Abstract
Stable quantitative trait loci (QTL) are important for deployment in marker assisted selection in wheat (Triticum aestivum L.) and other crops. We reported QTL discovery in wheat using a population of 217 recombinant inbred lines and multiple statistical approach including multi-environment, multi-trait and epistatic interactions analysis. We detected nine consistent QTL linked to different traits on chromosomes 1A, 2A, 2B, 5A, 5B, 6A, 6B and 7A. Grain yield QTL were detected on chromosomes 2B.1 and 5B across three or four models of GenStat, MapQTL, and QTLNetwork while the QTL on chromosomes 5A.1, 6A.2, and 7A.1 were only significant with yield from one or two models. The phenotypic variation explained (PVE) by the QTL on 2B.1 ranged from 3.3–25.1% based on single and multi-environment models in GenStat and was pleiotropic or co-located with maturity (days to heading) and yield related traits (test weight, thousand kernel weight, harvest index). The QTL on 5B at 211 cM had PVE range of 1.8–9.3% and had no significant pleiotropic effects. Other consistent QTL detected in this study were linked to yield related traits and agronomic traits. The QTL on 1A was consistent for the number of spikes m-2 across environments and all the four analysis models with a PVE range of 5.8–8.6%. QTL for kernels spike-1 were found in chromosomes 1A, 2A.1, 2B.1, 6A.2, and 7A.1 with PVE ranged from 5.6–12.8% while QTL for thousand kernel weight were located on chromosomes 1A, 2B.1, 5A.1, 6A.2, 6B.1 and 7A.1 with PVEranged from 2.7–19.5%. Among the consistent QTL, five QTL had significant epistatic interactions (additive × additive) at least for one trait and none revealed significant additive × additive × environment interactions. Comparative analysis revealed that the region within the confidence interval of the QTL on 5B from 211.4–244.2 cM is also linked to genes for aspartate-semialdehyde dehydrogenase, splicing regulatory glutamine/lysine-rich protein 1 isoform X1, and UDP-glucose 6-dehydrogenase 1-like isoform X1. The stable QTL could be important for further validation, high throughput SNP development, and marker-assisted selection (MAS) in wheat.
Introduction
Identification of consistent loci or haplotypes associated with traits is important in generation advancement decision, forward breeding, gene pyramiding and deployment recommendations in wheat and other crop breedings. These genomic regions can also play an important role during placement of new wheat variety into specific geographical footprints to maximize yield performance via genotype-environment matching. Several factors are considered prior to deployment of a QTL including the stability of the QTL across environment and genetic background, equivalency or effect of the QTL on other traits in elite germplasm, efficacy of the QTL if it is linked to disease or pest resistance, and availability of diagnostic marker in linkage disequilibrium with the QTL. In wheat, several strategies for MAS have been used to improve traits with majority focusing on simple traits [1, 2]. Notably, Rht-B1 and Rht-D1 linked to reduced stature have been deployed via MAS in wheat and in some environments a concomitant increase in yield have been observed although there seem to be a strong linkage disequilibrium between the two dwarfing genes and Fusarium head blight (FHB) susceptibility [2, 3]. A recently mapped QTL for reduced height, Rht24, on chromosome 6A has a potential for MAS and a diagnostic KASP marker has been developed [2, 4]. Our previous work using RIL population and 90K SNP array detected stable diagnostic marker tightly linked to Wsm2, a gene conferring resistance to wheat streak mosaic virus and is currently applied in MAS in winter wheat [5, 6]. Markers linked to Fhb1 and Qfhs.ifa5A, conferring resistance to Fusarium head blight, have also been applied in MAS in wheat breeding [7]. In addition, several other functional markers have been developed for agronomic traits, end-use quality and disease resistance [7, 8]
Detection of consistent QTL depends, in part, on the quality of phenotypic data, genotypic data, and statistical analysis. Typically, evaluation of mapping population is done over multiple seasons in different testing footprints that are representatives of target mega-environments. This approach facilitates detection of consistent QTL over environments and seasons by modeling multi-environment variance-covariance structures that account for heterogeneity of genetic variance. Recent advances in marker technology have led to development of dense and ultra-dense genetic maps providing a fairly good genome coverage [9]. In addition, advances in statistical modeling particularly application of linear mixed models (LMM) provides flexibility to include variance-covariance (VCOV) structure to account for heterogeneity in genetic variances and environmental correlation [10, 11]. LMM can account for both QTL-by-trait interactions (QTI) and QTL-by-environment interactions (QEI) including interaction with environmental covariables such as temperature, light duration and intensity, and moisture levels [10, 12].
Many genetic studies focusing on QTL discovery in wheat have utilized sparse genetic maps to tag QTL and rely on specific statistical method limiting the interpretation of the results to assumptions defined by the analysis approach. In addition, most studies assume the significance of epistatic and pleiotropic interactions underlying yield and yield components in wheat. Most of these QTL have been summarized and posted online on the catalogue of gene symbols. (https://shigen.nig.ac.jp/wheat/komugi/genes/symbolClassList.jsp). In the present study, we used high resolution genetic maps constructed using 90K array SNPs and implemented linear and linear mixed model QTL mapping approach in different genetic mapping software programs [13–16]. The SNP associated with significant QTL were used in combined analysis of pleiotropy and epistasis as outlined by Tyler et al. [13]. Our objectives were to map stable QTL for grain yield and yield components in one of the most popular hard red winter wheat, TAM 111, within the framework of single trait multi-environment and multi-trait analysis and examine marker-to-trait effects and marker-to-marker interactions through analysis of pleiotropy and epistasis [13, 16]. Consistent QTL detected from various statistical models were compared for their locations on consensus genetic and physical maps.
Materials and methods
Recombinant inbred line population and trial evaluation
A bi-parental mapping population was derived from a cross between CO960293-2 (PI 615160) [17] and TAM 111 (PI 631352) [18]. CO960293-2 was developed by Colorado Agricultural Experiment Station and co-released as a germplasm line by Colorado and Kansas Agricultural Experiment Stations primarily for resistance to wheat streak mosaic virus and Russian Wheat Aphid (Diuraphis noxi M.). TAM 111 is a popular cultivar developed and released by Texas A&M AgriLife Research in 2002. It has good performance in both low and high productivity environments [18, 19]. A trial comprising 217 recombinant inbred lines (RIL) plus three checks (four checks in 2012/13) was planted in eight environments from 2012 to 2014; each location-by-year combination was considered as an environment. The locations used in this study were in Texas at Texas AgriLife Research stations in Bushland (35° 06' N, 102° 27' W), Chillicothe (34° 07' N, 99° 18' W) and Etter (35° 59' N, 101° 59' W); Kansas State University Agricultural Research Center-Hays, Hays KS (38° 51' N, 99°20' W); University of Idaho Aberdeen Research and Extension Center, Aberdeen ID (42° 57' N, 112° 49' W); and Colorado State University Plainsman Research Center, Walsh CO (37° 25' N, 102° 18' W). The trials in Etter and Hays were evaluated in both the 2012/13 and 2013/14 crop seasons. The trials in Etter, Aberdeen and Walsh were under well-watered conditions whereas the remaining trials were under dryland conditions. The plot size was 4.645 m2 (4.459 m2 in Walsh) and all trials had two replications. Standard agronomic practices were carried out for each environment.
Trait measurements and data analyses
Grain yield was recorded in all the environments whereas yield components, plant height and days to heading were recorded in a subset of environments. Yield components were recorded in the Texas environments and Hays 2013 (HY13). Plant height was recorded in all the environments except Walsh 2014 (WA14) and Etter 2013 (ET13) whereas days to heading was recorded in ET13, HY13, Aberdeen 2013 (AB13) and Bushland 2014 (BS14). Days to heading was recorded at Feekes growth stage 10.1 as the number of days from January 1st to when 50% of the spikes had emerged from the boot. Percentage of green leaf area was visually rated in BS14 and Chillicothe 2014 (CH14) at Feekes growth stage 10.5 on a scale of 0–100%, where 0% = all the leaves senesced and 100% = all leaves green. Similarly, the greenness of the flag leaf was rated at Feekes growth stage 10.5 on a scale of 0–100% where 0% = whole flag leaf senesced and 100% = whole flag leaf green. At Feekes growth stage 11, plant height was measured in centimeters (cm) from representative plants in each plot as the distance from the base of the stem to the tip of the spike excluding awns. In addition, a half meter long sample from a uniformly filled and representative inner row was harvested from each plot and used for determination of biomass and yield components. The samples were oven-dried at 60°C for three days and the weight of each sample recorded. The total number of stems and the number of heads were counted for each biomass sample. The spikes m2, mean single head weight, kernels spike1, kernels m2, and thousand kernel weight were calculated from the plot sample. The spikes m2 was computed by dividing the number of heads by the sample plot area. The thousand kernel weight was determined by counting and weighing three sets of 100 kernels for each plot and multiplying the average weight by 10. Mean single head weight was calculated by dividing the total dry head weight per plot biomass sample by the number of heads. The kernels m-2 was estimated by dividing the weight of grain from the sample by single kernel weight. The harvest index was calculated as grain weight per sample divided by total weight of biomass. All trials were harvested using a combine harvester and the grain yield plot-1 was used to calculate yield in metric tons hectare-1. Test weight, in kg m-3, was measured using Seedburo equipment (www.seedburo.com, Des Plaines, IL, USA).
