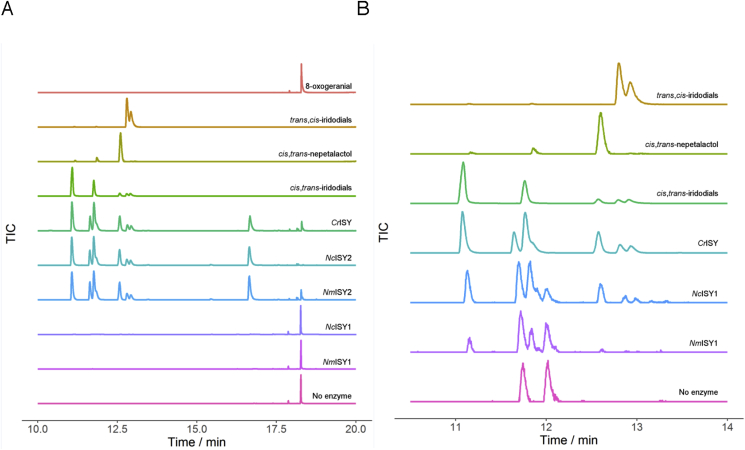
Fig. 5.
Biochemical assay of the four cloned Nepeta ISY enzymes as measured by GC-MS. A. GC-MS chromatograms showing authentic standards of substrate 8-oxogeranial, as well as cis-trans-nepetalactol, cis-trans-iridodial and trans-cis-iridodial. Enzymes from C. roseus, N. cataria (family 2) and N. mussinii (family 2) all have nearly identical product profiles. ISY from N. cataria (family 1) and N. mussinii (family 1) have very low catalytic activity. B. Product profiles of ISY from N. cataria (family 1) and N. mussinii (family 1) compared to that of C. roseus. To show trace levels of product, the chromatograms in this panel are normalized to their individual maximum signal intensity within the depicted range.
ISY from N. cataria and N. mussinii family 1 (NcISY1 and NmISY1) had very low catalytic enzymatic activity as evidenced by an end point assay (Fig. 5A), and could not subjected to steady state kinetic analysis. The activity was so low that these enzymes are most likely not physiologically relevant in iridoid biosynthesis. However, the trace amount of product that could be detected appeared to be primarily a mixture of cis-trans-nepetalactols and iridodials, similar to what is observed with the ISY from C. roseus (Fig. 5B). The family 2 enzymes (NcISY2 and NmISY2) showed substantially higher activity (Fig. 5). Notably, both NcISY2 and NmISY2 produced a mixture of products identical to the mixture of cis-trans-nepetalactol and iridodials produced by the C. roseus enzyme.
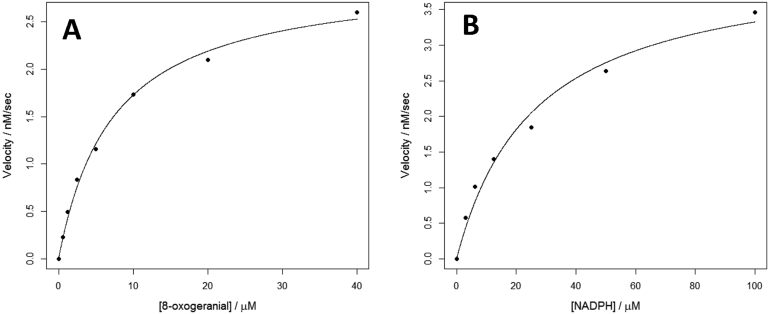
The steady-state kinetic constants for NmISY2 were obtained, using an assay based on consumption of NADPH (for 8-oxogeranial, Km = 7.3 ± 0.7 μM, kcat = 0.60 ± 0.02 s−1; for NADPH, Km = 26.2 ± 5.1 μM, kcat = 0.84 ± 0.06 s-1, all data mean±se.) (Fig. 6). Notably, the availability of the authentic trans-cis-iridodial standard demonstrated that small amounts of the trans-cis isomer were present in all of the enzymatically catalyzed reactions (Fig. 5A). However, due to the low levels of the trans-cis isomer observed in these assays, it seems unlikely that NcISY2 and NmISY2 provide a direct source of this diastereomer.
Fig. 6.
Kinetics of NmISY2. A. Varying 8-oxogeranial concentrations with NADPH concentration at 75 μM. B. Varying NADPH concentrations with 8-oxogeranial concentration at 50 μM.
