Significance
Flexibility in the endosymbiotic Symbiodinium community could provide reef-building corals with the capacity to survive environmental change, but this may be restricted to compatible host-symbiont combinations. Therefore, determining the underlying molecular, cellular, and physiological processes of symbiont compatibility is of critical importance for elucidating the resilience and adaptability of coral reefs. We coupled gene expression data with high-throughput metabolite profiling to compare the effects on the sea anemone Aiptasia when colonized by the thermally tolerant, opportunistic, but comparatively unproductive Symbiodinium trenchii vs. the regular symbiont species, Symbiodinium minutum. This powerful approach revealed strong evidence that optimal nutritional exchange and the response to intracellular oxidative stress are important determinants in the success of novel cnidarian-dinoflagellate symbioses.
Keywords: cnidarian, Aiptasia, Symbiodinium, metabolomics, transcriptomics
Abstract
The relationship between corals and dinoflagellates of the genus Symbiodinium is fundamental to the functioning of coral ecosystems. It has been suggested that reef corals may adapt to climate change by changing their dominant symbiont type to a more thermally tolerant one, although the capacity for such a shift is potentially hindered by the compatibility of different host-symbiont pairings. Here we combined transcriptomic and metabolomic analyses to characterize the molecular, cellular, and physiological processes that underlie this compatibility, with a particular focus on Symbiodinium trenchii, an opportunistic, thermally tolerant symbiont that flourishes in coral tissues after bleaching events. Symbiont-free individuals of the sea anemone Exaiptasia pallida (commonly referred to as Aiptasia), an established model system for the study of the cnidarian-dinoflagellate symbiosis, were colonized with the “normal” (homologous) symbiont Symbiodinium minutum and the heterologous S. trenchii. Analysis of the host gene and metabolite expression profiles revealed that heterologous symbionts induced an expression pattern intermediate between the typical symbiotic state and the aposymbiotic state. Furthermore, integrated pathway analysis revealed that increased catabolism of fixed carbon stores, metabolic signaling, and immune processes occurred in response to the heterologous symbiont type. Our data suggest that both nutritional provisioning and the immune response induced by the foreign “invader” are important factors in determining the capacity of corals to adapt to climate change through the establishment of novel symbioses.
Reef-building corals are dependent on an endosymbiosis with a highly diverse group of photosynthetic dinoflagellates of the genus Symbiodinium. Bidirectional exchange of organic and inorganic compounds is central to the ecological success and stability of the coral-dinoflagellate symbiosis and is critically important to the construction and persistence of coral reef ecosystems (reviewed in ref. 1). The algal symbiont receives dissolved inorganic nutrients (i.e., carbon, nitrogen, and phosphorus) from the host and also may receive host-derived amino acids, lipids, and fatty acids (2). In return, symbionts translocate products of carbon fixation and nitrogen assimilation to the host, including sugars (especially glucose), amino acids, and lipids (3, 4).
Disruption of the symbiosis (dysbiosis) due to the loss of Symbiodinium cells from coral tissues, known as coral bleaching, is due primarily to increasing sea surface temperatures from global warming and is occurring with increasing frequency and severity. Bleaching can cause decreased coral growth, fecundity, and fitness and dramatically increased mortality; therefore, it poses a major threat to the future of coral reefs (5). However, sublethal bleaching may present an opportunity for the existing mixed Symbiodinium community resident within a host to “shuffle” to an alternative dominant type or “switch” to a new type acquired from the surrounding environment (6, 7). Either mechanism could enable the holobiont (i.e., the coral, Symbiodinium, and other symbiotic microbes) to better withstand changing ocean conditions, such as elevated sea surface temperatures (6). These putative mechanisms are founded on the considerable molecular and physiological diversity within the genus Symbiodinium, with a single host species displaying different physiologies depending on the Symbiodinium type that it hosts (8).
Establishment of symbiosis between a host cnidarian and dinoflagellates from the surrounding environment includes mechanisms of interpartner recognition whereby compatible symbionts are retained within host cells and inappropriate invaders are removed (reviewed in ref. 1). The mechanisms responsible for the dynamic regulation and persistence of the symbiosis involve modulation of both the host immune response (9–11) and nutritional fluxes between the two partners (8). Different symbiont strains may release different types and quantities of photosynthate to the host and hence influence the nutritional potential of the symbiosis and its overall fitness (8, 12, 13). Therefore, unraveling these cellular and metabolic processes is imperative for understanding the potential for the evolution of novel host-symbiont pairings and the capacity of corals to withstand climate change.
