Abstract
Mapping population of recombinant inbred lines (RILs) representing 541 × Ot1-3 cross exhibited wide variations of benzoxazinoid (BX) content in leaves and roots, brown rust resistance, α-amylase activity in the grain, and resistance to preharvest sprouting. QTL mapping of major BX species using a DArT-based map revealed a complex genetic architecture underlying the production of these main secondary metabolites engaged in stress and allelopathy responses. The synthesis of BX in leaves and roots was found to be regulated by different QTL. The QTL for the BX content, rust resistance, α-amylase activity, and preharvest sprouting partially overlapped; this points to their common genetic regulation by a definite subset of genes. Only one QTL for BX located on chromosome 7R coincided with the loci of the ScBx genes, which were mapped as two clusters on chromosomes 5RS (Bx3-Bx5) and 7R (Bx1-Bx2). The QTL common for several BX species, rust resistance, preharvest sprouting, and α-amylase activity are interesting objects for further exploration aimed at developing common markers for these important agronomic traits.
Introduction
The most abundant secondary metabolites produced in rye leaves and roots are cyclic hydroxamic acids and their derivatives benzoxazinoids (BXs) playing various physiological roles such as defense against pests, allelopathic agents, inhibition of germination, and detoxification [1–4]. Rye contains the highest amounts of these compounds in the grain and green tissues, and there is a wide variation of the BX content among cultivars and breeding materials [4–7]. This points to a possibility for initiating breeding strategies aimed at increasing the BX content in rye cultivars, which then may be treated as a source of allelopathic compounds against many weed species [4]. Although the quantities of BX in rye grain were extensively studied, knowledge about the QTL underlying this variation is very limited. In general, this aspect is poorly recognized. According to the best of our knowledge, there are only two examples of research on QTL controlling the BX content, both concerning corn. In the first of these two studies [8] identified eight QTL stable across different environments and the position of the largest QTL co-localized with a majority of the structural genes of the DIMBOA pathway. Zheng et al. [9] using the NAM population located a major quantitative trait locus for a high DIMBOA content (concentrations of 1.5 mM or more) on the short arm of chromosome 4 where the BX gene cluster underlying the biosynthesis pathway of BX up to DIMBOA is mapped. Association of the BX content and SNP within the ScBx1-ScBx5 genes encoding the enzymes of the BX synthesis pathway was reported by Rakoczy-Trojanowska et al. [7]; this suggests that there is a functional allelic variation in these structural loci. SNP in the Bx genes were also found to be associated with variations of leaf rust resistance and preharvest sprouting (PHS) within a wide collection of genetically diverse rye materials [10]. This finding raises a question on the possible role of BX production in regulating such important agronomic traits as leaf rust resistance, PHS, and α-amylase activity in rye grain. Association mapping performed using DArT Seq markers revealed associated SNPs on chromosomes 1R (3) 2R (1), and 5R (1) for leaf rust resistance, and on chromosomes 1RL (1), 4RL (1), and 5RL (1) for the α-amylase activity along with numerous SNPs associated with PHS on each rye chromosome except 4R [10].
This paper presents a mapping analysis of Bx genes, QTL for BX content, leaf rust resistance, PHS, and α-amylase activity, performed in the 541 × Ot1-3 mapping population of recombinant inbred lines of rye.
Material and methods
Plant material
The plant material used in the experiments consisted of 98 recombinant inbred lines and parents for the 541 × Ot1-3 mapping population. This population was genotyped and previously used for the construction of an integrated genetic map of rye [11]. The experiment was conducted at the experimental plots in two growing seasons (2013 and 2014) and two locations: the West Pomeranian University of Technology in Szczecin, Poland (WPUT–N 53°26′51.291″, E 14°31′42.1278″) and Danko Plant Breeders Ltd. in Choryń, Poland (DANKO–N 52°02′19.7″, E 16°46′09.0″). Each RIL was represented by 33 plants in a given season, grown in randomized block design in three replications (11 plants per replicate). Further, observations from four experiments designated as Szczecin 2013, Szczecin 2014, Choryń 2013, and Choryń 2014 were scored.
Methods
Phenotyping
The following traits of RILs and parental lines were phenotyped: brown rust resistance (RR), PHS resistance (PHS-R), and α-amylase activity in grain (AMY).
RR
The reaction of RILs to Puccinia recondita was verified under field conditions where the source of inoculum was the uredospores of the local population of the fungus, naturally occurring in the environment and spontaneously infecting the plants. No special treatments were applied for enhancing the infection potential of the fungus. Rating was performed at the maximum brown rust epidemics intensity and was based on the average evaluation of all the plants in the plot.
The plant infection level was evaluated according to the following six-degree scale:
0 –resistant, no symptoms on plants
1 –resistant, chlorosis/necrosis, and/or single uredinia sporadically visible on leaves
2 –moderately resistant; single small uredinia regularly present on a majority of plants
3 –moderately susceptible; up to 40% of the leaf surface covered with uredinia
4 –susceptible; up to 70% of the leaf surface covered with uredinia
5 –very susceptible; over 70% of the leaf surface covered with uredinia
PHS
Three to six spikes from each RIL and each replicate representing individual plant were harvested at full ripeness and kept at room temperature for 3 days. Then, the spikes were sprayed with water for 5 min each day and kept in a moisture chamber at 20°C for 7 days for stimulated sprouting. Percentage of germinated kernels (the lower the resistance, the higher grain germination) was determined to assess PHS resistance (PHS).
AMY
The amount of 1.5 g of kernels from each RIL and each replicate showing no external signs of sprouting were milled into flour. Grain used for the α-amylase assay was stored at room temperature for the first 2 weeks after harvest and then kept at 4°C for the 3 months before the analysis. The α-amylase was extracted in 4 mL of distilled water mixed with 1 g of the flour, equilibrated for 10 min at room temperature, and finally, centrifuged at 8000 × rpm at 4°C for 20 min. The simple method of estimating the level of AMY [12] was used in which the diameters of the α-amylase diffusion circles (in millimeters) in the agarose gel containing starch stained with iodine show a linear relationship with the logarithm of the enzyme activity (in enzyme units per milliliter).
The highest values mean for RR, the lowest infection degree; PHS-R, the highest susceptibility; and AMY, the highest enzymatic activity.
Sample preparation and analysis of BX content
Plant samples, the above-ground parts (L) and roots (R), were collected two weeks after the start of the vegetation in the spring, when the plants were in the development stage 21–22 BBCH (20–24 Zadoks development stage), in the experiments performed at Choryń. The tissues after cutting were immediately frozen and lyophilized. Further, 100 mg d.w. of the plant material (L and R) was mixed with diatomaceous earth, placed in stainless steel extraction cells, and extracted with 70% methanol at an operating pressure of 10 MPa and at 40°C by using an accelerated solvent extraction system (ASE 200, Dionex, Sunnyvale, CA). The cells were filled with 250/750 mg of LiChroprep RP-18 (40–60 μm, Merck, Germany) for the roots and the above-ground parts, respectively. After evaporating to dryness under reduced pressure, extracts were reconstituted in 1 mL of methanol containing 0.1% (v/v) acetic acid and stored at −20°C. Prior to the analyzes, the extracts were centrifuged for 20 min at 23,000 × g at 4°C.
The quantitative analyzes of the BX content were described in detail by Rakoczy-Trojanowska et al. [7].
