ABSTRACT
While research on T cell exhaustion in context of cancer particularly focuses on CD8+ cytotoxic T cells, the role of inhibitory receptors on CD4+ T-helper cells have remained largely unexplored. TIGIT is a recently identified inhibitory receptor on T cells and natural killer (NK) cells. In this study, we examined TIGIT expression on T cell subsets from CLL patients. While we did not observe any differences in TIGIT expression in CD8+ T cells of healthy controls and CLL cells, we found an enrichment of TIGIT+ T cells in the CD4+ T cell compartment in CLL. Intriguingly, CLL patients with an advanced disease stage displayed elevated numbers of CD4+ TIGIT+ T cells compared to low risk patients. Autologous CLL-T cell co-culture assays revealed that depleting CD4+ TIGIT+ expressing T cells from co-cultures significantly decreased CLL viability. Accordingly, a supportive effect of TIGIT+CD4+ T cells on CLL cells in vitro could be recapitulated by blocking the interaction of TIGIT with its ligands using TIGIT-Fc molecules, which also impeded the T cell specific production of CLL-prosurvival cytokines. Our data reveal that TIGIT+CD4+T cells provide a supportive microenvironment for CLL cells, representing a potential therapeutic target for CLL treatment.
KEYWORDS: chronic lymphocytic leukemia, T cell exhaustion, TIGIT, PD-1, microenvironment
Introduction
Chronic lymphocytic leukemia (CLL) is a B cell malignancy, which is associated with substantial T subset skewing and T cell defects.1 In particular, we and others have previously shown that T cells from CLL patients show typical signs of T cell exhaustion, as they exhibit increased expression of inhibitory receptors and defects in proliferation, cytokine expression and synapse formation.2-5 In general, T cell exhaustion is the functional silencing of antigen experienced T cells, contributing to peripheral T cell tolerance to avoid excessive immune pathology during T cell responses,6 a process initially described during chronic viral infections7. However, T cell exhaustion was also found to occur in cancer and is supposed to significantly contribute to immune evasion of cancer cells by rendering cancer specific T cells non-functional.8 A key molecule in T cell exhaustion was found to be programmed death-1 (PD-1), which functionally impedes T cell receptor (TCR) mediated signaling on exhausted T cells.9 In addition to PD-1, a number of different inhibitory receptors were recently found to be associated with T cell exhaustion. The concept that the exhausted phenotype could be reversed by simply blocking these inhibitory receptors by monoclonal antibodies led to a renaissance of cancer immune therapy with specific immune checkpoint blockade using PD-1 antibodies being considered as a major breakthrough in cancer treatment.10 Considering the strength of checkpoint blockade, it is important to investigate further inhibitory receptors aside of PD-1 to increase efficacy and minimize side effects of this treatment approach, especially in light of the fact that not all cancer entities respond to PD-1 blockade. For CLL, no clear effects were noticed for single agent anti-PD-1 therapy (pembrolizumab) except for patients with Richter's syndrome, whereas at least partial responses were reported for CLL patients receiving anti-PD-1 therapy (nivolumab) combined with ibrutinib.11,12
TIGIT (T cell immunoreceptor with Ig and ITIM domains) is a recently identified inhibitory receptor which is expressed on T cells, natural killer (NK) and NKT cells.13,14 TIGIT has a cytoplasmic tail containing an immunoglobulin tail tyrosine (ITT)- like phosphorylation motif and an immunoreceptor tyrosine-based inhibitory motif (ITIM)(15). The natural ligands for TIGIT are the poliovirus receptors (PVR) CD155 and CD112. TIGIT signaling involves the recruitment of the phosphatase SHIP1 to the ITIM and downstream inhibition of NF-kB, PI3 K and MAPK pathways.16,17 PVRs are widely expressed on different cell types and were also found to be expressed on a number of cancer cells.18-21 In parallel, TIGIT+ tumor infiltrating CD8+ T cells could be detected in small-cell lung cancers, colorectal cancers and melanoma.22-24 Similar to the situation of CTLA-4 and CD28, which are sharing the same ligands25, TIGIT competes with CD226, a costimulatory T cell molecule for PVR binding and can directly prevent CD226 signaling by impeding its homodimerization.22 Hence, agonistic TIGIT antibodies could decrease T cell function similar to CD226 knockdown26,27 and TIGIT inhibition was recently shown to increase T cell functions of melanoma specific CD8+T cells.28
In this study, we investigated expression and function of TIGIT expressing T cells in CLL patients. We observed an increase in TIGIT expressing CD4+T cells in CLL compared to healthy controls and we provide evidence that TIGIT+CD4+T cells display a microenvironment important for CLL survival. Our data propose TIGIT to be a potential target for immune therapy in CLL.
Results
TIGIT expressing CD4+ T cells are elevated in patients with CLL
To evaluate TIGIT expression in comparison to other inhibitory receptors (PD-1 and 2B4/CD244) which we previously examined in CLL,2 we performed phenotypic characterization of T cells from peripheral blood samples of CLL patients and age-matched healthy controls using flow cytometry (Fig 1a and b). In line with our previous results,2 we found that the percentages of CD4+ or CD8+ T cells expressing 2B4 or PD-1 were not significantly different between patients and healthy controls (Fig 1b). However, within the CD4+ but not the CD8+ T cell subset, we observed a significant increase in the percentage of TIGIT+ cells in CLL patients compared to healthy controls (Fig 1b). Most of the TIGIT+CD4+T cells were also positive for PD-1 (Fig 1c), reflected in a high correlation of PD-1 and TIGIT expression in this T cell subset (Fig 1d). The percentage of TIGIT+CD4+ T, PD-1+CD4+ and PD-1+CD8+T cells was higher in bone marrow compared to peripheral blood of the same patient and we observed a high correlation of TIGIT, PD-1 and 2B4 expression in peripheral blood and bone marrow (supplementary figure S1). Notably, the inhibitory receptor TIM-3 was not detectable on T cells from peripheral blood of CLL patients (supplementary figure S2). Moreover, while TIGIT expression on CD8+T cells did not correlate with advanced disease stage or prior treatment, we found significantly increased TIGIT+CD4+T cells in patients with unmutated IgVH status, with RAI stage ≥1/≥ Binet B and in treated patients. No correlation with other prognostic factors (Zap70, CD38, CD49 d expression or chromosomal aberrations) was observed (Fig 1e and supplementary figure S3).