The sea anemone Exaiptasia pallida (14) (commonly referred to as Aiptasia) is a widely adopted model system for the study of coral symbiosis physiology and cell biology (15, 16). Its ease of culture, rapid growth rate, and accessible genomic resources make it a valuable resource for empirical studies of symbiosis that cannot be carried out in corals. Aiptasia most commonly forms an association with Symbiodinium minutum (ITS2 type B1) (13, 17, 18). Crucially, unlike corals, Aiptasia can be easily rendered aposymbiotic and recolonized with a range of different symbiont types, including types not found to be associated with Aiptasia in nature, making it an ideal system for the study of symbiosis establishment and host-symbiont regulation (e.g., ref. 13).
Here we used a combination of two high-throughput “omics” techniques (transcriptomics and metabolomics) to characterize gene expression and metabolite profiles of the cnidarian host in response to symbiotic state and symbiont type. Specifically, we compared aposymbiotic Aiptasia vs. those colonized by the homologous S. minutum vs. those colonized by the heterologous Symbiodinium trenchii (ITS2 type D1a) (19). The environmentally tolerant, opportunistic, but relatively unproductive S. trenchii has been shown to repopulate corals after bleaching events in numerous studies (6, 20); however, its role in overall coral survival and reef health is not fully understood (21, 22). These integrated analyses allow us to develop a complex account of the specific pathways and processes involved in the regulation and maintenance of cnidarian-dinoflagellate symbiosis, and of how the host responds to introduction of the novel heterologous S. trenchii.
Results
Colonization Success.
The two symbiont species, homologous S. minutum and heterologous S. trenchii, reached similar densities in Aiptasia (Mann–Whitney U; P = 0.589). Mean ± SEM cell densities of S. minutum and S. trenchii were 11.1 ± 2.8 × 106 and 10.5 ± 2.4 × 106 cells mg−1 protein, respectively. These values are consistent with full colonization (12, 23).
Transcriptomic Analysis.
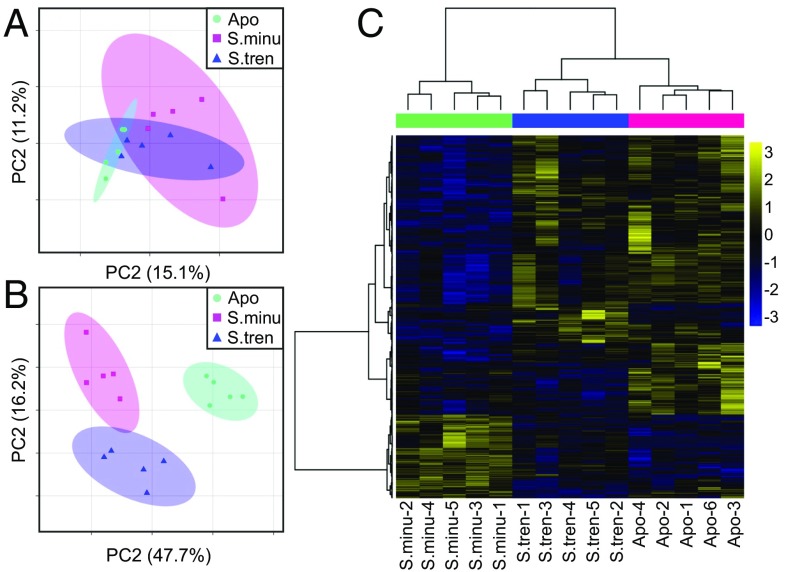
A reference transcriptome consisting of 35,516 cleaned, high-quality individual transcripts was tested for differential expression (Table S1 and Dataset S1). Principle component analysis (PCA) of the variance-stabilized data revealed expression differences among the three symbiotic states (Fig. 1A and Table S2). While there was some overlap among the three groups, the greatest difference was between aposymbiotic and S. minutum-colonized anemones; S. trenchii-colonized hosts showed intermediate expression. Statistical analysis of the variance-stabilized data (SI Materials and Methods) identified 686 differentially expressed (DE) genes according to symbiotic state (Dataset S2). PCA of the DE genes showed a clear separation between the symbiotic states (Fig. 1B). The gene expression of hosts with heterologous symbionts was closer to that of aposymbiotic anemones than to that of hosts with homologous symbionts. Likewise, the expression heatmap of the DE genes within the three symbiotic states showed close alignment between aposymbiotic and S. trenchii-colonized anemones, with S. minutum-colonized hosts the most different (Fig. 1C).