Individual and combined environment data was subjected to analysis of variance (ANOVA) in SAS (SAS Institute, 2013) to determine the significance of genotypic and other components of the model. The statistical model used for individual environment analysis was as follows:
Where Yik is the observed phenotypic value of the ith genotype in kth replicate, μ is the overall mean, Rk is the replication effect, Gi is the genetic effect of ith genotype and Ɛik is the residual. All components were considered fixed. The statistical model for combined analysis of variance was as follows:
Where Yijk is the observed value of the ith genotype in the jth environment and kth replicate, R(E) is replication nested within the environment, Ej is the effect of the environment, (GEI)ij is genotype-by-environment interaction, Ɛijk is the residual. To compute mean squares, all the components were considered as fixed whereas for variance components, all components in the model were considered as random. Best linear unbiased predictors (BLUP) and variance components were computed using residual maximum likelihood adapted to META-R program [20]. The BLUP were used for QTL analysis in GenStat version 17 [15]. For single trait multi-environment QTL analysis, the appropriate VCOV structure was modeled in GenStat to account for heterogeneity of genetic variances and correlation among environments [10, 12]. The best VCOV was selected based on the Schwarz information criterion. The genetic correlations (ρg) between pairs of traits were computed using the following formula in METAR:
Where COVx,y is the genetic covariance between trait x and y, σ2x is the variance of phenotype x and σ2y is the variance of phenotype y [20]. Entry-mean heritability estimates were computed using the formula:
Where r is the number of replication, t is the number of environments, σ2g is genotype variance, σ2ge is the GEI variance, and σ2e is the residual variance [21]
DNA extraction and genotyping
Total genomic DNA was extracted from leaf samples of 217 RIL using a CTAB method with minor modification [22, 23]. The RIL and four sets of each parent were genotyped using 90K SNP array based on the manufacturer’s protocol (www.illumina.com). The fluorescence signal was captured using an Illumina scanner and subsequently processed using GenomeStudio software (www.illumina.com). To adjust clusters with skewed cluster separation, manual curation of the data was done by examining the clusters in a Cartesian plot. Loci with low average normalized intensity and undefined clusters were excluded prior to downstream analysis. The genotype data set consisting of 8,819 high quality SNP was used for downstream statistical analysis [5, 24].
Linkage mapping and QTL analysis
We implemented linkage mapping in JoinMap version 4.0 [25]. Prior to linkage map construction, SNPs with identical loci scoring at 100% similarity were omitted to eliminate genetic redundancy and improve computation efficiency. In addition, all SNP with significant segregation distortion based on Chi square test (P < 0.05) were also omitted. We grouped loci into linkage groups based on Independence LOD with increasing stringency from 2.0 to 30.0 and the incremental step of 1.0. The Kosambi mapping function was used to convert recombination frequency into centiMorgans [26]. The pairwise recombination frequency was calculated based on a maximum likelihood (ML) algorithm with the default settings in JoinMap [25]. The final linkage map of 5,580 SNPs covering all of 21 chromosomes with 44 linkage groups was used for QTL analysis. Linkage groups were assigned to chromosomes based on loci information in the 9K and 40K genetic maps [27, 28].
Multi-environment and multi-trait QTL analyses were performed in GenStat based on a LMM framework as described by several authors [10, 12, 29]. In GenStat, QTL mapping was implemented in a stepwise manner commencing with simple interval mapping followed by at least two rounds of composite interval mapping using QTL identified to control the genetic background [30–32]. Backward selection was conducted on QTL detected and the final effects were estimated based on multi-QTL model [3032]. The markers associated with QTL were used for combined analysis of pleiotropy and epistasis using the CAPE package in R [13, 33, 34].
In addition, MapQTL 6.0 [25] was used to analyze and identify significant QTL for each trait based on single environment data. We used QTLNetwork sofware [16] to analyze the significant QTL additive effects, epistasis effects and their interactions with environments across data from multiple environments. The detected QTL results were compared within the three models (single-environment, multi-environment, and multi-trait) of GenStat. Comparative analyses were conducted based on the linked SNPs of unique and consistent QTL using databases from T3 (https://t3sandbox.org/t3/sandbox/wheat/, accessed on March 1, 2017) and the Chinese Spring reference genome from International Wheat Genome Sequence Consortium (IWGSC RefSeqv1.0, https://wheat-urgi.versailles.inra.fr/Seq-Repository/Assemblies, accessed on March 1, 2017).
Results
Phenotypic analysis of yield and its related traits
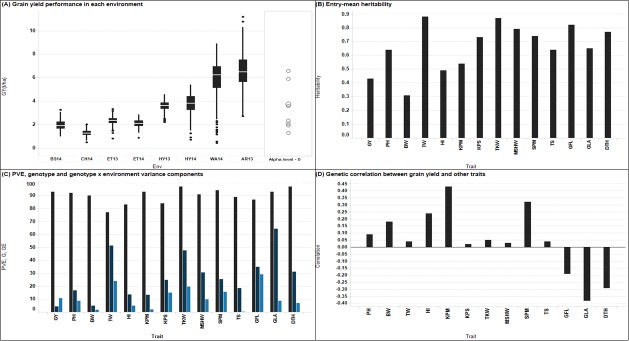
Combined ANOVA across environments revealed significant (P < 0.001) genotype and environment components for traits whereas the GEI was statistically significant for all traits except for biomass weight (S1 Table). The genotype × environment variance component for grain yield was higher than variance due to genotype. A reverse trend was observed for all the other traits where genotype variance components were greater than genotype × environment variance (Fig 1C). The highest variance component due to genotype were observed for green leaf area, thousand kernel weight, and test weight. The phenotypic variance explained (PVE) by the model fit was greater than 80% for all the traits except test weight which had PVE of 77% (Fig 1C). The entry-mean heritability ranged from moderate (0.40 to 0.60) to high (> 0.60) except for biomass weight which had heritability of 0.31 (Fig 1B). Moderate estimates of heritability (0.43–0.64) were observed for grain yield, harvest index, and total stems whereas the remaining traits had high heritability (0.65–0.88). The average grain yield across environments was 3.6 t ha-1 with a corresponding test weight of 750.4 kg m-3. On average, the population had 142 days to heading and the mean plant height was 63.2 cm. The average number of spikes m-2 was 376-. A single spike had an average of 28 kernels and weighed 0.7 g. The average thousand kernel weight was 26.2 g (S1 Table). We observed high yield variability in performance both within and across environments (Fig 1A). The highest range in yield was observed in AB13 and WA14 environments compared to dryland experiments. Dryland experiments showed differences in phenotypic expression with the trial in CH14 showing the least range in yield performance (0.5 to 2.0 t ha-1) attributed to a severe drought stress leading to a narrow window for the grain filling stage. Nonetheless, we still detected significant genotypic variation and the entry-mean heritability was 0.65 (Fig 1A and S1 Table). Comparison of average grain yield showed that environment AR13, WA14, and CH14 were significantly different (P < 0.01) from each other and from all other environments in the present study as indicated by nonoverlapping comparison circles (Fig 1A). BS14 and CH14 were not significant from each other. Similarly, the average grain yield in HY13 and HY14 was not significant from each other. Grain yield under drought stress condition ranged from 1.3 t ha-1 in CH14 to 3.9 t ha-1 in HY13 (Fig 1A).
Fig 1.
Visualization of phenotypic performance (A) Individual environment boxplot for grain yield. The y-axis is grain yield in t ha1 and the x-axis represents environments. The mid line in the box represent the median, the lower and upper horizontal lines of the box represent 25 and 75 percentiles, respectively. The lower whisker represents the 25th percentile minus 1.5 × inter-quartile range (IQR) and the upper whisker is the 75th percentile plus 1.5 × IQR. (B) Entry-mean heritability (C) Percentage of phenotypic variance explained (PVE), genotype and genotype x environment components (D) Genetic correlation between grain yield and agronomic traits.
Across environments, grain yield had significant genetic correlation (P < 0.01) with days to heading, harvest index, kernels m-2, biomass weight, spikes m-2, green leaf area, and greenness of the flag leaf (Fig 1D). The negative correlation observed for grain yield with both green leaf area and greenness of the flag leaf suggest that delayed chlorophyll decay may not necessarily translate to increased grain yield in the US High Plains. The significant correlation between grain yield and harvest index, biomass weight and spikes m-2 suggest that improvement in grain yield may be achievable through indirect selection of these traits. The highest significant correlation across environment (ρg = 0.43, P < 0.01) was observed between grain yield and spikes m-2 (Fig 1D). The genetic correlation between grain yield with biomass weight was moderate (ρg = 0.40 to 0.60) to high (ρg > 0.60) in all trials where the yield components were recorded (data not shown). Kernels m-2 was significantly correlated with grain yield in three environments but was not significantly correlated with grain yield in CH14 (data not shown). Test weight was positively correlated with grain yield in four environments although the correlations were less than 0.30 (data not shown).
Consistent QTL for grain yield, yield components and agronomic traits
We detected nine consistent QTL on chromosomes 1A, 2A, 2B, 5A, 5B, 6A, 6B, and 7A (Table 1, S2–S6 Tables). The QTL on chromosome 1A was linked to spikes m-2 across all the four analysis methods, henceforth referred to as M1, M2, M3 and M4 for method 1 (GenStat-multiEnv), method 2 (GenStat-SingleEnv), method 3 (MapQTL), and method 4 (QTLNetwork), respectively. The magnitude of additive effects and PVE varied from one method to the next. The PVE for spikes m-2 in M1 ranged from 5.8–7.9% with an additive effect range of 13.1–19.1 spikes. The PVE for spikes m-2 in M2 was 7.8% whereas the range in M3 (6.1–8.1%) was similar to M1. This same QTL was also linked to harvest index in M1 and M3; mean single head weight in M1 and M2; kernels spike-1 in M2, M3, and M4; thousand kernel weight in M3 and M4 and test weight in M2 (Table 1).
