Abstract
Background
Notch signaling is broadly required during embryogenesis, frequently activating the transcription of two bHLH transcription factors, Hes1 and Hes5. But, it remains unresolved when and where Hes1 and Hes5 act alone or together during development. Here, we analyzed a Hes5-GFP BAC transgenic mouse, as a proxy for endogenous Hes5. We directly compared transgenic GFP expression with Hes1, and particular markers of embryonic lens and retina development.
Results
Hes5-GFP is dynamic within subsets of retinal and lens progenitor cells, and differentiating retinal ganglion neurons, in contrast to Hes1 found in all progenitor cells. In the adult retina, only Müller glia express Hes5-GFP. Finally, Hes5-GFP is upregulated in Hes1 germline mutants, consistent with previous demonstration that Hes1 suppresses Hes5 transcription.
Conclusions
Hes5-GFP BAC transgenic mice are useful for identifying Hes5-expressing cells. Although Hes5-GFP and Hes1 are coexpressed in particular developmental contexts, we also noted cohorts of lens or retinal cells expressing just one factor. The dynamic Hes5-GFP expression pattern, coupled with its derepressed expression in Hes1 mutants, suggests that this transgene contains the relevant cis-regulatory elements that regulate endogenous Hes5 in the mouse lens and retina.
Keywords: Hes1, Hes5, lens development, retina, neurogenesis, Notch signaling
Introduction
The Notch signaling pathway maintains a balance between dividing and differentiating cells, and releases waves of progenitors to initiate differentiation at the right time, and in the correct proportions. A Notch signal initiated by ligand-receptor binding, invokes a series of proteolytic events, resulting in Notch intracellular protein domain (ICD) translocation into the nucleus. The Notch-ICD forms a complex with nuclear proteins RBPJ/Su(H) and MAML to activate target gene transcription, including the Hes1 and Hes5 transcriptional repressors (reviewed in Davis and Turner, 2001; Kageyama et al., 2007; Kopan and Ilagan, 2009). To fully understand Notch signaling mechanisms during development, it is necessary to define the contexts that require different combinations of downstream effector genes.
The rodent eye is an established model for investigating mechanisms of progenitor cell proliferation and cell fate specification, and the ocular expression patterns of Notch pathway genes have been described over the past decade (Lindsell et al., 1996; Bao and Cepko, 1997). Lens development initiates when an ectodermally derived lens placode evaginates to form a lens vesicle, from which differentiated fiber cells arise in two separate waves. Jag1, Notch1, Notch2, Rbpj, and Hes1 are more or less ubiquitous in early lens progenitor cells. However, once fiberogenesis initiates, the expression of both Notch receptors, Rbpj, Hes1 and Hes5 are all restricted to the anterior epithelial layer (AEL), whereas Jag1 is localized to the equatorial transition zone, where overt differentiation occurs (Jia et al., 2007; Rowan et al., 2008; Le et al., 2009; Le et al., 2012). Multiple loss-or-gain of function studies support the idea that a Jag1-Notch2-Rbpj mediated signal stimulates AEL cells to either proliferate or differentiate (Jia et al., 2007; Rowan et al., 2008; Le et al., 2009; Saravanamuthu et al., 2012).
Simultaneously, the optic vesicle, which is comprised of neuroepithelial cells, invaginates into the optic cup/retina. Four retinal cell types (RGCs, cones, amacrines and horizontal neurons) appear largely before birth, whereas rods, bipolar neurons and Müller glia appear after birth. In the mouse retina Dll1, Dll3, Dll4, Notch1, Notch3, Rbpj, Hes1 and Hes5 are all expressed during retinal development (Ishibashi et al., 1995; Lindsell et al., 1996; Bao and Cepko, 1997; Lee et al., 2005; Nelson et al., 2006; Nelson and Reh, 2008; Rocha et al., 2009). Numerous loss- and gain-of-function studies demonstrated that a Dll1-Notch1-Rbpj-Hes1 signal promotes retinal progenitor cell (RPC) proliferation and suppresses RGC neurogenesis, whereas a Dll4-Notch1-Rbpj signal stimulates progenitor cells but suppresses cone genesis (Takatsuka et al., 2004; Riesenberg et al., 2009; Rocha et al., 2009; Zheng et al., 2009; Maurer et al., 2014). The Notch pathway generally blocks neurogenesis by suppressing proneural bHLH factor expression. For example, Hes1 transcriptionally represses multiple proneural bHLH genes (reviewed in Ohtsuka et al., 1999; Kageyama et al., 2008; Ohsawa and Kageyama, 2008), including Atoh7, which is essential for RGC development (Takatsuka et al., 2004; Lee et al., 2005). Hes5 has been described to similarly suppress neurogenesis in the CNS, but its molecular mode of action during early retinal development and its roles within Müller glia of the postnatal retina remain poorly understood. Moreover, it is unclear when and where developing lens and retinal cells utilize Hes1 or Hes5 alone or together.
Here we evaluated Hes5-GFP BAC Tg mice for their recapitulation of endogenous Hes5 expression in the developing mouse eye. We found Hes5-GFP expression initiates at E10.5 in discrete subsets of lens pit and optic vesicle cells, but in the lens subsequently becomes expressed by all lens AEL and transition zone cells. Once retinal neurogenesis begins in the optic cup, we saw distinct cohorts of RPCs expressing Hes5-GFP, Hes1 or both. At E11.5, retinal cells expressing Hes5-GFP only coexpressed Pou4f+, a definitive RGC marker. Moreover, we found that nascent cone photoreceptors do not express Hes5-GFP. In the adult retina, Hes5-GFP is only present in Müller glia. The diversity of Hes5-GFP BAC expression by embryonic RGCs and adult Müllers is consistent with the dynamics of endogenous Hes5 mRNA expression (Hojo et al., 2000; Nelson et al., 2011). Finally, we observed Hes5-GFP is derepressed when Hes1 gene dosage was reduced.
Results
Although by no means the only readout of Notch signaling, cells expressing Hes1 and/or Hes5 are categorized as recipients of a canonical Notch signal (Kageyama et al., 2008; Kopan and Ilagan, 2009). In the mouse there are specific antibodies that label Hes1-expressing cells (Lee et al., 2005), but no Hes5 antibody of comparable quality and specificity. Therefore it has not been possible to visualize which ocular cells express Hes5 protein, alone or in combination with Hes1. In an effort to circumvent this issue, we evaluated a Hes5-GFP BAC transgenic (Tg) mouse, obtained from the Mutant Mouse Regional Resource. We directly compared GFP expression to a battery of early eye development markers, and several Notch pathway proteins, including Hes1.
