Abstract
The maintenance of normal heart function requires proper control of protein turnover. The ubiquitin-proteasome system is a principal regulator of protein degradation. Mdm2 is the main E3 ubiquitin ligase for p53 in mitotic cells thereby regulating cellular growth, DNA repair, oxidative stress and apoptosis. However, which of these Mdm2-related activities are preserved in differentiated cardiomyocytes has yet to be determined. We sought to elucidate the role of Mdm2 in the control of normal heart function. We observed markedly reduced Mdm2 mRNA levels accompanied by highly elevated p53 protein expression in the hearts of wild type mice subjected to myocardial infarction or trans-aortic banding. Accordingly, we generated conditional cardiac-specific Mdm2 gene knockout (Mdm2f/f;mcm) mice. In adulthood, Mdm2f/f;mcm mice developed spontaneous cardiac hypertrophy, left ventricular dysfunction with early mortality post-tamoxifen. A decreased polyubiquitination of myocardial p53 was observed, leading to its stabilization and activation, in the absence of acute stress. In addition, transcriptomic analysis of Mdm2-deficient hearts revealed that there is an induction of E2f1 and c-Myc mRNA levels with reduced expression of the Pgc-1a/Ppara/Esrrb/g axis and Pink1. This was associated with a significant degree of cardiomyocyte apoptosis, and an inhibition of redox homeostasis and mitochondrial bioenergetics. All these processes are early, Mdm2-associated events and contribute to the development of pathological hypertrophy. Our genetic and biochemical data support a role for Mdm2 in cardiac growth control through the regulation of p53, the Pgc-1 family of transcriptional coactivators and the pivotal antioxidant Pink1.
Introduction
Heart failure occurs when the heart is unable to adequately pump blood to meet the demands of the body at normal filling pressures. It is a clinical syndrome that is defined by symptoms of fatigue, shortness of breath and fluid retention [1–5] and is the leading cause of morbidity and mortality in North America. Heart failure has been identified as the second leading cause of extended hospital stays rendering this disease an ever-increasing socioeconomic burden [6–8]. The quality of life and the prognosis for this group of patients remains poor with one-year survival rates less than 40% [9].
In mammals, adult cardiomyocytes are terminally-differentiated cells with a rather limited capacity to proliferate to any appreciable extent for cardiac regeneration after injury [10–14]. This renders the heart particularly vulnerable to ischemic, toxic or biomechanical injury. To compensate for the irreversible damage, the left ventricle undergoes hypertrophy and remodeling. In mice, hypertrophic growth of cardiomyocytes is a result in reprogramming of fetal genes [15–17]. Although this response is considered initially compensatory, cardiomyocyte hypertrophy eventually leads to alterations in myocyte gene expression, metabolism, excitation contraction coupling, accompanied with enhanced apoptosis and fibrosis which is ultimately detrimental and progressive left ventricular dysfunction develops [18–22].
Cardiac remodeling is associated with protein synthesis and degradation. While elevated protein synthesis is a well-established process in cardiac hypertrophy, the link between proteasome-mediated protein degradation and cardiac hypertrophy is less well understood [23–26]. The Ubiquitin-Proteasome System (UPS) represents the major protein degradation pathway regulating a multitude of signaling pathways [27]. However, while inhibition of proteasomal function has proven beneficial towards maintaining cardiac homeostasis in vivo, this approach has suffered poor clinical translation [28–30]. Currently there is a great impetus to utilize downstream specificity of the UPS via the E3 ubiquitin ligases that ubiquitinate specific substrate proteins leading to their degradation. Indeed, harnessing E3 ubiquitin ligase specificity to regulate key factors governing cardiac homeostasis and hypertrophy holds potential for therapeutic intervention.
The E3 ubiquitin ligase Mdm2 is ubiquitously expressed as a multi-domain member of the RING finger domain family that attaches mono- and/or poly-ubiquitin chains onto its target substrates [31]. Mdm2 targets many proteins regulating pleiotropic biological processes including cell growth, proliferation and apoptosis, such as the tumor suppressors Trp53 (p53), retinoblastoma (Rb1) or the transcription factor E2f1 and targets them for proteasomal degradation [32–34].
The p53/Mdm2 pathway is one of the best characterized circuitries [35,36] that translates growth and survival signals into specific gene expression patterns, regulating tumor-free survival of an organism [37–39]. In normal cells, p53 expression is kept at low levels by Mdm2, through proteasomal degradation. In response to acute stress, Mdm2 is inactivated and increased p53 levels block cell division and induce apoptosis. Conversely, p53 can activate Mdm2 transcription, thereby forming a negative feedback loop that curtails p53 activity. Mdm2 is required for regulation of p53 in vivo since Mdm2 knockout mice die early in embryogenesis due to p53-dependent apoptosis [40].
Regulation of growth and apoptosis in mitotic cells and tissues by the p53/Mdm2 circuitry has been well established [41–43]. Intriguingly, Akt-dependent phosphorylation of Mdm2 increases its ubiquitination activity which results in downregulation of p53 thereby enhancing cellular survival and proliferation [44]. However, apart from the analysis of Mdm2 in transgenic mice or germline knockout models [45–47], the precise physiological function of Mdm2 in the adult heart has yet to be determined. Therefore, we recognized the importance of investigating the potential role of Mdm2 as a regulator of growth regulated processes.
In this study, we report significant decreases in Mdm2 mRNA levels in samples obtained from patients with end-stage heart failure. We also describe Mdm2 and p53 transcript and protein expression levels in murine models of heart failure, and evaluate the alterations in cardiac function, energy metabolism and reactive oxygen species (ROS) defense in the presence or absence of Mdm2 in the adult mouse heart. We show that p53 is a specific Mdm2 target and that ablation of Mdm2 leads to activation of p53 in the absence of acute stress. The loss of Mdm2 severely impacts redox homeostasis and mitochondrial function through activation of the transcription factors c-Myc and E2f1, both known for their ability to induce apoptosis and inhibit antioxidative systems. Under these conditions, negative regulation of the Pgc-1a/Ppar/Esrr axis and Pink1 evoked oxidative stress, energy deprivation and cardiac dysfunction. All these processes are Mdm2-dependent events, and can contribute to the development of pathological hypertrophy, left ventricular dysfunction and premature mortality. Our data identify Mdm2 as a regulator of cardiac homeostasis through the persistent upregulation of ROS-detoxifying systems and mitochondrial energy metabolism.
Materials and methods
Cardiac-specific Mdm2f/f;mcm conditional mutant mice
To determine if Tam and Cre expression is toxic to cardiomyocytes we included vehicle- and Tam-injected wild type C57BL/6J, MerCreMer (mcm) and Mdm2f/f mice in all initial analyses of the corresponding mutants. We found that mice of these experimental groups were phenotypically indistinguishable from vehicle-injected Mdm2f/f;mcm control animals used in this study, as judged by heart body weight ratios, ventricular fibrosis and fractional shortening. All animal usage in this study was in accordance with approved institutional animal care guidelines of the UHN (AUP 1815/1379, Canadian Council in Animal Care). All animals used in this study were 10 weeks old (22-26g) at the beginning of experimentation. All experiments used isogenic controls of matched age and sex.
The Mdm2f/f (01XH9) and p53f/f mice (strain number 01XC2) mice were obtained from Frederick (Rockville, MD 20852 USA). These strains were previously backcrossed into a C57BL/6J background for 7 generations. The mcm strain (Tg(Myh6-cre/Esr1*)1Jmk/J) was from Jackson (Bar Harbor, ME 04609 USA). In these mice, the cardiac muscle a-myosin heavy chain 6 promoter drives the expression of Cre (c) recombinase fused to two mutant (m) estrogen-receptor ligand-binding domains (mcm) when exposed to Tam [48]. We crossed mcm transgenic mice on a C57BL/6J background with mice carrying the conditional alleles Mdm2f/f to obtain Mdm2f/f;mcm animals. We crossed mcm transgenic mice on a C57BL/6J background with mice carrying the conditional alleles p53f/f to obtain p53f/f;mcm animals. We crossed the heterozygous offspring of p53f/f;mcm and Mdm2f/f;mcm breeders to obtain Mdm2f/f;p53f/f;mcm animals. The Mdm2f/+;mcm strain was obtained by crossing Mdm2f/f;mcm mice with wild type (C57BL/6J) animals. We obtained Mdm2f/f;p53f/+;mcm mice by breeding Mdm2f/f;p53f/f;mcm animals with Mdm2f/f;mcm mice. After weaning, experimental male mice were housed in groups of 3–5 animals in mechanically ventilated cages (600 cm2; changed fortnightly) environmentally enriched by bedding and nesting materials and crawl tubes. Animals were held in a temperature-controlled environment at 19–22°C on a diurnal 12h light cycle. Mice were provided free access to standard non-medicated pelleted laboratory rodent chow (Harlan) and tap water ad libitum from a portable water faucet.
DNA isolated from fresh tail snips was employed for genotyping. Samples were incubated in 300ul of 50 mM NaOH for 2h, 80°C while rocking. Then, samples were neutralized with 25 ul 1.0 M HCl, 700 ul H2O, vigorously vortexed, centrifuged for 13,000 rpm for 10 min and stored at 4°C. We used 0.5 ul of DNA solution per PCR reaction and the following primers: Mdm2 forward 5’-CTGTGTGAGCTGAGGGAGATGTG-3’; Mdm2 reverse 5’-CCTGGATTTAATCTGCAGC ACTC-3’. p53 forward 5’-CACAAAAACAGGTTAAACCCAG-3’; p53 reverse 5’-AGCACAT AGGAGGCAGAGAC-3’. Mcm forward 5’-AGGTGGACCTGATCATGGAG-3’; mcm reverse 5’-ATACCGGAGATCATGCAAGC-3. We performed PCR analysis with Quanta Accustart Geltrac with GelDye (no. 95136-04K; VWR) and Platinum Blue Supermix (no. 12580–023; Invitrogen): Mdm2 wild-type allele, 310 bp; Mdm2 floxed allele, 397 bp; mcm 440 bp. p53 wild type allele, 288 bp. p53 floxed allele 370 bp. Genomic recombination efficiency at the Mdm2 gene locus was measured by PCR employing the following primers spanning exon 7 to 9: forward 5’-CCTCCCCCACGTCTACTAAA-3’; reverse 5’-TGTTTTGACAGCAGGTCAGG -3’. Amplicon size: 590 bp.
An ethanol-peanut oil emulsion of 4-Hydroxytamoxifen (Tam; H6278, Sigma-Aldrich) was prepared by completely dissolving 100 mg Tam in 5 ml highly purified ethanol (ACS reagent grade, anhydrous, absolute; no. 6590–32; Ricca Chemical, Fisher) while vortexing vigorously for 5–8 min. Peanut oil (32 ml) (P2144; Sigma-Aldrich) was added and the emulsion was again vigorously vortexed for 2 min. Then, the emulsion was sonicated on ice at highest output for 10–30 sec until it became translucent, aliquoted, and stored at -20°C for several months. Shortly before usage, Tam was melted in a 37°C water bath, briefly vortexed, and 200–250 ul were immediately injected intraperitoneally into conscious mice. Animals were injected daily between 5–6 pm for four consecutive days (98–83 ug/g body weight cumulative dosage). Homologous recombination was completed at 7 days post-Tam. Vehicle control mice were intraperitoneally injected with an ethanol-peanut oil emulsion lacking Tam.
For 5-bromo-2´-deoxyuridine (BrdU) (B5002; Sigma-Aldrich) labeling in vivo, BrdU was dissolved in PBS (10 mg/ml), aliquoted and stored at -20°C. Conscious mice were intraperitoneally injected with 200 ul BrdU (cumulative dosage of 160 mg/kg) twice daily at 6d and 7d post-Tam, and sacrificed at 16 hours after the last injection. The membrane permeable proteasome inhibitor MG132 as ready-made solution in DMSO was purchased from Sigma (M7449). At 7 days post-Tam, mice were injected with MG132 (30 umol/kg body weight) and sacrificed 6 hours later [49,50].
Microarray analysis
All microarray data were submitted to the ArrayExpress database (Experiment ArrayExpress accession: E-MTAB-5441; http://www.ebi.ac.uk/arrayexpress).
Total RNA from mouse LV tissues was isolated with Trizol reagent (no. 15596026; Thermo Fisher Scientific). Phase lock Gels (no. 826754; VWR) were employed to eliminate interphase-protein contaminations. RNA quality was assessed by 260/280 and 260/230 absorption ratios employing a Nanodrop spectrophotometer (NanoDrop; Thermo Fisher Scientific), and an Agilent Bioanalyzer at the Microarray Facility, Centre for Applied Genomics, The Hospital for Sick Children (Toronto). RNA samples were processed for analysis by Affymetrix Mouse Gene 1.0 ST expression arrays at the Centre for Applied Genomics. Processing of probe level data and all subsequent analyses were performed using GeneSpring (Version 13.2; Agilent Technologies Inc.). Genome wide data from the gene expression microarrays were normalized and filtered for genes with significant expression levels (log2 fold change ±1.3; P < 0.05) in Tam-treated Mdm2f/f;mcm compared with vehicle injected control Mdm2f/f;mcm (n = 3) employing GeneSpring. Selection of top-ranked differentially expressed genes in the GO gene sets was performed on the bases of arbitrary threshold for fold changes plus statistical significance according to the t test with Benjamini-Hochberg correction (log2 fold change ±1.3; P < 0.01).
Statistical analyses
Data are means±s.e.m. as indicated at the bottom of each Figure legend. We used factorial design analysis of variance (ANOVA) or t-tests to analyze data as appropriate employing GraphPad InStat (version 3.1) and GraphPad Prism (GraphPad Software, version 7.0; La Jolla, CA 92037 USA). Significant ANOVA values were subsequently subjected to simple main effects analyses or post hoc comparisons of individual means using the Tukey method as appropriate. We considered P values of < 0.05 as significant.
Myocardial infarction, transaortic banding and echocardiography
For myocardial infarction, mice were subjected to ligation of the left anterior descending artery at the level of the left atrial appendage or sham operation. For transaortic banding (TAB), mice were anesthetized using 2% (vol/vol) isoflurane/95% (vol/vol) oxygen and ventilated. A left thoracotomy was performed and the pericardium opened. The transverse aortic arch was ligated using a 30-gauge needle with an overlying suture. Sham mice underwent a comparable operation in which a suture was passed without ligation.
Echocardiography on anesthetized mice (2.0% isoflurane, 98% oxygen) was performed using a 15-MHz linear ultrasound transducer (Vivid7; GE). Body temperature was maintained at 37°C. M-mode measurements of the LV end-diastolic diameter (LVEDD) and LV end-systolic diameter (LVESD) were made in triplicate from short-axis views at the level of the papillary muscle and averaged over three to six beats. LVEDD was measured at the time of the apparent maximal LV diastolic dimension, whereas LVESD was measured at the time of the most anterior systolic excursion of the posterior wall. LV fractional shortening (FS) was calculated as follows: FS = (LVEDD − LVESD)/LVEDD × 100%.
Immunofluorescence microscopy, morphometric analyses, apoptosis and DNA replication assays
Mice were euthanized without anesthesia by cervical dislocation between 9-11am. Anticoagulant was not administered. Hearts were quickly excised and rinsed in 20 ml ice-cold PBS, pH 7.4 without Ca2+/Mg2+ (no. 10010023; Thermo Fisher Scientific), fixed in 4% PBS-buffered formalin (10 ml) for 50 min at room temperature with constant agitation and incubated in 0.3 M glycine in PBS (pH 7.4; 10 ml) at 4°C for 3 to 5 days. After embedding hearts in Tissue-Tek OCT Compound (Sakura, Finetek; VWR) sections were cut at 10 um thickness using a cryostat (HM525 NX; Thermo Fisher Scientific), and mounted on histological slides (Superfrost Plus Microslides; no. 48311–703; VWR). For BrdU (5-Bromo-2′-deoxyuridine) labeling experiments, DNA was denatured in 2N HCl for 30 min post-fixation. Specimen were then permeabilized and stained with antibodies to cardiac-specific nuclear Mef2a and BrdU. Detection of fragmented genomic DNA was performed by terminal deoxynucleotidyl transferase-mediated dUTP nick-end-labeling (TUNEL) according to the manufacturer’s instructions (no. 1684795910; Roche). These samples were co-stained with cardiac-specific anti-sarcomeric a-actinin after permeabilization. Specimen were permeabilized in 1.0% Triton X-100 (X100; Sigma-Aldrich) in TBS (20 mM Tris, 150 mM NaCl), pH 7.6 for 60 minutes at room temperature. Samples were incubated with primary antibodies (listed in Table 1 in TBS/1.0% Triton X-100, for 16–20 hours at room temperature without agitation. Thereafter, specimen were briefly rinsed with TBS. For visualization, samples were incubated with secondary antibodies, diluted 150-fold in TBS/1.0% Triton X-100 for 45 min in the dark (Alexa Fluor 488-goat anti-rat IgG, A-11006; Alexa Fluor 555-goat anti-rabbit IgG, A-21429; Alexa Fluor 555-goat anti-mouse IgG, A-2142; Alexa Fluor 647-goat anti-mouse IgG, A-20990 (Thermo Fisher Scientific). Specimen were briefly rinsed with TBS, and nuclear DNA was visualized with 4,6-diamidino-2-phenylindole (Dapi; 1.0 ug/ml in PBS) (D9542; Sigma-Aldrich). ProLong Diamond antifade reagent (no. 36965; Thermo Fisher Scientific) was applied, and samples were sealed with a coverslip. Three-dimensional confocal laser scanning microscopy was performed on a Zeiss LSM700 confocal microscope and LSM Zen 2009 data acquisition software (AOMF-Advanced Optical Microcopy Facility, Ontario Cancer Institute, Toronto, ON Canada). For determination of cardiomyocyte cell size and ventricular remodeling, ventricular samples were stained with sarcomeric a-actinin and Alexa Fluor 488-conjugated wheat germ agglutinin (WGA) (W11261; Thermo Fisher Scientific). Cross-dimensions of adult cardiomyocytes and fibrotic area were determined by planimetry of immunofluorescence microphotographs using ImageJ (Version 1.51d; National Institutes of Health, Bethesda, MD, https://imagej.nih.gov/ij/). After recording, simple adjustments and assembly of entire and cropped microphotographs were performed employing Adobe Photoshop CS6.
