Abstract
In recent years, RNA has attracted widespread attention as a unique biomaterial with distinct biophysical properties for designing sophisticated architectures in the nanometer scale. RNA is much more versatile in structure and function with higher thermodynamic stability compared to its nucleic acid counterpart DNA. Larger RNA molecules can be viewed as a modular structure built from a combination of many ‘Lego’ building blocks connected via different linker sequences. By exploiting the diversity of RNA motifs and flexibility of structure, varieties of RNA architectures can be fabricated with precise control of shape, size, and stoichiometry. Many structural motifs have been discovered and characterized over the years and the crystal structures of many of these motifs are available for nanoparticle construction. For example, using the flexibility and versatility of RNA structure, RNA triangles, squares, pentagons and hexagons can be constructed from phi29 pRNA-three-way junction (3WJ) building block. This review will focus on 2D RNA triangles, squares and hexamers; 3D and 4D structures built from basic RNA building blocks; and their prospective applications in vivo as imaging or therapeutic agents via specific delivery and targeting. Methods for intracellular cloning and expression of RNA molecules and the in vivo assembly of RNA nanoparticles will also be reviewed.
Graphical/Visual Abstract
The 3WJ motif derived from the packaging RNA of bacteriophage phi29 DNA packaging motor is highly thermodynamically stable. The 3WJ can be tuned to construct RNA triangles and squares. Through intermolecular interaction RNA hexamer can be constructed. The RNA triangular units can be further assembled into RNA 2D triangle, square, pentamer, hexamer, and arrays as well as 3D structures including tetrahedron, prism and dendrimers. The multi-functional RNA nanoparticles have shown enormous potential as delivery vehicles for targeted cancer therapy.
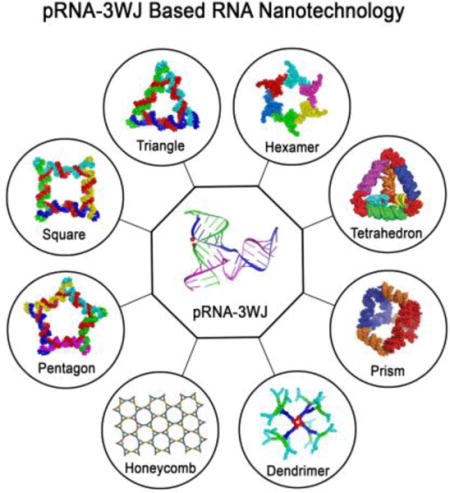
Introduction
A concept now widely accepted is that RNA is thermodynamically more stable than DNA and is much more versatile in structure and function compared to DNA, while in many cases display properties similar to proteins. Many secondary and tertiary structural RNA motifs have been revealed by biochemical, chemical and crystal analysis1,2. Several large structured RNA molecules such as, ribosomal RNA (rRNA) exist in nature, and these RNA complexes can be viewed as sophisticated architectures built from simple building blocks via modular assembly principles3. The diverse folding patterns lead to versatility in structure, which is necessary in order to interact with their specific substrates or targets, proteins, small ligands, DNA or other RNAs4. Genome sequencing revealed that only 1.5% of the human genomic dsDNA codes for proteins5. Subsequent evidence revealed that a substantial part of the rest 98.5% (used to be called “Junk DNA”) actually codes for small and, more recently, long noncoding RNA6. An increasing number of RNA molecules with new functions are being uncovered almost on a daily basis. Two significant hallmarks in drug development have been chemical drugs and protein-based drugs. A third milestone is projected to be RNA as drugs or drugs that target RNA7,8.
RNA nanotechnology involves bottom-up self-assembly of RNA nanoparticles in which the scaffold, targeting ligands, therapeutics agents, regulatory modules, and imaging agents can be exclusively made of RNA8–11. Varieties of RNA structural motifs such as three-way junction (3WJ)11,12, four-way junction (4WJ)13, kink-turn (helix–internal loop–helix motif with a 50° bend in the helical axis)14, hairpins1, pseudoknot (at least two helices and two loops crossing the grooves of the helices)15, C-loops (where helical twists are increased in RNA helices)1, right-angle motifs (internal helical angle of 90°)16, tetraloop-receptors (hairpin motif interacting with a structured internal loop)17, paranemic motifs (crossover motif of stacked helices)18 and kissing loops19,20 have been investigated as scaffolds for constructing compact and stable RNA nanostructures with various shapes and sizes. RNA exhibits phenomenal structural flexibility exemplified by packaging RNA (pRNA)21 derived from bacteriophage phi29 DNA packaging motor. The pRNA has several structural features that can be used as a building block for constructing elegant 2D, 3D, and 4D architectures11,19,22–27. Many crystal structures of other RNA motifs are available and several motifs have been catalogued in databases1,14,15,20,28,29, such as RNAJunction and RNA 3D Motif Atlas from which one can extract detailed information such as, sequence, geometry, and function to rationally design new nanostructures with a high degree of control and predictability for desired applications.
In addition to structural diversity, RNA molecules exhibit diverse functions and immense potential for therapeutics development. RNA aptamers are an emerging class of targeting ligands, which function similar to protein antibodies by binding to their target with high affinity and specificity30,31. When integrating aptamers to RNA nanoparticles, they can guide the RNA nanoparticles to their target receptor molecule overexpressing regions and facilitate cell entry via receptor-mediated endocytosis31,32. Some aptamers can also function as inhibitor of signalling pathways33,34. Ribozymes are RNA molecules that can catalyze biochemical reactions35–37 and thereby serve as anti-viral and anti-cancer therapeutics. Riboswitches are structured RNA molecules that can change their conformation in response to environmental stimuli35,38. Short interfering RNA (siRNA) and microRNA (miRNA) are short RNA fragments typically regulate post-transcriptional gene expression39,40. More non-coding RNAs are being discovered performing various regulatory roles in the cells. All these RNA-based modules have tremendous potentials to serve as novel drugs for disease treatment, but the bottleneck remains efficient delivery into specific diseased cells while minimizing damage to healthy cells and tissues. RNA nanotechnology has potential to overcome many of the challenges, as evidenced by series of publications showing that RNA nanoparticles display favourable attributes from a clinical translation perspective9,22,24,25,41–46: homogenous and controllable in size and shape, which is important from manufacturing view-point; display tunable thermodynamic stability25,26,47, chemical stability (ex. use of 2′-Fluoro modified pyrimidines)11,48 and mechanical stability49, which is unique from a biomaterials physiochemical property standpoint; can be chemically synthesized at large scale and self-assembled with high efficiency from modular units harboring functionalities, which is necessary for quality control for pre-clinical and clinical trials; display favorable biodistribution and pharmacological profiles, which is essential for transitioning to clinic; and finally, composed exclusively of RNA, which can facilitate regulatory approval of the biomaterial for clinical use.
This review aims to highlight the versatility of some of the RNA motifs used for constructing programmable 2D RNA triangles, squares and hexamers as well as 3D RNA prisms, tetrahedrons and dendrimers. The structural features and design parameters are discussed in the context of RNA nanoparticle production. Finally, some of their applications in nanomedicine with an emphasis on in vivo biodistribution and therapeutic delivery studies are discussed to demonstrate the tremendous potentials of RNA nanotechnology platform in the nanomedicine frontier.
TRIANGULAR RNA STRUCTURES CONSTRUCTED USING VARIOUS RNA MOTIFS
pRNA-3WJ based triangles
The pRNA is a naturally occurring RNA structure. Each pRNA monomer contains a 3WJ motif, which is the central domain of pRNA and can be assembled from three short strands ‘a’ (18 nt), ‘b’ (20 nt) and ‘c’ (16 nt). The 3WJ (PDB ID: 4KZ2)50 is composed of three helices: H1, H2 and H3. Helices H1 and H3 are coaxially stacked aligned along a common axis forming ~180° angle. Helices H1 and H2 form a 60° angle. Each helix can be extended by sticky ends and then linked together with a central common strand by hybridization to form a triangle26 (Fig. 1a). The final triangle contains four strands (three external and one internal) and has three 3WJ motifs at the vertices. The internal helical angles are all 60°. The smallest planar triangle is ∼12 nm in dimensions with 8-nt sticky ends that result in a 0° dihedral angle between each 3WJ motifs. The entire construct is resistant to boiling temperature. Moreover, the vertices can be functionalized easily and then used for construction of supramolecular assemblies as well as delivery of therapeutic modules26.
Figure 1. Design and construction of various RNA triangular nanostructures.
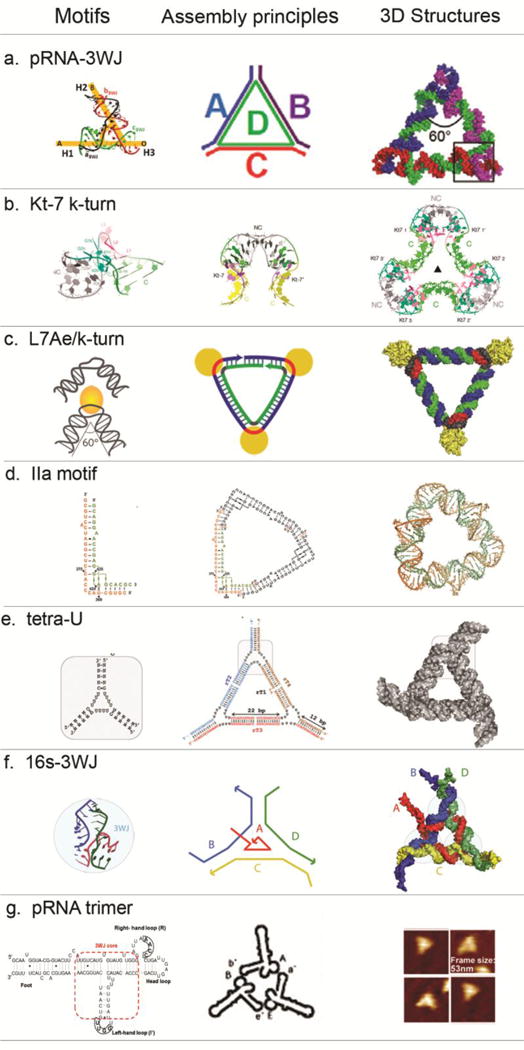
(a) RNA nanotriangle designed and formed from four RNA oligos with pRNA-3WJ motifs at the corners56. Adapted with permission from Ref.56 © 2014 Oxford University Press. (b) RNA nanotriangles designed and formed from two RNA oligos with two k-turn units at each corner (PDB: 4SC1)51. Adapted with permission from Ref.51 © 2016 The Royal Society of Chemistry. (c) RNA-protein complex nanotriangle designed and formed utilizing the interaction of k-turn motif and L7Ae protein52. Adapted with permission from Ref.52 © 2011 Nature Publishing Group. (d) RNA nanotriangles designed and formed from four RNA oligos with either short or long IIa motif (PDB: 4P97, 4PHY)53. Adapted with permission from Ref.53 © 2016 John Wiley & Sons, Inc. (e) RNA nanotriangle designed from three tetra-Us as building unit at the corner. Adapted with permission from Ref.54 © 2017 Elsevier Inc. (f) RNA nanotriangle designed and formed from four RNA oligos with 3WJ motifs (PDB:11836) at the corners15. Adapted with permission from Ref.15 © 2011 American Chemical Society. (g) RNA triangles designed using loop-loop interactions. Three pRNA monomers A-b’, B-e’ and E-a’ form interlocking loops to form the triangle. Adapted with permission from Ref.57 © 2004 American Chemical Society.
