Abstract
Sequence-specific peptides have been demonstrated to self-assemble into structurally defined nanoscale objects including nanofibers, nanotubes, and nanosheets. The latter structures display significant promise for the construction of hybrid materials for functional devices due to their extended planar geometry. Realization of this objective necessitates the ability to control the structural features of the resultant assemblies through the peptide sequence. The design of a amphiphilic peptide, 3FD-IL, is described that comprises two repeats of a canonical 18 amino acid sequence associated with straight α-helical structures. Peptide 3FD-IL displays 3-fold screw symmetry in a helical conformation and self-assembles into nanosheets based on hexagonal packing of helices. Biophysical evidence from TEM, cryo-TEM, SAXS, AFM, and STEM measurements on the 3FD-IL nanosheets support a structural model based on a honeycomb lattice, in which the length of the peptide determines the thickness of the nanosheet and the packing of helices defines the presence of nanoscale channels that permeate the sheet. The honeycomb structure can be rationalized on the basis of geometrical packing frustration in which the channels occupy defect sites that define a periodic superlattice. The resultant 2D materials may have potential as materials for nanoscale transport and controlled release applications.
Graphical Abstract
INTRODUCTION
The creation of functional materials using the basic principles of molecular design is a main objective of the emergent field of nanoarchitectonics.1,2 Peptides, proteins, and related foldamers represent useful substrates for the construction of functional materials in that intermolecular interactions can be encoded within the corresponding sequence to direct self-assembly into well-defined supramolecular structures. Two-dimensional nanoscale assemblies (i.e., nanosheets) represent particularly desirable targets, in that these materials can serve as scaffolds to spatially arrange exogenous substrates, potentially with precision, for integration into devices. To date, collagen-mimetic peptides,3,4 peptoids,5 β-sheet peptides,6,7 and globular proteins8,9 have been employed for the creation of two-dimensional materials through self-assembly in solution.
In contrast, the ubiquitous α-helical structural motif has been less utilized for fabrication of structurally defined two-dimensional assemblies. Often, however, the crystallographically determined structures of synthetic helical peptides reveal the presence of two-dimensional layers within three-dimensional crystals, indicating the potential for nanosheet formation from self-assembly of helices.10–12 Similarly, uniform-sized rods of α-helical poly(benzyl-l-glutamate) were observed to pack into layered structures of defined width in lyotropic mesophases and thin solid films.13 The thickness of the smectic layers could be directly correlated with the length of the peptide in an α-helical conformation, and, therefore, is amenable to sequence control.
Two-dimensional assemblies of helices may be fabricated from either coiled-coil structural motifs or straight α-helices. While interhelical packing interactions are reasonably well understood from structural analyses of native proteins,14 most helix-helix interactions involve crossing angles that deviate from a strictly parallel packing arrangement. Classical coiled-coils,15 which comprise discrete bundles of α-helices having a canonical heptad repeat sequence, display a crossing angle of approximately 20° that defines a left-handed superhelical twist. Coiled-coils have the advantage that the structural interactions that guide self-assembly into specific oligomerization states (n = 2–7)16–19 are well understood and amenable to computational design. However, the structural features that determine lateral packing of coiled-coils are less well understood. The packing of coiled-coils in two dimensions requires that adjacent bundles interact at interfaces that are inclined with respect to each other and continuously twist about the superhelical axis. While such interfaces have been designed,20,21 the degree of curvature limits contact between adjacent bundles such that self-assembly may be precluded except under conditions in which interbundle interactions can be precisely engineered. While the latter process remains a challenge, the computational design of coiled-coils that self-assemble into crystalline 1D,22,23 2D,24 and 3D20 assemblies has been reported.
In contrast, single-crystal structural analyses of straight α-helices indicate that these materials crystallize well in lattices consisting of packed layers of peptides. However, in most cases, the peptide sequences are relatively short, such that the helical conformation is thermodynamically unstable in the absence of helix-promoting solvents (e.g., trifluoroethanol) or without the incorporation of helicogenic noncanonical amino acids (e.g., 2-amino-isobutyric acid). Moreover, little information is available regarding the specificity of lateral interactions between adjacent helices within these layered structures. However, the computational design of a conformationally stable antiparallel trimeric helical bundle was recently reported, in which the α-helices pack in a stable arrangement with minimal superhelical twist.19 These results suggest that straight α-helices could form stable assemblies if the interacting helical faces were based on a repeat sequence that reinforced the canonical 18 amino acid repeat.25 We report herein the design of a synthetic peptide, 3FD-IL, that self-assembles from aqueous solution to form structurally defined two-dimensional assemblies that comprise a hexagonal honeycomb structure based on packing of aligned α-helices (Figure 1).
Figure 1.
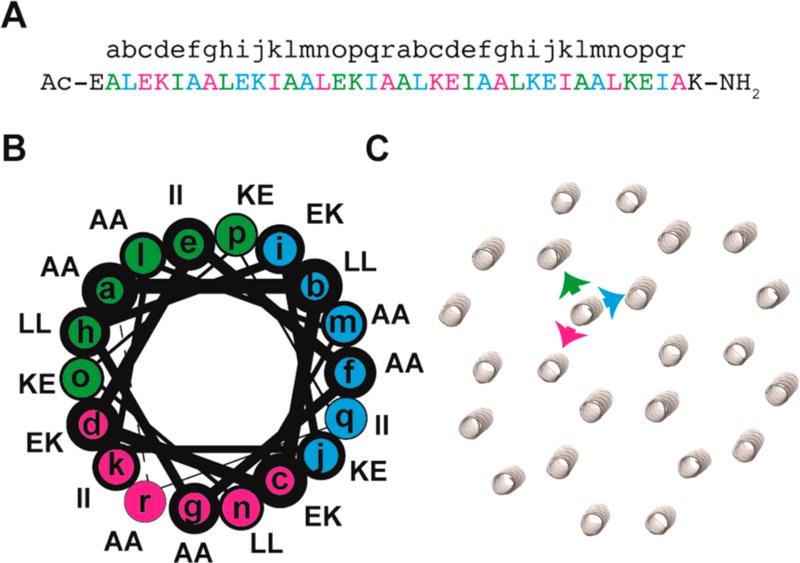
(A) Amino acid sequence of 3FD-IL indicating the register of residues within the 18 amino acid repeats. (B) Helical wheel representation of the repeat sequences of 3FD-IL. (C) Proposed packing of 3FD-IL helices within the hexagonal honeycomb model depicting the 3-fold symmetry of the sequence. The three faces are represented in cyan, green, and magenta.
RESULTS AND DISCUSSION
Peptide 3FD-IL, a 38 amino acid sequence, was designed to form a straight α-helix with 3-fold screw symmetry (Figure 1A). The core peptide sequence (Ala2-Ala37) consists of two successive 18 amino acid repeats; each of which defines three identical faces of six amino acids within the structural context of an α-helical conformation (Figure 1B). Based on the geometrical parameters of an α-helix, successive faces within the peptide sequence are angularly offset by −120° and axially offset by 9 Å (6 × 1.5 Å helical rise/residue).25 In addition, the charged amino acids in the N-terminal three faces are electrostatically complementary to the C-terminal three faces. Finally, a negatively charged glutamic acid residue and positively charged lysine residue were placed at the N-terminus and C-terminus of the capped peptide, respectively, to compensate for the structural influence of the helix macrodipole.26
Since detailed structural information was not available for the design of selectively interacting straight α-helices, the rationale for the design of the 3FD-IL peptide sequence was based on consideration of the 18 amino acid repeat as a sequential series of heptad (7 amino acid) and hendecad (11 amino acid) repeats.15 Sequence determinants are more readily available for the hendecad and, especially, the heptad repeat sequences. The sequence-preferences of the individual heptad and hendecad repeats are unlikely to perfectly capture the geometrical requirements of straight α-helices, but nonetheless provided a reasonable approximation for the initial design of the 3FD-IL sequence.
