Abstract
Background:
Thiopurine S-methyltransferase (TPMT) enzyme metabolizes thiopurine drugs which are widely used in various disciplines as well as in leukemias. Individual enzyme activity varies depending on the genetic polymorphisms of TPMT gene located at chromosome 6. Up to 14% of popu-lation is known to have a decreased enzyme activity, and if treated with standard doses of thiopurines, these individuals are at a high risk of severe Adverse Drug Reactions (ADR) as myelosuppression, gas-trointestinal intolerance, pancreatitis and hypersensitivity. However, TPMT-deficient patients can suc-cessfully be treated with decreased thiopurine doses if enzyme status is identified by a prior testing. TPMT status identification is a pioneering experience in application of pharmacogenetic testing in clini-cal settings. 4 TPMT (*2, *3A, *3B, *3C) alleles are known to account for 80-95% of a decreased en-zyme activity, and therefore, identifying the presence of these alleles supported by phenotypic measure-ment of the enzyme activity can reveal patient’s TPMT status. Evaluation of the levels of thiopurine me-tabolites further supports the practice of appropriate dose adjustment by providing the efficient monitor-ing of drug cytotoxicity.
Conclusion:
We hereby review the thiopurine pharmacogenetics and the methods applied in common practice to evaluate patient’s TPMT status.
Keywords: Thiopurine methyltransferase, pharmacogenetics, azathioprine, 6-mercapturine, 6-thioguanine, TPMT testing
1. INTRODUCTION: Pharmacogenetics
Individual responses to drugs can vary depending on the genetic variations in their metabolic pathways. Pharmacogenetics provides better prediction of drug efficacy and toxicity by linking differences in gene expression to drug response. Increasing knowledge on genome-disease and genome-drug interactions provides rationale for individualizing drug therapy by providing each patient with the right dose, ensuring improvement in an outcome of disease and decreasing Adverse Drug Reactions (ADR) [1-3].
Modern pharmacogenetics emerged in the 1950s with a series of discoveries revealing the influence of genetics on drug response [4-7]. With the increasing knowledge in genetics and development of molecular techniques in the last two decades, prior genotypic testing became a feasible option to predict drug response before the drug administration. Suxamethonium/mivacurium, warfarin, perhexiline and azathioprine/6-mercaptopurine/6-thioguanine are some of the examples of drugs with a strong evidence or a potential for the future application of a genetic testing prior to the dose adjustment [2].
The rationale behind pharmacogenetics is to study the influence of genetic polymorphisms on drug’s metabolic pathways. Many factors like age, concomitant diseases, substances influencing activity of drug-metabolizing enzymes are known to alter the drug efficacy.
Variations in the DNA sequences of genes encoding drug-metabolizing enzymes, drug target proteins and drug transporters can also cause variable drug responses in patients. Sequence variations can abolish, decrease, or increase the activity of a drug-metabolizing enzyme [8, 9]. Screening of the enzyme phenotype or genotype prior to treatment could predict the therapy outcome. Currently, the most successful example of a pharmacogenetic testing in a clinical setting is phenotypic and genotypic screening of TPMT enzyme status prior to the prescription of thiopurine drugs [10]. In this article, we review the significance of TPMT testing, supporting evidences, population studies of allelic frequencies, and major methods of patient’s TPMT status determination.
2. CLINICAL SIGNIFICANCE OF TPMT
TPMT is a cytosolic enzyme (E.C. 2.1.1.67) with the highest levels in heart and liver and relatively low levels in brain and lungs [11]. It catalyses the S-methylation of aromatic and heterocyclic sulphydryl compounds including anticancer and immunosuppressive thiopurine drugs, which are Azathioprine (AZA), 6-mercaptopurine (6MP) and 6-thioguanine (6TG) [12]. Erythrocytes TPMT activity in healthy males is 7% higher than in healthy females, and 4-6% higher in smokers compared to non-smokers. It is also higher in newborns (about 50%), while children demonstrate almost the same enzyme activity as adults [13].
Thiopurines are used in gastroenterology, dermatology, rheumatology and organ transplantation as immunosupressors, as well as in the management of Acute Lymphoblastic Leukaemia (ALL) as anti-cancer drugs [14-19]. 6MP and AZA are widely used immunosuppressants in the treatment of Inflammatory Bowel Diseases (IBD) such as Crohn’s disease and ulcerative colitis. Currently, about half of such patients receive thiopurine treatment. AZA and 6MP are also effective in autoimmune diseases and in preventing transplant graft rejection due to their effects on T-cells [20-22].
Although thiopurines are widely used, ADR like gastrointestinal intolerance, pancreatitis, hypersensitivity and myelosuppression are observed in up to 30% of patients [23, 24]. Thiopurines have a narrow therapeutic index. The difference between therapeutic and toxic doses is very little, which makes appropriate dose adjustment more complicated. Individuals with normal TPMT activity can be given full thiopurine doses, whereas TPMT-deficient patients are recommended to start with drastically reduced doses and to decrease the starting dose gradually. Those patients are at the high risk of severe ADR, as high levels of drug metabolites can accumulate if they are given standard thiopurine doses. Severe myelosuppression cases with a reduced blood cell production can occasionally be fatal, causing increased susceptibility to infections, clotting abnormalities and anaemia [25]. Varying TPMT enzyme activity has an influence on drug metabolism and cytotoxicity of thiopurines. That is why, the determination of TPMT enzyme status of a patient and appropriate dose adjustment is a matter of concern.
3. THIOPURINE DRUG METABOLISM
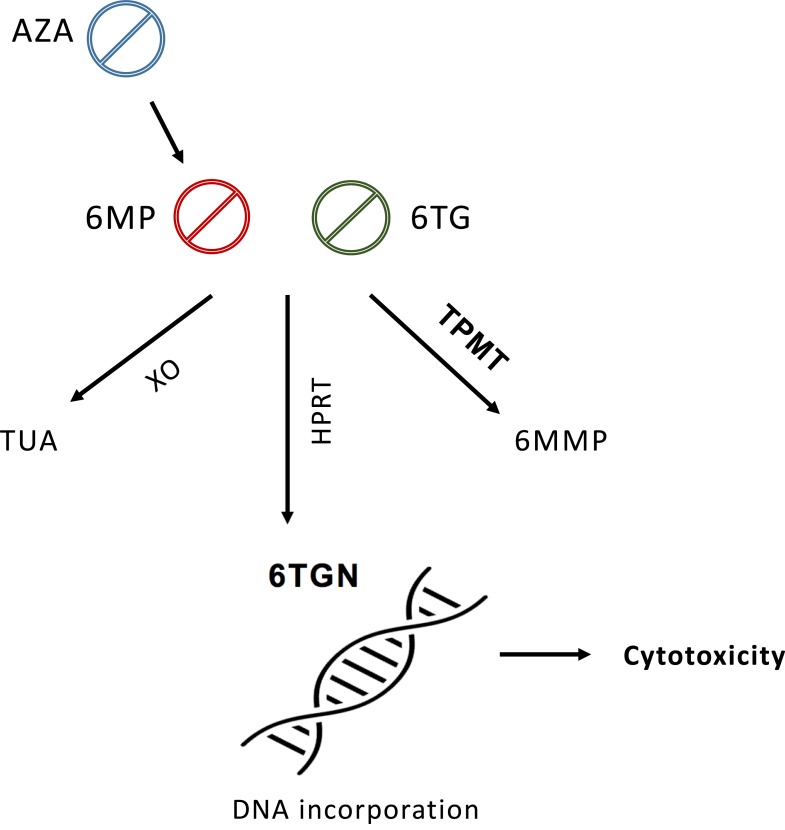
Thiopurines have a poor bioavailability and a short plasma half-life (6MP has a mean half-life of 1.9±0.6 hours, 6TG 1.5 hours, and AZA less than 2 hours) [26-28]. Thiopurine drugs are purine nucleoside analogues. AZA is the 1-methyl-4-nitro-imadazolyl of thioguanine. It is classed as a prodrug which is converted by the sulphydryl compounds, glutathione and cysteine to 6MP in the Red Blood Cells (RBC) [29]. This metabolite undergoes activation into thioguanine nucleotides (6TGN) via a multi-step enzymatic pathway. 6TGN’s therapeutic efficacy is primarily related to their cytotoxic action (Fig. 1). They distort the structure of the newly synthesized DNA and prevent proliferation of bone marrow and hematopoiesis. Intracellular 6TGN is incorporated into DNA instead of guanine nucleotides, and triggers cell-cycle arrest and apoptosis after a strand breakage [30-31].