Table 1. Summary of consistent QTL for yield, yield components and agronomic traits.
| Chrom | QTL ID | Peak (cM) |
M1b | M2 b | M3 b | M4 b | Favorable alleles e | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Traita | A c | PVE d | A c | PVE d | A c | PVE d | A c | ||||
| 1A | 1 | 170–179 | KPS | 0.82 | 7.9 | 0.5–0.8 | 5.6–7.5 | 0.6 | P2 | ||
| 173–186 | SPM | 13.1–19.1 | 5.8–7.9 | 14.0 | 7.8 | 11.8–18.7 | 6.1–8.6 | 9.8 | P1 | ||
| 179–181 | TKW | 0.6–0.7 | 9.0 | 0.3 | P2 | ||||||
| 201–204 | HI | 0.006–0.008 | 3.0–6.1 | 0.01 | 8.8 | P2 | |||||
| 173–179 | MSHW | 0.02 | 4.1–10.9 | 0.03 | 7.0–11.5 | P2 | |||||
| 196 | TW | 4.4 | 5.3 | P2 | |||||||
| 2A.1 | 3 | 46–68 | KPS | 0.83 | 5.1–8.1 | 0.8–1.3 | 8.1–12.2 | 0.8–1.5 | 5.6–12.8 | 0.8 | P2 |
| 46–52 | TW | 6.7–22.0 | 2.5–12.2 | 19.6 | 9.7 | P1 | |||||
| 2B.1.2 | 4 | 122–125 | SPM | 16.9 | 9.7 | 16.7 | 8.5 | 7.8 | P1 | ||
| 142 | PH | 0.5–0.9 | 2.2–7.4 | P2 | |||||||
| 142–152 | DTH | 0.4–0.7 | 8.3–21 | P1 | |||||||
| 2B.1 | 5 | 170–173 | GY | 0.05–0.25 | 3.3–25.1 | 0.12 | 13.4 | 0.11 | 8.6–20.7 | P2 | |
| 167 | KPS | 0.7 | 5.3 | P2 | |||||||
| 165 | SPM | 6.8–13.9 | 1.8–6.0 | P1 | |||||||
| 166–173 | TKW | 0.4–1.0 | 2.7–19.5 | 0.7 | 8.6 | 0.5–1.0 | 6.1–18.4 | P2 | |||
| 170–173 | HI | 0.004–0.016 | 3.3–22.9 | 0.01–0.02 | 5.6–7.0 | 0.01–0.02 | 9.8–25.9 | 0.013 | P2 | ||
| 164–168 | TW | 5.5–8.6 | 1.8–14.4 | 6.6 | 5.9 | P2 | |||||
| 167 | DTH | 0.3–0.9 | 2.5–30.6 | P1 | |||||||
| 168–173 | GFL | 0.1–0.2 | 10.6–36.4 | 0.01 | 21.2–39.5 | P1 | |||||
| 168–175 | GLA | 0.2–0.3 | 32.0–42.1 | 0.2 | 21.1–25.4 | P1 | |||||
| 5A.1 | 8 | 99 | GY | 0.04–0.25 | 2.2–5.2 | P2 | |||||
| 99 | SPM | 9.0 | P1 | ||||||||
| 86–117 | TKW | 0.5–0.8 | 6.1–8.6 | 0.52 | P2 | ||||||
| 100 | KPM | 126.1 | 9.0 | P1 | |||||||
| 5B | 11 | 211–243 | GY | 0.04–0.20 | 1.8–9.3 | 0.10 | 7.2 | 0.11 | 7.7–8.2 | 0.07 | P2 |
| 6A.2 | 14 | 131 | GY | 0.06 | 5.2 | P2 | |||||
| 129 | KPS | 0.3 | P2 | ||||||||
| 125–127 | SPM | 19.5 | 9.3 | 20.0 | 8.8 | 8.7 | P2 | ||||
| 129–133 | TKW | 0.8–1.1 | 11.8–17.5 | 0.7–1.0 | 12.8–14.0 | 0.7–1.0 | 9.3–13.4 | 0.7 | P1 | ||
| 115–122 | HI | 0.006 | 1.3–6.5 | 0.01 | 7.1–8.3 | 0.01 | P1 | ||||
| 129 | TW | 7.3 | 7.2 | P1 | |||||||
| 113 | TS | 2.7 | 7.2 | P2 | |||||||
| 130–131 | PH | 0.8 | 0.8–6.3 | 1.1 | 7.8 | P1 | |||||
| 107 | GLA | 0.1 | 4 | P2 | |||||||
| 6B.1 | 15 | 130–135 | SPM | 5.5–22.7 | 1.7–9.4 | 18.6 | 8.5 | 12.9–14.9 | 6.8–9.5 | P1 | |
| 133–166 | TKW | 0.4–0.6 | 2.5–4.5 | 0.3 | P2 | ||||||
| 127–151 | TW | 4.5–13.5 | 3.5–5.9 | 6.7 | 6.2 | P1 | |||||
| 146–148 | PH | 1.9–2.4 | 7.7–9.2 | P2 | |||||||
| 7A.1 | 16 | 20 | GY | 0.1 | 7.4 | 0.11 | 7.2 | P1 | |||
| 4–20 | KPS | 0.5–1.0 | 2.4–7.3 | 0.9 | 8.4 | 0.5–1.1 | 5.3–8.6 | 0.5 | P2 | ||
| 0–20 | TKW | 0.3 | 0.9–2.1 | 0.7 | 9.5 | 0.6–0.7 | 6.2–8.7 | 0.6 | P1 |
aAbbreviation of traits: GY grain yield, TW test weight, DTH days to heading, PH plant height, HI harvest index, SPM spike m2, KPS kernels spike1, KPM, kernels m2, MSHW mean single head weight, TKW, thousand kernel weight, TS total stems, GLA green leaf area, GFL greenness of flag leaf
b M1 Method 1, GenStat Multi-env model; M2 Method 2, GenStat Single-env model; M3 Method 3, MapQTL; M4 Method 4, QTLNetwork
c A Additive effect; Ignore the negative signs because the parental favorable alleles are labelled
d PVE Phenotypic variations explained
e P1 female parent, CO960293-2; P2 male parent, TAM 111
The interval region for this 1A QTL harbors seven protein coding gene with transcript ID TRIAE_CS42_1AL_TGACv1_001932_AA0036960, Traes_1AS_656CB2399, Traes_1AS_7B084FDFA, Traes_1AS_3160922E9, Traes_1AS_6CB929C18, Traes_1AS_1CBC21AE8, Traes_1AS_5317928F1. This transcript indicates that the 1A QTL is around the centromere given some genes are on the short arm and others on the long arm (http://plants.ensembl.org).
The QTL on chromosome 2A.1 was consistently linked to the number of kernels spike-1 with PVE of 5.1–8.1% in M1, 8.1% and 12.2% in M2, and 5.6–12.8% in M3. In all the QTL analysis approach, this 2A.1 QTL had an additive effect of about one kernel per spike. In addition to kernel spike-1, it was linked to test weight in M1 and M2 where it had PVE of 2.5–12.2% and 9.7% in M1 and M2, respectively. Comparative search showed that the gene, Traes_2AL_2EC344DEE, is within the region of 2A.1 QTL on the long arm of chromosome 2A (http://plants.ensembl.org).
Two QTL, repeatable across environment and statistical analysis methods were detected on chromosome 2B.1. The first one was at the region of 122 to 152 cM of 2B.1, was significantly associated with spikes m-2 in M2 and M3 with a corresponding PVE of 9.7% and 8.5%, respectively. The additive effect for this QTL was 16 spikes m-2 in M2 and M3 and 7.8 spikes m-2 in M4. Analysis using M2 showed that this 2B.1 QTL was also linked to day to heading with PVE range of 8.3–20.7%. BLAST search showed that 20 genes falls within the confidence interval of this first QTL on 2B.1 (http://plants.ensembl.org; https://urgi.versailles.inra.fr). The second QTL on chromosome 2B.1 was linked to harvest index in all the four methods but to yield and thousand kernel weight in the first three methods. We also detected significant statistical association of this QTL on test weight, greenness of the flag leaf and green leaf area in both M1 and M2. For grain yield, this 2nd QTL on 2B.1 had a PVE range of 3.3–25.1% in M1, 13.4% in M2, and 8.6–20.7% in M3. Overall, the additive effect for grain yield ranged from 0.05–0.25 t ha-1 depending on the environment and statistical analysis method. The PVE for this 2nd QTL on 2B.1 on harvest index was 3.3–22.9% in M1, 5.6% and 7.0% in M2, and 9.8–25.9% in M3. The corresponding additive effect for harvest index was 0.004–0.016 in M1, 0.01 and 0.02 in M2, and 0.01–0.02 in M3. This 2nd QTL on 2B.1 was also linked to days to heading in M1 with a PVE range of 2.5–30.6%. Based on the database search of the linked SNP markers, this QTL is very close to the PpdB1 gene, 1.9 Mb apart on the Chinese Spring reference genome (www.wheatgenome.org) and is associated with protein coding gene heat shock 70kDa protein TRIAE_CS42_2BS_TGACv1_146035_AA0453700, Traes_2BS_96D64756B, Traes_2BS_F3D1922E5 and Traes_2BS_B88CDE912 on the short arm of chromosome 2B (S8 Table) (http://plants.ensembl.org).
The QTL on 5A.1 was associated with yield based on M1 analyses, spikes m-2 from M4, and thousand kernel weight from M3 and M4 (Table 1). However, this is a QTL with small effect based on its additive effect and PVE. The peak SNP for 5A.1 is within TRIAE_CS42_5AL_TGACv1_375092_AA1215930 gene which has 7 exons and 20 variants (http://plants.ensembl.org).
The QTL on 5B at 211 cM was consistently linked to grain yield across environments and methods with PVE of 1.8–9.3%, 7.2%, and 7.7–8.2% in M1, M2, and M3, respectively. This QTL had an additive effect of 0.04–0.20 t ha-1 in M1, 0.1 t ha-1 in M2, 0.11 in M3, and 0.07 in M4. The region spanning 5B.1 on the long arm of chromosome 5B harbors TRIAE_CS42_5BL_TGACv1_408832_AA1364560 gene which has a single transcript coding of 146 amino acid. The QTL on 6A.2 was consistently linked to thousand kernel weight with a corresponding PVE of 11.8–17.5% in M1, 12.8–14.0% in M2, and 9.3–13.4% in M3. In addition, 6A.2 QTL was also linked to harvest index in M1, M3, M4; spikes m2 in M2, M3, M4; and plant height in M1 and M2. One of the peak SNP (IWB8924) for 6A.2 is within TRIAE_CS42_6AL_TGACv1_472318_AA1520780 gene on the long arm of chromosome 6A (http://plants.ensembl.org). The gene has four transcripts and codes Diacylglycerol kinase which play a role in signaling under biotic and abiotic stress [35]. Another peak marker, IWB67907, is within a predicted TRIAE_CS42_6AL_TGACv1_472781_AA1525920 gene that codes Alphamannosidase. In other plants, alphamannosidase has enzymatic function on Nglycans [36, 37]. IWB7004, also within confidence interval for 6A.2, is linked to TRIAE_CS42_1BS_TGACv1_051115_AA0177850 gene for uncharacterized protein in wheat (http://plants.ensembl.org). The QTL on 6B.1 was significantly associated with spikes m2 and had PVE of 1.7–9.4% in M1, 8.5% in M2, and 6.8–9.5% in M3. The additive effect of 6B.1 QTL for spikes m2 was 5.5–22.7, 18.6, and 12.9–14.9 spikes m2 in M1, M2, and M3, respectively. A search for genes within the QTL region revealed two protein coding genes TRIAE_CS42_6BL_TGACv1_501196_AA1614830 on 6BL, and TRIAE_CS42_6BS_TGACv1_514653_AA1662770 on 6BS indicating that the QTL is located near the centromeric region. On chromosome 7A.1, we detected a QTL consistently linked to the number of kernels spike1 and thousand kernel weight across environment and QTL analysis methods. The PVE for thousand kernel weight was 0.9–2.1%, 9.5%, and 6.2–8.7% in M1, M2, and M3, respectively. The corresponding additive effect was 0.3 g, 0.7 g, 0.6–0.7 g, respectively. For the number of kernels spike1, the PVE was 2.4–7.3%, 8.4%, and 5.3–8.6% in M1, M2, and M3, respectively. BLAST search of peak SNP showed that TRIAE_CS42_7AL_TGACv1_557259_AA1778880 gene is located within the region spanning 7A.1.
