Abstract
A comprehensive study on characterization and genetic diversity analysis was carried out in 16 ‘Ogura’-based ‘CMS’ lines of cabbage using 14 agro-morphological traits and 29 SSR markers. Agro-morphological characterization depicted considerable variations for different horticultural traits studied. The genotype, ZHA-2, performed better for most of the economically important quantitative traits. Further, gross head weight (0.76), head length (0.60) and head width (0.83) revealed significant positive correlation with net head weight. Dendrogram based on 10 quantitative traits exhibited considerable diversity among different CMS lines and principle component analysis (PCA) indicated that net and gross head weight, and head length and width are the main components of divergence between 16 CMS lines of cabbage. In molecular study, a total of 58 alleles were amplified by 29 SSR primers, averaging to 2.0 alleles in each locus. High mean values of Shannon’s Information index (0.62), expected (0.45) and observed (0.32) heterozygosity and polymorphic information content (0.35) depicted substantial polymorphism. Dendrogram based on Jaccard’s similarity coefficient constructed two major groups and eight sub-groups, which revealed substantial diversity among different CMS lines. In overall, based on agro-morphological and molecular studies genotype RRMA, ZHA-2 and RCA were found most divergent. Hence, they have immense potential in future breeding programs for the high-yielding hybrid development in cabbage.
Keywords: Cabbage, Dendrogram, PCA, Polymorphism, Simple sequence repeat (SSR)
Introduction
Cabbage (Brassica oleracea L. var. capitata) is one of the most important cole vegetable crops grown worldwide under temperate to sub-tropical climatic conditions (Singh et al. 2013). Today, most of the cabbage genotypes grown in the world are hybrids and they are expected to form heads as early as possible along with higher yield (Cervenski et al. 2012). In India, majority of the vegetable hybrids are sold by private sector at exorbitant price (Koundinya and Kumar 2014). Hence, there is an urgent need to make available high-yielding quality hybrid seeds from public sector to the farming community at reasonable price to keep the farming profitable and competitive. For successful F1 hybrid breeding in cabbage, an efficient, economic, reliable and stable method of hybrid seed production without adulteration of self-fertilized seeds from each parent is needed. Because of the small size and structure of flowers, it is very difficult to produce hybrid seeds through manual emasculation and pollination techniques in cabbage (Yamagishi and Bhat 2014). The self-incompatibility (SI) system as reviewed by Kitashiba and Nasrallah (2014) is effective in hybrid breeding of Brassica vegetables. However, SI system is not always stable, and may be broken down by elevated temperature or drought conditions. This insist on finding out the more effective and reliable system of economic hybrid seed production in Brassica vegetables.
Cytoplasmic male sterility (CMS) is an important mechanism, which is stable and widely applicable to F1 hybrid seed production in all Brassicaceae crops (Yamagishi and Bhat 2014). In CMS genotypes, pollen production is inhibited, whereas pistil functions normally. CMS is controlled by a gene located in the mitochondrial genome. The nuclear genome of brassica vegetables which lacks fertility restorer gene (Rf) and CMS-inducing mitochondrial gene leads to nuclear–cytoplasmic incompatibility which results in production of CMS plants. There are several sources of the CMS cytoplasm in brassica species viz, ogura, kosena, polima, hau, nap, Diplotaxis muralis, Diplotaxis catholica, B. tournefortii, etc., but ‘Ogura’ cytoplasm is the only source which is widely used in Brassica vegetables. Ogura CMS system was discovered in Japanese radish (Raphanus sativus) of an unknown cultivar by Ogura (1968). Later on, European scientists introduced Ogura CMS into Brassica oleracea by inter-generic hybridization and repeated back-crossing (Bannerot et al. 1974). Since then, Ogura’-based CMS lines are worldwide used for hybrid seed production in Brassica vegetables.
The estimates of genetic diversity are useful for germplasm characterization and help to identify suitable parents for hybrid breeding in cabbage. The parental lines with sufficient diversity/polymorphism are selected based on degree of genetic diversity (Louarn et al. 2007), which can be evaluated with the help of agro-morphological traits and biochemical markers. In any plant breeding program, agro-morphological traits have immense value for the selection of parents with maximum variation (Zhang et al. 2008). Nowadays, molecular markers are considered valuable than agro-morphological traits as these are devoid from the perplexing effect of environment (Pejic et al. 1998). Hence, marker-assisted breeding aids in the selection of breeding material in conventional breeding programs (Frey et al. 2004; Liu et al. 2004). In recent years, simple sequence repeats (SSRs) have become excellent tool for the plant breeders. These markers are highly valuable due to high reproducibility, polymorphism, co-dominance and transferability in allied plant species and genera (Rana et al. 2015). They also resolve the variations that are caused due to widespread crossing and phenotypic plasticity imposed by environmental fluctuations (Nybom and Weising 2010). Therefore, present investigation was aimed to characterize and estimate the extent of diversity in ‘Ogura’-based CMS lines of cabbage using agro-morphological traits and SSR markers for the selection of diverse and superior parental lines for high-yielding quality hybrid development in cabbage.
Materials and methods
Experimental material and site
The present investigation was conducted at Experimental Research Farm and Molecular Laboratory of ICAR-IARI Regional Station, Katrain, Kullu, HP, India, during the year 2015 and 2016. The experimental material for present study comprised of 16 CMS lines developed by the introgression of ‘Ogura’ cytoplasm from a CMS source line ‘EC-173419’ of Brassica oleracea L. (received from ICAR-NBPGR, New Delhi) into different nuclear backgrounds of cabbage through repeated backcrossing for more than six generations. For agro-morphological characterization, seeds of all the genotypes were sown in the well-prepared nursery beds during August 2015 and 2016. After 30 days, seedlings were transplanted in the experimental field in Randomized Complete Block design (RCBD) at a spacing of 45 cm × 45 cm in the plots having size 3.0 m × 3.0 m during, September 2015 and 2016 and replicated thrice. Data were recorded on different qualitative and quantitative traits periodically from arbitrarily selected 10 plants from all replications. All the standard cultural practices as necessary for raising healthy crop stand of cabbage have been followed according to ICAR-IARI, Regional Station guidelines (Sharma 2003).
Isolation, purification and quantification of genomic DNA
For molecular characterization, seeds of each genotype were sown in pro-trays during July 2016 and were kept in the polyhouse for further vegetative growth. Isolation and purification of genomic DNA was done from 100 mg fresh green leaves of 30-day-old seedlings of each cabbage genotype using CTAB (cetyltrimethyl ammonium bromide) methodology given by Doyle and Doyle (1990). For DNA quantification, 2 µl of each DNA sample along with the uncut lambda DNA (100 ng/µl) was run on 0.8 per cent agarose gel (Sigma-Aldrich, USA). Dilution of samples (25–50 ng DNA/µl) was done in Tris–EDTA buffer and then they were stored at − 80 °C for further analyses.