Fig. (1).
Simplified diagram of thiopurine drug metabolism. Thiopurines are metabolized into different products via multi-step enzymatic pathways. Their pharmacologically active metabolites 6TGN exhibit cytotoxic action by incorporating into DNA instead of guanine nucleotides. TPMT diverts substrates away from this pathway, therefore it is inversely related to 6TGN concentration and cytotoxicity. Abbvreviations: AZA, Azathioprine; 6MP, 6-Mercaptopurine; 6TG, 6-Thioguanine; XO, Xanthine oxidase; TUA, Thiouric acid; HPRT, hypoxanthine phosphoribosyltransferase; 6TGN, thioguanine nucleotides; TPMT, thiopurine methyltransferase; 6MMP, 6-Methyl mercaptopurine.
Three enzymes, Hypoxanthine Phosphoribosyltransferase (HPRT), Xanthine Oxidase (XO) and Thiopurine Methyltransferase (TPMT) compete to break down 6MP into various metabolites. HPRT is the primary anabolic route of 6MP, leading to pharmacologically active 6TGN. XO metabolizes 6MP into inactive thiouric acid which is excreted from the body [32]. This catabolic process takes place in the liver and intestinal mucosa.
On the other hand, TPMT methylation diverts a proportion of available substrate away from HPRT pathway to form 6-methylmercaptopurine (6MMP). Therefore, TPMT enzyme activity is inversely related to 6TGN levels, subsequently, to the drug toxicity. Deficiency in TPMT activity will result in more substrate following the HPRT route and the higher concentration of 6TGN produced [33]. On the contrary, a high TPMT activity will result in less 6TGN being produced, posing a potential treatment failure. Furthermore, high 6MMP levels in patients with an increased TPMT activity can inhibit purine biosynthesis and cause liver damage.
4. TPMT GENE
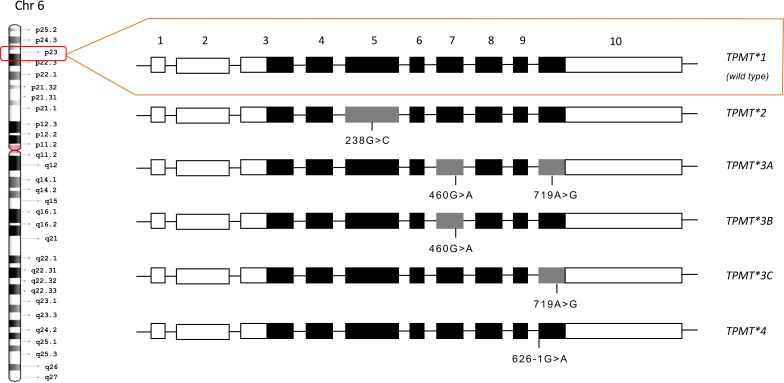
Activity of TPMT enzyme is related to Single Nucleotide Polymorphisms (SNPs) which are inherited co-dominantly [34]. It gives rise to the three patterns of the enzyme activity: homozygous TPMT-deficient patients with two mutant alleles (approx. 1 in 178 to 1 in 3736 individuals), heterozygous individuals with a decreased enzyme activity (approx. 3-14% of the population), and homozygous wild-type individuals (approx. 86-97% of the population) [35]. More than 30 allelic variants of TPMT gene, which is located in the short arm of 6th chromosome (6p22.3), have been reported to date [11]. The gene comprises 10 exons, 8 of them encode for 245 amino acid long protein, and common mutations are located in exon 5, 7 and 10 (Fig. 2).
Fig. (2).
Schematic diagram of the human thiopurine methyltransferase gene and its common mutant alleles. The gene is located in the short arm of the chromosome 6 and is comprised of 10 exons. Mutations of the common alleles are located in the V, VII and X exons. Gray rectangles represent exons that contain mutations. White rectangles are untranslated regions and black rectangles represent exons in the open reading frame.
TPMT alleles contain point mutations leading to amino acid substitutions (TPMT*2, -*3A, -*3B, -*3C, -*3D, -*5, and -*6), formation of a premature stop codon (TPMT*3D), or destruction of a splice site (TPMT*4). However, only 4 of these alleles (TPMT*2, -*3A, -*3B, -*3C) account for 80-95% of people with decreased TPMT activity, and the rest of the alleles are very rare [36-41].
5. ALLELE FREQUENCIES
Despite the known common alleles among the world population, frequencies of mutant alleles have been reported to vary between different ethnic groups (Table 1) [35, 42-55]. The most frequent allele in most populations is TPMT*3A, which is mainly followed by TPMT*3C. Europeans especially have been reported to have one of the highest frequencies of TPMT*3A, and all the mutant alleles in common [13, 42-45]. On the other hand, Asians and Africans show higher frequencies of TPMT*3C [35, 52-55]. TPMT*3C is mainly detected in these ethnicities, and the rest of the world shows a considerably lower frequency of it. It is also almost the only mutant allele observed in the Asians. The less common allele TPMT*2 is mainly seen in South American and Middle Eastern ethnicities, especially in Iranian population [35, 48]. Turkey, on the other hand, has been reported to have a lower frequency of mutant alleles compared to nearby countries [47].
Table 1.
Frequencies of TPMT mutant alleles in major ethnic groups and populations (%).
| Ethnicity | *2 | *3A | *3B | *3C | *4 | Reference |
|---|---|---|---|---|---|---|
| Caucasian | 0.2 | 3.5 | 0.04 | 0.4 | -1 | [35] |
| Mediterranean | 0.4 | 2.5 | 0.4 | 0.5 | - | [35] |
| Middle Eastern | 0.7 | 1.1 | 0.5 | 0.5 | - | [35] |
| Asian | 0 | 0.01 | 0 | 1.5 | - | [35] |
| African | 0.08 | 0.2 | 0 | 4.9 | - | [35] |
| South American | 0.8 | 2.8 | 0.05 | 0.9 | 0.05 | [35] |
| European | 0.5 | 5.7 | 0 | 0.8 | - | [42] |
| British | 0.5 | 4.5 | 0 | 0.3 | - | [43] |
| German | 0.2 | 4.5 | 0 | 0.4 | - | [13] |
| Russian | 0.1 | 2.3 | 0 | 0.4 | - | [44] |
| Italian-Caucasian | 0 | 2.2 | 0.3 | 0.3 | - | [45] |
| Spanish | 0 | 2.5 | 0.7 | 1 | - | [46] |
| Turkish | 0 | 0.9 | 0 | 0.9 | - | [47] |
| Iranian | 2.2 | 1.7 | 1.6 | 0.5 | - | [48] |
| Israeli | 0 | 1.2 | 0 | 0.4 | - | [49] |
| Kazak | 0 | 0.3 | 0 | 0.9 | - | [50] |
| Egyptian | 0 | 0.3 | 0 | 1.3 | - | [51] |
| Chinese | 0 | 0.1 | 0 | 1 | - | [52] |
| Japanese | 0 | 0 | 0 | 1.6 | - | [53] |
| Korean | 0 | 0 | 0 | 1 | - | [54] |
| Indian | 0 | 0.5 | 0 | 0.8 | - | [55] |
1frequency was not reported.