2.3. Sequence of Nepeta ISY
Structural and mechanistic studies of ISY from C. roseus have revealed that the key active site residue is Tyr178 (Kries et al., 2016, Hu et al., 2015). This residue is also conserved in all ISY homologs identified from Nepeta (Fig. 3). A lysine residue in the active site, Lys146, has been shown to play a catalytic role in progesterone 5-β-reductase, the short chain dehydrogenase most closely related to ISY (Thorn et al., 2008). While this lysine residue is conserved in ISY (C. roseus), mutational analysis demonstrated that this residue does not play an essential role in catalysis (Kries et al., 2016, Hu et al., 2015). Nevertheless, along with tyrosine, this lysine forms part of the core conserved residues of the short chain dehydrogenase protein family, of which ISY is a member. While family 1 NISY contained this lysine residue, this residue was replaced with a phenylalanine in members of the more catalytically active family 2. Moreover, previous work suggests that Gly150 of ISY (C. roseus) allows conformational flexibility of the active site, allowing the enzyme to assume both open and closed forms (Kries et al., 2016). This residue is mutated to an alanine in family 2 NISY, a mutation that has been shown to lock the active site in an open conformation (Kries et al., 2016). However, these sequence differences do not appear to alter either the catalytic efficiency or the product profile of these enzymes. This highlights the plasticity of the ISY enzyme responsible for the synthesis of the iridoid scaffold. As more sequence data becomes available for the Lamiaceae, we will be able to place the ISY enzymes within a phylogenetic context of the entire Lamiaceae family to better understand how the ISY enzyme evolved.
3. Discussion
Members of the Nepeta genus produce nepetalactones, which are iridoid-type monoterpenes that impact plant-insect interactions. The stereochemical variation found among the nepetalactones has a profound influence on the resulting biological activities. However, the biosynthetic pathway of the nepetalactones at the onset of this work was unknown.
Here, we report the transcriptomes of leaves from two Nepeta plants, N. cataria (catnip) and N. mussinii (catmint), plants that produce qualitatively different profiles of nepetalactone stereoisomers. Using this transcriptome database, we searched for homologs of ISY, a key iridoid biosynthetic enzyme that generates the iridoid bicyclic scaffold, along with the 4α and 7α carbon stereocenters, from the linear substrate 8-oxogeranial. After identification, cloning and heterologous expression of these homologs, in vitro biochemical assays indicate that both Nepeta species harbor two ISY homologs. Although one homolog (NcISY1 and NmISY1) has only trace catalytic activity, the other (NcISY2 and NmISY2) has robust catalytic activity toward the 8-oxogeranial substrate. Notably, this ISY in both N. cataria and N. mussinii displayed an unexpected mutation of the active site lysine, as well as mutation of a glycine known to impact the conformational flexibility of the active site.
Surprisingly, the Nepeta ISY enzymes assayed gave identical product profiles (Fig. 5), despite the fact that the stereochemistry of the iridoid scaffolds accumulated by these two species are different (Fig. 2). The product profile of the Nepeta enzymes was also very similar to the products produced by the previously reported ISY from C. roseus, a plant that produces iridoids exclusively from the cis-trans nepetalactol isomer.
This discovery highlights that the Nepeta ISY enzymes, along with the amino acid sequence differences observed in these enzymes, are not responsible for setting the stereochemistry at the 4α and 7α carbons. Downstream enzymes that catalyze isomerization of nepetalactol or nepetalactone, or alternative cyclization enzymes, could be required to yield the stereochemical variation that is observed in the Nepeta nepetalactones. The transcriptomic data reported here will facilitate the identification of these downstream enzymes and lead to a greater understanding of the factors influencing the stereochemical variation of these important iridoids produced in Nepeta.
4. Experimental
4.1. Plant material
Nepeta mussinii and N. cataria were obtained from Herbal Haven, Coldhams Farm, Rickling, Saffron Walden, CB11 3YL, UK. N. racemosa ‘Walker's Low’ was obtained from Notcutts Garden Center, Daniels Road, Norwich, Norfolk, NR4 6QP, UK. RNA was harvested from mature leaves of plants in a vegetative state using Qiagen RNeasy Plant Mini kit according to the manufacturer's instructions. For metabolic profiling, approximately 50 mg of frozen leaf tissue were homogenized with a tungsten bead in a ball mill (27 Hz, 30 s, 3 repetitions). MeOH (300 μL) was added, mixed vigorously and the resulting slurry was extracted with hexane (600 μL). The top hexane layer was removed, and applied to a Phenomenex Strata SI-1 Silica column (55 μM, 70 Å, 100 mg/mL). The hexane flow through was discarded. The compounds of interest were eluted from the column with 20% EtOAc/Hexane (500 μL), and this eluant was analysed by GC-MS. Three individual plants were analysed for each species; no significant metabolite variation between individuals was observed.