Kink-turn motif based triangles
Kink-turn (k-turn) motifs are observed in dsRNA where a short bulge is followed by G•A and A•G base-pairs. A kink is generated with an internal angle of 50° between the two helical axes. Lilley and colleagues developed a crystal lattice guided design of RNA nanoparticles using a double-k-turn motif, whereby two inverted k-turns (Kt-7; PDB: 4CS1) form a horse-shoe like structure by coaxial end-to-end stacking of the RNA helices51. The two loops are separated by 10 base pairs and lie on opposite strands with a two-fold rotational axis. Three two-k-turn units can associate to form a triangle with three-fold rotational symmetry (Fig. 1b). The triangle can also be formed in presence of L7Ae proteins that drive the folding of k-turns51 (Fig. 1c). Similarly, Saito’s group used the box C/D k-turn RNA and ribosomal protein L7Ae to generate synthetic equilateral shaped RNA-protein complex52. The proteins facilitate the formation of triangles by stabilizing the k-turn regions with 60° between the helices. The results demonstrate how versatile k-turn motifs are in nature for organizing long-range architectures and mediating tertiary contacts. Furthermore, RNA-protein complexes provide a new avenue of inducing structural changes (mimicking ribosomes) and for organizing proteins into larger machineries for nanotechnological applications.
Seneca Valley Virus (SVV)-Internal Ribosome Entry Site (IRES) RNA motif based triangles
Hermann and colleagues developed a crystal structure guided principle for self-assembling RNA nano-triangles53. The corner motifs were extracted from ligand-responsive RNA switch motifs located in subdomain IIa of IRES elements of SVV (Fig. 1d). The triangle is composed of three symmetrical identical corner motifs, each forming an inner and outer strand harboring 4-nt sticky end that base-pairs with neighboring corner strands. The overall structure is twisted to accommodate the 90° corner motifs. The edge lengths can be increased to form larger triangles. The 12 termini of the 6 short single strands are located on one face of the triangle and this arrangement offers opportunities for constructing complex shapes as well as functionalization for sensors and nanotechnology applications. Since the motifs are ligand-dependent switches, conformational changes can be also induced to tune the biophysical properties of the triangle.
Tetra-U helix based triangles
Khisamutdinov and colleagues used a computational approach to design an artificial 3WJ structure consisting of three dsRNA helices connected with a bulge of 4×4×4 single-stranded Us54 (Fig. 1e). The four Us provide greater flexibility for assembly of higher-ordered structures. Planar equilateral triangular geometries can be formed with the core 3WJ scaffold at the vertices which are connected by 22 bp duplexes (2 complete RNA turns). The triangles can be functionalized with imaging dyes and siRNA.
Thermus thermophiles 16S rRNA based triangles
Shapiro and colleagues searched through RNAJunction database20 using NanoTiller55 software and identified a 3WJ motif from 16S rRNA that has an intrinsic interhelical angle of ~60°, and two interhelical angles of ~150°15 (Fig. 1f). Triangular shaped RNA nanoparticle was then generated with the 3WJ at each vertex and assembled from four strands.
Hand-in-hand (loop–loop) interactions to produce RNA trimers
Phi29 pRNA contains two domains: a helical domain located at the paired 5′- and 3′- ends58 and an interlocking domain located at the central part of the pRNA structure that binds the connector58–61. The central domain of each pRNA subunit contains a right-hand loop and a left-hand loop, that can be re-engineered as needed to form higher order complexes. The right-hand loop is denoted with an uppercase letter (ex. A) and the left loop with a lowercase letter with a prime (ex. a′). When the two loops interact by complementation, a same letter pair with upper and lower cases respectively, is assigned. For instance, when pRNA A-b′ is mixed with pRNA B-e′ plus pRNA E-a′, hand-in-hand interactions will produce a trimer with A interacting with A′, B interacting with b′ and E interacting with e′ (Fig. 1g).
RNA SQUARE NANOSTRUCTURES CONSTRUCTED USING VERSATILE RNA MOTIFS
pRNA-3WJ based squares
The pRNA-3WJ (PDB ID: 4KZ2)50 scaffold is highly versatile. To form nanostructure with a square geometry, the native pRNA-3WJ internal angle of 60° can be stretched to 90°25. The final square consists of five strands (four short external strands and one complementary long internal strand) (Fig. 2a). The resulting nanoparticle contains a 3WJ at each vertex and A-type duplex RNA along each edge. The linking sequences can be shortened or extended to create squares with tunable sizes. Additional RNA sequences or motifs can be placed at each vertex for desired functionalities.
Figure 2. Design and construction of various RNA square nanostructures.
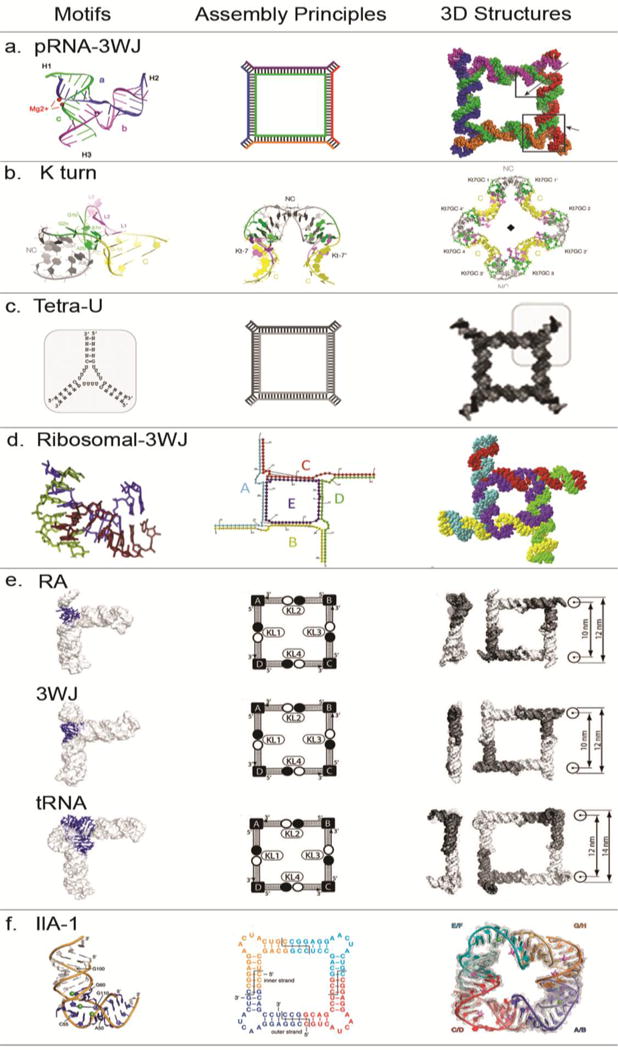
(a) RNA nanosquare designed and formed from five RNA oligos with pRNA-3WJ motifs at the corners25. Adapted with permission from Ref.25 © 2014 American Chemical Society. (b) RNA nanosquares designed and formed from two RNA oligos with two k-turn units at each corner (PDB: 4SC1)51. Adapted with permission from Ref.51 © 2016 The Royal Society of Chemistry. (c) RNA nanosquares designed from three-way junction formed by tetra-Us and helixes as building unit at the corner. Adapted with permission from Ref.54 © 2017 Elsevier Inc. (d) RNA nanosquares designed and formed from five RNA oligos with 3WJ motifs at each corner(PDB, 2OGM)28. Adapted with permission from Ref.28 © 2016 Elsevier. (e) Tectosqaure nanoparticles designed and formed from RA, 3WJ and tRNA motifs16. Adapted with permission from Ref.16 © 2009 American Chemical Society. (f) RNA nanosquare designed and formed from four piece of identical RNA oligos with IIa-1 motifs (PDB:3P59) at the corners62. Adapted with permission from Ref.62 © 2011 National Academy of Sciences.
Kink-turn motif based square
Lilley and colleagues used the same double k-turn motif (Kt-7; PDB: 4CS1) described earlier to form a square shaped geometry through coaxial end-to-end interactions51 (Fig. 2b). A four-fold axis of rotational symmetry is observed through the center of the square. As seen in triangles, the square-shaped structure can also be formed in presence of L7Ae proteins.
