ABSTRACT
Nine aph genes, including aph(2″)-Ib, aph(2″)-Ic, aph(2″)-Ig, aph(2″)-If, aph(2″)-If1, aph(2″)-If3, aph(2″)-Ih, aac(6′)-Ie–aph(2″)-Ia, and aac(6′)-Ie–aph(2″)-If2, were previously identified in Campylobacter. To measure the contribution of these alleles to aminoglycoside resistance, we cloned nine genes into the pBluescript and expressed them in Escherichia coli DH5α. The nine aph expressed in E. coli showed various levels of resistance to gentamicin, kanamycin, and tobramycin. Three genes, aac(6″)-Ie–aph(2″)-Ia, aph2″-If1, and aph2″-Ig, showed increased MICs to amikacin, and five aph genes were transferrable.
KEYWORDS: Campylobacter, gentamicin resistance, NARMS, cloning and expression
TEXT
The aminoglycoside 2″-phosphotransferase [APH(2″)] family is a major contributor to gentamicin resistance (Genr) in Campylobacter. Nine variants of aph genes were previously identified in Campylobacter isolated from human and retail chickens, including seven monofunctional aph(2″) genes, aph(2″)-Ib, aph(2″)-Ic, aph(2″)-Ig, aph(2″)-If, aph(2″)-If1, aph(2″)-If3, and aph(2″)-Ih, and two bifunctional aph(2″) genes, aac(6′)-Ie/aph(2″)-Ia and aac(6′)-Ie–aph(2″)-If2. Five of them, aph(2″)-Ig, aph(2″)-If1, aph(2″)-If3, aph(2″)-Ih, and aac(6′)-Ie–aph(2″)-If2, were novel genes (1).
Toth et al. showed that aph(2″)-If confers resistance to several aminoglycosides (2). Since the APH(2″) family is genetically diverse and the percentage of amino acid identity between subfamilies can be as low as 25.9% (1–3), it is important to investigate transferability and the attribution of variants of aph gene resistance to other aminoglycosides, in addition to gentamicin.
Eleven Campylobacter strains, five C. jejuni and six C. coli, obtained from the National Antimicrobial Resistance Monitoring System (NARMS) program were used in this study (Table 1). All isolates were previously sequenced using the Illumina MiSeq (Illumina, San Diego, CA, USA) (1, 4). The coding sequences of nine aph genes were synthesized and cloned in a pBluescript expression vector by GenScript (Piscataway, NJ). For two bifunctional aph(2″) genes, only the aph(2″)-Ia and aph(2″)-If2 genes were cloned into the pBluescript vector. The pBluescript::aph(2″) recombinant plasmids were transformed into competent Escherichia coli DH5α, according to the protocol provided by Thermo Fisher Scientific (Invitrogen, Carlsbad, CA). The clones were grown on LB agar plates containing 50 μg/ml ampicillin and 8 μg/ml gentamicin.
TABLE 1.
Campylobacter strains used in this study
| Strain IDa | Species | Source | Yr | Resistance phenotypeb | Resistance genesc | Mutation | Purpose |
|---|---|---|---|---|---|---|---|
| N29710d | C. coli | Chicken breast | 2011 | GEN TET | aadE aad9 aph(2″)-Ig aph(3′)-IIIa blaOXA-61 sat4 tetO | None | Cloning donor |
| N20344d | C. coli | Chicken breast | 2009 | GEN TET | aph(2″)-Ic aph(3′)-IIIa blaOXA-61 tetO | None | Cloning donor |
| 41912d | C. jejuni | Human | 2008 | CIP GEN NAL TET | aadE aac(6′)-Ie–aph(2″)-If2 blaOXA-61 tetO | GyrA86I | Cloning donor |
| 41921d | C. jejuni | Human | 2008 | CIP GEN NAL TET | aadE aad9 aac(6′)-Im aph(2″)-Ib tetO | GyrA86I | Cloning donor |
| 41945d | C. coli | Human | 2010 | CIP AZI CLI ERY TEL GEN NAL TET | aadE aad9 aac(6′)-Ie–aph(2″)-Ia aph(2″)-If1 aph(3′)-IIIa blaOXA-61 catA tetO | GyrA86I 23S rRNA 2075G | Cloning donor |
| 41898e | C. coli | Human | 2003 | CIP GEN NAL TET | aadE aac(6′)-Ie–aph(2″)-Ia blaOXA-61 tetO | GyrA86I | Donor |
| 41902e | C. jejuni | Human | 2005 | CIP GEN NAL TET | aph(2″)-If tetO blaOXA-61 | GyrA86I | Cloning donor |
| 41904e | C. coli | Human | 2006 | CIP AZI CLI ERY TEL GEN NAL TET | aadE aad9 aph(2″)-If3 lnuC aph(3′)-IIIa sat4 tetO | GyrA86I 23S rRNA 2075G | Cloning donor |
| 41905e | C. jejuni | Human | 2007 | CIP AZI CLI ERY TEL GEN NAL TET | aadE aad9 aph(2″)-Ih sat4 aph(3′)-IIIa blaOXA-61 tetO | GyrA86I 23S rRNA 2075G | Cloning donor |
| N18880 | C. jejuni | Chicken breast | 2008 | AZT ERY TEL | None | 23S rRNA 2075G | Recipient |
| N46788F | C. coli | Cattle | 2014 | CIP CLI FEN NAL TET | blaOXA-61 aph(3′)-IIIa tetO | GyrA86I | Recipient |
ID, identifier.