4.2. Compounds
Syntheses and isolation of key compounds are outlined below. All of the compounds have been described previously. 1H NMR spectra were used to demonstrate compound purity (Supporting Information). 8-oxogeranial was synthesized from geranyl acetate as previously described in Geu-Flores et al. (2012). Cis-trans-nepetalactol was synthesized by DIBAL-H reduction of cis-trans-nepetalactone as previously described in Geu-Flores et al. (2012).
Cis-trans-nepetalactone – Leaves and shoots (∼100 g) of Nepeta fassinii “Six Hills Giant” (Burncoose Nurseries, Gwennap, Redruth, Cornwall, TR16 6BJ, UK) and water (300 mL) were blended (3 × 1 min) in a kitchen blender. The dark green slurry was extracted with dichloromethane (10 × 100 mL). Fractions were analysed by GC-MS to determine nepetalactone presence. Combined fractions were filtered, washed with brine (200 mL), dried over anhydrous Na2SO4 and evaporated to dryness in vacuo yielding a yellow oil (1.098 g crude). The nepetalactone was purified by silica flash chromatography (60 g silica, 3 × 25 cm). The crude oil was loaded onto the column in hexane and eluted in hexane/EtOAc fractions (40 mL) with increasing concentrations of EtOAc. Fractions were analysed by GC-MS; those containing the desired nepetalactone isomer were combined and evaporated to dryness yielding the pure cis-trans-nepetalactone (155 mg). Product identity was verified by NMR spectroscopy.
Trans-cis-nepetalactone – Catnip oil (2 mL, Health and Herbs, 425 Ellsworth St. SW Albany, OR 97321, USA, https://betterhealthherbs.com/) was purified by silica flash chromatography. The crude oil was applied to the column and eluted with hexane/EtOAc with increasing concentrations of EtOAc. Fractions containing the same compounds (as analysed by TLC) were combined and evaporated to dryness. Pure β-caryophyllene (700 mg) and trans-cis-nepetalactone (441 mg) were isolated. Compound identity was determined by NMR spectroscopy and comparison to literature values.
Trans-cis-iridodials – Trans-cis-nepetalactone (212 mg) was dissolved in hexanes (30 mL) and cooled to −78 °C in an inert atmosphere. A solution of DIBAL-H in hexanes (213 mg, 5% v/v) was added dropwise over 10 min and left to stir for 1 h. Baeckstrom reagent (1.6 g, celite:Na2SO4 1:1) was added and the reaction was stirred for 1 h at −78 °C and then 2 h at room temperature. The mixture was filtered through a glass frit and concentrated in vacuo yielding the crude product. The compound was purified by silica flash chromatography and eluted in EtOAc/hexane with increasing concentrations of EtOAc. Fractions were combined and evaporated to dryness to yield a mixture of the two trans-cis-iridodials (64 mg). The identity of the compound was verified by 1H NMR.
Cis-trans-iridodials – Cis-trans-nepetalactol (0.6 mM) was incubated in dilute aqueous HCl (100 mM) overnight, extracted into EtOAc and analysed by GC-MS.
4.3. Transcriptome and bioinformatics
RNA-sequencing (RNA-seq) libraries were constructed using the Kappa stranded RNA-seq kit and libraries were sequenced on an Illumina HiSeq 2500 generating 150 nt paired-end reads. Read quality was assessed using FASTQC (v0.11.2; http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) with default parameters and adaptors and low quality sequences removed using Trimmomatic (Parameter: LEADING:10 TRAILING:10 SLIDINGWINDOW:4:15 MINLEN:30) (v0.32, (Bolger et al., 2014). Only surviving paired-end reads were used to generate de novo assemblies using Trinity (v2014-07-17) (Grabherr et al., 2011) and only transcripts >250 bp were reported (Table 1). The protein sequences of iridoid synthases from Catharanthus roseus (AFW98981.1) and Olea europaea L (ALV83438.1) were used to BLAST the predicted peptides of the N. mussinii and N. cataria transcriptomes (Camacho et al., 2009). Two ISY candidates from each transcriptome were identified.