Tetra-U helix based squares
The tetra-U helix described earlier is a highly flexible module and can be used a building block to build square geometries by stretching the intra-helical angle to 90°54 (Fig. 2c). The formation of a homogenous RNA band in native PAGE demonstrated that the square is stable. Furthermore, DNA/RNA hybrids can be formed to fine-tune the physiochemical properties of the complexes for controlled release of cargoes.
rRNA derived from bacterial ribosome
Following is an example of building a nanostructure using information available in database. Nanotiler is a software developed by Shapiro and colleagues to build RNA nanostructures55. One of its algorithm is a JunctionScanner than can scan through RNA Junction database20 and extract structural elements, like a 3WJ derived from large ribosomal unit of Deinococcus radiodurans (PDB: 2OGM)63. The core junction consists of three strands of length 4 nt, 5 nt and 7 nt forming approximately a 90° between the helical axes (Fig. 2d). In the final square configuration, the sides of the square are 47 nt in length, while the core strand in 58 nt in length. Each strand forms an intermolecular interaction with the other component strands.
90° RNA motifs derived from cellular translational machineries to generate squares
Jaeger and colleagues used three topologically different 90° motifs exhibiting different thermodynamic, kinetic and folding properties to construct RNA squares16 (Fig. 2e). The first is the RA-motif, a highly prevalent structure in rRNAs. It forms a L-shaped structure by packing two helical stems along their shallow-groove through ribose-zipper interactions. The second is the 3WJ motif derived from 23S rRNA in Haloarcula marismortui. It forms a T-shaped structure, where two helices are co-axially stacked and the third helix protrudes out at 90°. The third is the tRNA-motif derived from tRNA(qSer), hybrid class I/class II tRNA, which consists of 4- or 5-way junctions that form a L-shaped structure. The individual motifs have two kissing loops at two helical branches and these form loop-loop interactions with three other monomeric units to generate square shaped structures.
Hepatitis C virus (HCV) - Internal Ribosome Entry Site (IRES) RNA motif
Herman and colleagues used the 90° bend in the domain IIa bulge in the IRES of HCV genome to generate a double-stranded RNA square62 (Fig. 2f). Molecular modelling revealed that a 100 nt square can be self-assembled from four copies of two distinct strands, an inner strand of 10 nt and an outer strand of 15 nt. Each strand needs a four-base single stranded sticky-end for complementary hybridization. The square was assembled, crystallized and the final structure solved at 2.2 Å resolution. The sides of the square are ~6 nm and the inner cavity was ~1.8 nm. The results demonstrate the feasibility of sequence-dependent programmed self-assembly of structured RNA nanoparticles.
RNA HEXAMER NANOSTRUCTURE CONSTRUCTED USING VERSATILE RNA MOTIFS
Hexameric pRNA ring produced by hand-in-hand interaction of reengineered right and left-hand loops
In nature, the pRNA from bacteriophage phi29 DNA packing motor forms a hexameric ring to gear the motor via hand-in-hand or loop-loop interactions21,64,65,87. The 4-nt loop sequences of the individual monomers can be engineered to form dimer, trimer, and hexamer nanoparticles by bottom-up self-assembly of re-engineered RNA fragments64,65. However, the resulting constructs are not thermodynamically stable for in vivo applications. Stable interlocking loops can be generated by extending the native 4-nt to 7-nt complementary loop sequences19,66 (Fig. 3a). The resulting constructs can be further functionalized with desired functional modules for desired applications19,67. In this approach, all functionalities can be pre-designed in the primary sequence and then assembled, thus ensuring the production of homogeneous nanoparticles.
Figure 3. Design and construction of various RNA hexamer nanostructures.
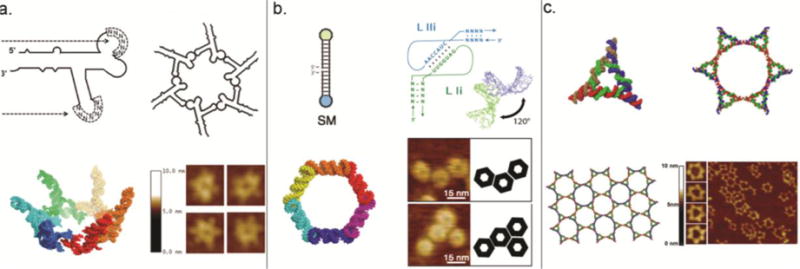
(a) pRNA hexamers designed and formed through intermolecular hand-in-hand interactions among 6 pieces of pRNAs19,21,61,64,65. Adapted with permission from Ref.19 © 2013 RNA Society. (b) RNA hexagonal nano-ring designed and formed through RNAI/II kissing complexes70. Adapted with permission from Ref.70 © 2011 American Chemical Society. (c) RNA hexamers designed and formed through linking six triangles formed from pRNA-3WJ with helixes, constructing a supramolecular pattern resembling honeycombs. Adapted with permission from Ref.26 © 2014 American Chemical Society.
Hexamer nanorings based on RNAI/IIi kissing complexes
Shapiro, Jeager and colleagues used a strategy based on the principle of RNA architectonics68,69, whereby ssRNAs harboring partially folded structural motifs form monomers followed by bottom-up assembly into nanocomplexes utilizing tertiary hydrogen bond formations. Hexamers based on the geometry of the RNAI/IIi kissing-loop complex can be assembled using two approaches70,71. Two analogous RNA monomers, each with a single loop sequence on either end of the central helix, can self-assemble through specific complementary kissing-loop interactions (Fig. 3b). Alternatively, a single monomer designed with both complementary loop sequences can self-assemble with other monomers. However, for complete control over assembly, six specific kissing-loop interactions are required and these monomers need to assemble selectively in concert. Accordingly, six distinct 7-nt sequences were designed computationally and assembled into hexameric rings with high yield70. These hexameric rings can be further generated co-transcriptionally in one pot, and also can be functionalized with siRNAs capable of being processed by Dicer71.
Hexamers formed by sticky end linkage of pRNA-3WJ triangles
The pRNA-3WJ triangles can be functionalized at the vertices with specific sticky ends. A triangular monomer unit is constructed with 6-nt sticky ends denoted A, b’ and C. When mixed in 1:1 ratio with another triangular monomer with 6-nt sticky ends denoted a’, b’ and c’, thermostable hexamers can be generated through sticky-end complementary binding of A-a’ and C-c’26 (Fig. 3c). The b’ corner ends remains open, if needed, for post-assembly functionalization with desired module, such as siRNA.
RNA 3D NANOSTRUCTURES CONSTRUCTED USING VERSATILE RNA MOTIFS
3D RNA nanostructures are particularly attractive for encapsulating drugs, imaging modules and other functionalities for not only shielding the functional modules from in vivo cellular environment, but also for controlled release of the modules at the specific site.
pRNA-based nanocages
Guo and colleagues designed tetrahedrons72 (Fig. 4a) and triangular nanoprisims73 (Fig. 4b) utilizing the pRNA-3WJ as a building block. First, triangular structures were assembled with a 3WJ motif at the vertices, which were then joined by duplexes. A 21 nt single-stranded RNA linker was then added to each vertex to link two triangles face-to-face through complementary hybridization. For added flexibility to generate the 90° bend, four Us were added between the linker sequence and core 3WJ sequence. The entire construct assembles from eight strands and displays a size of ~11 nm. The size is tunable by simply increasing or decreasing the sides of the prism by one helical turn. The nanoprism can encapsulate therapeutic cargoes and also harbor functionalities at the vertices.
Figure 4. Design and construction of various 3D RNA nanostructures.
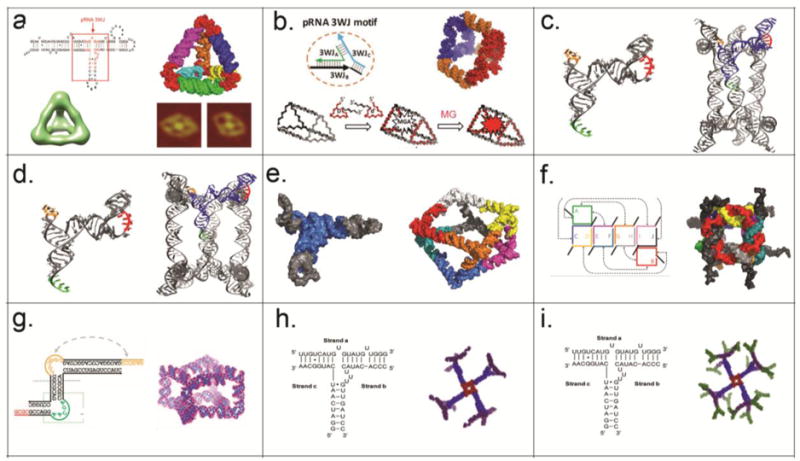
(a) RNA tetrahedron designed and formed by placing pRNA-3WJ at the four corners72 as shown by AFM and cryo-EM. Adapted with permission from Ref.72 © 2016 Wiley VCH Verlag GmbH and Co. KGaA, Weinheim. (b) RNA prism designed and formed by placing pRNA-3WJ at the six corners73. Small RNA molecules, such as MG aptamer can be encapsulated inside the prism. Adapted with permission from Ref.73 © 2016 Wiley VCH Verlag GmbH and Co. KGaA, Weinheim. (c–d) RNA triangular nanoprism I and tetragonal nanoprism II designed and self-assembled by re-engineered pRNA74. Adapted with permission from Ref.74 © 2014 Macmillan Publishers Limited. (e) Polyhedron made of tRNA subunits75. Adapted with permission from Ref.75 © 2010 Macmillan Publishers Limited. (f) Ten stranded RNA cube with dangling ends76. Adapted with permission from Ref.76 © 2010 Macmillan Publishers Limited. (g) Homo-octameric prism designed and formed with an RNA tile with T junction structure78. Adapted with permission from Ref.78 © 2015 Macmillan Publishers Limited. (h–i) RNA dendrimer G3 and G4 designed and formed by placing pRNA-square at the central and extended with multiple pRNA-3WJ structures27. Adapted with permission from Ref.27 © 2015 Elsevier Inc.
