ABSTRACT
Streptomycin, the first drug used for the treatment of tuberculosis, shows limited activity against the highly resistant pathogen Mycobacterium abscessus. We recently identified two aminoglycoside-acetylating genes [aac(2′) and eis2] which, however, do not affect susceptibility to streptomycin. This suggests the existence of a discrete mechanism of streptomycin resistance. M. abscessus BLASTP analysis identified MAB_2385 as a close homologue of the 3″-O-phosphotransferase [APH(3″)] from the opportunistic pathogen Mycobacterium fortuitum as a putative streptomycin resistance determinant. Heterologous expression of MAB_2385 in Mycobacterium smegmatis increased the streptomycin MIC, while the gene deletion mutant M. abscessus ΔMAB_2385 showed increased streptomycin susceptibility. The MICs of other aminoglycosides were not altered in M. abscessus ΔMAB_2385. This demonstrates that MAB_2385 encodes a specific and prime innate streptomycin resistance determinant in M. abscessus. We further explored the feasibility of applying rpsL-based streptomycin counterselection to generate gene deletion mutants in M. abscessus. Spontaneous streptomycin-resistant mutants of M. abscessus ΔMAB_2385 were selected, and we demonstrated that the wild-type rpsL is dominant over the mutated rpsLK43R in merodiploid strains. In a proof of concept study, we exploited this phenotype for construction of a targeted deletion mutant, thereby establishing an rpsL-based counterselection method in M. abscessus.
KEYWORDS: rapidly growing mycobacteria, streptomycin, aminoglycoside resistance, phosphotransferases, counterselection marker, rpsL
INTRODUCTION
In the past several decades, Mycobacterium abscessus has emerged as one of the most clinically relevant nontuberculous mycobacteria (1–3). M. abscessus belongs to the rapidly growing mycobacteria (RGM) and was first described in 1953, isolated from a patient suffering from a knee infection (4, 5). M. abscessus skin and soft tissue infections, posttraumatic wound infections, or disseminated disease occurs after tattooing, mesotherapy, or surgical intervention. Sources of these infections include contaminated ink, multidose vials, surgical instruments, and even tap water (6–13, 70). Outbreaks of disseminated M. abscessus infections are described mainly in immunocompromised patients (12, 13). Additionally, M. abscessus accounts for more than 80% of all pulmonary infections with RGM (14). The pathogen poses a major threat for patients suffering from pulmonary disorders, like bronchiectasis or cystic fibrosis (1, 14). Occasionally, these pulmonary infections are fatal (14, 15). M. abscessus infections are difficult to treat due to the high-level antibiotic resistance conferred by an almost impermeable cell wall, drug efflux pumps, or drug-modifying enzymes (16–18).
Recently, various enzymes responsible for resistance to macrolides (Erm41), rifamycins (Arr_Mab), β-lactams (BlaMab), capreomycin (Eis2), and several aminoglycosides, such as amikacin, tobramycin, or kanamycin B [AAC(2′) and Eis2], were described (19–23). M. abscessus is naturally resistant to the aminoglycoside streptomycin (24), while streptomycin susceptibility was not affected by deletion of aac(2′) and eis2, indicating a discrete streptomycin resistance mechanism (22). In total, 12 putative aminoglycoside phosphotransferases, potentially having overlapping functions, are encoded in the genome of M. abscessus (18). In Mycobacterium fortuitum, another member of the RGM, an aminoglycoside, 3″-O-phosphotransferase [encoded by aph(3″)] was described as a streptomycin resistance determinant (25), while no streptomycin-modifying enzymes have been identified in other mycobacteria, like Mycobacterium tuberculosis or Mycobacterium smegmatis (26). These organisms are naturally susceptible to streptomycin (27–29). Streptomycin was the first drug used to treat tuberculosis (30). Streptomycin binds to the small ribosomal subunit and induces miscoding; it inhibits the initiation of translation and interferes with proofreading (31). Clinically acquired streptomycin resistance in M. tuberculosis is associated with single-point mutations in genes encoding the S12 protein of the small ribosomal subunit (rpsL) or the 16S rRNA (rrs) (32–35). RpsL and rRNA alterations interfere with streptomycin binding and ameliorate streptomycin-induced disturbance of ribosome dynamics and thereby confer streptomycin resistance (36–38).
Tools for the genetic manipulation of M. abscessus are still limited (22, 39, 40), as most of the conventional genetic tools for mycobacteria are not very effective and require high antibiotic concentrations due to the high level of intrinsic resistance (40). An rpsL-based counterselection strategy was previously exploited for allelic-exchange experiments in mycobacteria (41). rpsL+ can be used as counterselectable marker, since the wild-type rpsL is dominant over the mutated rpsLmut allele in a merodiploid strain, resulting in a growth disadvantage in the presence of streptomycin, i.e., a streptomycin-susceptible phenotype (42). The rpsL+ counterselection strategy has been exploited to generate various allelic-exchange mutants for characterizing mechanisms of genome stability, protein homeostasis, posttranslational modification, virulence, and drug resistance in M. smegmatis, Mycobacterium bovis BCG, and M. tuberculosis (43–52), but also in other genera (53).
In this study, we identified the innate streptomycin resistance determinant in M. abscessus. We compared the susceptibilities of the wild type and the targeted gene deletion mutant devoid of the putative streptomycin resistance determinant to streptomycin and other aminoglycosides. In addition, we demonstrate that rpsL mutations confer streptomycin resistance and introduce rpsL+ as a novel counterselection marker for M. abscessus, based on streptomycin susceptibility.
RESULTS
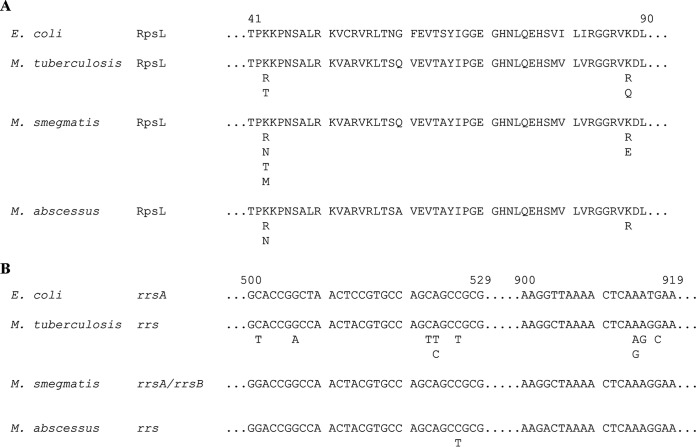
In silico identification of a putative streptomycin resistance determinant.
M. abscessus shows lower streptomycin susceptibility than M. tuberculosis. Streptomycin susceptibility in M. tuberculosis is affected by single nucleotide polymorphisms in either rpsL or rrs. Sequence comparison of M. abscessus ATCC 19977T RpsL (accession number YP_001704579) (18) and rrs (accession number NC_010397, region 1462398 to 1463901) (https://www.ncbi.nlm.nih.gov/nuccore/NC_010397.1?report=GenBank&from=1462398&to=1463901) (18) with homologous sequences in M. tuberculosis H37Rv (accession numbers: RpsL, NP_215196 [54]; rrs, NR_044826 [55]) and M. smegmatis mc2 155 (accession numbers: RpsL, YP_885784; rrsA, NC_008596, region 3823615 to 3825142 [https://www.ncbi.nlm.nih.gov/nuccore/NC_008596.1?from=3823615&to=3825142], and rrsB, NC_008596, region 5029475 to 5027948 [https://www.ncbi.nlm.nih.gov/nuccore/NC_008596.1?from=5027948&to=5029475]) did not reveal any alterations at codons 43 and 88 of RpsL or in relevant positions conferring streptomycin resistance in the 530 loop and the 915 region of 16S rRNA (Fig. 1). M. abscessus genome annotation, genome analysis, and aminoglycoside susceptibility testing suggested the presence of several putative aminoglycoside-modifying enzymes (18, 22). However, the already characterized aminoglycoside resistance genes aac(2′) and eis2 did not affect streptomycin resistance levels (22), indicating that another mechanism is responsible for the intrinsic streptomycin resistance in M. abscessus. In M. fortuitum, another RGM, a streptomycin resistance determinant, APH(3″), was identified (25). We used BLASTP to identify putative M. fortuitum FC1 APH(3″) (accession number ABC68330) (25) homologues in the genome of M. abscessus. Here, we identified MAB_2385 with an amino acid sequence identity of 55%.
