ABSTRACT
New fluoroquinolones (FQs) have been shown to be more active against drug-resistant Mycobacterium tuberculosis strains than early FQs, such as ofloxacin. Sitafloxacin (STFX) is a new fluoroquinolone with in vitro activity against a broad range of bacteria, including M. tuberculosis. This study aimed to determine the in vitro activity of STFX against all groups of drug-resistant strains, including multidrug-resistant M. tuberculosis (MDR M. tuberculosis), MDR M. tuberculosis with quinolone resistance (pre-XDR), and extensively drug-resistant (XDR) strains. A total of 374 drug-resistant M. tuberculosis strains were tested for drug susceptibility by the conventional proportion method, and 95 strains were randomly submitted for MIC determination using the microplate alamarBlue assay (MABA). The results revealed that all the drug-resistant strains were susceptible to STFX at a critical concentration of 2 μg/ml. Determination of the MIC90s of the strains showed different MIC levels; MDR M. tuberculosis strains had a MIC90 of 0.0625 μg/ml, whereas pre-XDR and XDR M. tuberculosis strains had identical MIC90s of 0.5 μg/ml. Common mutations within the quinolone resistance-determining region (QRDR) of gyrA and/or gyrB did not confer resistance to STFX, except that double mutations of GyrA at Ala90Val and Asp94Ala were found in strains with a MIC of 1.0 μg/ml. The results indicated that STFX had potent in vitro activity against all the groups of drug-resistant M. tuberculosis strains and should be considered a new repurposed drug for treatment of multidrug-resistant and extensively drug-resistant TB.
KEYWORDS: XDR M. tuberculosis, multidrug resistance, quinolones, sitafloxacin
INTRODUCTION
Drug-resistant tuberculosis (TB) is still one of the major public health threats globally. There are approximately 480,000 multidrug-resistant M. tuberculosis (MDR M. tuberculosis) (resistance to at least rifampin and isoniazid)-infected patients per year. Of the patients with MDR M. tuberculosis infections, 9.5% were found to have extensively drug-resistant M. tuberculosis (XDR M. tuberculosis) infections (1). Fluoroquinolones (FQs) are the most effective drugs when used in combination with injectable drugs, like amikacin, kanamycin, or capreomycin, for the treatment of MDR M. tuberculosis. MDR M. tuberculosis that resists any FQs and one of the injectable drugs is defined as XDR M. tuberculosis. FQs have been shown to be potent drugs against Mycobacterium tuberculosis, and later FQs, like moxifloxacin and gatifloxacin, have higher activity than the early drugs (ofloxacin and ciprofloxacin). Previous studies demonstrated the effectiveness of later FQs against MDR M. tuberculosis strains with resistance to early FQs (2–4). Sitafloxacin (STFX), a C-8 chloroquinolone, is a new fluoroquinolone with in vitro activity against a broad range of Gram-positive, Gram-negative, and anaerobic bacteria. STFX has also been shown to have potent activity against M. tuberculosis and Mycobacterium avium complex both in vitro and in macrophage or epithelial cell lines (5, 6). However, there has been limited information regarding the activity of STFX against drug-resistant M. tuberculosis. This study aimed to determine the in vitro activity of STFX against all groups of drug-resistant M. tuberculosis strains, namely, MDR, MDR with quinolone resistance (pre-XDR), and XDR tuberculosis strains. MICs and mutations associated with quinolone resistance were also determined in this study.
RESULTS
Drug susceptibility testing (DST).
Three hundred seventy-four M. tuberculosis clinical strains were tested for first-line and second-line anti M. tuberculosis drug susceptibility by the proportional method. Three hundred fifteen and 59 strains were identified as MDR M. tuberculosis and non-MDR M. tuberculosis strains, respectively. Of the 315 MDR M. tuberculosis strains, 73 and 41 were categorized as pre-XDR M. tuberculosis (MDR M. tuberculosis with resistance to any FQs) and XDR M. tuberculosis (MDR M. tuberculosis with resistance to ofloxacin [OFX] and amikacin and/or kanamycin) strains, respectively. The resistance patterns are summarized in Table 1. STFX showed the greatest in vitro activity; all the tested isolates were susceptible to STFX. For other FQs, later drugs showed better activity against both MDR and non-MDR M. tuberculosis strains than the early drugs. Of 274 MDR M. tuberculosis strains, 19%, 9.1%, 10.2%, and 1.4% were resistant to OFX, levofloxacin (LVFX), moxifloxacin (MOFX), and gatifloxacin (GTFX), respectively. Similar results were obtained with the XDR M. tuberculosis strains; STFX showed the greatest activity, whereas GTFX had greater activity than MOFX and LVFX, with 19.5%, 56.1%, and 80.1% resistance, respectively. Of the 59 non-MDR M. tuberculosis strains, only 5.1%, 3.4%, and 1.7% were resistant to OFX, LVFX, and MOFX, respectively. All of the non-MDR M. tuberculosis strains were susceptible to GTFX and STFX.