ScBx gene mapping
The sequences of the ScBx1–ScBx5 genes previously sequenced and characterized in the rye inbred line L318 were used [13]. Sequences of the ScBx1–ScBx5 genes in parental lines 541 and Ot1-3 were compared; this led to the detection of SNPs, which were used as a 3′ ends of the primers. Alternatively, primers were developed within consensus regions, and restriction enzymes were used for revealing the allelic polymorphisms in SNPs. Sequences of primers, PCR conditions, and restriction enzymes used were presented in S1 Protocol. Polymorphisms among parental lines were initially tested within a group of 16 lines of the 541 × Ot1-3 mapping population to check for an appropriate 1:1 Mendelian segregation. Then, all RILs of the mapping population were genotyped. A reference genetic map consisting mainly of DArT markers was used for mapping both ScBx genes and QTL [11]. Mapping was performed using the JoinMap 4.0 computer program [14].
Statistical methods
Observations of the BX content were transformed by log10(x+1). The analysis of variance (ANOVA) was performed in the mixed model with years and locations as fixed effects, and lines (RILs) as random effects. Variance components were compared to (3 × std. err.) to assess their magnitude. Broad sense heritability was calculated according to Cullis et al. [15]. Correlation coefficient was calculated on average data over years and locations. Computations were performed using Genstat 17 [16].
QTL mapping
QTL mapping was performed with the software package WinQTL Cartographer 2.5 [17] using the procedure for composite interval mapping (CIM), with the threshold value of LOD set at a score at 3.0. QTL with an LOD value between 2.5 and 3.0 was considered putative.
Results
QTL for benzoxazinoid (BX) content
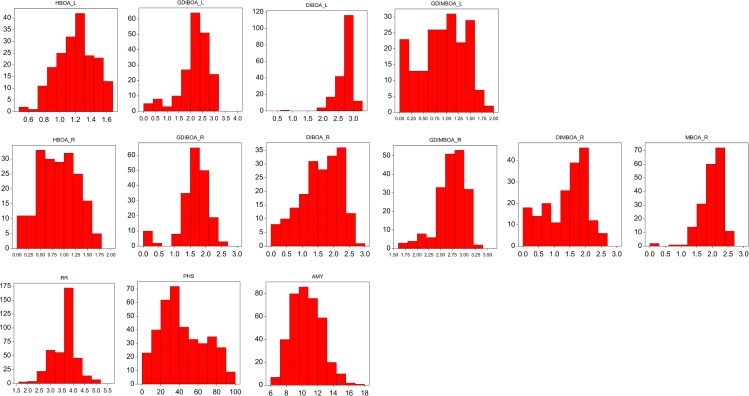
Differences in BX quantities between the parental lines were low (although statistically significant) and unstable in consecutive years (Table 1). Variation ranges found among RILs were much larger than those found in the parental generation in both years (from about 1.5 to 47 times), which suggests a substantial genetic transgression component in the observed variation. The prevailing BX species in rye leaves were DIBOA and GDIBOA. Lower amounts of GDIMBOA and HBOA were found in this tissue. Roots contained higher amounts of GDIMBOA, MBOA, and GDIBOA, and lower amounts of DIBOA, DIMBOA, and HBOA. Broad sense heritability of the BX species ranged between c. 30% and 40% with two exceptions (16.95% for GDIMBOA_R and 55.36% for MBOA_R, Table 2). But, the substantial lines × years interaction (Table 3) and apparent deviations from the normal distribution lowered the chances of finding stable QTL across different environments (Fig 1). A number of significant QTL for particular BX species were found (Tables 4 and 5, Fig 2). Depending on a year of study, 2 (2014) to 3 (2013) QTL were detected for HBOA in leaves, with one common on chromosome 6RL and three separate on chromosomes 1R, 4RS, and 7RS. QTL for HBOA in roots were found on chromosome 5R in 2013 and on chromosomes 5R (1) and 6R (2) in 2014. The QTL mapping of the GDIBOA amount in leaves revealed 1 QTL in 2014 and 4 QTL in 2013, located on the distal part of chromosome 1RS (1); distal part of chromosomes 2RS (1), 5R (1), and 7RL (2) coinciding with the locus for the ScBx1-2 genes. No QTL for GDIBOA in roots were found in 2013, but in 2014, there were 3 detected QTL on chromosomes 3RS, 6RS, and 6RL. QTL for DIBOA in leaves were dispersed on chromosomes 3R and 7RL (adjacent to the ScBx1 locus) in 2013, whereas in 2014, they were detected on chromosomes 1R, 6R, and 7R. The QTL for DIBOA in roots was found on chromosomes 3R, 6RS, and 7RL in 2013 and on chromosomes 1RL, 2RL, 4RL, and 6R (2) in 2014. Closely linked but not overlapping QTL on chromosomes 1R, 3R, and 6R were detected for DIBOA from both leaves and roots. The QTL for GDIMBOA from leaves were mapped on chromosomes 1R, 2RS, 5RS, and 7R in 2013 and only on 6R in 2014. The distribution of GDIMBOA QTL in leaves partially overlapped with that in roots where the QTL were located on chromosomes 1R, 2RS (coinciding QTL found in leaves and roots), and 6RS in 2014 and on chromosome 7R (2013) near the ScBx1 locus. DIMBOA QTL were detected only in roots. They were located on chromosomes 3R and 6R in 2013 and on chromosomes 1RL and 6RS in 2014. MBOA QTL were not detected in leaves. MBOA QTL in roots were represented in 2013 by two loci on chromosomes 1R and 4RL and by one locus on chromosome 2RL in 2014.
Table 1. Characteristics of BX contents (μg/g d.w.) in leaves (L) and roots (R).
| BX | Year | Parents | RILs | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P1 OT1-3 |
Std. err. | P2 541 |
Std. err. | P1-P2 | No. of lines | Mean | Std. error of mean | Min | Max | C.V. [%] | ||
| 1. HBOA_L | 2013 | 1.068 | 0.054 | 1.262 | 0.141 | -0.194* | 96 | 1.218 | 0.029 | 0.543 | 1.646 | 23.10 |
| 2014 | 1.198 | 0.062 | 1.032 | 0.047 | 0.166* | 96 | 1.194 | 0.018 | 0.752 | 1.611 | 15.10 | |
| 2. GDIBOA_L | 2013 | 2.459 | 0.018 | 1.882 | 0.215 | 0.577* | 96 | 1.719 | 0.062 | 0 | 2.383 | 35.20 |
| 2014 | 2.499 | 0.009 | 2.62 | 0.066 | -0.121* | 96 | 2.616 | 0.024 | 2.013 | 3.084 | 9.10 | |
| 3. DIBOA_L | 2013 | 2.774 | 0.036 | 2.725 | 0.023 | 0.049 | 96 | 2.618 | 0.034 | 0.725 | 3.049 | 12.60 |
| 2014 | 2.886 | 0.037 | 2.746 | 0.070 | 0.140* | 96 | 2.829 | 0.014 | 2.498 | 3.104 | 4.80 | |
| 4. GDIMBOA_L | 2013 | 1.756 | 0.096 | 0.504 | 0.275 | 1.252* | 96 | 0.967 | 0.058 | 0 | 1.837 | 59.00 |
| 2014 | 0.785 | 0.031 | 1.298 | 0.070 | -0.513* | 96 | 0.853 | 0.040 | 0 | 1.549 | 45.40 | |
| 5. HBOA_R | 2013 | 1.279 | 0.032 | 1.177 | 0.043 | 0.102 | 96 | 1.199 | 0.024 | 0.679 | 1.758 | 19.60 |
| 2014 | 0.788 | 0.009 | 0.261 | 0.043 | 0.527* | 96 | 0.556 | 0.026 | 0 | 1.244 | 45.40 | |
| 6. GDIBOA_R | 2013 | 1.950 | 0.045 | 1.559 | 0.106 | 0.391* | 96 | 1.494 | 0.06 | 0 | 2.23 | 39.40 |
| 2014 | 1.878 | 0.040 | 1.444 | 0.152 | 0.434* | 96 | 1.761 | 0.036 | 1.010 | 2.564 | 20.30 | |
| 7. DIBOA_R | 2013 | 2.361 | 0.071 | 2.467 | 0.026 | -0.106 | 96 | 2.050 | 0.035 | 1.014 | 2.773 | 16.50 |
| 2014 | 1.200 | 0.083 | 0.912 | 0.298 | 0.288 | 96 | 1.115 | 0.053 | 0 | 2.453 | 46.30 | |
| 8. GDIMBOA_R | 2013 | 2.948 | 0.035 | 2.811 | 0.032 | 0.137* | 96 | 2.674 | 0.03 | 1.726 | 3.123 | 11.10 |
| 2014 | 3.017 | 0.062 | 2.839 | 0.107 | 0.178* | 96 | 2.781 | 0.032 | 1.628 | 3.270 | 11.20 | |
| 9. DIMBOA_R | 2013 | 2.123 | 0.083 | 2.062 | 0.050 | 0.061 | 96 | 1.727 | 0.048 | 0 | 2.635 | 27.50 |
| 2014 | 1.217 | 0.109 | 1.903 | 0.072 | -0.686* | 96 | 1.056 | 0.068 | 0 | 2.531 | 63.50 | |
| 10. MBOA_R | 2013 | 2.347 | 0.073 | 2.528 | 0.029 | -0.181* | 96 | 2.126 | 0.025 | 1.441 | 2.673 | 11.30 |
| 2014 | 2.196 | 0.063 | 1.985 | 0.053 | 0.211* | 96 | 1.826 | 0.042 | 0 | 2.333 | 22.50 | |
*—difference between parents significant according to the LSD test at P < 0.05
Table 2. Characteristics of rust resistance (RR), preharvest sprouting (PHS) and alpha-amylase activity in rye grain (AMY).