In order to simplify the design parameters, we chose to restrict the mode of helical interaction to dimeric interfaces between straight α-helices. Heptad sequences tend to favor dimerization when isoleucine and leucine occupy the core a-and d-positions, respectively, of the heptad repeat.16,27 In comparison, the association between helical interfaces for hendecad repeats is mediated through interactions between residues at the a-, d-, e-, and h-positions. However, isolated dimers based on hendecad repeats have not been characterized at atomic-level resolution. Indeed, Lupas has postulated that dimers derived from hendecad repeats should place residues at the a- and h-positions in close proximity to the polypeptide backbone of the opposite helix such that only amino acids with small side chains could be tolerated at those sites.28 The b subunits of bacterial F0F1-ATPases contain a domain comprising a series of hendecad repeats that forms an extended dimer on the basis of biophysical measurements in solution.29,30 Alanine residues are strongly conserved at the a- and h-positions of these hendecad repeats, as expected based on Lupas’ hypothesis. The d- and e-positions of the hendecad repeats do not point directly toward the opposite helix and can tolerate amino acids with longer side-chains. In the design of 3FD-IL, we chose to employ combinations of amino acids with charged side-chains, i.e., glutamic acid and lysine, in order to promote solubility of the peptide and introduce complementary electrostatic interactions upon self-association. Thus, each interacting interface comprises six amino acid residues at the a- and d-positions of the heptad repeats and the a-, d-, e-, and h-positions of the hendecad repeats (i.e., the a-, d-, h-, k-, l-, and o-positions of an 18 amino acid repeat).
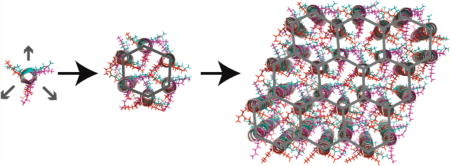
On the basis of this sequence design, we hypothesized that the straight helices of 3FD-IL would behave as 3-fold symmetric tectons that could self-assemble into a two-dimensional open framework structure having hexagonal symmetry. The 2D framework would be stabilized through a combination of hydrophobic interactions and electrostatic interactions between structurally adjacent helices (Figure 1B, 1C). This mode of assembly should result in the creation of a nanoporous two-dimensional structure in which the charged amino acid side-chains line the lumen of the channel (Figure 1C).
Peptide 3FD-IL could be readily prepared in high yield and purity via microwave assisted solid-phase peptide synthesis. The peptide was readily soluble in aqueous buffers within the pH range from 4 to 9. Circular dichroism spectropolarimetry indicated that 3FD-IL adopted a strongly helical conformation within this pH range. The circular dichroism spectra of 3FD-IL in aqueous buffer (10 mM TAPS, pH 8.5) are representative of its conformational properties and corresponded to conditions that promoted the formation of ordered assemblies (vide infra). CD spectra of nascent solutions of peptide 3FD-IL indicated the presence of α-helical secondary structure (Figure 2A).
Figure 2.
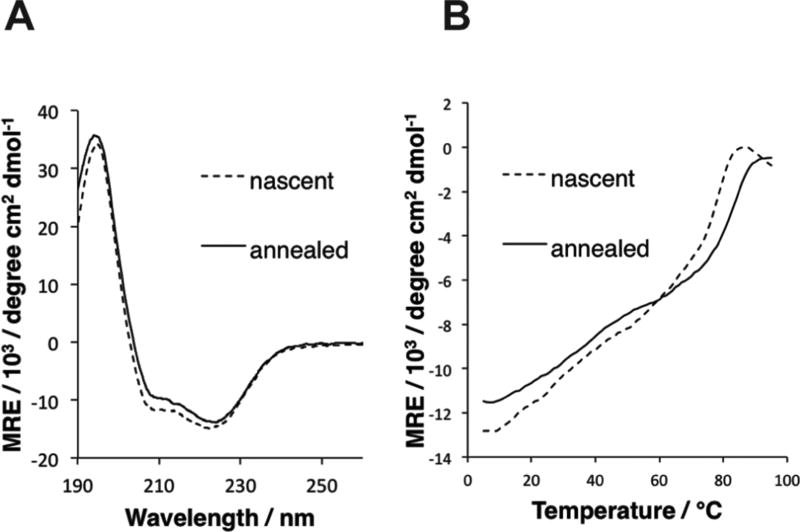
(A) Circular dichroism spectra of nascent and thermally annealed specimens of 3FD-IL in TAPS buffer (10 mM, pH 8.5). (B) CD melting curves for nascent and thermally annealed solutions of 3FD-IL in TAPS buffer (10 mM, pH 8.5).
Thermolysis and slow cooling from 90 °C to 40 °C (0.2 °C/ min) resulted in retention of the helical CD signature of 3FD-IL (Figure 2A). The first derivative of the CD melting curve of nascent and thermally annealed samples of 3FD-IL displayed a similar response, in which a melting transition (Tm) was observed at elevated temperature (Figure 2B and Supporting Figure S1). The Tm for the thermally annealed specimen of 3FD-IL occurred at ca. 84 °C, which is slightly higher than the corresponding Tm of 78 °C observed for the nascent sample. This observation may be attributable to the larger size of the self-assembled species after controlled thermal annealing (vide infra). The melting transitions are broadened to an extent, which may reflect the distribution of oligomeric assembly states within the specimens. A similar broadening of the CD melting transition has been observed for two-dimensional collagen assemblies that display polymorphism in lateral association.3
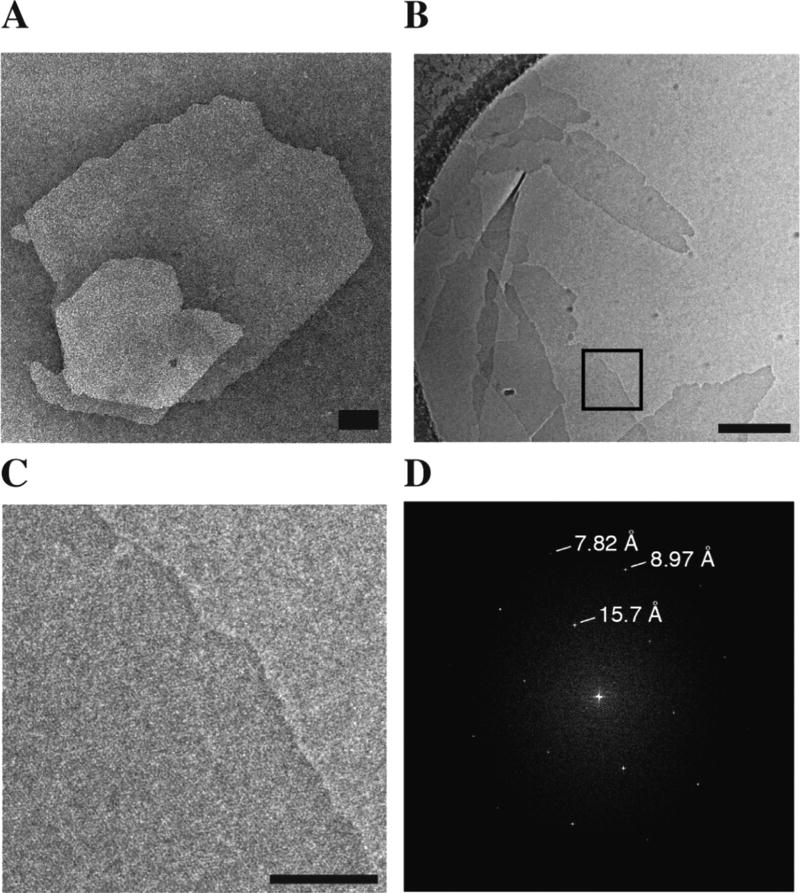
The self-assembly behavior of 3FD-IL was analyzed using transmission electron microscopy (TEM) and atomic force microscopy (AFM). TEM analysis of thermally annealed solutions of 3FD-IL indicated the presence of two-dimensional assemblies having a size distribution from a few hundred nanometers to several micrometers in apparent width. (Figure 3A). The nanosheets occasionally displayed sharp edges, but seemed susceptible to fragmentation from mechanical shearing due to deposition on the EM grid. Note that TEM analysis of nascent solutions of 3FD-IL indicated the presence of nanosheets, but these species were smaller than the corresponding assemblies from thermally annealed samples (Supporting Figure S2). The morphology of the 3FD-IL assemblies was confirmed using electron cryomicroscopy (Figure 3B), which demonstrated that the nanosheets remained largely intact under conditions of controlled vitrification. At higher magnification (boxed region), the presence of a periodic lattice within the nanosheet could be observed in which nanoscale pores could be discerned (Figure 3C and Supporting Figure S3). An FFT of the nanosheet displayed several lattice spacings, as well as an underlying hexagonal symmetry for the individual assemblies (Figure 3D). In contrast, samples consisting solely of vitrified buffer lacked this lattice structure and its associated periodicity (Supporting Figure S4).