Table 1.
Transcriptomic metrics.
| Metric | N. cataria | N. mussinii | N. racemosa |
|---|---|---|---|
| No. of transcripts | 77,879 | 69,775 | 78,057 |
| Maximum length of transcripts (bp) | 10,772 | 11,683 | 11,535 |
| N50 transcript size | 1490 | 1589 | 1330 |
| Average transcripts size | 998 | 1026 | 920 |
4.4. Cloning and protein expression
cDNA was prepared from leaf RNA using Invitrogen SuperScript III First-Strand Synthesis System kit following the manufacturer's instructions. Primers were designed based on the ISY transcript sequences and with 5′-overhangs for cloning (Table 2). These were used to PCR amplify ISY genes from the cDNA and clone directly into a pOPINF expression vector using an InFusion HD cloning kit. The pOPINF vector encodes an N-terminal His-tag. The gene sequences were verified by Sanger sequencing. Two ISY candidates from N. mussinii were obtained (NmISY1 and NmISY2). Sequencing revealed that three ISY candidates were cloned from N. cataria. Two were cloned from with same primer pair (NcISY1) and had 97% amino acid identity, and thus, only one was investigated further. The two ISY candidates from N. cataria investigated further were named NcISY1 and NcISY2. Proteins were subjected to a variety of expression conditions to maximize expression, with the most optimal conditions described below. All genes were expressed in strain soluBL21 (DE3) (Genlantis). Cells were pre-cultured overnight at 37 °C in LB medium containing 100 μg/mL carbenicillin. Aliquots of 200 μL each were used to inoculate two 2-L flasks containing 2YT medium and carbenicillin. When cultures reached an OD600 of 0.5–0.8 after ∼6 h shaking at 37 °C, protein production was induced by adding IPTG at a final concentration of 500 μM. Protein was expressed at 18 °C for ∼16 h. Cells were harvested by centrifugation, the supernatant was discarded and pellets were resuspended in 100 mL of 50 mM Tris-HCl buffer (pH 7.0) containing 300 mM NaCl, 1 mM DTT, one tablet of Complete EDTA free protease inhibitor (Roche), and 0.2 mg/mL lysozyme. The cells were disrupted by 7 min sonication on ice in cycles of 2 s sonication followed by a 3 s break. All subsequent steps were conducted at 4 °C. The lysate was centrifuged for 20 min at 35,000 g and the supernatant containing the soluble protein was applied on a 5 mL HisTrap column connected to an Äkta Xpress purifier (GE healthcare). His-tagged protein was eluted with a step gradient of 20 mM–500 mM imidazole in 50 mM Tris/glycine buffer adjusted to pH 8.0 with hydrochloric acid and supplemented with 5% (v/v) glycerol, 0.5 M NaCl, and 1 mM DTT. Fractions containing the protein of interest were collected and concentrated to 2–3 mL in an Amicon 10 kDa MWCO centrifugal filter (Millipore). The concentrated solution was further purified by size-exclusion chromatography on a Superdex 200 16/60 GF column (GE Healthcare). Fractions corresponding to the molecular weight of the ISY (C. roseus) dimer (83 kDa) were collected, combined, concentrated in an Amicon 10 kDa MWCO centrifugal filter and stored at −20 °C after flash freezing in liquid nitrogen. Protein concentrations were determined spectrophotometrically at 280 nm using extinction coefficients calculated with the protparam tool (http://web.expasy.org/protparam/). Despite extensive optimization of purification conditions, it was not possible to purify proteins to complete homogeneity as judged by SDS-PAGE (Fig. 4).
Table 2.