Fig. 1.
RNA-Seq gene expression of Aiptasia in response to symbiotic state. (A) PCA of all genes. (B) PCA of all DE genes; clusters replicate samples by symbiotic state (FDR 10%). (C) Heat map of all DE genes (P < 0.1). Rows are genes, and columns are biological replicate samples of the three symbiotic treatments —homologous S. minutum (S. minu), heterologous S. trenchii (S. tren), and aposymbiotic (Apo) (n = 5)—as ordered by hierarchical clustering of genes based on Pearson’s correlation of their expression across samples. The color scale is log2 fold change relative to the mean.
Of the 686 DE genes, 409 had protein annotations (UniProt) by homology to known genes and were assigned a Gene Ontology (GO) molecular function (Fig. S1 and Dataset S3). Sixteen percent of DE genes were grouped under stress response, composed primarily of genes involved in oxidative stress and apoptosis, two processes that play critical roles in animal innate immune responses to microbes (24). Redox genes included glutathione-handling enzymes and heat shock proteins. Apoptosis-related genes included caspase-8 and tumor necrosis factor receptors. Transport-related proteins composed 13% of the DE genes and included a large number involved in metabolite transport: ABC transporters, solute-carrier proteins, and monocarboxylate transporters. Proteins involved in oxygen transport and G protein receptor signal transduction also fell into this group. Fifteen percent of the DE genes were categorized under metabolism. Carbohydrate metabolism included membrane-associated glycosyl-phosphatidylinositol–linked carbonic anhydrase, implicated in inorganic carbon transport, and Niemann–Pick type C2. Multiple DE genes were grouped in the category of synthesis and degradation of lipids.
Metabolite Profiling.
A total of 89 compounds were identified in the polar and semipolar extracts of free metabolite pools from hosts, consisting primarily of amino acids, metabolic intermediates, organic acids, sugars, and fatty acids (Dataset S4). PCA showed distinct clustering of the three symbiotic states (Fig. S2). Similar to the PCA of expression profiles, the greatest difference was seen between aposymbiotic and homologous S. minutum-colonized anemones, while the heterologous S. trenchii-colonized hosts spanned the intermediate region.
Strikingly, of the 89 metabolites identified, only 8 were higher in relative abundance in S. minutum-colonized host pools compared with S. trenchii-colonized host pools (Fig. S3 and Dataset S5). The remaining 81 metabolites were in higher abundance in the S. trenchii-colonized host pools, including all nine detected unsaturated fatty acids. Univariate statistical analysis of the fold change in relative abundance identified 10 compounds that were of significantly greater abundance in S. trenchii-colonized host pools compared with S. minutum-colonized host pools. These compounds included two monounsaturated fatty acids [palmitoleic acid (C16:1) and oleic acid (C18:1)], three polyunsaturated fatty acids [PUFAs; linoleic acid (C18:2), linolenic acid (C18:3), and dihomo-γ-linolenic acid (C20:3)], saturated fatty acid butyric acid (C4:0), the amino acids methionine and tyrosine, the amino acid-derivative tyramine, the CoA precursor pantothenic acid, and the amino sugar glucosamine. No compounds were significantly lower in S. trenchii-colonized host pools compared with S. minutum-colonized host pools.
Joint Enrichment Analysis.
Joint enrichment analysis (SI Materials and Methods) of the transcriptomic and metabolomic data revealed 22 differentially affected processes in Aiptasia colonized with heterologous S. trenchii compared with those colonized with homologous S. minutum (Table 1). Five of the processes referred to broad categories in the Reactome database (signal transduction, diseases, gene expression, metabolism, and transport of small molecules), and 17 processes referred to specific pathways within these categories. Whereas differentially affected genes displayed a mixture of up- and down-regulation in expression, the vast majority of metabolites affected across all processes were up-regulated in S. trenchii-colonized hosts compared with S. minutum-colonized hosts.
Table 1.