SSR markers and PCR amplification
A total of 106 primer pairs (Integrated DNA technologies, New Delhi, India) were tested for molecular diversity analysis in 16 ‘Ogura’-based cytoplasmic male-sterile (CMS) lines of cabbage. Out of which only 29 primer pairs showed polymorphic bands in the different lines under study (Table 1). PCR amplification was performed using Eppendorf Mastercycler Nexus GSX1 in 10 μl reaction volume comprising: 5.00 µl premixed ready to use Go Taq® green master mix [DNA polymerase, 2× reaction buffer (pH 8.5), dATP (400 µM), dGTP (400 µM), dCTP (400 µM), dTTP (400 µM) and MgCl2 (3 mM)], primer pair (1.00 µl), template DNA (1.00 µl) and nuclease free water (3.00 µl). DNA amplification was carried out for 35 cycles with denaturation at 94 °C (1 min), annealing at 55 °C (1 min) and extension at 72 °C (2 min). The final extension was done at a temperature of 72 °C for 10 min.
Table 1.
List of SSR primers used in molecular characterization of different CMS lines of cabbage
| S. no. | LG | cM | Oligo name | Sequence (5′ to 3′) |
|---|---|---|---|---|
| 1. | C03 | 61.81 | BoSF042 | F-CGGCTTGACAGAATTGGACT R-TCCTATTCCACACCAAAGCC |
| 2. | C03 | 37.19 | BoSF062 | F-CTAGTGTTCGCCGAAGTGGT R-AAAAGGTGTCATGGAGTGCC |
| 3. | C03 | 92.42 | BoSF2985 | F-GGTTTCATAAAACATCTGTAGTTCGTC R-TGCAAGACATCTTTATTTCTTCCTC |
| 4. | N08 | 76.00 | 0112GO4A | F-CGAACATCTTAGGCCGAATC R-GGTTAACCTGCGGGATATTG |
| 5. | C01 | 47.66 | BoSF2345 | F-GCTCCGATGATCACGATTCT R-CTTCATCCCCTCACCACACT |
| 6. | C01 | 74.46 | CB10258 | F-ATGATGCCTAGCATGTCC R-AAGCTAAAGCGAAAGAAGC |
| 7. | C09 | 35.57 | BoGMS1498 | F-TCAACAGAACACATCCACAG R-TAGTGCCATAGAAACCATCTT |
| 8. | C01 | 54.72 | BoSF063 | F-GAATGTTTCCTCTGCTTGGC R-TCAAAATCAGGAGAATCGGG |
| 9. | CO2 | 125.93 | BRAS011 | F-TGGGACGTAGTCAGTCAACAA R-CCAAGTGCGAGAAGAGGAAG |
| 10. | C03 | 32.33 | BoSF966 | F-ATCCCATTGTCGTTATCCCA R-CGTCGTCTAGCGATGATGAA |
| 11. | C03 | 9.90 | BoSF1131 | F-GAAGTTTCACTGCCTCTCGG R-CTTCGTTAACCTCGCGAAAG |
| 12. | C06 | 67.96 | BoSF1215 | F-AGTATCAAACCCGCCTGTTG R-GGGTCGTATTAATCGCGTGT |
| 13. | C04 | 62.62 | BoE862 | F-AGCAAAGGCGGGGGAATGATAC R-ATGACAAAGACCACCCACACCAAT |
| 14. | CO7 | 75.51 | BoSF2313 | F-AAGGAGGATCACGAGGAGGT R-CATGGTAGCATCGAAAGCCT |
| 15. | C07 | 22.27 | BoE7830 | F-AATGGCGGTGGTGTTGG R-TTGGGCGACTAAAGAAAAAT |
| 16. | C08 | 90.92 | BoSF2680 | F-AAAGGTTAGGTGGTTGGATAAAGA R-TGTCTTCTGATGCCTTGGTCT |
| 17. | C09 | 45.88 | BoSF1640 | F-AGCACAACTACCTGAACCTCT R-TTTATCCTCGGTCTTCTCTCT |
| 18. | C07 | 30.78 | BoSF2860 | F-CATGCTTGCCTGAAAAGACA R-CCTTGTACTGCTCCTCTGCC |
| 19. | C05 | 70.88 | BoSF317 | F-CCAACTCCGGTCAATCATCT R-GCCCCTTTCTGTGTGACATT |
| 20. | C08 | 107.79 | BoSF2612 | F-CGTAGCCGTCTCTTACGCAT R-TTCAGTCCAGCGTTCAACAG |
| 21. | C06 | 82.37 | BoSF2054 | F-GAAGGAACAAGAGGATGGCA R-TCATGTTGTCGAGAATCCCA |
| 22. | C04 | 59.23 | BoSF184 | F-TTGCACGTACGTCTTTGAGG R-CTGCAACGAGGATGAAAACA |
| 23. | A10 | 78.65 | BoGMS0206 | F-TACTCCACGGTCTTCTACTTG R-GGGATAGTGATGTTGTTGATG |
| 24. | C03 | 20.89 | BrBAC214 | F-CGTATAATTTTCATAGGCGACG R-AGCATGCTTATGACTCTGGGA |
| 25. | C05 | 27.01 | BoSF2878 | F-CCTTGCGTCTGAAACATCAA R-TTACCGGGAGTAAATGCAGC |
| 26. | C08 | 80.34 | BoGMS1460 | F-CGAGAGGTGAAGAACAAGAG R-AAATAAGAGAAGAGAAACCGTC |
| 27. | C06 | 3.00 | BoGMS0952 | F-CAGTGAGTAACATTTGGCTG R-CGAGAGAGAAAGTGATGAGAG |
| 28. | C01 | 36.26 | BoSF1207 | F-CGACGCTAGACCAAGGTTTC R-GGAAAACCTTCTGCCAATGA |
| 29. | C02 | 77.19 | BoSF1167 | F-TTCGTTCCTTCGTTCATTCC R-AGGTAGTGGAGGAGGTCGGT |
Electrophoresis and gel documentation of amplified DNA
The amplified DNA products along with 1-kb DNA ladder (Fermentas, Lithuania) were separated by electrophoresis in agarose gel (2%) having ethidium bromide (10 mg/µl). The gel was run at 70 mA voltage for 2 h in 1× TBE buffer (pH 8.0) and it was picturized on a gel documentation system (BioSpectrum® Imaging System™, UK).