TABLE 1.
Resistance patterns of M. tuberculosis clinical strains used in this study
| Resistance patternb | No. of strains | No. resistant toa: |
||||||
|---|---|---|---|---|---|---|---|---|
| AK | K | OFX | LVFX | MOFX | GTFX | STFX | ||
| H | 5 | 0 | 0 | 1 | 1 | 0 | 0 | 0 |
| HS | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| R | 38 | 0 | 0 | 2 | 1 | 1 | 0 | 0 |
| RS | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| RE | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| RSE | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| HR | 64 | 2 | 2 | 7 | 6 | 7 | 2 | 0 |
| HRS | 88 | 14 | 16 | 27 | 19 | 13 | 6 | 0 |
| HRE | 52 | 8 | 9 | 16 | 10 | 9 | 2 | 0 |
| HRSE | 111 | 18 | 18 | 43 | 23 | 22 | 2 | 0 |
| Total | 374 | 42 | 45 | 96 | 60 | 52 | 12 | 0 |
Critical drug concentrations: amikacin (AK), 6.0 μg/ml; kanamycin (K), 6.0 μg/ml; OFX, 2.0 μg/ml, LVFX, 2.0 μg/ml; MOFX, 2.0 μg/ml; GTFX, 2.0 μg/ml; and STFX, 2.0 μg/ml.
H, isoniazid 0.2 μg/ml; R, rifampin 1.0 μg/ml; S, streptomycin 2.0 μg/ml; E, ethambutol 5.0 μg/ml.
MICs.
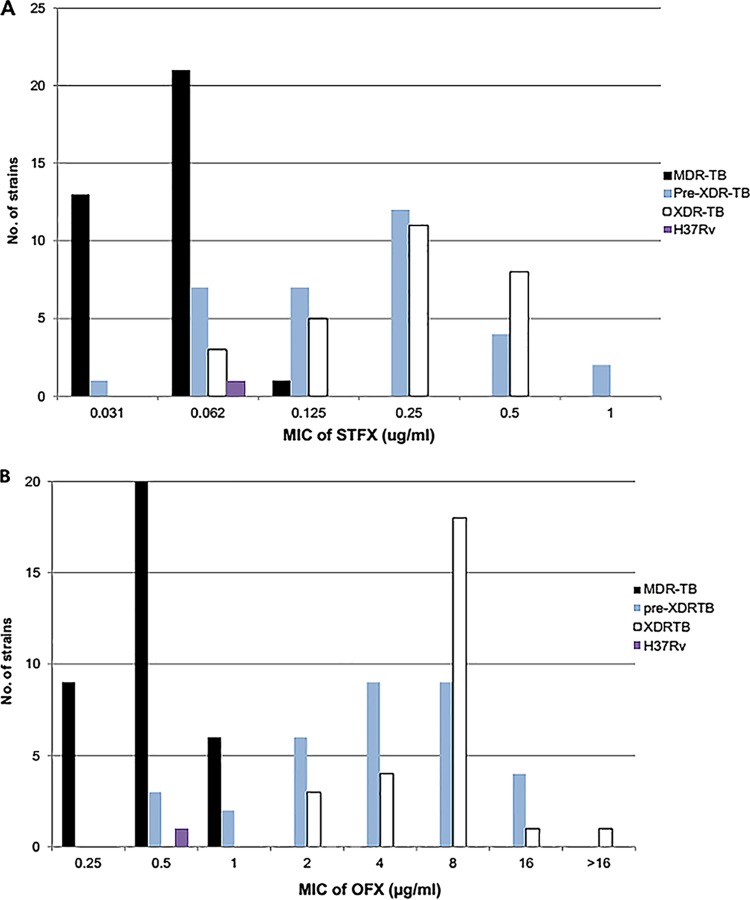
A total of 95 clinical strains, comprising 35 MDR, 33 pre-XDR, and 27 XDR M. tuberculosis strains, were randomly selected, and the MICs against STFX and OFX were determined by the alamarBlue assay. The majority of the strains (85%) had STFX MICs of ≤0.25 μg/ml (Fig. 1 and data not shown). Almost all MDR M. tuberculosis strains (34 of 35; 97%) had MICs of ≤0.0625 μg/ml, whereas pre-XDR and XDR M. tuberculosis showed slightly higher MICs; 81.8% and 70.4% of pre-XDR and XDR M. tuberculosis, respectively, had MICs of ≤0.25. Two pre-XDR M. tuberculosis strains showed MICs of 1.0 μg/ml. Determination of the MIC50s and MIC90s of the test strains showed different MIC levels among the resistance groups, as summarized in Table 2. In this experiment, we tested OFX in parallel with STFX. Unlike STFX, 58% of the test strains had MICs of ≥2 μg/ml; however, 29 of 35 (82.8%) MDR M. tuberculosis strains were still susceptible to OFX, with a MIC of ≤0.5 μg/ml (data not shown). M. tuberculosis H37Rv was used as a susceptible control and had STFX and OFX MICs of 0.0625 and 0.5 μg/ml, respectively.