Overlapping and distinct Hes1 and Hes5-GFP expression domains in the prenatal eye
This Hes5-GFP BAC transgene inserted onto the mouse X chromosome. Hemizygotes were propagated by mating transgenic females to wild type males, so that on average 50% of the progeny would be transgenic. Over the course of this study, we identified 92 transgenics of 198 total weanling mice (48%, n= 18 litters) using the MMRRC recommended genotyping protocol. Recently, we switched to assessing transgenic inheritance directly in our mouse colony, using a GFP flashlight and scoring expression in adult lenses (see Methods). To date, we have identified 27 GFP+ of 55 total weanling mice (49%, n=6 litters). In our embryonic analyses, we initially compared PCR genotyping to live GFP screening using a fluorescent microscope, UV light source and GFP filter. We saw 100% correlation between these methods, among 31 of 31 transgenic embryos, from 4 litters. As a consequence we switched exclusively to live embryo screening for the Hes5-GFP transgene. To date we identified 232 GFP+ out of 451 embryos (52%, n= 34 litters), including those with the transgene intercrossed with Hes1 KO/+ or Hes1KO/+;Atoh7LacZ/+ mice. We also observed 100% expressivity of the GFP reporter within cryosections (232 out of 232), all of which displayed the expected tissue-specific patterns. Overall we conclude that this Hes5-GFP BAC transgenic line is both stably inherited and a reliable reporter.
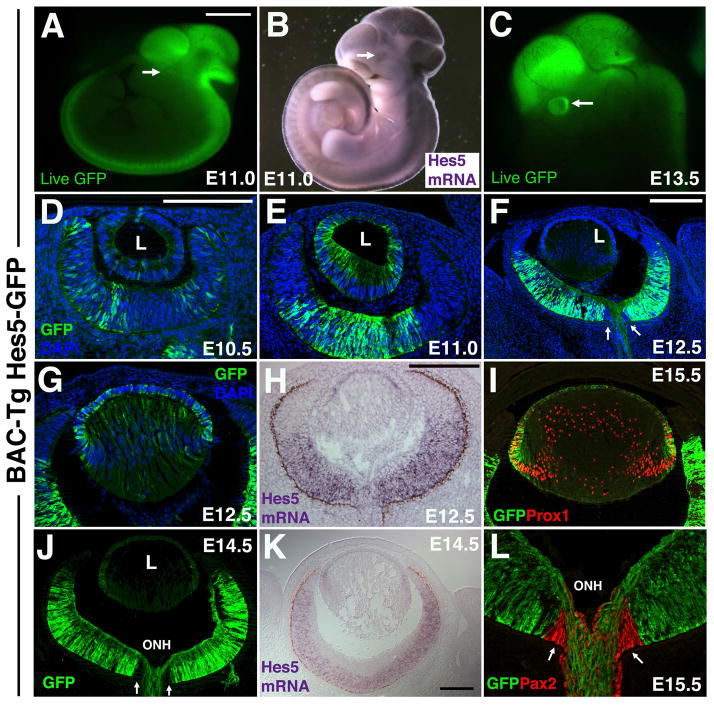
We first assessed the live fluorescence of Hes5-GFP BAC transgenic embryos from E9.5 through E16.5. No GFP expression was obvious at E9.5, but starting at E10.0 (Fig 5A) GFP expression was seen in restricted forebrain and midbrain domains, and more uniformly throughout the hindbrain and spinal cord (Fig 1A, 5A). Between E10.5 to E11, additional somite, olfactory epithelium and eye domains became apparent. At the gross level, Hes5-GFP mimics Hes5 mRNA expression (compare Fig 1A to 1B). By E13.5 Hes5-GFP was very obvious in the developing eye (arrow in Fig 1C). We next analyzed fixed cryosections, labeled with anti-GFP, and noted patchy Hes5-GFP expression in peripheral RPCs at E10.5 (Fig 1D), which was not maintained at E11, where Hes5-GFP-expressing cells were now restricted to the central cup (Fig 1E), a domain that had expanded peripherally at E12.5 (Fig 1F). These relatively rapid changes were unexpected, given that GFP transgenic lines are often prone to persistent GFP reporter expression. We also compared anti-GFP labeling of Hes5-GFP to Hes5 mRNA in situ expression (compare Figs 1F,1G to 1H, Fig 1J to 1K). We observed that Hes5 mRNA and Hes5-GFP are both present in lens AEL cells, but Hes5-GFP was retained in a subset of in central lens fiber cells (Fig 1G), whose nuclei are Prox1+ (Fig 1I). In the E12.5 central retina both Hes5 mRNA and Hes5-GFP are robustly expressed, but not at the periphery, where the ciliary body forms. At E14.5, Hes5 mRNA is restricted to subsets of RPCs, whereas both progenitor cells and nascent RGC neurons express Hes5-GFP, including their axons extending through the optic nerve head (ONH in Fig 1J,1L) and into the optic nerve. Intriguingly, optic nerve head cells, uniquely marked by Pax2 (red nuclei in Fig 1L), do not express Hes5-GFP (arrows in Fig 1F, 1J and 1L).
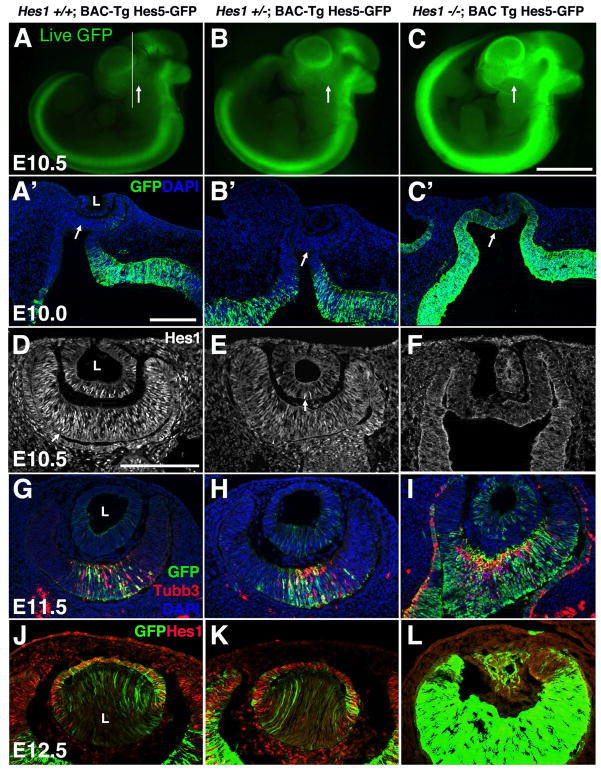
Figure 5. Hes5-GFP BAC Tg expression is regulated by Hes1.
In each experiment (row), although not quantitative, labeling conditions and imaging time/parameters were held constant among genotypes. A–C) Live GFP expression among littermates. Line in A indicates sectioning plane for A′–C′ and arrows point to forming eye. Hes5-GFP expression is upregulated in Hes1 mutants, consistent with Hes5 mRNA expression (ref). A′–C′) Anti-GFP labeling of fixed ocular sections also highlights changes in Hes5-GFP expression in Hes1 mutants. Arrows point to forming optic cup. D–F) Loss of Hes1 protein expression from E10.5 Hes1 mutants. Arrows in D,E point to Hes1 nuclear protein expression. G–I) In the absence of Hes1 activity, neuronal differentiation (Tubb3+ cells in red) is also derepressed in the optic cup and RPE. J–L) Removal of Hes1 results in lens microphthalmia, although some Hes5-GFP expression persists (L). Bar in C (for A–C) = 1 mm; in A′ (for A′–C′) = 10 microns and in D (for D–L) = 20 microns; n ≥ 3 embryos per age and genotype analyzed.