Statistical analysis
Agro-morphological data (pooled data of 2015 and 2016) were subjected to analysis of variance in OPSTAT software by following Gomez and Gomez (1984). The Pearson’s correlation coefficient, principle component analysis (PCA) and dendrogram based on single-linkage euclidean distance were calculated and constructed through SPSS 16.0 and Statistica software packages. Molecular data were analyzed only for 29 primers, which furnished scorable and polymorphic bands. Various genetic diversity estimates such as observed number of alleles (n a), effective number of alleles (n e), expected heterozygosity (H e), observed heterozygosity (H o) and Shannon information index (I) were estimated through POPGENE software (version 1.32) by following Yeh et al. (1997). The polymorphism information content (PIC) was computed through Cervus version 3.0 software as per the formulae given by Botstein et al. (1980). UPGMA (unweighted pair group method of arithmetic mean) dendrogram based on principle component analysis was constructed using NTSYSpc 2.0 (Rohlf 1998) software.
Results and discussion
Agro-morphological traits
Qualitatively assessed traits
Substantial variations were observed for different qualitative traits under study (Table 2). Head color was recorded as purple, light purple, reddish purple, green, dark green and purple green in four, three, one, six, one and one genotypes, respectively. Head compactness varied from compact in 15 genotypes to very compact in only 1 genotype, i.e., RCA. Among all the genotypes, eight, seven and one genotype had round, flat and oval shape heads, respectively. Further, leaf waxiness varied from highly waxy to medium waxy to less waxy in four, eight, and four genotypes, respectively. Hence, considerable variations observed for different qualitative traits in the available germplasm offer the chance for selection of suitable parental lines for quality hybrid development in cabbage. Earlier workers (Balkaya et al. 2005; Mohamed et al. 2012; Kibar et al. 2016) had correspondingly recorded wide variations in leaf color and head shape in cabbage.
Table 2.
Mean performance of 16 CMS lines of cabbage for different qualitative and quantitative traits (Pooled data of 2015 and 2016)
| Trait Line | Qualitative traits | Quantitative traits | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Head color | Head compactness | Head shape | Leaf waxiness | Plant height (cm) | Plant spread (cm) | Gross head weight (kg) | Number of non- wrapper leaves | Head length (cm) | Head width (cm) | Net head weight (kg) | Stalk length (cm) | Core length (cm) | Harvest index (%) | |
| RCA | Purple | Very compact | Round | Highly waxy | 18.00 | 53.00 | 1.56 | 12.00 | 14.50 | 15.00 | 1.30 | 1.00 | 8.50 | 83.75 |
| PMA | Purple | Compact | Round | Highly waxy | 20.00 | 60.00 | 1.98 | 11.67 | 14.33 | 15.33 | 1.38 | 1.50 | 10.50 | 69.41 |
| ZHA-3 | Purple | Compact | Flat | Medium waxy | 21.00 | 60.67 | 2.48 | 16.00 | 16.50 | 18.00 | 1.52 | 1.00 | 11.00 | 61.51 |
| ZHA-1 | Light purple | Compact | Flat | Medium waxy | 24.00 | 57.80 | 2.37 | 14.00 | 12.33 | 15.17 | 1.30 | 1.50 | 8.50 | 55.42 |
| ZHA-2 | Light purple | Compact | Flat | Medium waxy | 28.00 | 66.00 | 3.65 | 18.00 | 16.00 | 21.30 | 2.02 | 1.00 | 8.50 | 55.39 |
| RZA | Reddish purple | Compact | Flat | Highly waxy | 21.00 | 58.80 | 1.95 | 12.00 | 14.50 | 16.67 | 0.97 | 1.00 | 6.00 | 49.96 |
| RRMA | Light purple | Compact | Round | Highly waxy | 26.00 | 68.77 | 2.04 | 14.00 | 15.00 | 16.00 | 0.96 | 1.80 | 9.00 | 47.07 |
| 3-1A | Green | Compact | Round | Less waxy | 24.00 | 52.00 | 2.51 | 9.80 | 17.50 | 19.00 | 1.94 | 2.00 | 6.50 | 77.06 |
| 1-1A | Green | Compact | Flat | Less waxy | 19.00 | 46.00 | 1.88 | 11.00 | 14.50 | 17.00 | 1.67 | 1.00 | 5.00 | 89.17 |
| 2-1A | Green | Compact | Flat | Less waxy | 21.00 | 50.00 | 2.10 | 13.00 | 15.00 | 18.50 | 1.89 | 1.50 | 5.70 | 89.86 |
| 60-1A | Green | Compact | Round | Medium waxy | 20.00 | 48.00 | 2.36 | 14.00 | 15.50 | 17.00 | 1.90 | 0.50 | 8.00 | 80.40 |
| 924A | Green | Compact | Round | Medium waxy | 20.67 | 45.00 | 0.71 | 14.40 | 12.57 | 13.58 | 0.55 | 1.15 | 4.75 | 77.56 |
| 836A | Green | Compact | Round | Medium waxy | 19.00 | 63.00 | 2.35 | 12.00 | 17.00 | 18.00 | 1.32 | 1.00 | 5.00 | 56.32 |
| CH2A | Dark green | Compact | Flat | Medium waxy | 21.00 | 60.00 | 2.51 | 15.00 | 13.50 | 19.00 | 1.69 | 0.50 | 8.20 | 67.12 |
| KRA | Purple | Compact | Oval | Less waxy | 23.00 | 59.50 | 1.76 | 13.75 | 13.25 | 13.42 | 0.85 | 0.90 | 5.75 | 48.50 |
| RCGA | Purple green | Compact | Round | Medium waxy | 22.00 | 52.00 | 1.35 | 11.00 | 12.00 | 14.50 | 0.96 | 0.81 | 7.00 | 71.24 |
| CD (0.05) | – | – | – | – | 3.97 | 2.79 | 0.14 | 0.88 | 1.21 | 0.91 | 0.06 | 0.19 | 0.49 | 5.58 |
| ± SE (d) | – | – | – | – | 1.93 | 1.36 | 0.07 | 0.43 | 0.59 | 0.44 | 0.03 | 0.09 | 0.24 | 2.72 |
| CV (%) | – | – | – | – | 10.90 | 2.96 | 3.99 | 3.95 | 4.92 | 3.23 | 2.47 | 9.95 | 3.99 | 4.93 |
Quantitatively measured traits
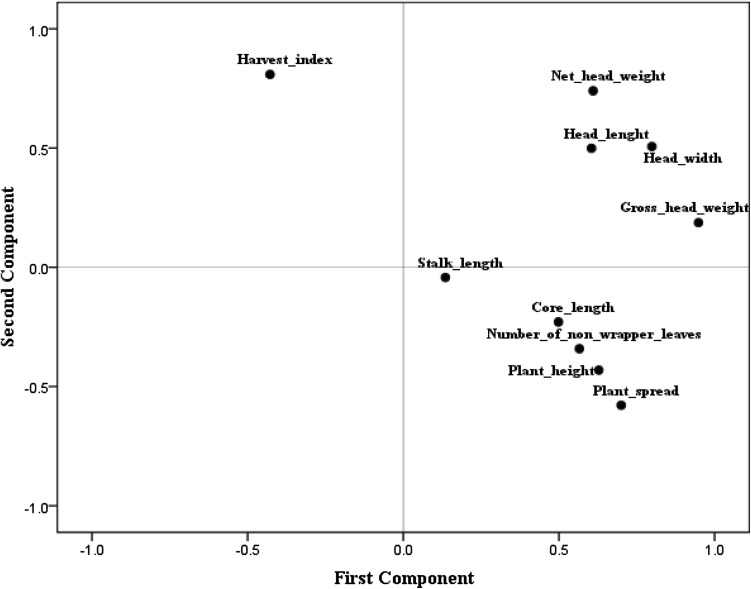
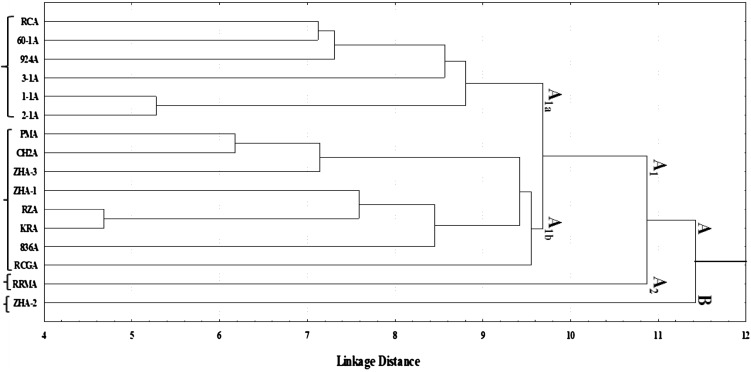