FIG 1.
Distribution of MICs of STFX (A) and OFX (B) among groups of resistant M. tuberculosis clinical strains.
TABLE 2.
MIC50s and MIC90s of sitafloxacin for M. tuberculosis clinical strainsa
| Clinical strain (n = 95) (no.) | MIC50 (μg/ml) | MIC90 (μg/ml) | Range (μg/ml) |
|---|---|---|---|
| MDR M. tuberculosis (35) | 0.0625 | 0.0625 | 0.0312–0.125 |
| Pre-XDR M. tuberculosis (33) | 0.25 | 0.5 | 0.0312–1.0 |
| XDR M. tuberculosis (27) | 0.25 | 0.5 | 0.0625–0.5 |
Range, 0.0312 to 4 μg/ml; MIC for M. tuberculosis H37Rv, 0.0625 μg/ml.
DNA sequencing.
Nucleotide sequence analysis of gyrA and gyrB was performed in 53 strains representing each of the STFX MIC levels. All the strains had mutations detected in either the gyrA or gyrB gene. Most of the mutations were located in the quinolone resistance-determining region (QRDR) of gyrA (90.7%); Asp94Gly was the most common mutation, representing 40.7% of the gyrA or gyrB mutations (Table 3). Neither the mutation site nor the type of mutation was significantly related to the STFX MIC level. Notably, double mutations of Ala90Val and Asp94Ala were found in strains with a MIC of 1.0 μg/ml. Mutations in gyrB were found in 5 strains without the gyrA mutations; 4 strains had Glu540Asp and one had Asn538Asp Asp656Tyr. Almost all the strains had STFX MICs of ≤0.125 μg/ml, except that the strains carrying double mutations had MICs of 0.25 μg/ml. Compared with the OFX MIC level, the Glu540Asp mutation did slightly increase the OFX MIC to 1 μg/ml, whereas Asn538Asp Asp656Tyr showed 4- to 8-fold higher MICs, with an OFX MIC of 4.0 μg/ml. In addition, multiple mutations within both gyrA and gyrB were found in 14 strains (Table S3). However, all the strains that carried mutations outside the QRDR had comutations with either codon 90 or 94, and 10 of 14 strains had a silent mutation at gyrB codon 87 (Glu87Glu).
TABLE 3.