GEN, gentamicin; TET, tetracycline; CIP, ciprofloxacin; NAL, nalidixic acid; AZI, azithromycin; CLI, clindamycin; ERY, erythromycin; TEL, telithromycin; FFN, florfenicol.
The genes in bold are aminoglycoside 2″-phosphotransferase [aph(2″)] genes.
The aph(2″) genes from these isolates were successfully transferred to Gens recipient strains.
The aph(2″) genes from these isolates were not successfully transferred to Gens recipient strains.
Expression of the cloned aph genes in E. coli DH5α was first determined by MICs of gentamicin using broth microdilution (CMV3AGNF; Thermo Fisher Scientific, Trek Diagnostics, Cleveland, OH), following standard protocols (5). E. coli DH5α carrying the pBluescript vector without the aph genes was used as a control. To measure the contribution of different aph genes to additional aminoglycoside resistance, agar dilution was performed, and the MICs of six aminoglycosides, including gentamicin, kanamycin, streptomycin, neomycin, tobramycin, and amikacin, were determined. Agar dilution plates were prepared with concentrations ranging from 0.125 μg/ml to 1,024 μg/ml. The MIC was recorded as the lowest concentration of antimicrobial agent that completely inhibited the growth of an organism after incubation at 35°C for 16 to 20 h, according to guidelines of the Clinical and Laboratory Standards Institute (CLSI) (6)
For conjugation, nine Genr Campylobacter strains that carried variants of aph(2″) genes were used as donor strains, including four C. jejuni (41912, 41921, 41902, and 41905) and five C. coli (41945, 41898, 41904, N29710, and N20344) strains. Two gentamicin-susceptible (Gens) strains, C. jejuni N18880 and C. coli N46788F, were used as recipient strains (Table 1). Conjugation was performed as described by Chen et al. (7). Successful transconjugants were then confirmed by antimicrobial susceptibility testing (AST) and whole-genome sequence (WGS) analysis (4, 8).
AST showed that the expression of the nine aph genes in E. coli resulted in various levels of resistance to gentamicin, kanamycin, and tobramycin. The MICs from different clones ranged from 16 to 64 μg/ml for gentamicin, 64 to 512 μg/ml for kanamycin, and 8 to 128 μg/ml for tobramycin. Comparing the MICs of E. coli that carry pBluescript::aph(2″) to the control strain, there were 32- to 128-, 64- to 512-, and 16- to 256-fold increases in the MICs to gentamicin, kanamycin, and tobramycin, respectively (Table 2). The MIC differences from nine clones to the same drug may explain the diversity of this family. Furthermore, three clones carrying aph(2″)-Ia, aph2″-If3, and aph2″-Ig showed 8- to 16-fold increases in their MICs to amikacin compared to those of the E. coli control strain. None of the nine aph gene clones showed resistance to streptomycin or neomycin (Table 2).
TABLE 2.