FIG 1.
Sequence comparisons of RpsL (A) and rrs (B), the relevant streptomycin resistance determinants in M. tuberculosis and M. smegmatis. Published polymorphisms, each singly conferring streptomycin resistance, are listed below the relevant positions. The numbering is based on E. coli. Reference sequences of the E. coli K-12 substrain MG1655 RpsL (accession number NP_417801) (63) and rrsA (NC_000913, regions 4035531 to 4037072) (https://www.ncbi.nlm.nih.gov/nuccore/NC_000913.3?report=GenBank&from=4035531&to=4037072) (63) are shown. Amino acid substitutions in RpsL were observed at positions 43 and 88. The 530 loop and the 915 region of rrs are predominant for resistance mutations in M. tuberculosis H37Rv. M. smegmatis mc2 155 has two rrs copies (rrsA and rrsB), and thus, no resistance mutations were observed (29). In rare cases, nucleotide substitutions outside the two regions have been associated with streptomycin resistance (29, 33, 64–67). Comparison of the streptomycin resistance-conferring regions showed that amino acids and nucleotides, respectively, in M. abscessus ATCC 19977T are identical to those in M. tuberculosis H37Rv and M. smegmatis mc2 155. Generated spontaneous streptomycin-resistant M. abscessus ATCC 19977T mutants revealed substitutions in RpsL at amino acids 43 and 88 and mutations in the 530 loop of the single rrs gene. These polymorphisms have been described in other mycobacteria.
Heterologous expression of MAB_2385 and generation of a MAB_2385 gene deletion mutant.
MAB_2385 was cloned into the single-copy integrative vector pMV361-aac(3)IV and transformed into M. smegmatis. Transformation of pMV361-aac(3)IV-MAB_2385 into M. smegmatis increased the streptomycin MIC 8-fold (median, 4 mg/liter) compared to the wild type (median, 0.5 mg/liter) or strains carrying the empty vector pMV361-aac(3)IV (median, 0.5 mg/liter).
The suicide allelic replacement vector pSE-ΔMAB_2385 targeting MAB_2385 carrying an apramycin resistance cassette and M. tuberculosis katG for isoniazid negative selection was transformed into M. abscessus. M. tuberculosis KatG converts the prodrug isoniazid into its active form and thereby confers isoniazid susceptibility. Selection on apramycin was applied, and single-crossover transformants resulting from homologous recombination were identified by Southern blotting (Fig. 2). Counterselection of single-crossover recombinants with isoniazid resulted in the M. abscessus MAB_2385 gene deletion mutant (M. abscessus ΔMAB_2385). The complementation vector pMV361-aac(3)IV-MAB_2385 was transformed in M. abscessus ΔMAB_2385. The deletion of MAB_2385 and the gene's complementation in the mutant strain were verified by Southern blotting (Fig. 2).
FIG 2.
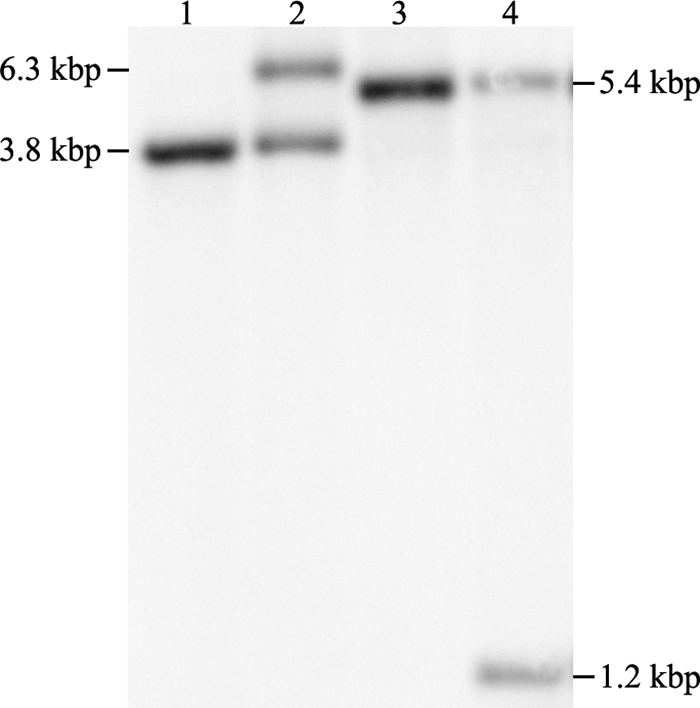
Southern blot analysis of the MAB_2385 gene deletion mutant and its complementation. Genomic DNA of the wild-type M. abscessus ATCC 19977T (lane 1), the pSE-MAB_2385 recombinant after apramycin selection for site-specific homologous recombination (single crossover) (lane 2), the MAB_2385 gene deletion mutant after counterselection with isoniazid (lane 3), and the complemented ΔMAB_2385 transformed with the vector pMV361-aac(3)IV-MAB_2385 (lane 4) was isolated, digested with Tth111I, and probed with a flanking region upstream of MAB_2385. The pattern is consistent with the predicted wild-type fragment at 3.8 kbp, single-crossover fragments at 3.8 kbp and 6.3 kbp, the gene deletion mutant fragment at 5.4 kbp, and gene complementation with fragments at 1.2 kbp and 5.4 kbp.
Antibiotic susceptibility testing of M. abscessus strains.
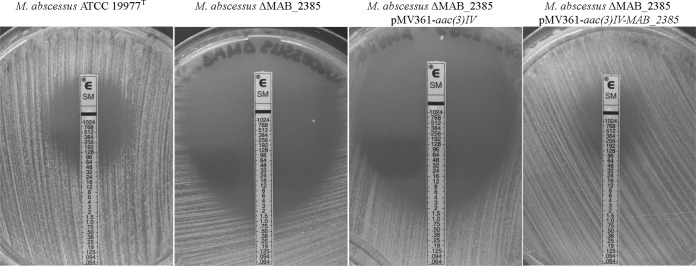
Initially, Etests were performed to compare streptomycin susceptibilities of the wild-type M. abscessus, M. abscessus ΔMAB_2385, M. abscessus ΔMAB_2385 pMV361-aac(3)IV, and M. abscessus ΔMAB_2385 pMV361-aac(3)IV-MAB_2385 strains. The M. abscessus wild type had a MIC of 64 mg/liter. The M. abscessus ΔMAB_2385 deletion mutant exhibited significantly increased susceptibility (1.5 mg/liter), while the complementation of MAB_2385 restored the wild-type phenotype (96 mg/liter). The vector pMV361-aac(3)IV did not affect the streptomycin susceptibility of M. abscessus ΔMAB_2385 (Fig. 3).
FIG 3.
Streptomycin Etest of M. abscessus ATCC 19977T, the MAB_2385 gene deletion mutant M. abscessus ΔMAB_2385, and transformants of the vector backbone pMV361-aac(3)IV and the complementation vector pMV361-aac(3)IV-MAB_2385. The bacterial suspension was evenly spread on the LB agar plate before the streptomycin Etest strip was placed on the surface of the plate. The plates were incubated for 5 days. The MIC of the wild type is 64 mg/liter, while the gene deletion mutant and the vector backbone transformant have a MIC of 1.5 mg/liter. The complemented mutant has a MIC similar that of the wild type, 96 mg/liter.