Gyrase mutations and MICs for 53 M. tuberculosis clinical strains
| Mutation | No. of strains | No. of resistant strainsa |
MIC (μg/ml) |
||
|---|---|---|---|---|---|
| MOFX | GTFX | STFX (mode) | OFX (mode) | ||
| GyrA | |||||
| Asp94Gly | 22 | 12 | 4 | 0.125–0.5 (0.25) | 2–8 (8) |
| Asp94Ala | 6 | 0 | 0 | 0.0625–0.25 (0.0625) | 2–8 (2) |
| Asp94Asn | 4 | 3 | 0 | 0.25–0.5 (0.5) | 8–16 (8) |
| Asp94His | 3 | 3 | 0 | 0.25–0.5 (0.25) | 4–8 (4) |
| Asp94Tyr | 2 | 1 | 0 | 0.0625–0.125 | 4–8 |
| Ala90Val | 8 | 4 | 2 | 0.125–0.5 (0.125) | 2–16 (2) |
| Ala90Val Asp94Ala | 2 | 2 | 1 | 0.5–1 | 16 |
| Ala90Val Asp94Asn | 1 | 1 | 0 | 0.625 | 4 |
| GyrB | |||||
| Glu540Asp | 4 | 4 | 0 | 0.031–0.125 (0.0625) | 0.5–1 (0.5) |
| Asn538Asp Asp656Tyr | 1 | 0 | 1 | 0.25 | 4 |
Critical concentration, 2 μg/ml.
DISCUSSION
Previous studies investigating the in vitro and in vivo activities of STFX compared with GTFX, LVFX, and the synthesized quinolone HRS-903 revealed that STFX had greater antimycobacterial activity than other quinolones and showed good activity against MDR M. tuberculosis (5, 6). A recent study also supported the potent in vitro activity of STFX, as well as GTFX and MOFX, against ciprofloxacin-resistant M. tuberculosis (7). Drug susceptibility testing of early and later fluoroquinolones using a critical concentration of 2 μg/ml on Middlebrook 7H10 medium in our study revealed greater in vitro activity of STFX against MDR, pre-XDR, and XDR M. tuberculosis strains than of LVFX, MOFX, and GTFX. No STFX-resistant strain was seen using this critical concentration of 2 μg/ml. More importantly, all XDR M. tuberculosis strains tested in this study were susceptible to STFX, whereas 19.5%, 56.1%, and 80.1% were resistant to GTFX, MOFX, and LVFX, respectively. In addition, STFX showed lower MIC50 and MIC90 values against the MDR M. tuberculosis strains, with a MIC50 and a MIC90 of 0.0625 μg/ml compared to the higher MIC50 and MIC90 of 0.25 and 0.5 μg/ml, respectively, against quinolone-resistant MDR M. tuberculosis (pre-XDR M. tuberculosis) and XDR M. tuberculosis. This is the first study to report the MIC50s and MIC90s of STFX against all types of resistant M. tuberculosis strains. The MIC50s of STFX in this study were 2-fold lower than that of GTFX (MIC50 = 0.5 μg/ml) reported previously, when 80 OFX-resistant M. tuberculosis clinical strains were evaluated (8). Our results supported the greater in vitro activity of STFX than of other, later quinolones, like MOFX and GTFX. Sitafloxacin is a promising drug that can inhibit OFX-, LVFX-, MOFX-, and GTFX-resistant M. tuberculosis, and it showed superior in vitro activity against all types of drug-resistant strains.
The molecular mechanism of quinolone resistance in M. tuberculosis is mostly related to mutations within the DNA gyrase-encoding genes (gyrA and gyrB), particularly at the QRDR of gyrA, and it is rarely caused by mutations in the gyrB gene (8–11). Different types of gyrase mutations have been reported to produce variable MIC levels in early and later quinolones (11). Most studies reported that the common mutations conferring quinolone resistance were located in the QRDR at codons 88, 90, 91, and 94 of gyrA. Asp94Gly was the predominant mutation commonly found in almost all studies and caused resistance to both early and later quinolones (9). In contrast, mutations in the QRDR of gyrB, although found much less frequently (≤2%) than the gyrA mutations, showed different MIC levels among the quinolones. The GyrB Glu540Asp mutation has been shown to mediate higher MICs of later quinolones, particularly 8-methoxy quinolones like MOFX, than of early quinolones (12). In this study, the gyrA and gyrB genes of 53 OFX-resistant strains were sequenced. The results revealed that 90.7% of the strains had mutations at gyrA in codon 90 or 94 or both. Asp94Gly was the most common codon, found in 40.7% of the resistant strains. Mutations at gyrB were found in 5 strains; 4 strains had Glu540Asp, and 1 had Asn538Asp. The results of the susceptibility testing of these 53 OFX-resistant strains using a critical drug concentration of 2 μg/ml revealed that 30 (57%) and 8 (15%) of the strains were resistant to MOFX and GTFX, respectively (Table 3). STFX showed greater activity; all 53 strains were