| Trait | Year | Location | Parents | RILs | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P1 OT1-3 |
Std. err. | P2 541 |
Std. err. | P1-P2 | No. of lines | Mean | Std. err. | Min | Max | C.V. (%) | |||
| 11. RR | 2013 | Choryn | 4.00 | 0.58 | 4.00 | 0.58 | 0 | 96 | 4.18 | 0.04 | 3.33 | 5.00 | 9.53 |
| Szczecin | 3.33 | 0.33 | 4.00 | 0.00 | -0.67 | 96 | 3.58 | 0.03 | 3.00 | 4.33 | 9.02 | ||
| 2014 | Choryn | 4.00 | 0.00 | 4.33 | 0.33 | -0.33 | 96 | 3.88 | 0.04 | 3.00 | 4.67 | 9.42 | |
| Szczecin | 3.33 | 0.33 | 3.33 | 0.88 | 0 | 96 | 2.97 | 0.04 | 2.00 | 4.00 | 12.71 | ||
| 12. PHS | 2013 | Choryn | 41.11 | 9.81 | 71.02 | 5.58 | -29.91* | 93 | 33.41 | 1.54 | 0 | 68.26 | 44.53 |
| Szczecin | 18.02 | 2.24 | 89.92 | 3.80 | -71.90* | 96 | 49.14 | 2.97 | 4.43 | 95.08 | 59.25 | ||
| 2014 | Choryn | 52.96 | 10.65 | 52.97 | 3.11 | -0.01 | 91 | 35.49 | 1.70 | 2.63 | 82.14 | 45.61 | |
| Szczecin | 50.46 | 10.16 | 82.58 | 0.76 | -32.12* | 93 | 55.96 | 2.55 | 4.55 | 100.00 | 44.01 | ||
| 13. AMY | 2013 | Choryn | 16.83 | 1.09 | 10.33 | 0.33 | 6.50* | 94 | 11.27 | 0.16 | 8.00 | 15.33 | 14.18 |
| Szczecin | 16.67 | 0.44 | 13.33 | 0.60 | 3.34* | 96 | 10.95 | 0.20 | 7.33 | 15.67 | 17.93 | ||
| 2014 | Choryn | 11.33 | 1.60 | 9.00 | 0.00 | 2.33 | 96 | 8.97 | 0.12 | 6.00 | 12.00 | 12.96 | |
| Szczecin | 16.67 | 0.93 | 12.67 | 0.88 | 4.00* | 95 | 11.29 | 0.18 | 7.00 | 17.00 | 15.54 | ||
*—difference between parents significant according to the LSD test at P < 0.05
Table 3. ANOVA results for BX quantities in rye tissues.
| Trait | P-value for Years | Variance component for Lines | Std. error | Variance component for interaction Lines x Years* | Std. error | Heritability (%) |
|---|---|---|---|---|---|---|
| HBOA_L | 0.430 | 0.011 | 0.006 | 0.040* | 0.006 | 34.41 |
| GDIBOA_L | < 0.001 | 0.055 | 0.022 | 0.143* | 0.022 | 41.33 |
| DIBOA_L | < 0.001 | 0.017 | 0.007 | 0.041* | 0.007 | 41.73 |
| GDIMBOA_L | 0.051 | 0.075 | 0.025 | 0.145* | 0.023 | 47.95 |
| HBOA_R | < 0.001 | 0.013 | 0.006 | 0.042* | 0.007 | 35.82 |
| GDIBOA_R | < 0.001 | 0 | - | 0.235* | 0.035 | - |
| DIBOA_R | < 0.001 | 0.029 | 0.019 | 0.133* | 0.023 | 26.84 |
| GDIMBOA_R | 0.011 | 0.008 | 0.009 | 0.078* | 0.012 | 16.95 |
| DIMBOA_R | < 0.001 | 0 | - | 0.322* | 0.049 | - |
| MBOA_R | < 0.001 | 0.043* | 0.012 | 0.063* | 0.010 | 55.36 |
*—significance of variance components assessed approximately by comparison with (3 x std. error)
Fig 1. Distribution of variation of BX, RR, PHS and AMY within the 541×Ot1-3 mapping population of RILs after log10(x+1) conversion.
Table 4. Characteristics of QTL detected in 2013.