MICs of six aminoglycosides for E. coli DH5α expressing the nine aph(2″) genes
| Bacterial strain name | aph genes in pBluescript | MIC (μg/ml) |
|||||
|---|---|---|---|---|---|---|---|
| Gentamicin | Kanamycin | Tobramycin | Amikacin | Neomycin | Streptomycin | ||
| DH5a | pBluescript | 0.5 | 1 | 0.5 | 1 | 1 | 2 |
| E. coli-Ia | aac(6′)-Ie–aph(2″)-Iaa | 64 | 512 | 128 | 8 | 0.5 | 2 |
| E. coli-Ib | aph(2″)Ib | 16 | 64 | 16 | 1 | 1 | 2 |
| E. coli-Ic | aph(2″)-Ic | 32 | 64 | 16 | 0.5 | 0.5 | 2 |
| E. coli-If | aph(2″)If | 32 | 128 | 16 | 2 | 1 | 4 |
| E. coli-If1 | aph(2″)-If1 | 32 | 128 | 16 | 2 | 1 | 2 |
| E. coli-If2 | aac(6′)-Ie–aph(2″)-If2a | 32 | 128 | 32 | 2 | 1 | 2 |
| E. coli-If3 | aph(2″)-If3 | 32 | 128 | 8 | 8 | 0.5 | 2 |
| E. coli-Ig | aph(2″)-Ig | 32 | 128 | 16 | 16 | 1 | 2 |
| E. coli-Ih | aph(2″)-Ih | 64 | 128 | 16 | 1 | 0.5 | 2 |
For bifunctional aminoglycoside resistance genes, only aminoglycoside phosphotransferase genes [aph(2″)-Ia and aph(2″)-If2] were cloned to the pBluescript vector.
A similar study conducted by Toth et al. showed that when aph(2″)-Ia and aph(2″)-If were cloned in pET22b(+) vector and expressed in E. coli BL21, the MICs increased 32- to 128-fold to kanamycin, tobramycin, and gentamicin. There was only a 1- to 2-fold increase in the MICs to neomycin and amikacin compared with control strain E. coli JM83 (2). Our results agreed with their findings for these two antimicrobial agents, except that pET22b::aph(2″)-Ia expressed in E. coli JM83 had a 4-fold lower MIC to tobramycin (32 μg/ml) than to pBluescript::aph(2″)-Ia expressed in E. coli DH5α (128 μg/ml). The difference in MICs to tobramycin between these two experiments could be due to the use of different cloning vectors and expression strains of E. coli, as well as the use of different AST methods.
The conjugation study showed that aph(2″)-Ib (41921), aph(2″)-Ic (N20344), aph(2″)-If1 (41945), aph(2″)-If2 (41912), and aph(2″)-Ig (N29710) were successfully transferred to Gens strains, either in C. jejuni N18880 or C. coli N46788. Strain 41945 carried two aph(2″) genes, bifunctional aac(6′)-Ie–aph(2″)-Ia and monofunctional aph(2″)-If1, and only aph(2″)-If1 was transferred based on the comparative genomic analysis. The other four aph(2″) genes, including three monofunctional aph(2″)-If (41902), aph(2″)-If3 (41904), aph(2″)-Ih (41905) genes and one bifunctional aac(6′)-Ie–aph(2″)-Ia (41898) gene, were not transferred to Gens strains (Table 1).
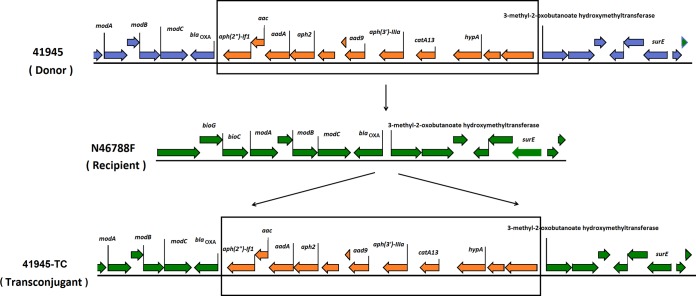
WGS analysis of donors, recipients, and transconjugants showed that transferred aph(2″) genes, including aph(2″)-Ib, aph(2″)-Ic, and aph(2″)-Ig, and aac(6′)-Ie–aph(2″)-If2, are located on plasmids. However, the aph(2″)-If1 gene from strain 41945, which is located on a chromosome, also was transferred. Comparative genomic analysis of donor (41945), recipient (N46788F), and transconjugant (41945-TC) strains showed that the aminoglycoside resistance island from the donor chromosome was integrated into the chromosome of recipient cells (41945-TC) through a recombination event (Fig. 1). WGS data showed that aadE, aad9, sat4, and aphA-3, one of the aph(2″) genes, and the tetO genes were often clustered together, forming a resistance island located either on the chromosome or the pTet plasmid (4, 7). Similar aminoglycoside resistance genomic islands were found either on the plasmid pCG8245 of C. jejuni or in the chromosome of C. coli SX81 (9, 10).