Figure 1.
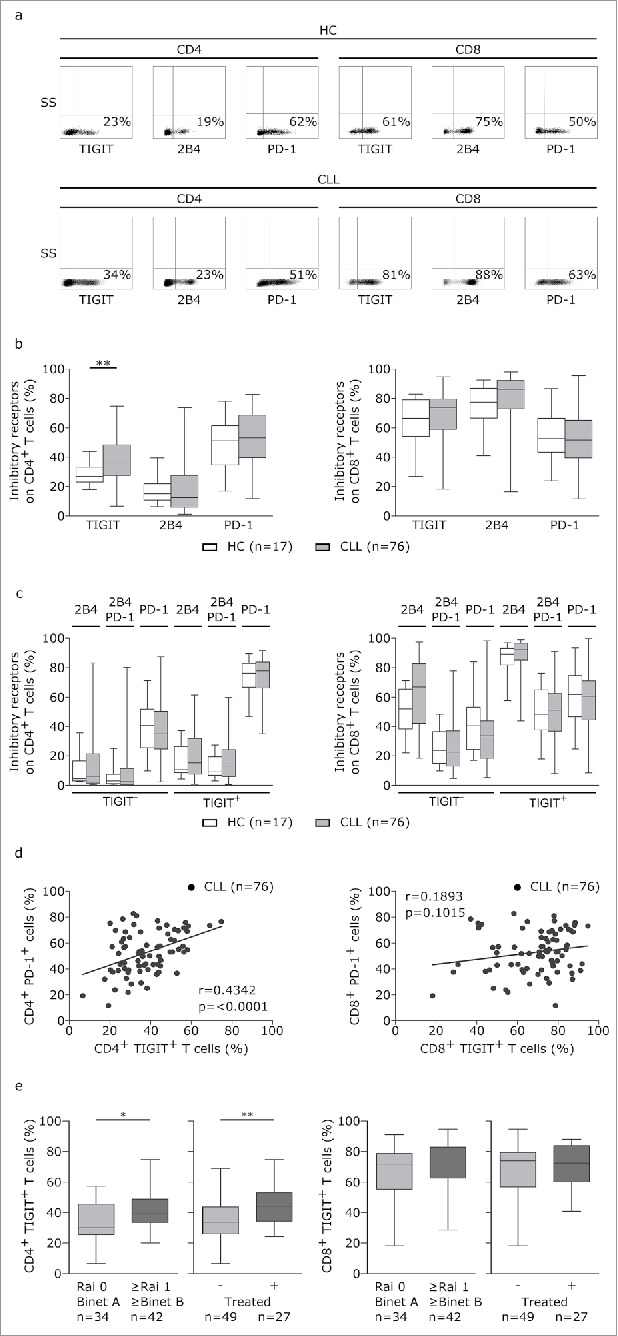
T cells from CLL patients display elevated TIGIT expression. (a-b) Peripheral blood samples from CLL patients (CLL) and age-matched healthy controls (HC) were analyzed by flow cytometry (FACS) with regard to TIGIT, 2B4 and PD-1 expression on CD4 or CD8 T cells. (c) Distribution of inhibitory receptors on TIGIT- or TIGIT+ T cells. (d) Correlation of TIGIT and PD-1 expression on CD4+ or CD8+ T cells. (e) Distribution of CD4+TIGIT+ cells in patients divided according to clinical markers of disease burden (Rai/ Binet stage or treatment status).
TIGIT is preferentially expressed on antigen experienced and Th1 polarized cells in CLL
To more thoroughly evaluate TIGIT expression on T cells, we analysed expression of TIGIT in combination with T cell subset defining markers. In line with previous reports, we observed a general shift towards effector memory T cell populations as defined by CD62 L/CD45RA expression in CLL (Fig 2a). By analyzing naïve (CD62 L+CD45RA+), central memory (TCM:CD62 L+CD45RA-), effector memory (TEM: CD62 L-CD45RA-), and terminally differentiated effector memory (TEMRA:CD62 L-CD45RA+) CD4+ and CD8+T cells with respect to their expression of TIGIT, we found that within the TIGIT+CD4+T cells, TEMRA and naïve cells were significantly enriched whereas the percentage of TCM cells was decreased in CLL (Fig 2b). In TIGIT+CD8+T cells, TEMRA subsets were expanded (Fig 2b). Counting the absolute number of cells within the respective T cell subsets revealed that TIGIT+CD4+T cells were enriched within the naïve, TEM and TEMRA subsets and CD8+TIGIT+T cells within the TEM subset in CLL (Fig 2c). We further characterized Th1 (CD45RA-CXCR3+CCR4-), Th2 (CD45RA-CCR3-CCR4+), regulatory T (Treg; CD127-CD25+) and follicular helper T (Tfh; CXCR5+PD-1+) cells and found a general skewing towards Th1, Treg and Tfh subsets in CLL compared to healthy controls (Fig 3a). While we observed a significant increase in the percentage of TIGIT+Th1 cells in peripheral blood from CLL patients compared to healthy controls, the absolute cell numbers of TIGIT+Th1, TIGIT+Treg as well as of TIGIT+Tfh cells were all significantly increased in peripheral blood from CLL patients compared to healthy controls (Fig 3b and c), with no apparent correlation with tumor load or treatment status (supplementary figure S4).In addition, we found that in CLL, a significantly smaller percentage of TIGIT+CD4+T cells expressed CD226, a costimulatory molecule competing with TIGIT for the same ligands (Fig 3d). Further analysis of the CD8 compartment revealed decreased percentage of CD226 expressing cells among TIGIT+T cells compared to TIGIT- CD8+T cells (Fig 3d) and a negative correlation of CD226 with TIGIT expression (Fig 3e).
Figure 2.