Primers used for cloning. Overhangs used for pOPINF InFusion are in bold.
| Gene | Direction | Sequence 5′-3′ |
|---|---|---|
| NmISY1 | Forward | AAGTTCTGTTTCAGGGCCCGAGCTGGTGGTGGG |
| NmISY1 | Reverse | ATGGTCTAGAAAGCTTTAGGGAACAATCTTGAAAGCTTTCAC |
| NmISY2 | Forward | AAGTTCTGTTTCAGGGCCCGAGCATGAGCTTGAGCTGG |
| NmISY2 | Reverse | ATGGTCTAGAAAGCTTTAGGTATAGGTAGAAGGAACAAGGTTGTAAG |
| NcISY1 | Forward | AAGTTCTGTTTCAGGGCCCGAGCTGGTGGTGGGC |
| NcISY1 | Reverse | ATGGTCTAGAAAGCTTTAGGGAACAATCTTGAAAGCTTTCAC |
| NcISY2 | Forward | AAGTTCTGTTTCAGGGCCCGAGCATGAACTGGTGGAGG |
| NcISY2 | Reverse | ATGGTCTAGAAAGCTTTAAGAAATAGTAGAGGAAGGAACAATCTTGTAAGC |
4.5. Assay conditions, end point reactions
Reactions were conducted with 8-oxogeranial (500 μM), NADPH (1 mM), MOPS pH 7.0 (200 mM), NaCl (100 mM), tetrahydrofuran (THF) (0.5% v/v) and Nepeta ISYs (0.5 μM). A negative control was conducted without enzyme, and a positive control was conducted with ISY (C. roseus). Reactions were incubated for 3 h at 30 °C before extraction with EtOAc (100 μL) and analysis on GC-MS.
4.6. Kinetic measurement conditions
Kinetics of NADPH consumption were determined spectrophotometrically on a PerkinElmer Lambda 35 instrument at a wavelength of 340 nm in cuvettes with 1 cm path length. Reactions contained 200 mM MOPS buffer pH 7.0, 100 mM sodium chloride and 5 nM NmISY2, in a total volume of 800 μL. 8-oxogeranial was added from a stock solution in inhibitor free THF. A THF co-solvent concentration of 1–1.4% was maintained in the assay to ensure substrate solubility. For determination of 8-oxogeranial parameters, 75 μM NADPH was used. For determination of NADPH kinetic parameters, 50 μM 8-oxogeranial was used. Cuvettes were equilibrated to 25 °C before the reaction was initiated by addition of enzyme. Absorbance values were recorded at a rate of 1 Hz. The R software environment was used to fit linear initial rates over 2–5 min of the enzyme reaction. Kinetic parameters are reported as a best fit estimate ± SE. Background NADPH consumption was subtracted from initial rates. The Michaelis-Menten equation was fitted to the data points in R by the nls function to obtain values for the kinetic parameters. The enzyme NmISY2 appeared to lose activity during storage at −20 °C, so kinetic parameters were measured with freshly purified enzyme.
4.7. GC-MS method
Samples were injected in split mode (2 μL, split ratio 20:1) at an inlet temperature of 220 °C on a Hewlett Packard 6890 GC-MS equipped with a 5973 mass selective detector (MSD), and an Agilent 7683B series injector and autosampler. Separation was performed on a Zebron ZB5-HT-INFERNO column (5% phenyl methyl siloxane; length: 35 m; diameter: 250 μm) with guard column. Helium was used as mobile phase at a constant flow rate of 1.2 mL/min and average velocity 37 cm/s. After 5 min at 80 °C, the column temperature was increased to 110 °C at a rate of 2.5 K/min, then to 280 °C at 120 K/min, and kept at 280 °C for another 4 min. A solvent delay of 5 min was allowed before collecting MS spectra at a fragmentation energy of 70 eV.
Author contributions
N.H.S. and S.E.O. conceived the study; N.H.S. identified plants, cloned ISY genes, prepared standards, obtained initial results, and generated conclusions; L.C. purified the protein; B.L. generated kinetic data; M.O.K. screened plants; C.R.B and D.Z. generated transcriptome; S.E.O. wrote the manuscript.
Conflict of interest
The authors declare no conflict of interest.