Summary of the significantly enriched pathways
| Pathway name | Genes | Metabolites |
| Signal transduction** | ||
| Signaling by GPCR** | 6/4 | 1/16 |
| GPCR ligand binding* | 3/3 | 1/13 |
| Class A/1 rhodopsin-like receptors* | 1/2 | 0/11 |
| GPCR downstream signaling* | 5/3 | 1/9 |
| Diseases** | ||
| Glycogen storage diseases* | 6/7 | 1/9 |
| Metabolic disorders of oxidation enzymes** | 3/6 | 1/33 |
| Gene expression** | 6/9 | 1/17 |
| Metabolism** | ||
| Biological oxidations** | 1/2 | 1/22 |
| Metabolism of amino acids, derivatives, and urea cycle** | 2/7 | 1/24 |
| Tyrosine metabolism* | 0/3 | 1/7 |
| Metabolism of vitamins and cofactors* | 2/1 | 1/8 |
| Metabolism of lipids and lipoproteins* | 7/5 | 2/16 |
| Protein digestion and absorption** | 2/2 | 1/19 |
| Fatty acid, TAG, and ketone metabolism** | 3/2 | 0/10 |
| Purine metabolism* | 3/1 | 1/8 |
| Transmembrane transport of small molecules** | ||
| SLC-mediated transport** | 5/4 | 2/38 |
| Transport of glucose/other sugars** | 3/1 | 0/22 |
| ABC transporters** | 3/1 | 1/25 |
Pathways were detected as significantly enriched (*P < 0.05; **P < 0.001) in S. trenchii-containing hosts vs. S. minutum-containing hosts when FDR <0.05 and at least three genes or metabolites were present. The numbers of down-regulated/up-regulated genes and metabolites in S. trenchii-colonized hosts are shown. A list of the genes/metabolites in each pathway is provided in Dataset S6.
Discussion
Host phenotypes of the three symbiotic states are distinctly different as measured by gene expression and metabolite profiles. Hosts fully colonized with S. trenchii either occupy an intermediate position between aposymbiotic and S. minutum-colonized hosts or are more closely aligned with aposymbiotic hosts (Fig. 1 and Fig. S2). Numerous gene expression studies have documented differences between the symbiotic and aposymbiotic state in corals and sea anemones (25), including recent high-throughput RNA-Seq studies (11, 26). Two microarray studies that examined gene expression of hosts colonized with homologous and heterologous symbionts also showed distinct host expression profiles as a function of symbiont type (9, 27).
We grouped information from the expression and metabolite profiling and integrated analysis into functional categories highlighting genes and metabolites likely involved in the cnidarian-Symbiodinium symbiosis. What emerged is a picture of dramatically different biological processes involved in immunity, cell signaling, and metabolism between hosts fully colonized by homologous symbionts and those colonized by heterologous symbionts (Fig. 2, Table 1, and Datasets S3, S5, and S6). Such differences could contribute to the relatively rapid rate of colonization by homologous symbionts compared with heterologous symbionts in Aiptasia (28). In what follows, we provide detailed descriptions of the biological patterns from integrated analysis.
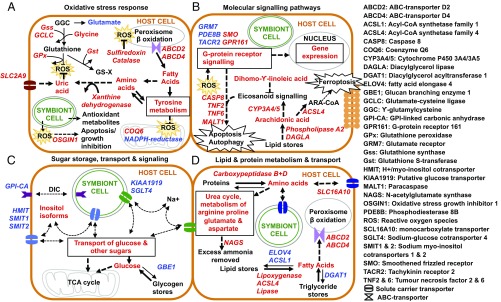
Fig. 2.
Cellular processes altered by colonization by a heterologous symbiont: (A) oxidative stress response; (B) molecular signaling pathways; (C) sugar storage, transport, and signaling; and (D) lipid and protein metabolism and transport. Type color indicates up-regulated (red) and down-regulated (blue) expression in the heterologous S. trenchii-colonized host tissues. Entities in black type were not measured in this study. Arrow thickness indicates direction of pathway activity, and dashed lines indicate putative activity. Only genes and metabolites likely involved in the symbiosis are shown. Italic type represents genes, and roman type represents metabolites. Data are from Datasets S3, S5, and S6.
Host Innate Immunity Processes Are Up-Regulated in Hosts Colonized with Heterologous Symbionts.
Evidence throughout the individual and linked datasets indicates different host innate immune responses to the two different symbiont species. The data point toward immunotolerance and immune quiescence in hosts containing homologous symbionts, and immunoresistance and a stress response in those containing heterologous symbionts. These processes are grouped into oxidative stress response and signaling pathways (Fig. 2 A and B, respectively).
Oxidative Stress Response.