6. GENOTYPE – PHENOTYPE CORRELATION
Numerous in vitro, preclinical and clinical studies demonstrated a significant concordance between TPMT genotype and thiopurine-related ADR. Homozygous mutant TPMT alleles have been associated with the absence of the thiopurine catabolism into 6MMP in human RBC, lymphocytes, liver and kidney [34, 56-58]. TPMT knock-out mice exhibited a higher thiopurine-related morbidity and mortality compared to heterozygous and wild-type mice [59]. Furthermore, clinical studies demonstrated a 60-70% concordance in patients with a low TPMT activity, and >90% overall [13]. Since TPMT enzyme diverts thiopurines away from the HPRT pathway preventing the formation of cytotoxic and anti-leukemic product 6TGN, TPMT wild-type genotype is associated with an increased risk of relapse and higher levels of residual leukaemia in ALL patients [60-62]. Homozygous and heterozygous individuals, on the other hand, demonstrated higher risks of life-threatening toxicity, myelosuppression, and leukopenia from standard thiopurine doses [15, 63-65]. Testing of the genotype is thus a potential diagnostic tool for identifying individuals prone to poor thiopurine metabolism, and preventing ADR. Specificity, sensitivity and the positive and negative predictive value of the genotype testing were reported to be higher than 90% [13].
7. METHODS OF TPMT STATUS DETERMINATION
Since thiopurine ADR has been shown to correlate with a TPMT activity, the determination of TPMT status by laboratory methods can predict the toxicity potential of patients in advance, as well as provide the monitoring of effective drug dosing. It can be done by measuring a TPMT activity or levels of thiopurine metabolites in erythrocytes (6TGN), and by an identification of TPMT genotype [10, 15, 66].
Since the first description of TPMT assay [57], it has gradually become an established clinical practice [67, 68], and is now recommended by most guidelines [35]. Nevertheless, TPMT testing does not completely exclude thiopurine ADR. Factors other than genetic predisposition can also influence cytotoxicity. Although a decreased TPMT activity is associated with thiopurine-related toxicity, not all ADRs are initiated due to a low enzyme activity. Approximately 70% of patients with adverse events have normal TPMT activity, and factors as concomitant diseases, drug therapies, and other impairments in the drug metabolic pathway can also have a role in initiating ADR. There are other genes like Inosine Triphosphate Pyrophosphohydrolase (ITPA) and recently discovered NUDT15 has been linked to the thiopurine metabolism [69-74]. However, its significance is weaker or there is a lack of sufficient evidence to support its clinical use for now.
TPMT screening via measuring erythrocyte enzyme activity or TPMT genotyping is ideally performed prior to treatment for an individual dose assignment, whereas monitoring 6TGN concentrations of a patient already under the thiopurine therapy provides useful information on patient’s response to thiopurines and enables adequate dose adjustment [10, 75].
7.1. TPMT Phenotyping
Human TPMT enzyme activity is measured in RBC. Erythrocyte TPMT activity has been demonstrated to correlate with other tissues, especially with liver and kidney [57, 76], and it is also easily accessible from peripheral blood. Measurement of TPMT activity is performed by incubating whole blood or erythrocyte lysate with either 6MP or 6TG, and the methyl donor S-Adenosylmethionine (SAM), yielding 6-Methylmercaptopurine (6MMP) or 6-methylthioguanine (6MTG). These products can then be measured by different methods, including radiochemical assay, High-Performance Liquid Chromatography (HPLC), Enzyme-Linked Immunosorbent Assay (ELISA), and mass spectrometry.
Radiochemical assay is based on the transfer of methyl groups from radio-labelled 14C-methyl-SAM to 6MP. The radioactivity of the reaction products is then measured by scintillation counter [77]. This is not commonly used due to the operational difficulties of working with radioactive isotopes, and its safety implications.
HPLC, on the other hand, is a commonly used method. Reaction products of the assay are measured by injecting them into a column with high pressure liquid, and after separation, they are quantified via absorbance or fluorescence detection [78, 79].
ELISA uses an antibody specific to the 6MMP. Antibodies binding to the metabolite and streptavidin-coated microplate well produce a colour signal which is detected via spectrophotometer [80].
Liquid Chromatography – Tandem Mass Spectrometry is now used to measure TPMT enzyme activity, as well as to quantify thiopurine metabolites for clinical monitoring of patients under the thiopurine therapy [81].
Despite the variety of methods available for phenotyping, some pre-analytical factors interfering test results should be carefully taken into consideration when choosing an appropriate method. Enzymatic reactions are easily affected by outside factors like incubation temperature, duration, presence of inhibitors and the source and concentration of a substrate [82]. TPMT activity results are also influenced by red cell transfusions [83], as well as leukaemia and treatment related factors in ALL patients [84]. Variety in the enzyme activity due to age, gender, ethnicity, erythrocyte lifespan, and all the factors mentioned above should be considered when interpreting TPMT activity results [85-87].
7.2. TPMT Genotyping
Since the 4 most common alleles (TPMT*2, TPMT*3A, TPMT*3B, and TPMT*3C) account for 80% to 95% of a decreased TPMT activity [13, 15, 35], most of the genotyping assays include these SNPs only. This can occasionally cause misdiagnosis in cases where the rare mutations are present, since the assay will not be able to detect them [35]. Genotyping assays are also not able to differentiate whether two mutations are located in the same allele, or in opposite alleles (e.g. *1/3A or *3B/3C). However, genotyping results are not influenced by outside factors and are relatively stable. Considering some weaknesses of both phenotyping and genotyping methods, it is recommended that their results are interpreted together [10, 75].
TPMT genotyping assays are mainly based on the techniques including Restriction Fragment Length Polymorphism (RFLP), Single Strand Conformational Polymorphism (SSCP), Amplification Refractory Mutation System (ARMS), Reverse Dot-Blot (RDB) hybridization, denaturing HPLC, pyrosequencing [88], Microchip Technology and Mass Spectrometry (MALDI-TOF).
RFLP was one of the early methods applied in TPMT genotyping using restriction endonucleases which cleave DNA molecules at specific nucleotide sequences. Its major drawback is possible erroneous readings due to incomplete restriction digestion [67].
SSCP is based on electrophoresis of the exons of TPMT gene, since different mutations cause different migratory patterns on a gel [89]. This method is not routinely used in clinical settings.
ARMS uses primers designed to function only when anneal to specific sequences. Consequently, PCR product can be detected if a target sequence is present. It can be performed in a multiplex, containing the several primers complementary for different mutations [90].
RDB hybridization is able to detect multiple mutations simultaneously. PCR amplification with biotinylated primers is followed by hybridization of amplification products onto the strip containing multiple allele-specific oligonucleotide probes. If mutant sequences are present, they are bound to oligonucleotides, and are detected via colour reaction [91].
Denaturing HPLC is particularly effective in analysing DNA polymorphisms. PCR products are denatured and reannealed to form hetero duplexes, which make extra peak in HPLC data and can be easily distinguished from a wild type [13].
DNA microchip technology similarly uses multiple complementary oligonucleotides attached to microchips. Fluorescently labelled oligonucleotides are then detected via lasers. This technique is also able to detect multiple polymorphisms simultaneously [92].
Mass spectrometry is another state of the art technique providing multiplex genotyping. Matrix-assisted laser desorption/ionization - time of flight (MALDI-TOF) mass spectrometry is able to detect minor differences in base pair mass, which is applied to differentiate SNPs [93].