Acknowledgments
We gratefully acknowledge funds from the US National Science Foundation Plant Genome Research Program to CRB and SOC (IOS-1444499) as well the UK Biotechnological and Biological Sciences Research Council (BBSRC) and Engineering and Physical Sciences Research Council (EPSRC) joint-funded OpenPlant Synthetic Biology Research Centre (BB/L014130/1). The raw RNA-sequencing reads used in the transcriptome assembly have been deposited in the National Center for Biotechnology Information Sequence Read Archive under BioProject ID (PRJNA379302). The assembled transcriptomes are available at the Dryad Digital Repository under doi (to be released upon publication). Sequences for NmISY1 (KY882235), NmISY2 (KY882236), NcISY1 (KY882233), NcISY2 (KY882234) have been deposited in GenBank.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.phytochem.2017.10.004.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- Alagna F., Geu-Flores F., Kries H., Panara F., Baldoni L., O'Connor S.E., Osbourn A. Identification and characterization of the iridoid synthase involved in oleuropein biosynthesis in olive (Olea europaea) Fruits. J. Biol. Chem. 2016;291:5542–5554. doi: 10.1074/jbc.M115.701276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldrich J.R., Chauhan K., Zhang Q.H. Pharmacophagy in green lacewings. Peer J. 2016;4 doi: 10.7717/peerj.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates R.B., Eisenbraun E.J., McElvain S.M. The configurations of the nepetalactones and related compounds. J. Am. Chem. Soc. 1958;80:3420–3424. [Google Scholar]
- Bellesia F., Grandi R., Pagnoni Ugo M., Pinetti A., Trave R. Biosynthesis of nepetalactone in Nepeta cataria. Phytochemistry. 1984;23:83–87. [Google Scholar]
- Birkett M.A., Pickett J.A. Aphid sex pheromones: from discovery to commercial production. Phytochemistry. 2003;62:651–656. doi: 10.1016/s0031-9422(02)00568-x. [DOI] [PubMed] [Google Scholar]
- Birkett M.A., Hassanali A., Hoglund S., Pettersson J., Pickett J.A. Repellent activity of catmint, Nepeta cataria, and iridoid nepetalactone isomers against Afro-tropical mosquitoes, ixodid ticks and red poultry mites. Phytochemistry. 2011;72:109–114. doi: 10.1016/j.phytochem.2010.09.016. [DOI] [PubMed] [Google Scholar]
- Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L. BLAST+: architecture and applications. BMC Bioinforma. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark L.J., Hamilton J.G.C., Chapman J.V., Rhodes M.J.C., Hallahan D.L. Analysis of monoterpenoids in glandular trichomes of the catmint Nepeta racemosa. Plant J. 1997;11:1387–1393. [Google Scholar]
- Collu G., Unver N., Peltenburg-Looman A.M., van der Heijden R., Verpoorte R., Memelink J. Geraniol 10-hydroxylase, a cytochrome P450 enzyme involved in terpenoid indole alkaloid biosynthesis. FEBS Lett. 2001;508:215–220. doi: 10.1016/s0014-5793(01)03045-9. [DOI] [PubMed] [Google Scholar]
- Dawson G.W., Griffiths D.C., Janes N.F., Mudd A., Pickett J.A., Wadhams L.J., Woodcock C.M. Identification of an aphid sex pheromone. Nature. 1987;325:614–616. [Google Scholar]
- Dawson G.W., Pickett J.A., Smiley D.W. The aphid sex pheromone cyclopentanoids: synthesis in the elucidation of structure and biosynthetic pathways. Bioorg. Med. Chem. 1996;4:351–361. doi: 10.1016/0968-0896(96)00012-0. [DOI] [PubMed] [Google Scholar]
- Eisenbraun E.J., Browne C.E., Irvin-Willis R.L., McGurk D.J., Eliel E.L., Harris D.L. Structure and stereochemistry 4aβ,7α,7aβ-nepetalactone from Nepeta mussini and its relationship to the 4aα,7α,7aα- and 4aα,7α,7aβ-nepetalactones from N. cataria. J. Org. Chem. 1980;45:3811–3814. [Google Scholar]
- Eisner T. Catnip: its raison d' etre. Science. 1964;146:1318–1320. doi: 10.1126/science.146.3649.1318. [DOI] [PubMed] [Google Scholar]