Figure 1. Hes5-GFP BAC-Tg reporter expression relative to Hes5 mRNA.
A,B) Whole embryo Hes5-GFP fluorescence, compared to Hes5 mRNA in situ expression. Arrows point to forming eye. C) At E13.5 GFP fluorescence in the developing eye is obvious (arrow). In all other panels, GFP expression was visualized by anti-GFP labeling of fixed cryosections, collected in the horizontal plane of sectioning. D–F) Dynamic changes in Hes5-GFP during early eye formation. F–H) Comparison of Hes5-GFP expression at E12.5 in the developing lens and retina. Arrows in F point to the optic nerve head, which lacks anti-GFP labeling. I) E15.5 Hes5-GFP expression in lens AEL and transition zone(bracket), compared to Prox1 nuclear expression (red) within transition zone and lens fiber cell nuclei. J,K) Comparison of Hes5-GFP and Hes5 mRNA expression at E14.5. Hes5-GFP is present in RPCs (like the endogenous gene), but also within RGCs, including their axons extending into the optic nerve head (ONH). J,L) Pax2+ optic nerve head (arrows) and optic stalk cells do not express GFP. Dorsal is up in A–C; Anterior is up, and nasal left in panels D–L. L = lens in D–F,J; ONH = optic nerve head in J,L. Scale bar in A=1mm (for A–C); in D (for D,E,G,L) or H=20 microns; in F for F,I,J) or K=10 microns.
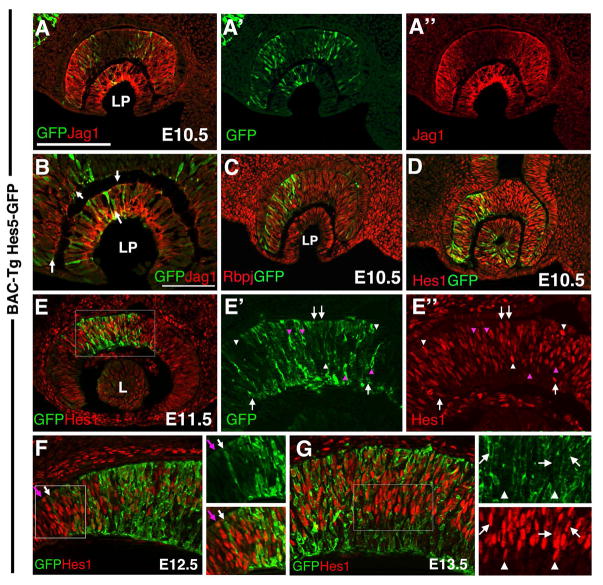
By antibody multiple labeling, we next compared Hes5-GFP expression with endogenous Jag1, Rbpj and Hes1 proteins, in the E10.5–E13.5 developing eye. In both the E10.5 lens pit and optic cup, Hes5-GFP expression is first seen within subsets of progenitor cells. Those lens progenitor cells expressing Hes5-GFP, co-expressed Jag1 at the cell membrane (Figs 2A, arrows in 2B), whereas other Jag1+ cells (negative for Hes5-GFP), express Hes1 (Fig 2C). All Hes5-GFP cells co-express the ubiquitous Rbpj protein throughout the developing eye (Fig 2C). We conclude that during the early phase of lens development, progenitor cells are actively signaling since they all express Jag1 and Rbpj. However, subpopulations could be identified by virtue of expression of Hes1 alone, or Hes1+Hes5GFP (Fig 2D).
Figure 2. Hes5-GFP expression in the early eye relative to other Notch signaling pathway components.
A,A′A″, B) Hes5-GFP and Jag1 protein overlap in subsets of distal optic cup and lens pit cells. Arrows in B point to cells with Jag1 expression (red) at the cell membrane, surrounding cytoplasmic GFP. Nasal is to the left here, and in C,D. C) All Hes5-GFP cells co-express Rbpj. D) At this same age, every Hes5-GFP lens, optic cup and stalk cell also coexpresses Hes1 protein. E, E′,E″) Starting at E11.5, some GFP + only (pink arrowhead) and some Hes1+ only (white arrowheads) cells can be identified, although the majority of Hes5-GFP+ cells coexpress Hes1 (white arrows). E′ and E″ are 8X zoomed magnifications of boxed area in E. F) Comparison of Hes5-GFP and Hes1 expression at the E12.5 ciliary margin. 4X zoom of boxed area is shown in adjacent panels. Hes1 is present in RPCs and throughout the forming ciliary margin (left boxed area), but, Hes5-GFP+ is confined to the retina, like Hes5 mRNA (Fig 1H). White arrow points to distal-most extent of the Hes5-GFP domain, and the pink arrow denotes adjacent distal row of Hes1+ cells. G) Comparison of Hes5-GFP and Hes1 protein in E13.5 central retina. Arrows denote coexpressing cells, whereas white arrowheads point to Hes1+ only progenitors. Nascent RGCs in the inner retina express Hes5-GFP+. All images are in the horizontal plane of section. Scleral/apical is up in all panels; LP = lens pit; L= lens vesicle; Mag bar in A (for A,B,C,E) = 20 microns; in D (for D,E′,E″,F,I) = 50 microns.
Meanwhile in the adjacent optic cup, distal optic cup cells coexpress Hes5-GFP at the boundaries of a Jag1 distal expression domain (Fig 2A). Although no function has been reported for this specific Jag1 domain, it does precede the advancing wave of neurogenesis from E9.5 to E12.5 (Le et al., 2009; Maurer et al., 2014), further suggesting the potential for a Jag1-mediated signal in the distal optic cup. It is also interesting that Hes5-GFP is more easily visualized within nasal RPCs (Figs 2C, 2D). When retinal neurogenesis initiates at E11.5, the majority of Hes5-GFP+ cells coexpress Hes1 (white arrows), but smaller cohorts of Hes1+ (white arrowheads in Fig 2E) or Hes5-GFP+ cells (pink arrowheads) became distinguishable. At E12.5 a boundary appears between the forming ciliary body and retina (Marcucci et al., 2016). Here too we noted differences between Hes5-GFP and Hes1 protein expression. Although Hes5-GFP is confined to retinal cells (white arrow in Fig 2F, Fig 1F, 1H), Hes1 is expressed by both RPCs and presumptive ciliary body cells. From E12.5 to E16.5, we found central RPCs were double positive for Hes1+Hes5-GFP (arrows in Fig 2G), although some Hes1+ RPCs (arrowheads) was also present. By contrast, Hes5-GFP was uniquely expressed by inner GCL neurons (Fig 2G), corresponding to nascent RGCs (Fig 3).