susceptible to STFX, with a MIC range of 0.0625 to 1.0 μg/ml, indicating that gyrA and gyrB mutations had less effect on STFX susceptibility than on MOFX and GTFX. Notably, one strain with double mutations at Ala90Val and Asp94Ala was resistant to all the test quinolones but still had an STFX MIC of 1.0 μg/ml. Although a previous study reported that either Gly88Cys, Ser91Pro, Asp94Gly, or Asp94Asn mutation caused a higher MIC50 of ≥1.56 μg/ml and that double mutations, namely, Ala90Val Ser91Pro, Ala90Val Asp94Gly, and Ala90Val Asp94Ala, contributed to a higher MIC than single mutations (7), our study showed different results; either Asp94Gly or Asp94Asn alone did not confer STFX resistance (MIC ≤ 0.5 μg/ml), except that the double mutations Ala90Val Asp94Ala resulted in a 2-fold-higher MIC (1.0 μg/ml). This might result from the difference in the study populations and the genetic backgrounds of the strains, which might have had other, unknown mechanisms (i.e., drug efflux pumps, cell wall permeability, or drug-inactivating/modifying enzymes) that had a synergistic effect on the STFX-resistant phenotype. One limitation of our study is that we did not sequence any FQ-susceptible strains to confirm the presence of wild-type gyrA or gyrB. Instead, we inferred that the MDR M. tuberculosis group that was susceptible to all the FQs tested should contain wild-type gyrA or gyrB. The MIC values of this group were lower than those of the pre-XDR M. tuberculosis and XDR M. tuberculosis groups (Table 2 and Fig. 1).
Sitafloxacin (Gracevit; Daiichi Sankyo, Tokyo, Japan) is a new oral fluoroquinolone and has been approved in Japan and Thailand since 2008 and 2011, respectively, for the treatment of lower respiratory tract and urinary tract infections. STFX has excellent distribution in various tissues, including bronchi, alveoli, and cavities in the lung caused by tuberculosis and tissue granuloma. The intracellular drug concentration is up to 4 times higher than the drug concentration in the blood. STFX is available in 50-mg tablets. The maximum concentrations of STFX in serum (Cmax) for 50-mg and 100-mg doses are 0.5 and 1.0 μg/ml, respectively (13), which is 8-fold more than the MIC90 (0.0625 μg/ml) against MDR M. tuberculosis and equal to the MIC90 (0.5 μg/ml) against XDR M. tuberculosis strains. The dose of STFX for MDR M. tuberculosis should be 50 mg twice a day, while the dose for pre-XDR M. tuberculosis and XDR M. tuberculosis should be higher, 100 mg twice a day. STFX has been found to be safe, with a low incidence of side effects, mainly involving the gastrointestinal system. Acceptable adverse events have been reported with long-term administration of STFX (12.1 ± 7.3 months) for treatment of pulmonary MAC (Mycobacterium avium complex) disease (14). Since data on the sitafloxacin breakpoint are limited, in this study, we used the same breakpoint as for ofloxacin, with 2 μg/ml as the critical concentration for performing the drug susceptibility testing (2, 3). This was a limitation of our study, since this critical concentration might be too high compared to the MIC values of STFX. In agreement with the most recent study, Yi et al. (15) demonstrated the in vitro activity of STFX compared with ciprofloxacin, levofloxacin, and moxifloxacin against drug-susceptible and drug-resistant M. tuberculosis strains and recommended an epidemiological cutoff (ECOFF) of 0.125 μg/ml for STFX. Considering the Cmax of 1.0 μg/ml for a STFX 100-mg dose and the results of our study, we therefore recommended using 0.5 μg/ml as the critical concentration for drug susceptibility testing of sitafloxacin.
In summary, our study showed potent in vitro activity of STFX against all types of resistance, including MDR, pre-XDR, and XDR M. tuberculosis strains. STFX showed greater activity than 8-methoxy fluoroquinolones like MOFX and GTFX; mutations of gyrA and gyrB had less effect on STFX susceptibility than on GTFX and MOFX. STFX should be considered a new repurposed drug for treatment of multidrug- and extensively drug-resistant M. tuberculosis. A prospective, multicenter, randomized, controlled clinical trial should be carried out to evaluate the effectiveness of STFX versus MOFX or GTFX against multidrug- and extensively drug-resistant M. tuberculosis.