- Fernández-Grandon G.M., Poppy G.M. Response of Aphidius colemani to aphid sex pheromone varies depending on plant synergy and prior experience. Bull. Entomol. Res. 2015;105:507–514. doi: 10.1017/S0007485315000371. [DOI] [PubMed] [Google Scholar]
- Fernández-Grandon G.M., Woodcock C.M., Poppy G.M. Do asexual morphs of the peach-potato aphid, Myzus persicae, utilise the aphid sex pheromone? Behavioural and electrophysiological responses of M. persicae virginoparae to (4aS,7S,7aR)-nepetalactone and its effect on aphid performance. Bull. Entomol. Res. 2013;103:466–472. doi: 10.1017/S0007485313000126. [DOI] [PubMed] [Google Scholar]
- Formisano C., Rigano D., Senatore F. Chemical constituents and biological activities of Nepeta species. Chem. Biodiv. 2011;8:1783–1818. doi: 10.1002/cbdv.201000191. [DOI] [PubMed] [Google Scholar]
- Geu-Flores F. An alternative route to cyclic terpenes by reductive cyclization in iridoid biosynthesis. Nature. 2012;492:138–142. doi: 10.1038/nature11692. [DOI] [PubMed] [Google Scholar]
- Glinwood R.T., Du Y.-J., Smiley D.W.M., Powell W. Comparative responses of parasitoids to synthetic and plant-extracted nepetalactone component of aphid sex pheromones. J. Chem. Ecol. 1999;25:1481–1488. [Google Scholar]
- Goldansaz S.H., Dewhirst S., Birkett M.A., Hooper A.M., Smiley D.W., Pickett J.A., Wadhams L., McNeil J.N. Identification of two sex pheromone components of the potato aphid, Macrosiphum euphorbiae (Thomas) J. Chem. Ecol. 2004;30:819–834. doi: 10.1023/b:joec.0000028434.19319.b4. [DOI] [PubMed] [Google Scholar]
- Grabherr M.G. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallahan D.L., Dawson G.W., West J.M., Wallsgrove R.M. Cytochrome P-450 catalyzed monoterpene hydroxylation in Nepeta mussinii. Plant Physiol. Biochem. 1992;30:435–443. [Google Scholar]
- Hallahan D.L., West J.M., Smiley D.W.M., Pickett J.A. Nepetalactol oxidoreductase in trichomes of the catmint Nepeta racemosa. Phytochemistry. 1998;48:421–427. [Google Scholar]
- Hardie J., Peace L., Pickett J.A., Smiley D.W.M., Storer J.R., Wadhams L.J. Sex pheromone stereochemistry and purity affect field catches of male aphids. J. Chem. Ecol. 1997;23:2547–2554. [Google Scholar]
- Hu Y., Liu W., Malwal S.R., Zheng Y., Feng X., Ko T.P., Chen C.C., Xu Z., Liu M., Han X., Gao J., Oldfield E., Guo R.T. Structures of iridoid synthase from Cantharanthus roseus with bound NAD(+), NADPH, or NAD(+)/10-oxogeranial: reaction mechanisms. Angew. Chem. Int. Ed. 2015;54:15478–15482. doi: 10.1002/anie.201508310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iijima Y., Gang D.R., Fridman E., Lewinsohn E., Pichersky E. Characterization of geraniol synthase from the peltate glands of sweet basil. Plant Physiol. 2004;134:370–379. doi: 10.1104/pp.103.032946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann M., Wink M. Molecular systematics of the Nepetoideae (family Labiatae): phylogenetic implications from rbcL gene sequences. Z. Naturforsch. C. 1994;49:635–645. doi: 10.1515/znc-1994-9-1015. [DOI] [PubMed] [Google Scholar]
- Khan M.A., Jones I., Loza-Reyes E., Cameron M.M., Pickett J.A., Birkett M.A. Interference in foraging behaviour of European and American house dust mites Dermatophagoides pteronyssinus and Dermatophagoides farinae (Acari: Pyroglyphidae) by catmint, Nepeta cataria (Lamiaceae) Exp. Appl. Acarol. 2012;57:65–74. doi: 10.1007/s10493-012-9532-2. [DOI] [PubMed] [Google Scholar]
- Koczor S., Szentkirályi F., Pickett J.A., Birkett M.A., Tóth M. Aphid sex pheromone compounds interfere with attraction of common green lacewings to floral bait. J. Chem. Ecol. 2015;41:550–556. doi: 10.1007/s10886-015-0585-7. [DOI] [PubMed] [Google Scholar]