Figure 3.
(A) TEM image of a representative nanosheet from self-assembly of annealed solutions of 3FD-IL (scale bar = 200 nm). (B) Cryo-TEM image of 3FD-IL nanosheets (scale bar = 200 nm). (C) Magnification of the boxed region displaying the periodic lattice of the 3FD-IL nanosheet (scale bar = 50 nm). (D) FFT indicating the hexagonal symmetry of the periodic lattice of a representative nanosheet and associated lattice spacings.
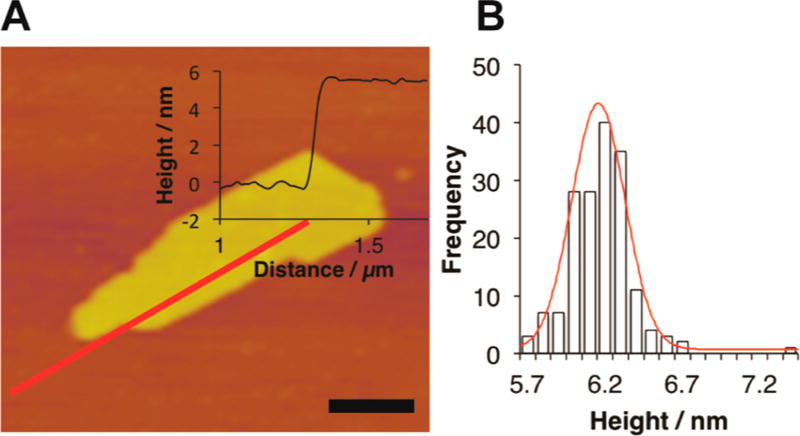
Tapping mode atomic force microscopy of the 3FD-IL assemblies confirmed the morphology observed from TEM measurements (Figure 4A). AFM measurements of the heights of the nanosheets indicated an apparent thickness of 62 ± 1.6 Å (Figure 4A and 4B) with a relatively narrow distribution of heights and no evidence for multilayer sheets. The length of the peptide was calculated to be 57 Å (38 residues × 1.5 Å rise/ residue), assuming a uniform α-helical conformation along the contour length of the peptide. The close correspondence between the measured height of the nanosheet and the calculated length of 3FD-IL suggests that the peptides pack in a perpendicular orientation with respect to the surface of the nanosheet. We have observed a similar packing arrangement for nanosheets derived from collagen-mimetic peptides, in which the triple helices are oriented perpendicular to the sheet surface.3 The small discrepancy between the calculated and measured sheet thickness may reflect a slight registry shift between adjacent helices within the structure. Axial offsets of 2–3 Å are typically observed between antiparallel helices in coiled-coil structures,31 and may be relevant to helix packing interactions within the structural model for the 3FD-IL nanosheets (vide infra).
Figure 4.
(A) Representative AFM image of 3FD-IL nanosheets with height profiles (inset). Scale bar = 200 nm. (B) AFM height histogram with fit (red curve) for 3FD-IL nanosheets.
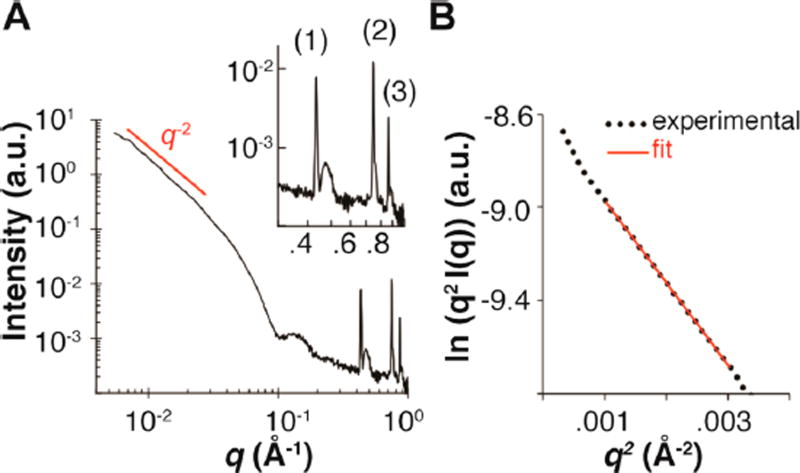
Small angle X-ray scattering (SAXS) experiments were employed to determine information about the shape and internal structure of the assemblies in solution. A plot of intensity versus momentum transfer (q) revealed several critical features related to the structure of the nanosheets. First, the scattering intensity in the low q (~ <0.05 Å−1) region followed a power law dependence of q−2, indicating the presence of sheet-like or disk-like assemblies in solution (Figure 5A and Supporting Figure S5). Second, fitting of the low q data using the modified Guinier equation for sheet-like forms32 indicated that the sheets have an average thickness of ca. 65 Å in solution (Figure 5B), which agrees well with the AFM height measurements.
Figure 5.
(A) Small-angle X-ray scattering (SAXS) curve for a solution of 3FD-IL (10 mg/mL) in TAPS buffer (10 mM, pH 8.5). Inset: expansion of the high q region depicting the positions of the three diffraction peaks. Peaks (d-spacing): (1), 14.6 Å; (2), 8.4 Å; (3), 7.3 Å. (B) Modified Guinier plot, ln(q2 I(q)) versus q2, of scattering data for 3FD-IL using an equation for sheet-like forms: , where It,0 is a constant, and thickness (T) can be calculated as .
Furthermore, Bragg reflections were observed in the high q region of the scattering curve (Figure 3A). This suggests that the nanosheets have a high degree of internal order. The Bragg reflections correspond to d-spacings of (1) 14.6 Å, (2) 8.4 Å, and (3) 7.3 Å. The relationship between these peaks is (1):(1)/ √3:(1)/√4, which is consistent with a hexagonal arrangement of helices within the nanosheets. FFTs obtained from individual nanosheets imaged using electron cryomicroscopy display the same mathematical relationship between lattice spacings observed at 15.7 Å, 9.0 Å, and 7.8 Å (Figure 3D). The slightly larger distances determined from cryo-EM image analysis may indicate that the lattice expands upon vitrification at low temperature. Nevertheless, the correspondence of structural parameters between measurements is consistent with formation of structurally defined populations of self-assembled nanosheets of 3FD-IL.
As the SAXS data were acquired at ambient temperature in buffered aqueous solution, we posited that these data were more representative of the structure under the conditions in which these materials might find application. Therefore, the distances from SAXS analysis were utilized to construct a structural model for the hexagonal unit cell. The two most probable packing arrangements of the 2D lattice would involve: (1) a hexagonal close packed (hcp) lattice of helices or (2) a hexagonal honeycomb (i.e., graphene-like) structure in which helices are positioned at 3-fold sites in the 2D lattice. Pseudohexagonal close packing of helices has been commonly observed in the crystal structures of synthetic helical peptides10 as well as thin solid films from dried down lamellar mesophases of monodisperse, α-helical PBLG.13 However, the 3-fold screw symmetry of 3FD-IL is more consistent with a honeycomb lattice (Figure 1). A detailed description of the two different models can be found in the Supporting Information (Supporting Figures S6 and S7).
The different structural models for the 3FD-IL nanosheets were constructed on the basis of antiparallel packing arrangements of adjacent helices based on several considerations. Foremost, an antiparallel orientation minimizes dipole-dipole repulsion between structurally adjacent α-helices.26 In the crystal structures of helical peptides, antiparallel arrangements seem more common than parallel arrangements, probably due to the aforementioned consideration.10 However, the pattern of charged amino acids in the sequence of 3FD-IL is compatible with the formation of charge-complementary interactions in either a parallel or an antiparallel alignment between helices.