Susceptibility to streptomycin and other aminoglycosides was subsequently determined by broth microdilutions. Table 1 shows the median MICs evaluated on day 5. M. abscessus ΔMAB_2385 is 16-fold more susceptible to streptomycin than the wild type and the complementation mutant. Susceptibility to other aminoglycosides was not altered. In M. abscessus ΔMAB_2385 pMV361-aac(3)IV-MAB_2385, the wild-type phenotype was restored, while the M. abscessus ΔMAB_2385 pMV361-aac(3)IV control strain remained susceptible to streptomycin. On days 3, 7, and 12, similar differences in streptomycin MICs were observed (see Table S1 in the supplemental material).
TABLE 1.
Median MIC values after 5 days
| Drug | MIC (mg/liter)a |
|||
|---|---|---|---|---|
| Wild type | ΔMAB_2385 |
|||
| No vector | pMV361-aac(3)IV | pMV361-aac(3)IV-MAB_2385 | ||
| Streptomycin | 32 | 2 | 2 | 32 |
| Spectinomycin | >256 | >256 | >256 | >256 |
| Apramycin | 0.5 | 0.5 | NAb | NA |
| Kanamycin A | 1 | 1 | NA | NA |
| Kanamycin B | 8 | 8 | NA | NA |
| Amikacin | 2 | 1 | 2 | 2 |
| Hygromycin B | 256 | 256 | 256 | 256 |
Broth microdilutions in cation-adjusted Mueller Hinton broth.
NA, the aac(3)IV gene confers resistance to several aminoglycosides, including kanamycin A and kanamycin B, and thus, drug susceptibility testing was not applicable.
Generation and characterization of spontaneous streptomycin-resistant mutants.
The streptomycin-susceptible M. abscessus ΔMAB_2385 was plated on agar plates containing high streptomycin concentrations (32 mg/liter and 128 mg/liter) to select for spontaneous streptomycin-resistant colonies. Streptomycin-resistant colonies appeared at frequencies of approximately 10−6 and 10−7 on 32-mg/liter and 128-mg/liter streptomycin LB agar, respectively. Colonies were picked and screened by Sanger sequencing for alterations in rpsL and rrs. We identified mutations resulting in amino acid substitutions at codon 43 (K43N and K43R) and codon 88 (K88R) in RpsL. Other streptomycin-resistant colonies without any mutation in rpsL carried C491T mutations in rrs (Fig. 1). For further studies and to establish an rpsL-based counterselection marker in M. abscessus, a strain carrying an RpsL K43R alteration was used.
Reversion of the streptomycin-resistant phenotype.
In several bacterial species, merodiploid strains carrying an rpsL+ and an rpsLmut allele are streptomycin susceptible, indicating that the wild-type allele is dominant (42). We transformed a single-copy integrating vector carrying the wild-type [pMV361-aac(3)IV-rpsL+] or the K43R [pMV361-aac(3)IV-rpsLK43R] mutated rpsL into the streptomycin-resistant M. abscessus ΔMAB_2385 rpsLK43R strain. As control, we transformed the empty backbone vector pMV361-aac(3)IV. The strains were plated on LB agar containing apramycin or a combination of apramycin and streptomycin and were incubated for 5 days before CFU were determined. Apramycin selection was applied to avoid vector loss through integrase-mediated excision (56). A one-tailed (HA<H0) two-sample Student's t test [t test with 4 degrees of freedom: t(4) = −7.3853; P < 0.001] revealed a significantly (104-fold) reduced number of CFU of the strain carrying the pMV361-aac(3)IV-rpsL+ vector when it was plated on apramycin and streptomycin compared to apramycin alone. Bacteria carrying the vectors pMV361-aac(3)IV [t(4) = −0.1261; P = 0.4529] and pMV361-aac(3)IV-rpsLK43R [t(4) = −1.3399; P = 0.1257] did not show reduced numbers of CFU when plated on apramycin and streptomycin compared to apramycin alone (Fig. 4). The data indicate that rpsL+ confers a streptomycin-susceptible phenotype on M. abscessus ΔMAB_2385 rpsLK43R.
FIG 4.
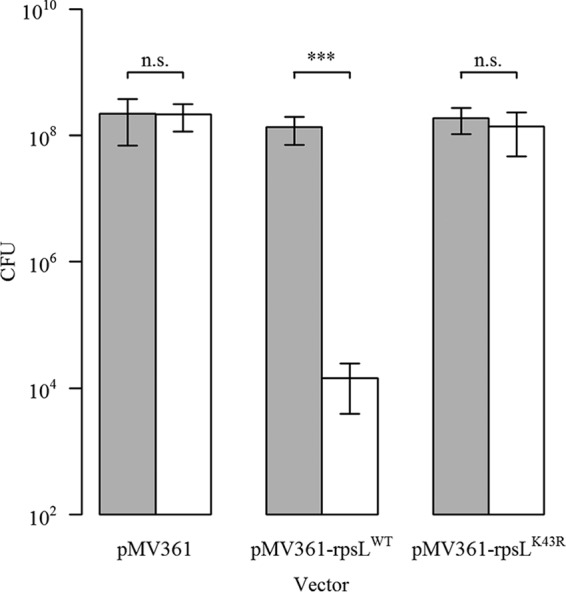
CFU obtained after plating M. abscessus ΔMAB_2385 rpsLK43R strains carrying pMV361-aac(3)IV (empty control), pMV361-aac(3)IV-rpsL+, or pMV361-aac(3)IV-rpsLK43R on apramycin (gray bars) or apramycin and streptomycin (white bars) selective LB agar plates. Each strain was diluted to an OD600 of 0.1, plated on apramycin- or apramycin- and streptomycin-containing LB agar plates, and incubated for 5 days. Bacterial counts on the different antibiotics were determined. The plot shows the means and standard deviations of the results of three independent experiments. One-tailed two-sample Student's t test: ***, P < 0.001 n.s., not significant.
Streptomycin as a counterselection marker.
Having shown that rpsL+ confers streptomycin susceptibility on M. abscessus ΔMAB_2385 rpsLK43R, we wanted to test whether the rpsL+ counterselection method could be used for the generation of targeted deletion mutants in M. abscessus. For a proof of concept study, we chose the eis2 (MAB_4532c) locus, which was previously deleted by Rominski et al. (22). We constructed a novel allelic replacement vector by cloning the rpsL+ allele and the DsRed2 fluorescence marker into the eis2 suicide vector pSE-Δeis2 to produce pSE-aac(3)IV:DsRed2-Δeis2-rpsL+. The vector was transformed in M. abscessus ΔMAB_2385 rpsLK43R. Transformants were selected on apramycin plates and identified by red fluorescence. Single-crossover recombinants identified by Southern blotting were plated on streptomycin LB agar to select for double-crossover recombinants. The CFU counts on streptomycin plates were compared to the CFU counts on apramycin-selective LB agar plates. In three independent experiments, the mean frequency was 8.25 × 10−4 based on CFU counts. Fluorescence imaging determined that on average 11.14% of the clones still carried the DsRed2 of the suicide vector, while almost 89% did not show red fluorescence. Twenty nonfluorescent colonies were picked and screened by Southern blotting for second intramolecular recombination. All the colonies had undergone this intramolecular recombination. Five were unmarked eis2 gene deletion mutants, while 15 were eis2 wild-type revertants. These results demonstrated that the streptomycin counterselection strategy can be applied to generate deletion mutants in the M. abscessus ΔMAB_2385 rpsLK43R strain (Fig. 5).
FIG 5.