Table 1. Antibodies employed for IF, IP and WBa.
| Gene name | Catalog No. | Vendor | Application | Dilution Factor |
|---|---|---|---|---|
| Aco2 | 6922 | Cell Signaling | WB | 1000 |
| a-actinin, sarcomeric | A7811 | Sigma | IF | 50 |
| Atp5a | ab14748 | Abcam | WB | 1000 |
| Bak | 3184 | Cell Signaling | WB | 1000 |
| Bax | 2772 | Cell Signaling | WB | 1000 |
| BrdU | MCA2060 | AbD Serotec | IF | 50 |
| Cat | 14097 | Cell Signaling | WB | 1000 |
| Complex I | ab109798 | Abcam | ELISA | 5 ug |
| Cox4i1 | ab16056 | Abcam | WB | 1000 |
| Cx43 | CBL171 | Sigma | WB | 1000 |
| CytC | ab13575 | Abcam | WB | 1000 |
| E2f1 | sc-193 | Santa Cruz | WB | 500 |
| Erk1/2 | 9102 | Cell Signaling | WB | 1000 |
| Erk1/2 Pi-Thr202/Tyr204 | 9101 | Cell Signaling | WB | 1000 |
| Esrra/b | NBP1-47254 | Novus | WB | 1000 |
| Esrrg | 27–388 | ProScience | WB | 1000 |
| Gpx4 | Ab125066 | Abcam | WB | 1000 |
| Gsta1 | sc-100546 | Santa Cruz | WB | 500 |
| Lam | L9393 | Sigma | WB | 1000 |
| Mdm2 | M8558 | Sigma | WB | 1000 |
| Mef2a | ab32866 | Abcam | IF | 50 |
| Myc | sc-764 | Santa Cruz | WB | 500 |
| Ndufa8 | BS3336 | BioWorld | WB | 1000 |
| normal rabbit IgG | 2729 | Cell Signaling | IP | 7.5 ug—1000 |
| Npm1 | B0556 | Sigma | WB | 1000 |
| Opa1 | ab42364 | Abcam | WB | 1000 |
| p38 | 9112 | Cell Signaling | WB | 1000 |
| p38 Pi-Thr180/Tyr182 | 9211 | Cell Signaling | WB | 1000 |
| p53 | BML-SA293 | Enzo Life Sci. | IP WB | 1000 |
| Pgc—1a | 3934–100 | BioVision | WB | 1000 |
| Pink1 | 3929–100 | BioVision | WB | 1000 |
| Pln | 8495 | Cell Signaling | WB | 1000 |
| Ppara | PAB11321 | Abnova | WB | 1000 |
| Sdhb | ab14714 | Abcam | WB | 1000 |
| SERCA2 | 9580 | Cell Signaling | WB | 1000 |
| Sirt3 | 5490 | Cell Signaling | WB | 1000 |
| Sod2 | 611580 | Becton Dickinson | WB | 4000 |
| Txnrd2 | ab16841 | Abcam | WB | 1000 |
| ubiquitin | 13–1600 | Thermo Fisher | WB | 7.5 ug |
aIF, immunofluorescence microscopy. IP, immunoprecipitation. WB, Western blot.
Cell fractionation, total heart tissue extracts and isolation of mitochondria
Subcellular cell fractions were prepared using the NE-PER Kit (no. 78833; Pierce) supplemented with phosphatase inhibitors (1.0 mM Na3VO4, 20 mM NaF, 10 mM b-glycerophosphate; Sigma) and with a protease inhibitor cocktail containing AEBSF, Aprotinin, Bestatin, E64, Leupeptin and Pepstatin (no. P2714; Sigma). Ventricular specimen were mechanically homogenized in RIPA Buffer (no. 9806; Cell Signaling) composed of 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1.0 mM EGTA, 1.0% NP-40, 1.0% sodium deoxy cholate, 2.5 mM sodium pyrophosphate, 1.0 mM b-glycerophosphate, 1.0 mM Na3VO4, 1.0 mg/ml leupeptin, phosphatase and protease inhibitors. Mechanochemical assisted isolation of mitochondria from adult mouse ventricular tissues was performed using extraction buffer A (10 mM HEPES, pH 7.5; 200 mM mannitol; 70 mM sucrose; 1.0 mM EGTA) supplied with the Mitochondrial Isolation Kit (MITOISO1; Sigma) according to the manufacturer’s instructions. Isolated mitochondria derived from one heart were resuspended in 200 ul storage buffer (10 mM HEPES, pH 7.5, containing 250 mM sucrose, 1.0 mM ATP, 80 uM ADP, 5.0 mM sodium succinate, 2.0 mM K2HPO4, 1.0 mM DTT) (MITOISO1; Sigma), and used directly in mitochondrial complex activity assays, or aliquoted and stored at -80°C for Western blot analysis. The protein concentration was approximately 3.0–4.0 ug/ul as determined by fluorometry (Qubit 2.0 Fluorometer; no. Q33217; Thermo Fisher Scientific).
Mitochondrial Complex I function
The relative activity of mitochondrial Complex I was determined employing the Complex I Enzyme Activity Assay kit (MS141; Abcam), according to the manufacturer’s procedures except that the supplied 96-well plates were re-coated with antibodies to Complex I or IV (5.0 ug/well) and freshly purified mitochondria were used (50 ul/well). The Complex I kit determines the diaphorase-type activity of Complex I. This activity is not dependent on the presence of ubiquinone and, therefore, inhibitors, such as rotenone, do not inhibit Complex I activity in this assay. Importantly, the activity assay is affected by assembly deficiencies, and, thus enabled us to specifically determine the impact of decreased Ndufa8 levels on Complex I activity. Colorimetric increases in absorbance at 450 nm for Complex I assays were recorded at room temperature for 30 min at 2 min intervals using a spectrophotometer (FlexStation 3; Molecular Devices).
Immunoprecipitation assays and immunoblotting
For immunoprecipitation (IP) assays, cellular extracts in supplemented RIPA Buffer were incubated with antibodies to ubiquitin (7.5 ug) or normal rabbit IgG (7.5 ug) covalently linked to protein A agarose beads (Seize X Protein A IP Kit; no. 26149; Pierce) for 3 hours at 4°C. Immunocomplexes were washed twice with lysis buffer and boiled in SDS sample buffer (no. 7722; Cell Signaling). Protein samples were resolved by SDS-PAGE employing 4–12% and 3–8% NuPAGE pre-cast gels (Life Technologies), and PVDF membranes (iBlot; Life Technologies). The presence of ubiquitinated p53 in the immunoprecipitate was determined by immunoblotting with anti-ubiquitin antibodies. The following secondary antibodies were employed for chemiluminescence detection of proteins: horseradish peroxidase (HRP)-conjugated anti-rabbit IgG (no. 7074; Cell Signaling), HRP-conjugated anti-mouse IgG (no. 7076; Cell Signaling), and Luminata Crescendo (WBLUR0100, Millipore). The complete list of antibodies employed in our study can be found in Table 1.
Detection of oxidative damage, antioxidants levels, and metabolism assays
Individual ventricular specimen were rapidly cut into 4–6 pieces, briefly rinsed in 40 ml ice-cold PBS, immediately snap frozen in liquid nitrogen, and stored at -80°C. Assays for determination of small metabolic molecules (4-HAE, ATP, GSH/GSSG) were deproteinized prior to analysis using the perchlorate-based precipitation based Deproteinizing Sample Preparation Kit according to the supplier’s instruction (no. K808-200; BioVision). All assays were performed according to the manufacturer’s instructions with minor modifications: 4-HAE/MDA (Bioxytech LPO-586 kit; Oxis), 8-OHdG (Bioxytech 8-OHdG-EIA kit; Oxis), ATP (ENLITEN ATP Assay System Bioluminescence no. FF200; Promega), ADP (ADP-Glo Kinase Assay V6930; Promega), aconitase (Bioxytech Aconitase-340 kit; Oxis), caspase-3 and 7 (Caspase-Glo 3/7 Assay; G8090; Promega), caspase 8 (Caspase-Glo 8 Assay; G8200; Promega) catalase (Bioxytech Catalase-520 kit; Oxis), and GSH/GSSG (Bioxytech GSH/GSSG-412 kit; Oxis). Each sample was measured in duplicate. Final values were normalized by the total protein concentration for each sample determined prior to precipitation.
Determination of the mitochondrial membrane potential (ΔΨm)
The membrane permeable fluorescence dye JC-1 (no. T3168; Thermo Fisher Scientific) is a mitochondria membrane potential (MMP) probe that exhibits potential-dependent accumulation in mitochondria, indicated by a fluorescence emission shift from green (525 nm) to red (590 nm). This depolarization of MMP occurs at early stages of oxidative stress and cell death. This potential-sensitive emission shift from green to red is due to concentration-dependent formation of red fluorescent J aggregates, which in turn is dependent on MMP. Thus, decreases in MMP are measured by decreases in the intensity of emitted red fluorescence. Isolated mitochondria (10 ul/well) in HBSS (without Ca2+/Mg2+, phenol red no.14185-052; Thermo Fisher Scientific) were incubated with JC-1 (5.0 ug/ml in NADH-containing Complex I assay buffer; MS141, Abcam) for 1–2 min and then treated with antimycin. JC-1 emission at 525/595 nm was recorded (2 readings/min for 30 min) using a fluorescence spectrophotometer (Flex Station 3; Molecular Devices). The rate between two time points (emission at 595 nm/min) was calculated in the most linear range of decline for JC-1 emission. ROS levels were determined employing the 2’,7’-dichlorofluorescin diacetate (DCFDA) Cellular Reactive Oxygen Species Detection Assay Kit (ab113851; Abcam) according to the manufacturer’s instructions.
Mitochondrial biogenesis and capacity
Impaired mitochondrial (mt) biogenesis is a key marker of physiologic stress or genetic mutations. Mt have multiple circular genomes (mtDNA) that are replicated independently from the nuclear genome (nDNA). The mtDNA encodes 13 core polypeptides involved in oxidative phosphorylation in addition to 22 tRNA and 12S and 16S rRNA genes for mt protein synthesis. We used the relative mtDNA copy as a surrogate for mitochondrial biogenesis. Therefore, the mtDNA copy number was determined by qPCR of the mitochondrial gene cytochrome b (mtCytb) normalized to levels of a nuclear (n) encoded single-copy gene, B2m, using DNA preparations from total ventricular lysates. The ratio of mtDNA to nDNA in ventricle-injected hearts was arbitrarily set to 1. We used mitochondrial gene transcription as a surrogate for mitochondrial capacity. Therefore, the mRNA expression levels of the mtCytb gene were measured by RT-qPCR and corrected for the transcript expression of nuclear B2m. The ratio of transcript levels of mtCytb to B2m in ventricle-injected hearts was set to 1.
Reverse transcription and quantitative real time PCR assays
We carried out two-step reverse transcriptase (RT) and quantitative real-time polymerase chain reactions (qPCR) on a LightCycler 480 (Roche; TMDT Core Facility) for mRNA analysis. Total RNA from mouse and human ventricular cardiac specimen was isolated with Trizol reagent (no. 15596026; Thermo Fisher Scientific), and Phase-lock Gels (no. 826754; VWR) were employed to eliminate interphase-protein contaminations. We used 500 ng total RNA in a 20 ul reaction for first-strand cDNA synthesis employing the SensiFast cDNA synthesis kit (BIO-65053; Bioline). For qPCR, we employed 4.0 ul first-strand synthesis product, diluted 5-fold with MilliQ-grade water and the Quanta Accustart II PCR Supermix (no, 95136–04; VWR) with EvaGreen dye (no. 31000; VWR). Mouse qPCR primers [51] used in this study are summarized in Table 2. Relative quantification of transcript levels was performed using the ddCt method with normalization to beta-2 microglobulin (B2m) employing the data analysis module. Mdm2 transcript levels were determined in human samples with Taqman assays employing standard human gene probes for B2m (no. HS99999907_m1) and Mdm2 (HS01066930_m1; Thermo Fisher Scientific). Relative quantification of Mdm2 transcript levels was performed using the ddCt method with normalization to B2m employing the data analysis module.
Table 2. Primer sequences used in RT-qPCR assays.
| Gene Symbol | Forward | Reverse |
|---|---|---|
| Adra1a | CTAAGGCCATTCTACTTGGGGT | CGAGTGCAGATGCCGATGA |
| Adra1b | CGGACGCCAACCAACTACTT | AACACAGGACATCAACCGCTG |
| Adrb1 | CTCATCGTGGTGGGTAACGTG | ACACACAGCACATCTACCGAA |
| Adrb2 | GGGAACGACAGCGACTTCTT | GCCAGGACGATAACCGACAT |
| Ace2 | TCCAGACTCCGATCATCAAGC | GCTCATGGTGTTCAGAATTGTGT |
| ANP | GCTTCCAGGCCATATTGGAG | GGGGGCATGACCTCATCTT |
| Apaf1 | AGTGGCAAGGACACAGATGG | GGCTTCCGCAGCTAACACA |
| ATPaf1 | CCCCTTCTACGACCGCTAC | CCACTGGCTGCTTTCGGAA |
| ATP5a1 | TCTCCATGCCTCTAACACTCG | CCAGGTCAACAGACGTGTCAG |
| B2m | TTCTGGTGCTTGTCTCACTGA | CAGTATGTTCGGCTTCCCATTC |
| Bak1 | CAACCCCGAGATGGACAACTT | CGTAGCGCCGGTTAATATCAT |
| Bax | TGAAGACAGGGGCCTTTTTG | AATTCGCCGGAGACACTCG |
| BNP | GAGGTCACTCCTATCCTCTGG | GCCATTTCCTCCGACTTTTCTC |
| Casp1 | ACAAGGCACGGGACCTATG | TCCCAGTCAGTCCTGGAAATG |
| Casp3 | TGGTGATGAAGGGGTCATTTATG | TTCGGCTTTCCAGTCAGACTC |
| Casp8 | TGCTTGGACTACATCCCACAC | TGCAGTCTAGGAAGTTGACCA |
| Col1a1 | GCTCCTCTTAGGGGCCACT | CCACGTCTCACCATTGGGG |
| Col1a2 | GTAACTTCGTGCCTAGCAACA | CCTTTGTCAGAATACTGAGCAGC |
| Col4a1 | CTGGCACAAAAGGGACGAG | ACGTGGCCGAGAATTTCACC |
| Col4a2 | GACCGAGTGCGGTTCAAAG | CGCAGGGCACATCCAACTT |
| Col5a1 | CTTCGCCGCTACTCCTGTTC | CCCTGAGGGCAAATTGTGAAAA |
| Col5a2 | TTGGAAACCTTCTCCATGTCAGA | TCCCCAGTGGGTGTTATAGGA |
| Col6a1 | CTGCTGCTACAAGCCTGCT | CCCCATAAGGTTTCAGCCTCA |
| Col6a2 | AAGGCCCCATTGGATTCCC | CTCCCTTCCGACCATCCGAT |
| Col6a3 | GCTGCGGAATCACTTTGTGC | CACCTTGACACCTTTCTGGGT |
| Col8a1 | ACTCTGTCAGACTCATTCAGGC | CAAAGGCATGTGAGGGACTTG |
| Col11a1 | ACAAAACCCCTCGATAGAAGTGA | CTCAGGTGCATACTCATCAATGT |
| Col12a1 | AAGTTGACCCACCTTCCGAC | GGTCCACTGTTATTCTGTAACCC |
| Col15a1 | CCCAGGGAAGAATGGAGAAGT | CCAGAGCCTTCAATCTCAAATCC |
| Cox4i1 | ATTGGCAAGAGAGCCATTTCTAC | CACGCCGATCAGCGTAAGT |
| Cyc1 | CAGCTTCCATTGCGGACAC | GGCACTCACGGCAGAATGAA |
| Cytb | TTCTGAGGTGCCACAGTTATT | GAAGGAAAGGTATTAGGGCTAAA |
| E2f1 | CTCGACTCCTCGCAGATCG | GATCCAGCCTCCGTTTCACC |
| Esrra | GCACCTGGGCTCTAGTTGC | TACAGTCCTCGTAGCTCTTGC |
| Esrrg | AAGATCGACACATTGATTCCAGC | CATGGTTGAACTGTAACTCCCAC |
| aMHC | GCCCAGTACCTCCGAAAGTC | GCCTTAACATACTCCTCCTTGTC |
| bMHC | ACTGTCAACACTAAGAGGGTCA | TTGGATGATTTGATCTTCCAGGG |
| Mdm2 | TGTCTGTGTCTACCGAGGGTG | TCCAACGGACTTTAACAACTTCA |
| Mfn1 | ATGGCAGAAACGGTATCTCCA | CTCGGATGCTATTCGATCAAGTT |
| Mfn2 | ACCCCGTTACCACAGAAGAAC | AAAGCCACTTTCATGTGCCTC |
| Myl2 | AGGACGAGTGAACGTGAAAAAT | GATTGCCGGTAACGTCAGGG |
| Myl3 | TGGGGAAGCCAAAACAGGAAG | AGCCATCAGTTTCTCTACCTCA |
| Mylk3 | GTGTGCCAAGGCATGGATCA | CCTCCTGTAGTCAGTACATGGT |
| Myc | ATGCCCCTCAACGTGAACTTC | CGCAACATAGGATGGAGAGCA |
| Ndufa5 | ATGGCGGGCTTGCTGAAAA | GCTGCATGTTTAGGAAAGTGCTT |
| Ndufa6 | TCGGTGAAGCCCATTTTCAGT | CTCGGACTTTATCCCGTCCTT |
| Ndufa8 | GGAGCTGCCAACTCTGGAAG | CCAGCGGCACAGCATAAAC |
| Npm1 | ATGGAAGACTCGATGGATATGGA | ACCGTTCTTAATGACAACTGGTG |
| Opa1 | CGACTTTGCCGAGGATAGCTT | CGTTGTGAACACACTGCTCTTG |
| p53 | CCCCTGTCATCTTTTGTCCCT | AGCTGGCAGAATAGCTTATTGAG |
| Pgc-1a | TATGGAGTGACATAGAGTGTGCT | CCACTTCAATCCACCCAGAAAG |
| Pink1 | TTCTTCCGCCAGTCGGTAG | CTGCTTCTCCTCGATCAGCC |
| Pln | AAAGTGCAATACCTCACTCGC | GGCATTTCAATAGTGGAGGCTC |
| Ppara | AGAGCCCCATCTGTCCTCTC | ACTGGTAGTCTGCAAAACCAAA |
| Ryr2 | ATGGCTTTAAGGCACAGCG | CAGAGCCCGAATCATCCAGC |
| Sdha | GGAACACTCCAAAAACAGACCT | CCACCACTGGGTATTGAGTAGAA |
| Sdhb | AATTTGCCATTTACCGATGGGA | AGCATCCAACACCATAGGTCC |
| Tcap | GATGCGCCTGGGTATCCTC | GATCGAGACAGGGTACGGC |
| Tnnc1 | GCGGTAGAACAGTTGACAGAG | CCAGCTCCTTGGTGCTGAT |
| Tnni3 | TCTGCCAACTACCGAGCCTAT | CTCTTCTGCCTCTCGTTCCAT |
| Tnnt2 | CAGAGGAGGCCAACGTAGAAG | CTCCATCGGGGATCTTGGGT |
| Tpm2 | AAGTCGCTGATAGCCTCAGAG | GGTCTGGTGTATCTCCACGTTC |
| Tpm3 | GTCCCGTTGCCGAGAGATG | CTCGTCGCTAATGGCCTTGTA |
| Tpm4 | CCGGAGTAGTGGGGCAAATAC | GGCGCGAAGAAGCAAAAGC |
| Ttn | GACACCACAAGGTGCAAAGTC | CCCACTGTTCTTGACCGTATCT |
| Uqcr11 | TGCTGCGCAGGTTTCTAGGC | TCCACAGGACTACGCTGTGT |
Human cardiac samples
Human studies were conducted with the approval of the University Health Network Research Ethics Board (protocol 10–0703-TE). Informed written consent was obtained from patients with end-stage heart failure before the insertion of a left ventricular assist device. We purchased certified total RNA from normal adult human hearts from BioChain (Newark, CA 94560, USA).