To test for relative helix orientation, three peptide variants of 3FD-IL were designed in which the sequences comprised different patterns of charged residues, but the overall amino acid composition and polar patterning were retained. Peptides 3FD-IL-EKEK, 3FD-IL-KEKE, and 3FD-IL-EEKK were designed such that the pattern of charged amino acids favored the parallel arrangement for the first two peptides and the antiparallel packing arrangements for the third peptide (Supporting Figure S8). In other words, the full complement of attractive Coulombic interactions would be observed if the peptide assembled into a 2D lattice based on either a parallel orientation (for 3FD-IL-EKEK or 3FD-IL-KEKE) or an antiparallel orientation (for 3FD-IL-EEKK), but the interactions would be completely repulsive in the opposite orientation for each peptide, respectively.
The CD spectra of all three peptides displayed a strongly α-helical signature (Supporting Figure S9). TEM image analysis of assemblies derived from either 3FD-IL-EKEK or 3FD-IL-KEKE indicated the presence of a heterogeneous mixture of fibrils, ribbons, sheets, and aggregates (Supporting Figures S10 and S11). In contrast, TEM image analysis demonstrated that aqueous solutions of 3FD-IL-EEKK formed an abundance of highly ordered nanosheets (Supporting Figure S12). In addition, synchrotron SAXS analysis of annealed nanosheet solutions of the antiparallel peptide, 3FD-IL-EEKK, displayed a similar scattering profile to that observed for nanosheets of 3FD-IL, including the position of the diffraction peaks in the high q region of the scattering curve (Supporting Figure S13).
These data indirectly suggested that adjacent helices within the 3FD-IL nanosheets are aligned in an antiparallel orientation. As a final test of this hypothesis, an approximately equimolar mixture of 3FD-IL-EKEK and 3FD-IL-KEKE peptides was prepared and thermally annealed. The mixture retained its α-helical CD signature after heat treatment (Supporting Figure S9). TEM analysis indicated the presence of nanosheets as the sole species present in solution (Supporting Figure S14). However, the nanosheets appeared to have a more irregular morphology than those derived from self-assembly of either 3FD-IL or 3FD-IL-EEKK. On the basis of the honeycomb structural model, heteromeric association between 3FD-IL-EKEK and 3FD-IL-KEKE could result in the formation of a 2D assembly but only if the respective peptides were aligned in an antiparallel orientation.
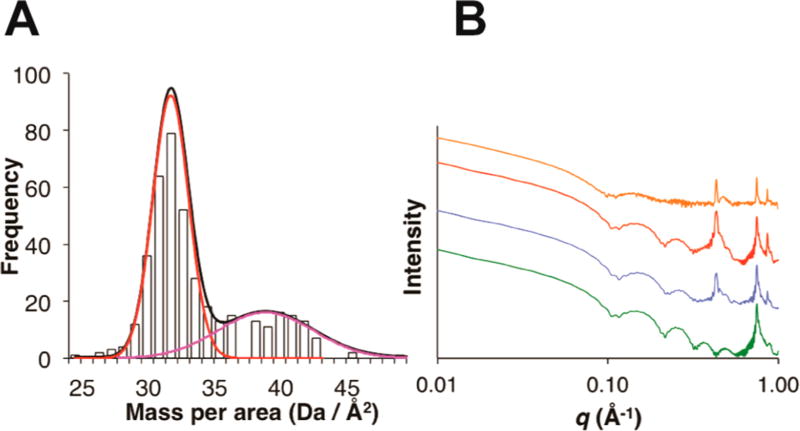
Two lines of evidence were employed to distinguish between the hexagonal honeycomb and hexagonal close-packed structural models. Scanning transmission electron microscopy (STEM) was utilized to determine the mass/area (M/A) values for freeze-dried nanosheets of 3FD-IL. The histogram of the mass/area plot indicates the presence of a major species with a mean M/A value of 31.7 ± 1.4 Da/Å2 (Figure 6A) Theoretical mass-per-area measurements can be calculated for the two model lattices, which affords values of 33.2 Da/Å2 and 50.1 Da/ Å2 for the hexagonal honeycomb and hexagonal close-packed lattices, respectively (Figure 7 and Supporting Figures S6 and S7). The experimental STEM mass/area data more closely correspond to the hexagonal honeycomb model (Figure 7). A minor species with a higher mass/area (38.8 ± 3.6 Da/Å2) is also observed, although its value is less than that expected for a hexagonal close-packed lattice. FFT of STEM images obtained on negatively stained nanosheets of 3FD-IL indicated the presence of a periodicity of 15.1 Å with hexagonal symmetry (Supporting Figure S15). The observed value from STEM data is slightly larger than the 14.6 Å spacing from the SAXS data, but not significant enough to skew the conclusions of the analysis. These observed differences in lattice parameters might result from slightly different methods for sample preparation between the different analytical methods, which become manifested due to the porosity of the honeycomb structure.
Figure 6.
(A) STEM mass/area histogram for freeze-dried specimens of the 3FD-IL nanosheets with Gaussian fits for a major (red) and minor (magenta) population of self-assembled species. (B) Comparison of the experimental scattering profile (orange curve) for 3FD-IL nanosheets to a series of calculated scattering profiles for a hexagonal honeycomb structure in which the channels are empty (red curve), filled with water (blue curve), or filled within an additional helix (green curve). Calculated scattering curves were based on a disk-like model with a radius of 250 Å using the lattice parameters derived from the original SAXS measurements.
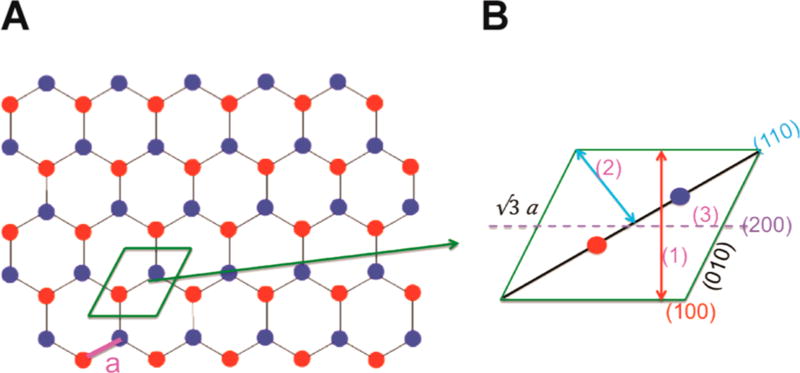
Figure 7.
(A) Top view of the proposed hexagonal honeycomb lattice model for the 3FD-IL nanosheets. Adjacent helices are aligned antiparallel to each other with oppositely oriented helices depicted as red and blue circles. The unit cell is depicted in green. The interhelical distance (a) is calculated to be 9.7 Å. (B) Expansion of the 2D unit cell of the 3FD-IL nanosheets. The lattice spacings corresponding to (1) d100 (14.6 Å), (2) d110 (8.4 Å), and (3) d200 (7.3 Å) are indicated. The length of the 2D unit cell is 16.8 Å (√3 × 9.7 Å). The area of the parallelogram that defines the 2D unit cell is 245 Å2 (base of 16.8 Å × height of 14.6 Å). The unit cell contains two peptides of mass 4070 Da. The calculated mass/area of the model structure is 33.2 Da/Å2 (2 × 4070 Da/245 Å2).
Theoretical modeling of the SAXS data, especially in the high q region of the scattering profile, provided additional support for the hexagonal honeycomb model for the 3FD-IL nanosheets. The latter model requires the presence of internal nanosized pores within the two-dimensional lattice (Figure 1C and Figure 7). In this model, the pores would be lined with polar amino acid residues. Theoretical modeling of the AXS profile of the honeycomb structure approximates quite well the experimental scattering curve. Modeling of water molecules into the channels does not significantly alter the theoretical scattering profile. However, additional peptides modeled into the channel (essentially equivalent to the hexagonal close-packed model) resulted in significant changes to the calculated scattering profile. Clear discrepancies can be distinguished between the calculated scattering profile for the hcp model and the experimental SAXS data for 3FD-IL in solution. In particular, the absence of the sharp peak at a q value of 0.431 Å−1 (corresponding to the d-spacing of 14.6 Å) in the calculated scattering curve for the hcp model is quite distinct from that of the experimental scattering profile. Moreover, the honeycomb lattice is qualitatively in agreement with structural data on helix-helix interactions. The spacing between helices was determined to be 9.7 Å, which is consistent with the centroid-to-centroid distances between helices within crystal structures of helical peptides10 and in α-helical coiled-coils.33
Collectively, these data support a model for the 3FD-IL nanosheets in which the helices are aligned perpendicular to the surface of the assembly and in which structurally adjacent helices are aligned in an antiparallel orientation (Figure 7). Recent reports of nanosheets based on collagen-mimetic peptides have provided evidence that the triple helices pack in an antiparallel manner in the 2D lattice due to charge complementary interactions.3 However, the 3FD-IL nanosheets differ from the collagen nanosheets in that an open framework structure based on hexagonal packing symmetry is observed upon self-assembly. In contrast, the structural evidence supports tetragonal packing of collagen triple helices as a recurrent feature within the corresponding 2D assemblies.