The S. trenchii-colonized hosts showed significant activation of processes involved in a homeostatic cellular oxidative stress response, including reactive oxygen species (ROS)- and reactive nitrogen species (RNS)-producing pathways, oxidative stress response proteins, and the synthesis of enzymatic and nonenzymatic antioxidants (Fig. 2A). This provides evidence that hosts were responding to an oxidative stress event. A number of NADPH reductases (ROS attenuators) were down-regulated, potentially resulting in ROS accumulation in S. trenchii-colonized host tissues. Coenzyme Q6, produced in response to mitochondrial hypoxia (29), and the antioxidant enzymes catalase and sulfiredoxin were up-regulated in S. trenchii-colonized hosts.
There were multiple examples of increased production of nonenzymatic antioxidants in S. trenchii-colonized hosts. Uric acid was significantly more abundant, and genes involved in uric acid production and transport were significantly up-regulated. Xanthine dehydrogenase, involved primarily in purine metabolism and uric acid formation, was up-regulated, as was the uric acid transporter SLC2A9. The amino acid tyrosine was also more abundant, and tyrosine metabolism was up-regulated in S. trenchii-colonized hosts. A role for tyrosine in defense against reactive oxygen stress has been suggested in the moon jellyfish Aurelia aurita (30). Finally, the glutathione (GSH) pathway was up-regulated in S. trenchii-colonized hosts. GSH detoxifies ROS directly or serves as an electron donor in a reaction catalyzed by glutathione peroxidase (31). Four GSH pathway enzymes were up-regulated: glutathione peroxidase, glutathione synthase, glutamate-cysteine ligase, and glutathione transferase. Crucially, the abundance of glutamate, a GSH precursor, was lower in S. trenchii-colonized hosts, corresponding to an up-regulation in GSH synthesis.
ROS regulation is a key component in the innate immune response to both harmful and beneficial invaders (32). It can play a signaling role and also participate in responses to destroy invaders, such as the microbial killing mechanism (24), and has key roles in the regulation of cnidarian-dinoflagellate mutualisms (1). In addition, ROS mitigation is a hallmark of cnidarian-dinoflagellate symbiosis physiology, given that hosts need mechanisms to manage hyperoxia and the ROS produced by their highly productive symbionts (33). Our results demonstrate a disruption of cellular oxidative homeostasis in the heterologous S. trenchii-colonized hosts. This up-regulation of an oxidative-stress response is consistent with the accumulation of ROS and RNS in the S. trenchii-colonized host tissues. This observation is compatible with the known roles of ROS and RNS in the cnidarian bleaching cascade (33), as well as with previous reports that colonization of Aiptasia with homologous symbionts results in a down-regulation of host nitric oxide (NO) production pathways (34), while inoculation with unsuitable or dysfunctional symbionts leads to heightened host NO production (35).
Elevated antioxidant defenses in S. trenchii-colonized hosts could enhance the thermotolerance and/or oxidative tolerance of the holobiont, perhaps helping to explain stress resilience conferred by this symbiont type (6, 19, 20). Elevation of host antioxidant pools in response to S. trenchii may explain the ability of this heterologous symbiont type to maintain a symbiosis with Aiptasia despite the putative functional consequences, including decreased productivity and growth.
Signaling Pathways.
Numerous differences in host cell signaling pathways were identified in the individual and integrated datasets (Fig. 2B). The integrated data indicate alterations in signaling driving membrane trafficking and oxidative stress, both possibly related to signaling across the symbiosome and/or regulation of apoptosis and autophagy. Membrane trafficking is likely a critical component of innate immune regulation and interpartner signaling in cnidarian-dinoflagellate symbiosis, but this remains very poorly understood and is a current area of interest in the field (1). The results show an up-regulation of G protein-associated stress signaling and lipid signaling pathways active in a stress response in the heterologous S. trenchii-colonized hosts (Fig. 2B).
G protein coupled receptor (GPCR) signaling was one of the major differentially enriched processes emerging from the integrated analysis (Fig. 2B), suggesting dramatic differences in signal transduction between hosts harboring the two different symbiont species. GPCRs are the most abundant and diverse group of membrane receptors in eukaryotes (36). They typically transduce inputs via cAMP or phosphatidylinositol signaling pathways (36) that in turn lead to an enormous diversity of cellular responses. The role of GPCRs in the context of cnidarian-dinoflagellate symbiosis is essentially unexplored to date; one study identified GPCRs in symbiosomes isolated from Aiptasia, suggesting a possible involvement in interpartner signaling (37).