QTLNetwork analyses for additive affects, epistasis, and their interactions revealed more significant effects (Table 1, S2 and S3 Tables). Epistasis was important for thousand kernel weight with seven significant additive × additive (A × A) interaction detected between QTL on chromosome 1A, 1B.1, 3A.3, 3B.1, 5B, 6B and QTL on 5D.1, 7A.3, and 7B. Three out of these seven were between Qtkw.tamu.7B.2 and a QTL on 1A, 3B.1, and 6B (S3 Table). There was no significant additive × additive × environment (A × A × E) interaction for thousand kernel weight but additive × environment (A × E) interaction was important. Among the ten QTL linked to thousand kernel weight, Qtkw.tamu.6A.2 showed significant A × E interaction at CH14 (S2 Table). The number of spikes m-2 had nine significant epistases detected with one showing significant A × A × E interaction. Five significant A × A interaction for spikes m-2 were between one major QTL and a new QTL that was not detected based method 1 to 4 while one epistasis was between two new QTL. We detected one significant epistasis for kernel spike1 and harvest index. The latter involved interaction between two new QTL while the former involved interaction between Qkps.tamu.3D.1 and a new QTL for kernel spike-1. Only the A × A × E for two major stable QTL Qhi.tamu.2B.1 and Qhi.tamu.6A.2 was significant (S3 Table).
The QTL on 1A, 1B.1, 1B.3, and 2B.1 at 170 cM as well as the QTL on 5B at 243 cM showed significant A × E for grain yield whereas two QTL on 1A and 2B.1 at 170 cM showed significant A × E for harvest index (S2 Table). An interaction between two QTL, Qgy.tamu.3D.1 and Qgy.tamu.5B.1, was not significant for A × A epistasis but significant for A × A × E interactions. Overall, the environment AB13 (Aberdeen, Idaho 2013) was different from other environments. Five out of the six significant A × A × E interactions involved AB13 (S3 Table).
Multi-trait QTL for yield and yield components
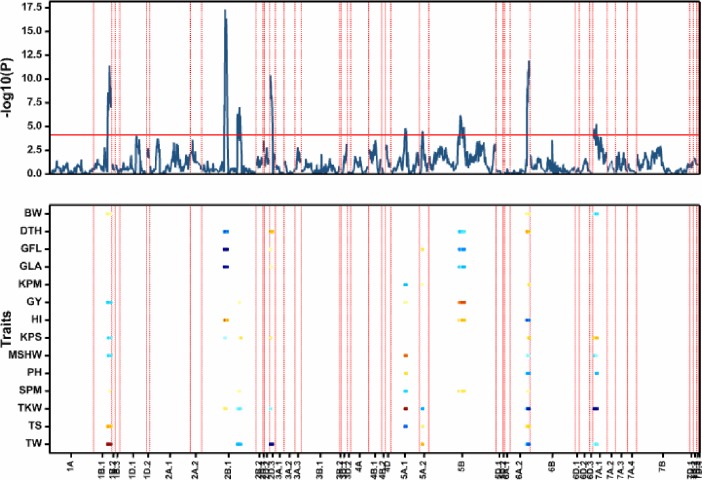
The multi-trait genetic model across seven environments revealed nine significant QTL with all QTL detected showing significant QTI as indicated by a color coded HVA (red color = HVA from TAM 111, blue = HVA from CO960293-2) [10, 12] (Table 2, Fig 2).Seven of the nine multi-trait QTL were also detected based on M1 and M2 approach (Table 1). Multi-trait QTL on chromosomes 2B (Qmt.tamu.2B.1) and 6A.2 (Qmt.tamu.6A.2) were associated with the highest number of traits suggesting that these genomic regions are essential in wheat breeding for higher yield (Fig 2).
Table 2. Multitrait QTL detected using data pooled across environments.
| QTL Namea | QTL ID | Chr | Position (cM) |
Peak SNP | CI_LLb | CI_ULc | -log10(P) | Traitsd | AEe | PVEf (%) | HVAg | Detected in single environment analysish |
Detected in multi- environment |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Qmt.tamu.1B.1 | 18 | 1B.1 | 111.4 | IWB37118 | 91.3 | 123.1 | 10.9 | BW, GY, KPS, MSHW, TS, TW |
0.17, 0.23, 0.18, 0.19, 0.24, 3.60 |
3.0, 5.4, 3.3, 3.7, 1.1, 8.0 |
P2, P1, P1, P1, P2, P2 |
Yes: for TW in HY13 and ET13 | No |
| Qmt.tamu.2B.1 | 5 | 2B.1 | 165.5 | IWB43273 | 163.5 | 167.5 | 17.3 | DTH, GFL, GLA, HI, TKW, TW |
0.43, 0.76, 0.63, 0.48, 0.35, 2.15 |
18.4, 57.7, 39.2, 22.9, 12.0, 2.8 |
P1, P1, P1, P2, P2, P2 |
Yes: for GFL and GLA in BS14; HI, GFL, GLA, and GY in CH14; DTH and HI in ET13; DTH, TW, and TKW in HY13 | Yes: for DTH, GFL, GLA, GY, HI, and SPM |
| Qmt.tamu.2B.1.1 | 19 | 2B.1 | 269.3 | IWB70591 | 221.3 | 317.3 | 7.6 | GY, SPM, TKW, TW | 0.19, 0.15, 0.16, 2.96 | 3.5, 2.2, 2.4, 5.2 | P2, P2, P1, P1 | Yes: for GY in ET13 | No |
| Qmt.tamu.2D.3 | 6 | 2D.3 | 1.2 | IWB26013 | 0.0 | 8.4 | 9.9 | DTH, GFL, GLA, TW | 0.27, 0.10, 0.11, 0.42 | 7.5, 1.0, 1.1, 17.7 | P2, P2, P2, P1 | Yes: for TW in ET13, ET14, and HY13; DTH in AB13 | Yes: for TW |
| Qmt.tamu.5A.1 | 8 | 5A.1 | 100.4 | IWB912 | 83.6 | 117.2 | 5.1 | KPM, GY, MSHW, PH, SPM, TKW, TS |
0.19, 0.20, 0.24, 0.15, 0.15, 0.27, 0.30 |
3.7, 4.0, 5.6, 2.2, 2.1, 7.3, 9.0 |
P1, P2, P2, P2, P1, P2, P1 |
Yes: for KPM in BS14 | Yes: for GY |
| Qmt.tamu.5A.2 | 9 | 5A.2 | 17.7 | IWB6809 | 0.0 | 56.4 | 4.9 | GFL, KPM, TKW, TW | 0.09, 0.18, 0.20, 2.19 | 0.8, 3.4, 3.9, 2.8 | P2, P2, P1, P2 | Yes: for TKW in BS14; KPM in CH14; TW in HY13 | Yes: for KPM, KPS, and TKW |
| Qmt.tamu.5B.1 | 11 | 5B | 226.5 | IWB21839 | 201.2 | 251.8 | 7.3 | DTH, GFL, GLA, GY, HI, SPM |
0.16, 0.15, 0.12, 0.26, 0.14, 0.15 |
2.7, 2.4, 1.4, 7.0, 2.0, 2.2 |
P1, P1, P1, P2, P2, P2 |
Yes: for GY in HY13 | Yes: for GY |
| Qmt.tamu.6A.2 | 14 | 6A.2 | 130.7 | IWB26244 | 111.6 | 133.5 | 11.8 | DTH, HI, KPS, MSHW, PH, SPM, TKW, TS, TW |
0.14, 0.23, 0.13, 0.15, 0.25, 0.13, 0.29, 0.17, 3.35 |
1.9, 5.1, 1.6, 2.2, 6.3, 1.7, 8.3, 2.9, 6.9 |
P2, P1, P2, P1, P1, P2, P1, P2, P1 |
Yes: for SPM and TKW in ET13; TKW in ET14; TKW, TW, and PH in HY13 | Yes: for HI, PH, and TKW |
| Qmt.tamu.7A.1 | 16 | 7A.1 | 20.6 | IWB7632 | 2.9 | 38.3 | 4.9 | BW, KPS, PH, TKW | 0.17, 0.18, 0.19, 0.30 | 3.0, 3.3, 3.7, 8.7 | P1, P2, P1, P1 | Yes: GY in ET14; KPS and TKW in HY13 | Yes: for KPS and TKW |
a QTL name including trait, institute, and chromosome location; the chromosome location part is unique for each numbered QTL if the peak positions were less than 40 cM; within each chromosome fragment, different numbered QTL will have various chromosome fragment parts starting from the fragment name, then adding “.1, .2, …”.
b CI_LL, lower limit of QTL confidence interval in centiMorgans.
c CI_UL, upper limit of QTL confidence interval in centiMorgans.
d Abbreviation of traits: GY grain yield, TW test weight, DTH days to heading, PH plant height, HI harvest index, KPM kernels m2, BW biomass weight, SPM spike m2, KPS kernels spike1, MSHW mean single head weight, TKW, thousand kernel weight, TS total stems, GLA green leaf area, GFL greenness of flag leaf
e Additive effects corresponding to each trait in the trait column.
f PVE, phenotypic variance explained (%) corresponding to each trait in the trait column.
g HVA, High value allele corresponding to each trait in the trait column. P1 = CO960293-2, P2 = TAM 111.
h ENV, environment, AB13 Aberdeen 2013, BS14 Bushland 2014, CH14 Chillicothe 2014, ET13 Etter 2013, ET14 Etter 2014, HY13 Hays 2013, HY14 Hays 2014.