The perusal of pooled data (2015 and 2016) in Table 2 revealed the considerable variations for different quantitative traits under study viz. plant height (18.00–28.00 cm), plant spread (45.00–68.77 cm), gross head weight (0.71–3.65 kg), number of non-wrapper leaves (9.80–18.00), head length (12.00–17.50 cm), head width (13.42–21.30 cm), net head weight (0.55–2.02 kg), stalk length (0.50–2.00 cm), core length (4.75–11.00 cm) and harvest index (47.07–89.86%). Among all the genotypes, ZHA-2 performed better for plant height (28.00 cm), gross head weight (3.65 kg), head width (21.30 cm) and net head weight (2.02 kg). While, the genotypes RRMA for plant spread (68.77 cm); 3-1A for number of non-wrapper leaves (9.80) and head length (17.50 cm); 2-1A for harvest index (89.86%); 60-1A and CH2A for stalk length (0.50 cm) and 924A for core length (4.75 cm), recorded significant desirable performance. Hence, these genotypes must be taken into consideration, while making the selection of parental lines for improvement in yield and its attributing traits in CMS lines of cabbage. Similarly, Atter et al. (2009), Cervenski et al. (2012) and Chura et al. (2016) had reported wide variations for different quantitative traits using different lines of cabbage. Further, Pearson’s correlation coefficient between different quantitative traits revealed significant positive correlation of plant height with plant spread (0.53) and gross head weight (0.51); plant spread with gross head weight (0.54); gross head weight with head length (0.59), head width (0.85) and net head weight (0.76); head length with head width (0.70) and net head weight (0.60) and head width with net head weight (0.83) (Table 3). Singh et al. (2010) and Kibar et al. (2014) had also reported significant positive association of head weight with plant height and head diameter with head length in cabbage. Significant negative correlation of plant height (−0.54) and plant spread (−0.84) was recorded with harvest index in 16 CMS lines of cabbage. Negative correlation of plant height with harvest index in cabbage was also reported earlier by Singh et al. (2010), but results were found non-significant. Principle component analysis (PCA) is used to detect more important traits, which helps the plant breeders to carry out trait-specific breeding programs (Yousuf et al. 2011). The outcome of PCA revealed that first three components having eigen values greater than one were retained in the analysis because of the substantial amount of the variations. They had the variance of 39.27, 24.34 and 13.79% and aggregating to 77.40% of total variations explained (Table 4). The first factor (PC1) had the highest positive values for plant height (0.63), plant spread (0.70), gross head weight (0.95), number of non-wrapper leaves (0.57), head length (0.60) and head width (0.80). While second factor (PC2) was found superior for net head weight (0.74) and harvest Index (0.81). The third factor (PC3) recorded highest positive values for stalk length (0.92). The positive values of different traits in three components indicated their importance for divergence among 16 CMS lines of cabbage, whereas negative values showed the lowest contribution to the divergence. Further, loading of different traits based on first two principle components indicates that net head weight, gross head weight, head length and head width are the main components of divergence between 16 CMS lines of cabbage, whereas contribution of rest of traits under study was found comparatively less in divergence (Fig. 1). Hence, main emphasis should be given on the net head weight, gross head weight, head length and head width for yield improvement in cabbage. In a study on PCA for 15 agro-morphological traits in cabbage, Cervenski et al. (2011) found that first three principal components having eigen values greater than one revealed 99.99% of total variations among the cabbage cultivars for different traits under study. While, Kibar et al. (2016) observed 45.34% of total variations explained by first three principle components in different genotypes of cabbage. Dendrogram constructed using single-linkage euclidean distance based on 10 agro-morphological traits divided the 16 CMS lines of cabbage into two distinct groups viz., A and B (Fig. 2). Group A was further bifurcated into two sub-groups viz, A1 and A2. Further, A1 was alienated into two sub-groups (A1a and A1b). A1a comprised of six genotypes viz, RCA, 60-1A, 924A, 3-1A, 1-1A and 2-1A, while A1b accommodated eight genotypes viz, PMA, CH2A, ZHA-3, ZHA-1, RZA, KRA, 836A and RCGA. On the other hand, sub-group A2 and group B, each consisted of single genotype RRMA and ZHA-2, respectively. It means that these are the most distant lines among all the genotypes studied. Hence, these novel genotypes can be used as female parent for making superior heterotic crosses with the maintainer lines of CMS genotypes of other groups (A1a and A1b) in cabbage. Cervenski et al. (2010) and Kibar et al. (2016) had also used hierarchical method of clustering to discriminate different cultivars of cabbage.