In a different approach, Mao and colleagues designed RNA nanoprsims using pRNA monomers as building blocks74. Two approaches, one relying on loop-loop interactions and the other on palindrome sequence mediated sticky-end hybridization was used. Triangular prisms were designed using three pRNA monomers with three different loop sequences (a-b’, b-c’, c-a’) (Fig. 4c) and a four-base palindrome sequence at the 3’-end. Upon annealing, the three complementary loop sequences hybridize (a/a’; b/b’ and c/c’) via loop-loop interactions to form a closed triangle while the sticky ends at the 3’-ends link to each other to connect the triangular faces. Similarly, using the same design principles, a tetragonal prism can be constructed from four pRNA monomers (a-b’, b-c’, c-d’, and d-a’) with a four-base palindrome sequence at the 3’-end (Fig. 4d).
Transfer RNA (tRNA) polyhedrons
Jaeger and colleagues used tRNA as a structural motif and spatial addressability feature to design well defined 3D polyhedral structures75. Class II tRNAs, such as tRNA(Ser) has a 5-way helix junction with three arms extending in the x-, y- and z-dimensions making angles of 120° 70° and 25°, respectively. By employing an inverse folding design strategy, tRNA(Ser) motif can be used to generate a square geometry through loop-loop interactions. Upon addition of a single-stranded sticky-end, two-squares can be connected through four sets of tail-tail interactions to generate a non-uniform square anti-prism (Fig. 4e). Due to the structural constraints imposed by tRNA motif, the squares are twisted 30° relative to each other, and thus the eight triangular sides are non-equilateral. As proof-of-concept, the tRNA motifs can be functionalized with biotin in order to conjugate streptavidin protein, thereby demonstrating its potential to organize molecules with high precision and as a potential carrier of drugs.
RNA cubes designed in silico
In contrast to aforementioned use of known structural elements with pre-folded RNAs, Shapiro and colleagues designed a 3D RNA cube computationally from short strands and then experimentally validated the designs76. The cubes are ~13 nm long, composed of 6 core strands or 10 oligos with single stranded corner linkers amenable for functionalization (Fig. 4f). The cubes can be self-assembled in one pot co-transcriptionally. The thermal stabilities can be fine-tuned by altering the composition of strands with DNA (low Tm) or 2’-F (high Tm). The kinetics of association and tunable thermal and chemical stabilities can be used for conditional activation of multiple functionalities to modulate biological pathways77.
Nanoprisms
Mao and colleagues designed RNA tiles that can self-assemble into a homo-octameric cube78. The RNA tile consists of two strands and contains three helical regions, two internal single-stranded loops and two single stranded tails (Fig. 4g). Three structural features are utilized in the design of the prism structure: a T-shaped junction (artificial), a 90° bend (derived from RNA element in HCV) and sticky-end hybridization. The tiles form homo-tetramers via T-junctions and the squares further homo-dimerize through sticky end cohesion to generate homo-octameric prisms in a cooperative assembly process.
RNA dendrimers
Dendrimers are particularly attractive for delivering multiple types of therapeutics, but several challenges remain for conventional chemical building units: (1) Scaffold assembly involves multistep chemical synthesis and purification which leads to low yield and structural defects for anything bigger than G2 dendrimers; (2) Site-specific functionalization for introducing diverse payloads is extremely challenging requiring several protection and de-protection steps; (3) Current platforms often lack targeting capabilities and rely mainly on EPR effects for tumor delivery; some use protein-based targeting ligands, which in turn, can induce immunogenicity; (4) Organic dendrimers require additional steps, such as cross-linking followed by PEGylation to increase their stability, and reduce cytotoxicity; (5) Organic dendrimers rely on their cationic nature to bind anionic RNAi and thus have difficulty in reaching a homogenous state important for drug quality control as well as in vivo toxicity. (6) Organic dendrimers generally suffer from low water-solubility and high aggregation propensity. Recently Guo and colleagues used pRNA-3WJ as a scaffold to construct G0‒G4 RNA dendrimers with precise control of size (20–50 nm), shape (3D globular) and stoichiometry (3–32 functionalities)27 (Fig. 4h, i). RNA dendrimers harboring imaging, targeting and therapeutic modules can be assembled with high efficiency by mixing the component strands in equimolar ratio. RNA dendrimers exhibit elasticity, which will facilitate trafficking across narrow cavities, as well as processing of RNAi. Compared to other polymers, the degradation of RNA can be timely controlled by adjusting the percent and location of 2'-F nucleotides. RNA dendrimers also retain all the favourable thermodynamic and pharmacological traits of the pRNA-3WJ.
APPLYING THE FLEXIBILITY AND VERSITILITY OF RNA PROPERTY FOR CONSTRUCTING RNA TRIANGLE, SQUARE, PENTAGON BY TUNING INTERIOR RNA 3WJ ANGLE FROM 60° TO 90° OR 108°
A simple method for the design and synthesis of RNA triangle, square, and pentagon have been developed by stretching RNA 3WJ native angle from 60° to 90° and 108°, using the 3WJ of the pRNA from bacteriophage phi29 dsDNA packaging motor56 (Fig. 5). This method for the construction of elegant polygons can be applied to other RNA building blocks including RNA 4-way, 5-way, and other multi-way junctions to construct higher order 3D structures with controllable size, shape and geometry.
Figure 5. Design and construction of RNA triangle, square and pentagon from pRNA-3WJ motif.
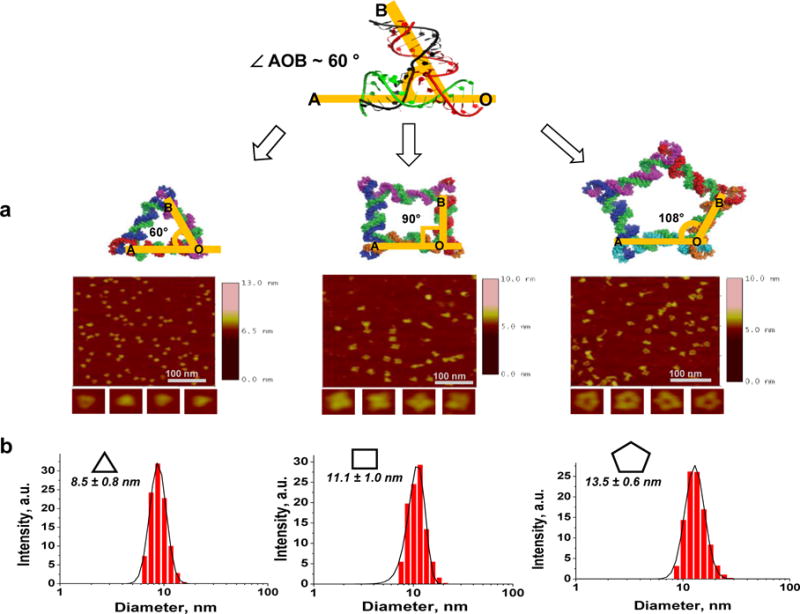
(a) 3D model and Atomic Force Microscopy (AFM) images, and (b) Dynamic Light Scattering (DLS) assay showing the size of RNA nanoparticles56. Adapted with permission from Ref.56 © 2014 Oxford University Press
DIVERSE RNA NANOSTRUCTURES WITH DISEASED CELL TARGETING AND THERAPEUTIC DELIVERY EFFECTS
RNA nanoparticles can be developed into targeted drug delivery systems. RNA based therapeutics (such as siRNA or miRNA) and targeting ligands (such as, aptamers) can be simply incorporated to the RNA nanoparticle scaffold by fusing the sequences. Chemical based ligands (such as, folate or galactose) or chemotherapeutic drugs can be conjugated to the terminal ends of the helices using standard conjugation methods. RNA nanoparticles can be delivered locally, as exemplified by ocular delivery and intra-tumoral delivery or more commonly through systemic injection, as discussed in detail below. By controlling the size, shape and physiochemical properties of RNA nanoparticles, the pharmacological profiles in vivo can be tuned to achieve optimal targeting and therapeutic efficacy.
Ocular delivery of RNA nanoparticles
Li and colleagues demonstrated that delivery of RNA nanoparticles to different ocular tissues (conjunctiva, cornea, retina, and sclera) is size and shape dependent79. Following non-invasive subconjunctival injection, ocular biodistribution was investigated for pRNA-3WJ (three-branched structure) and pRNA-X (four-branched structure extended from pRNA-3WJ) nanoparticles, which are similar in size, but different in shape. The RNA nanoparticles were distributed from the injection site to different ocular tissues and then cleared from the eyes rapidly via the lymph duct. Significant internalization of both the pRNA-3WJ and pRNA-X nanoparticles were observed in conjunctiva and cornea cells, but only the pRNA-X was able to penetrate the retina cells (Fig. 6a).
Figure 6. Application of multifunctional RNA nanoparticles in cancer research.