Fig 2. Genomewide scan for multi-trait QTL for grain yield and agronomic traits.
The upper graph is the QTL profile plot with the y-axis representing the log of likelihood, -log (P), for declaring significance of QTL. The red horizontal line represents the threshold corrected for the number of independent tests using Li and Ji [38]. The lower profile is the genomewide heat map of significant QTL across environments. The y-axis is the traits and the x-axis represents the chromosomes. The light blue to blue color indicates the high value allele originates from CO9602932 and the yellow-red color indicates the high value allele originates from TAM 111.
Grain yield was linked to multi-trait QTL on chromosome 1B.1, 2B.1, 5A.1, and 5B with all QTL except 1B.1 showing HVA from TAM 111 (Fig 2). Biomass weight was linked to multi-trait QTL on chromosome 1B.1 and 7A.1 (Table 2 and Fig 2). Multi-trait QTL associated with plant height were detected on chromosome 5A.1, 6A.2, and 7A.1 and the chromosomal location of multi-trait QTL for plant height on 6A.2 agreed with single trait multi-environment QTL model (Tables 1 and 2). Days to heading was linked to multi-trait QTL on 2B.1, 2D.3, 5B, and 6A.2 whereas green leaf area and greenness of the flag leaf were associated with multi-trait QTL on chromosome 2B.1, 2D.3, and 5B although the greenness of the flag leaf had an additional QTL on 5A.2 (Table 2). The multi-trait QTL with significant additive effect on kernels m2 were mapped on chromosome 5A.1 and 5A.2 (Table 2). Harvest index was associated with multi-trait QTL on 2B.1, 5B, and 6A.2 whereas kernel spike-1 QTL with significant additive effects were detected on 1B.1, 6A.2, and 7A.1 (Table 2). Mean single head weight was linked to multi-trait QTL on chromosomes 1B.1, 5A.1, and 6A.2. Thousand kernel weight QTL were mapped on chromosome 2B.1, 5A.1, 5A.2, 6A.2, and 7A.1 (Table 2). The QTL for thousand kernel weight detected on 2B.1, 5A.2, 6A.2, and 7A.1 were also detected in a single trait multi-environment model (Table 1). The multi-trait QTL linked to spikes m2 were detected on chromosome 2B.1, 5A.1, 5B, and 6A.2 with all QTL showing HVA from TAM 111 except for the QTL on 5A.1. Test weight was associated with multi-trait QTL on 1B.1, 2B.1, 2D.3, 5A.2, 6A.2, and 7A.1 whereas the multi-trait QTL with significant additive effect on total stems were detected on chromosome 1B.1, 5A.1, and 6A.2 (Table 2). The multi-trait QTL on 1B.1, 2B.1, 2D.3, 6A.2, and 7A.1 were associated with kernels spike1. However, a test for significance of additive revealed that only the QTL on 1B.1, the multi-trait QTL on 6A.2 and on 7A.1 had significant additive effect on kernels spike1. Mean single head weight was associated with multi-trait QTL on 1B.1, 5A.1, 6A.2, and 7A.1 although the latter had nonsignificant additive effect.
The co-location observed in multi-trait analysis was supported by results from previous analysis. In M1 analysis, mean single head weight, spikes m2 and harvest index were co-located on chromosome 1A from 173.0 cM to 202.9 cM whereas thousand kernel weight, kernels m2, and kernels spike1 were co-located on 5A.2 from 1.0 cM and 24.7 cM, respectively based on peak SNP position (S4 Table and S1–S3 Figs). Test weight and kernels spike1 were co-located on chromosome 2A.1 at 51.8 cM and 53.7 cM, respectively (S4 Table and S1–S3 Figs). QTL for days to heading, greenness of the flag leaf, green leaf area, grain yield, harvest index, spikes m2, thousand kernel weight, and test weight were co-located from 165.5 cM to 172.9 cM on chromosome 2B.1. In addition, thousand kernel weight, plant height, and harvest index were co-located on 6A.2 from 115.3 cM to 129.7 cM (S4 Table and S1–S3 Figs). In M2 QTL model showed colocation at 168–175 cM of 2B.1 for greenness of the flag leaf and green leaf area and in the same map position as M1 colocation (Table 1 and S4 and S5 Tables). Spikes m-2 and thousand kernel weight were co-located on chromosome 6A.2 in ET13. In HY13, colocation of QTL was observed for test weight and thousand kernel weight on 2B.1; thousand kernel weight, test weight, and plant height on 6A.2; and kernel spike-1 and thousand kernel weight on 7A.1 (S4 and S5 Tables). The colocation of QTL for different traits could partly explain the genetic correlation observed in this study (Fig 1D).
Combined analysis of pleiotropy and epistasis
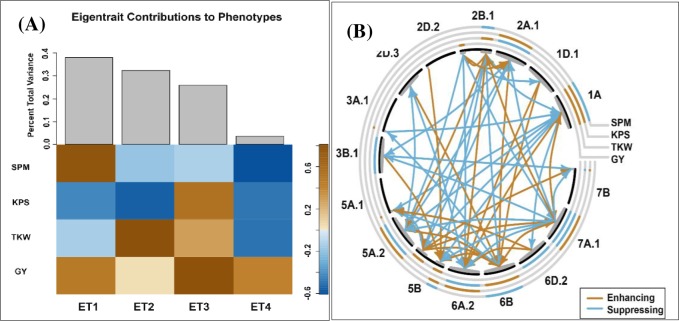
Combined analysis of pleiotropy and epistasis (CAPE) jointly uses multiple phenotype and genetic markers to model and define marker-to-trait effects and marker-to-marker interactions. The detected marker-to-trait effect and marker interactions are defined either as enhancing (allele from male parent) or repressing (allele from female parent). Typically, the linear independence of the phenotypic data is achieved by singular value decomposition (SVD) to extract composite traits (eigentraits) prior to analysis [13, 39, 40]. In this study, SVD of grain yield and three yield components (spikes m-2, kernels spike-1, and thousand kernel weight) generated four eigentraits (ET). The first three ET accounted for approximately 90% of the variation in the phenotype and were used for CAPE to elucidate interaction patterns underlying grain yield and yield components. The black arc lines represent linkage groups. Light grey concentric lines represent traits with the innermost concentric line representing grain yield followed sequentially by yield components namely thousand kernel weight, kernels spike-1, and spikes m-2 (Fig 3B). A network of marker-to-marker interaction is represented by color-coded arrow line depending on whether the interaction is enhancing (brown) or repressing (blue). The segment of the linkage group involved in the interaction is marked by grey color inside the black arc. The main effect calculated based on a subset of markers (markers associated with significant effect in the previous section) are mapped along the concentric lines. Both positive and negative pleiotropic effects were observed on chromosomes 1A, 2A.1, 2B.1, 5A.2, 5B, 6A.2, 6B, 7A.1, and 7B (Table 3 and Fig 3B). Markers on 2B.1, IWB70591 and IWB64246, had enhancing effect on grain yield. In addition, IWB23950 at 228.0 cM and IWB52093 at 402.6 cM on chromosome 5B had enhancing effect on grain yield. On the contrary, IWA3983 on 3B.1, IWB31561 on 6D.2, and IWB11000 on 7A.1 had a repression effect on grain yield (Table 3 and Fig 3B). Thousand kernel weight was enhanced by IWB46316 and IWB42357 on 1A; IWB16370 and IWB8143 on 2B.1; IWB47055 on 3A.1; IWB912 on 5A.1; IWB72333, IWB11040, IWB9108 on 6B. QTL that had repression effect on thousand kernel weight include IWB7015 on 2A.1; IWB74769 on 5A.2; IWB7004 on 6A.2; and IWA7406 and IWB11000 on 7A.1 (Table 3). The two markers, IWB46316 and IWB42357 on 1A, enhancing thousand kernel weight had positive pleiotropic effect on kernels spike1 but had repression effect on spikes m2. In addition, the IWB16370 on 2B.1 which had positive effect on thousand kernel weight showed negative pleiotropic effect on spikes m2 (Table 3; Fig 3B). Besides IWB46316 on 1A, other markers that had enhancing effect on kernels spike1 include IWB13287 and IWB42357 on 1A; IWB7015 and IWB28453 on 2A.1; IWB12338, IWB8687 and IWB14407 on 5A.2; IWB29746 on 5B; IWB7004 on 6A.2; IWB41660 and IWB7632 on 7A.1 (Table 3 and Fig 3B). IWB7015, IWB7004, and IWB10879 enhancing KPS had a negative pleiotropic effect on thousand kernel weight.
Fig 3. Interactions and pleiotropic patterns based on markers linked to significant QTL from multi-environment and multi-trait QTL analysis.
(A) Eigentraits (ET) generated through singular value decomposition of phenotypic data. The first three ET explained approximately 90% of the phenotypic variation and were used for combined analysis of pleiotropy and epistasis (CAPE). The legend indicates the direction of variance among traits. For instance, for ET1, kernel spike-1 (KPS) and thousand kernel weight (TKW) vary in opposite direction compared to grain yield (GY) and spike m-2 (SPM). (B) Interaction patterns from CAPE with main effects (brown and blue) depicted on the concentric rings and interactions depicted by color-coded arrow lines within the plot. Brown arrow line represents enhancing effect whereas blue arrow line indicates repressing effect.