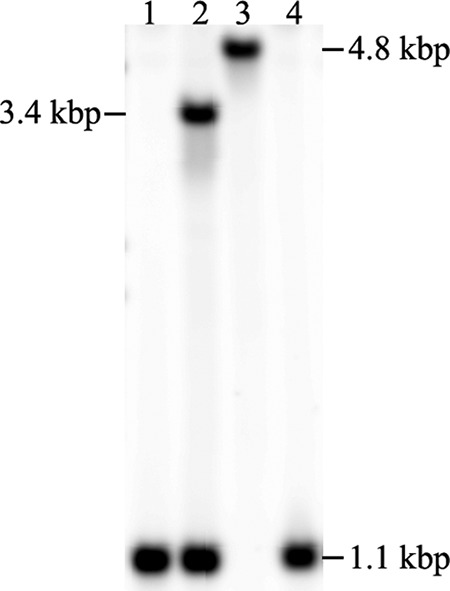
Southern blot analysis of the eis2 gene deletion mutant, with streptomycin used for counterselection. Genomic DNA of the streptomycin-resistant M. abscessus ΔMAB_2385 rpsLK43R (lane 1); after transformation with pSE-(aac3)IV:DsRed2-Δeis2-rpsL+ vector (lane 2); and each of the two possible outcomes after counterselection with streptomycin, the gene deletion mutant (lane 3) or the parental strain (lane 4), was isolated and digested with Van91I. The pattern obtained is in accordance with the pattern reported by Rominski et al. (22); a 1.1-kpb wild-type fragment, a 1.1-kbp and a 3.4-kbp fragment for single-crossover transformants, and a 4.8-kbp fragment for the eis2 gene deletion mutant.
DISCUSSION
MAB_2385 is the intrinsic streptomycin resistance determinant in M. abscessus.
M. abscessus is a clinically relevant pathogen with several intrinsic resistance mechanisms. Increased interest in this highly resistant mycobacterium resulted in the identification of resistance mechanisms for macrolides (19), rifamycins (20), β-lactams (21), and capreomycin and aminoglycosides (22). The molecular mechanism of M. abscessus streptomycin resistance has not been identified yet. Alterations in the rpsL or the rrs gene associated with clinically acquired streptomycin resistance in M. tuberculosis have not been found in M. abscessus (29). In the genome of M. abscessus, several putative aminoglycoside-modifying enzymes, mostly aminoglycoside phosphotransferases, were annotated (18). By scanning the genome of M. abscessus for the presence of the streptomycin resistance determinant APH(3″) of M. fortuitum, we identified MAB_2385 as a candidate resistance gene. Heterologous expression of MAB_2385 in M. smegmatis increased MIC levels. Subsequent drug susceptibility testing in M. abscessus ΔMAB_2385 showed increased susceptibility to streptomycin compared to the wild type and the complemented strain. The susceptibility of M. abscessus ΔMAB_2385 to other aminoglycosides was not affected. Heinzel et al. had previously demonstrated that 3″-phosphotransferases specifically phosphorylate streptomycin, but not kanamycin (57). These findings support the assumption that MAB_2385 is a streptomycin 3″-O-phosphotransferase. However, additional biochemical studies should be performed to directly demonstrate streptomycin-modifying activity of MAB_2385.
Streptomycin as a counterselection marker in M. abscessus.
So far, the genetic toolbox for M. abscessus is ill equipped, and only a few efficient positive and negative selectable markers have been described (22, 39, 40). Sensitizing M. abscessus to streptomycin by deletion of MAB_2385 and subsequent selection of streptomycin-resistant rpsL mutants offers the possibility to exploit the rpsL+ counterselection strategy described previously for M. tuberculosis, M. bovis BCG, and M. smegmatis (41, 58). Hence, we selected rpsL mutants resulting in K43R substitutions, since high-level resistance was observed in other mycobacteria carrying the corresponding allele (32).
The introduction of a vector with the wild-type small ribosomal protein S12 allele (rpsL+) into M. abscessus ΔMAB_2385 rpsLK43R significantly reduced the number of colonies grown on streptomycin-containing agar plates by a factor of approximately 10−4, indicating that rpsL+ is an efficient counterselection marker. Intramolecular recombination between the wild-type and the mutant rpsL most likely explains why a few bacteria still grew on streptomycin plates. The frequency is approximately 10−4 to 10−5 (59). Other events, such as the occurrence of spontaneous resistance mutations in the wild-type rpsL (10−6) or the loss of the vector catalyzed by the vector's integrase (8 × 10−5) (56) coupled with spontaneous apramycin resistance-conferring mutations (10−6) (in total, 8 × 10−11) are less frequent. Finally, in a proof of concept study, we generated a gene deletion mutant by introducing rpsL+ in a modified eis2 allelic replacement vector and subsequent streptomycin counterselection, thereby establishing a new negative selection marker for M. abscessus.
In summary, we demonstrated that MAB_2385 specifically confers streptomycin resistance but does not have an effect on susceptibility to other aminoglycosides. Further, we were able to generate spontaneous streptomycin-resistant mutants in the M. abscessus ΔMAB_2385 strain carrying mutations in rpsL. This enabled us to introduce an rpsL-based streptomycin counterselection system in M. abscessus by exploiting the fact that in merodiploid strains the wild-type rpsL confers a streptomycin-susceptible phenotype.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
M. abscessus ATCC 19977T was grown either in Middlebrook 7H9-OADC (oleic acid-albumin-dextrose-catalase) (BD Difco, Sparks, MD, USA) with 0.05% Tween 80 (7H9) (Hänsler AG, Switzerland) or on solid LB agar plates. Kanamycin A (50 mg/liter), apramycin (50 mg/liter), isoniazid (32 mg/liter), or streptomycin (32 mg/liter or 128 mg/liter) (Sigma-Aldrich, Switzerland) was added for primary selection of transformants and counterselection and isolation of spontaneous resistant mutants, respectively. Preparation of electrocompetent cells (22) and extraction of genomic DNA with phenol-chloroform-isoamyl alcohol (41) were described previously.
M. smegmatis mc2 155 was grown in LB with 0.05% Tween 80 or 7H9 broth and on Middlebrook 7H10-OADC or LB agar plates. For primary selection, kanamycin A (50 mg/liter) or apramycin (50 mg/liter) was supplemented. Electrocompetent cells were prepared as described previously, except that 7H9 was used as the growth medium (60). Escherichia coli XL1-Blue MRF′ was grown in LB broth or on LB agar plates containing either ampicillin (liquid, 100 mg/liter; solid, 120 mg/liter) or kanamycin A (50 mg/liter) (Sigma-Aldrich, Switzerland). All bacterial strains were grown at 37°C.
Vector construction and generation of strains.
The strains used in this study are listed in Table 2. Promega's (Switzerland) pGEM-T easy vector system was used for cloning of PCR fragments. The integrative single-copy vector pMV361-aac(3)IV and the suicide vector pSE-apr-katG (pSE) were described previously (22). For heterologous expression of MAB_2385 in M. smegmatis and for complementation, a 1,606-bp HindIII/NheI fragment of the MAB_2385 region was amplified from genomic DNA of M. abscessus ATCC 19977T by PCR with the primers 5′-CGGAAGCTTCAAGGATTACCCGACACCAC-3′ and 5′-CCGCTAGCTGGATCAGCTCCAGGATG-3′ (restriction enzyme recognition sites are underlined). The PCR fragment was ligated into pGEM-T Easy and subsequently cut out with HindIII and NheI. pMV361-aac(3)IV was also cut with HindIII and NheI, and the MAB_2385 fragment was ligated into the linearized backbone, resulting in pMV361-aac(3)IV-MAB_2385. The vectors pMV361-aac(3)IV and pMV361-aac(3)IV-MAB_2385 were transformed into M. smegmatis mc2 155.