Table 3.
Pearson’s correlation coefficients among quantitatively measured traits in 16 CMS lines of cabbage
| Traits | Plant height (cm) | Plant spread (cm) | Gross head weight (kg) | Number of non-wrapper leaves | Head length (cm) | Head width (cm) | Net head weight (kg) | Stalk length (cm) | Core length (cm) | Harvest index (%) |
|---|---|---|---|---|---|---|---|---|---|---|
| Plant height (cm) | 1.00 | 0.53* | 0.51* | 0.46 | 0.06 | 0.30 | 0.10 | 0.38 | 0.21 | − 0.54* |
| Plant spread (cm) | 1.00 | 0.54* | 0.42 | 0.25 | 0.28 | − 0.04 | 0.14 | 0.48 | − 0.84** | |
| Gross head weight (kg) | 1.00 | 0.46 | 0.59* | 0.85** | 0.76** | 0.02 | 0.38 | − 0.29 | ||
| Number of non-wrapper leaves | 1.00 | − 0.01 | 0.31 | 0.12 | − 0.30 | 0.38 | − 0.37 | |||
| Head length (cm) | 1.00 | 0.70** | 0.60* | 0.24 | 0.11 | 0.02 | ||||
| Head width (cm) | 1.00 | 0.83** | 0.01 | 0.13 | 0.04 | |||||
| Net head weight (kg) | 1.00 | 0.01 | 0.22 | 0.39 | ||||||
| Stalk length (cm) | 1.00 | 0.07 | − 0.05 | |||||||
| Core length (cm) | 1.00 | − 0.22 | ||||||||
| Harvest index (%) | 1.00 |
*p < 0.05, **p < 0.01
Table 4.
Eigen vectors for first three principal components of quantitatively measured traits in 16 CMS lines of cabbage
| Traits | Principle componenta | ||
|---|---|---|---|
| PC1 | PC2 | PC3 | |
| Plant height (cm) | 0.63 | − 0.43 | 0.26 |
| Plant spread (cm) | 0.70 | − 0.58 | 0.10 |
| Gross head weight (kg) | 0.95 | 0.19 | − 0.08 |
| Number of non-wrapper leaves | 0.57 | − 0.34 | − 0.56 |
| Head length (cm) | 0.60 | 0.50 | 0.29 |
| Head width (cm) | 0.80 | 0.51 | − 0.05 |
| Net head weight (kg) | 0.61 | 0.74 | − 0.09 |
| Stalk length (cm) | 0.13 | − 0.04 | 0.92 |
| Core length (cm) | 0.50 | − 0.23 | − 0.16 |
| Harvest index (%) | − 0.43 | 0.81 | − 0.07 |
| Eigen value | 3.93 | 2.43 | 1.38 |
| Percentage of variance | 39.27 | 24.34 | 13.79 |
| Cumulative per cent of variance | 39.27 | 63.61 | 77.40 |
PC Principal component. aExtracted through principle component analysis
Fig. 1.
Loading of different traits based on first two principal components
Fig. 2.
Dendrogram showing clustering pattern of 16 CMS lines based on 10 agro-morphological traits constructed using single-linkage Euclidean distance
SSR polymorphism and diversity analysis
In this study, 106 primer pairs were tested to estimate molecular diversity in 16 CMS lines of cabbage. Out of them, only 29 primer pairs showed reproducible and polymorphic bands and were selected for further studies (Table 1). The 29 SSR primers were found highly polymorphic and useful to differentiate different genotypes under study (Fig. 3). Raybould et al. (1999) and Louarn et al. (2007) had also revealed the usefulness of microsatellite markers to distinguish different genotypes of cabbage. In overall, 58 alleles were amplified through 29 SSR primers, averaging to two alleles in each locus. This average value agrees with the Cui et al. (2008) for Brassica rapa (2.91) suggesting appreciable allelic frequency among the genotypes studied, but reasonably lower than that as reported by Mohamed et al. (2016) in different Brassica species (3.92). This might be due to the use of genotypes belonging to single species. All the SSR markers were able to amplify only two alleles per locus among the tested genotypes. The maximum value (0.69) of Shannon’s Information Index (I) was exhibited by total 10 primers, while it was observed minimum (0.41) in two primers viz, BoGMS1460 and BoSF1167. In our study, mean value of ‘I’ was recorded as 0.62, which is greater than as observed earlier by Paulauskas et al. (2013) in Brassica napus (0.12). The expected heterozygosity (0.45) had higher mean values than observed heterozygosity (0.32). Prajapat et al. (2014) had also observed higher mean values of expected heterozygosity (0.56) than observed heterozygosity (0.30). Highest (0.75) observed heterozygosity (H o) was reported in the primer BoSF062, while lowest value (0.00) was recorded for three SSR primers viz. BoE862, BoSF1640 and BoGMS0206. The mean value of observed heterozygosity in this study was found greater than earlier reports by Pascher et al. (2010) in commercial varieties of Brassica napus (0.23) and Prajapat et al. (2014) in different Brassica species (0.30). In the meanwhile, expected heterozygosity (H e) was recorded maximum (0.57) and minimum (0.25) with the primer pairs BoSF1640 and BoGMS1460, respectively. In line with our study, Ofori and Becker (2008) had also reported similar mean value of expected heterozygosity in different cultivars of Brassica rapa. Polymorphic information content (PIC) was used to estimate allelic frequency and diversity among different CMS lines of cabbage (Table 5). PIC with a population mean of 0.34 was recorded highest in total six primers (0.38) and lowest value (0.22) was observed for the primers BoGMS1460 and BoSF1167. Naushad et al. (2012) had also reported varied values of PIC (0.17–0.75), with a mean value of 0.46 using SSR markers in different Brassica species. In the present study, different parameters of diversity exhibited high mean values, signifying allelic abundance in different CMS lines of cabbage. This allelic abundance might be attributed to diverse nuclear backgrounds of cabbage used for the development of ‘Ogura’-based CMS lines of cabbage under study.