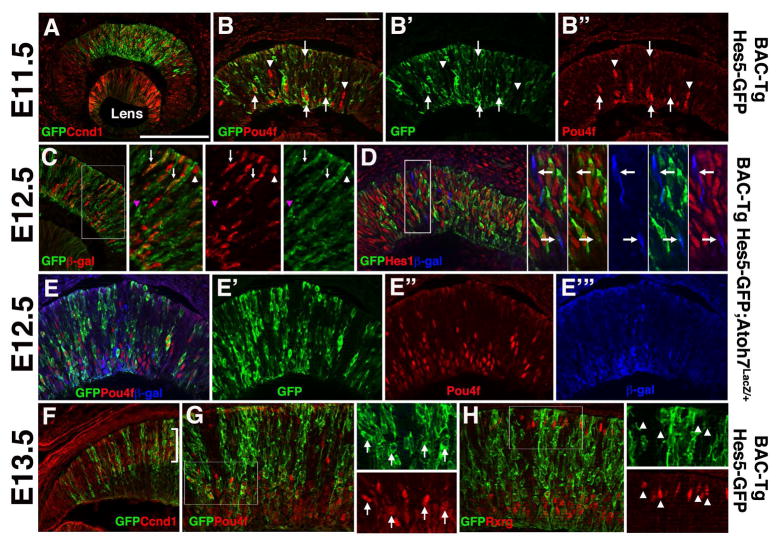
Figure 3. Hes5-GFP expression correlates with RGC but not cone neurogenesis.
A) Most Hes5-GFP cells in the E11.5 central optic cup coexpress the mitotic marker Ccnd1. B,B′B″) But a subset of nascent Pou4f+ RGCs also express Hes5-GFP (arrows). Arrowheads denote Pou4f+ nuclei that are Hes5-GFP-negative. C) In Hes5-GFP/+;Atoh7LacZ/+ eyes, anti-GFP and anti-βgal colabeling highlight three types of RPCs: double positive (white arrows), βgal-only (white arrowhead) GFP-only (pink arrowhead).
D) Anti-GFP, anti-Hes1 and anti-βgal triple labeling further distinguishes a subset of Atoh7LacZ/+ cells that do not express Hes1 or Hes5-GFP proteins (arrows in zoomed insets of boxed area). E,E′E″, E‴) Subpopulations of E12.5 RPCs express different combinations of GFP, Pou4f (red) and β-gal (blue). F) At E13.5, a proportion of Hes5-GFP cells reside in the outer, neuroblast layer of the retina (bracket), as marked by Ccnd1 expression. G) Nearly all Pou4f+ RGCs in the E13.5 central retina coexpress Hes5-GFP. Arrows in 4X zoomed inset point to double positive cells. H) The Rxrg transcription factor is expressed by early RGCs (inner retina) and cone photoreceptors (outer retina) (Roberts et al., 2005). Although inner Rxrg+ RGCs coexpress Hes5-GFP, Rxrg+ cones were devoid of Hes5-GFP. Arrowheads in 4X zoomed insets point to Rxrg+ cones. Scleral/apical is up in all panels; mag bars in A (for A,C,F) = 20 microns; in B (for B, D, E, G, H) = 50 microns.
Differential Hes expression during prenatal retinal neurogenesis
A long-term goal of our work is to uncover the distinct molecular mechanisms by which Notch signaling regulates RGC versus cone photoreceptor neurogenesis. So we compared Hes5-GFP expression to that of the cell cycle marker Ccnd1/CyclinD1, Atoh7LacZ (β-gal) or Pou4f. The Atoh7 lineage contains RPCs with competence to adopt the RGC fate (Yang et al., 2003; Brzezinski et al., 2012), whereas Pou4f expression indicates cells that differentiated as RGC neurons (Xiang et al., 1993). We noted that Hes5-GFP expression localizes to the central optic cup at E11.5, coincident with the initiation of neurogenesis (Figs 1E, 2E, 3A). While most central RPCs coexpressed Hes5-GFP and Ccnd1 (Fig 3A), we also saw sparse Hes5-GFP+ cells in the innermost optic cup (arrows 3B). With the expansion of neurogenesis between E11.5 and E13.5, many more Hes5-GFP cells coexpressed Pou4f+, although occasional Pou4f+ only RGCs were also observed (arrowheads in Fig 3B, Figs 3E, 3G). This is consistent with a recent study reporting that Hes5 RPCs give rise to all contralateral-projecting RGCs (Kuwajima et al., 2017). The subset of Pou4f+ only RGCs might correlate with intrinsic differences among RGC subtypes, or with some aspect of Hes5-GFP reporter kinetics. Overall, we conclude that this Hes5-GFP BAC transgene is a short-term lineage tracer for the majority of RGCs.
Next we analyzed Hes5-GFP expression in retinal sections from Atoh7LacZ/+;BAC Tg Hes5GFP hemizygous embryos. At E12.5 we noted mostly double-positive (GFP+ β-gal+) cells (Figs 3C–3E), but also observed occasional single-labeled cells, either GFP+ (pink arrowhead in 3C) or β-gal+ (white arrowhead in 3C, white arrow in 3D insets). We previously showed mutually exclusive expression of Hes1 and β-gal proteins in E11.5 Atoh7LacZ/+ retinas, consistent with Hes1 repression of Atoh7 expression (Takatsuka et al., 2004; Lee et al., 2005). Therefore, to further highlight the differences between Hes5-GFP and Hes1 expression at the cellular level, Atoh7LacZ/+;Hes5GFP sections were triple labeled with β-gal, GFP and Hes1 antibodies (Fig 3D). In addition to Hes1-only, Hes5-GFP only and Hes1+Hes5GFP+ cohorts, we also saw a minor population of β-gal (Atoh7LacZ/+) cells (arrows pointing to blue-only cells in 3D insets). Similarly, triple labeling of Atoh7LacZ/+;Hes5GFP sections with β-gal, GFP and Pou4f antibodies, showed many Hes5-GFP/Pou4f double positive cells, particularly in the inner retina, but also sparse GFP− or Pou4f− only cells(Fig 5E). With the onset of retinal lamination at E13.5, Hes5-GFP+ cells were present in the neuroblast layer, marked by Ccnd1 expression (bracket in Fig 3F), and in inner RGCs coexpressing Pou4f (Fig 3G). Importantly, Hes5-GFP expression was absent in Rxrg+ cone photoreceptors (Fig 3H). We conclude that when Hes1 expression is shut off by RPCs adopting the RGC fate, Hes5-GFP becomes activated within these differentiating neurons. Moreover, cone photoreceptors do not express Hes5-GFP.