Lanci et al., have observed a honeycomb structure in a computationally designed peptide crystal, in which 2D layers of triple helical coiled-coils stack along the z-direction (crystallo-graphic c-axis).20 Single layers of the corresponding structures have not been observed, thus far, as isolated nanosheets in solution, perhaps due to relatively weak lateral interactions between trimers. More recently, Zhang et al., reported the computational design of open framework 2D lattices derived from coiled-coil peptides.24 In this approach, the interfaces between component four-helix bundles were engineered to afford 2D lattices displaying different underlying plane symmetries. The self-assembly of 3FD-IL does not require prior design of a folded protein motif such as a coiled-coil subunit. The design is quite robust in that it can accept variations in the identity of amino acid residues as long as the underlying sequence pattern is maintained in terms of positional preferences for small hydrophobic, large hydrophobic, and charged/polar residues. However, this approach is likely to be far less flexible than that of Zhang et al., in that the underlying symmetry of the 2D lattice will be less amenable to rational variation.
The question arises as to the driving force for selective formation of the honeycomb lattice with respect to the hexagonal close-packed lattice for 2D assemblies derived from self-assembly of 3FD-IL and its structurally related congeners. Crystallographic studies of polymers34 (including polypeptides35–38) have demonstrated that geometrical frustration can occur within trigonal lattices such as the hexagonal close-packed lattice. In a hexagonal close-packed lattice, it is impossible for every helix to pack into an arrangement in which all nearest neighbor interactions involve the lowest energy, antiparallel orientation. This geometrical frustration results in distortion of the lattice to form an ordered structure that can be considered as a superlattice of defect sites. This situation is analogous to that of spin glasses, in which the arrangement of anti-ferromagnetically coupled Ising spins on a trigonal lattice results in an obligate frustrated state.39 In helical polymer crystals, a structural distortion can occur to alleviate the geometrical frustration, which results in a honeycomb lattice in which one polymer chain occupies defect sites that the define the superlattice.34
In the case of the 3FD-IL nanosheets, the peptide sequence could promote the formation of a “geometrically frustrated” honeycomb lattice. Rather than undergo a structural distortion, the 3FD-IL helices form an open framework structure in which two subunits pack in the energetically preferred antiiparallel orientation within the unit cell (Figure 7). We hypothesize that the channel occupies the defect site and defines the superlattice of the honeycomb structure (Figure 7). The periodicity of the polar groups and the complementary interactions between charges stabilize the pore structure and occlude the presence of an additional helix. The absence of a helix in the channel, as would be present in a hexagonal close-packed structure, ensures that the two antiparallel helices are energetically equivalent and experience identical interhelical interactions.
The hexagonal honeycomb morphology has been observed in layered structures corresponding to metal–organic frameworks (MOFs),40 covalent organic frameworks (COFs),41 and supramolecular organic frameworks (SOFs).42 However, the isolation of freely standing nanosheets of uniform thickness and internal structure from these materials represents a significant challenge. Supramolecular self-assembly in solution holds significant promise for fabrication of freely standing, structurally defined nanosheets of uniform thickness.42–47 The 3FD-IL nanosheets more closely resemble these supramolecular organic frameworks (SOFs) than two-dimensional assemblies derived from either MOFs or COFs. The lamellar thickness, the internal structure, and the surface chemistry of these peptide nanosheets can be controlled through the composition, sequence, and length of the protomer. Recent progress in the de novo design of synthetic peptide and protein assemblies has demonstrated that ordered structures approaching the complexity of structures derived from DNA nanotechnology48 can be assembled from relatively simple components under environmentally benign conditions.20,23,24,49–51 In particular, a broad range of peptide- and protein-based subunits have now been described that self-assemble into structurally defined two-dimensional assemblies with the potential to encode function.52
CONCLUSIONS
The formation of the 3FD-IL nanosheets indicates that robust two-dimensional structures can result from self-assembly of simple α-helical protomers. The corresponding nanosheets display significant long-range order and thermal stability at relatively low concentration (ca. 0.1–2.5 mM) in solution. The sequence of 3FD-IL directs the formation of a hexagonal honeycomb structure that defines nanosized pores, which suggests that these materials may find application in transport and encapsulation of appropriately sized ions and small molecules. Further development of these materials for applications will require the availability of higher resolution (i.e., atomic-level) structural information. Recently, direct electron detection devices, in combination, with electron cryomicroscopy have enabled reconstruction at near atomic-resolution structures of natural53 and synthetic54 protein- and peptide-based helical assemblies and of 2D crystals of membrane proteins, such as aquaporin-0 (AQP0).55 The extended structural order of the 3FD-IL assemblies suggests that they might serve as useful substrates in this type of structural analysis, which would provide insights that could facilitate the design of more structurally complex and operationally functional 2D assemblies.
METHODS
Materials
Chemical reagents were purchased from Sigma-Aldrich Chemical Co. (St. Louis, MO) or Anaspec, Inc. (Fremont, CA), unless otherwise noted. Fmoc-PEG-PAL-PS resin for solid-phase peptide synthesis was purchased from Applied Biosystems, Inc. (Foster City, CA).
Peptide Synthesis
The 3FD-IL peptide and the 3FD-IL-EKEK, 3FD-IL-KEKE, and 3FD-IL-EEKK variants were either purchased from GenScript USA, Inc. (Piscataway, NJ) or synthesized in house. In the latter case, peptides were prepared using microwave-assisted synthesis on a CEM Liberty solid-phase peptide synthesizer. Fmoc-PEG-PAL-PS resin was used for synthesis, which affords the C-terminal amide derivatives. Standard Fmoc protection chemistry was used for coupling, which was promoted using standard activation protocols based on DIEA/HBTU and base-induced deprotection of the Fmoc group (20% piperidine in dimethylformamide). All peptides were acetylated at the N-terminus using 20% acetic anhydride. The crude peptides were purified using reverse phase high performance liquid chromatography (HPLC) on a C18 column using a water-acetonitrile (0.1% trifluoroacetic acid) gradient. The target HPLC fractions were collected and lyophilized. The purity of the peptides was analyzed using electrospray ionization mass spectrometry and analytical HPLC (see Supporting Figures S16–S23). Peptides 3FD- IL, 3FD-IL-EKEK, 3FD-IL-KEKE, and 3FD-IL-EEKK were dissolved in aqueous buffer (10 mM TAPS, pH 8.5) at an approximate concentrations of either 3 or 10 mg/mL. Thermal annealing of the peptide samples was performed using the following protocol in a programmable thermal cycler: (1) peptide solutions were initially heated to 90 °C for 10 min and (2) peptide solutions were cooled to a final temperature of 40 °C at a cooling rate of 0.2 °C/min. The resultant specimens were stored at ambient temperature prior to analysis.
Circular Dichroism Spectropolarimetry
CD measurements were performed on a Jasco J-810 CD spectropolarimeter in 0.10 mm quartz cells (Hellma Analytics) at a peptide concentration of 1 mg/mL in 10 mM TAPS buffer (pH 8.5). Spectra were recorded from 260 to 190 nm at a scanning rate of 50 nm/min and a resolution of 0.1 nm. CD melting experiments were performed in the temperature range from 5 to 95 °C at a heating rate of 40 °C/hour. The intensity of the CD signal at 222 nm was monitored as a function of temperature. Melting temperatures were obtained from the first derivative of the melting curves.