Results
Acute genetic ablation of Mdm2 increases p53 protein stability
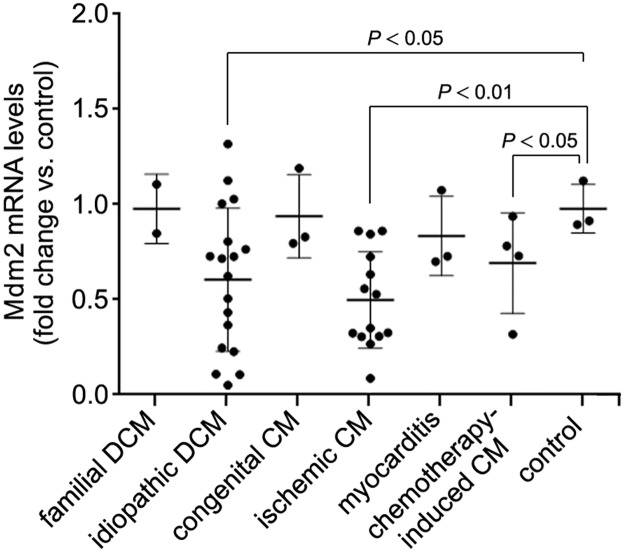
We analyzed mRNA isolated from LV samples of patients with end-stage HF of varying etiologies. When compared to mRNA isolated from healthy hearts, Mdm2 gene expression was significantly decreased in samples from patients with idiopathic dilated cardiomyopathy (DCM), ischemic cardiomyopathy and chemotherapy-induced cardiomyopathy (P < 0.05) (Fig 1), suggesting a correlation between Mdm2 expression and end-stage HF of these etiologies. Hence, to elucidate the potential function of the p53/Mdm2 circuitry in cardiomyocytes, we examined their protein expression and mRNA levels in left ventricular samples derived from C57BL/6J wild-type mice subjected to myocardial infarction (MI) or trans-aortic banding (TAB). We observed that Mdm2 protein levels were significantly decreased after MI (P < 0.001) (Fig 2A and 2B) and markedly downregulated after TAB in wild-type mice (P < 0.05) (Fig 2C and 2D). In contrast, p53 protein contents were significantly increased in both heart failure models (Fig 2A–2D). However, the same was not observed in p53 mRNA levels post-MI (Fig 2E) and post-TAB (Fig 2F) (P > 0.05) demonstrating that p53 protein expression is regulated post-transcriptionally. These data illustrate that when chronically exposed to ischemic or mechanical stress, the murine heart undergoes maladaptive changes including impaired Mdm2 expression that is inversely correlated to p53 protein levels. These findings are compatible with the view that Mdm2 exerts an activity in adult cardiomyocytes that maintains p53 at low levels and inhibits its activation. However, it can be robustly activated in response to ischemic and biomechanical stress, contributing to ventricular remodeling and the development of heart failure [52–57].
Fig 1. Down-regulation of Mdm2 mRNA levels in human end-stage heart failure determined by RT-qPCR.
DCM, dilated cardiomyopathy. CM, cardiomyopathy. Control, normal human heart. Data are means±s.e.m.
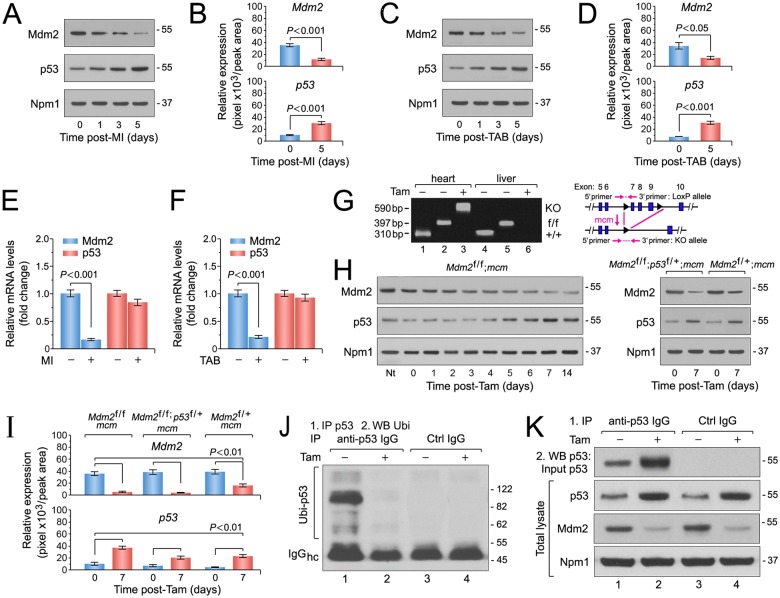
Fig 2. The E3 ubiquitin ligase Mdm2 is indispensable for the negative regulation of p53 protein stability in adult cardiomyocytes in vivo.
Ctrl, control. Hc, heavy chains. IgG, immunoglobulin G. IP, immunoprecipitation. Tam, 4-hydroxytamoxifen. Ubi, ubiquitin. WB, Western blot. Numbers on the right indicate the relative molecular protein weight in kilodalton. (A) Mdm2 and p53 protein levels after ischemic injury (myocardial infarction, MI). Immunoblot analysis of left ventricular extracts (60 ug total protein/lane) of C57BL/6J wild type mice at the indicated time points was performed employing anti-Mdm2 and anti-p53 antibodies as indicated on the left. Animals were 13 weeks old at the time of analysis. For normalization, Western blots were probed with anti-nucleophosmin (Npm1). One representative immunoblot of 3 independent experiments is shown. (B) Protein levels shown in Fig 2A were quantified with ImageJ software. n = 3. (C) Mdm2 and p53 protein levels after acute pressure overload (TAB). Immunoblot analysis of left ventricular extracts (60 ug total protein/lane) of C57BL/6J wild-type mice at the indicated time points was performed employing anti-Mdm2 and anti-p53 antibodies as indicated on the left. Animals were 13 weeks old at the time of analysis. One representative immunoblot of 3 independent experiments is shown. (D) Quantification of protein levels shown in Fig 2C. n = 3. (E) Transcript levels of endogenous Mdm2 and p53 in 10-week-old wild-type mice post-MI as analyzed by RT-qPCR. n = 4. (F) RT-qPCR analysis of endogenous Mdm2 and p53 transcript levels in 10-week-old wild-type mice post-TAB as analyzed by RT-qPCR. n = 4. (G) Heart-specific deletion of Mdm2. Schematic structure of the floxed alleles of Mdm2 (right panel). Genomic PCR results (left panel) of DNA isolated from LV tissue or liver control samples of wild-type (wt), vehicle injected control Mdm2f/f;mcm (-Tam) and Mdm2f/f;mcm mice at 7 days post-Tam (+Tam). Animals were 12 weeks old at the time of analysis. Numbers on the left refer to amplicon sizes in base pairs (bp). lane 1: Mdm2+/+;mcm, LV, -Tam. lane 2: Mdm2f/f;mcm, LV, -Tam. Lane 3: Mdm2f/f;mcm, LV +Tam. Lane 4: Mdm2+/+;mcm, liver, -Tam. lane 5: Mdm2f/f;mcm, liver, -Tam. Lane 6: Mdm2f/f;mcm, liver, +Tam. One representative result of 3 independent experiments is shown. (H) Immunoblot analysis of Mdm2 and p53 levels in left ventricular extracts (60 ug total protein/lane) of Mdm2f/f;mcm (left panel), Mdm2f/+;mcm and Mdm2f/f;p53f/+;mcm mice (right panel) employing specific antibodies as indicated on the left. Animals were 13 weeks old at the time of analysis. One representative immunoblot of 3 independent experiments is shown. (I) Quantification of protein levels of endogenous Mdm2 and p53 in the indicated strains shown in Fig 2H. n = 3. (J and K) Mdm2 regulates p53 protein stability in the adult mouse heart by regulation of its ubiquitin-mediated proteasomal degradation. (J) At 7d post-Tam, Mdm2f/f;mcm mice were intraperitoneally injected with the proteasomal inhibitor MG132 (30 mmol/kg body weight) for 6 hours. Left ventricular lysates were immunoprecipitated (IP) with anti-p53 antibodies or normal rabbit IgG. Ubiquitinated p53 proteins in the immunoprecipitates were identified by immunoblotting with antibodies to ubiquitin. One representative immunoblot of 3 independent experiments is shown. IgG, immunoglobulin G. IP, immunoprecipitation. Ubi, ubiquitin. WB, Western blot. (K) Levels of endogenous Mdm2 and p53 proteins in total left ventricular extracts prepared from Mdm2f/f;mcm mice in the presence and absence of Tam (middle and bottom panels). Samples were subjected to anti-p53 immunoprecipitations and Western blots were probed with anti-p53 antibodies (top panel). The same samples as in Fig 3J were analyzed. The mice were 12 weeks old at the end of the experiment. One representative immunoblot of 3 independent experiments is shown. Fig 2 data are means±s.e.m.
To explore the reciprocal regulation of p53 by Mdm2 mechanistically in vivo, we crossed transgenic mice expressing Cre recombinase flanked by mutated estrogen receptors (MerCreMer; mcm) with mice carrying loxP flanked alleles (f/f) of Mdm2 [58] to obtain Mdm2f/f;mcm animals. The day of the last injection was arbitrarily set to 0. Four consecutive daily intraperitoneal Tam injections induced genetic ablation of Mdm2 at 7 days post-Tam (Fig 2G). We observed a marked increase in p53 protein levels with an overlapping decrease in Mdm2 expression in left ventricular extracts from Mdm2f/f;mcm mice at 7 days post-Tam (Fig 2H) indicating a high recombination efficiency (> 90%) as determined by densitometric immunoblot analysis (Fig 2I). As expected, we detected approximately 50% less p53 protein in LV extracts of Tam-treated Mdm2f/+;mcm and Mdm2f/f;p53f/+;mcm mice (Fig 2H and 2I). Based on these findings, we surmised that Mdm2 regulates p53 protein stability in differentiated cardiomyocytes. To investigate this further, endogenous p53 was immunoprecipitated with anti-p53 antibodies from left ventricular lysates of Mdm2f/f;mcm that were intraperitoneally injected with the proteasome inhibitor MG132 (Fig 2J) [49,50]. Western blots were subsequently reacted with antibodies specifically recognizing ubiquitin (Fig 2K). The immunoblot analysis clearly demonstrates the presence of ubiquitinated p53 in vehicle-injected Mdm2f/f;mcm animals that is absent in Mdm2-deficient hearts. These findings imply that Mdm2 is a major E3-ubiquitinase for p53 in the adult heart at baseline.
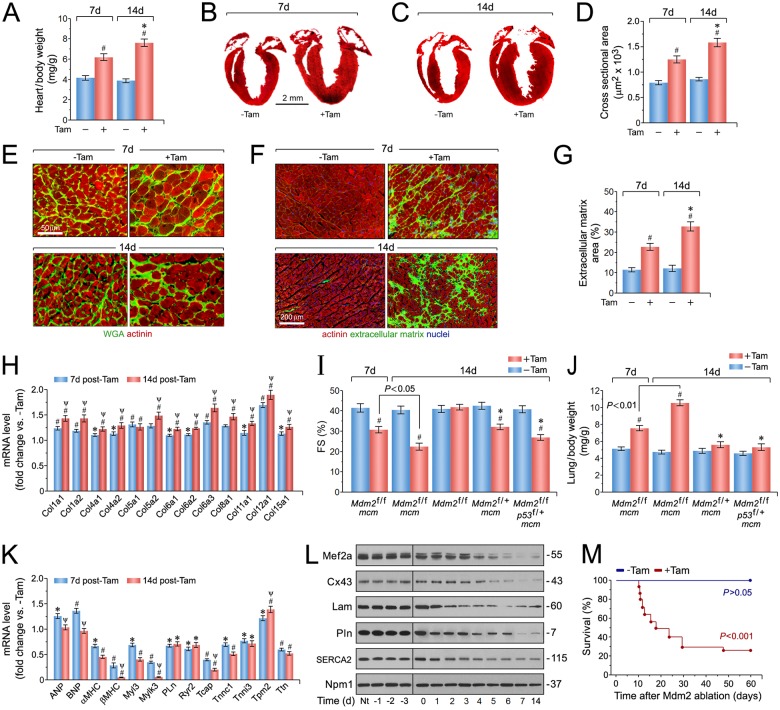
Acute cardiac-specific ablation of Mdm2 is associated with pathological cardiac hypertrophy and early death
To determine the physiological consequence of Mdm2 ablation, cardiac morphology, and function in Mdm2f/f;mcm mice were assessed in the presence and absence of Tam. We found that Mdm2f/f;mcm developed normally without an obvious cardiac phenotype in the absence of Tam (Fig 3A to 3C). In contrast, Mdm2f/f;mcm mice developed concentric cardiac hypertrophy as early as 7 days post-Tam, with significant wall thickening (Fig 3A and 3B), and an average increase of 98% (P < 0.001) (Fig 3A) in heart weight/body weight (HBW) ratios at 14 days post-Tam in comparison to vehicle controls (Fig 3A and 3C). To determine whether the increased HBW ratio in Mdm2f/f;mcm hearts was due to an increase in cell size, the width of cardiomyocytes was measured in wheat germ agglutinin (WGA)-stained specimen of the left ventricular free wall in these animals. In these samples, the cross-sectional area of Mdm2-deficient cardiomyocytes was already significantly elevated after 7 days (P < 0.01) (Fig 3D and 3E). At 14 days after Tam administration, there was a 1.9-fold increase in cardiomyocyte width in Mdm2f/f;mcm mice (P < 0.01) compared with vehicle injected controls (Fig 3D and 3E). Mdm2f/f;mcm hearts also exhibited extensive interstitial fibrosis over controls at 7 days and 14 days post-Tam (2.7-fold higher) (P < 0.01) (Fig 3F and 3G). We then measured mRNA levels of different collagens in left ventricular samples and found the transcript levels of several collagen (Col) classes including Col type I, Col IV and Col VI, were significantly increased in Mdm2f/f;mcm at 7 days and 14 days post-Tam when compared to vehicle-treated controls (P < 0.01) (Fig 3H). Examination of cardiac function by echocardiography on viable adult Mdm2f/f;mcm mice post-Tam revealed significant decreases in fractional shortening as early as 7 days post-Tam, that deteriorated further by 14 days (23 + 2.4%), in comparison to vehicle-injected Mdm2f/f;mcm (41 + 4.3%; P < 0.01) and Tam-treated Mdm2f/f control groups (Fig 3I). Importantly, FS was significantly higher in hearts of Tam-treated Mdm2f/+;mcm and Mdm2f/f;p53f/+;mcm mice when compared to Mdm2f/f;mcm animals post-Tam (P < 0.05) (Fig 3I). Moreover, lung/body weight ratios were significantly increased in Mdm2f/f;mcm mice at 7 days and 14 days post-Tam (P < 0.01) (Fig 3J). This was not observed in the Mdm2f/+;mcm and Mdm2f/f;p53f/+;mcm animals (P > 0.05). Thus, Mdm2f/f;mcm mice developed congestive heart failure after Tam administration, that appeared to be due to a higher p53 activity.
Fig 3. Ablation of Mdm2 is associated with the development of concentric hypertrophy and cardiac dysfunction.
(A) Heart-weight corrected for body weight in Mdm2f/f;mcm mice at 7d and 14 days post-Tam. n = 24. #P < 0.01 vs. -Tam. *P < 0.01 vs. Mdm2f/f;mcm at 7 days post-Tam. (B) Masson staining of longitudinal cardiac sections of Mdm2f/f;mcm mice at 7 days post-Tam. (C) Masson staining of longitudinal cardiac sections of Mdm2f/f;mcm mice at 14 days post-Tam. (D) Quantification of cross-sectional area of adult cardiomyocytes in Mdm2f/f;mcm mice shown in Fig 2E. n = 6–8. #P < 0.01 vs. -Tam. *P < 0.05 vs. Mdm2f/f;mcm at 7 days post-Tam. (E) Immunofluorescence microscopy of wheat germ agglutinin (WGA, green; top panel) and cardiomyocyte-specific anti-actinin (red; bottom panel) stained left ventricular sections from Mdm2f/f;mcm mice. (F) Immunofluorescence microscopy employing WGA staining (green) of the extracellular matrix, cardiomyocyte-specific anti-actinin (red), and Dapi (blue) to visualize nuclear DNA in left ventricular sections from Mdm2f/f;mcm mice. (G) Quantification of extracellular matrix area indicative of left ventricular fibrosis in Mdm2f/f;mcm mice shown in Fig 2F. n = 4. #P < 0.01 vs. -Tam. *P < 0.05 vs. Mdm2f/f;mcm at 7 days post-Tam. (H) Transcript levels of differentially expressed collagen types in Tam-treated Mdm2f/f;mcm mice as determined by RT-qPCR at 7 days and 14 days post-Tam. n = 4. #P < 0.01 vs. -Tam. *P < 0.05 vs. -Tam. ΨP < 0.05 vs. 7d +Tam. (I) Fractional shortening (FS) determined by M-mode echocardiography of the indicated strains at 7 and 14 days post-treatment with Tam or vehicle. n = 6. #P < 0.01 vs. -Tam. *P < 0.05 vs. Mdm2f/f;mcm at 14 days post-Tam. (J) Lung/body weight ratios in various Mdm2/p53 mutant mice at 7 and 14 days after Tam treatment. n = 6. #P < 0.01 vs. -Tam. *P < 0.05 vs. Mdm2f/f;mcm at 14 days post-Tam. (K) Levels of hypertrophic marker genes in Mdm2f/f;mcm mice as analyzed by qRT-PCR at 7 days and 14 days post-Tam. n = 4. #P < 0.01 vs. -Tam. *P < 0.05 vs. -Tam. ΨP < 0.05 vs. 7d +Tam. (L) Immunoblot analysis of cardiac-specific gene expression in left ventricular samples from Mdm2f/f;mcm mice. Nt, no treatment. Western blots were repeated at least once with similar results. (M) Acute genetic ablation of Mdm2 evokes premature death. Kaplan-Meier survival curves of conditional Mdm2f/f;mcm mice. n = 20. Fig 3A to Fig 3M: 12-week-old mice (7 days post-Tam) and 13-week-old mice (14 days post-Tam) were analyzed. Fig 3 data are means±s.e.m.
To explore the molecular basis for the cardiac abnormalities in Mdm2f/f;mcm mice exposed to Tam, RT-qPCR was carried out on total RNA isolated from left ventricular tissue samples at 7 days and 14 days post-Tam. Notably, we detected altered transcript levels of structural and stress-responsive cardiac genes as early as 7 days post-Tam. In particular, significant decreases in expression of genes regulating Ca2+-handling (Pln, Ryr2), and contractile proteins (for example, Mylk3, Tnnc1, Tnni3, Ttn) were observed and in keeping with the cardiac dysfunction of Mdm2-deficient hearts (P < 0.01) (Fig 3K). Intriguingly, transcripts for atrial natriuretic factor (ANF), and brain natriuretic peptide (BNP), canonical markers of cardiac hypertrophy and heart failure, were initially increased by 7 days, but dropped to control levels at 14 days (P < 0.05) in Mdm2f/f;mcm mice after Tam-administration, whereas alpha and beta myosin heavy chain (a/bMHC) mRNA were significantly downregulated by -39% and -47% (P < 0.001) (Fig 3K). All these effects were further corroborated by immunoblot analysis of selected cardiac genes in Mdm2f/f;mcm mice over a time course of 14 days in the presence and absence of Tam (Fig 3L). Importantly, 50% of Mdm2f/f;mcm mice died unexpectedly by 18 days post-Tam, and more than 75% of animals succumbed within 30 days that was not observed in vehicle-treated Mdm2f/f;mcm controls (P < 0.001) (Fig 3M). In contrast, heterozygosity of p53 was sufficient to rescue the lethality of Mdm2f/f;mcm mice exposed to Tam in the same observation period (Fig 3M). Early death of Mdm2f/f;mcm mice could also be rescued by heterozygosity of Mdm2, demonstrating a specific gene dose effect for Mdm2 in the adult heart.