| Trait | QTL | Chromosome | Marker interval | QTL position (cM) | LOD | Additive effect | R2 (%) |
|---|---|---|---|---|---|---|---|
| HBOA_L | HBOA_AG/13_1 | 1R | XrPt390335—XrPt411480 | 155.3 | 2.8 | -3.19 | 6.7 |
| HBOA_AG/13_2 | 4R | XrPt398853—XrPt506862 | 45.5 | 3.9 | -4.11 | 10.7 | |
| HBOA_AG/13_3 | 6R | XrPt508835—XrPt410804 | 176.4 | 4.5 | -4.40 | 14.4 | |
| GDIBOA_L | GDIBOA_AG/13_1 | 2R | XrPt505615—XrPt507714 | 25.8 | 8.6 | 41.42 | 34.0 |
| GDIBOA_AG/13_2 | 5R | XrPt398734—XrPt507068 | 99.5 | 2.9 | -20.28 | 7.6 | |
| GDIBOA_AG/13_3 | 7R | XrPt508579 –ScBx2 | 128.2 | 5.9 | 34.46 | 16.5 | |
| GDIBOA_AG/13_4* | 7R | ScBx1 –XrPt509722 | 139.8 | 2.6 | 21.05 | 8.2 | |
| DIBOA_L | DIBOA_AG/13_1 | 3R | XrPt399424—Xpsr754L218 | 91.3 | 3.7 | -97.35 | 12.9 |
| DIBOA_AG/13_2 | 7R | XrPt508579 –ScBx2 | 128.2 | 3.5 | 87.84 | 12.1 | |
| GDIMBOA_L | GDIMBOA_AG/13_1 | 1R | XrPt399269—XrPt509253 | 142.4 | 3.4 | 5.46 | 10.5 |
| GDIMBOA_AG/13_2 | 2R | XrPt505615—XrPt507714 | 23.6 | 4.5 | 6.12 | 14.2 | |
| GDIMBOA_AG/13_3* | 5R | XrPt398734—XrPt509607 | 98.8 | 2.6 | -4.64 | 7.8 | |
| GDIMBOA_AG/13_4 | 7R | XrPt508579 –ScBx2 | 128.2 | 3.9 | 5.82 | 12.3 | |
| HBOA_R | HBOA_R/13_1 | 5R | XrPt508373—XrPt507068 | 98.9 | 3.2 | -4.77 | 10.8 |
| DIBOA_R | DIBOA_R/13_1 | 3R | XrPt66766—XrPt116220 | 104.7 | 3.4 | -40.95 | 9.7 |
| DIBOA_R/13_2 | 6R | Xpsr106.1L350—XrPt401117 | 72.6 | 2.9 | -37.88 | 8.3 | |
| DIBOA_R/13_3 | 7R | XrPt507302—XrPt508293 | 158.3 | 4.5 | -46.37 | 13.2 | |
| GDIMBOA_R | GDIMBOA_R/13_1* | 7R | XrPt401834—XrPt506935 | 9 | 2.6 | -104.20 | 14.7 |
| DIMBOA_R | DIMBOA_R/13_1 | 3R | XrPt398766—XrPt507717 | 109 | 3.3 | -27.41 | 10.0 |
| DIMBOA_R/13_2 | 6R | XrPt347114—XrPt508051 | 129.6 | 3.6 | 28.46 | 11.1 | |
| MBOA_R | MBOA_R/13_1 | 1R | XrPt399269—XrPt401024 | 140 | 3.2 | 32.58 | 10.7 |
| MBOA_R/13_2 | 4R | Xpsr119L1000—XrPt505236 | 187.2 | 5.1 | -44.84 | 17.0 | |
| RR_DANKO | RR_DANKO/13_1 | 1R | XrPt390549—XrPt505946 | 54.8 | 4.9 | 0.16 | 15.6 |
| RR_DANKO/13_2 | 5R | XrPt508794—XrPt4100863 | 94 | 4.2 | -0.15 | 13.3 | |
| RR_WPUT | RR_WPUT/13_1 | 4R | XrPt508495—XrPt400917 | 130.5 | 3.1 | 0.12 | 10.5 |
| RR_WPUT/13_2 | 5R | XrPt401573—XrPt346094 | 135.8 | 4.3 | -0.17 | 25.5 | |
| PHS_DANKO | PHS_DANKO/13_1 | 2R | Xscm0188—XrPt508521 | 97.5 | 5.0 | -7.76 | 22.2 |
| PHS_WPUT | PHS_WPUT/13_1 | 5R | XrPt401573—XrPt346094 | 123.4 | 4.0 | -12.63 | 17.7 |
| PHS_WPUT/13_2 | 6R | XrPt100398 -XrPt401305 | 21.4 | 3.0 | 23.44 | 45.5 | |
| PHS_WPUT/13_3 | 7R | XrPt506902—XrPt508061 | 102.1 | 3.8 | -10.92 | 12.2 | |
| AMY_WPUT | AMY_WPUT/13_1 | 1R | XrPt400138—Xbcd304 | 235.8 | 5.0 | -0.77 | 13.7 |
| AMY_WPUT/13_2 | 2R | XrPt120224—XrPt399788 | 125.2 | 3.6 | 0.69 | 10.3 | |
| AMY_WPUT/13_3 | 3R | XrPt399830—XrPt398766 | 90.8 | 5.3 | -0.78 | 14.5 | |
| AMY_WPUT/13_4* | 5R | XrPt508373—XrPt508518 | 98.9 | 2.7 | -0.54 | 6.9 | |
| AMY_DANKO | AMY_DANKO/13_1* | 4R | XrPt509036—XrPt506651 | 206.9 | 2.6 | -0.70 | 8.7 |
| AMY_DANKO/13_2 | 6R | XrPt100398 -XrPt401305 | 1.4 | 7.9 | 1.20 | 38.8 | |
| AMY_DANKO/13_3 | 6R | XrPt410804—XrPt411086 | 187.8 | 2.9 | -0.80 | 7.6 |
Table 5. Characteristics of QTL detected in 2014.
| Trait | QTL | Chromosome | Marker interval | QTL position (cM) | LOD | Additive effect | R2 (%) |
|---|---|---|---|---|---|---|---|
| HBOA_L | HBOA_AG/14_1 | 6R | XrPt346701-XrPt410804 | 170.3 | 4.9 | -2.87 | 16.1 |
| HBOA_AG/14_2 | 7R | XrPt505150-XrPt402431 | 15 | 3.5 | 2.30 | 9.9 | |
| GDIBOA_L | GDIBOA_AG/14_1 | 1R | XrPt390516-XrPt347149 | 0 | 5.2 | 111.87 | 19.2 |
| DIBOA_L | DIBOA_AG/14_1 | 1R | XrPt509253-XrPt411480 | 154.7 | 3.1 | -65.95 | 9.3 |
| DIBOA_AG/14_2 | 6R | XrPt401725-XrPt508436 | 147.5 | 3.4 | -69.48 | 9.9 | |
| DIBOA_AG/14_3 | 7R | XrPt505150-XrPt402431 | 16 | 4.5 | 83.18 | 13.7 | |
| GDIMBOA_L | GDIMBOA_AG/14_1 | 6R | XrPt508562-Xpsr454L1000 | 212.4 | 4.4 | 3.62 | 15.3 |
| HBOA_R | HBOA_R/14_1 | 5R | XrPt389838-XrPt399887 | 122.4 | 3.5 | 0.95 | 12.9 |
| HBOA_R/14_2 | 6R | XrPt402654-XrPt508054 | 157 | 5.3 | -1.12 | 17.5 | |
| HBOA_R/14_3* | 6R | XrPt509216-XrPt410804 | 165.3 | 2.6 | -0.81 | 9.3 | |
| GDIBOA_R | GDIBOA_R/14_1* | 3R | XrPt410966-XrPt505403 | 49.9 | 2.7 | 19.83 | 7.3 |
| GDIBOA_R/14_2 | 6R | Xpsr106.1L350-XrPt401117 | 72.4 | 3.7 | -24.61 | 10.7 | |
| GDIBOA_R/14_3 | 6R | XrPt507940-XrPt506539 | 150.8 | 5.0 | -27.97 | 14.6 | |
| DIBOA_R | DIBOA_R/14_1 | 1R | XrPt399269-XrPt401024 | 138 | 2.8 | -8.42 | 5.2 |
| DIBOA_R/14_2 | 2R | XrPt398489-XrPt402116 | 159.1 | 3.6 | -10.39 | 7.9 | |
| DIBOA_R/14_3 | 4R | XrPt508265-XrPt402590 | 195.4 | 3.0 | -12.16 | 5.9 | |
| DIBOA_R/14_4* | 6R | XrPt401305-Xpsr106.1L350 | 40.9 | 2.7 | -8.79 | 5.7 | |
| DIBOA_R/14_5 | 6R | XrPt398480-XrPt400777 | 157 | 3.1 | -12.07 | 10.4 | |
| GDIMBOA_R | GDIMBOA_R/14_1 | 1R | XrPt505839-XrPt410988 | 233.5 | 3.6 | 157.57 | 12.1 |
| GDIMBOA_R/14_2 | 2R | XrPt389332-XrPt402421 | 23.4 | 3.8 | 147.16 | 11.7 | |
| GDIMBOA_R/14_3* | 6R | XrPt100398-XrPt401305 | 19.4 | 2.7 | -274.70 | 39.3 | |