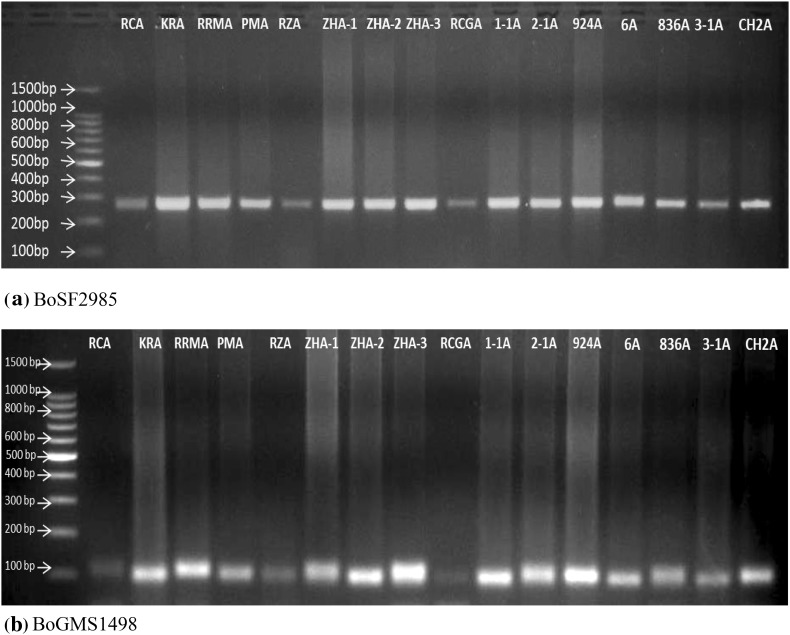
Fig. 3.
PCR amplification profile of 16 CMS lines of cabbage using SSR primer a BoSF2985 and b BoGMS1498 where, M molecular size marker (1 Kb ladder). Molecular sizes (in bp) are given on left
Table 5.
Genetic diversity statistics for 29 SSR loci studied in 16 CMS lines of cabbage
| S. no. | Locus | n a | n e | I | H o | H e | PIC |
|---|---|---|---|---|---|---|---|
| 1. | BoSF042 | 2 | 1.54 | 0.54 | 0.46 | 0.37 | 0.29 |
| 2. | BoSF062 | 2 | 1.97 | 0.69 | 0.75 | 0.51 | 0.37 |
| 3. | BoSF2985 | 2 | 1.97 | 0.68 | 0.20 | 0.51 | 0.37 |
| 4. | 0112GO4A | 2 | 1.38 | 0.45 | 0.33 | 0.29 | 0.24 |
| 5. | BoSF2345 | 2 | 1.77 | 0.63 | 0.36 | 0.45 | 0.34 |
| 6. | CB10258 | 2 | 2.00 | 0.69 | 0.29 | 0.52 | 0.38 |
| 7. | BoGMS1498 | 2 | 1.36 | 0.43 | 0.19 | 0.27 | 0.23 |
| 8. | BoSF63 | 2 | 2.00 | 0.69 | 0.67 | 0.52 | 0.38 |
| 9. | BRAS011 | 2 | 1.87 | 0.66 | 0.47 | 0.48 | 0.36 |
| 10. | BoSF966 | 2 | 1.92 | 0.67 | 0.40 | 0.50 | 0.37 |
| 11. | BoSF1131 | 2 | 1.77 | 0.63 | 0.07 | 0.45 | 0.34 |
| 12. | BoSF1215 | 2 | 1.47 | 0.50 | 0.27 | 0.33 | 0.27 |
| 13. | BoE862 | 2 | 1.64 | 0.58 | 0.00 | 0.41 | 0.32 |
| 14. | BoSF2313 | 2 | 1.70 | 0.60 | 0.42 | 0.43 | 0.33 |
| 15. | BoE7830 | 2 | 2.00 | 0.69 | 0.29 | 0.52 | 0.38 |
| 16. | BoSF2680 | 2 | 1.99 | 0.69 | 0.46 | 0.52 | 0.37 |
| 17. | BoSF1640 | 2 | 2.00 | 0.69 | 0.00 | 0.57 | 0.38 |
| 18. | BoSF2860 | 2 | 1.55 | 0.54 | 0.46 | 0.37 | 0.29 |
| 19. | BoSF317 | 2 | 1.99 | 0.69 | 0.46 | 0.52 | 0.37 |
| 20. | BoSF2612 | 2 | 1.91 | 0.67 | 0.21 | 0.50 | 0.36 |
| 21. | BoSF2054 | 2 | 1.97 | 0.69 | 0.13 | 0.53 | 0.37 |
| 22. | BoSF184 | 2 | 2.00 | 0.69 | 0.39 | 0.52 | 0.38 |
| 23. | BoGMS0206 | 2 | 1.96 | 0.68 | 0.00 | 0.51 | 0.37 |
| 24. | BrBAC214 | 2 | 1.97 | 0.69 | 0.38 | 0.53 | 0.37 |
| 25. | BoSF2878 | 2 | 2.00 | 0.69 | 0.08 | 0.52 | 0.38 |
| 26. | BoGMS1460 | 2 | 1.32 | 0.41 | 0.29 | 0.25 | 0.22 |
| 27. | BoGMS0952 | 2 | 1.96 | 0.68 | 0.29 | 0.51 | 0.37 |
| 28. | BoSF1207 | 2 | 1.87 | 0.66 | 0.60 | 0.48 | 0.36 |
| 29. | BoSF1167 | 2 | 1.32 | 0.41 | 0.29 | 0.26 | 0.22 |
| Mean | 2 | 1.80 | 0.62 | 0.32 | 0.45 | 0.34 |
n a Observed number of alleles, n e effective number of alleles, I Shannon’s Information index, H o observed heterozygosity, H e expected heterozygosity and PIC polymorphic information content
Cluster and principal component analysis
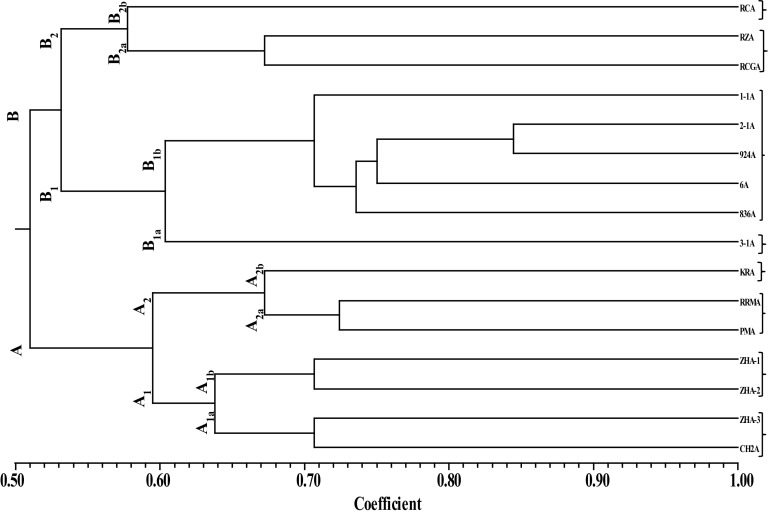
Dendrogram constructed through Jaccard’s similarity coefficient and UPGMA method exhibited the similarity coefficient of 0.51 and allocated the 16 CMS lines of cabbage into two major groups, i.e., A and B (Fig. 4). Group A was further divided into two sub-groups viz, A1 and A2. Further, A1 was bifurcated into two sub-groups viz, A1a (ZHA-3 and CH2A) and A1b (ZHA-1 and ZHA-2) and A2 was also alienated into two sub-groups viz, A2a (PMA and RRMA) and A2b (KRA). Group B was also divided into two sub-groups viz, B1 and B2. Further, B1 was bifurcated into two groups viz, B1a (3-1A) and B1b (1-1A, 2-1A, 924A, 6A and 836A). The sub-group B2 was also alienated into two sub-groups viz, B2a (RZA and RCGA) and B2b (RCA). Here in this study, sub-group B2b comprised of a single genotype, i.e. RCA with least similarity coefficient of 0.58, designating this to be most distant line among all the genotypes studied. This elite genotype can be used as female parent for making superior heterotic crosses with the maintainers of CMS genotypes of other groups. In the present studies, two genotypes of group B1b viz, 2-1A and 924A due to highest similarity index (0.85) were found genetically most identical among all the tested CMS lines of cabbage. Hence, hybridization between these genotypes using their maintainers as pollen parents will not prove effective to yield superior hybrid combination. On the other hand, crossing between the CMS genotypes and maintainers of sub-groups A1, A2, B1 and B2, might have the opportunity to get superior heterotic combinations. Saxena et al. (2011) using RAPD and SSR markers had also reported that UPGMA dendrogram give similar pattern of genetic diversity among different cabbage cultivars. Likewise, Mohamed et al. (2016) based on SSR data, had clustered different genotypes of Brassica oleracea into distinct groups, indicating considerable level of genetic variations among different Brassica Spp.
Fig. 4.
UPGMA dendrogram showing clustering pattern of 16 CMS lines based on 58 alleles constructed using Jaccard’s similarity coefficient
Conclusion