TABLE 2.
Strains used in this study
| Strain | Vector | Description | Source |
|---|---|---|---|
| E. coli XL1-Blue MRF′ | Cloning | Stratagene | |
| M. smegmatis mc2 155 | Wild type | 68 | |
| pMV361-aac(3)IV | Backbone control | This study | |
| pMV361-aac(3)IV-MAB_2385 | Expression of MAB_2385 | This study | |
| M. abscessus ATCC 19977T | Wild type | 69 | |
| M. abscessus ΔMAB_2385a | Gene deletion mutant | This study | |
| pMV361-aac(3)IV | Backbone control | This study | |
| pMV361-aac(3)IV-MAB_2385 | Gene complementation | This study | |
| M. abscessus ΔMAB_2385 rpsLK43Rb | K43R alteration in RpsL | This study | |
| pMV361-aac(3)IV | Backbone control | This study | |
| pMV361-aac(3)IV-rpsL+ | Expression of rpsL+ | This study | |
| pMV361-aac(3)IV-rpsLK43R | Expression of rpsLK43R | This study |
M. abscessus ΔMAB_2385 was derived from M. abscessus ATCC 19977T.
M. abscessus ΔMAB_2385 rpsLK43R was derived from M. abscessus ΔMAB_2385.
The gene deletion suicide vector pSE-ΔMAB_2385 was generated by cloning two regions flanking MAB_2385. A 1,548-bp PscI/KpnI upstream fragment and a 1,484-bp KpnI/HindIII downstream fragment were amplified by PCR using genomic DNA of M. abscessus ATCC 19977T. The primers for the amplification of the upstream fragment were 5′-GGGACATGTCGGGGATTTCTACACGCTTA-3′ and 5′-CGGGGTACCTTTCGCATAGCGAGAACC-3′, while the primers for the amplification of the downstream fragment were 5′-CGGGGTACCAACCGGTACGGAGCGTCT-3′ and 5′-CCCAAGCTTACACACACGTGATGCAGACC-3′. Both fragments were separately cloned into the pGEM-T Easy vector, subsequently cut with the appropriate enzymes, and stepwise ligated into the linearized pSE vector. The vector pSE-ΔMAB_2385 was transformed in M. abscessus ATCC 19977T. Apramycin was used for positive selection and isoniazid for counterselection. Genomic DNA of M. abscessus transformants was isolated and cut with Tth111I to confirm gene deletion by Southern blotting. The vector pMV361-aac(3)IV-MAB_2385 was transformed into the M. abscessus ΔMAB_2385 deletion mutant to complement the strain. As a control, the vector backbone pMV361-aac(3)IV was transformed in parallel.
A suspension of mid-log-phase M. abscessus ΔMAB_2385 was plated on LB agar containing streptomycin (32 mg/liter and 128 mg/liter) and incubated for 5 days. Colonies were picked and screened for alterations in rpsL by Sanger sequencing with the primers 5′-GTTACCAGCTGCGAACCGTA-3′ and 5′-GAAAAACGCAGGACAACAGG-3′. A clone carrying a K43R amino acid substitution (M. abscessus ΔMAB_2385 rpsLK43R) was used for further studies. Mutations in rrs were detected by sequencing with the primers 5′-ACCGGGAATTTGACTCAGGT-3′ and 5′-GAAAACGAGCGAGGCTATGT-3′.
The wild-type (rpsL+) and the mutated (rpsLK43R) rpsL were PCR amplified with the primers 5′-CGGAAGCTTCTCCAGAGCGCCGTACAC-3′ and 5′-CCCGCTAGCCGCACTGTCGTTCCTGGAT-3′. This resulted in 1,375-bp HindIII/NheI fragments, which were cloned into pGEM-T Easy, cut with HindIII and NheI, and subsequently ligated into the HindIII/NheI-linearized pMV361-aac(3)IV vector. The resulting vectors, pMV361-aac(3)IV-rpsL+ and pMV361-aac(3)IV-rpsLK43R, as well as the backbone vector pMV361-aac(3)IV, were transformed into M. abscessus ΔMAB_2385 rpsLK43R.
A wild-type rpsL fragment (1,375 bp) with MunI (instead of HindIII/NheI) restriction sites was amplified by PCR with the primers 5′-CGGCAATTGCTCCAGAGCGCCGTACAC-3′ and 5′-CCCCAATTGCGCACTGTCGTTCCTGGAT-3′. The fragment was cloned into pGEM-T Easy, reisolated by digestion with MunI, and inserted into the MunI-linearized eis2 suicide targeting vector pSE-Δeis2 (22), resulting in pSE-Δeis2-rpsL+. This vector was further modified to facilitate identification of transformants (which are mostly single-crossover transformants) and double-crossover recombinants resulting from counterselection of single-crossover transformants: the apramycin cassette was replaced by an apramycin (aac3)IV:DsRed2 fusion cassette to produce pSE-(aac3)IV:DsRed2-Δeis2-rpsL+. This vector was transformed in M. abscessus ΔMAB_2385 rpsLK43R, and positive selection was performed with apramycin to select transformants resulting from intermolecular homologous recombination between cloned eis2 sequences and the target gene. Subsequent counterselection of single-crossover transformants was performed with streptomycin (32 mg/liter). The second (intramolecular) recombination events resulted in the deletion of eis2 or reversion to wild-type eis2. This was confirmed by Southern blotting as described by Rominski et al. (22).
Etest.
Bacteria were grown on LB agar plates. After incubation, a bacterial suspension was adjusted to a McFarland standard of 0.50. Therefore, 1 to 10 colonies were picked with a cotton swab and transferred to a glass tube containing 0.9% NaCl. A small volume of the suspension was evenly spread on an LB agar plate with a cotton swab. A streptomycin Etest strip (concentration range, 0.064 to 1,024 mg/liter) was placed in the middle of the plate and incubated for 5 days until growth at the border was well defined before the MIC was determined.
MIC assays.
Streptomycin, spectinomycin, apramycin, amikacin, kanamycin A, and kanamycin B (all from Sigma-Aldrich, Switzerland) were dissolved in water and filter sterilized. Hygromycin B (Invivogen, San Diego, CA, USA) was delivered as a 100-g/liter solution. The MIC assays were in principle performed according to CLSI guidelines for antimicrobial susceptibility testing, with slight modifications, and were conducted essentially as described previously in 96-well microtiter plates (Corning Inc., NY, USA) (20, 61). For preparation of the inocula, M. abscessus colonies were picked from LB plates as described previously, while M. smegmatis was grown in liquid cultures. Growth of M. abscessus was visually inspected on days 3, 5, 7, and 12, while growth of M. smegmatis was visually inspected on day 3.
Selectivity in merodiploid rpsL strains.
M. abscessus ΔMAB_2385 rpsLK43R carrying any of the three vectors pMV361-aac(3)IV, pMV361-aac(3)IV-rpsL+, and pMV361-aac(3)IV-rpsLK43R was inoculated in 10 ml 7H9 with apramycin (50 mg/liter) and grown for 5 days. The optical density at 600 nm (OD600) was adjusted to 0.1. Subsequently, 10-fold serial dilutions of the bacterial suspension were prepared, and 100 μl was spread on LB agar plates with apramycin (50 mg/liter) or a combination of apramycin (50 mg/liter) and streptomycin (32 mg/liter). After 5 days of incubation, CFU were counted. Statistical analysis was performed with the statistical software R (version 3.1.1) (62).
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by a grant from the Swiss National Science Foundation (grant no. 31003 A_153349), as well as by the Institute of Medical Microbiology and the University of Zurich.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.01427-17.