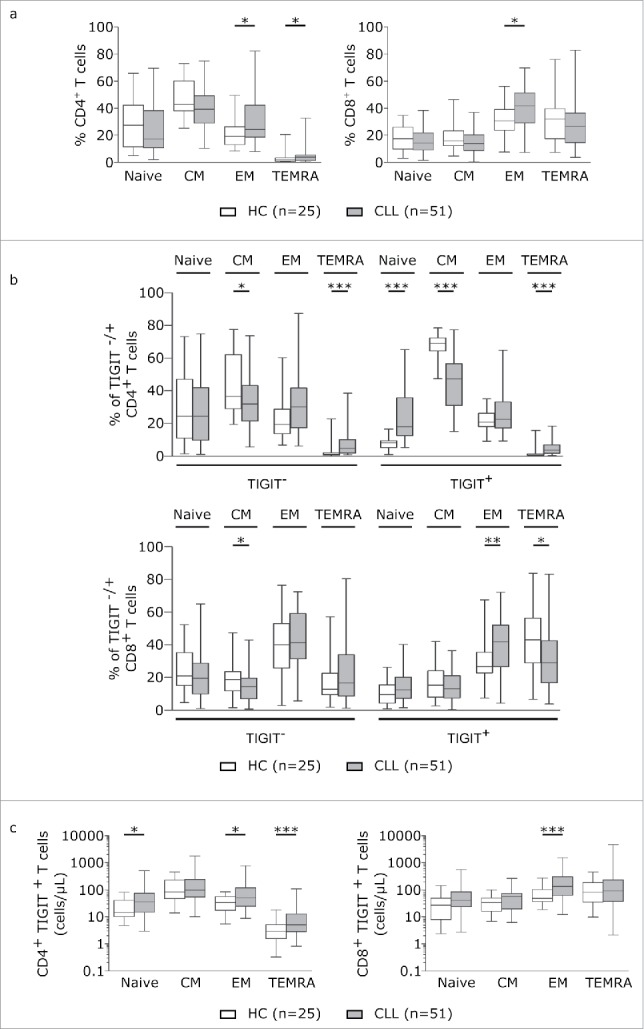
TIGIT is particularly expressed on antigen experienced T cells. (a) T cell subsets in peripheral blood samples from CLL patients and age-matched healthy controls (HC) were measured by FACS analysis defined by CD62 L and CD45RA. Plots represent naïve (Tnaive: CD62 L+CD45RA+), central memory (TCM: CD62 L+CD45RA-), effector memory (TEM: CD62 L-CD45RA-) and terminally differentiated effector memory (TEMRA: CD62 L-CD45RA+). (b) T cell subset distribution in the TIGIT- and TIGIT+ T cell compartment. (c) Absolute cell counts (cells/µL blood) of CD4+ and CD8+ subsets expressing TIGIT.
Figure 3.

Absolute cell counts of TIGIT+Th1, TIGIT+Treg and TIGIT+Tfh cells are significantly increased in CLL. (a) Plot of percentages of Th1, Th2, Treg and Tfh among CD4+ T cells from controls and CLL patients. (b) Percentage of TIGIT expressing Th1, Th2, Treg and Tfh cells. (c) Absolute cell counts (cells/µL). (d) Percentage of CD226+ cells among TIGIT- versus TIGIT+ CD4+ or CD8+ cells in HCs or CLL patients.(e) Correlation of TIGIT and CD226 expression on CD4+ or CD8+ T cells.
TIGIT+CD4+ but not TIGIT+CD8+ T cells affect in vitro survival of autologous CLL cells
Next, we wanted to assess CLL prosurvival or proapoptotic capacities of TIGIT+ CD4+ and CD8+ T cells. To this end, we performed autologous CLL/T cell co-culture assays using CD3/CD28 activated T cells which were either depleted of TIGIT or PD-1 expressing T cells using flow cytometric cell sorting (as outlined in the methods section; Fig 4a). Thereby we found that absence of TIGIT+ cells from all T cells in our co-culture setting resulted in significant decrease in the percentage of viable CLL cells (Fig 4b). While absence of PD-1+ T cells had no significant effect on CLL cells, absence of both PD-1+ and TIGIT+ T cells again resulted in decreased CLL viability (Fig 4b). To elucidate whether the prosurvival impact of TIGIT+ T cells depends on CD4+ or CD8+ T cells, we further depleted T cells from CD4+TIGIT+ or CD8+TIGIT+ cells prior to co-culture with CLL cells (Fig 4a). Thereby we observed that only absence of TIGIT+CD4+ but not TIGIT+CD8+T cells decreased CLL cell survival (Fig 4b). In these assays, also absence of PD-1+CD4+ T cells resulted in decreased CLL cell survival (Fig 4b). Of note, this effect on CLL survival in these co-culture assays was not based on reduced overall numbers of T cells, as the T/CLL cell ratio was not significantly different in the respective assays (Fig 4c).
Figure 4.

CD4+ TIGIT+ cells provide a supportive microenvironment for CLL cells. (a) Representative dot plots showing gating strategy for flow cytometric cell sorting. (b) PBMCs have been depleted of TIGIT+, PD-1+ or TIGIT+PD-1+ CD4+ or CD8+ cells followed by incubation with CD3/CD28 activating beads. After 5 days in culture, CLL viability was measured and corresponding T/ CLL ratios were analysed (n = 6) (c).
Blocking TIGIT interactions decreases CLL viability and interferes with production of prosurvival cytokines
To further examine the CLL-supportive function of TIGIT+CD4+T cells, we analyzed the cytokine expression profile of TIGIT+ and TIGIT- CD4+T cells using intracellular cytokine staining (Fig 5a). We observed a significantly higher percentage of IFNγ and IL-10 producing CD4+T cells within the TIGIT+ population while the percentage of IL-21 and IL-4 producing cells was comparable in TIGIT+ and TIGIT- subpopulations (Fig 5b). In addition, low but measurable expression levels of the ligands for TIGIT (CD112 and CD155) could be detected on the surface of primary CLL samples as well as on T cells (Fig 5c).
Figure 5.