| DIMBOA_R | DIMBOA_R/14_1 | 1R | XrPt389926-XrPt508678 | 268.1 | 3.3 | 30.01 | 11.6 |
| DIMBOA_R/14_2 | 6R | XrPt506337-Xpsr106.1L350 | 40.9 | 3.1 | -15.70 | 9.9 | |
| MBOA_R | MBOA_R/14_1 | 2R | XrPt505891-XrPt401684 | 152.7 | 4.1 | -22.42 | 13.5 |
| RR_DANKO | RR_DANKO/14_1 | 1R | XrPt400461-XrPt411480 | 154.7 | 3.9 | 0.13 | 12.1 |
| RR_DANKO/14_2 | 1R | XrPt507632-Xbcd304 | 240.1 | 3.0 | -0.11 | 8.0 | |
| RR_DANKO/14_3 | 3R | XrPt505403-XrPt508819 | 57.4 | 3.3 | -0.11 | 8.7 | |
| RR_DANKO/14_4 | 3R | XrPt66766-XrPt390684 | 105.5 | 4.8 | 0.14 | 13.3 | |
| RR_DANKO/14_5* | 5R | XrPt508071-XrPt506821 | 224.9 | 2.6 | -0.10 | 7.0 | |
| RR_WPUT | RR_WPUT/14_1 | 1R | XrPt508703-XrPt344175 | 39.4 | 3.0 | 0.12 | 10.0 |
| RR_WPUT/14_2 | 3R | XrPt507316-XrPt508407 | 187.3 | 3.0 | -0.12 | 10.4 | |
| PHS_DANKO |
PHS_DANKO/14_1 | 1R | XrPt390321-XrPt506100 | 54.8 | 2.9 | -5.29 | 8.9 |
| PHS_DANKO/14_2 | 1R | XrPt400239-XrPt506887 | 217.9 | 2.8 | -5.29 | 8.7 | |
| PHS_WPUT | PHS_WPUT/14_1 | 5R | XrPt506718-XrPt508861 | 286.9 | 4.0 | -10.22 | 14.5 |
| AMY_DANKO | AMY_DANKO/14_1 | 2R | XrPt505534-XrPt346940 | 128.3 | 2.9 | 0.38 | 9.9 |
| AMY_DANKO/14_2 | 3R | XrPt400319-XrPt401081 | 109 | 4.0 | -0.42 | 12.2 | |
| AMY_DANKO/14_3 | 5R | XrPt390157-XrPt508794 | 70.8 | 3.0 | -0.47 | 15.5 | |
| AMY_WPUT | AMY_WPUT/14_1 | 3R | XrPt398786-XrPt505593 | 103.6 | 3.4 | -0.74 | 11.3 |
| AMY_WPUT/14_2 | 4R | XrPt509488-XrPt401194 | 68.2 | 3.6 | -0.78 | 12.6 |
Fig 2. Localization ScBx1 –ScBx5 genes and QTL of BX, RR, PHS and AMY on the 541×Ot1-3 genetic map.
The R2 value for most of the QTL for BX was close to 10%. The greatest impact on the BX content in leaves was exerted by GDIBOA_L/13_1 (R2 = 34%) on chromosome 2R (2013), and in roots, by GDIMBOA_R/14_3 (R2 = 39.3%) on chromosome 6R (2014). QTL were significant only in one year of the study (with one exception on 6RL), which illustrates their instability due to the significant environmental component of variation including lines versus years relationship.
Except for the QTL on the distal part of chromosome 2RS, there were no coinciding QTL for the same BX species in leaves and roots, which strongly supports the hypothesis of the differences in the regulation of BX production in these two plant tissues. In contrast, there are several coinciding QTL for different BX species like those for leaf GDIBOA, GDIMBOA, and HBOA on 5RS; leaf GDIBOA and GDIMBOA on 2RS; and leaf GDIBOA, GDIMBOA, and DIBOA on 7R. Similarly, coinciding QTL for different types of BX in roots were found on chromosomes 2RL (DIBOA and MBOA), 3R and 6RS (DIMBOA and DIBOA), 4RL (MBOA and DIBOA), and 6RS (GDIBOA and DIBOA). They may represent pleiotropic genes regulating the production of more than one BX species or clusters of tightly linked genes with specificity for a given species of BX. Coinciding QTL for BX in both leaves and roots may constitute the genetic background for the significant correlation of their amounts detected in this study (Table 6).
Table 6. Significant correlation coefficients between studied traits (P < 0.01).
| HBOA_L | GDIBOA_L | DIBOA_L | GDIMBOA_L | HBOA_R | GDIBOA_R | DIBOA_R | GDIMBOA_R | DIMBOA_R | MBOA_R | RR | PHS | AMY | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HBOA_L | 1.00 | ||||||||||||
| GDIBOA_L | 0.52 | 1.00 | |||||||||||
| DIBOA_L | 0.49 | 0.46 | 1.00 | ||||||||||
| GDIMBOA_L | 0.29 | 0.77 | 1.00 | ||||||||||
| HBOA_R | 0.47 | 1.00 | |||||||||||
| GDIBOA_R | 0.38 | 0.30 | 0.65 | 1.00 | |||||||||
| DIBOA_R | -0.25 | 0.75 | 0.61 | 1.00 | |||||||||
| GDIMBOA_R | 0.33 | 0.40 | 1.00 | ||||||||||
| DIMBOA_R | 0.31 | 0.47 | 0.31 | 0.80 | 1.00 | ||||||||
| MBOA_R | -0.33 | 0.39 | 1.00 | ||||||||||
| RR | 1.00 | ||||||||||||
| PHS | 1.00 | ||||||||||||
| AMY | 1.00 |
Structural loci encoding enzymes of the BX biosynthesis pathway were mapped on the distal part of chromosome 5RS (a cluster of the ScBx3, ScBx4, and ScBx5 genes) and on the central part of the 7R chromosome (a cluster of the ScBx1 and ScBx2 genes) (Fig 2). The QTL for BX in leaves and roots did not coincide with these structural genes except for the QTL for GDIMBOA in roots, which was mapped near the ScBx1 and ScBx2 gene cluster on chromosome 7R.
QTL for resistance to preharvest sprouting (PHS)
In each experiment with one exception (Choryń 2014), differences between parental lines were substantial, showing that line Ot1-3 represents resistant and line 541 susceptible phenotypes (Table 2). The RIL variation ranges were large in each experiment overcoming those of parental lines, which led to high coefficients of variation. But, because of a very high line × location and line × year interactions, the heritability of the trait calculated for the whole experiment was very low (2.18%) (Table 7). Only four QTL for PHS were detected in 2013 and three others in 2014 (Tables 4 and 5, Fig 2). The highest determination coefficient has PHS_WPUT/13_2 QTL from the distal part of chromosome 6RS. The QTL for PHS were distributed on 1RS (1), 1RL (1), 2R (2), 5R (2), and 6RS (1).