Rare alleles which cannot be detected by routine genotyping are investigated by Sequence-based techniques. Conventional Sanger sequencing as well as the less time consuming and labour intensive High-Resolution Melting Analysis (HRMA) are applied to analyze complete coding sequence of the TPMT gene and to determine the spectrum and the prevalence of the TPMT gene sequence variations [94, 95]. However, sequence-based techniques are not used as first-line diagnostic methods due to their expensiveness and not being readily available in most diagnostic facilities. Therefore, initial genotyping is usually performed by aforementioned methods in a multiplex, including most of the clinically significant alleles which account for up to 95% of a decreased enzyme activity. An unexpected negative result, which is not in concordance with a decreased enzyme activity or patient’s thiopurine-related ADR, is then followed by a complete TPMT sequencing to investigate the possibility of rare alleles.
The genotype-phenotype concordance of TPMT has been reported to be 60-70% in patients with a low TPMT activity, and >90% overall [96]. Although genotyping methods are more reliable and are not influenced by red cell transfusions, drug interactions and sample age, they are relatively costly and time-consuming. Since most commercial methods are able to detect only 4-5 most common alleles, genotyping can also miss the rest alleles, identifying these patients as a wild type. Neither measurement of TPMT enzyme activity nor identifying TPMT genotype is sufficient by itself to be applied as a single method to reveal patient’s thiopurine status. Phenotyping results can be influenced by aforementioned outside factors, whereas genotyping is comparably time-consuming and more complicated to perform. Most laboratories initially perform phenotyping as a main TPMT screening method and use genotyping to confirm controversial cases. However, most recent guidelines recommend thiopurine dosage according to the genotype, which makes genotyping of all patients necessary [35].
7.3. Measurement of Thiopurine Metabolites
6TGN are thiopurine metabolites, and their levels are measured in patients already receiving a thiopurine therapy in order to monitor individual drug metabolism and provide adequate dose adjustment. 6TGN levels have been demonstrated to correlate with a disease remission and leukopenia [97, 98], making it a successful indicative for therapeutic drug monitoring. Additionally, TPMT activity has been reported to be induced in patients receiving thiopurine therapy [99]. Therefore, measuring metabolite levels is considered a comparably reliable method for monitoring thiopurine metabolism.
CONCLUSION
ADR attributed to thiopurines is known to be dose-dependent, and especially myelosuppression, which is more common in childhood ALL compared to patients receiving thiopurines as immunosupressors, requires a special attention of clinicians and can be prevented by an individual dose adjustment. This is possible by a prior testing and identification of patient’s TPMT status in advance. Considering the limitations of phenotyping and genotyping of TPMT activity, both methods are recommended to be applied in a combination supported by the measurement of thiopurine metabolite levels. The variety of techniques enables to choose the appropriate one for clinical facilities with different recources.
CONSENT FOR PUBLICATION
Not applicable.
ACKNOWLEDGEMENTS
We would like to express our acknowledgements to Ulviyya Bayramova for her guidance on language editing.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
REFERENCES
- 1.Roses A.D. Pharmacogenetics and drug development: the path to safer and more effective drugs. Nat. Rev. Genet. 2004;5(9):645–656. doi: 10.1038/nrg1432. [DOI] [PubMed] [Google Scholar]
- 2.Gardiner S.J. Pharmacogenetics, drug-metabolizing enzymes, and clinical practice. Pharmacol. Rev. 2006;58(3):521–590. doi: 10.1124/pr.58.3.6. [DOI] [PubMed] [Google Scholar]
- 3.Ma Q., Lu A.Y. Pharmacogenetics, pharmacogenomics, and individualized medicine. Pharmacol. Rev. 2011;63(2):437–459. doi: 10.1124/pr.110.003533. [DOI] [PubMed] [Google Scholar]
- 4.Hughes H.B., Biehl J.P., Jones A.P., Schmidt L.H. Metabolism if isoniazid in man as related to the occurrence of peripheral neuri-tis. Am. Rev. Respir. Dis. 1954;70:266–273. doi: 10.1164/art.1954.70.2.266. [DOI] [PubMed] [Google Scholar]
- 5.Alving A.S., Carson P.E., Flanagan C.L., Ickes C.E. Enzymatic deficiency in primaquine-sensitive erythrocytes. Science. 1956;124:484–485. doi: 10.1126/science.124.3220.484-a. [DOI] [PubMed] [Google Scholar]
- 6.Kalow W. Familial incidence of low pseudocholinesterase level. Lancet. 1956;2:576–577. doi: 10.1016/s0140-6736(56)90869-8. [DOI] [PubMed] [Google Scholar]
- 7.Motulsky A.G. Drug reactions enzymes, and biochemical genetics. J. Am. Med. Assoc. 1957;1165(7):835–837. doi: 10.1001/jama.1957.72980250010016. [DOI] [PubMed] [Google Scholar]
- 8.Evans W.E., Relling M.V. Moving towards individualized medicine with pharmacogenomics. Nature. 2004;429:464–468. doi: 10.1038/nature02626. [DOI] [PubMed] [Google Scholar]
- 9.Lin J.H. Pharmacokinetic and pharmacodynamic variability: a daunting challenge in drug therapy. Curr. Drug Metab. 2007;8:109–136. doi: 10.2174/138920007779816002. [DOI] [PubMed] [Google Scholar]
- 10.Ford L.T., Berg J.D. Thiopurine S-methyltransferase (TPMT) assessment prior to starting thiopurine drug treatment; a Phar-macogenomic test whose time has come. J. Clin. Pathol. 2010;63(4):288–295. doi: 10.1136/jcp.2009.069252. [DOI] [PubMed] [Google Scholar]
- 11.GeneCards Database TPMT Gene. GCID: GC06M018072. http://Genecards.org
- 12.Fotoohi A.K., Coulthard S.A., Albertioni F. Thiopurines: factors influencing toxicity and response. Biochem. Pharmacol. 2010;79(9):1211–1220. doi: 10.1016/j.bcp.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 13.Schaeffeler E., Fischer C., Brockmeier D., Wernet D., Moerike K., Eichelbaum M., Zanger U.M., Schwab M. Comprehensive analysis of thiopurine S-methyltransferase phenotype-genotype correlation in a large population of German-Caucasians and identi-fication of novel TPMT variants. Pharmacogenetics. 2004;14(7):407–417. doi: 10.1097/01.fpc.0000114745.08559.db. [DOI] [PubMed] [Google Scholar]