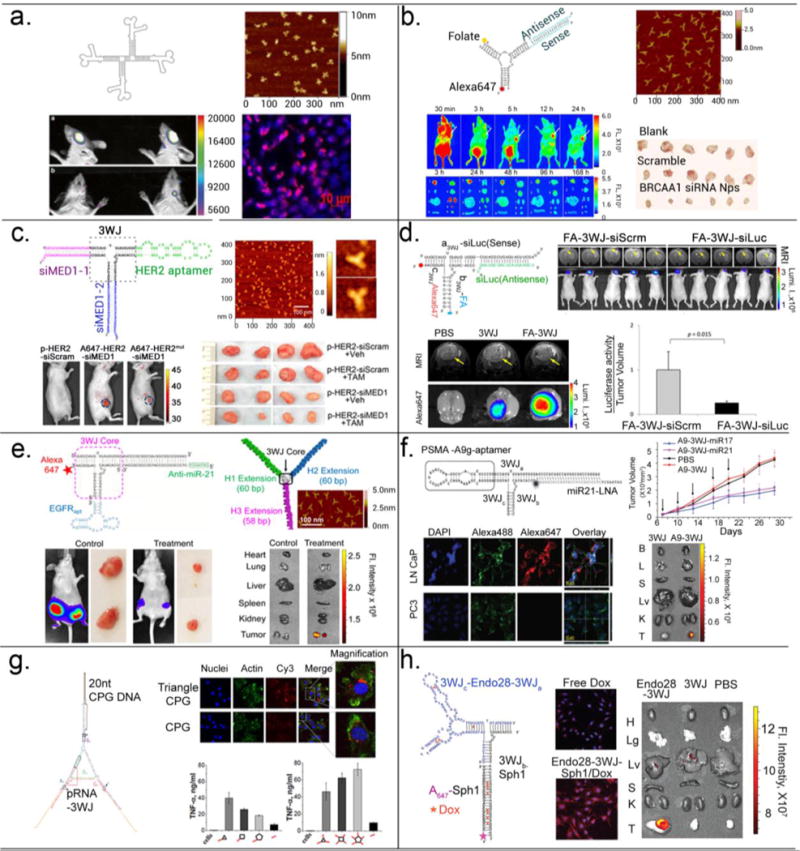
(a) pRNA-X nanoparticle labelled with Alexa647 distributed to retinal cells after subconjunctival injection79. Adapted with permission from Ref.79 © 2014 Springer International Publishing Group. (b) Folate directed delivery of 3WJ-BRCAA1 siRNA to gastric cancers. Tumor inhibition observed in a gastric cancer xenograft mice model after systemic injection46. Adapted with permission from Ref.46 © 2014 Macmillan Publishers Limited. (c) Her2 aptamer directed delivery of MED siRNA to breast cancers and effect in overcoming the tamoxifen resistance of xenograft human breast cancer43. Adapted with permission from Ref.43 © 2016 American Chemical Society. (d) Folate directed delivery of 3WJ-luciferase siRNA to glioblastoma and effect in silencing luciferase gene expression in glioblastoma mice model after systemic injection80. Adapted with permission from Ref.80 © 2015 Impact Journals, LLC. (e) EGFR aptamer directed delivery of 3WJ-anti-miR21 to breast cancer cells and effect in inhibition of cancer growth in orthotopic mice model after systemic injection24. Adapted with permission from Ref.24 © 2015 American Chemical Society. (f) PSMA aptamer directed delivery of 3WJ-anti-miR21 and 3WJ-anti-miR17 to prostate cancer cells and effect in inhibition of cancer growth in mice model after systemic injection41. Adapted with permission from Ref.41 © 2016 The American Society of Gene and Cell Therapy. (g) RNA-CpG polygons induced strong cytokine induction in vivo56. Adapted with permission from Ref.56 © 2014 Oxford University Press. (h) Annexin A2 aptamer directed delivery of 3WJ-doxorubicin to ovarian cancer cells, and its effect in targeting to ovarian cancer in mice model after systemic injection81. Adapted with permission from Ref.81 © 2017 Elsevier Inc.
Systemic delivery of siRNA to treat gastric cancer and breast cancer
Therapeutic pRNA-3WJ nanoparticles were constructed harboring with Folate as a targeting ligand, Alexa-647 as an imaging module and an siRNA targeting BRCAA1 (breast cancer-associated antigen 1) that is over-expressed in gastric cancer. Upon systemic injection, RNA nanoparticles targeted subcutaneous MGC803 xenografts with little or no accumulation in healthy vital organs and tissues (Fig. 6b). The RNA nanoparticles were internalized into the cells via Folate receptor mediated endocytosis. The double-stranded BRCAA1 siRNA region is processed by RISC complex in the cytoplasm resulting in silencing of BRCAA1 gene and subsequent down-regulation of Bcl-2 gene, and further up-regulation of Rb and Bax gene, inducing cell apoptosis and inhibiting tumor growth46. Similaly, Zhang and colleagues used pRNA-3WJ based nanoparticles to tackle tamoxifen-resistant breast cancer43. They constructed pRNA−3WJ-HER2apt (HER2 targeting RNA aptamer)−siMED1 (ER coactivator Mediator Subunit 1) nanoparticle to deliver siRNA to HER2-overexpressing human breast cancer cells. Upon cellular internalization via HER2 aptamer, the siRNA reduced MED1 expression, and significantly decreased ERα-mediated gene transcription. Following i.v. treatment in an orthotopic xenograft model, tumor growth and metastasis were inhibited and the cancer cells become sensitized to tamoxifen (Fig. 6c). In addition, RNA nanoparticles also reduced the cancer stem cell content of breast tumors when combined with tamoxifen treatment in vivo. In a more challenging orthotopic glioblastoma mouse model, pRNA-3WJ nanoparticles harboring folate as a targeting ligand and luciferase siRNA was able to target glioblasoma cells specifically and knockdown luciferase expression after systemic injection80 (Fig. 6d). The results highlight the potential to treat glioblastoma using RNA nanoparticles.
Systemic delivery of anti-miRNA to treat breast, prostate and brain cancer
Targeted delivery of anti-miRNA to knock-down oncogenic miRNA is exemplified by the pRNA-3WJ nanoparticles in three different tumor models. As anti-miRNA module, an 8-mer Locked Nucleic Acid (LNA, conformationally restricted nucleotide analogs) was fused to the pRNA-3WJ scaffold. LNAs are resistant to nucleases and upon binding to their complementary miRNA seed region82 with very high affinity and specificity, they can trigger miRNA inhibition in a dose dependent manner. In an orthotopic Triple Negative Breast Cancer model, pRNA-3WJ nanoparticles harboring EGFR targeting RNA aptamers and anti-miR-21 suppressed tumor growth efficiently without collateral damage to healthy cells24 (Fig. 6e). In a similar manner, pRNA-3WJ nanoparticles were administered systemically to a subcutaneous prostate tumor xenograft bearing mice, pRNA-3WJ nanoparticle harboring PSMA targeting RNA aptamers could deliver anti-miR-21 or anti-miR-17 to inhibit tumor growth at low doses (Fig. 6f). More impressively, in an orthotopic patient-derived glioblastoma model, cancer cell apoptosis and tumor growth regression were observed after systemic injection of pRNA-3WJ/Folate/anti-miR-21 nanoparticles44. The specific knock-down of miR-21 in the aforementioned tumor models were validated by assaying the downstream target genes by RT-PCR at mRNA levels and Western blot at the protein levels.
Local delivery of siRNA using hexameric RNA nanoring
Hexameric RNA nanorings can be functionalized with 6 double-stranded siRNAs (ex. siGFP) at the arms83. Athymic nude mice were subcutaneously grafted with MDA-MB-231 cells stably expressing GFP. For in vivo delivery, the nanorings were associated with bolaamphiphilic cationic carriers and injected intratumorally. The silencing of GFP gene was analyzed by measuring the fluorescent intensities in treated vs control tumors ex vivo. A ~90% decrease in GFP fluorescence intensity was observed for the nanorings indicating that the nanorings can be efficiently processed by Dicer to release the siRNA and trigger gene knockdown.
RNA nanostructures for immunotherapy
The immune responses of RNA nanoparticles is tunable and depends on the sequence, size and shape of the RNA nanoparticle. The pRNA-based nanoparticles by themselves do not induce release of pro-inflammatory cytokines and interferons in nature, which is an attractive feature of a therapeutic delivery vector. Upon incorporating CpG (short sequences containing unmethylated C and G nucleotides) DNA motifs, pRNA nanoparticles can become highly immuno-stimulatory. In both pRNA-polygons56 and pRNA nanoprisms73 fused CpG DNA induced strong induction of TNF-α and IL-6 depending on the shape of polygons and number of CpG per nanoparticle, compared to CpG alone (Fig. 6g). The results demonstrate the potentials of RNA nanoparticles to serve as vaccine adjuvant and as an immunotherapeutic reagent.
RNA nanostructures for chemotherapy
Chemical drugs remain the first line of treatment for cancer. Various nanodrug delivery systems have been explored to reduce its systemic toxicity during treatment. Compared to traditional lipid vesicle based nano-delivery systems, RNA nanoparticles have a great advantage of sub-nanometer scale size controllability, and thus greatly reduce nonspecific accumulation of drugs in liver and lung, to eventually reduce its side effect. Annexin A2-pRNA-3WJ nanoparticles harboring ovarian cancer targeting aptamer can specifically target ovarian cancer xenograft tumor in mice after tail vein injection and showed great potential for targeted doxorubicin delivery with enhanced efficacy81 (Fig. 6h).
Cloning and expression of RNA molecules and assembly of RNA nanoparticles in the cell or in vivo
Cloning and expression of proteins in the cell or in the body have been investigated extensively for many decades. However, there are limited information regarding cloning and expression of RNA molecules in the cell or in vivo. The unusual modularity of phi29 motor pRNA made it possible to fabricate and assemble functional RNA nanoparticles including dimers, trimers, tetramers and hexamers in vitro or in the cell via hand-in-hand interactions by bottom up self-assembly64. The thermostable transfer RNA (tRNA) motif has been used as a scaffold to escort and express varieties of small RNA motifs, including phi29 pRNA, Sephadex aptamer and Streptavidin aptamer84,85.
Instability after expession in the cell and misfolding resulting in loss of function are frequently encountered during constructing of fusion RNA complexes due to changes in energy landscapes and the nearest-neighbour principles. The pRNA-3WJ motif (Fig. 1a) exhibits unusually robust properties as a scaffold with a low folding energy11,47. It can be assembled from three pieces of RNA strands resulting in a structure with high thermodynamic stability, can assemble with high efficiency even in the absence of metal salts, is resistance to denaturation even in the presence of 8 M urea, and does not dissociate at ultra-low concentrations11,50. The motif can provide a leading core and drive the correct folding of other functional RNA molecules fused to the RNA complex with controllable and predictable outcome86. The three component strands of 3WJ motif can be cloned into DNA and displayed as three discontinuous nucleotide fragments. After expression into RNA, the three sections can fold spontaneously into a tight core that promotes the correct folding of other RNA functional groups fused to the RNA sequence. Three individual fragments dispersed at any location within the sequence would allow the other RNA functional modules to fold into their authentic structures with functions, as tested using HBV ribozyme, siRNA, and aptamers of Spinach, Malachite Green (MG), and Streptavidin as model functional groups (Fig. 7). Only 9 complementary nucleotides are required for any two of the three 18-nt 3WJ components. The three 9-bp segments can override other double-stranded segments with more than 15-bp within the fusion RNA. This system enables the production of fusion RNA complexes harbouring multiple functional RNA molecules with correct folding for many potential applications in nanobiotechnology. This system has also been used to investigate the principles governing the folding of RNA molecules in vivo and in vitro. It was found that temporal production of RNA sequences during in vivo transcription caused RNA to fold into different conformations that could not be predicted with the routine principles derived from in vitro studies.