MATERIALS AND METHODS
Bacterial strains and antibiotics.
Overall, 374 drug-resistant M. tuberculosis strains isolated from July 2013 to July 2016 from the Drug-Resistant Tuberculosis Research Laboratory, Drug-Resistant Tuberculosis Research Fund, Faculty of Medicine Siriraj Hospital, Mahidol University, Bangkok, Thailand were investigated in this study. Mycobacteria were cultivated on Löwenstein-Jensen (LJ) medium and incubated at 37°C for 4 weeks.
All drug discs containing OFX, LVFX, MOFX, and GTFX were purchased from BBL (Difco, Detroit, MI, USA). OFX powder was purchased from Sigma-Aldrich (St. Louis, MO, USA). STFX discs and powder were kindly provided by Daiichi Sankyo Co., Ltd.
Drug susceptibility testing.
DST was carried out using the disc elution method on Middlebrook 7H10 (Difco, Detroit, MI, USA) supplemented with 10% oleic acid-albumin dextrose-catalase (OADC) (BBL, Becton Dickinson, USA) as recommended by the CLSI (16). Briefly, the appropriate drug discs were dispensed aseptically into the centers of individual quadrants of sterile plastic dishes. Exactly 5.0 ml each of sterile, tempered (52°C), complete M7H10 medium was pipetted over the discs, and the plates were left overnight at room temperature to permit the drug to diffuse uniformly. The inoculum was prepared by suspending M. tuberculosis cells in Middlebrook 7H9 with the turbidity adjusted to match that of MacFarland no. 1 standard. The cell suspension was diluted to 10−2 (approximately 105 CFU/ml) and 10−4 (approximately 103 CFU/ml) in sterile distilled water; these two dilutions were inoculated onto each quadrant and onto a quadrant containing drug-free medium, which was used as a control. The plate was incubated at 37°C until colonies appeared on the control quadrant (approximately 2 to 4 weeks). Resistance was reported when the colonies on the drug-containing quadrant appeared ≥1% compared to the drug-free control quadrant. For quality control, M. tuberculosis H37Rv ATCC 27294 was used as a control for DST. This strain was used at each time when a batch of DST was set up. If any resistance was observed in the control strain, all the results in that batch were not interpreted.
Determination of the MIC.
The MIC was determined using the Microplate alamarBlue assay (MABA) as previously described (17). Briefly, all of the outer wells of the test plates were filled with sterile water to minimize medium evaporation during incubation. The wells in rows B to G of columns 3 to 11 were filled with 100 μl of Middlebrook 7H9 broth. One hundred microliters of the drug solutions was added to each well in rows B to G of columns 2 and 3. Then, 100 μl of the mixture was transferred from column 3 to column 4, and the contents of the wells were mixed well. Serial 1:2 dilutions were continued through column 10 (STFX, 0.03125 to 4.0 μg/ml; OFX, 0.125 to 16.0 μg/ml), and 100 μl of excess medium was discarded from the wells in column 10. One hundred microliters of M. tuberculosis inoculum (approximately 105 CFU/ml) was added to each well in rows B to G of columns 2 to 11. Thus, the wells in column 11 served as drug-free (inoculum-only) controls. The plates were incubated at 37°C for 5 days. Twenty microliters of alamarBlue dye (Accumed International, Westlake, OH, USA) and 12.5 μl of 20% sterilized Tween 80 reagent were added to well B11. The plates were reincubated at 37°C for 24 h. If the solution in well B11 turned pink, the reagents were added to all of the wells in the plate (if it remained blue, the dye-reagent mixture would be added to another control well, and the result would be read the next day). The plates were incubated for an additional 24 h at 37°C, and the colors of all the wells were recorded. The MIC was defined as the lowest drug concentration that prevented a color change from blue to pink.
DNA isolation.
Two or three loops of mycobacterial culture were suspended in Tris-EDTA buffer (10 mM Tris-HCl, pH 8.0, 1 mM EDTA), and the suspension was heated at 80°C for 20 min to kill the mycobacterial cells. The heat-killed mycobacterial suspension was used for DNA extraction via an enzymatic method using lysozyme, and the DNA was then purified with cetyltrimethylammonium bromide-NaCl and chloroform-isoamyl alcohol (18). The DNA purities and concentrations were measured using a NanoDrop 1000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA).