Table 7. ANOVA results for traits.
| Trait | P-value for Years (Y) | P-value for Locations (P) | P-value for interaction Y x P | Variance component for Lines (L) | Std. error | Variance component for interaction L x Y | Std. error | Variance component for interaction L x P | Std. error | Heritability (%) |
|---|---|---|---|---|---|---|---|---|---|---|
| RR | < 0.001 | < 0.001 | < 0.001 | 0.026 | 0.009 | 0.008 | 0.008 | 0 | - | 52.14 |
| PHS | 0.005 | < 0.001 | 0.175 | 4.40 | 38.50 | 67.47 | 18.88 | 264.38 | 46.99 | 2.18 |
| AMY | < 0.001 | < 0.001 | < 0.001 | 1.43 | 0.30 | 0.20 | 0.08 | 0.55 | 0.132 | 71.85 |
QTL for α-amylase activity (AMY)
The grains of parental lines exhibited large differences in the α-amylase activity, and a large variation range was detected among RILs in each experiment (Table 2).
The environmental component of variation due to the line × location interaction was significant (Table 7). In contrast to the other traits, the heritability of the α-amylase activity proved to be very high (71.85%), and its distribution was close to normal (Fig 1). Among the seven QTL detected in 2013, the one on chromosome 6R was the most efficient (R2 = 38.8%) (Table 4, Fig 2). Five QTL detected in 2014 had similar low determination coefficients (c. 10%) (Table 5, Fig 2). QTL were found on distal parts of chromosome 1RL, on the proximal part of chromosome 2RL (present in two years), on the proximal part of chromosomes 2RL and 3R in three adjacent positions), and on chromosomes 4RS, 4RL, 5RS, 6RS, 6RL, and 7RS.
QTL for leaf rust resistance (RR)
Parental lines did not differ significantly with respect to rust resistance (Table 7). But, the variation ranges among RILs were substantial because of transgression effects, which assured that the genetic variation in a mapping population although not high (CV close to 10%) was sufficient for successful QTL mapping. In addition, the results of ANOVA showed that the environmental component of variation was low and the heritability was high (Table 4).
Chromosome 1R carried four QTL for leaf rust resistance. They were distributed on the distal part of the 1RS arm (2) and on the 1RL arm (2) (Tables 4 and 5, Fig 2). Three QTL for rust resistance were detected on chromosome 3R: the first on the short arm, the second in the centromeric region 3Rc, and the third on the long arm. Chromosome 4R contained one Lr QTL on the 4Rc. Two other QTL for rust resistance were in the 5Rc region. A majority of the RR QTL determined the trait variation in c. 10%, while the RR QTL on chromosome 5R (RR_WPUT/13_2) had a much higher impact (25.5%). Each of the RR QTL was detected only in one year and in one location, which shows that their effectiveness was highly dependent on environmental changes.
Coinciding QTLs for different traits
The most interesting was the coinciding of QTL for rust resistance affecting leaves and the α-amylase activity in rye grain. Such cases were found on chromosomes 1RL and 3R (Fig 2), but they did not result in a significant correlation between these traits (Table 6). The two coinciding QTL for PHS and rust resistance were detected on chromosomes 1RS and 5R (Tables 4 and 5, Fig 2). Except for the distal region on chromosome 1RL, the proximal region of 2RL, and the distal region of 6RS, there were no genomic locations with coinciding PHS and AMY QTL, which may partially confirm the lack of correlation between these two traits (Table 6).
The QTL for BX in leaves or roots were found in several locations accompanied by AMY QTL as on chromosomes 1RL, 3R, 5R, 6RS, and 6RL. The most apparent coincidence for PHS and GDIMBOA QTL from roots was on the distal part of chromosome 6RS. A common position of QTL for the HBOA content in roots and the PHS QTL was also detected on chromosome 5R. The detected similarities between BX and AMY or PHS QTL distribution did not result in a significant correlation between these traits (Table 6). Coinciding QTL for different traits raise a question on the pleiotropic effects of the underlying genes. They also provide strong support for the validity of the QTL detection.
Discussion
Results of mapping the ScBx genes were in agreement with the earlier reports on their distribution in the contigs of the rye BAC library [13]. The ScBx3, ScBx4, and ScBx5 genes were mapped as a one gene cluster on the distal part of chromosome 5RS and ScBx1 with the ScBx2 gene as another cluster on chromosome 7R. This result clearly shows that both groups of genes are not linked to other BX genes: ScGT encoding UDP-glucosyltransferase and ScGlu encoding beta-glucosidase which were shown to be located on chromosomes 4R and 2R, respectively [18]. The map positions of the ScBx1-5 cluster on 5RS and the ScBx1-2 cluster on 7R, determined for the first time in this study, confirm the earlier results attributing the Bx genes to rye chromosomes [18,19]. The map position of Bx genes in the rye genome is thus not syntenious to that found in corn where they are all clustered on the distal part of the short arm of chromosome 4 [20]. The two clusters of the BX genes were also assigned to chromosomes in wheat: Bx1 and Bx2 to chromosome group 4 and Bx3, Bx4, and Bx5 to the short arm of the chromosomes of homoeologous group 5 [18]. Since a segment of chromosome 4L is located on rye chromosome 7RS because of ancient translocation [21], it may be inferred that the Bx1-2 cluster from wheat chromosome 4DL and that from rye chromosome 7R are located synteniously.
The studied complex QTL system for BX in the rye genome constitutes a network of regulatory genes affecting the expression of the structural Bx genes encoding the enzymes of the BX biosynthesis pathway. The Bx genes located in two clusters on chromosomes 5RS and 7R have no or a slight effect on the variation of BX production. Only one QTL for GDIBOA_L with a determination coefficient of 16.5% and additive value of 34.5 coincided in one year with the Bx1-2 genes on chromosome 7R. A similar situation was found for the α-amylase activity in rye where none of the QTL detected coincided with the structural genes encoding the α-amylase isozymes playing a major role in the catabolism of the endosperm starch [22,23]. Allelic forms of the Bx structural genes have little effect on their expression. Instead, a high level of genetic variation in regulatory genes enables a fast and specific adjustment of enzyme synthesis in response to environmental conditions. Another observation supporting this hypothesis is that of the completely different sets of QTL regulating the production of a particular BX species in roots and leaves. These organ-specific regulatory networks probably reflect different defensive mechanisms against biotic and abiotic stresses acting upon roots and leaves or may result from the activation of different members of the multigene families in each tissue. A second possible explanation of the apparently small impact of polymorphisms in the Bx structural loci on the BX content, found in this study, is the lack of functional polymorphism in the Bx genes within the mapping population. This hypothesis is supported by earlier results of association mapping, which showed the presence of SNPs in the Bx genes associated with the BX content within a wide range of rye genotypes [7]. Similarly, the results reported by [8] and [9] support the significant role of polymorphisms within the Bx1 sequences in controlling the BX content in corn.
Results of the QTL mapping revealed that although the heritability of the BX content in rye is promising for breeding purposes, individual QTL are not stable across environments and years. This is a common conclusion from many QTL studies based on the whole mapping populations where phenotypic differences between lines are most often small and prone to environmental factors. This problem is overcome in bidirectional selective genotyping (BSG) studies where only extreme phenotypes, derived through divergent selection and much more independent of environmental changes, are analyzed [22–24]. These studies show that the individual effects of QTL are usually low, but their impact on the trait due to their interactions with other QTL may be substantial.