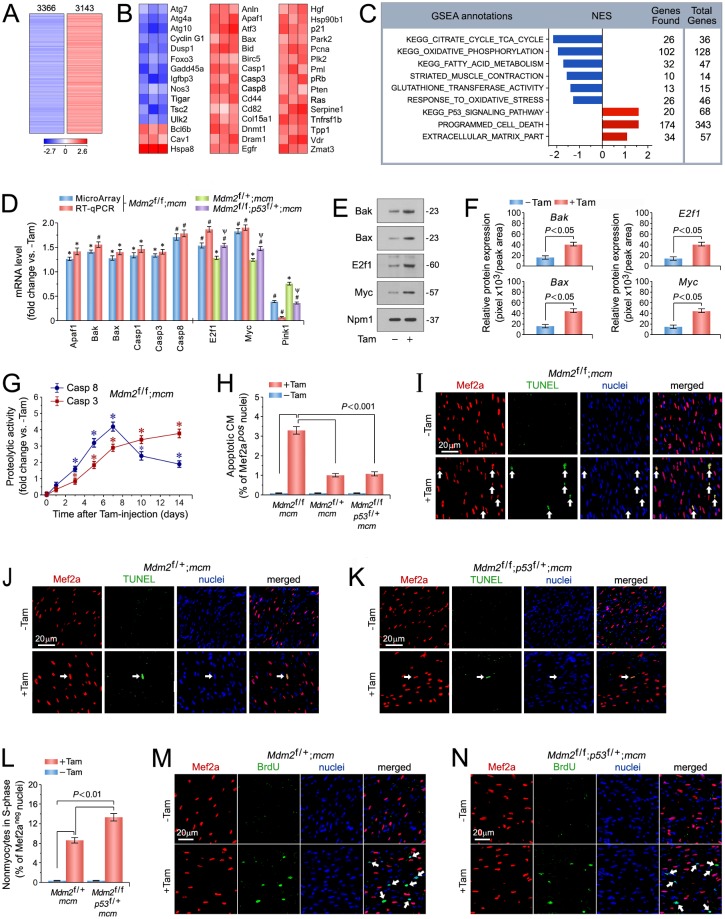
Genome-wide expression profiling of Mdm2-regulated transcripts
To uncover mechanisms responsible for the cardiac phenotype induced by Mdm2-deficiency at the transcriptome level, genome-wide RNA expression profiling was performed on left ventricular samples from Mdm2f/f;mcm mice in the presence and absence of Tam. Acute ablation of Mdm2 induced a broad derangement of the cardiac transcriptome (Fig 4A). At 14 days after Tam administration, 6,509 of 26,166 individual genes were differentially regulated in the hearts of Mdm2f/f;mcm mice (false discovery rate < 0.01%) in comparison to vehicle-injected Mdm2f/f;mcm mice. Moreover, significant enrichment of canonical p53 target genes [59] in Mdm2-deficient hearts indicates an association between activated p53 and Mdm2 ablation (P < 0.01) (Fig 4B).
Fig 4. p53 is spontaneously active in Mdm2-deficient cardiomyocytes and induces apoptosis.
(A) Genome-wide messenger RNA (mRNA) microarray profiling reveals that conditional genetic ablation of Mdm2 induces profound alterations in the cardiac transcriptome indicating a high degree of complexity associated with Mdm2-deficiency. Heat map of unsupervised hierarchical cluster analysis at a high confidence threshold identifies 6,509 transcripts (rows) out of 26,166 individual genes (columns) that were enriched in the hearts of Mdm2f/f;mcm mice post-Tam relative to vehicle-injected controls. Values (log2 expression) are shown by color and intensity of shading. Blue, repressed. Red, induced. n = 3 biological replicates. P < 0.01. Fold change >1.3. Mice (13 weeks old) were analyzed at 14 days post-Tam. (B) Heat maps examining the impact of genomic modifications in Mdm2-deficient hearts (columns) on previously validated p53 target genes (rows). Values, log2 expression. n = 3. P < 0.01. Fold change >1.3. (C) Gene Set Enrichment Analysis (GSEA) of biological processes among all differentially expressed transcripts, assessed by overrepresentation of GSEA terms for the biological function of each transcript in hearts of Mdm2f/f;mcm mice post-Tam. NES, normalized enrichment score. NES have False Discovery Rate (FDR) q-values <0.1. Blue, inhibited processes. Red, activated processes. (D) Expression levels of key genes selected from top-ranked gene sets identified in Fig 4A, involved in the regulation of cardiomyocyte apoptosis in the indicated Mdm2 and p53 mutant mice as analyzed by RT-qPCR at 7 days post-Tam. n = 3. *P < 0.05 vs. -Tam. #P < 0.01 vs. -Tam. Ψ < 0.05 vs. Mdm2f/+;mcm and Mdm2f/f;mcm. (E) Immunoblot analysis of master regulators of apoptosis in Mdm2f/f;mcm mice at 7 days post-Tam employing specific antibodies as indicated on the left. One representative immunoblot of 3 independent experiments is shown. (F) Protein levels shown in Fig 4E were quantified with ImageJ software. n = 3. (G) Time course of caspase 3/8 activation in Mdm2f/f;mcm mice. n = 4. *P < 0.05 vs. -Tam. (H) Quantification of apoptotic cardiomyocytes shown in Fig 4I-K. n = 4. (I to K) Analysis of apoptosis in left ventricular cardiomyocytes (white arrows) in various Mdm2 and p53 mutant strains at 7 days post-Tam by immunofluorescence microscopy and terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay. Red, anti-Mef2a. Green, TUNEL. Blue, DAPI stain of nuclear genomic DNA. (L) Induction of cell cycle entry and DNA synthesis is restricted to non-myocytes in the indicated Mdm2 and p53 mutant strains. Quantification of non-myocytes in S phase of the cell cycle shown in Fig 4M and N. n = 4. (M, N) Analysis of DNA replication in left ventricular non-myocytes (white arrows) in the indicated Mdm2 and p53 mutant strains at 7 days post-Tam by immunofluorescence microscopy. Red, anti-Mef2a. Green, BrdU. Blue, DAPI stain of nuclear genomic DNA. Mice were 13 weeks old at 14 days post-Tam (Fig 4A to C). Animals were 12 weeks old mice at 7 days post-Tam (Fig 4C to N). Abbreviations of gene names in Fig 4B: Anln, anillin actin binding protein; Apaf1, apoptotic peptidase activating factor 1; Atf3, activating transcription factor 3; Atg7, autophagy related 7; Atg4a, autophagy related 4A cysteine peptidase; Atg10, autophagy related 10; Bax, BCL2-associated X protein; Bcl6b, B cell CLL/lymphoma 6, member B; Bid, BH3 interacting domain death agonist; Birc5, baculoviral IAP repeat-containing 5; Casp1, caspase 1; Cav1, caveolae protein caveolin 1; Cd44, CD44 antigen; Cd82, CD82 antigen; Col18a1, collagen type XVIII alpha 1; Cyclin G1 (Ccng1); Dnmt1, DNA methyltransferase (cytosine-5) 1; Dram1, DNA-damage regulated autophagy modulator 1; Dusp1, dual specificity phosphatase 1; Egfr, epidermal growth factor receptor; Foxo3, forkhead box O3; Gadd45a, growth arrest and DNA-damage-inducible 45 alpha; Hgf, hepatocyte growth factor; Hspa8, heat shock protein 8; Hsp90b1, heat shock protein 90 beta (Grp94) member 1; Igfbp3, insulin-like growth factor binding protein 3; Nos3, endothelial cell nitric oxide synthase 3; p21, cyclin-dependent kinase inhibitor 1A (Cdkn1a); Park2 (parkin), Parkinson disease autosomal recessive, juvenile 2; Pcna, proliferating cell nuclear antigen; Plk2, polo-like kinase 2; Pml, promyelocytic leukemia; pRb, retinoblastoma 1 (Rb1); Pten, PTEN induced putative kinase 1; Ras, Harvey rat sarcoma virus oncogene (Hras); Serpine1, serine (or cysteine) peptidase inhibitor clade E member 1; Tigar, Trp53 induced glycolysis regulatory phosphatase; Tnfrs1b, TNF receptor superfamily member 1B; Tpp1, tripeptidyl peptidase I; Tsc2, tuberous sclerosis 2; Ulk2, unc-51 like kinase 2; Vdr, vitamin D receptor; Zmat3, zinc finger matrin-type 3. Fig 4 data are means±s.e.m.
To identify biological processes underlying the development of the cardiac phenotype in the absence of Mdm2 at 14 days post-Tam, Gene set enrichment analysis (GSEA) analysis was performed. GSEA identified several highly significantly gene clusters with similar biological functions in the mutant Mdm2 transcriptome that were known to be critically involved in the regulation of normal heart function (P < 0.01) (Fig 4C). The top ranked GSEA terms (Fig 4C) with negative (downregulated genes, blue) normalized enrichment scores (NES) were the p53 signaling pathway, tricarboxylic acid cycle (TCA), oxidative phosphorylation, fatty acid metabolism, striated muscle contraction (as analyzed in Fig 3J and 3K), glutathione transferase, and response to oxidative stress. Programmed cell death and extracellular matrix (as analyzed in Fig 3F, 3G and 3H) were highly enriched gene sets containing upregulated transcripts (positive NES, red) (Fig 4C). The development of a cardiac phenotype occurred as early as 7 days in these animals (Fig 3A–3K). Therefore, we decided to test the physiological relevancy of main regulators in these gene sets for their ability to drive the development of heart failure associated with Mdm2 loss at an earlier time point at 7 days post-Tam.
Spontaneous activation of p53 in Mdm2-deficient cardiomyocytes induces apoptosis
With the observation of elevated p53 protein levels in hearts of Mdm2f/f;mcm mice exposed to Tam (Fig 2H), we initially investigated the sensitivity of this strain to p53-triggered oxidative stress and apoptosis. Top ranked genes included direct proapoptotic p53 target genes Apaf1 [60–62], caspases 1 [63], 3 [64,65] and 8 [66–68], E2f1 [69–72], and c-Myc (Myc) [71,73] as well as Bax and Bak, key p53-interacting factors during transcription-independent p53 cell death [74,75]. E2f1 [72] and Myc [73] are ubiquitously expressed, highly pleiotropic transcription factors. In mitotic cells, they promote proliferation, apoptosis, DNA damage, sensitivity to reactive oxygen species (ROS), and the oxidative stress response [70,71,73]. Significant upregulation (P < 0.05) of all of these factors in Tam-injected Mdm2f/f;mcm was confirmed by real time PCR (RT-qPCR) (Fig 4D) and immunoblot analysis (Fig 4E and 4F) at 7 days post-Tam.
Next, we analyzed whether mRNA expression of E2f1, Myc and Pink1 are regulated by p53 or by Mdm2 independently of p53. Thus, RT-qPCR was carried out on total RNA isolated from Tam-treated Mdm2f/+;mcm and Mdm2f/f;p53f/+;mcm mice. We observed, that transcript levels of E2f1 and Myc were significantly higher in hearts of Mdm2f/f;p53f/+;mcm mice as compared to Mdm2f/+;mcm animals post-Tam (P < 0.05) (Fig 4D). Moreover, Pink1 mRNA expression was lower in LV samples from Mdm2f/f;p53f/+;mcm mice in comparison to Mdm2f/+;mcm animals (P < 0.05) (Fig 4D). These findings are consistent with the hypothesis that gene expression of E2f1, Myc and Pink1 regulated by p53, and occurs in a Mdm2-independent manner.
Transcriptional activation of caspase-3 and 8 in a p53-dependent fashion has been well established in previous studies [65,67]. Thus, we examined the kinetics of caspase-8 (extrinsic cell death pathway) and caspase-3 (intrinsic apoptosis) [76] activation to determine their relationship to the development of heart failure in the absence of Mdm2. Spectrophotometric measurement of caspase-dependent proteolytic activities in left ventricular lysates were shown to be significantly induced in the presence of Tam within 3 days (caspase-8) and 5 days (caspase-3) (P < 0.05), and reached their maximum at 7 days (Fig 4G). Significant levels of caspase-3 and 8 activities were not detected in control mice (Fig 4G). Notably, we observed a marked increase in the number of TUNEL-positive cardiomyocyte nuclei in fixed left ventricular specimen from Tam-injected Mdm2f/f;mcm mice (3.3 ± 0.46%; P < 0.001) as compared to controls (0.1 ± 0.02%; P < 0.001) (Fig 4H and 4I). Importantly, reduction of p53 levels in Tam-treated Mdm2f/+;mcm (Fig 4H and 4J), and Mdm2f/f;p53f/+;mcm mice (Fig 4H and 4K) significantly reduced the degree of cardiomyocyte apoptosis in comparison to the Mdm2f/f;mcm strain (P < 0.001). We infer from this data, that p53 is a physiological relevant target of Mdm2. These findings suggest that induction of cardiomyocyte apoptosis by p53 due to loss of Mdm2, plays a role in the initiation phase of a process that culminates in left ventricular dysfunction and premature death.
We have previously reported, that homozygous genetic deletion of both p53 and Mdm2 in adult mice significant increases adult cardiomyocyte proliferation [77]. In contrast, CM in Mdm2f/f;p53f/+;mcm mice exhibited a markedly lower extent of cell cycle re-entry post-Tam. Notably, deficiency of p53 alone was insufficient to induce CM DNA replication [77]. To investigate whether heterozygosity for p53 in Mdm2-deficient mice induces DNA synthesis in adult CM, we intraperitoneally injected Mdm2f/f;p53f/+;mcm mice with BrdU, and analyzed fixed LV cryosections for evidence of CMs in S phase of the cell cycle at 7d post-Tam. Mdm2f/+;mcm mice were also included in this analysis, because CM cell cycle activity in this strain had not been analyzed in previous studies [77]. Immunofluorescence imaging repeatedly showed significant DNA synthesis of non-CM cell populations (Fig 4L). There was no evidence of CMs in S phase in Mdm2f/f;p53f/+;mcm and Mdm2f/+;mcm mice with or without Tam injection (Fig 4M and 4N). Thus, heterozygosity for p53 on an Mdm2 negative genetic background is insufficient to induce cell cycle re-entry.
Mdm2-deficiency enhances oxidative stress in cardiomyocytes
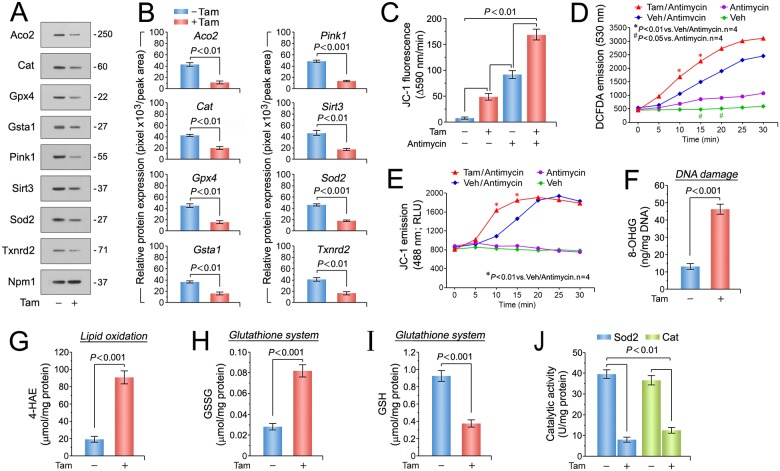
Enhanced oxidative stress associated with mitochondrial dysfunction has been well documented in patients with heart failure [78–80]. Since p53 [81], E2f1 [82], and Myc [83] play important roles in redox homeostasis and ROS generation, we analyzed the expression of the top-ranked antioxidant factors contained in the GSEA term “response to oxidative stress” (Fig 4C) by immunoblotting of left ventricular lysates. Pink1, mitochondrial Aco2, Cat, Gpx4, Gsta1, mitochondrial Sirt3, Sod2, and Txnrd2 were markedly downregulated in Mdm2f/f;mcm post-Tam, but not in control mice (Fig 5A and 5B). Since mitochondrial complex I (CI) activity and mitochondrial membrane potential is impaired in the hearts of systemic Pink1 knockout mice [84,85], we evaluated the consequence of Mdm2 ablation on mitochondrial function. Production of reactive oxygen species (ROS) by the mitochondrial respiratory chain is a main contributor to the development of heart failure associated with oxidative stress [86]. Two production sites for ROS have been identified at mt C1 and mt C3 [87,88]. The production of ROS at these sites can be greatly enhanced by specific inhibitor, rotenone for C1 and antimycin for C3 [89]. Both compounds stimulate ROS generation by induction of a reverse-electron flux [90]. To analyze the integrity of the mitochondrial transmembrane potential (MMP), mitochondrial isolated from left ventricles derived from Tam-treated Mdm2f/f;mcm mice were labeled with the fluorochrome JC-1 (2.0 uM), and then exposed to antimycin (50 uM). As expected, we observed a ROS-dependent impairment of MMP in mitochondrial from Mdm2-deficient hearts compared relative to vehicle controls at baseline and in the presence of antimycin that indicates enhanced ROS production (P < 0.01) (Fig 5C to 5E). In agreement with these findings, we found 3.5-fold higher levels of oxidative stress indicators 8-hydroxy-2′-deoxyguanosine (8-OHdG) (Fig 5F), and 4.5-fold higher levels of 4-hydroxyalkenals (4-HAE) (P < 0.001) (Fig 5G) in Tam-treated Mdm2f/f;mcm. This data suggest that Mdm2 participates in the negative regulation of Pink1 protein expression to contribute to oxidative stress. GSH (-59%)/GSSG (+283%) ratios, key components in the cellular anti-oxidation, were also significantly decreased in Tam injected Mdm2f/f;mcm hearts over controls (P < 0.001) (Fig 5H and 5I). Finally, administration of Tam to Mdm2f/f;mcm mice markedly downregulated activities of ROS detoxifying enzymes Sod2 (-80%) and Cat (-65%; P < 0.01) (Fig 5J). Taken together, our results demonstrate that Mdm2-deficiency is accompanied by elevated oxidative stress within the mitochondria.
Fig 5. Enhanced oxidative stress and impaired antioxidant systems in cardiomyocytes lacking Mdm2.
(A) Immunoblot analysis of key genes selected from top-ranked gene sets identified in Fig 3C involved in detoxification processes of reactive oxygen species (ROS). One representative immunoblot of 3 independent experiments is shown. Left ventricular samples from Mdm2f/f;mcm mice were analyzed at 7 days post-Tam. (B) Densitometric analysis of Western blot results shown in Fig 5A employing ImageJ software. Numbers at the bottom of each panel indicate fold-change. n = 3 (C) Isolated cardiac mitochondria from Tam-treated Mdm2f/f;mcm have a membrane potential (MMP) that is susceptible to ROS-induced depolarization. Mitochondria were mechanically isolated, incubated with JC-1 (5.0 ug/mL), and were then treated with antimycin (50 uM). JC-1 emission at 535/595 nm was recorded at 1 reading/min for 30 min using a fluorescence spectrophotometer. The rate between two time points (emission at 595 nm/min) was calculated in the most linear range of decline for JC-1 fluorescence intensity. n = 4. (D) DCFAD (dichlorofluorescin diacetate) emission data in isolated cardiac mitochondria from Tam-treated Mdm2f/f;mcm mice. n = 4. (E) JC-1 green emission data in isolated cardiac mitochondria from Tam-treated Mdm2f/f;mcm mice. RLU, relative light units. n = 4. (F) Higher oxidative genomic DNA damage in Mdm2f/f;mcm mice post-Tam when compared to vehicle-injected controls. Concentrations of 8-hydroxy-2’-deoxyguanosine (8-OHdG), a biomarker for oxidative DNA damage in the indicated strains was determined by a competitive enzyme-linked ELISA employing 8-OHdG antibodies. n = 4. (G) Significantly enhanced levels of 4-hydroxyalkenals (4-HAE), an indicator of ROS-dependent lipid peroxidation in Mdm2f/f;mcm mice post-Tam compared with control injected animals. n = 4. (H and I) Markedly reduced glutathione/oxidized glutathione (GSH/GSSG) ratios, an indicator of cardiac oxidative stress, in Mdm2f/f;mcm mice post-Tam compared to controls. n = 4. (J) Significantly downregulated mitochondrial superoxide dismutase (Sod2) and catalase (Cat) activities as determined spectrophotometrically in Mdm2f/f;mcm mice post-Tam. n = 4. Fig 5A to J: Left ventricular samples from 12-week-old mice were analyzed at 7 days post-Tam. Fig 5 data are means±s.e.m.