The experimental results on morphogenetic characterization and diversity analysis conclude that 16 CMS lines of cabbage have appreciable genetic variations. Based on agro-morphological and molecular studies, the genotype RRMA, ZHA-2 and RCA were found most distinct and divergent. Hence, these genotypes have immense potential in future breeding programs for the development of high-yielding economic hybrids. The estimates of Pearson’s correlation coefficient and PCA studies revealed that main emphasis should be given on gross head weight, head length and head width for yield improvement in different CMS lines of cabbage. Further, 29 SSR loci recorded high polymorphism and were found effective for differentiating different CMS lines under study. Hence, SSR markers can be utilized for germplasm characterization and association mapping for future breeding programs in ‘Ogura’-based CMS lines of cabbage.
Compliance with ethical standards
Conflict of interest
The authors declare that there is no conflict of interest.
References
- Atter RS, Sharma KC, Sundouri AS. Genetic variability for head yield and component traits in cabbage (Brassica oleracea var. capitata L.). Ind. J Ecol. 2009;36(1):88–90. [Google Scholar]
- Balkaya A, Yanmaz R, Apaydin A, Kar H. Morphological characterization of white head cabbage (Brassica oleracea var. capitata sub var. alba) genotypes in Turkey. N Z J Crop Hort Sci. 2005;33(4):333–341. doi: 10.1080/01140671.2005.9514367. [DOI] [Google Scholar]
- Bannerot H, Boulidard L, Cauderon Y Temp J (1974) Transfer of cytoplasmic male sterility from Raphanus sativus to Brassica oleracea. Eucarpia Meeting Cruciferae, Dundee, Scotland, pp 52–54
- Botstein D, White RL, Skolmick H, Davis RW. Construction of a genetic linkage map in man using restriction fragment length polymorphism. Am J Hum Genet. 1980;32:314–331. [PMC free article] [PubMed] [Google Scholar]
- Cervenski J, Gvozdanovic-Varga J, Vasic M, Glogovac S. Multivariate analysis for head weight and yield performance of experimental cabbage hybrids (Brassica oleracea var. capitata L.) Genetika. 2010;42(2):259–266. doi: 10.2298/GENSR1002259C. [DOI] [Google Scholar]
- Cervenski J, Gvozdanovic-Varga J, Glogovac S. Local cabbage (Brassica oleracea var. capitata L.) populations from Serbian Province of Vojvodina. Afr J Biotechnol. 2011;10(27):5281–5285. [Google Scholar]
- Cervenski J, Gvozdanovic-Varga J, Glogovac S. Variance components and correlations of agronomic traits among cabbage (Brassica oleracea var. capitata L.) maturity groups. Genetika. 2012;44(1):55–68. doi: 10.2298/GENSR1201055C. [DOI] [Google Scholar]
- Chura A, Negi PS, Pandey P. Assessment of heritability and genetic advancement for yield and yield attributing traits in Cabbage (Brassica oleracea var. Capitata L.) Int J Agri Innov Res. 2016;5(1):76–78. [Google Scholar]
- Cui XM, Dong YX, Hou XL, Cheng Y, Zhang JY, Jin MF. Development and characterization of microsatellite markers in Brassica rapa spp. chinensis and transferability among related species. Agric Sci China. 2008;7(1):19–31. doi: 10.1016/S1671-2927(08)60018-8. [DOI] [Google Scholar]
- Doyle JJ, Doyle JL. Isolation of plant DNA from fresh tissue. Focus. 1990;12:13–15. [Google Scholar]
- Frey JE, Frey B, Sauer C, Kellerhals M. Efficient low-cost DNA extraction and multiplex fluorescent PCR method for marker assisted selection in breeding. Plt Breed. 2004;123:554–557. doi: 10.1111/j.1439-0523.2004.01033.x. [DOI] [Google Scholar]
- Gomez KA, Gomez AA. Statistical procedure for agricultural research. 2. New York: Wiley; 1984. [Google Scholar]
- Kibar B, Karaagaç O, Kar H. Correlation and path coefficient analysis of yield and yield components in cabbage (Brassica oleracea var. capitata L.) Acta Sci Pol Hortorum Cultus. 2014;13(6):87–97. [Google Scholar]
- Kibar B, Karaagaç O, Kar H. Determination of morphological variability among cabbage (Brassica oleracea var. capitata L.) hybrids and their parents. Univ J Inst Sci Technol. 2016;6(1):31–44. doi: 10.21597/jist.2016119273. [DOI] [Google Scholar]
- Kitashiba H, Nasrallah JB. Self-incompatibility in Brassicaceae crops: lessons for interspecific incompatibility. Breed Sci. 2014;64:23–37. doi: 10.1270/jsbbs.64.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koundinya AVV, Kumar PP. Indian vegetable seeds industry: status and challenges. Int J Plt Animal Environ Sci. 2014;4(4):62–69. [Google Scholar]
- Liu P, Zhu J, Lu Y. Marker-assisted selection in segregating generations of self-fertilizing crops. Theor Appl Genet. 2004;109(2):370–376. doi: 10.1007/s00122-004-1636-6. [DOI] [PubMed] [Google Scholar]
- Louarn S, Trop AM, Holme IB. Database derived microsatellite markers (SSRs) for cultivar differentiation in Brassica oleracea. Genet Resour Crop Evol. 2007;54:1717–1725. doi: 10.1007/s10722-006-9181-6. [DOI] [Google Scholar]