Table 3. Significant marker-to-trait influences based on a subset of markers in combined analysis of pleiotropy and epitasis.
| Markers | QTL ID | Source linkage group | Source position | Target traitsa | Effect on target | Source of alleles | Effect | SE | |Effect|/SEb | P-value | q-value |
|---|---|---|---|---|---|---|---|---|---|---|---|
| IWB70591 | 19 | 2B.1 | 269.3 | GY | Enhancing | TAM 111 | 0.59 | 0.18 | 3.23 | 0.0013 | 0.0392 |
| IWB64246 | 19 | 2B.1 | 289.6 | GY | Enhancing | TAM 111 | 0.66 | 0.19 | 3.47 | 0.0005 | 0.0270 |
| IWB23950 | 11 | 5B | 228.0 | GY | Enhancing | TAM 111 | 0.84 | 0.20 | 4.23 | 0.0000 | 0.0008 |
| IWB52093 | 12 | 5B | 402.6 | GY | Enhancing | TAM 111 | 0.96 | 0.32 | 3.02 | 0.0025 | 0.0360 |
| IWB46316 | 21 | 1A | 148.1 | TKW | Enhancing | TAM 111 | 0.63 | 0.19 | 3.27 | 0.0013 | 0.0440 |
| IWB42357 | 1 | 1A | 196.0 | TKW | Enhancing | TAM 111 | 0.78 | 0.19 | 4.08 | 0.0001 | 0.0015 |
| IWB16370 | 4 | 2B.1 | 122.7 | TKW | Enhancing | TAM 111 | 0.79 | 0.20 | 3.86 | 0.0002 | 0.0045 |
| IWB8143 | 5 | 2B.1 | 166.7 | TKW | Enhancing | TAM 111 | 0.81 | 0.20 | 4.10 | 0.0000 | 0.0022 |
| IWB47055 | 25 | 3A.1 | 44.0 | TKW | Enhancing | TAM 111 | 0.85 | 0.21 | 4.05 | 0.0001 | 0.0031 |
| IWB912 | 8 | 5A.1 | 97.1 | TKW | Enhancing | TAM 111 | 0.67 | 0.19 | 3.52 | 0.0006 | 0.0104 |
| IWB72333 | 15 | 6B | 127.5 | TKW | Enhancing | TAM 111 | 0.79 | 0.21 | 3.76 | 0.0002 | 0.0061 |
| IWB11040 | 15 | 6B | 145.7 | TKW | Enhancing | TAM 111 | 0.78 | 0.21 | 3.67 | 0.0003 | 0.0068 |
| IWB9108 | 26 | 6B | 230.7 | TKW | Enhancing | TAM 111 | 0.67 | 0.18 | 3.71 | 0.0003 | 0.0081 |
| IWB46316 | 21 | 1A | 148.1 | KPS | Enhancing | TAM 111 | 0.58 | 0.18 | 3.18 | 0.0017 | 0.0298 |
| IWB13287 | 1 | 1A | 169.9 | KPS | Enhancing | TAM 111 | 0.76 | 0.21 | 3.57 | 0.0005 | 0.0096 |
| IWB42357 | 1 | 1A | 196.0 | KPS | Enhancing | TAM 111 | 0.70 | 0.20 | 3.52 | 0.0005 | 0.0136 |
| IWB7015 | 3 | 2A.1 | 43.9 | KPS | Enhancing | TAM 111 | 0.97 | 0.21 | 4.51 | 0.0000 | 0.0001 |
| IWB28453 | 3 | 2A.1 | 67.1 | KPS | Enhancing | TAM 111 | 0.65 | 0.19 | 3.47 | 0.0006 | 0.0073 |
| IWB12338 | 9 | 5A.2 | 1.0 | KPS | Enhancing | TAM 111 | 0.61 | 0.20 | 3.14 | 0.0019 | 0.0243 |
| IWB8687 | 9 | 5A.2 | 13.2 | KPS | Enhancing | TAM 111 | 0.73 | 0.20 | 3.57 | 0.0005 | 0.0215 |
| IWB14407 | 9 | 5A.2 | 24.7 | KPS | Enhancing | TAM 111 | 0.70 | 0.20 | 3.44 | 0.0007 | 0.0342 |
| IWB29746 | 12 | 5B | 384.8 | KPS | Enhancing | TAM 111 | 0.67 | 0.19 | 3.52 | 0.0005 | 0.0165 |
| IWB7004 | 14 | 6A.2 | 128.7 | KPS | Enhancing | TAM 111 | 0.63 | 0.19 | 3.40 | 0.0008 | 0.0312 |
| IWB41660 | 16 | 7A.1 | 11.2 | KPS | Enhancing | TAM 111 | 0.60 | 0.18 | 3.40 | 0.0008 | 0.0165 |
| IWB7632 | 16 | 7A.1 | 20.6 | KPS | Enhancing | TAM 111 | 0.71 | 0.21 | 3.34 | 0.0010 | 0.0190 |
| IWB10879 | 17 | 7B | 12.7 | KPS | Enhancing | TAM 111 | 0.59 | 0.19 | 3.06 | 0.0025 | 0.0443 |
| IWA3983 | 7 | 3B.1 | 107.9 | GY | Repressing | CO960293-2 | -0.69 | 0.19 | 3.52 | 0.0005 | 0.0164 |
| IWB31561 | 27 | 6D.2 | 41.1 | GY | Repressing | CO960293-2 | -0.69 | 0.19 | 3.55 | 0.0004 | 0.0217 |
| IWB11000 | 43 | 7A.1 | 57.8 | GY | Repressing | CO960293-2 | -0.61 | 0.20 | 3.11 | 0.0019 | 0.0298 |
| IWB7015 | 3 | 2A.1 | 43.9 | TKW | Repressing | CO960293-2 | -0.71 | 0.20 | 3.58 | 0.0005 | 0.0108 |
| IWB74769 | 9 | 5A.2 | 24.3 | TKW | Repressing | CO960293-2 | -0.93 | 0.18 | 5.10 | 0.0000 | 0.0000 |
| IWB7004 | 14 | 6A.2 | 128.7 | TKW | Repressing | CO960293-2 | -0.97 | 0.19 | 5.08 | 0.0000 | 0.0000 |
| IWA7406 | 16 | 7A.1 | 5.7 | TKW | Repressing | CO960293-2 | -0.74 | 0.17 | 4.31 | 0.0000 | 0.0008 |
| IWB11000 | 43 | 7A.1 | 57.8 | TKW | Repressing | CO960293-2 | -0.66 | 0.17 | 3.87 | 0.0002 | 0.0055 |
| IWB10879 | 17 | 7B | 12.7 | TKW | Repressing | CO960293-2 | -0.67 | 0.17 | 3.88 | 0.0001 | 0.0034 |
| IWB46316 | 21 | 1A | 148.1 | SPM | Repressing | CO960293-2 | -0.96 | 0.21 | 4.47 | 0.0000 | 0.0006 |
| IWB13287 | 1 | 1A | 169.9 | SPM | Repressing | CO960293-2 | -0.74 | 0.18 | 4.04 | 0.0001 | 0.0033 |
| IWB42357 | 1 | 1A | 196.0 | SPM | Repressing | CO960293-2 | -0.87 | 0.19 | 4.59 | 0.0000 | 0.0003 |
| IWB16370 | 4 | 2B.1 | 122.7 | SPM | Repressing | CO960293-2 | -0.69 | 0.20 | 3.46 | 0.0007 | 0.0343 |
| IWA4416 | 12 | 5B | 478.6 | SPM | Repressing | CO960293-2 | -0.59 | 0.17 | 3.35 | 0.0010 | 0.0420 |
| IWB72333 | 15 | 6B | 127.5 | SPM | Repressing | CO960293-2 | -0.76 | 0.19 | 4.05 | 0.0001 | 0.0016 |
| IWB8809 | 15 | 6B | 166.4 | SPM | Repressing | CO960293-2 | -0.70 | 0.18 | 3.83 | 0.0002 | 0.0025 |
| IWB9108 | 26 | 6B | 230.7 | SPM | Repressing | CO960293-2 | -0.82 | 0.18 | 4.63 | 0.0000 | 0.0002 |
a Abbreviation for traits: GY grain yield, TKW thousand kernel weight, KPS Kernels spike1, SPM Spikes m2
b The test statistic was computed by dividing the absolute effect by standard error (SE). The likelihood test for chance association was calculated using 500,000 permutations. The test statistic was compared with null distribution generated through permutation. To minimize inflation of false positives due to multiple tests, the false discovery rate (FDR) corrected P-values (q-values) were computed
Marker-to-marker interaction patterns were depicted using color-coded arrowed lines with direction of the arrow indicating the target markers enhanced or repressed (Fig 3B). Similar to marker-to-trait effect, the brown arrow lines indicate favorable interactions (enhancing) whereas blue lines indicate unfavorable interactions (suppressing). The effect of markers on 1A was enhanced by interaction with markers on 7A.1. The marker IWB7015 on 2A.1 was enhanced by interaction with IWB7053 and IWB64246 on 2B.1. Markers on 2B.1 were enhanced by interaction with markers on 2A.1, 2B.1, 6A.2, and 7A.1. IWB912 on 5A.1 was enhanced by IWB6263 whereas a set of markers mapped from 1.0 to 17.7 cM on 5A.2 were enhanced by interacting with several markers on 1A, 2B.1, 5A.1, and 5B (Fig 3B and S7 Table). On chromosome 5B, IWB21839, IWB59433, IWB29746, and IWB52093 were enhanced by several markers on 5A.2, 6B, 2B.1, 2D.2, and 3B.1 (Fig 3B and S7 Table). Several markers on 6A.2 were enhanced by interaction with markers on 1A, 1D.1, 2B.1, and 6D.2. In addition, IWB43368 mapped at 112.7 cM on 6A.2 had enhancing effect on IWB1295 mapped at 129.7 cM on the same chromosome. A number of markers detected on 1D.1, 2A.1, 2D.3, 5A.2, 5B, and 7A.1 enhanced markers detected on 6B. IWB7632 on 7A.1 was enhanced by IWB46316, IWB13287, and IWB42357 on 1A. IWB41660 showed positive interaction with IWB65641 on 5B whereas IWA7406 on 7A.1 showed positive interaction with IWB72333 and IWB8809 on 6B. Marker IWB65641 on 5B enhanced IWB10879 on 7B (Fig 3B and S7 Table). Negative marker-to-marker interactions with sources of alleles from CO9602932 were detected between 10 source chromosomes including 1A, 2A, 2B, 3A, 3B, 5A, 5B, 6A, 6B, and 7A (Fig 3B and S7 Table).