Mdm2 ablation inhibits the Pgc-1/Ppar/Esrr axis and leads to broad mitochondrial dysfunction
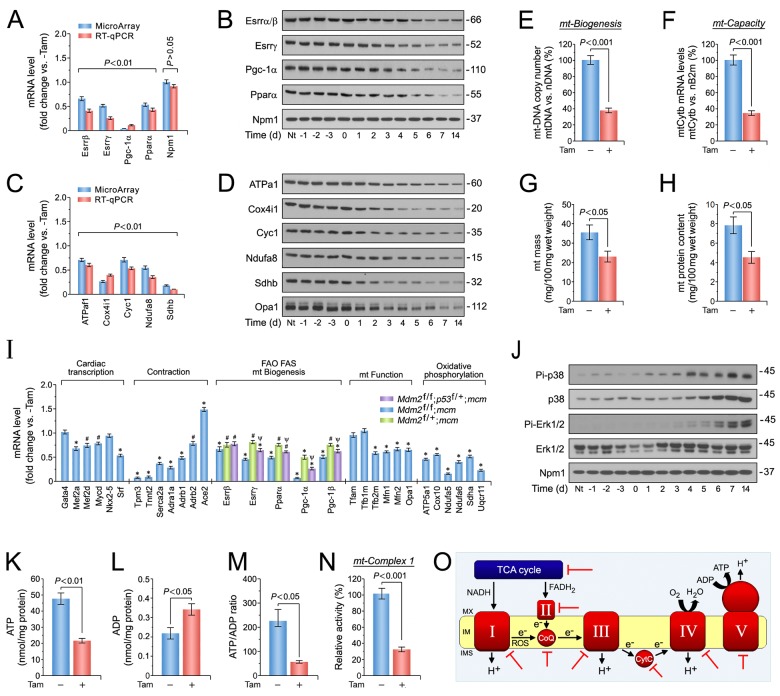
To determine whether activated p53, E2f1 and Myc in the absence of Mdm2 induces downregulation of major regulators of mitochondrial function and fatty acid oxidation, as predicted by our GSEA analysis (Fig 4C), we performed RT-qPCR and immunoblot analyses of left ventricular tissue samples from Mdm2f/f;mcm mice at 7 days post-Tam. Significant decreases in the mRNA levels of Esrrb/g Pgc-1a and Ppara/g in Mdm2f/f;mcm mice post-Tam were detected by RT-qPCR (P < 0.01) (Fig 6A), and confirmed by immunoblot analysis (Fig 6B).
Fig 6. Mdm2 is involved in the maintenance of mitochondrial biogenesis and bioenergetics.
(A) Expression levels of selected factors from top-ranked gene sets identified in Fig 4A that play a crucial role in the regulation of mitochondrial function and fatty acid oxidation as analyzed by RT-qPCR. n = 4. Left ventricular tissue samples from Mdm2f/f;mcm mice were analyzed at 7 days post-Tam. (B) Immunoblot analysis of key factors regulating mitochondrial biogenesis and fatty acid oxidation from Mdm2f/f;mcm mice. Western blots were repeated at least once with similar results. (C) Transcript levels of important constituents of the mitochondrial electron transport chain participating in oxidative phosphorylation in Mdm2f/f;mcm mice. n = 4. (D) Immunoblot analysis of important components of the mitochondrial electron transport chain complexes in Mdm2f/f;mcm mice. Western blots were repeated at least once with similar results. (E) Mitochondrial biogenesis, defined as relative DNA copy number of mitochondrial encoded Cytb gene normalized to the copy number of the nuclear gene B2m, was determined by qPCR in Mdm2f/f;mcm mice. n = 4. (F) Mitochondrial capacity, defined as relative mRNA levels of the nuclear gene cytochrome b (Cytb), a constituent of oxidative phosphorylation (OxPhos) complex III, normalized to B2m transcript expression, was determined by RT-qPCR in Mdm2f/f;mcm mice. (G) Decreased mitochondrial wet weight in ventricular tissue of Mdm2f/f;mcm mice post-Tam. n = 4. (H) Downregulation of total mitochondrial protein content in Mdm2-deficient ventricles. n = 4. (I) Cardiac-specific gene expression in the indicated Mdm2 and p53 mutant mice post-Tam as analyzed by RT-qPCR. n = 4. *P < 0.01 vs. -Tam. #P < 0.05 vs. -Tam. Ψ < 0.05 vs. Mdm2f/+;mcm and Mdm2f/f;mcm. (J) Activation of p38 and Erk1/2 signal transduction pathways is associated with the development of decompensated cardiac hypertrophy in Mdm2f/f;mcm mice exposed to Tam. LV extracts were prepared at the indicated time points, and were immunoblotted with the antibodies as indicated on the left. The amount of phosphorylation was determined using phosphorylation site-specific antibodies to p38 and Erk1/2. Western blots were repeated at least once with similar results. (K) Markedly impaired ATP production indicates mitochondrial dysfunction in Mdm2f/f;mcm mice post-Tam. ATP levels were analyzed with a bioluminescence-based assay. n = 4. (L) Acute genetic deletion of Mdm2 evokes increases in ADP levels in Mdm2f/f;mcm mice post-Tam. n = 4. (M) Mdm2 is indispensable for the maintenance of proper energy metabolism as indicated by a significantly lower ATP/ADP ratio in Mdm2f/f;mcm mice post-Tam. n = 4. (N) Mitochondrial complex I activity in Mdm2f/f;mcm mice was determined with an ELISA assay employing isolated mitochondria and complex I-specific antibodies. The NADH dehydrogenase activity of complex I was determined colorimetrical by monitoring the oxidation of NADH to NAD+. n = 4. (O) Potential Mdm2-regulated genes in a well-established mt interaction network. Red block arrows indicate significantly downregulated genes in Mdm2f/f;mcm mice post-Tam. CoQ, coenzyme Q10. CytC, cytochrome c. IMM, inner mt membrane. IMS, inner membrane space. MMX, mt matrix. ROS, reactive oxygen species. (A to M) Tam- and vehicle-injected Mdm2f/f;mcm animals were investigated. Unless indicated otherwise left ventricular samples from 12-week-old mice were analyzed at 7 days post-Tam. Fig 6 data are means±s.e.m.
The majority of the 1,500 mitochondrial proteins are encoded by nuclear genes. In contrast, the mitochondrial genome contains 13 protein-coding genes, 2 rRNAs and 22 tRNAs [91]. Since it has been firmly established that ATP levels are tightly correlated to contractility [92], we determined the expression levels of the top-ranked factors contained in the KEGG gene set oxidative phosphorylation (Fig 4C). Mitochondrial encoded genes all have the potential to modulate oxidative phosphorylation, albeit at differing levels, thus, it is crucial that mitochondrial transcription be coordinated with the other cellular signaling pathways that impose their own energy demands upon the cell [91,92]. Notably, we observed significant decreases in the expression of crucial components of respiratory chain complexes, including Atp5af1 (complex V, CV), Cox4i (CIV), somatic Cyc1 (CIII), Ndufa8 (CI) and Sdhb (CII), in RT-qPCR (P < 0.01) (Fig 6C) and Western blot analyses (Fig 6D) of left ventricular tissue samples derived from Mdm2f/f;mcm mice post-Tam. All these effects were not observed in vehicle-treated controls (Fig 6C).
Next, we investigated whether a global decline in mitochondrial biogenesis underlies the observed reduction in mitochondrial factors involved in OxPhos, because, for example, nuclear encoded proteins are actively transported into these organelles [91]. Therefore, mitochondrial DNA copy number was determined by qPCR of the mitochondrial gene cytochrome b (Cytb), normalized to levels of nuclear encoded B2m (beta-2 microglobulin). We noted a dramatically lower (-61%) mitochondrial copy number in Mdm2f/f;mcm post-Tam in comparison to untreated Mdm2f/f;mcm (P < 0.001) (Fig 6E). As expected from this result, mitochondrial respiratory capacity was also critically lower (-76%) in Mdm2f/f;mcm hearts post-Tam, while normal in controls (P < 0.001) (Fig 6F). We then analyzed the possibility, whether the observed decrease in mitochondrial Cytb transcription reflects a decrease in mitochondrial mass. Therefore, we directly measured mitochondrial wet weight and mitochondrial protein content in ventricular tissue of Mdm2f/f;mcm hearts post-Tam. Indeed, significant (P < 0.05) decreases in mitochondrial wet weight (Fig 6G) and mitochondrial protein content (Fig 6H) in Mdm2f/f;mcm exposed to Tam demonstrate that Mdm2 is involved in the maintenance of mitochondrial function.
We employed a RT-qPCR based screen to investigate transcriptional changes of gene sets that exert distinct function during cardiac hypertrophy. Intriguingly, we observed a significant downregulation (P < 0 .05 to 0.01) of key heart-specific transcription factors (Mef2a/d, Myocd, Srf) in Tam-treated Mdm2f/f;mcm when compared to control animals (Fig 6I). Expression of genes encoding important regulators of cardiac contractility (Adra1a, Adrb1, Serca2a, Ace2) were also significant lower in Mdm2f/f;mcm mice post-Tam (Fig 6I). Sarcomere organization profoundly influences cardiac function with abnormalities in this structure commonly observed in human HF. Strikingly, important contractile proteins (Myl2, Tpm3/4, Tnnt2) were downregulated in Mdm2f/f;mcm animals post-Tam (Fig 6I), demonstrating that Mdm2 is important for proper maintenance of the contractile apparatus. Beyond these genes, we also found key regulators of fatty acid metabolism (Esrrb/g, Ppara/g, Pgc-1a/b, Tfb1/2), mitochondrial fusion/fission (Mfn1/2, Opa1), and mitochondrial respiration (Atp5a1, Cox10, Ndufa5, Ndufa6, Sdha, Uqcr11) to be transcriptionally downregulated in Mdm2-mutant hearts (Fig 6I).
Next, we analyzed whether mRNA expression of Esrrb, Esrrg, Ppara, Pgc-1a, and Pgc-1b is subjected to regulation by p53, or occurs through Mdm2, independently of p53. Thus, RT-qPCR was carried out on total RNA isolated from Tam-treated Mdm2f/+;mcm and Mdm2f/f;p53f/+;mcm mice (Fig 6I). We observed, that transcript levels of Esrrg, Ppara and Pgc-1a were significantly higher in hearts of Mdm2f/f;p53f/+;mcm mice as compared to Mdm2f/+;mcm animals showing that these effects are regulated by p53, independent of Mdm2 (P < 0.05) (Fig 6I). Thus, it appears that Mdm2-dependent regulation of p53 transcriptional activity is necessary to maintain normal heart function.
It is well established, that the development of maladaptive cardiac hypertrophy involves the activation of distinct signal transduction pathways, including the mitogen-activated protein kinase (Mapk) signaling cascade [93]. Main Mapk signaling cascades consist of terminal effector kinases p38 and extracellular signal-regulated kinases (Erks). Both kinase families are each regulated by specific phosphorylation of adjacent Tyr and Thr recognition motifs [94]. Since hypertrophic growth of cardiomyocytes is associated with phosphorylation-dependent activation of p38 [95] and Erk1/2 [96], the phosphotransferase activity of these Mapks was investigated by immunoblotting employing antibodies that specifically recognize their phosphorylated active isoforms. Thus, we evaluated p38 activation by phosphorylation on Thr180/Tyr182, and found an increase in Mdm2f/f;mcm animals as early as 1d post-Tam (Fig 6J). Since the pro-hypertrophic function of Erk1/2 is associated with phosphorylation at Thr202/Tyr204, this Erk1/2 modification was analyzed by Western blotting employing phosphorylation site-specific antibodies. Notably, Tam-treatment triggered phosphorylation of Erk1/2 at Thr202/Tyr204 as early as day 4 in Mdm2f/f;mcm mice (Fig 6J). Taking these findings into consideration, we surmise that observed increases in p38 and Erk1/2 activation are a consequence of Mdm2 ablation. We also conclude that both signal transduction pathways are critical for the development of decompensated hypertrophy.
Mitochondrial dysfunction leads to lower production of mitochondrial ATP [97], and decreases in cytosolic ATP/ADP ratios are indicative of enhanced glycolytic ATP production, as recognized in heart failure. To gain a more complete understanding of the energy status in Mdm2-deficient cardiomyocytes, ADP levels were assessed. ATP levels were significantly lower (-54%) in the Mdm2-deficient hearts compared to vehicle controls (P < 0.001) (Fig 6K). We found higher ADP levels in the Mdm2f/f;mcm strain post-Tam (P < 0.05) in comparison to vehicle-injected controls (Fig 6L) and, thus, elevated ATP/ADP ratios (P < 0.05) (Fig 6M).
Immunocomplex assays confirmed a detrimental impact of Mdm2 on mitochondrial CI activity (-68%) in Mdm2f/f;mcm post-Tam, while such an effect was absent in vehicle-control animals (P < 0.001) (Fig 6N). This supports the hypothesis that negative regulation of crucial factors in the mitochondrial electron transport chain by inhibition of Pgc-1a, Ppara/g and Esrrb/g evokes severe mitochondrial deficiencies including fatty acid oxidation. Fig 6O illustrates the consequences of impaired Pgc-1a, Ppara/g and Esrrb/g axes on mitochondrial energy metabolism in the absence of Mdm2.
Discussion
Here we report a role for the E3 ubiquitin ligase Mdm2 in differentiated cardiomyocytes where it supresses pathological cardiac growth through the promotion of p53 degradation. Mechanistically, we found that genetic ablation Mdm2 lead to increased levels of p53, in the absence of any acute stress. This effect sufficed to induce global alterations of the cardiac transcriptome and is reflective of ROS generation, apoptosis, fibrosis, broad mt deficiencies and hypertrophy in the Mdm2-deficient hearts. These processes culminated in the development of LV dysfunction and early death. This observation led us to realize that basal Mdm2 activity is indispensable for the control of proper heart function.
Oxidative stress plays an important role in the development of cardiac remodeling and HF. At higher levels, ROS causes direct contractile dysfunction by directly modifying proteins central to excitation-contraction coupling, and induces p53-mediated apoptosis [21,78–80]. ROS also stimulates proliferation of cardiac fibroblasts, and activates metalloproteinases leading to remodeling of the extracellular matrix [22]. Mdm2-deficient hearts develop early signs of ROS-induced oxidative damage. It appears reasonable to assume, therefore, that the majority of ROS derived from dysfunctional mt is due to p53 instigated changes in gene transcription. We have previously shown, that at low levels, p53 activity is an important regulator of normal heart function [98]. In the presence of acute stress, however, hyper-activated p53 induces predominantly apoptosis, as shown in this and other studies [37,38]. Thus, the second mechanism by which ablation of Mdm2 caused pathological cardiac hypertrophy is through the induction of p53-dependent apoptosis. The third, less characterized, pathway of maladaptive hypertrophy in the absence of Mdm2 probably involves downregulation of cardiac transcription factors, including Mef2a and Pgc-1a, key regulators of mt physiology. In a mouse model of telomere dysfunction, Sahin et al. demonstrated that telomeric dysfunction activated p53, which in turn can represses the Pgc-1a gene promoter causing HF [99]. Thus, the authors were the first who established a direct link between ROS-activated p53 and mt function.
It has been firmly established that p53 is spontaneously active in all tissues in mice with reduced Mdm2 expression [100,101]. Our previous analysis of a cardiac-specific p53 knockout mouse revealed that p53 regulates >1,000 differentially expressed transcripts in more than 20 gene sets relevant to cardiac architecture, excitation-contraction coupling, mt biogenesis and oxidative phosphorylation capacity [98]. This study defined a new role for p53 as a novel master regulator of the cardiac transcriptome, and suggests a central role for the Mdm2/p53 pathway in the maintenance of cardiac homeostasis that extends mechanistically far beyond the regulation of apoptosis and hypertrophy, as previously reported [46]. In fact, we are the first to report the fundamental importance of Mdm2 for the p53 pathway with respect to its strong impact on a wide spectrum of essential processes including cardiac gene transcription, ROS defence, and mt energy metabolism.
In the classical model, p53 is activated in mitotic cells by oncogenic stress, DNA damage and other stress signals, triggering apoptosis and proliferative arrest [35,37]. It has been well established that two classes of caspases, initiator caspases including caspase 8, and effector caspases, such as caspase 3, are instrumental in the execution of apoptosis [102]. In addition, two apoptotic signaling pathways have been identified. Caspase 8 controls the death receptor pathway whereas caspase 3 activation is dependent on mt cytochrome c release and caspase 9 function. However, independent studies have confirmed that caspase 8 is frequently activated in response to apoptotic signals in a death receptor independent manner, but is dependent on the participation of activated caspase 3 [103]. This might explain the co-activation of caspase 3/8 in Mdm2-deficient cardiomyocytes as observed in our study.
Our study provides genetic proof that p53 is spontaneously active in Mdm2-deficient hearts, corroborating the prevailing view, that inhibition of p53 is the primary function of Mdm2. An increase in p53 activity, in response to Mdm2 loss, triggered a phenotype that alters normal heart function. What is the role of p53 in the regulation of all these responses? Our analysis of various Mdm2- and p53-mutant mice demonstrates, that a rise in p53 levels is responsible for the initial transcriptional changes. However, it appears very likely that p53 is not the sole cause that drives the development of the cardiac phenotype in Mdm2f/f;mcm mice.
We observed that the pleiotropic transcription factors E2f1 [72] and Myc [73], both playing important roles in the initiation phase of cell death, are co-induced by p53 (Fig 4D and 4E). It has been shown previously that endogenous Myc is important for p53-induced apoptosis in response to DNA damage in the adult mouse intestine [104]. The proapoptotic activities of E2f1 and p53 are also co-regulated [105]. For example, p53 and E2f1 are stabilized and activated in response to various stresses through phosphorylation by ataxia telangiectasia mutated (ATM) [106,107] and the checkpoint kinases Chk1/2 [108,109]. E2f1 and p53 also cooperate [110] to transactivate many proapoptotic genes, including Bax, caspases, and cytochrome c [111,112]. Moreover, the expression of key apoptotic genes, including Apaf-1, Puma, and Siva, are reportedly co-regulated by both factors [10]. In isolated cardiomyocytes and adult mouse heart, ectopic overexpression of E2f1 evoked apoptosis [113–118].
It is also noteworthy that Mdm2 exerts p53-independent activities that are at least partially mediated by the E2f1 pathway [119]. For example, Mdm2 can physically interact with the retinoblastoma protein [120–122], E2F1 itself, and Dp1, a transcriptional co-activator of E2f1. All these interactions can stimulate transcriptional transactivation of E2f1 target genes [123,124].
Despite the negative regulation of p53 by Mdm2, physiological stress can induce low levels of p53 activity that is involved in the regulation of glycolysis, oxidative phosphorylation, and oxidative stress in mitotic cells [28, 98,125–128] and in the adult mouse heart [129,130]. Seminal studies have firmly established that p53-mediated apoptosis generates ROS [131–133]. Based on this work, several independent investigations revealed a primary role for p53 in the generation of oxidative stress and mt dysfunction in mouse models of HF [10,101].