Hes5-GFP and adult Müller glia
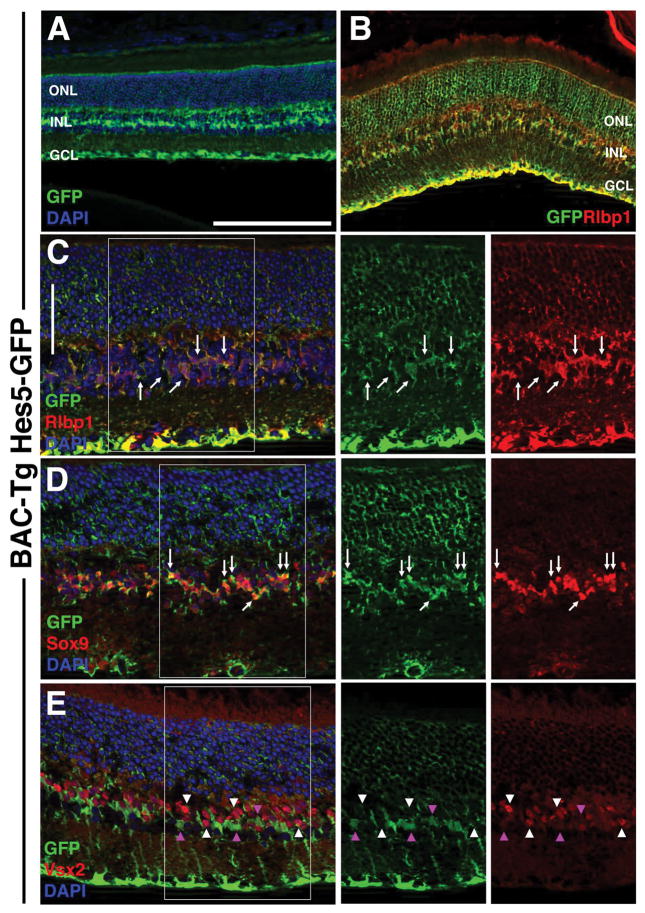
Throughout the developing CNS, glial cells are generated at the end of histogenesis. This also occurs in the retina where Müller glia are among the last cell types to develop (reviewed in Jadhav et al., 2009). Here too, Notch signaling and its downstream effectors, Hes1 and Hes5 are required for Müller glial differentiation (Hojo et al., 2000; Bernardos et al., 2005; Muto et al., 2009; Nelson et al., 2011). Hes5 mRNA was been previously shown to be restricted to Müller glia beginning at postnatal day 7 (P7) through adulthood (Nelson et al., 2011). Anti-GFP labeling of P21 BAC Tg Hes5GFP hemizygous cryosections showed localization to cells whose somas reside in the INL and processes span the entire retinal width (Figs 4A,B). This is consistent with Müller cell morphology. To validate the specificity of Hes5-GFP expression, we performed double immunofluorescent labeling with Müller markers Rlbp1/CRALBP and Sox9, as well as the bipolar neuronal marker Vsx2/Chx10. Essentially complete coexpression between Rlbp1 and GFP was observed (Figs 4B, arrows in 4C) and between GFP and nuclear Sox9 in the INL (Fig 4D arrows). Hes5-GFP expression was mutually exclusive to that of Vsx2 in bipolar neuron nuclei (arrowheads in Fig 4E). Overall we conclude that Hes5-GFP accurately reports Müller-specific Hes5 expression in the adult mouse retina.
Figure 4. Specific Hes5-GFP expression in adult Müller glia.
A) Anti-GFP labeling of P21 transgene hemizygous cryosection showing Hes5-GFP localization within particular INL cells and their cellular processes spanning the retina (from outer limiting membrane to nerve fiber layer). B) GFP coexpression with Rlbp1/Cralbp (arrows) in Müller cell bodies. C) Arrows point to coexpressing Müller cell bodies at higher magnification. D) Hes5-GFP was also co-expressed with Sox9 (arrows), another Müller cell nuclear marker. The circular structure within the GCL of boxed area is a blood vessel. E) No GFP+ INL cells (pink arrowheads) coexpress Vsx2, a bipolar marker (white arrowheads). Boxed areas in C,D,E are shown at same magnification at right, with marker channels separated. Scleral is up in all panels; bar in A,B = 20 microns; C–E = 50 microns, n ≥ 3 adult eyes analyzed per marker. ONL = outer nuclear layer, INL = inner nuclear layer, GCL = ganglion cell layer.
Hes5-GFP transgenic expression is modulated by Hes1 activity
Hes1 and Hes5 mRNA are each widely expressed in the developing nervous system, but the patterns are fairly complementary to one another (Hatakeyama et al., 2004). For example Hes1 is expressed in the anterior neural folds and optic vesicles, whereas Hes5 normally does not appear in the eye until after E10.5 (Hatakeyama et al., 2004; Lee et al., 2005). These differences in spatiotemporal expression are suggestive of compensatory gene activates, which was demonstrated in double and triple Hes mutant embryos (Hatakeyama et al., 2004; Baek et al., 2006). However, Hes genes also suppress one another in particular developmental contexts, including in the optic vesicle, where Hes5 mRNA is upregulated in Hes1−/− embryos (Hatakeyama et al., 2004). Whether this suppression occurs via direct transcriptional repression or indirectly through canonical Notch signaling has not been addressed in vivo. Therefore, we tested the possibility that Hes1 regulates Hes5-GFP BAC Tg expression by crossing this transgene into a Hes1 germline mutant background.
With the caveat that our data are not quantitative, we saw that GFP live fluorescence and anti-GFP staining of fixed sections strongly suggest derepression of transgenic expression in Hes1 mutants (Figs 5A–5C). Antibody labeling of tissue sections verified that there is a complete loss of Hes1 nuclear protein (Figs 5D–5F). Derepression of Hes5-GFP in ocular progenitor cells (lens and retina) persisted through E11.5 (Figs 5G–I) and E12.5 (Figs 5J–5L), when additional eye phenotypes of Hes1 mutants became manifested. These included premature neuronal differentiation, as marked by Tubb3 expression (Fig 5I), a morphologically abnormal optic cup (Fig 5I), ectopic neurons in the RPE (Fig 5I) and lens microphthalmia (Fig 5L) (Tomita et al., 1996; Takatsuka et al., 2004; Lee et al., 2005). Hes1+/− eyes were indistinguishable from wild type littermate controls. These findings are consistent with previous demonstration of Hes5 mRNA upregulation in Hes1 mutant ocular progenitor cells (Hatakeyama et al., 2004). Our data favor the idea that Hes1 normally suppresses Hes5, via a Hes1-responsive element contained in Hes5 noncoding DNA contained within the BAC transgene.
Discussion
The goal of this study was to evaluate a Hes5-GFP BAC transgenic mouse line as a reporter of Hes5 expression in the developing mouse eye. Hes5-GFP expression is first observable in E10.5 ocular sections, in both lens pit and optic vesicle cells. Once lens fiber cells appear, Hes5-GFP expression became largely localized to progenitor cells in the AEL and transition zone, although transient GFP expression in nascent fiber cells was noted. At the initiation of retinal neurogenesis particular subsets of RPCs expressed Hes5-GFP, Hes1, or both. The Hes5-GFP only retinal cells were most often located in the inner retina and coexpressed the RGC marker Pou4f. Yet in the adult retina, Hes5-GFP is specifically expressed by Müller glia (Fig 4 and Nelson et al., 2011). These retinal expression domains are consistent, but not identical, to those of another Hes5-GFP transgene, containing 3KB of genomic DNA from the Hes5 locus (Basak and Taylor, 2007; Nelson et al., 2011). This conventional transgenic line contains 1.6Kb of 5′ and 1.4Kb of intronic and 3′ noncoding DNA driving GFP expression from the endogenous promoter. These DNA segments contain 5 putative Rbpj and 1 Smad binding site, with a single Rbpj consensus site sufficient for all Notch1-inducible activity (Basak and Taylor, 2007). In addition, Hes5 3′ sequences have a negative effect on transgenic expression (Basak and Taylor, 2007). However, it remains unknown if this transgene contains all Hes5 cis-regulatory elements. Here we not only analyzed the ocular expression profile of a Hes5-GFP BAC transgenic mouse line, but compared GFP and Hes1 protein expression on a cell-by cell-basis, during key stages of early eye development.