Figure 7. Functional assays of fusion RNA complexes harboring multiple functionalities expressed in vivo.
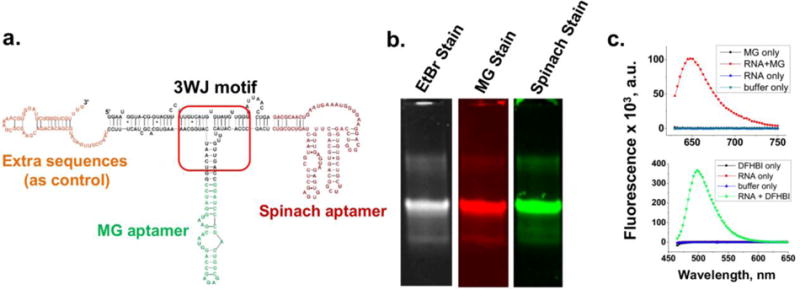
(a) Construction of RNA complex harboring Malachite Green binding and Spinach aptamer. (b) 8% native PAGE verifying the fluorogenic properties of the two RNA aptamers. (c) Fluorescence spectra of MG aptamer (top) and Spinach aptamer (bottom) in solution. Figure reproduced with permission from Ref.86 © 2013 Oxford University Press.
Perspectives
RNA as a biocompatible material is highly advantageous for nanotechnology due to its structural and functional attributes. RNA nanotechnology has provided a new means of fabricating varieties of RNA nanoparticles with exquisite control of size and shape along with tunable thermodynamic, chemical and mechanical properties. The RNA scaffolds can be functionalized with relative ease for RNA-based drugs such as siRNA, anti-miRNA, miRNA, riboswitch and ribozymes, and then self-assembled with high efficiency from modular building blocks and in the process, accelerate the progression of pure RNA-based drugs from the bench to the clinic. The use of different size, shape and physiochemical properties of RNA nanoparticles depends on the desired application and also on attaining optimal pharmacological profile with specific targeting, negligible off-target effects and toxicity, and optimal therapeutic efficacy.
Over the last decade, major challenges have been overcome to a large extent, particularly with regards to thermodynamic instability, chemical instability, toxicity and immunogenicity, but challenges still remain for widespread use of this promising platform in the research community: (1) RNA nanoparticles can get trapped in endosomes following receptor-mediated endocytosis, which can diminish their therapeutic efficacy to some extent. (2) User-friendly software need to be developed for designing RNA nanoparticles from simple set of building blocks for desired applications in a fairly quick time frame. RNA can fold into complex structures mediated by canonical and non-canonical base pairing, base stacking, and tertiary interactions and therefore computational methods for predicting the correct folding of RNA nanoparticles built from inter-molecular interactions are needed. Predicting kinetically trapped undesirable conformations of complex structures is a grand challenge and computational methods to balance kinetic aspects of RNA folding and thermodynamic control mediated by rigid motifs need to be developed. Exploiting the conformational dynamics of topologically equivalent motifs can lend further control of the thermodynamic and kinetic properties of the designed RNA nanoparticles. De novo design of RNA nanoparticles could perhaps offer more versatility in structure and function than what is available in nature for our desired applications and also offer the possibility of building complex RNA machineries. Furthermore, design of synthetic RNA nanostructures that are responsive of physiological stimuli such as protein and ligand binding and conditionally activate is largely an unexplored area. (3) RNA motif databases are relatively much smaller compared to small molecule and protein databases. Non-coding RNAs can adopt complex 3D structures that can possibly be used to build RNA nanomachines mimicking in vivo systems. As more papers in RNA biology and RNA structures are published, these novel structures need to be catalogued with available thermodynamic, kinetics and folding properties. (4) Advances in RNA chemistry are needed for large scale production of RNA strands (modified and longer than 80 bases) by chemical synthesis with high yield. (5) Following large scale synthesis, large-scale purification methods need to be developed. (6) One of the major advantages of using RNA is the feasibility of RNA nanoparticle production in vivo. While some progress has been made in this regard using the thermodynamically stable pRNA-3WJ11,47,86,88–90, endogenous production of programable functional RNA nanoparticles in mammalian cells to modulate biological pathways will be tremendous leap in synthetic biology. (7) CRISPR (Clustered Regularly Interspaced Short Palindromic repeats)- Cas9 has emerged as a versatile genome-editing technology91. RNA nanotechnology can possibly be integrated into the CRISPR tool to develop robust scaffolds for reprograming cells in an efficient manner.
Acknowledgments
The research in P.G.’s lab was supported by NIH Grants R01EB019036 and U01CA207946. The authors declare the following competing financial interest(s): P.G.’s Sylvan G. Frank Endowed Chair position in Pharmaceutics and Drug Delivery is funded by the CM Chen Foundation. P.G. is a consultant of Oxford Nanopore Technologies and Nanobio Delivery Pharmaceutical Co. Ltd. He is also the co-founder of Shenzhen P&Z Biomedical Co. Ltd and its subsidiary P&Z Biological Technology. F.H., F.P., Z.Z., S.G., H.H, and H.Y. work for Nanobio Delivery Pharmaceutical Company specializing in RNA Nanoparticles. The content is solely the responsibility of the authors and does not represent the official views of NIH.
Footnotes
The pRNA-3WJ is a thermodynamically stable motif, which was used to construct multiple structure 2D, 3D and 4D RNA nanostructures. The RNA triangle, square, hexamer, dendrimer, tetrahedron, and prism were constructed with pRNA-3WJ motif and discussed in this review.11,47,50,88–90
The pRNA-3WJ motif is an ideal scaffold to serve as a targeted drug delivery platform for clinical applications. RNA nanoparticles are negatively charged, which can reduce nonspecific binding to negatively charged cell membrane; they can be prepared by controlled synthesis with defined structure and stoichiometry; their multivalent nature provides a tool for combining therapy with targeting, delivery and detection functions. Targeted drug delivery could enhance therapeutic effects while minimizing side effects and toxicity.11,22–27,41–46
References
- 1.Leontis NB, Lescoute A, Westhof E. The building blocks and motifs of RNA architecture. Curr Opin Struct Biol. 2006;16:279–287. doi: 10.1016/j.sbi.2006.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leontis NB, Westhof E. Analysis of RNA motifs. Curr Opin Struct Biol. 2003;13(3):300–308. doi: 10.1016/s0959-440x(03)00076-9. [DOI] [PubMed] [Google Scholar]
- 3.Cruz JA, Westhof E. The Dynamic Landscapes of RNA Architecture. Cell. 2009;136(4):604–609. doi: 10.1016/j.cell.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 4.Brion P, Westhof E. Hierarchy and dynamics of RNA folding. Ann Rev Biophys Biomol Struct. 1997;26:113–137. doi: 10.1146/annurev.biophys.26.1.113. [DOI] [PubMed] [Google Scholar]
- 5.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 6.Claverie JM. Fewer genes, more noncoding RNA. Science. 2005;309(5740):1529–1530. doi: 10.1126/science.1116800. [DOI] [PubMed] [Google Scholar]
- 7.Shu Y, Pi F, Sharma A, Rajabi M, Haque F, Shu D, Leggas M, Evers BM, Guo P. Stable RNA nanoparticles as potential new generation drugs for cancer therapy. Adv Drug Deliv Rev. 2014;66C:74–89. doi: 10.1016/j.addr.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jasinski D, Haque F, Binzel DW, Guo P. Advancement of the Emerging Field of RNA Nanotechnology. ACS Nano. 2017;11(2):1142–1164. doi: 10.1021/acsnano.6b05737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo P. The emerging field of RNA nanotechnology. Nature Nanotechnology. 2010;5(12):833–842. doi: 10.1038/nnano.2010.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo P, Haque F, editors. RNA Nanotechnology and Therapeutics. CRC Press; 2013. pp. 1–593. [Google Scholar]
- 11.Shu D, Shu Y, Haque F, Abdelmawla S, Guo P. Thermodynamically stable RNA three-way junctions for constructing multifuntional nanoparticles for delivery of therapeutics. Nat Nanotechnol. 2011;6:658–667. doi: 10.1038/nnano.2011.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lescoute A, Westhof E. Topology of three-way junctions in folded RNAs. RNA. 2006;12(1):83–93. doi: 10.1261/rna.2208106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laing C, Schlick T. Analysis of four-way junctions in RNA structures. J Mol Biol. 2009;390(3):547–559. doi: 10.1016/j.jmb.2009.04.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schroeder KT, McPhee SA, Ouellet J, Lilley DM. A structural database for k-turn motifs in RNA. RNA. 2010;16(8):1463–1468. doi: 10.1261/rna.2207910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bindewald E, Afonin K, Jaeger L, Shapiro BA. Multistrand RNA secondary structure prediction and nanostructure design including pseudoknots. ACS Nano. 2011;5(12):9542–9551. doi: 10.1021/nn202666w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Severcan I, Geary C, V E, C A, Jaeger L. Square-shaped RNA particles from different RNA folds. Nano Lett. 2009;9:1270–1277. doi: 10.1021/nl900261h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Afonin KA, Lin YP, Calkins ER, Jaeger L. Attenuation of loop-receptor interactions with pseudoknot formation. Nucleic Acids Res. 2012;40(5):2168–2180. doi: 10.1093/nar/gkr926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Afonin KA, Cieply DJ, Leontis NB. Specific RNA self-assembly with minimal paranemic motifs. J Am Chem Soc. 2008;130:93–102. doi: 10.1021/ja071516m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shu Y, Haque F, Shu D, Li W, Zhu Z, Kotb M, Lyubchenko Y, Guo P. Fabrication of 14 Different RNA Nanoparticles for Specific Tumor Targeting without Accumulation in Normal Organs. RNA. 2013;19:766–777. doi: 10.1261/rna.037002.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bindewald E, Hayes R, Yingling YG, Kasprzak W, Shapiro BA. RNAJunction: a database of RNA junctions and kissing loops for three-dimensional structural analysis and nanodesign. Nucleic Acids Res. 2008;36:D392–D397. doi: 10.1093/nar/gkm842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo P, Erickson S, Anderson D. A small viral RNA is required for in vitro packaging of bacteriophage phi29 DNA. Science. 1987;236:690–694. doi: 10.1126/science.3107124. [DOI] [PubMed] [Google Scholar]