- 14.Mcleod H.L., Krynetski E.Y., Relling M.V., Evans W.E. Genetic polymorphism of thiopurine methyltransferase and its clinical relevance for childhood acute lymphoblastic leukemia. Leukemia. 2000;14(4):567–572. doi: 10.1038/sj.leu.2401723. [DOI] [PubMed] [Google Scholar]
- 15.Booth R.A., Ansari M.T., Loit E., Tricco A.C., Weeks L., Doucette S., Skidmore B., Sears M., Richmond S., Karsh J. Assessment of thiopurine S-methyltransferase activity in patients prescribed thiopurines: A systematic review. Ann. Intern. Med. 2011;154(12):814–823. doi: 10.7326/0003-4819-154-12-201106210-00009. [DOI] [PubMed] [Google Scholar]
- 16.Dong X-W. Thiopurine S-methyltransferase polymorphisms and thiopurine toxicity in treatment of inflammatory bowel dis-ease. World J. Gastroenterol. 2010;16(25):3187–3195. doi: 10.3748/wjg.v16.i25.3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moon W., Loftus E.V. Review Article: Recent advances in pharmacogenetics and pharmacokinetics for safe and effective thiopurine therapy in inflammatory bowel disease. Aliment. Pharmacol. Ther. 2016;43(8):863–883. doi: 10.1111/apt.13559. [DOI] [PubMed] [Google Scholar]
- 18.Benkov K., Lu Y., Patel A., Rahhal R., Russell G., Teitelbaum J. Role of Thiopurine metabolite testing and thiopurine methyl-transferase determination in pediatric IBD. J. Pediatr. Gastroenterol. Nutr. 2013;56(3):333–340. doi: 10.1097/MPG.0b013e3182844705. [DOI] [PubMed] [Google Scholar]
- 19.Fong S.C., Blaker P.A., Arenas-Hernandez M., Marinaki A.M., Sanderson J.D. Getting the best out of thiopurine therapy: Thiopurine s -methyltransferase and beyond. Biomarkers Med. 2015;9(1):51–65. doi: 10.2217/bmm.14.97. [DOI] [PubMed] [Google Scholar]
- 20.Cuffari C., Hunt S., Bayless T.M. Enhanced bioavailability of azathioprine compared to 6-mercaptopurine therapy in inflammatory bowel disease: correlation with treatment efficacy. Aliment. Pharmacol. Ther. 2000;14(8):1009–1014. doi: 10.1046/j.1365-2036.2000.00812.x. [DOI] [PubMed] [Google Scholar]
- 21.Chouchana L., Narjoz C., Beaune P., Loriot M.A., Roblin X. Review article: the benefits of pharmacogenetics for improving thi-opurine therapy in inflammatory bowel disease. Aliment. Pharmacol. Ther. 2012;35(1):15–36. doi: 10.1111/j.1365-2036.2011.04905.x. [DOI] [PubMed] [Google Scholar]
- 22.Deshpande A.R., Abreu M.T. Optimizing therapy with 6-mercaptopurine and azathioprine: to measure or not to measure? Therap. Adv. Gastroenterol. 2010;3(5):275–279. doi: 10.1177/1756283X10376121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lennard L., Lilleyman J.S. Individualizing therapy with 6-Mercaptopurine and 6-Thioguanine related to the thiopurine methyltransferase genetic polymorphism. Ther. Drug Monit. 1996;18(4):328–334. doi: 10.1097/00007691-199608000-00003. [DOI] [PubMed] [Google Scholar]
- 24.Anstey A., Lennard L., Mayou S.C., Kirby J.D. Pancytopenia related to azathioprine - an enzyme deficiency caused by a com-mon genetic polymorphism. J. R. Soc. Med. 1992;85:752–756. doi: 10.1177/014107689208501213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Slanar O., Chalupná P., Novotný A., Bortlík M., Krska Z., Lukás M. Fatal myelotoxicity after azathioprine treatment. Nucleosides Nucleotides Nucleic Acids. 2008;27(6):661–665. doi: 10.1080/15257770802143905. [DOI] [PubMed] [Google Scholar]
- 26.Cuffari C., Hunt S., Bayless T. Utilisation of erythrocyte 6-thioguanine metabolite levels to optimise azathioprine therapy in patients with inflammatory bowel disease. Gut. 2001;48(5):642–646. doi: 10.1136/gut.48.5.642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chan G.L., Erdmann G.R., Gruber S.A., Matas A.J., Canafax D.M. Azathioprine metabolism: pharmacokinetics of 6-mercaptopurine, 6-thiouric acid and 6-thioguanine nucleotides in renal transplant patients. J. Clin. Pharmacol. 1990;30(4):358–363. doi: 10.1002/j.1552-4604.1990.tb03606.x. [DOI] [PubMed] [Google Scholar]
- 28.Coulthard S., Hogarth L. The thiopurines: an update. Invest. New Drugs. 2005;23(6):523–532. doi: 10.1007/s10637-005-4020-8. [DOI] [PubMed] [Google Scholar]
- 29.Eklund B.I., Moberg M., Bergquist J., Mannervik B. Divergent activities of human glutathione transferases in the bioactivation of azathioprine. Mol. Pharmacol. 2006;70(2):747–754. doi: 10.1124/mol.106.025288. [DOI] [PubMed] [Google Scholar]
- 30.Fairchild C.R., Maybaum J., Kennedy K.A. Concurrent unilateral chromatid damage and DNA strand breakage in response to 6-thioguanine treatment. Biochem. Pharmacol. 1986;35(20):3533–3541. doi: 10.1016/0006-2952(86)90623-4. [DOI] [PubMed] [Google Scholar]
- 31.Somerville L., Krynetski E.Y., Krynetskaia N.F., Beger R.D., Zhang W., Marhefka C.A., Evans W.E., Kriwacki R.W. Struc-ture and dynamics of thioguanine-modified duplex DNA. J. Biol. Chem. 2003;278(2):1005–1011. doi: 10.1074/jbc.M204243200. [DOI] [PubMed] [Google Scholar]
- 32.Wong D.R., Derijks L.J., den Dulk M.O., Gemmeke E.H., Hooymans P.M. The role of xanthine oxidase in thiopurine metab-olism: a case report. Ther. Drug Monit. 2007;29(6):845–848. doi: 10.1097/FTD.0b013e31815bf4dc. [DOI] [PubMed] [Google Scholar]
- 33.Osterman M.T., Kundu R., Lichtenstein G.R., Lewis J.D. Asso-ciation of 6-thioguanine nucleotide levels and inflammatory bowel disease activity: a meta-analysis. Gastroenterology. 2006;130(4):1047–1053. doi: 10.1053/j.gastro.2006.01.046. [DOI] [PubMed] [Google Scholar]
- 34.Weinshilboum R.M., Sladek S.L. Mercaptopurine pharmaco-genetics: monogenic inheritance of erythrocyte thiopurine methyl-transferase activity. Am. J. Hum. Genet. 1980;32(5):651–662. [PMC free article] [PubMed] [Google Scholar]
- 35.Relling M.V., Gardner E.E., Sandborn W.J., Schmiegelow K., Pui C.H., Yee S.W., Klein T.E. Pharmacogenetics implementation consortium guidelines for thiopurine methyltransferase gen-otype and thiopurine dosing: 2013 Update. Clin. Pharmacol. Ther. 2013;93(4):324–325. doi: 10.1038/clpt.2013.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Appell M.L., Berg J., Duley J., Evans W.E., Kennedy M.A., Lennard L., Marinaki T., McLeod H.L., Relling M.V., Schaeffeler E., Schwab M. Nomenclature for alleles of the thiopu-rine methyltransferase gene. Pharmacogenet. Genomics. 2013;23(4):242–248. doi: 10.1097/FPC.0b013e32835f1cc0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garat A., Cauffiez C., Renault N., Lo-Guidice J.M., Allorge D., Chevalier D., Houdret N., Chavatte P., Loriot M.A., Gala J.L., Broly F. Characterisation of novel defective thiopurine S-methyltransferase allelic variants. Biochem. Pharmacol. 2008;76(3):404–415. doi: 10.1016/j.bcp.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 38.Ujiie S., Sasaki T., Mizugaki M., Ishikawa M., Hiratsuka M. Functional characterization of 23 allelic variants of thiopurine S-methyltransferase gene (TPMT* 2-* 24). Pharmacogenet. Genomics. 2008;18(10):887–893. doi: 10.1097/FPC.0b013e3283097328. [DOI] [PubMed] [Google Scholar]