During eye morphogenesis, Hes5-GFP labeled subsets of lens pit and optic vesicle progenitor cells, which along with Jag1 ligand expression in the optic vesicle (Le et al., 2009), is suggestive of differential Notch signaling. Notch1, Rbpj and Hes1 exhibit uniform mRNA and/or protein expression patterns at this age (Lindsell et al., 1996; Bao and Cepko, 1997; Lee et al., 2005; Riesenberg et al., 2009), and alpha-Cre;Jag1 conditional or Hes5 germline mutants display no optic cup phenotypes (Hojo et al., 2000; Zhou et al., 2013). Therefore, to test the biologic significance of these restricted patterns will necessitate laborious multiple ligand, receptor or effector mutant combinations. Hes1 and Hes5 can act redundantly to suppress neurogenesis, including within the embryonic retina. But, the compensatory mechanisms remains incompletely defined, in large part because the types of cells and developmental ages when one or both are expressed are still unknown. The most stringent method for addressing this issue would be to assess protein colocalization, but a specific Hes5 antibody is not available. Most transgenes are imperfect reporters, largely because their dynamics rarely match the endogenous protein. With this limitation in mind, the Hes5-GFP BAC transgenic mouse displayed relatively rapid changes in gene expression, particularly during early stages of eye development. This reporter was excluded from the developing ciliary body, RPE and optic nerve head, which all express Hes1. We also found that unlike Hes5 mRNA, Hes5-GFP expression persists in differentiating RGCs. This last observation confirms a recent study of contralateral-projecting RGCs, which suggested that the major class of RGCs derive from Hes5 mRNA-expressing progenitor cells (Kuwajima et al., 2017). Finally, we also observed that Hes5-GFP is expressed by a fraction of Atoh7LacZ/+ cells. The Atoh7 retinal lineage gives rise to all 7 cell major cell classes, with ~10% differentiating as RGCs (Brzezinski et al., 2012). Thus it is provocative to think that the GFP/β-gal+ population identifies those progenitors cells differentiating into RGCs.
Basak and Taylor previously demonstrated that Hes5 responsiveness to activated-Notch1 is mediated through a single Rbpj binding site situated near the 5′ promoter (Basak and Taylor, 2007), but the factors required to shut Hes5 expression off are largely unknown. This same study showed that 3′ noncoding sequences reduce Hes5 transcriptional levels, but the underlying basis, for this was not explored further. We found that a loss of Hes1 activity derepresses Hes5-GFP BAC transgenic expression, thereby extending the established idea of Hes1 repression of Hes5, to further support the possibility that it is direct at the Hes5 locus. In the future, it will be interesting to ask whether this regulatory relationship might be mediated by a particular Notch pathway signal, which stimulates Hes1 to block Hes5 transcription and how this correlates with the timing of neuronal and glial development.
Experimental Procedures
Animals
BAC Tg(Hes5-EGFP)CV50Gsat/Mmmh mice (stock strain 000316-MU) were generated in the GENSAT/NINDS Project by recombineering mouse BAC clone BX480 (RP24-341I10) to contain a GFP expression cassette (Gong et al., 2003). The resulting mouse transgenic line was maintained as a hemizygous stock on a CD-1 background, then cryoarchived at the MMRRC. After resuscitation this transgenic line, inserted on the X chromosome, was maintained by mating Hes5-GFP hemizygous females to CD-1 males, and PCR genotyping with recommended primers and protocol (https://www.mmrrc.org/catalog/sds.php?mmrrc_id=316), or by visualizing live GFP fluorescence in adult mouse lenses with a GFP flashlight and Royal Blue filter (NightSea). Transgenic embryos were similarly PCR genotyped and/or scored for live GFP expression using an MZ12 Leica dissection scope, equipped with an LED UV light source and GFP filter set.
Hes1 germline mutant mice (Hes1tm1Fgu) were maintained on an ICR background and genotyped using published protocols (Ishibashi et al., 1995). Atoh7LacZ/+ mice (Math5LacZ = Atoh7tm1Gla) were maintained on a CD-1 background and genotyped using published protocols (Brown et al., 2001). In some experiments, BAC Tg Hes5-GFP hemizyous females were mated to Hes1+/− males, creating compound heterozygous females, which were subsequently timed mated with Hes1+/− male mice for embryonic litters. BAC-Tg Hes5-GFP; Atoh7LacZ/+ embryos were similarly generated from timed matings between BAC Tg Hes5-GFP hemizyous females and Atoh7LacZ/LacZ males.
The date a vaginal plug was noted was assigned as embryonic age E0.5. A minimum of three embryos per age and genotype, from ≥2 litters were analyzed in each experiment. All mice were housed and cared for in accordance with guidelines provided by the National Institutes of Health, Bethesda, MD; Association for Research in Vision and Ophthalmology; and conducted with approval and oversight from the Cincinnati Children’s Hospital Research Foundation and UC Davis Institutional Animal Care and Use Committees.
Immunohistochemical analyses
Embryonic and adult eye tissues were fixed in 4% paraformaldehyde/PBS for 40–60 minutes at 4°C, processed through a sucrose/PBS series and cryoembedded. Antibody labeling of 10 micron cryosections used rat anti-βgal (gift from Tom Glaser, 1:1000)(Saul et al., 2008), rabbit anti-Ccnd1/CyclinD1 (Neomarkers clone SP4 1:100); chick anti-GFP (Invitrogen or Abcam 1:1000), rabbit anti-Hes1 (1:1000)(Lee et al., 2005), goat anti-Jag1 (Santa Cruz Biotechnology C-20 1:1000), rabbit anti-Ki67 (Vector Labs 1:1000), goat anti-pan Pou4f (Santa Cruz sc6026 1:50); rabbit anti-Prox1 (Biolegend/Covance 1:6000), rabbit anti-PAX2 (Biolegend/Covance 1:1000); rat anti-Rbpj (CosmoBio USA Clone T6709 1:100), mouse anti-Rlbp1/CRALBP (Affinity Bioreagents 1:100); rabbit anti-Rxrg (Santa Cruz Biotech 1:200); rabbit anti-Sox9 (Chemicon/Millipore 1:200); rabbit anti-Tubb3 (Biolegend/Covance 1:1000); sheep anti-Vsx2/Chx10 (Exalpha Biologicals N-terminus 1:1000); DAPI staining (Sigma Chemical 1:1000 dilution of 1mg/ml stock). Secondary antibodies were directly conjugated to Alexa Fluor350, Alexa Fluor488, or Alexa Fluor594 (Invitrogen), or were biotinylated (Jackson ImmunoResearch) and then sequentially labeled with Alexa488- or Alexa594-Streptavidin (Jackson ImmunoResearch).