REFERENCES
- 1.Griffith DE. 2003. Emergence of nontuberculous Mycobacteria as pathogens in cystic fibrosis. Am J Respir Crit Care Med 167:810–812. doi: 10.1164/rccm.2301001. [DOI] [PubMed] [Google Scholar]
- 2.Varghese B, Memish Z, Abuljadayel N, Al-Hakeem R, Alrabiah F, Al-Hajoj SA. 2013. Emergence of clinically relevant non-tuberculous mycobacterial infections in Saudi Arabia. PLoS Negl Trop Dis 7:e2234. doi: 10.1371/journal.pntd.0002234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bryant JM, Grogono DM, Rodriguez-Rincon D, Everall I, Brown KP, Moreno P, Verma D, Hill E, Drijkoningen J, Gilligan P, Esther CR, Noone PG, Giddings O, Bell SC, Thomson R, Wainwright CE, Coulter C, Pandey S, Wood ME, Stockwell RE, Ramsay KA, Sherrard LJ, Kidd TJ, Jabbour N, Johnson GR, Knibbs LD, Morawska L, Sly PD, Jones A, Bilton D, Laurenson I, Ruddy M, Bourke S, Bowler IC, Chapman SJ, Clayton A, Cullen M, Dempsey O, Denton M, Desai M, Drew RJ, Edenborough F, Evans J, Folb J, Daniels T, Humphrey H, Isalska B, Jensen-Fangel S, Jönsson B, Jones AM, Katzenstein TL, Lillebaek T, MacGregor G, Mayell S, Millar M, Modha D, Nash EF, O'Brien C, O'Brien D, Ohri C, Pao CS, Peckham D, Perrin F, Perry A, Pressler T, Prtak L, Qvist T, Robb A, Rodgers H, Schaffer K, Shafi N, van Ingen J, Walshaw M, Watson D, West N, Whitehouse J, Haworth CS, Harris SR, Ordway D, Parkhill J, Floto RA. 2016. Emergence and spread of a human-transmissible multidrug-resistant nontuberculous mycobacterium. Science 354:751–757. doi: 10.1126/science.aaf8156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shinnick TM, Good RC. 1994. Mycobacterial taxonomy. Eur J Clin Microbiol Infect Dis 13:884–901. doi: 10.1007/BF02111489. [DOI] [PubMed] [Google Scholar]
- 5.Moore M, Frerichs JB. 1953. An unusual acid-fast infection of the knee with subcutaneous, abscess-like lesions of the gluteal region: report of a case with a study of the organism, Mycobacterium abscessus, n. sp. J Investig Dermatol 20:133–169. doi: 10.1038/jid.1953.18. [DOI] [PubMed] [Google Scholar]
- 6.Falsey RR, Kinzer MH, Hurst S, Kalus A, Pottinger PS, Duchin JS, Zhang J, Noble-Wang J, Shinohara MM. 2013. Cutaneous inoculation of nontuberculous mycobacteria during professional tattooing: a case series and epidemiologic study. Clin Infect Dis 57:e143-7. doi: 10.1093/cid/cit347. [DOI] [PubMed] [Google Scholar]
- 7.Bechara C, Macheras E, Heym B, Pages A, Auffret N. 2010. Mycobacterium abscessus skin infection after tattooing: first case report and review of the literature. Dermatology 221:1–4. doi: 10.1159/000313974. [DOI] [PubMed] [Google Scholar]
- 8.Newman MI, Camberos AE, Ascherman J. 2005. Mycobacteria abscessus outbreak in US patients linked to offshore surgicenter. Ann Plast Surg 55:107–110. doi: 10.1097/01.sap.0000168030.87804.93. [DOI] [PubMed] [Google Scholar]
- 9.Maurer FP, Castelberg C, Von Braun A, Wolfensberger A, Bloemberg GV, Böttger EC, Somoskovi A. 2014. Postsurgical wound infections due to rapidly growing mycobacteria in Swiss medical tourists following cosmetic surgery in Latin America between 2012 and 2014. Euro Surveill 19:20905. doi: 10.2807/1560-7917.ES2014.19.37.20905. [DOI] [PubMed] [Google Scholar]
- 10.Furuya EY, Paez A, Srinivasan A, Cooksey R, Augenbraun M, Baron M, Brudney K, Della-Latta P, Estivariz C, Fischer S, Flood M, Kellner P, Roman C, Yakrus M, Weiss D, Granowitz EV. 2008. Outbreak of Mycobacterium abscessus wound infections among “lipotourists” from the United States who underwent abdominoplasty in the Dominican Republic. Clin Infect Dis 46:1181–1188. doi: 10.1086/529191. [DOI] [PubMed] [Google Scholar]
- 11.Falkinham JO., III 2011. Nontuberculous mycobacteria from household plumbing of patients with nontuberculous mycobacteria disease. Emerg Infect Dis 17:419–424. doi: 10.3201/eid1703.101510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morales P, Gil A, Santos M. 2010. Mycobacterium abscessus infection in transplant recipients. Transplant Proc 42:3058–3060. doi: 10.1016/j.transproceed.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 13.Fukui S, Sekiya N, Takizawa Y, Morioka H, Kato H, Aono A, Chikamatsu K, Mitarai S, Kobayashi S, Kamei S, Setoguchi K. 2015. Disseminated Mycobacterium abscessus Infection following septic arthritis: a case report and review of the literature. Medicine 94:e861. doi: 10.1097/MD.0000000000000861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Griffith DE, Girard WM, Wallace RJ Jr. 1993. Clinical features of pulmonary disease caused by rapidly growing mycobacteria. An analysis of 154 patients. Am Rev Respir Dis 147:1271–1278. doi: 10.1164/ajrccm/147.5.1271. [DOI] [PubMed] [Google Scholar]
- 15.Sermet-Gaudelus I, Le Bourgeois M, Pierre-Audigier C, Offredo C, Guillemot D, Halley S, Akoua-Koffi C, Vincent V, Sivadon-Tardy V, Ferroni A, Berche P, Scheinmann P, Lenoir G, Gaillard JL. 2003. Mycobacterium abscessus and children with cystic fibrosis. Emerg Infect Dis 9:1587–1591. doi: 10.3201/eid0912.020774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nessar R, Cambau E, Reyrat JM, Murray A, Gicquel B. 2012. Mycobacterium abscessus: a new antibiotic nightmare. J Antimicrob Chemother 67:810–818. doi: 10.1093/jac/dkr578. [DOI] [PubMed] [Google Scholar]
- 17.Brown-Elliott BA, Wallace RJ Jr. 2002. Clinical and taxonomic status of pathogenic nonpigmented or late-pigmenting rapidly growing mycobacteria. Clin Microbiol Rev 15:716–746. doi: 10.1128/CMR.15.4.716-746.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ripoll F, Pasek S, Schenowitz C, Dossat C, Barbe V, Rottman M, Macheras E, Heym B, Herrmann JL, Daffé M, Brosch R, Risler JL, Gaillard JL. 2009. Non mycobacterial virulence genes in the genome of the emerging pathogen Mycobacterium abscessus. PLoS One 4:e5660. doi: 10.1371/journal.pone.0005660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nash KA, Brown-Elliott BA, Wallace RJ Jr. 2009. A novel gene, erm(41), confers inducible macrolide resistance to clinical isolates of Mycobacterium abscessus but is absent from Mycobacterium chelonae. Antimicrob Agents Chemother 53:1367–1376. doi: 10.1128/AAC.01275-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rominski A, Roditscheff A, Selchow P, Böttger EC, Sander P. 2017. Intrinsic rifamycin resistance of Mycobacterium abscessus is mediated by ADP-ribosyltransferase MAB_0591. J Antimicrob Chemother 72:376–384. doi: 10.1093/jac/dkw466. [DOI] [PubMed] [Google Scholar]