- 39.Hamdan-Khalil R., Gala J.L., Allorge D., Lo-Guidice J.M., Horsmans Y., Houdret N., Broly F. Identification and functional analysis of two rare allelic variants of the thiopurine S-methyltransferase gene, TPMT* 16 and TPMT* 19. Biochem. Pharmacol. 2005;69(3):525–529. doi: 10.1016/j.bcp.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 40.Hamdan-Khalil R., Allorge D., Lo-Guidice J.M., Cauffiez C., Chevalier D., Spire C., Houdret N., Libersa C., Lhermitte M., Colombel J.F., Gala J.L. In vitro characterization of four novel non-functional variants of the thiopurine S-methyltransferase. Bio-chem. Bioph. Res. Co. 2003;309(4):1005–1010. doi: 10.1016/j.bbrc.2003.08.103. [DOI] [PubMed] [Google Scholar]
- 41.Salavaggione O.E., Wang L., Wiepert M., Yee V.C., Weinshil-boum R.M. Thiopurine S-methyltransferase pharmacogenetics: variant allele functional and comparative genomics. Pharmacogenet. Genomics. 2005;15(11):801–815. doi: 10.1097/01.fpc.0000174788.69991.6b. [DOI] [PubMed] [Google Scholar]
- 42.de la Moureyre C., Debuysere H., Mastain B., Vinner E., Marez D., Lo Guidice J.M., Chevalier D., Brique S., Motte K., Colombel J.F., Turck D. Genotypic and phenotypic analysis of the polymorphic thiopurine S‐methyltransferase gene (TPMT) in a Eu-ropean population. Br. J. Pharmacol. 1998;125(4):879–887. doi: 10.1038/sj.bjp.0702152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Collie-Duguid E.S., Sludden J., Li T., McLeod H.L. The fre-quency and distribution of thiopurine methyltransferase alleles in Caucasian and Asian populations. Pharmacogenet. Genomics. 1999;9(1):37–42. doi: 10.1097/00008571-199902000-00006. [DOI] [PubMed] [Google Scholar]
- 44.Samochatova E.V., Chupova N.V., Rudneva A., Makarova O., Nasedkina T.V., Fedorova O.E., Glotov A.S., Kozhekbaeva Z., Maiorova O.A., Roumyantsev A.G., Krynetski E.Y. TPMT genet-ic variations in populations of the Russian Federation. Pediatr. Blood Cancer. 2009;52(2):203–208. doi: 10.1002/pbc.21837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Serpe L., Calvo P.L., Muntoni E., D’Antico S., Giaccone M., Avagnina A., Baldi M., Barbera C., Curti F., Pera A., Eandi M. Thiopurine S-methyltransferase pharmacogenetics in a large-scale healthy Italian-Caucasian population: differences in enzyme activi-ty. Pharmacogenomics. 2009;10(11):1753–1765. doi: 10.2217/pgs.09.103. [DOI] [PubMed] [Google Scholar]
- 46.Corominas H., Domenech M., Laiz A., Gich I., Geli C., Diaz C., De Cuevillas F., Moreno M., Vazquez G., Baiget M. Is thio-purine methyltransferase genetic polymorphism a major factor for withdrawal of azathioprine in rheumatoid arthritis patients? Rheumatology. 2003;42(1):40–45. doi: 10.1093/rheumatology/keg028. [DOI] [PubMed] [Google Scholar]
- 47.Tumer T.B., Ulusoy G., Adali O., Sahin G., Gozdasoglu S., Arinç E. The low frequency of defective TPMT alleles in Turkish population: a study on pediatric patients with acute lymphoblastic leukemia. Am. J. Hematol. 2007;82(10):906–910. doi: 10.1002/ajh.20947. [DOI] [PubMed] [Google Scholar]
- 48.Bahari A., Hashemi M., Bari Z., Moazeni-Roodi A., Kaykhaei M.A., Narouie B. Frequency of thiopurine S-methyltransferase (TPMT) alleles in southeast Iranian population. Nucleosides Nucleotides Nucleic Acids. 2010;29(3):237–244. doi: 10.1080/15257771003720418. [DOI] [PubMed] [Google Scholar]
- 49.Efrati E., Adler L., Krivoy N., Sprecher E. Distribution of TPMT risk alleles for thioupurine toxicity in the Israeli population. Eur. J. Clin. Pharmacol. 2009;65(3):257–262. doi: 10.1007/s00228-008-0590-7. [DOI] [PubMed] [Google Scholar]
- 50.Wei H., Zhou S., Li C., Zhang J., Wu J., Huang M. Phenotyp-ing and genotyping studies of thiopurine S-methyltransferase in Kazaks. Pharm. Res. 2005;22(10):1762–1766. doi: 10.1007/s11095-005-7095-1. [DOI] [PubMed] [Google Scholar]
- 51.Hamdy S.I., Hiratsuka M., Narahara K., Endo N., El‐Enany M., Moursi N., Ahmed M.S., Mizugaki M. Genotype and allele frequencies of TPMT, NAT2, GST, SULT1A1 and MDR‐1 in the Egyptian population. Br. J. Clin. Pharmacol. 2003;55(6):560–569. doi: 10.1046/j.1365-2125.2003.01786.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang J.P., Zhou S.F., Chen X., Huang M. Determination of intra-ethnic differences in the polymorphisms of thiopurine S-methyltransferase in Chinese. Clin. Chim. Acta. 2006;365(1):337–341. doi: 10.1016/j.cca.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 53.Kumagai K., Hiyama K., Ishioka S., Sato H., Yamanishi Y., McLeod H.L., Konishi F., Maeda H., Yamakido M. Allelotype frequency of the thiopurine methyltransferase (TPMT) gene in Jap-anese. Pharmacogenet. Genomics. 2001;11(3):275–278. doi: 10.1097/00008571-200104000-00012. [DOI] [PubMed] [Google Scholar]
- 54.Cheon J.H., Kim J.H., Kim B.Y., Kim S.W., Hong S.Y., Eun C.S., Hong S.S., Byeon J.S., Kim T.I., Han D.S., Yang S.K. Al-lele frequency of thiopurine methyltransferase and inosine triphos-phate pyrophosphatase gene polymorphisms in Korean patients with inflammatory bowel diseases. Hepato-gastroenterol. 2008;56(90):421–423. [PubMed] [Google Scholar]
- 55.Kham S.K., Tan P.L., Tay A.H., Heng C.K., Yeoh A.E., Quah T.C. Thiopurine methyltransferase polymorphisms in a mul-tiracial Asian population and children with acute lymphoblastic leukemia. J. Pediatr. Hematol. Oncol. 2002;24(5):353–359. doi: 10.1097/00043426-200206000-00006. [DOI] [PubMed] [Google Scholar]
- 56.Van Loon J.A., Weinshilboum R.M. Thiopurine methyltransfer-ase biochemical genetics: human lymphocyte activity. Biochem. Genet. 1982;20:637–658. doi: 10.1007/BF00483962. [DOI] [PubMed] [Google Scholar]
- 57.Szumlanski C.L., Honchel R., Scott M.C., Weinshilboum R.M. Human liver thiopurine methyltransferase pharmacogenetics: bio-chemical properties, liver-erythrocyte correlation and presence of isozymes. Pharmacogenet. Genomics. 1992;2(4):148–159. [PubMed] [Google Scholar]
- 58.Van Loon J.A., Weinshilboum R.M. Thiopurine methyltransfer-ase isozymes in human renal tissue. Drug Metab. Dispos. 1990;18:632–638. [PubMed] [Google Scholar]