Transmission Electron Microscopy
TEM specimens were prepared from aqueous solutions of 3FD-IL (10 mg/mL) and the 3FD-IL-EKEK, 3FD-IL-KEKE, and 3FD-IL-EEKK variants (3 mg/ mL) in TAPS buffer (10 mM, pH 8.5) at the same concentration. The samples were deposited onto 200 mesh carbon-coated copper grids from Electron Microscopy Sciences (Hatfield, PA). After a 1 min incubation period, the samples were washed with ddH2O and stained with an aqueous solution of phosphotungstate stain (0.1%). Excess stain was wicked away from the grids after incubation on the grid for 30 s. TEM measurements were acquired on a JEOL JEM-1400 transmission electron microscope at an accelerating voltage of 120 kV.
Cryoelectron Microscopy
An aqueous solution of thermally annealed 3FD-IL nanosheets (10 mg/mL) in TAPS buffer (10 mM, pH 8.5) was diluted 1:10 with TAPS buffer and utilized for cryoelectron microscopy. The nanosheets were diluted to an approximate concentration of 1 mg/mL using TAPS buffer (10 mM, pH 8.5). Cryo-EM specimens were prepared by applying aliquots (4 µL) of the diluted solution of nanosheets to glow-discharged 200 mesh copper Quantifoil grids (Quantifoil Micro Tools GMBH; Groβlöbichau, Germany) and plunge freezing in liquid ethane using a Vitrobot Mark III (FEI, Hillsboro, Oregon). Cryo-EM data was collected using a JEOL JEM-2200FS 200 kV FEG-TEM with an in-column Omega energy filter (slit width 20 eV). Images were acquired on a Direct Electron, LP DE-20 direct electron detector (San Diego, CA) at a nominal magnification of 20 000× for a pixel size of 2.94 Å. Contrast was enhanced using a hole-free phase plate for phase imaging of cryoimmobilized specimens.
Atomic Force Microscopy
Specimens for AFM measurements were prepared from aqueous solutions of 3FD-IL nanosheets that had been previously assembled in TAPS buffer (10 mM, pH 8.5) at a peptide concentration of 10 mg/mL solutions. The nanosheet solution was diluted to a peptide concentration of 0.25 mg/mL using TAPS buffer (0.5 mM, pH 8.5) immediately before deposition onto a freshly cleaved mica substrate. The sample was spin-coated at a rate of 1500 rpm for 1 min. The AFM experiments were performed using a MFP-3D–BIO from Asylum Research. Silicon AFM tips (MikroMasch U.S.A.; Watsonville, CA) with a force constant (5.4–16 N/m) were used to image the specimens in tapping mode at a scan rate of 1 Hz with 256 points and lines.
Small- and Wide-Angle X-ray Scattering Measurements
Synchrotron SAXS/WAXS measurements were performed at the 12-ID-B beamline of Advanced Photon Sources at Argonne National Laboratory, using the methods described previously.3 Thermally annealed specimens of 3FD-IL nanosheets were assembled in TAPS buffer (10 mM, pH 8.5) at a peptide concentration of 10 mg/mL. The initial specimens were dialyzed against a solution of freshly prepared TAPS buffer (10 mM, pH 8.5) to remove residual trifluoroacetic acid. SAXS/WAXS data were acquired on the dialyzed solutions of peptide nanosheets in TAPS buffer (10 mM, pH 8.5) at 25 °C. A quartz capillary flow cell (1.5 mm diameter) was employed to prevent radiation damage. Twenty images were collected for each sample and buffer. The 2-D scattering images were converted to 1-D SAXS curves through azimuthally averaging after solid angle correction and then normalizing with the intensity of the transmitted X-ray beam, using the software package at beamline 12ID-B. The 1-D curves of the samples were averaged and subtracted with the background measured from the corresponding buffers.
Scanning Transmission Electron Microscopy
STEM data were acquired at Brookhaven National Laboratory (BNL). The STEM instrument operates at 40 keV with a scanning probe of <0.3 nm diameter produced from a cold field-emission source. Every electron emerging from the specimen is detected by one of the scintillator-photomultiplier detectors collecting 0- 15 mRadian (bright field), 15–40 mRadian (small-angle dark field) and 40–200 mRadian (large-angle dark field). The large-angle signal is proportional to the mass of atoms in the path of the beam. Specimen quality and mass calibration are checked by detailed comparison of the image to the known structure of tobacco mosaic virus (TMV). For mass-per-area (M/A) measurements, TMV rafts at a theoretical M/A value of 81.9 Da/Å2 were employed for calibration.
Thermally annealed specimens of 3FD-IL nanosheets were assembled in TAPS buffer (10 mM, pH 8.5) at a peptide concentration of 10 mg/mL. The initial specimens were diluted to a concentration of 3 mg/mL and dialyzed against a solution of freshly prepared TAPS buffer (10 mM, pH 8.5). Specimens are deposited on thin carbon (ca. 2 nm thick) supported on a thicker holey carbon film mounted on a titanium grid using the wet-film, hanging-drop method. TMV is added to the grid first as an internal control, followed by injection buffer, then specimen solution (in 20 mM MOPS buffer, pH 7.0) for 1 min, then 10 washes of 20 mM ammonium acetate pH 7.0. Excess solution is wicked from the edge with filter paper between each injection. After the last wash the grid is wicked to a thin layer (ca. 1 mm), fast frozen by plunging into liquid nitrogen slush and stored under liquid nitrogen. Grids are freeze-dried overnight in an ion-pumped chamber with an efficient cold trap and transferred under vacuum to the STEM cold stage (−160 °C). Imaging typically uses a dose of 20 el/Å2 (causing <5% mass loss, corrected by comparison to TMV). Mass measurements were performed off-line with the customized software PCMass56,57.
Supplementary Material
Acknowledgments
The electron microscopy data described here were gathered on either a JEOL JEM-2200FS 200 kV TEM (supported by a National Science Foundation Major Research Instrumentation Grant 0923395) or on a JEOL JEM-1400 120 kV TEM (supported by a National Institutes of Health Grant S10 RR025679). This study was supported in part by the Robert P. Apkarian Integrated Electron Microscopy Core (RPAIEMC), which is subsidized by the Emory College of Arts and Sciences and the Emory University School of Medicine and is one of the Emory Integrated Core Facilities. Additional support was provided by the National Center for Advancing Translational Sciences of the National Institutes of Health under award number UL1TR000454. The content is solely the responsibility of the authors and does not necessarily reflect the official views of the National Institutes of Health. This work benefited from the use of the A.P.S. funded by U.S. D.O.E. Office of Basic Energy Sciences, Division of Material Sciences, under contract W-31-109-Eng-38. We acknowledge the assistance of Dr. Anil Mehta for the construction of the molecular model of the honeycomb structure. V.P.C. acknowledges financial support from NSF grants CHE-1012620 and CHE-1412580. T.L. acknowledges the financial support from the National Science Foundation (CHE-1309817). E.R.W. acknowledges support from Emory University, Children’s Healthcare of Atlanta, the Georgia Research Alliance, the Center for AIDS Research at Emory University (P30 AI050409), the James B. Pendleton Charitable Trust, and NIH grant R01GM104540.
Footnotes
ASSOCIATED CONTENT
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.6b06592.
Additional experimental characterization of peptides and peptide assemblies (PDF)
The authors declare no competing financial interest.