- Kries H., Caputi L., Stevenson C.E., Kamileen M.O., Sherden N.H., Geu-Flores F., Lawson D.M., O'Connor S.E. Structural determinants of reductive terpene cyclization in iridoid biosynthesis. Nat. Chem. Biol. 2016;12:6–8. doi: 10.1038/nchembio.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liblikas I., Santangelo E.M., Sandell J., Baeckström P., Svensson M., Jacobsson U., Unelius C.R. Simplified isolation procedure and interconversion of the diastereomers of nepetalactone and nepetalactol. J. Nat. Prod. 2005;68:886–890. doi: 10.1021/np049647d. [DOI] [PubMed] [Google Scholar]
- McElvain S.M., Bright R.D., Johnson P.R. Constituents of the volatile oil of catnip. I. Nepetalic acid, nepetalactone and related compounds. J. Am. Chem. Soc. 1941;63:1558–1563. [Google Scholar]
- Meinwald J., Happ G.M., Labows J., Eisner T. Cyclopentanoid terpene biosynthesis in a phasmid insect and in catmint. Science. 1966;151:79–80. doi: 10.1126/science.151.3706.79. [DOI] [PubMed] [Google Scholar]
- Miettinen K., Dong L., Navrot N., Schneider T., Burlat V., Pollier J., Woittiez L., van der Krol S., Lugan R., Ilc T., Verpoorte R., Oksman-Caldentey K.M., Martinoia E., Bouwmeester H., Goossens A., Memelink J., Werck-Reichhart D. The seco-iridoid pathway from Catharanthus roseus. Nat. Commun. 2014;5:3606. doi: 10.1038/ncomms4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regnier F.E., Waller G.R., Eisenbraun E.J., Auda H. Biosynthesis of methylcyclopentane monoterpenoids. II. Nepetalactone. Phytochemistry. 1968;7:221–230. [Google Scholar]
- Sakurai K., Ikeda K., Mori K. Synthesis of mono- and sesquiterpenoids. Part 12. Both (4aS,7S,7aR)-(+)-nepetalactone and its antipode are powerful attractants for cats. Agric. Biol. Chem. 1988;52:2369–2371. [Google Scholar]
- Smith R.M., Brophy J.J., Cavill G.W.K., Davies N.W. Iridodials and nepetalactone in the defensive secretion of the coconut stick insects, Graeffea crouani. J. Chem. Ecol. 1979;5:727–735. [Google Scholar]
- Stewart-Jones A., Dewhirst S.Y., Durrant L., Fitzgerald J.D., Hardie J., Hooper A.M., Pickett J.A., Poppy G.M. Structure, ratios and patterns of release in the sex pheromone of an aphid, Dysaphis plantaginea. J. Exp. Bot. 2007;210:4335–4344. doi: 10.1242/jeb.009944. [DOI] [PubMed] [Google Scholar]
- Symmes E.J., Dewhirst S.Y., Birkett M.A., Campbell C.A., Chamberlain K., Pickett J.A., Zalom F.G. The sex pheromones of mealy plum (Hyalopterus pruni) and leaf-curl plum (Brachycaudus helichrysi) aphids: identification and field trapping of male and gynoparous aphids in prune orchards. J. Chem. Ecol. 2012;38:576–583. doi: 10.1007/s10886-012-0121-y. [DOI] [PubMed] [Google Scholar]
- Thorn A., Egerer-Sieber C., Jäger C.M., Herl V., Müller-Uri F., Kreis W., Muller Y.A. The crystal structure of progesterone 5 beta-reductase from Digitalis lanata defines a novel class of short chain dehydrogenases/reductases. J. Biol. Chem. 2008;283:17260–17269. doi: 10.1074/jbc.M706185200. [DOI] [PubMed] [Google Scholar]
- van Tol R.W., Helsen H.H., Griepink F.C., de Kogel W.J. Female-induced increase of host-plant volatiles enhance specific attraction of aphid male Dysaphis plantaginea (Homoptera: Aphididae) to the sex pheromone. Bull. Entomol. Res. 2009;99:593–602. doi: 10.1017/S0007485309006634. [DOI] [PubMed] [Google Scholar]
- Waller G.R., Price G.H., Mitchell E.D. Feline attractant, cis,trans-nepetalactone: metabolism in the domestic cat. Science. 1969;64:1281–1282. doi: 10.1126/science.164.3885.1281. [DOI] [PubMed] [Google Scholar]
- Zhu J.J., Berkebile D.R., Dunlap C.A., Zhang A., Boxler D., Tangtrakulwanich K., Behle R.W., Baxendale F., Brewer G. Nepetalactones from essential oil of Nepeta cataria represent a stable fly feeding and oviposition repellent. Med. Vet. Entomol. 2012;26:131–138. doi: 10.1111/j.1365-2915.2011.00972.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.