In a recent study, we were able to demonstrate that Myc is a specific target of the E3 ubiquitin ligase Mule, in the adult mouse heart [10]. Loss of Mule, with activation of Myc, severely impacts redox homeostasis and mt function through negative regulation of Pgc-1a and Pink1, leading to oxidative stress, energy deprivation and cardiac dysfunction. Based on these findings, it appears highly likely that transcriptional activation of Myc by p53 participates in the development of HF in Mdm2f/f;mcm mice. The role of E2f1 in the generation of oxidative stress has remained unclear. However, ROS production is essential for the apoptotic function of E2f1 [134]. Surprisingly, E2f1 can repress mt respiration at baseline in mammalian cells [129], whereas loss of drosophila dE2f compromises mt function [135]. Cardiac ischemia-reperfusion injury in mice led to an activation of E2f1 [136]. In addition, infarct size was decreased by 40% in E2f1 knockout mice, as would be expected if E2f1 stimulated apoptosis.
Maladaptive cardiac hypertrophy can be characterized by activation of a fetal gene program [137–139]. This group of genes includes ANP, BNP, fetal isoforms of contractile proteins (bMHC, skeletal a-actin), and smooth muscle genes (a-actin, SM22a) [140,141]. While all these factors are highly expressed in fetal hearts, they are transcriptionally silenced in the early neonatal period. Their re-expression under conditions of pathological stress is considered to play a mechanistic key role development of decompensated cardiac hypertrophy [142]. Intriguingly, we observed that ANP, BNP, and bMHC transcript levels were downregulated at 14d post-Tam (Fig 3K). Reportedly, the ANP gene promoter contains DNA-recognition sequences and low affinity binding sites for Gata4/6, Mef2, Nkx2.5, serum response factor (Srf) and Tbx5, that alone or cooperatively regulate ANP expression in cardiomyocytes [138;143–145]. Moreover, The BNP promoter contains Gata4/6, Srf, nuclear factor of activated T-cells (Nfat), and thyroid hormone responsive DNA binding site, all of which participate in the activation of BNP gene transcription [145,146]. In fetal cardiomyocytes, and under pathological conditions in the adult heart, bMHC expression is directly regulated by Mef2, Nfat4/3c and Nkx2.5 [147–149]. The Gata4 transcription has previously been shown to transactivate the promoters of many cardiac genes, including ANP, BNP, and bMHC [150]. Also, Morin et al. demonstrated, that Mef2 physically interacts with Gata4, and these Mef2/Gata4 protein complexes are then recruited to target promoters that containing Gata4 but not Mef2-specific DNA interaction sites [151]. The physiological function of Mef2 in this cooperation is to potentiate the transcriptional activity of Gata4 [151]. Based on these findings, it appears that the decrease in ANP, BNP, and bMHC expression observed in our study is caused by a marked downregulation of Mef2 family members and Srf transcription factors (Fig 6I).
An important question to answer is what is p53’s role in the down-regulation of cardiac-specific gene transcription in Mdm2f/f;mcm mice? Work from our group has shown that cardiac-specific conditional co-deletion of p53 and Mdm2 (DKO) in adult mice led to downregulation of cardiac gene expression and the development of a dilated cardiomyopathy, in the absence of cardiac hypertrophy [77] with alterations in miRNA expression. In the present study, we describe a broad dysregulation in the control of cardiac-specific gene expression in Mdm2-deficient hearts, whereas the conditional ablation of p53 in adult mice provided a cardioprotective effect through the increased expression of Ca2+ handling and contractile proteins [77]. The transcriptomic signature in p53-deficient hearts not only comprised genes important for cardiac architecture and function, but also mt biogenesis and bioenergetics, glucose and fatty acid metabolism, and transcription factors including Esrrg, Gata4, Mef2a, Myod, Pgc-1a and Tfam. How can these disparate findings be reconciled? One plausible explanation for this relies on the supposition that the default position for the p53 network in the normal heart is “on” wherein p53 is active, broadly regulating the cardiac transcriptome, albeit at low levels. This is in contrast to the classical model, where p53 is “off” until induced by acute stress [37,38]. In this model, ‘hyperactive’ p53 is responsible for the induction of apoptosis and oxidative stress. Thus, we conclude from all these findings that the Mdm2/p53 and Myc/E2f1 pathways, with crosstalk between them, explains the observed phenotype in Mdm2f/f;mcm mice.
In post-mitotic cardiomyocytes, the auto-regulatory feedback loop between p53 and Mdm2 [152] appears to be important to maintain normal heart function by restraining aberrant p53 activation. However, the individual contribution of Mdm2 to accomplish this remains unclear [153]. Firstly, it is highly unlikely that p53 is the only substrate of Mdm2 in differentiated cardiomyocytes in vivo [34]. Experimental evidence also suggests that other E3 ligases can regulate p53 stability. Secondly, other endogenous p53 inhibitors including Mdm4, and the ubiquitin ligases Cop1, Huwe1 (Mule) [154], and Prh2 quench p53-dependent transcriptionally activation in proliferating cells, in vitro [35,36,155]. This can potentially influence the contribution of Mdm2 in p53 regulation. Intriguingly, cardiac-specific loss of Mule activated Myc and p53, leading to the development of a phenotype similar to the Mdm2-deficient mice. This current study provides genetic proof for the view that the extent to which Mdm2 participates in buffering p53’s activity is significant. Indeed, further studies are needed to ascertain whether reduced Mdm2 expression in human HF represents a novel genetic abnormality that predisposes patients to HF, or is instead a consequence of HF itself.
The protein kinase Akt plays an important role in the regulation of survival [156] and hypertrophic growth [157]. Various growth factors promote their effect through activation of the phosphatidylinositide-3-kinase (Pi3K) signaling cascade. Mdm2 is one of several Akt substrates whose activation mediates the anti-apoptotic activity of Akt. Independent studies demonstrated, that Pi3K signaling induces Akt-dependent phosphorylation and activation of Mdm2 that eventually inhibits p53-dependent apoptosis [44,156]. Since Mdm2 inhibits cardiac hypertrophy, whereas Akt promotes this process, the existence of a negative feedback loop has been postulated, enabling Mdm2 to restrain Akt action [46].
In our study, transcriptomic analysis of Mdm2-mutant hearts revealed inhibition of known Myc-and E2f-target genes that were activated by p53 and are crucial for the regulation of mt energy metabolism and ROS detoxification under conditions of cellular stress. This is consistent with studies demonstrating that Myc and E2f transcription factor complexes bound to target promoters that shift from trans-activating to trans-repressing [45,158]. The questions remain as to how are E2f1 and Myc activated in Mdm2f/f;mcm mice post-Tam, and how are the transcriptional activities of p53, E2f1 and Myc interconnected to evoke such a profound cardiac phenotype in these mice? Intriguingly, the p53/Mdm2 [159] and Rb/E2f1 [70] pathways are both defective in most human cancers, underlining the importance of both pathways in regulating apoptosis and proliferation. In addition, E2f1 has been shown to induce cardiomyocyte apoptosis [160]. Mechanistically, p53 and E2f1 are stabilized in response to DNA damage [70] by enhanced oxidative stress in the hearts of Mdm2f/f;mcm mice post-Tam.
We have previously shown that Pink1-deficient hearts exhibit decreased CI activity, directly leading to a drop in mt membrane potential [84]. Indeed, defective OxPhos through defects in complex 1 (CI) (via Pink1) and CIV (via Myc-mediated transrepression of Ndufa4) paired with sustained fatty acid oxidation (FAO) in the Mdm2-deficient myocardium signifies the uncoupling of FAO and OxPhos, which leads to ROS generation [104]. The coincident downregulation of important redox regulators (Pink1, Cat, Sirt3, Sod2), and antioxidants (Gpx4, Gsta1, Txnrd2) in a p53/E2f1/Myc-dependent manner, renders the Mdm2-deficient heart sufficiently incapable to mobilize a defence against ROS, thereby leading to DNA damage. Clearly, downregulation of Pgc-1a and Pink1 in the absence of Mdm2 represents the loss of a central homeostatic pillar supporting substrate utilization and ROS defense in the myocardium, which ultimately has a severe impact on cardiomyocyte function.
Work from our own lab has shown that Pink1-deficient hearts are hypertrophic and have higher degrees of cardiomyocyte apoptosis with fibrosis [84]. In addition, these mice have greater levels of oxidative stress as reflected by higher levels of lipid peroxidation, DNA damage, and decreased aconitase activity in conjunction with reduced activity of several cytoplasmic and mt antioxidative systems. In response to biomechanical stress, the Pink1 knockout mice developed a further exaggeration in the degree of cardiac hypertrophy. Recently, a link between Pink1 dysfunction and the electron transport chain was provided. Under physiological conditions, Pink1 maintains CI activity by phosphorylation of its subunit Ndufa10, critical for ubiquinone reduction through the holo-complex [161]. Ectopic expression of the phospho-mimetic mutant Ndufa10 rescued mt depolarization defects in Pink1 knockout MEFs. CI dysfunction was also reversed by expression of the phospho-mimetic Ndufa10 in cells from Parkinson’s patients carrying Pink1 mutations. Whether Ndufa10 phosphorylation is mediated directly by Pink1 or is an indirect effect of Pink1, remains to be determined. Pink1 loss constitutes a mt defect that establishes a pathological imbalance in ROS production, leading to derangements in mt bioenergetics [84]. With the generation of abnormal ROS levels within the mt, a cascade of events is initiated, including a decrease in MMP, activation of the mt permeability transition pore (mPTP), and efflux of protons causing a decline in mt bioenergetics. This supports the clinical relevance of our finding that protein levels of Pink1 in end-stage human HF were also diminished [84], supporting a reciprocal relationship between Pink1 activity and HF.
In this study, we identified several genes including Pgc-1a and Pink1, downregulated following Mdm2 ablation, as principle factors mediating p53/Myc/E2f1-mediated decreases in mt dysfunction and altered energy metabolism. Pgc-1a is a central effector of mt biogenesis and FAO through transcriptional co-activation of Ppara [162]. Previously, Pgc-1a expression was shown to be inversely regulated in Myc-induced cardiac hypertrophy in transgenic mice [163]. This finding closely mirrors the outcome of our study in which Myc overexpression, driven by p53/E2f1 activation, downregulates Pgc-1a and, consequently, decreases expression of the nuclear receptors Ppara and Esrrb/g. The combined activities of both transcription factor complexes oversee the principle mechanism of ATP production in the heart through transcriptional regulation of fatty acid uptake and FAO factors [164]. Thus, their absence in Mdm2-deficient heart leaves the heart vulnerable to impaired ATP production, which even takes on greater impact when the heart is stressed. Fig 6N illustrates the consequences of impaired Pgc-1a, Ppara/g and Esrrb/g axes on mt energy metabolism in the absence of Mdm2. In conclusion, our study provides a new level of understanding of how heart function is maintained at baseline, and how Mdm2 participates in the mediation of this process.
Abbreviations
- Ace2
angiotensin I converting enzyme (peptidyl-dipeptidase A) 2
- Aco2
mitochondrial aconitase 2
- Adra1a
adrenergic receptor, alpha 1a
- Adrb2
adrenergic receptor, beta 2
- aMHC
myosin, heavy polypeptide 6 cardiac muscle alpha (Myh6)
- ANP (Nppa)
natriuretic peptide type A
- Apaf1
apoptotic peptidase activating factor 1
- ATP5a1
ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1
- ATPaf1
ATP synthase mitochondrial F1 complex assembly factor 1
- Bak
BCL2-antagonist/killer 1
- Bax
BCL2-associated X protein
- bMHC
myosin heavy polypeptide 7 cardiac muscle beta (Myh7)
- BNP (Nppb)
natriuretic peptide type B
- Casp1
caspase 1
- Casp3
caspase 3
- Casp8
caspase 8
- Cat
catalase
- Col
collagen (for example: Col1a1, collagen type I alpha 1)
- Cox10
cytochrome c oxidase assembly protein 10
- Cox4i1
cytochrome c oxidase subunit IV isoform 1
- Cx43
connexin 43 (Gja1, gap junction protein, alpha 1)
- E2f1
E2F transcription factor 1
- Esrrb
estrogen related receptor beta
- Esrrg
estrogen related receptor gamma
- Gata4
GATA binding protein 4
- Gpx4
glutathione peroxidase 4
- Gsta1
glutathione S-transferase, alpha 1
- Lam
laminin alpha 1 (Lama1)
- Mdm2
murine double-minute 2
- Mef2a
myocyte enhancer factor 2A
- Mef2d
myocyte enhancer factor 2D
- Mfn1
mitofusin 1
- Mfn2
mitofusin 2
- Myc
myelocytomatosis oncogene
- Myl3
myosin, light polypeptide 3
- Mylk3
myosin light chain kinase 3
- Ndufa5
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex 5
- Ndufa6
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex 6 (B14)
- Ndufa8
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex 8
- Nkx2-5
NK2 homeobox 5
- Npm1
nucleophosmin 1
- Opa1
optic atrophy 1
- p53
transformation related protein 53 (Trp53)
- Pgc-1a
peroxisome proliferative activated receptor gamma coactivator 1 alpha (Ppargc1a)
- Pgc-1b
peroxisome proliferative activated receptor gamma coactivator 1 beta (Ppargc1b)
- Pink1
PTEN induced putative kinase 1
- Pln
phospholamban
- Ppara
peroxisome proliferator activated receptor alpha
- Ryr2
cardiac ryanodine receptor 2
- Sdha
succinate dehydrogenase complex subunit A
- Sdhb
succinate dehydrogenase complex subunit B
- SERCA2
ATPase Ca2+ transporting cardiac muscle slow twitch 2 (Atp2a2)
- Sirt3
sirtuin 3
- Sod2
mitochondrial superoxide dismutase 2
- Srf
serum response factor
- Tcap
titin-cap
- Tfam
transcription factor A, mitochondrial
- Tfb1m
transcription factor B1, mitochondrial
- Tfb2m
transcription factor B2, mitochondrial
- Tnnc1
cardiac/slow skeletal troponin C
- Tnni3
troponin I cardiac 3
- Tnnt2
troponin T2, cardiac type
- Tpm2
tropomyosin 2 beta
- Tpm3
tropomyosin 3
- Ttn
titan
- Txnrd2
thioredoxin reductase 2
- Uqcr11
ubiquinol-cytochrome c reductase, complex III subunit XI
All annotations use the mouse gene nomenclature. Designations in brackets refer to the official gene symbol.
Data Availability
Data are available from Experiment ArrayExpress accession: E-MTAB-5441; http://www.ebi.ac.uk/arrayexpress.
Funding Statement
This work was funded by the Canadian Institutes for Health Research in the form of an operating grant (#299339) and salary support provided by the Canadian Institute for Health Research, Clinician-Scientist award for Dr. Filio Billia (#314888).