References
- 1.Ariga K, Li J, Fei J, Ji Q, Hill JP. Adv. Mater. 2016;28:1251–1256. doi: 10.1002/adma.201502545. [DOI] [PubMed] [Google Scholar]
- 2.Govindaraju T, Avinash MB. Nanoscale. 2012;4:6102–6117. doi: 10.1039/c2nr31167d. [DOI] [PubMed] [Google Scholar]
- 3.(a) Jiang T, Xu C, Liu Y, Liu Z, Wall JS, Zuo X, Lian T, Salaita K, Ni C, Pochan D, Conticello VPJ. Am. Chem. Soc. 2015;136:4300–4308. doi: 10.1021/ja412867z. [DOI] [PubMed] [Google Scholar]; (b) Jiang T, Xu C, Zuo X, Conticello VP. Angew. Chem. Int. Ed. 2015;53:8367–8371. doi: 10.1002/anie.201403780. [DOI] [PubMed] [Google Scholar]; (c) Jiang T, Vail OA, Jiang Z, Zuo X, Conticello VPJ. Am. Chem. Soc. 2015;137:7793–7802. doi: 10.1021/jacs.5b03326. [DOI] [PubMed] [Google Scholar]
- 4.(a) Xu F, Khan IJ, McGuinness K, Parmar AS, Silva T, Murthy NS, Nanda VJ. Am. Chem. Soc. 2016;135:18762–18765. doi: 10.1021/ja4106545. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Parmar AS, James JK, Grisham DR, Pike DH, Nanda VJ. Am. Chem. Soc. 2016;138:4362–4367. doi: 10.1021/jacs.5b10304. [DOI] [PubMed] [Google Scholar]
- 5.(a) Nam KT, Shelby SA, Choi PH, Marciel AB, Chen R, Tan L, Chu TK, Mesch RA, Lee BC, Connolly MD, Kisielowski C, Zuckermann RN. Nat. Mater. 2014;9:454–460. doi: 10.1038/nmat2742. [DOI] [PubMed] [Google Scholar]; (b) Sanii B, Kudirka R, Cho A, Venkateswaran N, Olivier GK, Olson AM, Tran H, Harada RM, Tan L, Zuckermann RNJ. Am. Chem. Soc. 2014;133:20808–20815. doi: 10.1021/ja206199d. [DOI] [PubMed] [Google Scholar]; (c) Kudirka R, Tran H, Sanii B, Nam KT, Choi PH, Venkateswaran N, Chen R, Whitelam S, Zuckermann RN. Biopolymers. 2014;96:586–595. doi: 10.1002/bip.21590. [DOI] [PubMed] [Google Scholar]; (d) Robertson EJ, Olivier GK, Qian M, Proulx C, Zuckermann RN, Richmond GL. Proc. Natl. Acad. Sci. U. S. A. 2014;111:13284–13289. doi: 10.1073/pnas.1414843111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hamley IW, Dehsorkhi A, Castelletto V. Chem. Commun. 2013;49:1850–1852. doi: 10.1039/c3cc39057h. [DOI] [PubMed] [Google Scholar]
- 7.Dai B, Li D, Xi W, Luo F, Zhang X, Zou M, Cao M, Hu J, Wang W, Wei G, Zhang Y, Liu C. Proc. Natl. Acad. Sci. U. S. A. 2015;112:2996–3001. doi: 10.1073/pnas.1416690112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suzuki Y, Cardone G, Restrepo D, Zavattieri PD, Baker TS, Tezcan FA. Nature. 2016;533:369–373. doi: 10.1038/nature17633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.(a) Brodin JD, Ambroggio XI, Tang C, Parent KN, Baker TS, Tezcan FA. Nat. Chem. 2014;4:375–382. doi: 10.1038/nchem.1290. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Brodin JD, Carr JR, Sontz PA, Tezcan FA. Proc. Natl. Acad. Sci. U. S. A. 2014;111:2897–2902. doi: 10.1073/pnas.1319866111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.(a) Chothia C, Levitt M, Richardson DJ. Mol. Biol. 1998;145:215–250. doi: 10.1016/0022-2836(81)90341-7. [DOI] [PubMed] [Google Scholar]; (b) Bowie JU. Nat. Struct. Biol. 1998;4:915–917. doi: 10.1038/nsb1197-915. [DOI] [PubMed] [Google Scholar]; (c) Walther D, Springer C, Cohen FE. Proteins: Struct., Funct., Genet. 1998;33:457–459. [PubMed] [Google Scholar]
- 11.(a) Karle IL. Acta Crystallogr., Sect. B: Struct. Sci. 2008;48:341–356. doi: 10.1107/s0108768192000673. [DOI] [PubMed] [Google Scholar]; (b) Vasudev PG, Shamala N, Balaram PGJ. Phys. Chem. B. 2008;112:1308–1314. doi: 10.1021/jp077231d. [DOI] [PubMed] [Google Scholar]
- 12.(a) Prive GG, Anderson DH, Wesson L, Cascio D, Eisenberg D. Protein Sci. 1999;8:1400–1409. doi: 10.1110/ps.8.7.1400. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Patterson WR, Anderson DH, DeGrado WF, Cascio D, Eisenberg D. Protein Sci. 1999;8:1410–1422. doi: 10.1110/ps.8.7.1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taylor KS, Lou MZ, Chin TM, Yang NC, Garavito RM. Protein Sci. 1996;5:414–421. doi: 10.1002/pro.5560050302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.(a) Yu SM, Conticello VP, Zhang G, Kayser C, Fournier MJ, Mason TL, Tirrell DA. Nature. 2000;389:167–170. doi: 10.1038/38254. [DOI] [PubMed] [Google Scholar]; (b) Yu SM, Soto C, Tirrell DAJ. Am. Chem. Soc. 2000;122:6552–6559. [Google Scholar]
- 15.(a) Lupas AN, Gruber M. Adv. Protein Chem. 2012;70:37–78. doi: 10.1016/S0065-3233(05)70003-6. [DOI] [PubMed] [Google Scholar]; (b) Woolfson DN, Bartlett GJ, Bruning M, Thomson AR. Curr. Opin. Struct. Biol. 2012;22:432–441. doi: 10.1016/j.sbi.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 16.Fletcher JM, Boyle AL, Bruning M, Bartlett GJ, Vincent TL, Zaccai NR, Armstrong CT, Bromley EH, Booth PJ, Brady RL, Thomson AR, Woolfson DN. ACS Synth. Biol. 2012;1:240–250. doi: 10.1021/sb300028q. [DOI] [PubMed] [Google Scholar]
- 17.Zaccai NR, Chi B, Thomson AR, Boyle AL, Bartlett GJ, Bruning M, Linden N, Sessions RB, Booth PJ, Brady RL, Woolfson DN. Nat. Chem. Biol. 2011;7:935–941. doi: 10.1038/nchembio.692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thomson AR, Wood CW, Burton AJ, Bartlett GJ, Sessions RB, Brady RL, Woolfson DN. Science. 2014;346:485–488. doi: 10.1126/science.1257452. [DOI] [PubMed] [Google Scholar]
- 19.Huang PS, Oberdorfer G, Xu C, Pei XY, Nannenga BL, Rogers JM, DiMaio F, Gonen T, Luisi B, Baker D. Science. 2014;346:481–485. doi: 10.1126/science.1257481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lanci CJ, MacDermaid CM, Kang SG, Acharya R, North B, Yang X, Qiu XJ, DeGrado WF, Saven JG. Proc. Natl. Acad. Sci. U. S. A. 2012;109:7304–7309. doi: 10.1073/pnas.1112595109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ogihara NL, Weiss MS, Degrado WF, Eisenberg D. Protein Sci. 1997;6:80–88. doi: 10.1002/pro.5560060109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.(a) Papapostolou D, Smith AM, Atkins ED, Oliver SJ, Ryadnov MG, Serpell LC, Woolfson DN. Proc. Natl. Acad. Sci. U. S. A. 2012;104:10853–10858. doi: 10.1073/pnas.0700801104. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Sharp TH, Bruning M, Mantell J, Sessions RB, Thomson AR, Zaccai NR, Brady RL, Verkade P, Woolfson DN. Proc. Natl. Acad. Sci. U. S. A. 2012;109:13266–13271. doi: 10.1073/pnas.1118622109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burgess NC, Sharp TH, Thomas F, Wood CW, Thomson AR, Zaccai NR, Brady RL, Serpell LC, Woolfson DN. J. Am. Chem. Soc. 2015;137:10554–10562. doi: 10.1021/jacs.5b03973. [DOI] [PubMed] [Google Scholar]
- 24.Zhang HV, Polzer F, Haider MJ, Tian Y, Villegas JA, Kiick KL, Pochan DJ, Saven JG. Sci. Adv. 2016;2:e1600307. doi: 10.1126/sciadv.1600307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pauling L, Corey RB, Branson HR. Proc. Natl. Acad. Sci. U. S. A. 1951;37:205–211. doi: 10.1073/pnas.37.4.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hol WJG. Prog. Biophys. Mol. Biol. 1985;45:149–195. doi: 10.1016/0079-6107(85)90001-x. [DOI] [PubMed] [Google Scholar]