- 22.Haque F, Shu D, Shu Y, Shlyakhtenko L, Rychahou P, Evers M, Guo P. Ultrastable synergistic tetravalent RNA nanoparticles for targeting to cancers. Nano Today. 2012;7:245–257. doi: 10.1016/j.nantod.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo P, Haque F. RNA Nanotechnology and Therapeutics—Methods in Molecular Biology. Vol. 1297. New York: Human Press; 2015. pp. 1–239. [DOI] [PubMed] [Google Scholar]
- 24.Shu D, Li H, Shu Y, Xiong G, Carson WE, Haque F, Xu R, Guo P. Systemic delivery of anti-miRNA for suppression of triple negative breast cancer utilizing RNA nanotechnology. ACS Nano. 2015;9:9731–9740. doi: 10.1021/acsnano.5b02471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jasinski D, Khisamutdinov EF, Lyubchenko YL, Guo P. Physicochemically Tunable Poly-Functionalized RNA Square Architecture with Fluorogenic and Ribozymatic Properties. ACS Nano. 2014;8:7620–7629. doi: 10.1021/nn502160s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khisamutdinov EF, Jasinski DL, Guo P. RNA as a boiling-resistant anionic polymer material to build robust structures with defined shape and stoichiometry. ACS Nano. 2014;8:4771–4781. doi: 10.1021/nn5006254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharma A, Haque F, Pi F, Shlyakhtenko L, Evers BM, Guo P. Controllable Self-assembly of RNA Dendrimers. Nanomedicine-Nanotechnology Biology and Medicine. 2015;12:835–844. doi: 10.1016/j.nano.2015.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parlea L, Bindewald E, Sharan R, Bartlett N, Moriarty D, Oliver J, Afonin KA, Shapiro BA. Ring Catalog: A resource for designing self-assembling RNA nanostructures. Methods. 2016;103:128–137. doi: 10.1016/j.ymeth.2016.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sarver M, Zirbel CL, Stombaugh J, Mokdad A, Leontis NB. FR3D: finding local and composite recurrent structural motifs in RNA 3D structures. Journal of Mathematical Biology. 2008;56(1–2):215–252. doi: 10.1007/s00285-007-0110-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bunka DH, Stockley PG. Aptamers come of age - at last. Nat Rev Microbiol. 2006;4(8):588–596. doi: 10.1038/nrmicro1458. [DOI] [PubMed] [Google Scholar]
- 31.Zhu H, Li J, Zhang XB, Ye M, Tan W. Nucleic acid aptamer-mediated drug delivery for targeted cancer therapy. ChemMedChem. 2015;10(1):39–45. doi: 10.1002/cmdc.201402312. [DOI] [PubMed] [Google Scholar]
- 32.Zhou J, Bobbin ML, Burnett JC, Rossi JJ. Current progress of RNA aptamer-based therapeutics. Front Genet. 2012;3:234. doi: 10.3389/fgene.2012.00234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keefe AD, Pai S, Ellington A. Aptamers as therapeutics. Nat Rev Drug Discov. 2010;9(7):537–550. doi: 10.1038/nrd3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sundaram P, Kurniawan H, Byrne ME, Wower J. Therapeutic RNA aptamers in clinical trials. Eur J Pharm Sci. 2013;48(1–2):259–271. doi: 10.1016/j.ejps.2012.10.014. [DOI] [PubMed] [Google Scholar]
- 35.Mulhbacher J, St-Pierre P, Lafontaine DA. Therapeutic applications of ribozymes and riboswitches. Curr Opin Pharmacol. 2010;10(5):551–556. doi: 10.1016/j.coph.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 36.Strobel SA, Cochrane JC. RNA catalysis: ribozymes, ribosomes, and riboswitches. Curr Opin Chem Biol. 2007;11(6):636–643. doi: 10.1016/j.cbpa.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scott WG. Ribozymes. Curr Opin Struct Biol. 2007;17(3):280–286. doi: 10.1016/j.sbi.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 38.Tucker BJ, Breaker RR. Riboswitches as versatile gene control elements. Curr Opin Struct Biol. 2005;15:342–348. doi: 10.1016/j.sbi.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 39.Hannon GJ. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 40.Pecot CV, Calin GA, Coleman RL, Lopez-Berestein G, Sood AK. RNA interference in the clinic: challenges and future directions. Nat Rev Cancer. 2011;11(1):59–67. doi: 10.1038/nrc2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Binzel D, Shu Y, Li H, Sun M, Zhang Q, Shu D, Guo B, Guo P. Specific Delivery of MiRNA for High Efficient Inhibition of Prostate Cancer by RNA Nanotechnology. Molecular Therapy. 2016;24:1267–1277. doi: 10.1038/mt.2016.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shu Y, Cinier M, Shu D, Guo P. Assembly of multifunctional phi29 pRNA nanoparticles for specific delivery of siRNA and other therapeutics to targeted cells. Methods. 2011;54:204–214. doi: 10.1016/j.ymeth.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y, Leonard M, Shu Y, Yang Y, Shu D, Guo P, Zhang X. Overcoming Tamoxifen Resistance of Human Breast Cancer by Targeted Gene Silencing Using Multifunctional pRNA Nanoparticles. ACS Nano. 2017;11(1):335–346. doi: 10.1021/acsnano.6b05910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee TJ, Yoo JY, Shu D, Li H, Zhang J, Yu JG, Jaime-Ramirez AC, Acunzo M, Romano G, Cui R, Sun HL, Luo Z, Old M, Kaur B, Guo P, Croce CM. RNA Nanoparticle-Based Targeted Therapy for Glioblastoma through Inhibition of Oncogenic miR-21. Mol Ther. 2017 doi: 10.1016/j.ymthe.2016.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee TJ, Haque F, Vieweger M, Yoo JY, Kaur B, Guo P, Croce CM. Functional assays for specific targeting and delivery of RNA nanoparticles to brain tumor. Methods Mol Biol. 2015;1297:137–152. doi: 10.1007/978-1-4939-2562-9_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cui D, Zhang C, Liu B, Shu Y, Du T, Shu D, Wang K, Dai F, Liu Y, Li C, Pan F, Yang Y, Ni J, Li H, Brand-Saberi B, Guo P. Regression of gastric cancer by systemic injection of RNA nanoparticles carrying both ligand and siRNA. Scientific reports. 2015;5:10726. doi: 10.1038/srep10726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Binzel DW, Khisamutdinov EF, Guo P. Entropy-driven one-step formation of Phi29 pRNA 3WJ from three RNA fragments. Biochemistry. 2014;53(14):2221–2231. doi: 10.1021/bi4017022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu J, Guo S, Cinier M, Shlyakhtenko LS, Shu Y, Chen C, Shen G, Guo P. Fabrication of stable and RNase-resistant RNA nanoparticles active in gearing the nanomotors for viral DNA packaging. ACS Nano. 2011;5(1):237–246. doi: 10.1021/nn1024658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu Z, Sun Y, Weber JK, Cao Y, Wang W, Jasinski D, Guo P, Zhou R, Li J. Directional mechanical stability of Bacteriophage phi29 motor's 3WJ-pRNA: Extraordinary robustness along portal axis. Sci Adv. 2017;3(5):e1601684. doi: 10.1126/sciadv.1601684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang H, Endrizzi JA, Shu Y, Haque F, Sauter C, Shlyakhtenko LS, Lyubchenko Y, Guo P, Chi YI. Crystal Structure of 3WJ Core Revealing Divalent Ion-promoted Thermostability and Assembly of the Phi29 Hexameric Motor pRNA. RNA. 2013;19:1226–1237. doi: 10.1261/rna.037077.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang L, Lilley DM. A quasi-cyclic RNA nano-scale molecular object constructed using kink turns. Nanoscale. 2016;8(33):15189–15195. doi: 10.1039/c6nr05186c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ohno H, Kobayashi T, Kabata R, Endo K, Iwasa T, Yoshimura SH, Takeyasu K, Inoue T, Saito H. Synthetic RNA-protein complex shaped like an equilateral triangle. Nat Nanotechnol. 2011;6(2):116–120. doi: 10.1038/nnano.2010.268. [DOI] [PubMed] [Google Scholar]