- Mohamed AEE, Bourke P, Germaine K, Malone R. Assessment of morphological variation in Irish Brassica oleracea species. J Agri Sci. 2012;4(10):20–34. [Google Scholar]
- Mohamed A, Sawi ELE, Germaine K, Baurke P, Malone R. Genetic diversity and population structure of Brassica oleracea germplasm in Ireland using SSR markers. CR Biol. 2016;339:130–140. doi: 10.1016/j.crvi.2016.02.002. [DOI] [PubMed] [Google Scholar]
- Naushad AT, Malik AR, Farhatullah D, Zabta KS. Genetic diversity in the locally collected Brassica species of Pakistan based on microsatellite markers. Pak J Bot. 2012;44(3):1029–1035. [Google Scholar]
- Nybom H, Weising K. DNA-based identification of clonally propagated cultivars. Plt Breed Rev. 2010;34:221–295. [Google Scholar]
- Ofori A, Becker HC. Breeding of Brassica rapa for biogas production: heterosis and combining ability of biomass yield. BioEnergy Res. 2008;1(1):98–104. doi: 10.1007/s12155-008-9001-2. [DOI] [Google Scholar]
- Ogura H. Studies on the new male-sterility in Japanese radish, with special reference to the utilization of this sterility towards the practical raising of hybrid seeds. Mem Fac Agric Kagoshima Univ. 1968;6:39–78. [Google Scholar]
- Pascher K, Macalka S, Rau D, Gollmann G, Reiner H, Glossl J, Grabherr G. Molecular differentiation of commercial varieties and feral populations of oilseed rape (Brassica napus L.) BMC Evol Biol. 2010;10(63):1–13. doi: 10.1186/1471-2148-10-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulauskas A, Jodinskienė M, Griciuvienė I, Zukauskienė J, Petraitienė E, Brazauskienė I. Morphological traits and genetic diversity of differently overwintered oilseed rape (Brassica napus L.) cultivars. Zemdirb Agric. 2013;100(4):409–416. doi: 10.13080/z-a.2013.100.052. [DOI] [Google Scholar]
- Pejic I, Ajmone-Marsan P, Morgante M, Kozumplick V, Castiglioni P, Taramino G, Motto M. Comparative analysis of genetic similarity among maize inbred lines detected by RFLPs, RAPDs, SSRs and AFLPs. Theor Appl Genet. 1998;97(8):1248–1255. doi: 10.1007/s001220051017. [DOI] [Google Scholar]
- Prajapat P, Sasidharan N, Kumar M, Prajapati V. Molecular characterization and genetic diversity analysis in four Brassica species using microsatellite markers. Bioscan. 2014;9(4):1521–1527. [Google Scholar]
- Rana JC, Chahota RK, Sharma V, Rana M, Verma N, Verma B, Sharma TR. Genetic diversity and structure of Pyrus accessions of Indian Himalayan region based on morphological and SSR markers. Tree Genet Genom. 2015;11(1):1–14. doi: 10.1007/s11295-014-0821-2. [DOI] [Google Scholar]
- Raybould AF, Mogg RJ, Clarke RT, Gliddon CJ, Gray AJ. Variation and population structure at microsatellite and isozyme loci in wild cabbage (Brassica oleracea L.) in Dorset (UK) Genet Resour Crop Evol. 1999;46:351–360. doi: 10.1023/A:1008658630440. [DOI] [Google Scholar]
- Rohlf FJ. NTSYSpc numerical taxonomy and multivariate analysis system version 2.0 user guide. Setauket: Applied Biostatistics Inc.; 1998. [Google Scholar]
- Saxena B, Kaur R, Bhardwaj SV. Assessment of genetic diversity in cabbage cultivars using RAPD and SSR markers. J Crop Sci Biotechnol. 2011;14(3):191–196. doi: 10.1007/s12892-011-0018-2. [DOI] [Google Scholar]
- Sharma SR. Cabbage. In: Sirohi PS, editor. Vegetable crop production. New Delhi: Division of Vegetable Science, IARI; 2003. [Google Scholar]
- Singh BK, Sharma SR, Kalia P, Singh B. Character association and path analysis of morphological and economic traits in cabbage (Brassica oleracea var. capitata) Ind J Agric Sci. 2010;80(2):116–118. [Google Scholar]
- Singh BK, Sharma SR, Singh B. Genetic variability, inheritance and correlation for mineral contents in cabbage (Brassica oleracea var. capitata L.) J Hortic Res. 2013;21(1):91–97. doi: 10.2478/johr-2013-0013. [DOI] [Google Scholar]
- Yamagishi H, Bhat SR. Cytoplasmic male sterility in Brassicaceae crops. Breed Sci. 2014;64:38–47. doi: 10.1270/jsbbs.64.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh FC, Yang RC, Boyle TB, Ye ZH, Mao JX. POPGENE, the user-friendly shareware for population genetic analysis. Canada: Molecular Biology and Biotechnology Centre, University of Alberta; 1997. [Google Scholar]
- Yousuf M, Ajmal SU, Munir M, Ghafoor A. Genetic diversity analysis for agro-morphological and seed quality traits in rapeseed (Brassica campestris L.) Pak J Bot. 2011;43(2):1195–1203. [Google Scholar]
- Zhang X, Blair MW, Wang S. Genetic diversity of Chinese common bean (Phaseolus vulgaris L.) landraces assessed with simple sequence repeat markers. Theor Appl Genet. 2008;17(4):629–640. doi: 10.1007/s00122-008-0807-2. [DOI] [PubMed] [Google Scholar]