Discussion
We applied different genetic models to define the underlying genetic basis of quantitative traits in winter wheat evaluated in different geographical testing footprints in the US Great Plains. A single trait multi-environment QTL analysis revealed that most traits had significant QEI, underscoring the need of multi-environment phenotyping to account for this variation. Across all the models and traits, we detected 43 unique QTL with most QTL showing colocation or pleiotropic effect. Nine of these QTL were consistent across environment and QTL analysis methods and seven of the nine QTL were also detected based on multi-trait QTL analysis approach. Stable grain yield QTL were detected on chromosomes 2B (Qgy.tamu.2B.1) and 5B (Qgy.tamu.5B.1) based on M1, M2, M3, and M4. The multi-trait QTL on 2B.1 was mapped at position 165.5 cM (172.9 cM in M1 analysis) and had a significant effect on grain yield and other traits (Tables 1 and 2, S2–S7 Tables, S1–S3 Figs). Based on genetic map position, Qgy.tamu.2B.1 was either pleiotropic or co-located with QTL for days to heading, test weight, kernel weight, spikes m2, greenness of the flag leaf, green leaf area, and harvest index. BLAST search using peak marker for Qgy.tamu.2B.1 revealed that it is associated with several genes including NAD-dependent histone deacetylase domain containing protein and heat shock 70 kDa protein (S8 Table). Four other multi-trait QTL (Qmt.tamu.1B.1, Qmt.tamu.2B.1.1, Qmt.tamu.5A.1, and Qmt.tamu.5B.1) were also linked to grain yield and other traits with Qmt.tamu.5B.1 showing map position consistency with QTL analysis in M1. Comparison of genetic maps in Wu et al. [41] and the present study, both constructed using 90K SNP, showed that IWA4518 (BobWhite_c8113_532) was mapped at 219.6 cM in the present work and falls within the confidence interval for the multi-trait QTL Qmt.tamu.2B.1.1. The base sequence for IWA4518 is within 1181 bp mRNA for uncharacterized protein in Aegilops tauschii subsp. tauschii and 1264 bp mRNA from Triticum aestivum cDNA clone WT006_C07 (www.ncbi.nlm.nih.gov). This region has also been associated with starch gelatinization in a study using 90K and 660K SNP array [42]. In their study, the linked SNP to the QTL for starch gelatinization was RAC875_c56101_368, which was mapped near Qmt.tamu.2B.1.1 in the present study.
We detected other stable QTL for thousand kernel weight on 6A.2, 6B, and 7A.1. A recent study using 90K SNP array reported kernel weight QTL on 3D, 4A, 5A, 6A, 7A, and 7B based on multi-environment data from Gao [43]. The map position of QTL on 6A, and 7A based on interval SNP were different from the present study suggesting that these could be new QTL for thousand kernel weight. A consistent QTL for mean single head weight (Qmshw.tamu.1A) was detected on 1A explaining 4.1–10.1% of the phenotypic variation. This region was linked to spike compactness and sterile spikelet number in a RIL population genotyped using 90K SNP array and SSR markers [44]. Plant height was linked to two stable QTL, one at 142 cM on 2B.1 and one on 6A.2 clustered or had pleiotropic effect on kernel weight, green leaf area, harvest index, and spikes m2. Recent work by Wurschum et al [2] and Tian et al [45] showed that chromosome 6A harbors Rht24 gene for reduced plant and that the gene is sensitive to gibberellic acid. These studies used SNP, GBS and SSR making it difficult to make direct comparison with the stable QTL for plant height detected in the present work although both QTL were mapped on the long arm of chromosome 6A. A summary of QTL by Triticeae Coordinated Agricultural Project (TCAP) reported. that IWB6528 (BS00012081_51), IWB56597 (RAC875_c31358_214), IWB4233 (BobWhite_c6771_697), and IWB54163 (RAC875_c15844_348) were associated with PH QTL on chromosome 2B (https://triticeaetoolbox.org/wheat/qtl/qtl_report.php). In the present work, the four SNP were mapped within the confidence interval (119.4–165 cM) for plant height QTL on chromosome 2B suggesting that this could be same QTL for plant height. The peak region for 2B.1.2 (IWB62653) was mapped at 142.2 cM compared to 144.6 cM, 144.7 cM, 144.0 cM and 145.1 for IWB6528, WB56597, IWB4233, and IWB54163, respectively. Megablast search of the IWB62653 sequence revealed that it is within 1656 bp mRNA of wre1n.pk0115.e3:fis clone (www.ncbi.nlm.nih.gov).
We detected stable QTL for test weight on chromosome 2A, 2B.1 at 164 cM and 6A.2. Both test weight QTL on 2B and 6A were also detected when data was analyzed using multi-trait QTL model with additional QTL Qmt.tamu.1B.1 on 1B, Qmt.tamu.2D on chromosome 2D, and Qmt.tamu.5A.1 on chromosome 5A (Table 3). In a previous study, IWB47726 (Kukri_c7770_176) and IWB73106 (Tdurum_contig68343_339) were associated with fertile spikelet number and spikelet compactness, respectively [44]. These two SNPs were mapped within the confidence interval for mult-trait QTL Qmt.tamu.1B.1 in the present study.
The significant QTI observed in multi-trait model suggests that pleiotropy and epistatic interactions can contribute to significant variations. Empirical studies on epistasis have reported varying results with some reporting significant contribution of epistasis in the modulation of quantitative traits while other studies have shown nonsignificant contribution [46]. Results of the multi-trait model were supported by further genetic analysis based on combined analysis of pleiotropy and epistasis which showed various patterns of genetic interactions among markers and traits. The marker-to-trait effect revealed that grain yield was enhanced by markers on 2B.1 and 5B with favorable alleles from TAM 111 but was repressed by a set of markers detected on 3B.1, 6D.2, and 7A.1 with alleles from CO9602932. Two markers, IWB46316 and IWB42357 on 1A enhancing thousand kernel weight had positive pleiotropic effect on kernels spike-1 but had negative pleiotropic effects on spikes m2 (Fig 3B and Table 3). Markers enhancing kernels spike-1 on 2A.1, 6A.2, and 7A.1 had a negative pleiotropic effect on thousand kernel weight. These results suggest the importance of epistasis and pleiotropy in the genetic architecture of yield components in winter wheat.
QTLNetwork analyses confirmed some results of epistasis from CAPE analyses and provided information on the importance QTL by environment and epistasis by environment interactions. A × A, A × E, and A × A × E interactions were identified for several traits across environments in this study. These interactions could be helpful in evaluation of the major QTL and potential candidate genes in marker-assisted breeding and genomic prediction. Validation and conversion to KASP of SNPs tightly linked to yield and its components will be useful for their utility in marker-assisted breeding.
Conclusions
We have provided QTL discovery results using both multi-environment, individual environment, and multi-trait analysis models of GenStat as well as using MapQTL and QTLNetwork. The multi-environment QTL for grain yield on 5B was repeatable in six out of eight environments. This could be an important target region for fine mapping and validation to identify SNPs associated with grain yield in wheat. A potential target region linked to mean single head weight was detected on 1A. The QTL for mean single head weight on 1A was constitutively expressed in all environments where data was recorded. Several QTL associated with multiple traits were detected through multi-trait QTL analysis approach. Multi-trait QTL on 2B.1 and 6A.2 were associated with the highest number of traits suggesting essential function of these genomic regions. The colocation observed in multi-environment QTL analysis and the results of multi-trait agreed in part with the genetic correlation observed in this study. Beyond QTL discovery, we defined both positive and negative marker-to-marker and marker-to-trait influences detected through joint analysis of pleiotropy and epistasis. Grain yield was enhanced by markers on chromosome 2B.1 and 5B with favorable alleles from TAM 111 but was repressed by several markers on 3B.1, 6D.2, and 7A.1 with favorable alleles from CO9602932. Other traits showed similar patterns of interactions. The results from combined analysis of pleiotropy and epistasis together with the standard QTL analysis provides a platform to build a set of QTL with enhancing effects on the trait of interest. The information could be useful in identification of markers for validation and subsequent deployment to improve grain yield as well as other traits in wheat.
Supporting information
(DOCX)
(DOCX)
(DOCX)
(XLSX)
(XLSX)
(XLSX)
Only QTL with significant (P < 0.05) additive effect are shown.
(XLSX)
Only QTL with significant additive effect are shown.