- 53.Boerneke MA, Dibrov SM, Hermann T. Crystal-Structure-Guided Design of Self-Assembling RNA Nanotriangles. Angew Chem Int Ed Engl. 2016;55(12):4097–4100. doi: 10.1002/anie.201600233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bui MN, Brittany JM, Viard M, Satterwhite E, Martins AN, Li Z, Marriott I, Afonin KA, Khisamutdinov EF. Versatile RNA tetra-U helix linking motif as a toolkit for nucleic acid nanotechnology. Nanomedicine. 2017;13(3):1137–1146. doi: 10.1016/j.nano.2016.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bindewald E, Grunewald C, Boyle B, O'Connor M, Shapiro B. A Computational strategies for the automated design of RNA nanoscale structures from building blocks using NanoTiler. Journal of Molecular Graphics & Modelling. 2008;27(3):299–308. doi: 10.1016/j.jmgm.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Khisamutdinov E, Li H, Jasinski D, Chen J, Fu J, Guo P. Enhancing immunomodulation on innate immunity by shape transition among RNA triangle, square, and pentagon nanovehicles. Nucleic Acids Res. 2014;42:9996–10004. doi: 10.1093/nar/gku516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shu D, Moll WD, Deng Z, Mao C, Guo P. Bottom-up assembly of RNA arrays and superstructures as potential parts in nanotechnology. Nano Lett. 2004;4:1717–1723. doi: 10.1021/nl0494497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang CL, Lee C-S, Guo P. The proximate 5' and 3' ends of the 120-base viral RNA (pRNA) are crucial for the packaging of bacteriophage f29 DNA. Virology. 1994;201:77–85. doi: 10.1006/viro.1994.1267. [DOI] [PubMed] [Google Scholar]
- 59.Reid RJD, Bodley JW, Anderson D. Characterization of the prohead-pRNA interaction of bacteriophage phi29. J Biol Chem. 1994;269:5157–5162. [PubMed] [Google Scholar]
- 60.Zhang CL, Trottier M, Guo PX. Circularly permuted viral pRNA active and specific in the packaging of bacteriophage Phi29 DNA. Virology. 1995;207:442–451. doi: 10.1006/viro.1995.1103. [DOI] [PubMed] [Google Scholar]
- 61.Chen C, Sheng S, Shao Z, Guo P. A dimer as a building block in assembling RNA: A hexamer that gears bacterial virus phi29 DNA-translocating machinery. J Biol Chem. 2000;275(23):17510–17516. doi: 10.1074/jbc.M909662199. [DOI] [PubMed] [Google Scholar]
- 62.Dibrov SM, McLean J, Parsons J, Hermann T. Self-assembling RNA square. Proc Natl Acad Sci USA. 2011;108(16):6405–6408. doi: 10.1073/pnas.1017999108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Davidovich C, Bashan A, uerbach-Nevo T, Yaggie RD, Gontarek RR, Yonath A. Induced-fit tightens pleuromutilins binding to ribosomes and remote interactions enable their selectivity. Proc Natl Acad Sci USA. 2007;104(11):4291–4296. doi: 10.1073/pnas.0700041104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guo P, Zhang C, Chen C, Trottier M, Garver K. Inter-RNA interaction of phage phi29 pRNA to form a hexameric complex for viral DNA transportation. Mol Cell. 1998;2:149–155. doi: 10.1016/s1097-2765(00)80124-0. [DOI] [PubMed] [Google Scholar]
- 65.Chen C, Zhang C, Guo P. Sequence requirement for hand-in-hand interaction in formation of pRNA dimers and hexamers to gear phi29 DNA translocation motor. RNA. 1999;5:805–818. doi: 10.1017/s1355838299990350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shu Y, Shu D, Haque F, Guo P. Fabrication of pRNA nanoparticles to deliver therapeutic RNAs and bioactive compounds into tumor cells. Nat Protoc. 2013;8(9):1635–1659. doi: 10.1038/nprot.2013.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guo S, Tschammer N, Mohammed S, Guo P. Specific delivery of therapeutic RNAs to cancer cells via the dimerization mechanism of phi29 motor pRNA. Hum Gene Ther. 2005;16:1097–1109. doi: 10.1089/hum.2005.16.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chworos A, Severcan I, Koyfman AY, Weinkam P, Oroudjev E, Hansma HG, Jaeger L. Building programmable jigsaw puzzles with RNA. Science. 2004;306(5704):2068–2072. doi: 10.1126/science.1104686. [DOI] [PubMed] [Google Scholar]
- 69.Afonin KA, Danilov EO, Novikova IV, Leontis NB. TokenRNA: A new type of sequence-specific, label-free fluorescent biosensor for folded RNA molecules. Chembiochem. 2008;9:1902–1905. doi: 10.1002/cbic.200800183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Grabow WW, Zakrevsky P, Afonin KA, Chworos A, Shapiro BA, Jaeger L. Self-Assembling RNA Nanorings Based on RNAI/II Inverse Kissing Complexes. Nano Lett. 2011;11(2):878–887. doi: 10.1021/nl104271s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Afonin KA, Kireeva M, Grabow WW, Kashlev M, Jaeger L, Shapiro BA. Co-transcriptional assembly of chemically modified RNA nanoparticles functionalized with siRNAs. Nano Lett. 2012;12(10):5192–5195. doi: 10.1021/nl302302e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li H, Zhang K, Pi F, Guo S, Shlyakhtenko L, Chiu W, Shu D, Guo P. Controllable Self-Assembly of RNA Tetrahedrons with Precise Shape and Size for Cancer Targeting. Adv Mater. 2016;28:7501–7507. doi: 10.1002/adma.201601976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Khisamutdinov EF, Jasinski DL, Li H, Zhang K, Chiu W, Guo P. Fabrication of RNA 3D Nanoprism for Loading and Protection of Small RNAs and Model Drugs. Advanced Materials. 2016;28:100079–100087. doi: 10.1002/adma.201603180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hao C, Li X, Tian C, Jiang W, Wang G, Mao C. Construction of RNA nanocages by re-engineering the packaging RNA of Phi29 bacteriophage. Nat Commun. 2014;5:3890. doi: 10.1038/ncomms4890. [DOI] [PubMed] [Google Scholar]
- 75.Severcan I, Geary C, Chworos A, Voss N, Jacovetty E, Jaeger L. A polyhedron made of tRNAs. Nat Chem. 2010;2(9):772–779. doi: 10.1038/nchem.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Afonin KA, Bindewald E, Yaghoubian AJ, Voss N, Jacovetty E, Shapiro BA, Jaeger L. In vitro assembly of cubic RNA-based scaffolds designed in silico. Nat Nanotechnol. 2010;5(9):676–682. doi: 10.1038/nnano.2010.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Halman JR, Satterwhite E, Roark B, Chandler M, Viard M, Ivanina A, Bindewald E, Kasprzak WK, Panigaj M, Bui MN, Lu JS, Miller J, Khisamutdinov EF, Shapiro BA, Dobrovolskaia MA, Afonin KA. Functionally-interdependent shape-switching nanoparticles with controllable properties. Nucleic Acids Res. 2017;45(4):2210–2220. doi: 10.1093/nar/gkx008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yu JW, Liu ZY, Jiang W, Wang GS, Mao CD. De novo design of an RNA tile that self-assembles into a homo-octameric nanoprism. Nature Communications. 2015;6:5724–5729. doi: 10.1038/ncomms6724. [DOI] [PubMed] [Google Scholar]
- 79.Feng L, Li SK, Liu H, Liu CY, LaSance K, Haque F, Shu D, Guo P. Ocular delivery of pRNA nanoparticles: distribution and clearance after subconjunctival injection. Pharmaceutical Research. 2014;31:1046–1058. doi: 10.1007/s11095-013-1226-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lee TJ, Haque F, Shu D, Yoo JY, Li H, Yokel RA, Horbinski C, Kim TH, Kim S-H, Nakano I, Kaur B, Croce CM, Guo P. RNA nanoparticles as a vector for targeted siRNA delivery into glioblastoma mouse model. Oncotarget. 2015;6:14766–14776. doi: 10.18632/oncotarget.3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pi F, Zhang H, Li H, Thiviyanathan V, Gorenstein DG, Sood AK, Guo P. RNA nanoparticles harboring annexin A2 aptamer can target ovarian cancer for tumor-specific doxorubicin delivery. Nanomedicine. 2016;13(3):1183–1193. doi: 10.1016/j.nano.2016.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Obad S, dos Santos CO, Petri A, Heidenblad M, Broom O, Ruse C, Fu C, Lindow M, Stenvang J, Straarup EM, Hansen HF, Koch T, Pappin D, Hannon GJ, Kauppinen S. Silencing of microRNA families by seed-targeting tiny LNAs. Nat Genet. 2011;43(4):371–378. doi: 10.1038/ng.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Afonin KA, Viard M, Koyfman AY, Martins AN, Kasprzak WK, Panigaj M, Desai R, Santhanam A, Grabow WW, Jaeger L, Heldman E, Reiser J, Chiu W, Freed EO, Shapiro BA. Multifunctional RNA nanoparticles. Nano Lett. 2014;14(10):5662–5671. doi: 10.1021/nl502385k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ponchon L, Dardel F. Large scale expression and purification of recombinant RNA in Escherichia coli. Methods. 2011;54:267–273. doi: 10.1016/j.ymeth.2011.02.007. [DOI] [PubMed] [Google Scholar]
- 85.Ponchon L, Dardel F. Recombinant RNA technology: the tRNA scaffold. Nat Methods. 2007;4(7):571–576. doi: 10.1038/nmeth1058. [DOI] [PubMed] [Google Scholar]
- 86.Shu D, Khisamutdinov E, Zhang L, Guo P. Programmable folding of fusion RNA complex driven by the 3WJ motif of phi29 motor pRNA. Nucleic Acids Res. 2013;42:e10. doi: 10.1093/nar/gkt885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jones CP, FerreDAmare AR. RNA quaternary structure and global symmetry. Trends in Biochemical Sciences. 2015;40(4):211–220. doi: 10.1016/j.tibs.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Binzel DW, Khisamutdinov E, Vieweger M, Ortega J, Li J, Guo P. Mechanism of three-component collision to produce ultrastable pRNA three-way junction of Phi29 DNA-packaging motor by kinetic assessment. RNA. 2016 Nov;22(11):1710–1718. doi: 10.1261/rna.057646.116. Epub 2016 Sep 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hao Y, Kieft JS. Three-way junction conformation dictates self-association of phage packaging RNAs. RNA Biol. 2016;13:635–45. doi: 10.1080/15476286.2016.1190075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hill AC, Schroeder SJ. Thermodynamic stabilities of three-way junction nanomotifs in prohead RNA. RNA. 2017;23:521–529. doi: 10.1261/rna.059220.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Charpentier E, Doudna JA. Biotechnology: Rewriting a genome. Nature. 2013;495(7439):50–51. doi: 10.1038/495050a. [DOI] [PubMed] [Google Scholar]