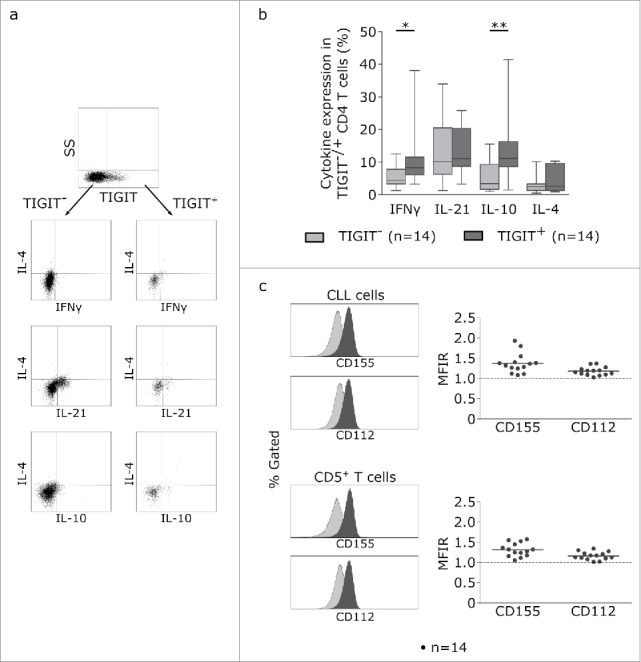
TIGIT+ cells display a distinct cytokine profile. (a) Representative dot plots showing intracellular cytokine production after cultivating CLL PBMCs for 24 h with CD3/CD28 activating beads. (b) Cytokine production of TIGIT- or TIGIT+CD4+ T cells in 14 samples. (c) Mean fluorescence intensity ratio (MFIR) of CD155 and CD112 on CD5+CD19+ CLL (top) or CD5+ T cells (bottom). The histograms show representative FACS plots of CLL cells (gated for CD5+CD19+cells) and T cells (CD5+ cells) stained with isotype controls (in gray) and CD112/CD155 specific antibodies (in black). The dot plots show results from n = 14 samples.
We next analyzed whether cytokine production could be modulated by blocking TIGIT/ligand interactions using recombinant TIGIT-Fc molecules. As shown in Fig 6a, presence of TIGIT-Fc resulted in impaired IFNγ and IL-10 production in T cells (Fig 6a). Notably, this effect was dependent on the presence of CLL cells, as in T cell solo cultures, cytokine production was not significantly affected by addition of TIGIT-Fc (Fig 6b).
Figure 6.

TIGIT blockade decreases CLL viability and interferes with production of prosurvival cytokines. (a, b) Impact of TIGIT blockade on cytokine production by CD4+ T cells. PBMCs (upper panel; n = 12) or purified T cells (bottom panel; n = 10) were activated with CD3/CD28 beads for 24 h in the presence of recombinant TIGIT-Fc protein (rhTIGIT Fc) or corresponding isotype control and cytokines were quantifiued by intracellular FACS staining. (b) FACS analysis of surface expression of TIGIT was performed on peripheral blood samples from CLL patients (n = 12). Plot of percentages of CD4+TIGIT+ T cells are shown, discriminating between TIGITlow (<52.6% CD4+TIGIT+cells) and TIGIThigh (>52.6% CD4+TIGIT+cells). (c) Plot represents difference in CLL viability between TIGIT-Fc or isotype control treated samples. Viability was determined by 7AAD/Annexin V staining after stimulating T/CLL co-cultures from TIGITlow (left) and TIGIThigh (right) patients shown in (b) using anti-CD3/CD28 beads for 5 days in the presence of recombinant TIGIT-Fc or isotype control.
Finally, we tested whether the CLL-supportive function of TIGIT+T cells could also be recapitulated by blocking TIGIT/ligand interactions using recombinant TIGIT-Fc molecules. Therefore, we assessed CLL viability in co-culture assays in response to 5 day anti-CD3/CD28 stimulation in presence or absence of recombinant TIGIT-Fc. In experiments on 12 CLL samples, we did not observe a significant impact on CLL survival, however, dividing samples in high and low expressing TIGIT+CD4+T cells based on ROC curve analysis using a cutoff of 52.6% TIGIT+cells within the CD4+T cell subset (Fig 6c), we found a significant impact on CLL viability in the TIGIThigh sample group (Fig 6c).
Discussion
In CLL, many alterations within the T cell compartment, such as skewing towards effector and Treg subsets,31 abnormal CD4/CD8 ratios,32 increased sensitivity of CD4+ T cells towards Fas-mediated apoptosis33 and the occurrence of expanded monoclonal CD4+ T cell populations have been observed.34,35 Also, an aberrant expression profile of inhibitory receptors associated with T cell exhaustion has been noticed in CD4+ and CD8+ T cells from CLL patients2 and within the CD8+ T cells, the expansion of PD-1 expressing cells featuring defects in proliferation and cytotoxicity have been described.2,3 Furthermore, in contrast to many aggressive tumors, CLL cells are strongly depending on the microenvironment for proliferation and survival1. In this regard, T cells were previously shown to execute a substantial CLL-supportive function.29,36 Hence, a more precise characterization of the CLL supportive T cell subset would allow to specifically target this CLL/T cell interaction, which would not only directly affect CLL viability but likely also render CLL cells more vulnerable to conventional treatment options. In this study, we found that particularly CD4+ T cells expressing the inhibitory receptor TIGIT were not only increased in patients with an advanced disease stage but also potent in increasing viability of autologous CLL cells in in vitro co-culture assays. Moreover, we could show that this supportive impact on CLL viability of TIGIT expressing T cells is dependent on TIGIT/ligand interactions, as this effect was targetable by recombinant TIGIT-Fc molecules. In line with this, we observed low but measurable levels of CD112 and CD155 on the surface of CLL cells, the ligands for TIGIT. Moreover, we demonstrated that IFNγ and IL-10 producing T cells were enriched in the TIGIT+ compartment and blocking TIGIT/ligand interactions suppressed the production of both IL-10 and IFNγ. This effect was dependent on the presence of CLL cells, further supporting a direct TIGIT mediated CLL/T cell crosstalk, consistent with recent observations that expression of these cytokines in T cells was increased upon TIGIT dependent interaction with antigen-presenting cells.13,37 The prosurvival effect of TIGIT+ T cells could well be based on secretion of these cytokines. Alhough the proinflammatory cytokine IFNγ has been associated with cytotoxic and antitumor mechanisms,38 IFNγ can also exert protumorigenic effects. IFNγ attenuates programmed cell death in CLL cells in vitro,39 and CLL T cells produce increased levels of IFNγ, while CLL cells show high expression of IFNγ-receptors.40 These data support an ambiguous role for IFNγ, which depends on the cellular, microenvironmental and molecular context.38
Recently, Joller et al. reported that TIGIT+ Tregs exhibit superior immunosuppressive functions compared to the TIGIT- counterpart.41 In this study, we observed a general increase of Tregs and we found that more Tregs were expressing TIGIT in CLL patients compared to healthy controls. This fits to the substantial IL-10 production in TIGIT+ CD4+ T cells from CLL patients as IL-10 not only dampens anti-tumor responses in vivo, but also has a direct protective and prosurvival effect on CLL cells.42 In line with our data, it has been recently reported that a subset of TIGIT+ circulating Tfh cells have a strong B cell-supportive effect due to protective cytokine secretion and high expression of costimulatory molecules.43 Additionally, the occurence of Tfh cells is significantly increased in CLL, particularly in high risk patients.44
Our results are of particular interest in light of recent approaches for anticancer immune therapies aiming at blocking TIGIT to reinvigorate anticancer immune responses (NCT02794571, NCT02913313; clinicaltrials.gov). Similar to therapeutic antibodies for CTLA-4 (ipilimumab) and PD-1 (nivolumab and pembrolizumab) or its ligand PD-L1 (atezolizumab, durvalumab), which have already been approved for multiple hematological malignancies, antibodies blocking TIGIT were developed to impede inhibitory signals on cytotoxic T cells in order to regain anti-tumor immunity.