FIG 1.
The chromosome-carried Genr island integrated into the chromosome of Gens isolate through conjugation.
In summary, all nine variants of aph(2″) genes are responsible for resistance to gentamicin, kanamycin, and tobramycin but not to neomycin or streptomycin. Three variants, including aph(2″)-Ia, aph(2″)-If3, and aph(2″)-Ig, showed decreased susceptibility to amikacin. Both the plasmid- and chromosome-carried aph(2″) gene can be transferred by conjugation. This study highlights the need for continuous monitoring of emergent resistance genes in foodborne pathogens.
ACKNOWLEDGMENTS
We are grateful to Maureen Davidson and Chris Whitehouse for helpful comments and manuscript review.
This work was supported by the U.S. Food and Drug Administration with internal funds as part of routine work.
The views expressed in this article are those of the authors and do not necessarily reflect the official policy of the Department of Health and Human Services, the U.S. Food and Drug Administration, the Centers for Disease Control and Prevention, or the U.S. Government. The mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture or Food and Drug Administration.
REFERENCES
- 1.Zhao S, Mukherjee S, Chen Y, Li C, Young S, Warren M, Abbott J, Friedman S, Kabera C, Karlsson M, McDermott PF. 2015. Novel gentamicin resistance genes in Campylobacter isolated from humans and retail meats in the U S A. J Antimicrob Chemother 70:1314–1321. doi: 10.1093/jac/dkv001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Toth M, Frase H, Antunes NT, Vakulenko SB. 2013. Novel aminoglycoside 2″-phosphotransferase identified in a Gram-negative pathogen. Antimicrob Agents Chemother 57:452–457. doi: 10.1128/AAC.02049-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chow JW. 2000. Aminoglycoside resistance in enterococci. Clin Infect Dis 31:586–589. doi: 10.1086/313949. [DOI] [PubMed] [Google Scholar]
- 4.Zhao S, Tyson GH, Chen Y, Li C, Mukherjee S, Young S, Lam C, Folster JP, Whichard JM, McDermott PF. 2015. Whole-genome sequencing analysis accurately predicts antimicrobial resistance phenotypes in Campylobacter spp. Appl Environ Microbiol 82:459–466. doi: 10.1128/AEM.02873-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Food and Drug Administration. 2014. National Antimicrobial Resistance Monitoring System retail meat annual report U.S. Food and Drug Administration, Washington, DC: https://www.fda.gov/AnimalVeterinary/SafetyHealth/AntimicrobialResistance/NationalAntimicrobialResistanceMonitoringSystem/ucm059103.htm. [Google Scholar]
- 6.CLSI. 2016. Performance standards for antimicrobial susceptibility testing, 26th ed CLSI supplement M100-S26. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 7.Chen Y, Mukherjee S, Hoffmann M, Kotewicz ML, Young S, Abbott J, Luo Y, Davidson MK, Allard M, McDermott P, Zhao S. 2013. Whole-genome sequencing of gentamicin-resistant Campylobacter coli isolated from U.S. retail meats reveals novel plasmid-mediated aminoglycoside resistance genes. Antimicrob Agents Chemother 57:5398–5405. doi: 10.1128/AAC.00669-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Food and Drug Administration. 2011. National Antimicrobial Resistance Monitoring System retail meat annual report. U.S. Food and Drug Administration, Washington, DC. [Google Scholar]
- 9.Nirdnoy W, Mason CJ, Guerry P. 2005. Mosaic structure of a multiple-drug-resistant, conjugative plasmid from Campylobacter jejuni. Antimicrob Agents Chemother 49:2454–2459. doi: 10.1128/AAC.49.6.2454-2459.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qin S, Wang Y, Zhang Q, Chen X, Shen Z, Deng F, Wu C, Shen J. 2012. Identification of a novel genomic island conferring resistance to multiple aminoglycoside antibiotics in Campylobacter coli. Antimicrob Agents Chemother 56:5332–5339. doi: 10.1128/AAC.00809-12. [DOI] [PMC free article] [PubMed] [Google Scholar]