- 21.Dubée V, Soroka D, Cortes M, Lefebvre AL, Gutmann L, Hugonnet JE, Arthur M, Mainardi JL. 2015. Impact of β-lactamase inhibition on the activity of ceftaroline against Mycobacterium tuberculosis and Mycobacterium abscessus. Antimicrob Agents Chemother 59:2938–2941. doi: 10.1128/AAC.05080-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rominski A, Selchow P, Becker K, Brülle JK, Dal Molin M, Sander P. 2017. Elucidation of Mycobacterium abscessus aminoglycoside and capreomycin resistance by targeted deletion of three putative resistance genes. J Antimicrob Chemother 72:2191–2200. doi: 10.1093/jac/dkx125. [DOI] [PubMed] [Google Scholar]
- 23.Rominski A, Schultess B, Müller DM, Keller PM, Sander P. 2017. Effect of β-lactamase production and β-lactam instability on MIC testing results for Mycobacterium abscessus. J Antimicrob Chemother 72:3070–3078. doi: 10.1093/jac/dkx284. [DOI] [PubMed] [Google Scholar]
- 24.Li G, Lian LL, Wan L, Zhang J, Zhao X, Jiang Y, Zhao LL, Liu H, Wan K. 2013. Antimicrobial susceptibility of standard strains of nontuberculous mycobacteria by microplate Alamar Blue assay. PLoS One 8:e84065. doi: 10.1371/journal.pone.0084065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramón-García S, Otal I, Martín C, Gómez-Lus R, Aínsa JA. 2006. Novel streptomycin resistance gene from Mycobacterium fortuitum. Antimicrob Agents Chemother 50:3920–3922. doi: 10.1128/AAC.00223-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramirez MS, Tolmasky ME. 2010. Aminoglycoside modifying enzymes. Drug Resist Updat 13:151–171. doi: 10.1016/j.drup.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wanger A, Mills K. 1996. Testing of Mycobacterium tuberculosis susceptibility to ethambutol, isoniazid, rifampin, and streptomycin by using Etest. J Clin Microbiol 34:1672–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ritz N, Tebruegge M, Connell TG, Sievers A, Robins-Browne R, Curtis N. 2009. Susceptibility of Mycobacterium bovis BCG vaccine strains to antituberculous antibiotics. Antimicrob Agents Chemother 53:316–318. doi: 10.1128/AAC.01302-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Springer B, Kidan YG, Prammananan T, Ellrott K, Böttger EC, Sander P. 2001. Mechanisms of streptomycin resistance: selection of mutations in the 16S rRNA gene conferring resistance. Antimicrob Agents Chemother 45:2877–2884. doi: 10.1128/AAC.45.10.2877-2884.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marshall G, Blacklock JWS, Cameron C, Capon NB, Cruickshank R, Gaddum JH, Heaf FRG, Hill AB, Houghton LE, Hoyle JC, Raistrick H, Scadding JG, Tytler WH, Wilson GS, Hart PD. 1948. Streptomycin treatment of pulmonary tuberculosis. Br Med J 2:769–782. doi: 10.1136/bmj.2.4582.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moazed D, Noller HF. 1987. Interaction of antibiotics with functional sites in 16S ribosomal RNA. Nature 327:389–394. doi: 10.1038/327389a0. [DOI] [PubMed] [Google Scholar]
- 32.Meier A, Sander P, Schaper KJ, Scholz M, Böttger EC. 1996. Correlation of molecular resistance mechanisms and phenotypic resistance levels in streptomycin-resistant Mycobacterium tuberculosis. Antimicrob Agents Chemother 40:2452–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sreevatsan S, Pan X, Stockbauer KE, Williams DL, Kreiswirth BN, Musser JM. 1996. Characterization of rpsL and rrs mutations in streptomycin-resistant Mycobacterium tuberculosis isolates from diverse geographic localities. Antimicrob Agents Chemother 40:1024–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Honoré N, Cole ST. 1994. Streptomycin resistance in mycobacteria. Antimicrob Agents Chemother 38:238–242. doi: 10.1128/AAC.38.2.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Musser JM. 1995. Antimicrobial agent resistance in mycobacteria: Molecular genetic insights. Clin Microbiol Rev 8:496–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Allen PN, Noller HF. 1989. Mutations in ribosomal proteins S4 and S12 influence the higher order structure of 16S ribosomal RNA. J Mol Biol 208:457–468. doi: 10.1016/0022-2836(89)90509-3. [DOI] [PubMed] [Google Scholar]
- 37.De Stasio EA, Moazed D, Noller HF, Dahlberg AE. 1989. Mutations in 16S ribosomal RNA disrupt antibiotic-RNA interactions. EMBO J 8:1213–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carter AP, Clemons WM, Brodersen DE, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V. 2000. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature 407:340–348. doi: 10.1038/35030019. [DOI] [PubMed] [Google Scholar]
- 39.Medjahed H, Gaillard JL, Reyrat JM. 2010. Mycobacterium abscessus: a new player in the mycobacterial field. Trends Microbiol 18:117–123. doi: 10.1016/j.tim.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 40.Cortes M, Singh AK, Reyrat JM, Gaillard JL, Nassif X, Herrmann J-L. 2011. Conditional gene expression in Mycobacterium abscessus. PLoS One 6:e29306. doi: 10.1371/journal.pone.0029306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sander P, Meier A, Böttger EC. 1995. rpsL+: a dominant selectable marker for gene replacement in mycobacteria. Mol Microbiol 16:991–1000. doi: 10.1111/j.1365-2958.1995.tb02324.x. [DOI] [PubMed] [Google Scholar]
- 42.Lederberg J. 1951. Streptomycin resistance; a genetically recessive mutation. J Bacteriol 61:549–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frischkorn K, Sander P, Scholz M, Teschner K, Prammananan T, Böttger EC. 1998. Investigation of mycobacterial recA function: protein introns in the RecA of pathogenic mycobacteria do not affect competency for homologous recombination. Mol Microbiol 29:1203–1214. doi: 10.1046/j.1365-2958.1998.01003.x. [DOI] [PubMed] [Google Scholar]
- 44.Sander P, De Rossi E, Böddinghaus B, Cantoni R, Branzoni M, Böttger EC, Takiff H, Rodriquez R, Lopez G, Riccardi G. 2000. Contribution of the multidrug efflux pump LfrA to innate mycobacterial drug resistance. FEMS Microbiol Lett 193:19–23. doi: 10.1111/j.1574-6968.2000.tb09396.x. [DOI] [PubMed] [Google Scholar]
- 45.Sander P, Papavinasasundaram KG, Dick T, Stavropoulos E, Ellrott K, Springer B, Colston MJ, Böttger EC. 2001. Mycobacterium bovis BCG recA deletion mutant shows increased susceptibility to DNA-damaging agents but wild-type survival in a mouse infection model. Infect Immun 69:3562–3568. doi: 10.1128/IAI.69.6.3562-3568.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sander P, Rezwan M, Walker B, Rampini SK, Kroppenstedt RM, Ehlers S, Keller C, Keeble JR, Hagemeier M, Colston MJ, Springer B, Böttger EC. 2004. Lipoprotein processing is required for virulence of Mycobacterium tuberculosis. Mol Microbiol 52:1543–1552. doi: 10.1111/j.1365-2958.2004.04041.x. [DOI] [PubMed] [Google Scholar]
- 47.Rampini SK, Selchow P, Keller C, Ehlers S, Böttger EC, Sander P. 2008. LspA inactivation in Mycobacterium tuberculosis results in attenuation without affecting phagosome maturation arrest. Microbiology 154:2991–3001. doi: 10.1099/mic.0.2008/018895-0. [DOI] [PubMed] [Google Scholar]