In this regard, our own results suggest that anti-TIGIT antibodies could specifically target the CLL-supportive TIGIT+CD4+T cell compartment, defining an alternative rationale for using these antibodies especially in combination with other treatment options.
Materials and methods
Patients
Peripheral blood samples were obtained from patients with confirmed diagnosis of CLL (Department of Haematology and Oncology, University Hospital Salzburg, Austria) on basis of >5000 light chain restricted B cells and CD5, CD19, CD20low and CD79blow co-expression per µl blood and twenty-nine age-matched healthy control samples (Stroke Prevention Center, Department of Neurology). Monoclonal antibodies for flow cytometric analysis were provided by Beckman coulter: CD5-PC7 (A21690), CD19-APC- Alexa Fluor 750 (A94681), CD23-APC (A69964), CD20-Pacific Blue (B49208)and CD79b- PE (IM1612). Of 114 measured patients, 36 received treatment and 78 were chemonaïve. Analysis of the mutational status of immunoglobulin heavy chain variable region (IgVH), expression of CD38, ZAP70, CD49d and assessment of genomic aberrations (Tri12, delChr11q, delChr17p and delChr13q) was performed as described previously(2). Overall 73.7% of patients showed genomic aberrations. For detailed patient information and treatment status see supplementary table 1 (samples used for respective experiments are given on individual excel spreadsheets within supplementary table 1). Full informed consent was obtained from all CLL patients and healthy volunteers and the study was conducted in accordance with institutional Guidelines and the Declaration of Helsinki and under approval of the Salzburg Ethics committee (no. 415-E/1287/4–2011 and 415-E/1287/13–2016). Peripheral blood mononuclear cells (PBMCs) were collected in heparinized or EDTA-coated tubes during routine examinations and separated by density centrifugation using Biocoll (Biochrom AG).
Immunofluorescence staining and flow cytometric analysis
PBMCs were separated by density centrifugation of fresh CLL blood samples. PBMCs were incubated with directly conjugated mAbs for 20 minutes at room temperature. Flow cytometric analysis was performed after erythrolysis with IOTest® 3 Lysing Solution (Beckman Coulter, IM3514) and washing steps. Monoclonal antibodies for analysis were provided by Biolegend: anti-human PD-1-Brilliant Violet 421 (329920),CXCR3-APC (353708), CXCR5-PerCP-Cy5.5 (356910), CCR4-PE-Cy7 (359410), CD62L-PerCP/Cy5.5 (304824) and CD226-PerCP/Cy5.5 (338314). Monoclonal antibodies provided by ThermoFisher included TIGIT-PE (12–9500-42), CD4-PE-Cyanine7 (25–0048-42), CD4-eFluor450 (48-0048-42), CD4-APC (MHCD0405), 2B4-APC (17-5837-42), CD3-Alexa Fluor 700 (56-0032-82), CD3-FITC (11-0039-42), CD45RA-APC-eFluor780 (47-0458-42), CD25-PE-Cyanine7 (25-0259-42), CD127-APC-eFluor780 (47-1271-82), CD155-PE (12-1550-41), CD112-APC (17-1128-42), CD19-FITC (11-0199-42), rhAnnexin V-FITC (BMS147FI), 7-AAD Viability Staining Solution (00-6993-50), TIM-3-PerCP-eFluor 710 (46-3109-42) and CD8-Pacific Orange (MCD0830). For viability assays, rhAnnexin V/FITC, 7-AAD Viability Staining Solution was used. Data acquisition was performed on Gallios™ Flow Cytometer research system (Beckman Coulter) and data analysis was performed using Kaluza 1.2 Flow Cytometry Analysis Software (Beckman Coulter).
Intracellular staining of cytokines
PBMCs were activated for 24 hours with CD3/CD28 coated beads (ThermoFisher, 111.31D) with or without recombinant human TIGIT Fc chimera protein, or the respective recombinant human IgG1 Fc control (100 µg/mL, R&D Systems, 7898-TG-050 and 110-HG). Monensin (ThermoFisher, 00-4505-51) was added during the final four hours of culture. Intracellular cytokines were stained with anti-human IFNγ-FITC (ThermoFisher,11-7319-82), IL-21-PE (ThermoFisher, 12-7219-42), IL-10-PerCP-eFluor710 (ThermoFisher, 46-7108-42) and IL-4-APC (ThermoFisher, 17-7049-42). For surface marker expression TIGIT-PE-Cyanine7 (ThermoFisher, 25-9500-42), CD3-AF700 (ThermoFisher, 56-0038-42) and CD4-Brilliant Violet 650 (Biolegend, 300536) were used.