Whole mount or cryosection in situ hybridization were performed as described in (Brown et al., 1998), using a Hes5 cDNA plasmid as a template for a digoxygenin-labeled antisense riboprobe.
Imaging
Digital Tiff images of live whole embryo GFP expression or embryos post whole mount in situ hybridization were captured using a Leica MZ12 dissecting microscope equipped with a UV light source, Optronics Magnafire digital B&W and color cameras and software. Digital Tiff images of embryonic eye cryosections, either post in situ or post- antibody labeling were collected using a Zeiss microscope equipped with color and black and white cameras and an Apotome deconvolution device, Axiovision (v7.0). The Adobe Photoshop (CS4) software program was used to convert Tiff images to Photoshop format and equivalently adjust their contrast.
Key Findings.
Dynamic Hes5-GFP expression during mouse eye development
Hes1 and Hes5-GFP delineate subpopulations of lens and retinal progenitor cells
Hes5-GFP labels embryonic RGCs and their axons
Hes5-GFP is a specifically expressed by adult Müller glia
Acknowledgments
Grant Sponsor:NIH/NEI; Grant Numbers: R01EY013612, R01EY018097
The authors thank Ryiochiro Kageyama for Hes1 germline mutant mice; the Mutant Mouse Regional Resource Center U42OD010918 for BAC Tg(Hes5-EGFP) mice; Kenny Campbell for a Hes5 mouse cDNA plasmid; Richard Lang for use of an Apotome deconvolution imaging system; Ashley Riesenberg for excellent technical assistance; and Tom Glaser, Kate Maurer, Anna LaTorre and Joel Miesfeld for valuable discussion.
References
- Baek JH, Hatakeyama J, Sakamoto S, Ohtsuka T, Kageyama R. Persistent and high levels of Hes1 expression regulate boundary formation in the developing central nervous system. Development. 2006;133:2467–2476. doi: 10.1242/dev.02403. [DOI] [PubMed] [Google Scholar]
- Bao ZZ, Cepko CL. The expression and function of Notch pathway genes in the developing rat eye. J Neurosci. 1997;17:1425–1434. doi: 10.1523/JNEUROSCI.17-04-01425.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basak O, Taylor V. Identification of self-replicating multipotent progenitors in the embryonic nervous system by high Notch activity and Hes5 expression. Eur J Neurosci. 2007;25:1006–1022. doi: 10.1111/j.1460-9568.2007.05370.x. [DOI] [PubMed] [Google Scholar]
- Bernardos RL, Lentz SI, Wolfe MS, Raymond PA. Notch-Delta signaling is required for spatial patterning and Muller glia differentiation in the zebrafish retina. Dev Biol. 2005;278:381–395. doi: 10.1016/j.ydbio.2004.11.018. [DOI] [PubMed] [Google Scholar]
- Brown NL, Kanekar S, Vetter ML, Tucker PK, Gemza DL, Glaser T. Math5 encodes a murine basic helix-loop-helix transcription factor expressed during early stages of retinal neurogenesis. Development. 1998;125:4821–4833. doi: 10.1242/dev.125.23.4821. [DOI] [PubMed] [Google Scholar]
- Brown NL, Patel S, Brzezinski J, Glaser T. Math5 is required for retinal ganglion cell and optic nerve formation. Development. 2001;128:2497–2508. doi: 10.1242/dev.128.13.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzezinski JAt, Prasov L, Glaser T. Math5 defines the ganglion cell competence state in a subpopulation of retinal progenitor cells exiting the cell cycle. Dev Biol. 2012;365:395–413. doi: 10.1016/j.ydbio.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R, Turner DL. Vertebrate hairy and Enhancer of split related proteins: transcriptional repressor regulating celluar differentiation and embryonic patterning. Oncogene. 2001;20:8342–8357. doi: 10.1038/sj.onc.1205094. [DOI] [PubMed] [Google Scholar]
- Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, Nowak NJ, Joyner A, Leblanc G, Hatten ME, Heintz N. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425:917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- Hatakeyama J, Bessho Y, Katoh K, Ookawara S, Fujioka M, Guillemot F, Kageyama R. Hes genes regulate size, shape and histogenesis of the nervous system by control of the timing of neural stem cell differentiation. Development. 2004;131:5539–5550. doi: 10.1242/dev.01436. [DOI] [PubMed] [Google Scholar]
- Hojo M, Ohtsuka T, Hashimoto N, Gradwohl G, Guillemot F, Kageyama R. Glial cell fate specification modulated by the bHLH gene Hes5 in mouse retina. Development. 2000;127:2515–2522. doi: 10.1242/dev.127.12.2515. [DOI] [PubMed] [Google Scholar]
- Ishibashi M, Ang S-L, Shiota K, Nakanishi S, Kageyama R, Guillemot F. Targeted disruption of mammalian hairy and Enhancer of split homolog-1 (HES-1) leads to up-regulation of neural helix-lop-helix factors, premature neurogenesis, and severe neural tube defects. Genes and Development. 1995;9:9136–9148. doi: 10.1101/gad.9.24.3136. [DOI] [PubMed] [Google Scholar]
- Jadhav AP, Roesch K, Cepko CL. Development and neurogenic potential of Muller glial cells in the vertebrate retina. Prog Retin Eye Res. 2009;28:249–262. doi: 10.1016/j.preteyeres.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia J, Lin M, Zhang L, York JP, Zhang P. The Notch signaling pathway controls the size of the ocular lens by directly suppressing p57Kip2 expression. Mol Cell Biol. 2007;27:7236–7247. doi: 10.1128/MCB.00780-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama R, Ohtsuka T, Kobayashi T. The Hes gene family: repressors and oscillators that orchestrate embryogenesis. Development. 2007;134:1243–1251. doi: 10.1242/dev.000786. [DOI] [PubMed] [Google Scholar]
- Kageyama R, Ohtsuka T, Kobayashi T. Roles of Hes genes in neural development. Dev Growth Differ. 2008;50(Suppl 1):S97–103. doi: 10.1111/j.1440-169X.2008.00993.x. [DOI] [PubMed] [Google Scholar]
- Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwajima T, Soares CA, Sitko AA, Lefebvre V, Mason C. SoxC Transcription Factors Promote Contralateral Retinal Ganglion Cell Differentiation and Axon Guidance in the Mouse Visual System. Neuron. 2017;93:1110–1125. e1115. doi: 10.1016/j.neuron.2017.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le TT, Conley KW, Brown NL. Jagged 1 is necessary for normal mouse lens formation. Dev Biol. 2009;328:118–126. doi: 10.1016/j.ydbio.2009.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le TT, Conley KW, Mead TJ, Rowan S, Yutzey KE, Brown NL. Requirements for Jag1-Rbpj mediated Notch signaling during early mouse lens development. Dev Dyn. 2012;241:493–504. doi: 10.1002/dvdy.23739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HY, Wroblewski E, Philips GT, Stair CN, Conley K, Reedy M, Mastick GS, Brown NL. Multiple requirements for Hes 1 during early eye formation. Dev Biol. 2005;284:464–478. doi: 10.1016/j.ydbio.2005.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsell CE, Boulter J, diSibio G, Gossler A, Weinmaster G. Expression patterns of Jagged, Delta1, Notch1, Notch2, and Notch3 genes identify ligand-receptor pairs that may function in neural development. Mol Cell Neurosci. 1996;8:14–27. doi: 10.1006/mcne.1996.0040. [DOI] [PubMed] [Google Scholar]
- Marcucci F, Murcia-Belmonte V, Wang Q, Coca Y, Ferreiro-Galve S, Kuwajima T, Khalid S, Ross ME, Mason C, Herrera E. The Ciliary Margin Zone of the Mammalian Retina Generates Retinal Ganglion Cells. Cell Rep. 2016;17:3153–3164. doi: 10.1016/j.celrep.2016.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer KA, Riesenberg AN, Brown NL. Notch signaling differentially regulates Atoh7 and Neurog2 in the distal mouse retina. Development. 2014 doi: 10.1242/dev.106245. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto A, Iida A, Satoh S, Watanabe S. The group E Sox genes Sox8 and Sox9 are regulated by Notch signaling and are required for Muller glial cell development in mouse retina. Exp Eye Res. 2009;89:549–558. doi: 10.1016/j.exer.2009.05.006. [DOI] [PubMed] [Google Scholar]
- Nelson BR, Gumuscu B, Hartman BH, Reh TA. Notch activity is downregulated just prior to retinal ganglion cell differentiation. Dev Neurosci. 2006;28:128–141. doi: 10.1159/000090759. [DOI] [PubMed] [Google Scholar]
- Nelson BR, Reh TA. Relationship between Delta-like and proneural bHLH genes during chick retinal development. Dev Dyn. 2008;237:1565–1580. doi: 10.1002/dvdy.21550. [DOI] [PubMed] [Google Scholar]
- Nelson BR, Ueki Y, Reardon S, Karl MO, Georgi S, Hartman BH, Lamba DA, Reh TA. Genome-wide analysis of Muller glial differentiation reveals a requirement for Notch signaling in postmitotic cells to maintain the glial fate. PLoS One. 2011;6:e22817. doi: 10.1371/journal.pone.0022817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsawa R, Kageyama R. Regulation of retinal cell fate specification by multiple transcription factors. Brain Res. 2008;1192:90–98. doi: 10.1016/j.brainres.2007.04.014. [DOI] [PubMed] [Google Scholar]
- Ohtsuka T, Ishibashi M, Gradwohl G, Nakanishi S, Guillemot F, Kageyama R. Hes1 and Hes5 as Notch effectors in mammalian neuronal differentiation. The EMBO Journal. 1999;18:2196–2207. doi: 10.1093/emboj/18.8.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riesenberg AN, Liu Z, Kopan R, Brown NL. Rbpj cell autonomous regulation of retinal ganglion cell and cone photoreceptor fates in the mouse retina. J Neurosci. 2009;29:12865–12877. doi: 10.1523/JNEUROSCI.3382-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts MR, Hendrickson A, McGuire CR, Reh TA. Retinoid X receptor (gamma) is necessary to establish the S-opsin gradient in cone photoreceptors of the developing mouse retina. Invest Ophthalmol Vis Sci. 2005;46:2897–2904. doi: 10.1167/iovs.05-0093. [DOI] [PubMed] [Google Scholar]
- Rocha SF, Lopes SS, Gossler A, Henrique D. Dll1 and Dll4 function sequentially in the retina and pV2 domain of the spinal cord to regulate neurogenesis and create cell diversity. Dev Biol. 2009;328:54–65. doi: 10.1016/j.ydbio.2009.01.011. [DOI] [PubMed] [Google Scholar]
- Rowan S, Conley KW, Le TT, Donner AL, Maas RL, Brown NL. Notch signaling regulates growth and differentiation in the mammalian lens. Dev Biol. 2008;321:111–122. doi: 10.1016/j.ydbio.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saravanamuthu SS, Le TT, Gao CY, Cojocaru RI, Pandiyan P, Liu C, Zhang J, Zelenka PS, Brown NL. Conditional ablation of the Notch2 receptor in the ocular lens. Dev Biol. 2012;362:219–229. doi: 10.1016/j.ydbio.2011.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saul SM, Brzezinski JAt, Altschuler RA, Shore SE, Rudolph DD, Kabara LL, Halsey KE, Hufnagel RB, Zhou J, Dolan DF, Glaser T. Math5 expression and function in the central auditory system. Mol Cell Neurosci. 2008;37:153–169. doi: 10.1016/j.mcn.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatsuka K, Hatakeyama J, Bessho Y, Kageyama R. Roles of the bHLH gene Hes1 in retinal morphogenesis. Brain Res. 2004;1004:148–155. doi: 10.1016/j.brainres.2004.01.045. [DOI] [PubMed] [Google Scholar]
- Tomita K, Ishibashi M, Nakahara K, Ang S-L, Nakanishi S, Guillemot F, Kageyama R. Mammalian hairy and Enhancer of split homolog 1 regulates differentiation of retinal neurons and is essential for eye morphogenesis. Neuron. 1996;16:723–734. doi: 10.1016/s0896-6273(00)80093-8. [DOI] [PubMed] [Google Scholar]
- Xiang M, Zhou L, Peng YW, Eddy RL, Shows TB, Nathans J. Brn-3b: a POU domain gene expressed in a subset of retinal ganglion cells. Neuron. 1993;11:689–701. doi: 10.1016/0896-6273(93)90079-7. [DOI] [PubMed] [Google Scholar]
- Yang Z, Ding K, Pan L, Deng M, Gan L. Math5 determines the competence state of retinal ganglion cell progenitors. Dev Biol. 2003;264:240–254. doi: 10.1016/j.ydbio.2003.08.005. [DOI] [PubMed] [Google Scholar]
- Zheng MH, Shi M, Pei Z, Gao F, Han H, Ding YQ. The transcription factor RBP-J is essential for retinal cell differentiation and lamination. Mol Brain. 2009;2:38. doi: 10.1186/1756-6606-2-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Tanzie C, Yan Z, Chen S, Duncan M, Gaudenz K, Li H, Seidel C, Lewis B, Moran A, Libby RT, Kiernan AE, Xie T. Notch2 regulates BMP signaling and epithelial morphogenesis in the ciliary body of the mouse eye. Proc Natl Acad Sci U S A. 2013;110:8966–8971. doi: 10.1073/pnas.1218145110. [DOI] [PMC free article] [PubMed] [Google Scholar]