- 27.Harbury PB, Zhang T, Kim PS, Alber T. Science. 1993;262:1401–1407. doi: 10.1126/science.8248779. [DOI] [PubMed] [Google Scholar]
- 28.Lupas A. Trends Biochem. Sci. 1996;21:375–382. [PubMed] [Google Scholar]
- 29.Del Rizzo PA, Bi Y, Dunn SD. J. Mol. Biol. 2006;364:735–746. doi: 10.1016/j.jmb.2006.09.028. [DOI] [PubMed] [Google Scholar]
- 30.Del Rizzo PA, Bi Y, Dunn SD, Shilton BH. Biochemistry. 2002;41:6875–6884. doi: 10.1021/bi025736i. [DOI] [PubMed] [Google Scholar]
- 31.Grigoryan G, DeGrado WFJ. Mol. Biol. 2011;405:1079–1100. doi: 10.1016/j.jmb.2010.08.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Porod G. In: Small Angle X-ray Scattering. Glatter O, Kratky O, editors. Academic Press; New York: 1982. pp. 17–51. [Google Scholar]
- 33.Harbury PB, Tidor B, Kim PS. Proc. Natl. Acad. Sci. U. S. A. 1995;92:8408–8412. doi: 10.1073/pnas.92.18.8408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lotz B. Macromolecules. 2012;45:2175–2189. [Google Scholar]
- 35.Watanabe J, Ono H, Uematsu I, Abe A. Macromolecules. 1985;18:2141–2148. [Google Scholar]
- 36.Watanabe J, Takashina Y. Macromolecules. 1991;24:3423–3426. [Google Scholar]
- 37.Sasaki SJ. Polym. Sci. Part B: Polym. Phys. 1991;29:527–535. [Google Scholar]
- 38.Sasaki S, Uematsu IJ. J. Polym. Sci. Polym. Phys. Ed. 1985;23:263–273. [Google Scholar]
- 39.Toulouse G. Commun. Phys. 1977;3:115–119. [Google Scholar]
- 40.Peng Y, Li Y, Ban Y, Jin H, Jiao W, Liu X, Yang W. Science. 2014;346:1356–1359. doi: 10.1126/science.1254227. [DOI] [PubMed] [Google Scholar]
- 41.Bunck DN, Dichtel WRJ. Am. Chem. Soc. 2013;135:14952–14955. doi: 10.1021/ja408243n. [DOI] [PubMed] [Google Scholar]
- 42.(a) Zhang KD, Tian J, Hanifi D, Zhang Y, Sue AC, Zhou TY, Zhang L, Zhao X, Liu Y, Li ZTJ. Am. Chem. Soc. 2015;135:17913–17918. doi: 10.1021/ja4086935. [DOI] [PubMed] [Google Scholar]; (b) Pfeffermann M, Dong R, Graf R, Zajaczkowski W, Gorelik T, Pisula W, Narita A, Mullen K, Feng XJ. Am. Chem. Soc. 2015;137:14525–14532. doi: 10.1021/jacs.5b09638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mitra S, Kandambeth S, Biswal BP, Khayum MA, Choudhury CK, Mehta M, Kaur G, Banerjee S, Prabhune A, Verma S, Roy S, Kharul UK, Banerjee RJ. Am. Chem. Soc. 2016;138:2823–2828. doi: 10.1021/jacs.5b13533. [DOI] [PubMed] [Google Scholar]
- 44.Kambe T, Sakamoto R, Hoshiko K, Takada K, Miyachi M, Ryu J, Sasaki S, Kim J, Nakazato K, Takata M, Nishihara HJ. Am. Chem. Soc. 2013;135:2462–2465. doi: 10.1021/ja312380b. [DOI] [PubMed] [Google Scholar]
- 45.(a) Bauer T, Zheng Z, Renn A, Enning R, Stemmer A, Sakamoto J, Schluter AD. Angew. Chem., Int. Ed. 2015;50:7879–7884. doi: 10.1002/anie.201100669. [DOI] [PubMed] [Google Scholar]; (b) Zheng Z, Opilik L, Schiffmann F, Liu W, Bergamini G, Ceroni P, Lee L, Schutz A, Sakamoto J, Zenobi R, VandeVondele J, Schluter AD. J. Am. Chem. Soc. 2015;136:6103–6110. doi: 10.1021/ja501849y. [DOI] [PubMed] [Google Scholar]; (c) Sakamoto R, Hoshiko K, Liu Q, Yagi T, Nagayama T, Kusaka S, Tsuchiya M, Kitagawa Y, Wong W, Nishihara H. Nat. Commun. 2015;6:6713. doi: 10.1038/ncomms7713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.(a) Zheng Y, Zhou H, Liu D, Floudas G, Wagner M, Koynov K, Mezger M, Butt H, Ikeda T. Angew. Chem., Int. Ed. 2009;52:4845–4848. doi: 10.1002/anie.201210090. [DOI] [PubMed] [Google Scholar]; (b) Lee E, Kim J, Lee M. Angew. Chem., Int. Ed. 2009;48:3657–3660. doi: 10.1002/anie.200900079. [DOI] [PubMed] [Google Scholar]
- 47.Dong R, Pfeffermann M, Liang H, Zheng Z, Zhu X, Zhang J, Feng X. Angew. Chem. Int. Ed. 2015;54:12058–12063. doi: 10.1002/anie.201506048. [DOI] [PubMed] [Google Scholar]
- 48.Rothemund PWK. Nature. 2006;440:297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 49.Gradisar H, Bozic S, Doles T, Vengust D, Hafner-Bratkovic I, Mertelj A, Webb B, Sali A, Klavzar S, Jerala R. Nat. Chem. Biol. 2013;9:362–366. doi: 10.1038/nchembio.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.(a) King NP, Sheffler W, Sawaya MR, Vollmar BS, Sumida JP, Andre I, Gonen T, Yeates TO, Baker D. Science. 2015;336:1171–1174. doi: 10.1126/science.1219364. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) King NP, Bale JB, Sheffler W, McNamara DE, Gonen S, Gonen T, Yeates TO, Baker D. Nature. 2015;510:103–108. doi: 10.1038/nature13404. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Bale JB, Park RU, Liu Y, Gonen S, Gonen T, Cascio D, King NP, Yeates TO, Baker D. Protein Sci. 2015;24:1695–1701. doi: 10.1002/pro.2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fletcher JM, Harniman RL, Barnes FR, Boyle AL, Collins A, Mantell J, Sharp TH, Antognozzi M, Booth PJ, Linden N, Miles MJ, Sessions RB, Verkade P, Woolfson DN. Science. 2013;340:595–599. doi: 10.1126/science.1233936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Magnotti E, Conticello V. Adv. Exp. Med. Biol. 2016;940:29–60. doi: 10.1007/978-3-319-39196-0_3. [DOI] [PubMed] [Google Scholar]
- 53.(a) Hospenthal MK, Redzej A, Dodson K, Ukleja M, Frenz B, Rodrigues C, Hultgren SJ, DiMaio F, Egelman EH, Waksman G. Cell. 2015;164:269–278. doi: 10.1016/j.cell.2015.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Sborgi L, Ravotti F, Dandey VP, Dick MS, Mazur A, Reckel S, Chami M, Scherer S, Huber M, Bockmann A, Egelman EH, Stahlberg H, Broz P, Meier BH, Hiller S. Proc. Natl. Acad. Sci. U. S. A. 2015;112:13237–13242. doi: 10.1073/pnas.1507579112. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Kudryashev M, Wang RY, Brackmann M, Scherer S, Maier T, Baker D, DiMaio F, Stahlberg H, Egelman EH, Basler M. Cell. 2015;160:952–962. doi: 10.1016/j.cell.2015.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.(a) Egelman EH, Xu C, DiMaio F, Magnotti E, Modlin C, Yu X, Wright E, Baker D, Conticello VP. Structure. 2015;23:280–289. doi: 10.1016/j.str.2014.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Wang RY, Kudryashev M, Li X, Egelman EH, Basler M, Cheng Y, Baker D, DiMaio F. Nat. Methods. 2015;12:335–338. doi: 10.1038/nmeth.3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stahlberg H, Biyani N, Engel A. Arch. Biochem. Biophys. 2015;581:68–77. doi: 10.1016/j.abb.2015.06.006. [DOI] [PubMed] [Google Scholar]
- 56.Wall JS, Simon MN. Methods Mol. Biol. 2001;148:589–601. doi: 10.1385/1-59259-208-2:589. [DOI] [PubMed] [Google Scholar]
- 57.PCMass is available at ftp.stem.bnl.gov
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.