- 59.Hartford C., Vasquez E., Schwab M., Edick M.J., Rehg J.E., Grosveld G., Pui C., Evans W.E., Relling M.V. Differential ef-fects of targeted disruption of thiopurine methyltransferase on mer-captopurine and thioguanine pharmacodynamics. Cancer Res. 2007;67:4965–4972. doi: 10.1158/0008-5472.CAN-06-3508. [DOI] [PubMed] [Google Scholar]
- 60.Stanulla M., Schaeffeler E., Flohr T., Cario G., Sahrauder A., Zimmermann M., Welte K., Ludwig W.D., Bartram C.R., Zanger U.M., Eichelbaum M., Schrappe M., Schwab M. Thiopurine methyltransferase (TPMT) genotype and early treatment response to mercaptopurine in childhood acute lymphoblastic leuke-mia. JAMA. 2005;293:1485–1489. doi: 10.1001/jama.293.12.1485. [DOI] [PubMed] [Google Scholar]
- 61.Lennard L., Lilleyman J.S., Van Loon J., Weinshilboum R.M. Genetic variation in response to 6-mercaptopurine for childhood acute lymphoblastic leukaemia. Lancet. 1990;336:225–229. doi: 10.1016/0140-6736(90)91745-v. [DOI] [PubMed] [Google Scholar]
- 62.Ansari A., Hassan C., Duley J., Marinaki A., Shobowale-Bakre M., Seed P., Meenan J., Yim A., Sanderson J. Thiopurine methyltransferase activity and the use of azathioprine in inflammatory bowel disease. Aliment. Pharmacol. Ther. 2002;16:1743–1750. doi: 10.1046/j.1365-2036.2002.01353.x. [DOI] [PubMed] [Google Scholar]
- 63.Relling M.V., Hancock M.L., Rivera G.K., Sandlund J.T., Ribei-ro R.C., Krynetski E.Y., Pui C.H., Evans W.E. Mercaptopurine therapy intolerance and heterozygosity at the thiopurine S-methyltransferase gene locus. J. Natl. Cancer Inst. 1999;91:2001–2008. doi: 10.1093/jnci/91.23.2001. [DOI] [PubMed] [Google Scholar]
- 64.McLeod H., Coulthard S., Thomas A.E., Pritchard S.C., King D.J., Richards S.M., Eden O.B., Hall A.G., Gibson B.E. Anal-ysis of thiopurine methyltransferase variant alleles in childhood acute lymphoblastic leukaemia. Br. J. Haematol. 1999;105:696–700. doi: 10.1046/j.1365-2141.1999.01416.x. [DOI] [PubMed] [Google Scholar]
- 65.Black A.J., McLeod H.L., Capel H.A., Powrie R.H., Matowe L.K., Pritchard S.C., Collie-Duguid E.S., Reid D.M. Thiopurine methyltransferase genotype predicts therapy-limiting severe toxici-ty from azathioprine. Ann. Intern. Med. 1998;129:716–718. doi: 10.7326/0003-4819-129-9-199811010-00007. [DOI] [PubMed] [Google Scholar]
- 66.Graham V. Thiopurine methyltransferase Phenotyping and Geno-typing in Clinical Practice.M.Phil. Thesis. The University of Birmingham. 2009. [Google Scholar]
- 67.Yates C.R., Krynetski E.Y., Loennechen T., Fessing M.Y., Tai H.L., Pui C.H., Relling M.V., Evans W.E. Molecular diagnosis of thiopurine S-methyltransferase deficiency: genetic basis for azathi-oprine and mercaptopurine intolerance. Ann. Intern. Med. 1997;126(8):608–614. doi: 10.7326/0003-4819-126-8-199704150-00003. [DOI] [PubMed] [Google Scholar]
- 68.Holme S.A., Duley J.A., Sanderson J., Routledge P.A., Anstey A.V. Erythrocyte thiopurine methyl transferase assessment prior to azathioprine use in the UK. QJM. 2002;95(7):439–444. doi: 10.1093/qjmed/95.7.439. [DOI] [PubMed] [Google Scholar]
- 69.Marinaki A.M., Ansari A., Duley J.A., Arenas M., Sumi S., Lewis C.M., Shobowale-Bakre E.M., Escuredo E., Fairbanks L.D., Sanderson J.D. Adverse drug reactions to azathioprine thera-py are associated with polymorphism in the gene encoding inosine triphosphate pyrophosphatase (ITPase). Pharmacogenet. Genomics. 2004;14(3):181–187. doi: 10.1097/00008571-200403000-00006. [DOI] [PubMed] [Google Scholar]
- 70.Marinaki A.M., Duley J.A., Arenas M., Ansari A., Sumi S., Lewis C.M., Shobowale‐Bakre M., Fairbanks L.D., Sanderson J. Mutation in the ITPA gene predicts intolerance to azathioprine. Nucleosides Nucleotides Nucleic Acids. 2004;23(8-9):1393–1397. doi: 10.1081/NCN-200027639. [DOI] [PubMed] [Google Scholar]
- 71.Von Ahsen N., Armstrong V.W., Behrens C., Von Tirpitz C., Stallmach A., Herfarth H., Stein J., Bias P., Adler G., Shipkova M., Oellerich M. Association of inosine triphosphatase 94C> A and thiopurine S-methyltransferase deficiency with adverse events and study drop-outs under azathioprine therapy in a prospective Crohn disease study. Clin. Chem. 2005;51(12):2282–2288. doi: 10.1373/clinchem.2005.057158. [DOI] [PubMed] [Google Scholar]
- 72.Yang S.K., Hong M., Baek J., Choi H., Zhao W., Jung Y., Haritunians T., Ye B.D., Kim K.J., Park S.H., Park S.K. A common missense variant in NUDT15 confers susceptibility to thi-opurine-induced leukopenia. Nat. Genet. 2014;46(9):1017–1020. doi: 10.1038/ng.3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang J.J., Landier W., Yang W., Liu C., Hageman L., Cheng C., Pei D., Chen Y., Crews K.R., Kornegay N., Wong F.L. In-herited NUDT15 variant is a genetic determinant of mercaptopu-rine intolerance in children with acute lymphoblastic leukemia. J. Clin. Oncol. 2015;33(11):1235–1242. doi: 10.1200/JCO.2014.59.4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Moriyama T., Nishii R., Perez-Andreu V., Yang W., Klussmann F.A., Zhao X., Lin T.N., Hoshitsuki K., Nersting J., Kihira K., Hofmann U. NUDT15 polymorphisms alter thiopurine metabolism and hematopoietic toxicity. Nat. Genet. 2016;48(4):367–373. doi: 10.1038/ng.3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lennard L. Implementation of TPMT testing. Br. J. C. Pharma-col. 2014;77(4):704–714. doi: 10.1111/bcp.12226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Woodson L.C., Dunnette J.H., Weinshilboum R.M. Pharmaco-genetics of human thiopurine methyltransferase: kidney-erythrocyte correlation and immunotitration studies. J. Pharmacol. Exp. Ther. 1982;222(1):174–181. [PubMed] [Google Scholar]
- 77.Weinshilboum R.M., Raymond F.A., Pazmino P.A. Human erythrocyte thiopurine methyltransferase: radiochemical microas-say and biochemical properties. Clin. Chim. Acta. 1978;85(3):323–333. doi: 10.1016/0009-8981(78)90311-x. [DOI] [PubMed] [Google Scholar]
- 78.Kröplin T., Weyer N., Gutsche S., Iven H. Thiopurine S-methyltransferase activity in human erythrocytes: a new HPLC method using 6-thioguanine as substrate. Eur. J. Clin. Pharmacol. 1998;54(3):265–271. doi: 10.1007/s002280050457. [DOI] [PubMed] [Google Scholar]
- 79.Ford L., Graham V., Berg J. Whole-blood thiopurine S-methyltransferase activity with genotype concordance: a new, sim-plified phenotyping assay. Ann. Clin. Biochem. 2006;43(5):354–360. doi: 10.1258/000456306778520070. [DOI] [PubMed] [Google Scholar]