References
- 1.Akazawa H. Mechanisms of cardiovascular homeostasis and pathophysiology—from gene expression, signal transduction to cellular communication. Circ J. 2015;79: 2529–2536. doi: 10.1253/circj.CJ-15-0818 [DOI] [PubMed] [Google Scholar]
- 2.Borlaug BA, Paulus WJ. Heart failure with preserved ejection fraction: pathophysiology, diagnosis, and treatment. Eur Heart J. 2011;32: 670–679. doi: 10.1093/eurheartj/ehq426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jessup M, Konstam MA, Mancini DM, Michl K, Oates JA, Rahko PS, et al. ACC/AHA 2005 Guideline Update for the Diagnosis and Management of Chronic Heart Failure in the Adult: a report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines (Writing Committee to Update the 2001 Guidelines for the Evaluation and Management of Heart Failure): developed in collaboration with the American College of Chest Physicians and the International Society for Heart and Lung Transplantation: endorsed by the Heart Rhythm Society. Circulation. 2005;112: e154–e235. doi: 10.1161/CIRCULATIONAHA.105.167586 [DOI] [PubMed] [Google Scholar]
- 4.Kapiloff MS, Emter CA. The cardiac enigma: current conundrums in heart failure research. F1000Res. 2016;5 doi: 10.12688/f1000research.7278.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mann DL, Bristow MR. Mechanisms and models in heart failure: The biomechanical model and beyond. Circulation. 2005;111: 2837–2849. doi: 10.1161/CIRCULATIONAHA.104.500546 [DOI] [PubMed] [Google Scholar]
- 6.Braunwald E, Bristow MR. Congestive heart failure: fifty years of progress. Circulation. 2000;102: IV14–23. [DOI] [PubMed] [Google Scholar]
- 7.Cohn JN. The management of chronic heart failure. N Engl J Med. 1996;335: 490–498. doi: 10.1056/NEJM199608153350707 [DOI] [PubMed] [Google Scholar]
- 8.Jessup M, Brozena S. Heart failure. N Engl J Med. 2003;348: 2007–2018. doi: 10.1056/NEJMra021498 [DOI] [PubMed] [Google Scholar]
- 9.Heidenreich PA, Albert NM, Allen LA, Bluemke DA, Butler J, Fonarow GC, et al. Forecasting the impact of heart failure in the United States: a policy statement from the American Heart Association. Circ Heart Fail. 2013;6: 606–619. doi: 10.1161/HHF.0b013e318291329a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dadson K, Hauck L, Hao Z, Grothe D, Rao V, Mak TW, et al. The E3 ligase Mule protects the heart against oxidative stress and mitochondrial dysfunction through Myc-dependent inactivation of Pgc-1α and Pink1. Sci Rep. 2017;7 doi: 10.1038/srep41490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pasumarthi KB, Field LJ. Cardiomyocyte cell cycle regulation. Circ Res. 2002;90: 1044–1054. [DOI] [PubMed] [Google Scholar]
- 12.Senyo SE, Steinhauser ML, Pizzimenti CL, Yang VK, Cai L, Wang M, et al. Mammalian heart renewal by pre-existing cardiomyocytes. Nature. 2012;493: 433–436. doi: 10.1038/nature11682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sutton MG, Sharpe N. Left ventricular remodeling after myocardial infarction: pathophysiology and therapy. Circulation. 2000;101: 2981–2988. [DOI] [PubMed] [Google Scholar]
- 14.Zebrowski DC, Becker R, Engel FB. Towards regenerating the mammalian heart: challenges in evaluating experimentally induced adult mammalian cardiomyocyte proliferation. Am J Physiol Heart Circ Physiol. 2016;310: H1045–H1054. doi: 10.1152/ajpheart.00697.2015 [DOI] [PubMed] [Google Scholar]
- 15.Maillet M, van Berlo JH, Molkentin JD. Molecular basis of physiological heart growth: fundamental concepts and new players. Nat Rev Mol Cell Biol. 2013;14: 38–48. doi: 10.1038/nrm3495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schoenfeld JR, Vasser M, Jhurani P, Ng P, Hunter JJ, Ross J Jr, et al. Distinct molecular phenotypes in murine cardiac muscle development, growth, and hypertrophy. J Mol Cell Cardiol. 1998;30: 2269–2280. [DOI] [PubMed] [Google Scholar]
- 17.van Berlo JH, Maillet M, Molkentin JD. Signaling effectors underlying pathologic growth and remodeling of the heart. J Clin Invest. 2013;123: 37–45. doi: 10.1172/JCI62839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gajarsa JJ, Kloner RA. Left ventricular remodeling in the postinfarction heart: a review of cellular, molecular mechanisms, and therapeutic modalities. Heart Fail Rev. 2011;16: 13–21. doi: 10.1007/s10741-010-9181-7 [DOI] [PubMed] [Google Scholar]
- 19.González A, López B, Ravassa S, Beaumont J, Arias T, Hermida N, et al. Biochemical markers of myocardial remodelling in hypertensive heart disease. Cardiovasc Res. 2009;81: 509–518. doi: 10.1093/cvr/cvn235 [DOI] [PubMed] [Google Scholar]
- 20.Lorell BH, Carabello BA. Left ventricular hypertrophy: pathogenesis, detection, and prognosis. Circulation. 2000;102: 470–479. [DOI] [PubMed] [Google Scholar]
- 21.Tsutsui H, Kinugawa S, Matsushima S. Oxidative stress and heart failure. Am J Physiol Heart Circ Physiol. 2011;301: H2181–H2190. doi: 10.1152/ajpheart.00554.2011 [DOI] [PubMed] [Google Scholar]
- 22.Travers JG, Kamal FA, Robbins J, Yutzey KE, Blaxall BC. Cardiac fibrosis: The fibroblast awakens. Circ Res. 2016;118: 1021–1040. doi: 10.1161/CIRCRESAHA.115.306565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Calise J, Powell SR. The ubiquitin proteasome system and myocardial ischemia. Am J Physiol Heart Circ Physiol. 2013;304: H337–H349. doi: 10.1152/ajpheart.00604.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Day SM. The ubiquitin proteasome system in human cardiomyopathies and heart failure. Am J Physiol Heart Circ Physiol. 2013;304: H1283–H1293. doi: 10.1152/ajpheart.00249.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X, Li J, Zheng H, Su H, Powell SR. Proteasome functional insufficiency in cardiac pathogenesis. Am J Physiol Heart Circ Physiol. 2011;301: H2207–H2219. doi: 10.1152/ajpheart.00714.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Willis MS, Bevilacqua A, Pulinilkunnil T, Kienesberger P, Tannu M, Patterson C. The role of ubiquitin ligases in cardiac disease. J Mol Cell Cardiol. 2014;71: 43–53. doi: 10.1016/j.yjmcc.2013.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Portbury AL, Ronnebaum SM, Zungu M, Patterson C, Willis MS. Back to your heart: ubiquitin proteasome system-regulated signal transduction. J Mol Cell Cardiol. 2012;52: 526–537. doi: 10.1016/j.yjmcc.2011.10.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hedhli N, Lizano P, Hong C, Fritzky LF, Dhar SK, Liu H, et al. Proteasome inhibition decreases cardiac remodeling after initiation of pressure overload. Am J Physiol Heart Circ Physiol. 2008;295: H1385–H1393. doi: 10.1152/ajpheart.00532.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Powell SR, Wang P, Katzeff H, Shringarpure R, Teoh C, Khaliulin I, et al. Oxidized and ubiquitinated proteins may predict recovery of postischemic cardiac function: essential role of the proteasome. Antioxid. Redox Signal. 2005;7: 538–546. doi: 10.1089/ars.2005.7.538 [DOI] [PubMed] [Google Scholar]
- 30.Pye J, Ardeshirpour F, McCain A, Bellinger DA, Merricks E, Adams J, et al. Proteasome inhibition ablates activation of NF-kappa B in myocardial reperfusion and reduces reperfusion injury. Am J Physiol Heart Circ Physiol. 2003;284: H919–926. doi: 10.1152/ajpheart.00851.2002 [DOI] [PubMed] [Google Scholar]
- 31.Iwakuma T, Lozano G. MDM2, an introduction. Mol Cancer Res. 2003;1: 993–1000. [PubMed] [Google Scholar]
- 32.Eischen CM, Lozano G. The Mdm network and its regulation of p53 activities: a rheostat of cancer risk. Hum Mutat. 2014;35: 728–737. doi: 10.1002/humu.22524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marine JC, Lozano G. Mdm2-mediated ubiquitylation: p53 and beyond. Cell Death Differ. 2010;17: 93–102. doi: 10.1038/cdd.2009.68 [DOI] [PubMed] [Google Scholar]
- 34.Riley MF, Lozano G. The Many Faces of MDM2 Binding Partners. Genes Cancer. 2012;3: 226–239. doi: 10.1177/1947601912455322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harris SL, Levine AJ. The p53 pathway: positive and negative feedback loops. Oncogene. 2005;24: 2899–2908. doi: 10.1038/sj.onc.1208615 [DOI] [PubMed] [Google Scholar]
- 36.Pant V, Lozano G. Dissecting the p53-Mdm2 feedback loop in vivo: uncoupling the role in p53 stability and activity. Oncotarget. 2014;5: 1149–1156. doi: 10.18632/oncotarget.1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vousden KH, Lu X. Live or let die: the cell’s response to p53. Nat Rev Cancer. 2002;2: 594–604. doi: 10.1038/nrc864 [DOI] [PubMed] [Google Scholar]
- 38.Vousden KH, Lane DP. p53 in health and disease. Nat Rev Mol Cell Biol. 2007;8: 275–283. doi: 10.1038/nrm2147 [DOI] [PubMed] [Google Scholar]
- 39.Vousden KH, Prives C. Blinded by the light: The growing complexity of p53. Cell. 2009;137: 413–431. doi: 10.1016/j.cell.2009.04.037 [DOI] [PubMed] [Google Scholar]
- 40.Donehower LA, Lozano G. 20 years studying p53 functions in genetically engineered mice. Nat Rev Cancer. 2009;9: 831–841. doi: 10.1038/nrc2731 [DOI] [PubMed] [Google Scholar]
- 41.Jones SN, Roe AE, Donehower LA, Bradley A. Rescue of embryonic lethality in mdm2-deficient mice by absence of p53. Nature. 1995;378: 206–208. doi: 10.1038/378206a0 [DOI] [PubMed] [Google Scholar]
- 42.Jones SN, Hancock AR, Vogel H, Donehower LA, Bradley A. Overexpression of Mdm2 in mice reveals a p53-independent role for Mdm2 in tumorigenesis. Proc Natl Acad Sci USA. 1998;95: 15608–15612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Montes de Oca Luna R, Wagner DS, Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature. 1995;378: 203–206. doi: 10.1038/378203a0 [DOI] [PubMed] [Google Scholar]
- 44.Ogawara Y, Kishishita S, Obata T, Isazawa Y, Suzuki T, Tanaka K, et al. Akt enhances Mdm2-mediated ubiquitination and degradation of p53. J Biol Chem. 2002;277: 21843–21850. doi: 10.1074/jbc.M109745200 [DOI] [PubMed] [Google Scholar]
- 45.Grier JD, Xiong S, Elizondo-Fraire AC, Parant JM, Lozano G. Tissue-specific differences of p53 inhibition by Mdm2 and Mdm4. Mol Cell Biol. 2006;26: 192–198. doi: 10.1128/MCB.26.1.192-198.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Toth A, Nickson P, Qin LL, Erhardt P. Differential regulation of cardiomyocyte survival and hypertrophy by MDM2, an E3 ubiquitin ligase. J Biol Chem. 2006;281: 3679–3689. doi: 10.1074/jbc.M509630200 [DOI] [PubMed] [Google Scholar]
- 47.Xiong S. Mouse models of Mdm2 and Mdm4 and their clinical implications. Chin J Cancer. 2013;32: 371–375. doi: 10.5732/cjc.012.10286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sohal DS, Nghiem M, Crackower MA, Witt SA, Kimball TR, Tymitz KM, et al. Temporally regulated and tissue-specific gene manipulations in the adult and embryonic heart using a tamoxifen-inducible Cre protein. Circ Res. 2001;89: 20–25. [DOI] [PubMed] [Google Scholar]
- 49.Luker GD, Pica CM, Song J, Luker KE, Piwnica-Worms D. Imaging 26S proteasome activity and inhibition in living mice. Nat Med. 2003;9: 969–973. doi: 10.1038/nm894 [DOI] [PubMed] [Google Scholar]
- 50.Potthoff MJ, Wu H, Arnold MA, Shelton JM, Backs J, McAnally J, et al. Histone deacetylase degradation and MEF2 activation promote the formation of slow-twitch myofibers. J Clin Invest. 2007;117: 2459–2467. doi: 10.1172/JCI31960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Spandidos A, Wang X, Wang H, Dragnev S, Thurber T, Seed B. A comprehensive collection of experimentally validated primers for Polymerase Chain Reaction quantitation of murine transcript abundance. BMC Genomics. 2008;9 doi: 10.1186/1471-2164-9-633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Matsusaka H, Ide T, Matsushima S, Ikeuchi M, Kubota T, Sunagawa K. Targeted deletion of p53 prevents cardiac rupture after myocardial infarction in mice. Cardiovasc Res. 2006;70: 457–465. doi: 10.1016/j.cardiores.2006.02.001 [DOI] [PubMed] [Google Scholar]
- 53.Shukla PC, Singh KK, Quan A, Al-Omran M, Teoh H, Lovren F, et al. BRCA1 is an essential regulator of heart function and survival following myocardial infarction. Nat Commun. 2011;2 doi: 10.1038/ncomms1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Toko H, Takahashi H, Kayama Y, Oka T, Minamino T, Okada S, et al. Ca2+/calmodulin-dependent kinase II delta causes heart failure by accumulation of p53 in dilated cardiomyopathy. Circulation. 2010;122: 891–899. doi: 10.1161/CIRCULATIONAHA.109.935296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Toth A, Jeffers JR, Nickson P, Min JY, Morgan JP, Zambetti GP, et al. Targeted deletion of Puma attenuates cardiomyocyte death and improves cardiac function during ischemia-reperfusion. Am J Physiol Heart Circ Physiol. 2006;291: H52–H60. doi: 10.1152/ajpheart.01046.2005 [DOI] [PubMed] [Google Scholar]
- 56.Wang EY, Gang H, Aviv Y, Dhingra R, Margulets V, Kirshenbaum LA. p53 mediates autophagy and cell death by a mechanism contingent on Bnip3. Hypertension. 2013;62: 70–77. doi: 10.1161/HYPERTENSIONAHA.113.01028 [DOI] [PubMed] [Google Scholar]
- 57.Zhang GX, Kimura S, Murao K, Obata K, Matsuyoshi H, Takaki M. Inhibition of cytochrome c release by 10-N-nonyl acridine orange, a cardiolipin-specific dye, during myocardial ischemia-reperfusion in the rat. Am J Physiol Heart Circ Physiol. 2010;298: H433–H439. doi: 10.1152/ajpheart.00938.2009 [DOI] [PubMed] [Google Scholar]
- 58.Mendrysa SM, McElwee MK, Michalowski J, O’Leary KA, Young KM, Perry ME. Mdm2 is critical for inhibition of p53 during lymphopoiesis and the response to ionizing irradiation. Mol Cell Biol. 2003;23: 462–72. doi: 10.1128/MCB.23.2.462-473.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9: 402–412. doi: 10.1038/nrm2395 [DOI] [PubMed] [Google Scholar]
- 60.Moroni MC, Hickman ES, Lazzerini Denchi E, Caprara G, Colli E, Cecconi F, et al. Apaf-1 is a transcriptional target for E2F and p53. Nat Cell Biol. 2001;3: 552–558. doi: 10.1038/35078527 [DOI] [PubMed] [Google Scholar]
- 61.Soengas MS, Alarcón RM, Yoshida H, Giaccia AJ, Hakem R, Mak TW, et al. Apaf-1 and caspase-9 in p53-dependent apoptosis and tumor inhibition. Science. 1999;284: 156–159. [DOI] [PubMed] [Google Scholar]
- 62.Fortin A, Cregan SP, MacLaurin JG, Kushwaha N, Hickman ES, Thompson CS, et al. APAF1 is a key transcriptional target for p53 in the regulation of neuronal cell death. J Cell Biol. 2001;155: 207–216. doi: 10.1083/jcb.200105137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gupta S, Radha V, Furukawa Y, Swarup G. Direct transcriptional activation of human caspase-1 by tumor suppressor p53. J Biol Chem. 2001;276: 10585–10588. doi: 10.1074/jbc.C100025200 [DOI] [PubMed] [Google Scholar]
- 64.Dumont P, Leu JI, Della Pietra AC 3rd, George DL, Murphy M. The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nat Genet. 2003;33: 357–365. doi: 10.1038/ng1093 [DOI] [PubMed] [Google Scholar]
- 65.Cregan SP, MacLaurin JG, Craig CG, Robertson GS, Nicholson DW, Park DS, et al. Bax-dependent caspase-3 activation is a key determinant in p53-induced apoptosis in neurons. J Neurosci. 1999;19: 7860–7889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Erster S, Mihara M, Kim RH, Petrenko O, Moll UM. In vivo mitochondrial p53 translocation triggers a rapid first wave of cell death in response to DNA damage that can precede p53 target gene activation. Mol Cell Biol. 2004;24: 6728–6741. doi: 10.1128/MCB.24.15.6728-6741.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ehrhardt H, Häcker S, Wittmann S, Maurer M, Borkhardt A, Toloczko A, et al. Cytotoxic drug-induced, p53-mediated upregulation of caspase-8 in tumor cells. Oncogene. 2008;27: 783–793. doi: 10.1038/sj.onc.1210666 [DOI] [PubMed] [Google Scholar]
- 68.Henry RE, Andrysik Z, París R, Galbraith MD, Espinosa JM. A DR4:tBID axis drives the p53 apoptotic response by promoting oligomerization of poised BAX. EMBO J. 2012;31: 1266–1278. doi: 10.1038/emboj.2011.498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chipuk JE, Green DR. Dissecting p53-dependent apoptosis. Cell Death Differ. 2006;13: 994–1002. doi: 10.1038/sj.cdd.4401908 [DOI] [PubMed] [Google Scholar]
- 70.Polager S, Ginsberg D. p53 and E2f: partners in life and death. Nat Rev Cancer. 2009;9: 738–748. doi: 10.1038/nrc2718 [DOI] [PubMed] [Google Scholar]
- 71.Tanaka H, Matsumura I, Ezoe S, Satoh Y, Sakamaki T, Albanese C, et al. E2F1 and c-Myc potentiate apoptosis through inhibition of NF-kappaB activity that facilitates MnSOD-mediated ROS elimination. Mol Cell. 2002;9: 1017–1029. [DOI] [PubMed] [Google Scholar]
- 72.Trimarchi JM, Lees JA. Sibling rivalry in the E2F family. Nat Rev Mol Cell Biol. 2002;3: 11–20. doi: 10.1038/nrm714 [DOI] [PubMed] [Google Scholar]
- 73.Dang CV. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol Cell Biol. 1999;19: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Leu JI, Dumont P, Hafey M, Murphy ME, George DL. Mitochondrial p53 activates Bak and causes disruption of a Bak-Mcl1 complex. Nat Cell Biol. 2004;6: 443–450. doi: 10.1038/ncb1123 [DOI] [PubMed] [Google Scholar]
- 75.Perfettini JL, Kroemer RT, Kroemer G. Fatal liaisons of p53 with Bax and Bak. Nat Cell Biol. 2004;6: 38638–38638. [DOI] [PubMed] [Google Scholar]
- 76.Fulda S, Debatin KM. Extrinsic versus intrinsic apoptosis pathways in anticancer chemotherapy. Oncogene. 2006;25: 4798–4811. doi: 10.1038/sj.onc.1209608 [DOI] [PubMed] [Google Scholar]
- 77.Stanley-Hasnain S, Hauck L, Grothe D, Aschar-Sobbi R, Beca S, Butany J, et al. p53 and Mdm2 act synergistically to maintain cardiac homeostasis and mediate cardiomyocyte cell cycle arrest through a network of microRNAs. Cell Cycle. 2017;26: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Christians ES, Benjamin IJ. Proteostasis and REDOX state in the heart. Am J Physiol Heart Circ Physiol. 2012;302: H24–H37. doi: 10.1152/ajpheart.00903.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.McMurray J, Chopra M, Abdullah I, Smith WE, Dargie HJ. Evidence of oxidative stress in chronic heart failure in humans. Eur Heart J. 1993;14: 1493–1498. [DOI] [PubMed] [Google Scholar]
- 80.Tsutsui H, Kinugawa S, Matsushima S. Oxidative stress and heart failure. Am J Physiol Heart Circ Physiol. 2011;301: H2181–H2190. doi: 10.1152/ajpheart.00554.2011 [DOI] [PubMed] [Google Scholar]
- 81.Sablina AA, Budanov AV, Ilyinskaya GV, Agapova LS, Kravchenko JE, Chumakov PM. The antioxidant function of the p53 tumor suppressor. Nat Med. 2005;11: 1306–1313. doi: 10.1038/nm1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Macleod KF. The role of the RB tumour suppressor pathway in oxidative stress responses in the haematopoietic system. Nat Rev Cancer. 2008;8: 769–781. doi: 10.1038/nrc2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Vafa O, Wade M, Kern S, Beeche M, Pandita TK, Hampton GM, et al. c-Myc can induce DNA damage, increase reactive oxygen species, and mitigate p53 function: a mechanism for oncogene-induced genetic instability. Mol Cell. 2002;9: 1031–1044. [DOI] [PubMed] [Google Scholar]