Depletion of T cells for in vitro co-culture experiments
The impact of specific T cells on the viability of autologous CLL cells was assessed in in vitro experiments. To deplete PBMCs from TIGIT+ and/or PD1+ cells, we stained PBMCs using TIGIT or PD1 antibodies and depleted positive cells by flow cytometric cell sorting (FACS Aria III, Becton Dickinson). To deplete PBMCs from all CD8+ T cells together with TIGIT+ or PD-1+ cells (to keep CD4+TIGIT- or PD-1- T cells in co-culture with CLL cells) we stained PBMCs using TIGIT or PD-1 and CD8 antibodies and depleted positive cells by flow cytometric cell sorting. To deplete PBMCs from all CD4+ T cells together with TIGIT+ or PD-1+ cells (to keep CD8+TIGIT- or PD-1- T cells in co-culture with CLL cells) we stained PBMCs using TIGIT and CD4 antibodies and depleted positive cells by flow cytometric cell sorting Purity of sorted cells was >92% for all experiments. Similar to Chauvin et al.,24 sorted PBMCs were stimulated in vitro with anti-CD3/CD28 coated beads (ThermoFisher). After five days CLL and T cells were stained with rhAnnexin V-FITC and 7-AAD Viability Staining Solution (ThermoFisher) and analyzed on a Gallios™ Flow Cytometer research system.
Blockade of TIGIT/PVRp
PBMCs were activated for five days with CD3/CD28 coated beads (ThermoFisher) in presence of recombinant human TIGIT Fc chimera protein or recombinant human IgG1 Fc control (100 µg/mL, R&D System). Antibodies used for viability stain were anti-CD5-AF700 (Beckman Coulter, A78835), CD19-PE, rhAnnexin V-FITC and 7-AAD Viability Staining Solution (ThermoFisher).
Statistical analysis
Statistical analyses were performed using Graph Pad Prism Version 5.02 (GraphPad Software, Inc.). Data are presented either as dot plots or as box plots showing 25th and 75th percentile and median inside the box. Whiskers represent minimum to maximum of all data.
Correlations were performed using the parametric pearson or nonparameric spearman test
Data was compared depending on Gaussian distribution (unpaired t test/ paired t test or Mann-Whitney test/ Wilcoxon matched pairs test). P values of less than 0.05 were considered statistically significant (*p < 0.05, **p < 0.01, ***p < 0.001).
Supplementary Material
Conflict of interest
The authors declare no conflict of interest.
Acknowledgments
The authors thank the stroke prevention center of the Christian-Doppler-Klinik Salzburg and the team of the Paracelsus 10,000 study for providing blood samples from healthy volunteers upon informed consent. This work was supported by the SCRI-LIMCR, the Province of Salzburg, the City of Salzburg, and grants from the Austrian Science Fund FWF (Projects P24100 to R.Gr., P28201 to R.Ge., I 2795-B28 to A.E., P25015 to T.N.H. and T 516-B13 to N.Z.).
References
- 1.Pleyer L, Egle A, Hartmann TN, Greil R. Molecular and cellular mechanisms of CLL: novel therapeutic approaches. Nat Rev Clin Oncol. 2009;6(7):405-18. doi: 10.1038/nrclinonc.2009.72. PMID:19488076 [DOI] [PubMed] [Google Scholar]
- 2.Gassner FJ, Zaborsky N, Neureiter D, Huemer M, Melchardt T, Egle A, Rebhandl S, Catakovic K, Hartmann TN, Greil R. Chemotherapy-induced augmentation of T cells expressing inhibitory receptors is reversed by treatment with lenalidomide in chronic lymphocytic leukemia. Haematologica. 2014;99(5):67-9. doi: 10.3324/haematol.2013.098459. PMID:24561794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Riches JC, Davies JK, McClanahan F, Fatah R, Iqbal S, Agrawal S, Ramsay AG, Gribben JG. T cells from CLL patients exhibit features of T-cell exhaustion but retain capacity for cytokine production. Blood. 2013;121(9):1612-21. doi: 10.1182/blood-2012-09-457531. PMID:23247726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramsay AG, Johnson AJ, Lee AM, Gorgun G, Le Dieu R, Blum W, Byrd JC, Gribben JG. Chronic lymphocytic leukemia T cells show impaired immunological synapse formation that can be reversed with an immunomodulating drug. J Clin Invest. 2008;118(7):2427-37. PMID:18551193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramsay AG, Clear AJ, Fatah R, Gribben JG. Multiple inhibitory ligands induce impaired T-cell immunologic synapse function in chronic lymphocytic leukemia that can be blocked with lenalidomide: eslishing a reversible immune evasion mechanism in human cancer. Blood. 2012;120(7):1412-21. doi: 10.1182/blood-2012-02-411678. PMID:22547582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frebel H, Nindl V, Schuepbach RA, Braunschweiler T, Richter K, Vogel J, Wagner CA, Loffing-Cueni D, Kurrer M, Ludewig B. Programmed death 1 protects from fatal circulatory failure during systemic virus infection of mice. J Exp Med. 2012;209(13):2485-99. doi: 10.1084/jem.20121015. PMID:23230000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brooks DG, McGavern DB, Oldstone MB. Reprogramming of antiviral T cells prevents inactivation and restores T cell activity during persistent viral infection. J Clin Invest. 2006;116(6):1675-85. doi: 10.1172/JCI26856. PMID:16710479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Catakovic K, Klieser E, Neureiter D, Geisberger R. T cell exhaustion: from pathophysiological basics to tumor immunotherapy. Cell Commun Signal. 2017;15(1):1. doi: 10.1186/s12964-016-0160-z. PMID:28073373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barber DL, Wherry EJ, Masopust D, Zhu B, Allison JP, Sharpe AH, Freeman GJ, Ahmed R. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature. 2006;439(7077):682-7. doi: 10.1038/nature04444. PMID:16382236 [DOI] [PubMed] [Google Scholar]