The complex genetic architecture of BX in rye makes it difficult to develop an efficient strategy for increasing the level of these important metabolites in both leaves and roots. From the QTLs for GDIBOA in leaves, the most promising for selection purposes might be those on chromosomes 1RS, 2RS, and 7R (coinciding with the ScBx1 gene). Both QTL for DIBOA in leaves on chromosome 7R seem to also be valuable for such selection. Roots can be enriched in BX by applying the QTL for GDIMBOA on chromosomes 1R, 2R, 6RS, and 7R.
The set of QTL for PHS revealed in this study constitutes only a part of the genetic architecture detected in the 541 × Ot1-3 intercross in earlier studies [22,25,26]. The QTL on chromosomes 1RL, 2R, 3R, 5R (3), and 6RS are located on the dense DArT map of the RIL population applied in this study in similar positions as those found on the F2 map consisting mainly of the RAPD and SSR markers [27] and agree with the results of association mapping performed within a wide range of rye genotypes [10]. The present experiment however did not identify the QTL for PHS on chromosomes 3RS, 6RL, and 7RS detected earlier by using both QTL mapping and BSG analysis. A proteomic analysis of lines representing the resistant and susceptible lines of the 541 × Ot1-3 cross showed that many identified proteins represented defensive functions against microbial and insect pests [28,29]. This should not be surprising as only the intact grain surface without any destructive effects of pets’ activities can resist water infiltration, which directly induces sprouting and the α-amylase production. This observation leads to the hypothesis about the possible role of BX in the resistance to PHS, which can be additionally supported by data about the inhibitory effect of BX on the germination ability [4]. The results of this study suggest that the coinciding positions of QTL for PHS and BX may reflect the common regulation of BX production and PHS in rye grain.
One of the first processes connected with the water uptake by the grain is the increase in the α-amylase activity in the endosperm. Therefore, all factors reducing the water permeability of the grain surface, including resistance against microbial infections and pest infestation should hinder the α-amylase production in aleurone cells. BXs are among metabolites that play a defensive role, and their relationship with the α-amylase activity is an important question. The negative effect of BX on a plant’s α-amylase activity was reported by Kato-Noguchi et al. [30]. As shown in this paper, several QTL for AMY coincide with those for BX. This observation opens up an opportunity for their common influence on both traits in the rye grain. Future studies should verify this hypothesis, which may play a positive role in finding valuable genes for rye breeding.
The α-amylase activity in the rye grain is a very variable trait, sensitive to environmental conditions, mainly the precipitation and temperature profiles during the period of grain maturation. Levels of this enzyme in the maturing grain were found to be not correlated with its predisposition to PHS [10,31]. Further, results reported in this paper are in agreement with the QTL mapping performed earlier within the 541×Ot1-3 mapping population [23,26,32]. The nine positions of QTL for AMY found here represent regions similar to those detected in previous reports, including an association mapping study [10]. The AMY and PHS QTL occupy similar positions in three chromosomal regions. This finding to some extent confirms earlier results showing that the genetic architecture of these traits is partially overlapping on chromosomes 1RL, 2R, 3R, and 6RL [23,26,33].
Despite several genetic, physiological, and breeding-directed studies, leaf rust caused by Puccinia recondita f. sp. secalis still constitutes a big challenge for rye breeders [34–36]. The SNPs detected for leaf rust resistance using association mapping on chromosome 1R [10] are in a region of RR QTL found in this study. But, no QTL on chromosome 2RS and 5RL, corresponding to the SNPs detected through association mapping, were found. RR QTL were also not found within regions of chromosomes 6RL and 7RL, where the Pr1 and Pr2 genes for leaf rust resistance were mapped [36]. QTL for rust resistance coincide in a majority of cases with the BX QTL and the QTL for the α-amylase activity, along with those for PHS on chromosome 5R. This finding is very important for the possibility of selecting molecular markers aimed at improving AMY, PHS, BX content, and rust resistance in rye. Such markers should be searched for in chromosome regions with coinciding QTL.
The physiological aspect of the common positions of QTL for defensive traits (BX content and rust resistance) and water stress-dependent traits such as PHS and AMY is very interesting. Further, the synthesis of the BX species as defensive metabolites is probably a part of a wider mechanism of mobilizing all necessary plant resources to eliminate the negative aspects of biotic and/or abiotic stresses.
Supporting information
(DOCX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was financially supported by the National Centre for Research and Development, Project No. PBS1/A8/12/2012 (http://www.ncbr.gov.pl/programy-krajowe/program-badan-stosowanych/). Danko Plant Breeders LTD provided partial funding for this project as a commercial partner. Danko Plant Breeders LTD also provided support in the form of salaries for Zofia Banaszak and Katarzyna Banaszak, but had no further role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The specific roles of these authors are articulated in the 'author contributions' section.
References
- 1.Niemeyer HM. Hydroxamic acids (4-hydroxy-1,4-benzoxazin-3-ones) defense chemicals in the Gramineae. Phytochemistry. 1988;27: 3349–3358. [Google Scholar]
- 2.Pérez FJ, Ormeño-Nuñez J. Difference in hydroxamic acid content in roots and root exudates of wheat (Triticum-aestivum L.) and rye (Secale cereale L.)–Possible role in allelopathy. J Chem Ecol. 1991;17: 1037–1043. doi: 10.1007/BF01402932 [DOI] [PubMed] [Google Scholar]
- 3.Poschenrieder C, Torra RP, Barcelo J. A role for cyclic hydroxamates in aluminium resistance in maize. J Inorg Biochem. 2005;99: 1830–1836. doi: 10.1016/j.jinorgbio.2005.05.017 [DOI] [PubMed] [Google Scholar]
- 4.Schulz M, Marocco A, Tabaglio V, Macias FA, Molinillo JMG. Benzoxazinoids in rye alllopathy- from discovery to application in sustainable weed control and organic farming. J Chem Ecol. 2013;39: 154–174. doi: 10.1007/s10886-013-0235-x [DOI] [PubMed] [Google Scholar]
- 5.Tanwir F, Fredholm M, Gregersen PL, Fomsgaard IS. Comparison of the levels of bioactive benzoxazinoids in different wheat and rye fractions and the transformation of these compounds in homemade foods. Food Chemistry. 2013;141: 444–450. doi: 10.1016/j.foodchem.2013.02.109 [DOI] [PubMed] [Google Scholar]
- 6.Brooks AM, Danehower DA, Murphy JP, Reberg-Horton C, Burton JD. Estimation of heritability of benzoxazinoid production in rye (Secale cereale L.) using gas chromatographic analysis. Plant Breed. 2012;131: 104–109. [Google Scholar]
- 7.Rakoczy-Trojanowska M, Orczyk W, Krajewski P, Bocianowski J, Stochmal A, Kowalczyk M. ScBx gene based association analysis of hydroxamate content in rye (Secale cereale L.). J Appl Genet. 2017. a;58: 1–9. doi: 10.1007/s13353-016-0356-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Butrón A, Chen YC, Rottinghaus GE, Mullen MD Genetic variation at bx controls DIMBOA content in maize. Theor Appl Genet. 2010;120: 721–734. doi: 10.1007/s00122-009-1192-1 [DOI] [PubMed] [Google Scholar]