There was up-regulation of multiple genes in S. trenchii-colonized hosts that are involved in apoptotic and autophagic processes, including tumor necrosis factor receptors, the paracaspase MALT1, and caspase-8 (Dataset S3). These processes are widely considered critical mechanisms in cnidarian-dinoflagellate dysbiosis (33) and are also hypothesized to play a role in immune recognition during the onset of symbiosis (38). Our data indicate signaling for an elevated immune response in S. trenchii-colonized animals and a corresponding attenuated immune response in homologous S. minutum-containing hosts, supporting previous proposals for immunotolerance as a part of cnidarian-dinoflagellate symbiosis recognition (10, 11, 26).
There are several indications of differences in lipid signaling in S. trenchii-colonized hosts compared with S. minutum-colonized hosts, suggesting a stress response (Fig. 2B). The lipid hydrolyzers diacylglycerol lipase and phospholipase A2 were up-regulated in S. trenchii-colonized hosts. Likewise, there was higher abundance of arachidonic acid (ARA; C20:4); a downstream product of hydrolysis and an essential PUFA. There was also an up-regulation of two cytochrome P450 monooxygenases (CYP3A4/5), which catalyze ARA and other PUFAs to biologically active, intercellular signaling molecules (eicosanoids) (39). These lipids participate in an oxidative stress response and are hypothesized to play a role in the heat stress response in symbiotic cnidarians (40). Finally, acyl-CoA synthetase long-chain family member 4 was up-regulated. This enzyme converts ARA to arachidonoyl-CoA, which is an important player in cellular oxidative stress and ferroptosis, a form of regulated cell death (41).
There is also evidence of stress modulation via lipid signaling in S. trenchii-colonized hosts. Three closely related long-chain fatty acids—C18:2n-6, C18:3n-6, and C20:3n-6 (dihomo-γ-linolenic acid)—were significantly elevated in S. trenchii-colonized hosts (Fig. 2B). Dihomo-γ-linolenic acid can act as an antagonist for the ARA eicosanoid signaling cascade to counter the inflammatory effects of ARA (42). Taken together, these data suggest that lipid signaling related to a stress response is occurring in S. trenchii-colonized hosts. These differentially produced stress-related metabolites provide potential biomarkers for future investigation into cellular signaling mechanisms in the cnidarian-dinoflagellate symbiosis.
S. trenchii-Colonized Host Metabolic Profiles Show a Strong Shift Toward Carbohydrate and Lipid Catabolism and Mobilization.
The separate datasets and integrated analysis show consistent patterns of elevated host catabolism of carbohydrate and lipid in S. trenchii-colonized hosts compared with S. minutum-colonized hosts. These data suggest that although the novel S. trenchii symbionts have colonized host tissues, the host is not increasing its energy reserves, but instead is catabolizing and mobilizing these stores to satisfy its energy needs.
Carbohydrate Transport and Storage.
Pathways involved in glycogen storage disorders in other organisms were identified in the integrated pathway results, indicating reduced glycogen storage in S. trenchii-colonized host tissues (Fig. 2C). Ten genes and 10 metabolites involved in these disorders were detected, including the glycogen synthesis gene glucan branching enzyme (GBE1), which was down-regulated in S. trenchii-colonized hosts. Similar reductions in GBE1 expression involved in glycogen synthesis have been observed during dysbiosis in the coral Acropora palmata under thermal stress conditions (43). We also observed a down-regulation of multiple sugar transporters, including the Na+/glucose cotransporter SGLT4 (16) and a putative Na+-dependent glucose transporter (KIAA1919), as well as multiple sugar-inositol transporters. The proton myo-inositol cotransporter HMIT is a member of the facilitative glucose transporter family that has been recently identified in the Aiptasia genome (16), and was down-regulated in the S. trenchii-colonized hosts. Also down-regulated were the myo-inositol/Na+ cotransporter SMIT1, involved in sugar transport and signaling (44), and the myo/chiro-inositol/glucose transporter SMIT2, previously found to be highly up-regulated in symbiotic host cnidarians (26).