Amplification and sequencing of the gyrA and gyrB genes.
The entire nucleotide sequence of the DNA gyrase operon of the gyrB and gyrA genes was amplified in 50-μl PCR mixtures consisting of 2.5 mM MgSO4, 0.25 mM (each) the four deoxynucleoside triphosphates (Thermo Fisher Scientific, Vilnius, Lithuania), 2 units of Platinum Taq DNA Polymerase High Fidelity (Invitrogen, Carlsbad, CA, USA), 20 pmol of primers (gyrB_Fw, 5′-GCACCAGGAAGAAAGATGTCC-3′, and gyrA-Rv, 5′-TTCCTCCTCAGATCGCTACG-3′), and 200 ng of template DNA under the following conditions: 2 min of initial denaturation at 94°C, 35 amplification cycles of 1 min at 94°C for denaturation, 1 min at 56°C for annealing, and 4 min at 68°C for extension, with a final extension at 68°C for 10 min (12). The entire gyrA and gyrB genes were sequenced in both directions using an ABI Prism 3730XL analyzer (Applied Biosystems, Foster City, CA, USA) by Macrogen (Seoul, South Korea).
The gyrB-gyrA sequences obtained were compared to those of M. tuberculosis H37Rv (GenBank accession number NC_000962.3) using ClustalW Multiple Alignment in BioEdit v7.2.0 software (19).
The study protocol was approved by the institutional review board of the Faculty of Medicine Siriraj Hospital, Mahidol University (EC no. 029/2557).
ACKNOWLEDGMENTS
We thank all staff of the Drug-Resistant Tuberculosis Research Laboratory, Drug-Resistant Tuberculosis Research Fund, Department of Microbiology, Faculty of Medicine Siriraj Hospital, Mahidol University, for their technical assistance.
This study was supported by the Drug-Resistant Tuberculosis Research Fund, Siriraj Foundation, Faculty of Medicine Siriraj Hospital, Mahidol University. A.C. was supported by a Chalermprakiat Grant, Faculty of Medicine Siriraj Hospital, Mahidol University.
We declare no competing interests.
This work is dedicated to the late Her Royal Highness Princess Galyani Vadhana Krom Luang Naradhiwas Rajanagarindra, patron of the Drug-Resistant Tuberculosis Research Fund.
REFERENCES
- 1.World Health Organization. 2016. Global tuberculosis report. WHO/HTM/TB/2016.13. WHO, Geneva, Switzerland. [Google Scholar]
- 2.Prammananan T, Arjratanakool W, Chaiprasert A, Leechawengwong M, Asawapokee N, Leelarasamee A, Dhiraputra C. 2005. Second-line drug susceptibilities of Thai multidrug-resistant Mycobacterium tuberculosis isolates. Int J Tuberc Lung Dis 9:216–219. [PubMed] [Google Scholar]
- 3.Prammananan T, Chaiprasert A, Leechawengwongs M. 2011. Eight-years experience of fluoroquinolone susceptibility testing of multidrug-resistant Mycobacterium tuberculosis isolates from Siriraj Hospital, Thailand. Int J Antimicrob Agents 37:84–85. doi: 10.1016/j.ijantimicag.2010.09.005. [DOI] [PubMed] [Google Scholar]
- 4.Chaiprasert A, Srimuang S, Tingtoy N, Makhao N, Sirirudeeporn P, Tomnongdee N, Theankeaw O, Charoensook S, Leechawengwongs M, Prammananan T. 2014. Second-line drug susceptibilities of multidrug-resistant tuberculosis strains isolated in Thailand: an update. Int J Tuberc Lung Dis 18:961–963. doi: 10.5588/ijtld.13.0197. [DOI] [PubMed] [Google Scholar]
- 5.Tomioka H, Sato K, Akaki T, Kajitani H, Kawahara S, Sakatani M. 1999. Comparative in vitro antimicrobial activities of the newly synthesized quinolone HSR-903, sitafloxacin (DU-6859a), gatifloxacin (AM-1155), and levofloxacin against Mycobacterium tuberculosis and Mycobacterium avium complex. Antimicrob Agents Chemother 43:3001–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sato K, Tomioka H, Sano C, Shimizu T, Sano K, Ogasawara K, Cai S, Kamei T. 2003. Comparative antimicrobial activities of gatifloxacin, sitafloxacin and levofloxacin against Mycobacterium tuberculosis replicating within Mono Mac 6 human macrophage and A-549 type II alveolar cell lines. J Antimicrob Chemother 52:199–203. doi: 10.1093/jac/dkg343. [DOI] [PubMed] [Google Scholar]