- 10.Intlekofer AM, Thompson CB. At the bench: preclinical rationale for CTLA-4 and PD-1 blockade as cancer immunotherapy. J Leukoc Biol. 2013;94(1):25-39. doi: 10.1189/jlb.1212621. PMID:23625198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.N Jain SB, Thompson PA, Ohanian M, Ferrajoli A, Pemmaraju N et al.. Nivolumab Combined with Ibrutinib for CLL and Richter Transformation: A Phase II Trial ASH. 2016. [Google Scholar]
- 12.W Ding JL-R, Call TG, Parikh SA, Leis JF, Shanafelt TD et al.. PD-1 Blockade with Pembrolizumab in Relapsed CLL Including Richter's Transformation: An Updated Report from a Phase 2 Trial (MC1485) ASH. 2016. https://ash.confex.com/ash/2016/webprogram/Paper95426.html. [Google Scholar]
- 13.Yu X, Harden K, Gonzalez LC, Francesco M, Chiang E, Irving B, Tom I, Ivelja S, Refino CJ, Clark H. The surface protein TIGIT suppresses T cell activation by promoting the generation of mature immunoregulatory dendritic cells. Nat Immunol. 2009;10(1):48-57. doi: 10.1038/ni.1674. PMID:19011627 [DOI] [PubMed] [Google Scholar]
- 14.Stanietsky N, Simic H, Arapovic J, Toporik A, Levy O, Novik A, Levine Z, Beiman M, Dassa L, Achdout H. The interaction of TIGIT with PVR and PVRL2 inhibits human NK cell cytotoxicity. Proc Natl Acad Sci U S A. 2009;106(42):17858-63. doi: 10.1073/pnas.0903474106. PMID:19815499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Le Mercier I Lines JL, Noelle RJ. Beyond CTLA-4 and PD-1, the Generation Z of Negative Checkpoint Regulators. Front Immunol. 2015;6:418. doi: 10.3389/fimmu.2015.00418. PMID:26347741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li M, Xia P, Du Y, Liu S, Huang G, Chen J, Zhang H, Hou N, Cheng X, Zhou L. T-cell immunoglobulin and ITIM domain (TIGIT) receptor/poliovirus receptor (PVR) ligand engagement suppresses interferon-gamma production of natural killer cells via beta-arrestin 2-mediated negative signaling. J Biol Chem. 2014;289(25):17647-57. doi: 10.1074/jbc.M114.572420. PMID:24817116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu S, Zhang H, Li M, Hu D, Li C, Ge B, Jin B, Fan Z. Recruitment of Grb2 and SHIP1 by the ITT-like motif of TIGIT suppresses granule polarization and cytotoxicity of NK cells. Cell Death Differ. 2013;20(3):456-64. doi: 10.1038/cdd.2012.141. PMID:23154388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kong Y, Zhu L, Schell TD, Zhang J, Claxton DF, Ehmann WC, Rybka WB, George MR, Zeng H, Zheng H. T-Cell Immunoglobulin and ITIM Domain (TIGIT) Associates with CD8+ T-Cell Exhaustion and Poor Clinical Outcome in AML Patients. Clin Cancer Res. 2016;22(12):3057-66. doi: 10.1158/1078-0432.CCR-15-2626. PMID:26763253 [DOI] [PubMed] [Google Scholar]
- 19.Nishiwada S, Sho M, Yasuda S, Shimada K, Yamato I, Akahori T, Kinoshita S, Nagai M, Konishi N, Nakajima Y. Clinical significance of CD155 expression in human pancreatic cancer. Anticancer Res. 2015;35(4):2287-97. PMID:25862891 [PubMed] [Google Scholar]
- 20.Iguchi-Manaka A, Okumura G, Kojima H, Cho Y, Hirochika R, Bando H, Sato T, Yoshikawa H, Hara H, Shibuya A. Increased Soluble CD155 in the Serum of Cancer Patients. PLoS One. 2016;11(4):e0152982. doi: 10.1371/journal.pone.0152982. PMID:27049654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carlsten M, Norell H, Bryceson YT, Poschke I, Schedvins K, Ljunggren HG, Kiessling R, Malmberg KJ. Primary human tumor cells expressing CD155 impair tumor targeting by down-regulating DNAM-1 on NK cells. J Immunol. 2009;183(8):4921-30. doi: 10.4049/jimmunol.0901226. PMID:19801517 [DOI] [PubMed] [Google Scholar]
- 22.Johnston RJ, Comps-Agrar L, Hackney J, Yu X, Huseni M, Yang Y, Park S, Javinal V, Chiu H, Irving B. The immunoreceptor TIGIT regulates antitumor and antiviral CD8(+) T cell effector function. Cancer Cell. 2014;26(6):923-37. doi: 10.1016/j.ccell.2014.10.018. PMID:25465800 [DOI] [PubMed] [Google Scholar]
- 23.Kurtulus S, Sakuishi K, Ngiow SF, Joller N, Tan DJ, Teng MW, Teng MW, Smyth MJ, Kuchroo VK, Anderson AC. TIGIT predominantly regulates the immune response via regulatory T cells. J Clin Invest. 2015;125(11):4053-62. doi: 10.1172/JCI81187. PMID:26413872 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Chauvin JM, Pagliano O, Fourcade J, Sun Z, Wang H, Sander C, Kirkwood JM, Chen TH, Maurer M, Korman AJ. TIGIT and PD-1 impair tumor antigen-specific CD8(+) T cells in melanoma patients. J Clin Invest. 2015;125(5):2046-58. doi: 10.1172/JCI80445. PMID:25866972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sansom DM. CD28, CTLA-4 and their ligands: who does what and to whom? Immunology. 2000;101(2):169-77. doi: 10.1046/j.1365-2567.2000.00121.x. PMID:11012769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramsbottom KM, Hawkins ED, Shimoni R, McGrath M, Chan CJ, Russell SM, Smyth MJ, Oliaro J. Cutting edge: DNAX accessory molecule 1-deficient CD8+ T cells display immunological synapse defects that impair antitumor immunity. J Immunol. 2014;192(2):553-7. doi: 10.4049/jimmunol.1302197. PMID:24337740 [DOI] [PubMed] [Google Scholar]