- 48.Tschumi A, Nai C, Auchli Y, Hunziker P, Gehrig P, Keller P, Grau T, Sander P. 2009. Identification of apolipoprotein N-acyltransferase (Lnt) in mycobacteria. J Biol Chem 284:27146–27156. doi: 10.1074/jbc.M109.022715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Imkamp F, Striebel F, Sutter M, Ozcelik D, Zimmermann N, Sander P, Weber-Ban E. 2010. Dop functions as a depupylase in the prokaryotic ubiquitin-like modification pathway. EMBO Rep 11:791–797. doi: 10.1038/embor.2010.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johansen P, Fettelschoss A, Amstutz B, Selchow P, Waeckerle-Men Y, Keller P, Deretic V, Held L, Kündig TM, Böttger EC, Sander P. 2011. Relief from Zmp1-mediated arrest of phagosome maturation is associated with facilitated presentation and enhanced immunogenicity of mycobacterial antigens. Clin Vaccine Immunol 18:907–913. doi: 10.1128/CVI.00015-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brülle JK, Tschumi A, Sander P. 2013. Lipoproteins of slow-growing Mycobacteria carry three fatty acids and are N-acylated by apolipoprotein N-acyltransferase BCG_2070c. BMC Microbiol 13:223. doi: 10.1186/1471-2180-13-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sander P, Clark S, Petrera A, Vilaplana C, Meuli M, Selchow P, Zelmer A, Mohanan D, Andreu N, Rayner E, Dal Molin M, Bancroft GJ, Johansen P, Cardona PJ, Williams A, Böttger EC. 2015. Deletion of zmp1 improves Mycobacterium bovis BCG-mediated protection in a guinea pig model of tuberculosis. Vaccine 33:1353–1359. doi: 10.1016/j.vaccine.2015.01.058. [DOI] [PubMed] [Google Scholar]
- 53.Meyer F, Pupkes H, Steinbüchel A. 2017. Development of an improved system for the generation of knockout mutants of Amycolatopsis sp. strain ATCC 39116. Appl Environ Microbiol 83:e02660-16. doi: 10.1128/AEM.02660-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE III, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail MA, Rajandream MA, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston JE, Taylor K, Whitehead S, Barrell BG. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 55.Kempsell KE, Ji YE, Estrada IC, Colston MJ, Cox RA. 1992. The nucleotide sequence of the promoter, 16S rRNA and spacer region of the ribosomal RNA operon of Mycobacterium tuberculosis and comparison with Mycobacterium leprae precursor rRNA. J Gen Microbiol 138:1717–1727. doi: 10.1099/00221287-138-8-1717. [DOI] [PubMed] [Google Scholar]
- 56.Springer B, Sander P, Sedlacek L, Ellrott K, Böttger EC. 2001. Instability and site-specific excision of integration-proficient mycobacteriophage L5 plasmids: development of stably maintained integrative vectors. Int J Med Microbiol 290:669–675. doi: 10.1016/S1438-4221(01)80004-7. [DOI] [PubMed] [Google Scholar]
- 57.Heinzel P, Werbitzky O, Distler J, Piepersberg W. 1988. A second streptomycin resistance gene from Streptomyces griseus codes for streptomycin-3″-phosphotransferase. Relationships between antibiotic and protein kinases. Arch Microbiol 150:184–192. [DOI] [PubMed] [Google Scholar]
- 58.Sander P, Springer B, Böttger EC. 2001. Gene replacement in Mycobacterium tuberculosis and Mycobacterium bovis BCG using rpsL+ as a dominant negative selectable marker. Methods Mol Biol 54:93–104. doi: 10.1385/1-59259-147-7:093. [DOI] [PubMed] [Google Scholar]
- 59.Pavelka MS Jr, Jacobs WR Jr. 1999. Comparison of the construction of unmarked deletion mutations in Mycobacterium smegmatis, Mycobacterium bovis bacillus Calmette-Guérin, and Mycobacterium tuberculosis H37Rv by allelic exchange. J Bacteriol 181:4780–4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jacobs WR Jr, Kalpana GV, Cirillo JD, Pascopella L, Snapper SB, Udani RA, Jones W, Barletta RG, Bloom BR. 1991. Genetic systems for mycobacteria. Methods Enzymol 204:537–555. doi: 10.1016/0076-6879(91)04027-L. [DOI] [PubMed] [Google Scholar]
- 61.Clinical and Laboratory Standards Institute 2011. Susceptibility testing of mycobacteria, nocardiae, and other aerobic actinomycetes, 2nd ed Approved standard M24-A2 CLSI, Wayne, PA, USA. [PubMed] [Google Scholar]
- 62.R Core Team 2014. R: a language and environment for statistical computing. 3.0.3. R Foundation for Statistical Computing, Vienna, VA. [Google Scholar]
- 63.Riley M, Abe T, Arnaud MB, Berlyn MKB, Blattner FR, Chaudhuri RR, Glasner JD, Horiuchi T, Keseler IM, Kosuge T, Mori H, Perna NT, Plunkett G, Rudd KE, Serres MH, Thomas GH, Thomson NR, Wishart D, Wanner BL. 2006. Escherichia coli K-12: a cooperatively developed annotation snapshot—2005. Nucleic Acids Res 34:1–9. doi: 10.1093/nar/gkj405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ramaswamy S, Musser JM. 1998. Molecular genetic basis of antimicrobial agent resistance in Mycobacterium tuberculosis: 1998 update. Tuber Lung Dis 79:3–29. doi: 10.1054/tuld.1998.0002. [DOI] [PubMed] [Google Scholar]
- 65.Finken M, Kirschner P, Meier A, Wrede A, Böttger EC. 1993. Molecular basis of streptomycin resistance in Mycobacterium tuberculosis: alterations of the ribosomal protein S12 gene and point mutations within a functional 16S ribosomal RNA pseudoknot. Mol Microbiol 9:1239–1246. doi: 10.1111/j.1365-2958.1993.tb01253.x. [DOI] [PubMed] [Google Scholar]
- 66.Meier A, Kirschner P, Bange FC, Vogel U, Böttger EC. 1994. Genetic alterations in streptomycin-resistant Mycobacterium tuberculosis: mapping of mutations conferring resistance. Antimicrob Agents Chemother 38:228–233. doi: 10.1128/AAC.38.2.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cuevas-Córdoba B, Cuellar-Sánchez A, Pasissi-Crivelli A, Santana-Álvarez CA, Hernández-Illezcas J, Zenteno-Cuevas R. 2013. rrs and rpsL mutations in streptomycin-resistant isolates of Mycobacterium tuberculosis from Mexico. J Microbiol Immunol Infect 46:30–34. doi: 10.1016/j.jmii.2012.08.020. [DOI] [PubMed] [Google Scholar]
- 68.Snapper SB, Melton RE, Mustafa S, Kieser T, Jacobs WR Jr. 1990. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol Microbiol 4:1911–1919. doi: 10.1111/j.1365-2958.1990.tb02040.x. [DOI] [PubMed] [Google Scholar]
- 69.Kusunoki S, Ezaki T. 1992. Proposal of Mycobacterium peregrinum sp. nov., nom. rev., and elevation of Mycobacterium chelonae subsp. abscessus (Kubica et al.) to species status: Mycobacterium abscessus comb. nov. Int J Syst Bacteriol 42:240–245. doi: 10.1099/00207713-42-2-240. [DOI] [PubMed] [Google Scholar]
- 70.Galmés-Truyols A, Giménez-Duran J, Bosch-Isabel C, Nicolau-Riutort A, Vanrell-Berga J, Portell Arbona M, Seguí-Prat B, Gumá-Torá M, Martí-Alomar I, Rojo-Arias MÁ, Ruiz-Veramendi M. 2011. An outbreak of cutaneous infection due to Mycobacterium abscessus associated to mesotherapy. Enferm Infecc Microbiol Clin 29:510–514. doi: 10.1016/j.eimc.2011.03.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.