- 84.Billia F, Hauck L, Konecny F, Rao V, Shen J, Mak TW. PTEN-inducible kinase 1 (PINK1)/Park6 is indispensable for normal heart function. Proc Natl Acad Sci USA. 2011;108: 9572–9577. doi: 10.1073/pnas.1106291108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Maejima Y, Chen Y, Isobe M, Gustafsson ÅB, Kitsis RN, Sadoshima J. Recent progress in research on molecular mechanisms of autophagy in the heart. Am J Physiol Heart Circ Physiol. 2015;308: H259–H268. doi: 10.1152/ajpheart.00711.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sadoshima J, Izumo S. The cellular and molecular response of cardiac myocytes to mechanical stress. Annu Rev Physiol. 1997;59: 551–571. doi: 10.1146/annurev.physiol.59.1.551 [DOI] [PubMed] [Google Scholar]
- 87.Ide T, Tsutsui H, Kinugawa S, Utsumi H, Kang D, Hattori N, et al. Mitochondrial electron transport complex I is a potential source of oxygen free radicals in the failing myocardium. Circ Res. 1999;85: 357–363. [DOI] [PubMed] [Google Scholar]
- 88.Petrosillo G, Ruggiero FM, Di Venosa N, Paradies G. Decreased complex III activity in mitochondria isolated from rat heart subjected to ischemia and reperfusion: role of reactive oxygen species and cardiolipin. FASEB J. 2003;17: 714–716. doi: 10.1096/fj.02-0729fje [DOI] [PubMed] [Google Scholar]
- 89.Chen YR, Zweier JL. Cardiac mitochondria and reactive oxygen species generation. Circ Res. 2014;114: 524–537. doi: 10.1161/CIRCRESAHA.114.300559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chen Q, Vazquez EJ, Moghaddas S, Hoppel CL, Lesnefsky EJ. Production of reactive oxygen species by mitochondria: central role of complex III. J Biol Chem. 2003;278: 36027–36031. doi: 10.1074/jbc.M304854200 [DOI] [PubMed] [Google Scholar]
- 91.Falkenberg M, Larsson NG, Gustafsson CM. DNA replication and transcription in mammalian mitochondria. Annu Rev Biochem. 2007;76: 679–699. doi: 10.1146/annurev.biochem.76.060305.152028 [DOI] [PubMed] [Google Scholar]
- 92.Kolwicz SC, Purohit S, Tian R. Cardiac metabolism and its interactions with contraction, growth, and survival of cardiomyocytes. Circ Res. 2013;113: 603–616. doi: 10.1161/CIRCRESAHA.113.302095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hunter JJ, Chien KR. Signaling pathways for cardiac hypertrophy and failure. N Engl J Med. 1999;341: 1276–1283. doi: 10.1056/NEJM199910213411706 [DOI] [PubMed] [Google Scholar]
- 94.Widmann C, Gibson S, Jarpe MB, Johnson GL. Mitogen-activated protein kinase: conservation of a three-kinase module from yeast to human. Physiol Rev. 1999;79: 143–180. [DOI] [PubMed] [Google Scholar]
- 95.Schulz R, Aker S, Belosjorow S, Konietzka I, Rauen U, Heusch G. Stress kinase phosphorylation is increased in pacing-induced heart failure in rabbits. Am J Physiol Heart Circ Physiol. 2003;285: H2084–H2090. doi: 10.1152/ajpheart.01038.2002 [DOI] [PubMed] [Google Scholar]
- 96.Takeishi Y, Huang Q, Abe J, Che W, Lee JD, Kawakatsu H, et al. Activation of mitogen-activated protein kinases and p90 ribosomal S6 kinase in failing human hearts with dilated cardiomyopathy. Cardiovasc Res. 2002;53: 131–137. [DOI] [PubMed] [Google Scholar]
- 97.Lombardi AA, Elrod JW. mtDNA damage in the development of heart failure. Am J Physiol Heart Circ Physiol. 2015;309: H393–H395. doi: 10.1152/ajpheart.00475.2015 [DOI] [PubMed] [Google Scholar]
- 98.Mak TW, Hauck L, Grothe D, Billia F. p53 regulates the cardiac transcriptome. Proc Natl Acad Sci USA. 2017;114: 2331–2336. doi: 10.1073/pnas.1621436114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sahin E, Colla S, Liesa M, Moslehi J, Müller FL, Guo M, et al. Telomere dysfunction induces metabolic and mitochondrial compromise. Nature. 2011;470: 359–365. doi: 10.1038/nature09787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.de Rozieres S, Maya R, Oren M, Lozano G. The loss of mdm2 induces p53-mediated apoptosis. Oncogene. 2000;19: 1691–1697. doi: 10.1038/sj.onc.1203468 [DOI] [PubMed] [Google Scholar]
- 101.Ringshausen I, O’Shea CC, Finch AJ, Swigart LB, Evan GI. Mdm2 is critically and continuously required to suppress lethal p53 activity in vivo. Cancer Cell. 2006;10: 501–514. doi: 10.1016/j.ccr.2006.10.010 [DOI] [PubMed] [Google Scholar]
- 102.Elmore S. Apoptosis: A Review of Programmed Cell Death. Toxicol Pathol. 2007;35: 495–516. doi: 10.1080/01926230701320337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tang D, Lahti JM, Kidd VJ. Caspase-8 activation and bid cleavage contribute to MCF7 cellular execution in a caspase-3-dependent manner during staurosporine-mediated apoptosis. J Biol Chem. 2000;275: 9303–9307. [DOI] [PubMed] [Google Scholar]
- 104.Phesse TJ, Myant KB, Cole AM, Ridgway RA, Pearson H, Muncan V, et al. Endogenous c-Myc is essential for p53-induced apoptosis in response to DNA damage in vivo. Cell Death Differ. 2014;21: 956–966. doi: 10.1038/cdd.2014.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Polager S, Ginsberg D. p53 and E2f: partners in life and death. Nat Rev Cancer. 2009;9: 738–748. doi: 10.1038/nrc2718 [DOI] [PubMed] [Google Scholar]
- 106.Canman CE, Lim DS, Cimprich KA, Taya Y, Tamai K, Sakaguchi K, et al. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science. 1998;281: 1677–1679. [DOI] [PubMed] [Google Scholar]
- 107.Lin WC, Lin FT, Nevins JR. Selective induction of E2F1 in response to DNA damage, mediated by ATM-dependent phosphorylation. Genes Dev. 2001;15: 1833–1844. [PMC free article] [PubMed] [Google Scholar]
- 108.Hirao A, Kong YY, Matsuoka S, Wakeham A, Ruland J, Yoshida H, et al. DNA damage induced activation of p53 by the checkpoint kinase Chk2. Science. 2000;287: 1824–1827. [DOI] [PubMed] [Google Scholar]
- 109.Stevens C, Smith L, La Thangue NB. Chk2 activates E2F-1 in response to DNA damage. Nature Cell Biol. 2003;5: 401–409. doi: 10.1038/ncb974 [DOI] [PubMed] [Google Scholar]
- 110.Wu X, Levine AJ. p53 and E2F-1 cooperate to mediate apoptosis. Proc Natl Acad Sci USA. 1994;91: 3602–3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Miyashita T, Reed JC. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell. 1995;80: 293–299. [DOI] [PubMed] [Google Scholar]
- 112.Nahle Z, Polakoff J, Davuluri RV, McCurrach ME, Jacobson MD, Narita M, et al. Direct coupling of the cell cycle and cell death machinery by E2F. Nat Cell Biol. 2002;4: 859–864. doi: 10.1038/ncb868 [DOI] [PubMed] [Google Scholar]
- 113.Hershko T, Chaussepied M, Oren M, Ginsberg D. Novel link between E2F and p53: proapoptotic cofactors of p53 are transcriptionally upregulated by E2F. Cell Death Differ. 2005;12: 377–383. doi: 10.1038/sj.cdd.4401575 [DOI] [PubMed] [Google Scholar]
- 114.Fortin A, MacLaurin JG, Arbour N, Cregan SP, Kushwaha N, Callaghan SM, et al. The proapoptotic gene SIVA is a direct transcriptional target for the tumor suppressors p53 and E2F1. J Biol Chem. 2004;279: 28706–28714. doi: 10.1074/jbc.M400376200 [DOI] [PubMed] [Google Scholar]
- 115.Kirshenbaum LA, Abdellatif M, Chakraborty S, Schneider MD. Human E2F-1 reactivates cell cycle progression in ventricular myocytes and represses cardiac gene transcription. Dev Biol. 1996;179: 402–411. doi: 10.1006/dbio.1996.0270 [DOI] [PubMed] [Google Scholar]
- 116.Agah R, Kirshenbaum LA, Abdellatif M, Truong LD, Chakraborty S, Michael LH, et al. Adenoviral delivery of E2F-1 directs cell cycle reentry and p53-independent apoptosis in postmitotic adult myocardium in vivo. J Clin Invest. 1997;100: 2722–2728. doi: 10.1172/JCI119817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.von Harsdorf R, Hauck L, Mehrhof F, Wegenka U, Cardoso MC, Dietz R. E2F-1 overexpression in cardiomyocytes induces downregulation of p21CIP1 and p27KIP1 and release of active cyclin-dependent kinases in the presence of insulin-like growth factor I. Circ Res. 1999;85: 128–136. [DOI] [PubMed] [Google Scholar]
- 118.Loughran O, La Thangue NB. Apoptotic and growth-promoting activity of E2F modulated by MDM2. Mol Cell Biol. 2000;20: 2186–2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sdek P, Ying H, Chang DL, Qiu W, Zheng H, Touitou R, et al. MDM2 promotes proteasome dependent ubiquitin-independent degradation of retinoblastoma protein. Mol Cell. 2005;20: 699–708. doi: 10.1016/j.molcel.2005.10.017 [DOI] [PubMed] [Google Scholar]
- 120.Xiao ZX, Chen J, Levine AJ, Modjtahedi N, Xing J, Sellers WR, et al. Interaction between the retinoblastoma protein and the oncoprotein MDM2. Nature. 1995;375: 694–698. doi: 10.1038/375694a0 [DOI] [PubMed] [Google Scholar]
- 121.Hsieh JK, Chan FS, O’Connor DJ, Mittnacht S, Zhong S, Lu X. RB regulates the stability and the apoptotic function of p53 via MDM2. Mol Cell. 1999;3: 181–193. [DOI] [PubMed] [Google Scholar]
- 122.Sdek P, Ying H, Zheng H, Margulis A, Tang X, Tian K, et al. The central acidic domain of MDM2 is critical in inhibition of retinoblastoma-mediated suppression of E2F and cell growth. J Biol Chem. 2004;279: 53317–53322. doi: 10.1074/jbc.M406062200 [DOI] [PubMed] [Google Scholar]
- 123.Martin K. et al. Stimulation of E2F1/DP1 transcriptional activity by MDM2 oncoprotein. Nature. 1995;375: 691–694. doi: 10.1038/375691a0 [DOI] [PubMed] [Google Scholar]
- 124.Jones RG, Plas DR, Kubek S, Buzzai M, Mu J, Xu Y, et al. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol Cell. 2005;18: 283–293. doi: 10.1016/j.molcel.2005.03.027 [DOI] [PubMed] [Google Scholar]
- 125.Matoba S, Kang JG, Patino WD, Wragg A, Boehm M, Gavrilova O, et al. p53 regulates mitochondrial respiration. Science. 2006; 312:1650–1653. doi: 10.1126/science.1126863 [DOI] [PubMed] [Google Scholar]
- 126.Bensaad K, Tsuruta A, Selak MA, Vidal MN, Nakano K, Bartrons R, et al. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126: 107–20. doi: 10.1016/j.cell.2006.05.036 [DOI] [PubMed] [Google Scholar]
- 127.Budanov AV, Sablina AA, Feinstein E, Koonin EV, Chumakov PM. Regeneration of peroxiredoxins by p53-regulated sestrins, homologs of bacterial AhpD. Science. 2004;304: 596–600. doi: 10.1126/science.1095569 [DOI] [PubMed] [Google Scholar]
- 128.Yoon KA, Nakamura Y, Arakawa H. Identification of ALDH4 as a p53-inducible gene and its protective role in cellular stresses. J Hum Genet. 2004;49: 134–140. doi: 10.1007/s10038-003-0122-3 [DOI] [PubMed] [Google Scholar]
- 129.Kimata M, Matoba S, Iwai-Kanai E, Nakamura H, Hoshino A, Nakaoka M, et al. p53 and TIGAR regulate cardiac myocyte energy homeostasis under hypoxic stress. Am J Physiol Heart Circ Physiol. 2010;299: H1908–H1916. doi: 10.1152/ajpheart.00250.2010 [DOI] [PubMed] [Google Scholar]
- 130.Nakamura H, Matoba S, Iwai-Kanai E, Kimata M, Hoshino A, Nakaoka M, et al. p53 promotes cardiac dysfunction in diabetic mellitus caused by excessive mitochondrial respiration-mediated reactive oxygen species generation and lipid accumulation. Circ Heart Fail. 2012;5: 106–115. doi: 10.1161/CIRCHEARTFAILURE.111.961565 [DOI] [PubMed] [Google Scholar]
- 131.Johnson TM, Yu ZX, Ferrans VJ, Lowenstein RA, Finkel T. Reactive oxygen species are downstream mediators of p53-dependent apoptosis. Proc Natl Acad Sci USA. 1996;93: 11848–11852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Polyak K, Xia Y, Zweier JL, Kinzler KW, Vogelstein B. A model for p53-induced apoptosis. Nature. 1997;389: 300–305. doi: 10.1038/38525 [DOI] [PubMed] [Google Scholar]
- 133.Hoshino A, Matoba S, Iwai-Kanai E, Nakamura H, Kimata M, Nakaoka M, et al. p53-TIGAR axis attenuates mitophagy to exacerbate cardiac damage after ischemia. J Mol Cell Cardiol. 2012;52: 175–184. doi: 10.1016/j.yjmcc.2011.10.008 [DOI] [PubMed] [Google Scholar]
- 134.Espada L, Meo-Evoli N, Sancho P, Real S, Fabregat I, Ambrosio S, et al. ROS production is essential for the apoptotic function of E2F1 in pheochromocytoma and neuroblastoma cell lines. PLoS One. 2012;7: e51544 doi: 10.1371/journal.pone.0051544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Blanchet E, Annicotte JS, Lagarrigue S, Aguilar V, Clapé C, Chavey C, et al. E2F transcription factor-1 regulates oxidative metabolism. Nat Cell Biol. 2011;13: 1146–1152. doi: 10.1038/ncb2309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ambrus AM, Islam AB, Holmes KB, Moon NS, Lopez-Bigas N, Benevolenskaya EV, et al. Loss of dE2F compromises mitochondrial function. Dev Cell. 2013;27: 438–451. doi: 10.1016/j.devcel.2013.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Angelis E, Zhao P, Zhang R, Goldhaber JI, Maclellan WR. The role of E2F-1 and downstream target genes in mediating ischemia/reperfusion injury in vivo. J Mol Cell Cardiol. 2011;51: 919–926. doi: 10.1016/j.yjmcc.2011.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kuwahara K, Nishikimi T, Nakao K. Transcriptional regulation of the fetal cardiac gene program. J Pharmacol Sci. 2012; 119: 198–203. [DOI] [PubMed] [Google Scholar]
- 139.Zarain-Herzberg A, Fragoso-Medina J, Estrada-Avilés R. Calcium-regulated transcriptional pathways in the normal and pathologic heart. IUBMB Life. 2011;63: 847–855. doi: 10.1002/iub.545 [DOI] [PubMed] [Google Scholar]
- 140.Black FM, Packer SE, Parker TG, Michael LH, Roberts R, Schwartz RJ, et al. The vascular smooth muscle alpha-actin gene is reactivated during cardiac hypertrophy provoked by load. J Clin Invest. 1991;88: 1581–1588. doi: 10.1172/JCI115470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Li L, Miano JM, Cserjesi P, Olson EN. SM22 alpha, a marker of adult smooth muscle, is expressed in multiple myogenic lineages during embryogenesis. Circ Res. 1996;78: 188–195. [DOI] [PubMed] [Google Scholar]
- 142.Olson EN. A decade of discoveries in cardiac biology. Nat Med. 2004;10: 467–474. doi: 10.1038/nm0504-467 [DOI] [PubMed] [Google Scholar]
- 143.Knowlton KU, Rockman HA, Itani M, Vovan A, Seidman CE, Chien KR. Divergent pathways mediate the induction of ANF transgenes in neonatal and hypertrophic ventricular myocardium. J Clin Invest. 1995;96: 1311–1318. doi: 10.1172/JCI118166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Houweling AC, van Borren MM, Moorman AF, Christoffels VM. Expression and regulation of the atrial natriuretic factor encoding gene Nppa during development and disease. Cardiovasc Res. 2005;67: 583–593. doi: 10.1016/j.cardiores.2005.06.013 [DOI] [PubMed] [Google Scholar]
- 145.Kuwahara K, Nakao K. Regulation and significance of atrial and brain natriuretic peptides as cardiac hormones. Endocr J. 2010;57: 555–565. [DOI] [PubMed] [Google Scholar]
- 146.LaPointe MC. Molecular regulation of the brain natriuretic peptide gene. Peptides. 2005;26: 944–956. doi: 10.1016/j.peptides.2004.08.028 [DOI] [PubMed] [Google Scholar]
- 147.Molkentin JD, Lu JR, Antos CL, Markham B, Richardson J, Robbins J, et al. A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell. 1998;93: 215–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.van Rooij E, Quiat D, Johnson BA, Sutherland LB, Qi X, Richardson JA, et al. A family of micrornas encoded by myosin genes governs myosin expression and muscle performance. Dev Cell. 2009;17: 662–673. doi: 10.1016/j.devcel.2009.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Control of stress-dependent cardiac growth and gene expression by a microrna. Science. 2007;316: 575–579. doi: 10.1126/science.1139089 [DOI] [PubMed] [Google Scholar]
- 150.Liang Q, De Windt LJ, Witt SA, Kimball TR, Markham BE, Molkentin JD. The transcription factors GATA4 and GATA6 regulate cardiomyocyte hypertrophy in vitro and in vivo. J Biol Chem. 2001;276: 30245–30253. doi: 10.1074/jbc.M102174200 [DOI] [PubMed] [Google Scholar]
- 151.Morin S, Charron F, Robitaille L, Nemer M. GATA-dependent recruitment of MEF2 proteins to target promoters. EMBO J. 2000;19: 2046–2055. doi: 10.1093/emboj/19.9.2046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Xiong S, Van Pelt CS, Elizondo-Fraire AC, Liu G, Lozano G. Synergistic roles of Mdm2 and Mdm4 for p53 inhibition in central nervous system development. Proc Natl Acad Sci USA. 2006;103: 3226–3231. doi: 10.1073/pnas.0508500103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Clegg HV, Zhang Y. Regulation of the p53 tumor suppressor pathway: the problems and promises of studying Mdm2’s E3 ligase function. Crit Rev Eukaryot Gene Expr. 2010;20: 77–86. [DOI] [PubMed] [Google Scholar]
- 154.Hao Z, Duncan GS, Su YW, Li WY, Silvester J, Hong C, et al. The E3 ubiquitin ligase Mule acts through the ATM-p53 axis to maintain B lymphocyte homeostasis. J Exp Med. 2012;209: 173–186. doi: 10.1084/jem.20111363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Naito AT, Okada S, Minamino T, Iwanaga K, Liu ML, Sumida T, et al. Promotion of CHIP-mediated p53 degradation protects the heart from ischemic injury. Circ Res. 2010;106: 1692–1702. doi: 10.1161/CIRCRESAHA.109.214346 [DOI] [PubMed] [Google Scholar]
- 156.Manfredi JJ. The Mdm2-p53 relationship evolves: Mdm2 swings both ways as an oncogene and a tumor suppressor. Genes Dev. 2010;24: 1580–1589. doi: 10.1101/gad.1941710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Ikeda M, Ide T, Fujino T, Matsuo Y, Arai S, Saku K, et al. The Akt-mTOR axis is a pivotal regulator of eccentric hypertrophy during volume overload. Sci Rep. 2015;5 doi: 10.1038/srep15881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Dang CV, O’Donnell KA, Zeller KI, Nguyen T, Osthus RC, Li F. The c-myc target gene network. Semin Cancer Biol. 2006;16: 253–264. doi: 10.1016/j.semcancer.2006.07.014 [DOI] [PubMed] [Google Scholar]
- 159.Liu G, Terzian T, Xiong S, Van Pelt CS, Audiffred A, Box NF, et al. The p53-Mdm2 network in progenitor cell expansion during mouse postnatal development. J Pathol. 2007;213: 360–368. doi: 10.1002/path.2238 [DOI] [PubMed] [Google Scholar]
- 160.Kirshenbaum LA, Abdellatif M, Chakraborty S, Schneider MD. Human E2F-1 reactivates cell cycle progression in ventricular myocytes and represses cardiac gene transcription. Dev Biol. 1996;179: 402–411. doi: 10.1006/dbio.1996.0270 [DOI] [PubMed] [Google Scholar]
- 161.Morais VA, Haddad D, Craessaerts K, De Bock PJ, Swerts J, Vilain S, et al. PINK1 loss-of-function mutations affect mitochondrial complex I activity via NdufA10 ubiquinone uncoupling. Science. 2014;344: 203–207. doi: 10.1126/science.1249161 [DOI] [PubMed] [Google Scholar]
- 162.Riehle C, Abel ED. Pgc-1 proteins and heart failure. Trends Cardiovasc Med. 2012;22: 98–105. doi: 10.1016/j.tcm.2012.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Ahuja P, Zhao P, Angelis E, Ruan H, Korge P, Olson A, et al. Myc controls transcriptional regulation of cardiac metabolism and mitochondrial biogenesis in response to pathological stress in mice. J Clin Invest. 2010;120: 1494–1505. doi: 10.1172/JCI38331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Watanabe K, Fujii H, Takahashi T, Kodama M, Aizawa Y, Ohta Y, et al. Constitutive regulation of cardiac fatty acid metabolism through peroxisome proliferator-activated receptor alpha associated with age-dependent cardiac toxicity. J Biol Chem. 2000;275: 22293–22299. doi: 10.1074/jbc.M000248200 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data are available from Experiment ArrayExpress accession: E-MTAB-5441; http://www.ebi.ac.uk/arrayexpress.