- 7.Suzuki Y, Nakalima C, Tamaru A, Kim H, Matsuba T, Saito H. 2012. Sensitivities of ciprofloxacin-resistant Mycobacterium tuberculosis clinical isolates to fluoroquinolones: role of mutant DNA gyrase subunits in drug resistance. Int J Antimicrob Agents 39:435–439. doi: 10.1016/j.ijantimicag.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 8.Li J, Gao X, Luo T, Wu J, Sun G, Liu Q, Jiang Y, Zhang Y, Mei J, Gao Q. 2014. Association of gyrA/B mutations and resistance levels to fluoroquinolones in clinical isolates of Mycobacterium tuberculosis. Emerg Microbes Infect 3:e19. doi: 10.1038/emi.2014.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Avalos E, Catanzaro D, Catanzaro A, Ganiats T, Brodine S, Alcaraz J, Rodwell T. 2015. Frequency and geographic distribution of gyrA and gyrB mutations associated with fluoroquinolone resistance in clinical Mycobacterium tuberculosis isolates: a systematic review. PLoS One 10:e0120470. doi: 10.1371/journal.pone.0120470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chien JY, Chiu WY, Chien ST, Chiang CJ, Yu CJ, Hsueh PR. 2016. Mutations in gyrA and gyrB among fluoroquinolone- and multidrug-resistant Mycobacterium tuberculosis isolates. Antimicrob Agents Chemother 60:2090–2096. doi: 10.1128/AAC.01049-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farhat MR, Jacobson KR, Franke MF, Kaur D, Sloutsky Mitnick ACD, Murray M. 2016. Gyrase mutations are associated with variable levels of fluoroquinolone resistance in Mycobacterium tuberculosis. J Clin Microbiol 54:727–733. doi: 10.1128/JCM.02775-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Disratthakit A, Prammananan T, Tribuddharat C, Thaipisuttikul I, Doi N, Leechawengwongs M, Chaiprasert A. 2016. Role of gyrB mutations in preextensively and extensively drug-resistant tuberculosis in Thai clinical isolates. Antimicrob Agents Chemother 60:5189–5197. doi: 10.1128/AAC.00539-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakashima M, Uematsu T, Kosuge K, Umemura K, Hakusui H, Tanaka M. 1995. Pharmacokinetics and tolerance of DU-6859a, a new fluoroquinolone, after single and multiple oral doses in healthy volunteers. Antimicrob Agents Chemother 39:170–174. doi: 10.1128/AAC.39.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujita K, Fujita M, Ito Y, Hirai T, Mio T, Watanabe K, Mishima M. 2016. Preliminary evaluation of a sitafloxacin-containing regimen for relapsed or refractory pulmonary Mycobacterium avium complex. Dis Open Forum Infect Dis 3:ofw147. doi: 10.1093/ofid/ofw147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yi L, Aono A, Chikamatsu K, Igarashi Y, Yamada H, Takaki A, Mitarai S. 2017. In vitro activity of sitafloxacin against Mycobacterium tuberculosis with gyrA/B mutations isolated in Japan. J Med Microbiol 66:770–776. doi: 10.1099/jmm.0.000493. [DOI] [PubMed] [Google Scholar]
- 16.CLSI. 2003. Susceptibility testing of mycobacteria, nocardiae, and other aerobic actinomycetes; approved standard. CLSI document M24-A. Clinical and Laboratory Standards Institute, Wayne, PA. [PubMed] [Google Scholar]
- 17.Collins L, Franzblau SG. 1997. Microplate Alamar blue assay versus BACTEC 460 system for high-throughput screening of compounds against Mycobacterium tuberculosis and Mycobacterium avium. Antimicrob Agents Chemother 41:1004–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Helden PD, Victor TC, Warren RM, van Helden EG. 2001. Isolation of DNA from Mycobacterium tuberculosis, p 19–30. In Parish T, Stoker NG (ed), Mycobacterium tuberculosis protocols. Humana Press, Totowa, NJ. [DOI] [PubMed] [Google Scholar]
- 19.Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98. [Google Scholar]