- 27.Zhang T, Wang J, Zhou X, Liang R, Bai Q, Yang L, Gu H, Gao G, Dong B, Zhu H. Increased expression of TIGIT on CD4+ T cells ameliorates immune-mediated bone marrow failure of aplastic anemia. J Cell Biochem. 2014;115(11):1918-27. PMID:24905442 [DOI] [PubMed] [Google Scholar]
- 28.Inozume T, Yaguchi T, Furuta J, Harada K, Kawakami Y, Shimada S. Melanoma Cells Control Antimelanoma CTL Responses via Interaction between TIGIT and CD155 in the Effector Phase. J Invest Dermatol. 2016;136(1):255-63. [DOI] [PubMed] [Google Scholar]
- 29.Tinhofer I, Weiss L, Gassner F, Rubenzer G, Holler C, Greil R. Difference in the relative distribution of CD4+ T-cell subsets in B-CLL with mutated and unmutated immunoglobulin (Ig) VH genes: implication for the course of disease. J Immunother. 2009;32(3):302-9. doi: 10.1097/CJI.0b013e318197b5e4. PMID:19242370 [DOI] [PubMed] [Google Scholar]
- 30.Weiss L, Melchardt T, Egle A, Grabmer C, Greil R, Tinhofer I. Regulatory T cells predict the time to initial treatment in early stage chronic lymphocytic leukemia. Cancer. 2011;117(10):2163-9. doi: 10.1002/cncr.25752. PMID:21523729 [DOI] [PubMed] [Google Scholar]
- 31.Piper KP, Karanth M, McLarnon A, Kalk E, Khan N, Murray J, Pratt G, Moss PA. Chronic lymphocytic leukaemia cells drive the global CD4+ T cell repertoire towards a regulatory phenotype and leads to the accumulation of CD4+ forkhead box P3+ T cells. Clin Exp Immunol. 2011;166(2):154-63. doi: 10.1111/j.1365-2249.2011.04466.x. PMID:21985361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bartik MM, Welker D, Kay NE. Impairments in immune cell function in B cell chronic lymphocytic leukemia. Semin Oncol. 1998;25(1):27-33. PMID:9482524 [PubMed] [Google Scholar]
- 33.Tinhofer I, Marschitz I, Kos M, Henn T, Egle A, Villunger A, Greil R. Differential sensitivity of CD4+ and CD8+ T lymphocytes to the killing efficacy of Fas (Apo-1/CD95) ligand+ tumor cells in B chronic lymphocytic leukemia. Blood. 1998;91(11):4273-81. PMID:9596676 [PubMed] [Google Scholar]
- 34.Zaborsky N, Holler C, Geisberger R, Asslaber D, Gassner FJ, Egger V, Piñón-Hofbauer J, Kocher T, Hartmann TN, Greil R. B-cell receptor usage correlates with the sensitivity to CD40 stimulation and the occurrence of CD4+ T-cell clonality in chronic lymphocytic leukemia. Haematologica. 2015;100(8):e307-10. PMID:25911550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rezvany MR, Jeddi-Tehrani M, Wigzell H, Osterborg A, Mellstedt H. Leukemia-associated monoclonal and oligoclonal TCR-BV use in patients with B-cell chronic lymphocytic leukemia. Blood. 2003;101(3):1063-70. doi: 10.1182/blood-2002-03-0746. PMID:12393705 [DOI] [PubMed] [Google Scholar]
- 36.Os A, Burgler S, Ribes AP, Funderud A, Wang D, Thompson KM, Tjønnfjord GE, Bogen B, Munthe LA. Chronic lymphocytic leukemia cells are activated and proliferate in response to specific T helper cells. Cell Rep. 2013;4(3):566-77. doi: 10.1016/j.celrep.2013.07.011. PMID:23933259 [DOI] [PubMed] [Google Scholar]
- 37.Zhao W, Dong Y, Wu C, Ma Y, Jin Y, Ji Y. TIGIT overexpression diminishes the function of CD4 T cells and ameliorates the severity of rheumatoid arthritis in mouse models. Exp Cell Res. 2016;340(1):132-8. doi: 10.1016/j.yexcr.2015.12.002. PMID:26683997 [DOI] [PubMed] [Google Scholar]
- 38.Zaidi MR, Merlino G. The two faces of interferon-gamma in cancer. Clin Cancer Res. 2011;17(19):6118-24. doi: 10.1158/1078-0432.CCR-11-0482. PMID:21705455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buschle M, Campana D, Carding SR, Richard C, Hoffbrand AV, Brenner MK. Interferon gamma inhibits apoptotic cell death in B cell chronic lymphocytic leukemia. J Exp Med. 1993;177(1):213-8. doi: 10.1084/jem.177.1.213. PMID:7678114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zaki M, Douglas R, Patten N, Bachinsky M, Lamb R, Nowell P, Nowell P, Moore J. Disruption of the IFN-gamma cytokine network in chronic lymphocytic leukemia contributes to resistance of leukemic B cells to apoptosis. Leuk Res. 2000;24(7):611-21. doi: 10.1016/S0145-2126(00)00022-9. PMID:10867137 [DOI] [PubMed] [Google Scholar]
- 41.Joller N, Lozano E, Burkett PR, Patel B, Xiao S, Zhu C, Xia J, Tan TG, Sefik E, Yajnik V. Treg cells expressing the coinhibitory molecule TIGIT selectively inhibit proinflammatory Th1 and Th17 cell responses. Immunity. 2014;40(4):569-81. doi: 10.1016/j.immuni.2014.02.012. PMID:24745333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fiorcari S, Maffei R, Audrito V, Martinelli S, Ten Hacken E, Zucchini P, Grisendi G, Potenza L, Luppi M, Burger JA. Ibrutinib modifies the function of monocyte/macrophage population in chronic lymphocytic leukemia. Oncotarget. 2016;7(40):65968-81. PMID:27602755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Godefroy E, Zhong H, Pham P, Friedman D, Yazdanbakhsh K. TIGIT-positive circulating follicular helper T cells display robust B-cell help functions: potential role in sickle cell alloimmunization. Haematologica. 2015;100(11):1415-25. doi: 10.3324/haematol.2015.132738. PMID:26250578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cha Z, Zang Y, Guo H, Rechlic JR, Olasnova LM, Gu H, Tu X, Song H, Qian B. Association of peripheral CD4+ CXCR5+ T cells with chronic lymphocytic leukemia. Tumour Biol. 2013;34(6):3579-85. doi: 10.1007/s13277-013-0937-2. PMID:23807677 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