- 80.Kharitonov A., Shchukina O., Lapin S., Bulgakova T., Bara-novsky A. P541 Can we predict myelotoxicity development during thiopurine therapy for inflammatory bowel disease? J. Crohn’s Colitis. 2014;8(Suppl. 1):S293. [Google Scholar]
- 81.Dervieux T., Meyer G., Barham R., Matsutani M., Barry M., Boulieu R., Neri B., Seidman E. Liquid chromatography-tandem mass spectrometry analysis of erythrocyte thiopurine nucleotides and effect of thiopurine methyltransferase gene variants on these metabolites in patients receiving azathioprine/6-mercaptopurine therapy. Clin. Chem. 2005;51(11):2074–2084. doi: 10.1373/clinchem.2005.050831. [DOI] [PubMed] [Google Scholar]
- 82.Loit E., Tricco A.C., Tsouros S., Sears M., Ansari M.T., Booth R.A. Pre-analytic and analytic sources of variations in thiopurine methyltransferase activity measurement in patients prescribed thio-purine-based drugs: A systematic review. Clin. Biochem. 2011;44(10):751–757. doi: 10.1016/j.clinbiochem.2011.03.022. [DOI] [PubMed] [Google Scholar]
- 83.Ford L., Prout C., Gaffney D., Berg J. Whose TPMT activity is it anyway? Ann. Clin. Biochem. 2004;41(6):498–500. doi: 10.1258/0004563042466866. [DOI] [PubMed] [Google Scholar]
- 84.Brouwer C., De Abreu R.A., Keizer-Garritsen J.J., Lambooy L.H., Ament K., ter Riet P.G., van Wering E.R., Trijbels F.J., Veerman A.J., Hoogerbrugge P.M., Bökkerink J.P. Thiopurine methyltransferase in acute lymphoblastic leukaemia: biochemical and molecular biological aspects. Eur. J. Cancer. 2005;41(4):613–623. doi: 10.1016/j.ejca.2004.10.027. [DOI] [PubMed] [Google Scholar]
- 85.McLeod H.L., Krynetski E.Y., Wilimas J.A., Evans W.E. Higher activity of polymorphic thiopurine S-methyltransferase in erythro-cytes from neonates compared to adults. Pharmacogenet. Genomics. 1995;5(5):281–286. doi: 10.1097/00008571-199510000-00003. [DOI] [PubMed] [Google Scholar]
- 86.Lennard L., Chew T.S., Lilleyman J.S. Human thiopurine me-thyltransferase activity varies with red blood cell age. Br. J. Clin. Pharmacol. 2001;52(5):539–546. doi: 10.1046/j.0306-5251.2001.01497.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.de Boer N.K., van Bodegraven A.A., de Graaf P., van der Hulst R.W., Zoetekouw L., van Kuilenburg A.B. Paradoxical elevated thiopurine S-methyltransferase activity after pancytopenia during azathioprine therapy: potential influence of red blood cell age. Ther. Drug Monit. 2008;30(3):390–393. doi: 10.1097/FTD.0b013e31816c20b3. [DOI] [PubMed] [Google Scholar]
- 88.Haglund S., Lindqvist M., Almer S., Peterson C., Taipalensuu J. Pyrosequencing of TPMT alleles in a general Swedish population and in patients with inflammatory bowel disease. Clin. Chem. 2004;50(2):288–295. doi: 10.1373/clinchem.2003.023846. [DOI] [PubMed] [Google Scholar]
- 89.de la Moureyre C.S., Debuysère H., Sabbagh N., Marez D., Vinner E., Chevalier E.D., Guidice J.M., Broly F. Detection of known and new mutations in the thiopurine S-methyltransferase gene by single-strand conformation polymorphism analysis. Hum. Mutat. 1998;12(3):177–185. doi: 10.1002/(SICI)1098-1004(1998)12:3<177::AID-HUMU5>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 90.Roberts R.L., Barclay M.L., Gearry R.B., Kennedy M.A. A multiplexed allele-specific polymerase chain reaction assay for the detection of common thiopurine S-methyltransferase (TPMT) mu-tations. Clin. Chim. Acta. 2004;341(1):49–53. doi: 10.1016/j.cccn.2003.10.029. [DOI] [PubMed] [Google Scholar]
- 91.Ganiere-Monteil C., Medard Y., Lejus C., Bruneau B., Pineau A., Fenneteau O., Bourin M., Jacqz-Aigrain E. Phenotype and genotype for thiopurine methyltransferase activity in the French Caucasian population: impact of age. Eur. J. Clin. Pharmacol. 2004;60(2):89–96. doi: 10.1007/s00228-004-0732-5. [DOI] [PubMed] [Google Scholar]
- 92.Nasedkina T.V., Fedorova O.E., Glotov A.S., Chupova N.V., Samochatova E.V., Maiorova O.A., Zemlyakova V.V., Roudne-va A.E., Chudinov A.V., Yurasov R.A., Kozhekbaeva J.M. Rap-id genotyping of common deficient thiopurine S-methyltransferase alleles using the DNA-microchip technique. Eur. J. Hum. Genet. 2006;14(9):991–998. doi: 10.1038/sj.ejhg.5201647. [DOI] [PubMed] [Google Scholar]
- 93.Schaeffeler E., Zanger U.M., Eichelbaum M., Asante-Poku S., Shin J.G., Schwab M. Highly multiplexed genotyping of thiopu-rine S-methyltransferase variants using MALDI-TOF mass spec-trometry: reliable genotyping in different ethnic groups. Clin. Chem. 2008;54(10):1637–1647. doi: 10.1373/clinchem.2008.103457. [DOI] [PubMed] [Google Scholar]
- 94.Kim H.Y., Lee S.H., Lee M.N., Kim J.W., Kim Y.H., Kim M.J., Lee Y.M., Kang B., Choe Y.H., Lee N.H., Kim D.H., Yoo K.H., Sung K.W., Lee S.Y., Koo H.H. Complete sequence-based screening of TPMT variants in the Korean population. Pharmacogenet. Genomics. 2015;25(3):143–146. doi: 10.1097/FPC.0000000000000117. [DOI] [PubMed] [Google Scholar]
- 95.Skrzypczak-Zielinska M., Borun P., Milanowska K., Jakubow-ska-Burek L., Zakerska O. , ; , ; Dobrowolska-Zachwieja, A.; Plawski, A.; Froster, U.G.; Szalata, M.; Slomski, R. High-Resolution melt-ing analysis of the TPMT gene: A study in the polish population. Genet. Test. Mol. Biomarkers. 2013;17(2):153–159. doi: 10.1089/gtmb.2012.0192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ford L., Kampanis P., Berg J. Thiopurine S-methyltransferase genotype-phenotype concordance: used as a quality assurance tool to help control the phenotype assay. Ann. Clin. Biochem. 2009;46(2):152–154. doi: 10.1258/acb.2008.008167. [DOI] [PubMed] [Google Scholar]
- 97.Gearry R.B., Barclay M.L., Roberts R.L., Harraway J., Zhang M., Pike L.S., George P.M., Florkowski C.M. Thiopurine me-thyltransferase and 6‐thioguanine nucleotide measurement: early experience of use in clinical practice. Intern. Med. J. 2005;35(10):580–585. doi: 10.1111/j.1445-5994.2005.00904.x. [DOI] [PubMed] [Google Scholar]
- 98.Lennard L., Rees C.A., Lilleyman J.S., Maddocks J.L. Child-hood leukaemia: a relationship between intracellular 6‐mercaptopurine metabolites and neutropenia. Br. J. Clin. Pharmacol. 1983;16(4):359–363. doi: 10.1111/j.1365-2125.1983.tb02178.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Thervet E., Anglicheau D., Toledano N., Houllier A.M., Noel L.H., Kreis H., Beaune P., Legendre C. Long-term results of TPMT activity monitoring in azathioprine-treated renal allograft recipients. J. Am. Soc. Nephrol. 2001;12(1):170–171. doi: 10.1681/ASN.V121170. [DOI] [PubMed] [Google Scholar]