- 9.Zheng LL, McMullen MD, Bauer E, Schon CC, Gierl A, Frey M Prolonged expression of the BX1 signature enzyme is associated with a recombination hotspot in the benzoxazinoid gene cluster in Zea mays. J Exp Botany. 2015;66: 3917–3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rakoczy-Trojanowska M, Krajewski P, Bocianowski J, Schollenberger M, Wakuliński W, Milczarski P, Masojć P, Targońska-Karasek M, Banaszak Z, Brukwiński W, Banaszak K, Orczyk W, Kilian A. Identification of single nucleotide polymorphisms associated with leaf rust resistance, alpha-amylase activity and preharvest sprouting in rye. Plant Mol Biol Rep. 2017. b;35(3): 366–378. doi: 10.1007/s11105-017-1030-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Milczarski P, Bolibok-Brągoszewska H, Myśków B, Stojałowski S, Heller-Uszyńska K, Góralska M, Brągoszewski P, Uszyński P, Kilian A, Rakoczy-Trojanowska M. A high density consensus map of rye (Secale cereale L.) based on DArT markers. PloS ONE. 2011;6(12): e28495 doi: 10.1371/journal.pone.0028495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Masojć P, Larsson-Raźnikiewicz M. Genetic variation of α-amylase levels among rye (Secale cereale L.) kernels, tested by gel diffusion technique. Swedish J Agric Res. 1991;21: 141–145. [Google Scholar]
- 13.Bakera B, Makowska B, Groszyk J, Niziołek M, Orczyk W, Bolibok-Brągoszewska H, Hromada-Judycka A, Rakoczy-Trojanowska M. Structural characteristics of ScBx genes controlling the biosynthesis of hydroxamic acids in rye (Secale cereale L.). J Appl Genet. 2015;56: 287–298. doi: 10.1007/s13353-015-0271-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stam P Construction of integrated genetic linkage maps by means of a new computer package: JoinMap. The Plant Journal. 1993;3: 739–744. [Google Scholar]
- 15.Cullis BR, Smith A, Coombes N. On the design of early generation variety trials with correlated data. JABES. 2006;11: 381–393. [Google Scholar]
- 16.International VSN. GenStat for Windows 16th Edition VSN International, Hemel Hempstead, UK: Web page: GenStat.co.uk. 2013. [Google Scholar]
- 17.Wang S, Basten CJ, Zeng ZB. Windows QTL Cartographer 2.5. Department of Statistics, North Carolina State University, Raleigh, NC: 2012. (http://statgen.ncsu.edu/qtlcart/WQTLCart.htm) [Google Scholar]
- 18.Sue M, Nakamura C, Nomura T. Dispersed benzoxazinone gene cluster: molecular characterization and chromosomal localization of Glucosyltransferase and Glucosidase Genes in wheat and rye. Plant Physiology. 2011;157: 985–997. doi: 10.1104/pp.111.182378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nomura T, Ishihara A, Imaishi H, Ohkawa H, Endo TR, Iwamura H. Rearrangement of the genes for the biosynthesis of benzoxazinones in the evolution of Triticeae species. Planta 2003;217: 776–782. doi: 10.1007/s00425-003-1040-5 [DOI] [PubMed] [Google Scholar]
- 20.Dutartre L, Hilliou F, Feyereisen R. Phylogenomics of the benzoxazinoid biosynthetic pathway of Poaceae: gene duplication and origin of the Bx cluster. BMC Evolutionary Biology. 2012;12: 64 doi: 10.1186/1471-2148-12-64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Devos KM, Atkinson MD, Chinoy CN, Francis HA, Harcourt RL, Koebner RMD, Liu CJ, Masojć P, Xie DX, Gale MD. Chromosomal rearrangements in the rye genome relative to that of wheat. Theor Appl Genet 1993;85: 673–680. doi: 10.1007/BF00225004 [DOI] [PubMed] [Google Scholar]
- 22.Masojć P, Lebiecka K, Milczarski P, Wiśniewska M, Łań A, Owsianicki R. Three classes of loci controlling preharvest sprouting in rye (Secale cereale L.) discerned by means of bidirectional selective genotyping (BSG). Euphytica. 2009;170: 123–129. [Google Scholar]
- 23.Masojć P, Wiśniewska M, Łań A, Milczarski P, Berdzik M, Pędziwiatr D, Pol-Szyszko M, Gałęza M, Owsianicki R. Genomic architecture of alpha-amylase activity in mature rye grain relative to that of preharvest sprouting. J Appl Genet. 2011;52: 153–160. doi: 10.1007/s13353-010-0025-x [DOI] [PubMed] [Google Scholar]
- 24.Masojć P, Bienias A, Berdzik M, Kruszona P. Genetic analysis carried out in population tails reveals diverse two-loci interactions as a basic factor of quantitative traits variation in rye. J Appl Genet. 2016;57: 165 https://doi.org/10.1007/s13353-015-0321-6. doi: 10.1007/s13353-015-0321-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Masojć P, Banek-Tabor A, Milczarski P, Twardowska M. QTLs for resistance to preharvest sprouting in rye (Secale cereale L.). J Appl Genet. 2007;48: 211–217. doi: 10.1007/BF03195215 [DOI] [PubMed] [Google Scholar]
- 26.Masojć P, Milczarski P. Relationship between QTLs for preharvest sprouting and alpha-amylase activity in rye grain. Mol Breed. 2009;23: 75–84. [Google Scholar]
- 27.Milczarski P, Banek-Tabor A, Lebiecka K, Stojałowski S, Myśków B, Masojć P. New genetic map of rye composed of PCR-based molecular markers and its alignment with the reference map of the DS2 × RXL10 intercross. J Appl Genet. 2007;48: 11–24. [DOI] [PubMed] [Google Scholar]
- 28.Masojć P, Kosmala A. Proteomic analysis of preharvest sprouting in rye using two-dimensional electrophoresis and mass spectrometry. Mol Breed. 2012;30: 1355–1361. doi: 10.1007/s11032-012-9721-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Masojć P, Kosmala A, Perlikowski D. Proteomic analysis of developing rye grain with contrasting resistance to preharvest sprouting. J Appl Genet. 2013;54: 11–19. doi: 10.1007/s13353-012-0127-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kato-Noguchi H, Macias FA, Molinillo JMG. Structure-activity relationship of benzoxazinones and related compounds with respect to the growth inhibition and alpha-amylase activity in cress seedlings. J Plant Physiol. 2010;167: 1221–1225. doi: 10.1016/j.jplph.2010.04.006 [DOI] [PubMed] [Google Scholar]
- 31.Twardowska M, Masojć P, Milczarski P. Pyramiding genes affecting sprouting resistance in rye by means of marker assisted selection. Euphytica. 2005;143: 257–260. [Google Scholar]
- 32.Masojć P, Milczarski P. Mapping QTLs for alpha-amylase activity in rye grain. J Appl Genet. 2005;46: 115–123. [PubMed] [Google Scholar]
- 33.Myśków B, Stojałowski S, Łań A, Bolibok-Brągoszewska H, Rakoczy Trojanowska M, Kilian A. Detection of the quantitative trait loci for α-amylase activity on a high-density genetic map of rye and comparison of their localization to loci controlling preharvest sprouting and earliness. Mol Breed. 2012;30(1): 367–376. doi: 10.1007/s11032-011-9627-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miedaner T, Sperling U. Effect of leaf rust on yield components of winter rye hybrids and assessment of quantitative resistance. J. Phytopathology. 1995;143: 725–730. [Google Scholar]
- 35.Miedaner T, Gey A, Sperling U, Gieger H. Quantitative-genetic analysis of leaf-rust resistance in seedling and adult-plant stages of inbred lines and their testcrosses in winter rye. Plant Breed. 2002;121: 475–479. [Google Scholar]
- 36.Wehling P, Linz A, Hackauf B, Roux SR, Ruge B, Klocke B. Leaf-rust resistance in rye (Secale cereale L.) 1. Genetic analysis and mapping of resistance genes Pr1 and Pr2. Theor Appl Genet. 2003;107: 432–438. doi: 10.1007/s00122-003-1263-7 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.