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
We thank all the staff at USDAARS genotyping center in Fargo, ND for their help in genotyping. We are grateful to the team that helped in phenotyping at KSU in Hays, KS; University of Idaho in Aberdeen, ID; and Colorado State University in Walsh, CO. We acknowledge the help of Anna Tyler from The Jackson Laboratory, Bar Harbor, ME for her help in CAPE analysis. We also acknowledge the help from undergraduate students namely Ashley Holmes, Jay Martin, Cody Shachter, and Brad Parker at Texas A&M AgriLife Research, Amarillo, Texas and graduate student Bharath Reddy from Department of Soil and Crop Sciences of TAMU at College Station.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by Monsanto Beachell-Borlaug International Scholars (MBBIS) program, Texas Wheat Producers Board, and Texas A&M AgriLife Research, National Institute of Food and Agriculture (US), funding number 2011-68002-30029 to Shuyu Liu; National Institute of Food and Agriculture (US), funding number 2017-67007-25939 to Shuyu Liu. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Gupta PK, Langridge P, Mir RR. Marker-assisted wheat breeding: present status and future possibilities. Molecular Breeding. 2010;26(2):145–61. doi: 10.1007/s11032-009-9359-7 [Google Scholar]
- 2.Würschum T, Langer SM, Longin CFH, Tucker MR, Leiser WL. A modern Green Revolution gene for reduced height in wheat. The Plant Journal. 2017. doi: 10.1111/tpj.13726 [DOI] [PubMed] [Google Scholar]
- 3.He X, Singh PK, Dreisigacker S, Singh S, Lillemo M, Duveiller E. Dwarfing genes RhtB1b and RhtD1b are associated with both type I FHB susceptibility and low anther extrusion in two bread wheat populations. PLoS ONE. 2016;11(9):e0162499 doi: 10.1371/journal.pone.0162499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Würschum T, Langer SM, Longin CFH. Genetic control of plant height in European winter wheat cultivars. Theoretical and Applied Genetics. 2015;128(5):865–74. doi: 10.1007/s00122-015-2476-2 [DOI] [PubMed] [Google Scholar]
- 5.Assanga S, Zhang G, Tan C-T, Rudd JC, Ibrahim A, Xue Q, et al. Saturated genetic mapping of wheat streak mosaic virus resistance gene Wsm2 in wheat. Crop Sci. 2017;57(1):332–9. doi: 10.2135/cropsci2016.04.0233 [Google Scholar]
- 6.Tan C-T, Assanga S, Zhang G, Rudd JC, Haley SD, Xue Q, et al. Development and validation of KASP markers for wheat streak mosaic virus resistance gene Wsm2. Crop Science. 2017;57(1):340–9. doi: 10.2135/cropsci2016.04.0234 [Google Scholar]
- 7.Liu S, Rudd JC, Guihua B, Haley SD, Ibrahim AMH, Qingwu X, et al. Molecular markers linked to important genes in hard winter wheat. Crop Sci. 2014;54(4):1304–21. doi: 10.2135/cropsci2013.08.0564 . [Google Scholar]
- 8.Liu Y, He Z, Appels R, Xia X. Functional markers in wheat: current status and future prospects. Theoretical and Applied Genetics. 2012;125(1):1–10. doi: 10.1007/s00122-012-1829-3 [DOI] [PubMed] [Google Scholar]
- 9.Semagn K, Babu R, Hearne S, Olsen M. Single nucleotide polymorphism genotyping using Kompetitive Allele Specific PCR (KASP): overview of the technology and its application in crop improvement. Mol Breeding. 2014;33(1):1–14. doi: 10.1007/s11032-013-9917-x. [Google Scholar]
- 10.Malosetti M, Ribaut J-M, van Eeuwijk FA. The statistical analysis of multi-environment data: modeling genotype-by-environment interaction and its genetic basis. Frontiers in Physiol. 2013;4:1–17. doi: 10.3389/fphys.2013.00044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith AB, Cullis BR, Thompson R. Analysis of crop cultivar breeding and evaluation trials: an overview of current mixed model approaches. J Agr Sci. 2005;143:449–62. [Google Scholar]
- 12.Boer MP, Wright D, Feng L, Podlich DW, Cooper M, van Eeuwijk FA. Mixed-model quantitative trait loci (QTL) analysis for multiple-environment trial data using environmental covariables for QTL-by-environment interactions, with an example in maize. Genetics. 2007;177(3):1801–13. doi: 10.1534/genetics.107.071068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tyler AL, Lu W, Hendrick JJ, Philip VM, Carter GW. CAPE: an R package for combined analysis of pleiotropy and epistasis. Plos Comput Biol. 2013;9(10):e1003270 doi: 10.1371/journal.pcbi.1003270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Ooijen JW. MapQTL 6, Software for the mapping of quantita tive trait loci in experimental populations of diploid species Kyazma B.V., Wageningen, Netherlands; 2009. [Google Scholar]
- 15.VSN International. GenStat for Windows 18th edition VSN International, Hemel Hempstead, UK: www.GenStat.co.uk. 2015. [Google Scholar]
- 16.Yang J, Hu H, Xia Z, Zhu J, Hu C, Yu R, et al. QTLNetwork: Mapping and visualizing genetic architecture of complex traits in experimental populations. Bioinformatics. 2008;24(5):721–3. doi: 10.1093/bioinformatics/btm494 [DOI] [PubMed] [Google Scholar]
- 17.Haley SD, Martin TJ, Quick JS, Seifers DL, Stromberger JA, Clayshulte SR, et al. Registration of CO960293-2 wheat germplasm resistant to wheat streak mosaic virus and Russian wheat aphid. Crop Sci. 2002;42(4):1381–2. doi: 10.2135/cropsci2002.1381 [Google Scholar]
- 18.Lazar MD, Worrall WD, Peterson GL, Fritz AK, Marshall D, Nelson LR, et al. Registration of 'TAM 111' wheat. Crop Sci. 2004;44(1):355–6. doi: 10.2135/cropsci2004.3550 [Google Scholar]
- 19.Xue Q, Rudd J, Liu S, Jessep K, Devkota R, Mahan J. Yield determination and water-use efficiency of wheat under water-limited conditions in the US Southern High Plains. Crop Sci. 2014;54(1):34–47. [Google Scholar]
- 20.Gregorio A, López M, Vargas M, Pacheco Á, Rodríguez F, Burgueño J, et al. META-R (Multi Environment Trial Analysis with R for Windows) Version 5.0, http://hdl.handle.net/11529/10201 International Maize and Wheat Improvement Center (CIMMYT). 2015.
- 21.Fehr WR, Fehr EL, Jessen HJ. Principles of cultivar development: New York: Macmillan; London: Collier Macmillan, c1987.; 1987. [Google Scholar]
- 22.Doyle JJ. Isolation of plant DNA from fresh tissue. Focus. 1990;12:13–5. [Google Scholar]
- 23.Liu S, Griffey C, Hall M, McKendry A, Chen J, Brooks W, et al. Molecular characterization of field resistance to Fusarium head blight in two US soft red winter wheat cultivars. Theoret Appl Genetics. 2013;126(10):2485–98. doi: 10.1007/s00122-013-2149-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu S, Assanga SO, Dhakal S, Gu X, Tan C-T, Yang Y, et al. Validation of chromosomal locations of 90K array single nucleotide polymorphisms in US wheat. Crop Sci. 2016;56(1):364–73. doi: 10.2135/cropsci2015.03.0194 [Google Scholar]
- 25.Van Ooijen JW. 2006. JoinMap ® 4, Software for the calculation of genetic linkage maps in experimental populations Kyazma B.V., Wageningen, Netherlands: 2006. [Google Scholar]
- 26.Kosambi DD. The esimation of map distances from recombination values. Ann Hum Genet. 1943;12(1):172–5. [Google Scholar]
- 27.Cavanagh C, Chao S, Wang S, Huang B, Stephen S, Kiani S, et al. Genome-wide comparative diversity uncovers multiple targets of selection for improvement in hexaploid wheat landraces and cultivars. Proc Nat Acad Sci. 2013;110(20):8057–62. doi: 10.1073/pnas.1217133110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang S, Wong D, Forrest K, Allen A, Chao S, Huang BE, et al. Characterization of polyploid wheat genomic diversity using a high-density 90 000 single nucleotide polymorphism array. Plant Biotechnol J. 2014;12(6):787–96. doi: 10.1111/pbi.12183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van Eeuwijk FA, Bink MCAM, Chenu K, Chapman SC. Detection and use of QTL for complex traits in multiple environments. Current opinion in plant biology. 2010;13(2):193–205. doi: 10.1016/j.pbi.2010.01.001 [DOI] [PubMed] [Google Scholar]
- 30.Bernardo R. Genomewide markers as cofactors for precision mapping of quantitative trait loci. Theor Appl Genet. 2013;126(4):999–1009. doi: 10.1007/s00122-012-2032-2 [DOI] [PubMed] [Google Scholar]
- 31.Zeng Z. Precision mapping of quantitative trait loci. Genetics. 1994;136:1457–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jansen RC, Stam P. High resolution of quantitative traits into multiple loci via interval mapping. Genetics. 1994;136(4):1447–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.R Core Team. R: A language and environment for statistical computing R foundation for statistical computing, Vienna, Austria: https://www.R-project.org/ 2015. [Google Scholar]
- 34.Tyler LA, Wei L, Justin JH, Vivek MP, Greg WC. cape: Combined analysis of pleiotropy and epistasis. R package version 1.3. http://CRAN.R-project.org/package=cape. 2014. [Google Scholar]
- 35.Escobar-Sepúlveda HF, Trejo-Téllez LI, Pérez-Rodríguez P, Hidalgo-Contreras JV, Gómez-Merino FC. Diacylglycerol kinases are widespread in higher plants and display inducible gene expression in response to beneficial elements, metal, and metalloid ions. Frontiers in Plant Science. 2017;8:129 doi: 10.3389/fpls.2017.00129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hossain MA, Nakano R, Nakamura K, Hossain MT, Kimura Y. Molecular characterization of plant acidic alpha-mannosidase, a member of glycosylhydrolase family 38, involved in the turnover of N-glycans during tomato fruit ripening. Journal of biochemistry. 2010;148(5):603–16. Epub 2010/08/28. doi: 10.1093/jb/mvq094 . [DOI] [PubMed] [Google Scholar]
- 37.Strasser R. Biological significance of complex N-glycans in plants and their impact on plant physiology. Frontiers in Plant Science. 2014;5:363 doi: 10.3389/fpls.2014.00363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li J, Ji L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity. 2005;95(3):221–7. doi: 10.1038/sj.hdy.6800717 [DOI] [PubMed] [Google Scholar]
- 39.Carter GW, Hays M, Sherman A, Galitski T. Use of pleiotropy to model genetic interactions in a population. Plos Genetics. 2012;8(10):e1003010 doi: 10.1371/journal.pgen.1003010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tyler AL, McGarr TC, Beyer BJ, Frankel WN, Carter GW. A genetic interaction network model of a complex neurological disease. Genes, Brain, and Behavior. 2014;13(8):831–40. doi: 10.1111/gbb.12178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu J, Wang Q, Liu S, Huang S, Mu J, Zeng Q, et al. Saturation mapping of a major effect QTL for stripe rust resistance on wheat chromosome 2B in cultivar Napo 63 using SNP genotyping arrays. Frontiers in Plant Science. 2017;8(653). doi: 10.3389/fpls.2017.00653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jin H, Wen W, Liu J, Zhai S, Zhang Y, Yan J, et al. Genome-wide QTL mapping for wheat processing quality parameters in a Gaocheng 8901/Zhoumai 16 recombinant inbred line population. Frontiers in Plant Science. 2016;7:1032 doi: 10.3389/fpls.2016.01032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gao F, Wen W, Liu J, Rasheed A, Yin G, Xia X, et al. Genome-wide linkage mapping of QTL for yield components, plant height and yield-related physiological traits in the Chinese wheat cross Zhou 8425B/Chinese spring. Frontiers in Plant Science. 2015;6:1099 doi: 10.3389/fpls.2015.01099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhai H, Feng Z, Li J, Liu X, Xiao S, Ni Z, et al. QTL analysis of spike morphological traits and plant height in winter wheat (Triticum aestivum L.) using a high-density SNP and SSR-based linkage map. Frontiers in Plant Science. 2016;7(1617). doi: 10.3389/fpls.2016.01617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tian X, Wen W, Xie L, Fu L, Xu D, Fu C, et al. Molecular mapping of reduced plant height gene Rht24 in bread wheat. Frontiers in Plant Science. 2017;8:1379 doi: 10.3389/fpls.2017.01379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carlborg O, Haley CS. Epistasis: too often neglected in complex trait studies? Nat Rev Genet. 2004;5(8):618–25. doi: 10.1038/nrg1407 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
(XLSX)
(XLSX)
(XLSX)
Only QTL with significant (P < 0.05) additive effect are shown.
(XLSX)
Only QTL with significant additive effect are shown.
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.