The decreased activity of these transporters would putatively result in a reduction in the intercellular and/or intracellular movement of sugars, yet we observed a high abundance of free pools of carbohydrates in the S. trenchii-colonized host tissues (Fig. S3). Taken together, these results indicate alterations in the processing of glycogen synthesis and an increase in sugar catabolism in S. trenchii-colonized anemones. Corals have been shown to form storage deposits of glycogen, suggesting that glycogen metabolism is indicative of a productive symbiosis (4). Our results suggest that S. trenchii-colonized anemones, in contrast, catabolize their sugars and do not accumulate glycogen deposits compared with S. minutum-colonized hosts.
Our results are consistent with previous studies showing that Symbiodinium genotypic diversity affects the quality and quantity of material translocated to the host (8, 13, 45), particularly those studies examining the performance of hosts harboring S. trenchii. Physiological experiments with S. trenchii-colonized Aiptasia revealed lower rates of carbon translocation than seen in hosts harboring homologous Symbiodinium (12). Studies in corals showed that S. trenchii-colonized corals experience reduced carbon translocation (46) or altered photosynthetic performance and energetics (47), leading to slower coral growth (21).
Amino Acid Metabolism and the Urea Cycle.
Symbiosis with the heterologous S. trenchii resulted in shifts in the metabolism and transport of host protein and amino acids (Fig. 2D). Multiple enzymes involved in the catabolism of proteins and amino acids, along with three monocarboxylate transporters, SLC16A10s, involved in the translocation of aromatic amino acids, were up-regulated in S. trenchii-colonized hosts (Dataset S3). The relative abundance of all detected amino acids was higher in S. trenchii-colonized tissues compared with S. minutum-colonized tissues (Fig. S2). Furthermore, pathway analysis revealed up-regulation of the urea cycle and of the metabolism of arginine, proline, glutamate, aspartate, and tyrosine (Fig. 2D). There was also an up-regulation of N-acetylglutamate synthase, which participates in the removal of excess ammonia (48). This is of particular interest given that nitrogen cycling within the symbiosis includes ammonium produced via the urea cycle, and that control of nitrogen provision is important for the regulation of host-symbiont biomass (reviewed in ref. 1).
Lipid Metabolism and Transport.
The datasets show strikingly different patterns in lipid metabolism between hosts harboring the two different symbiont species. Overall, S. trenchii-colonized hosts display strong evidence of lipid catabolism, reduced lipid stores and mobilization, and transport of lipids and fatty acids compared with hosts colonized with homologous S. minutum. Pathways involved in lipid and fatty acid degradation and transport were up-regulated in S. trenchii-colonized hosts, and conversely, enzymes responsible for fatty acid synthesis and elongation were down-regulated (Fig. 2D). In the expression analysis, lipase, lipoxygenase and two triglyceride lipases were up-regulated, whereas DGAT1, an enzyme involved in triglyceride synthesis, and the lipid biosynthesis enzymes ACSL1 and ELOVL4 were down-regulated (Dataset S3). These expression data are consistent with a concomitant higher abundance of the saturated fatty acid butyric acid and five unsaturated fatty acids in S. trenchii-colonized host metabolite pools (Fig. S3 and Dataset S5). Finally, ABCD2 and ABCD4, two transporters with peroxisomal fatty acid import functions, were up-regulated in the S. trenchii-colonized hosts (Dataset S3).
These results indicate that the mobilization of stored lipids and fatty acids is required by the host to support central metabolism when harboring S. trenchii, presumably due to a reduction of photosynthate (either as carbohydrate or lipid) from the S. trenchii symbiont. This complements evidence from other cnidarian-dinoflagellate symbioses showing that host lipid quantity and quality vary with symbiont type (49). Thus, our data not only support the importance of trophic plasticity for the survival of these associations (12), but also suggest that once host energy stores are exhausted, inadequate nutritional exchange between host and symbiont is likely to be a limiting factor. Given that changes in lipid and fatty acid composition have been shown to reflect changes in the nutrition, ecology, and ultimately health of corals (50), endosymbiont type and novel associations thus may affect the long-term survival of the holobiont.
Ecological Implications.
The idea that the capacity to change host-symbiont combinations could provide a punctuated and accelerated ability to adapt to changing environmental conditions has been a controversial and much-discussed area of cnidarian symbiosis for several decades (e.g., ref. 51). Overall, animals harboring the novel symbiont S. trenchii show evidence of immunoresistance and heterotrophy, in contrast to the immunotolerance and symbiont-derived nutrition displayed in S. minutum-colonized animals. These patterns are consistent with previous studies of host cnidarians colonized with S. trenchii that have shown attenuated productivity and slower growth of the association and, in the case of calcifying organisms, reduced coral reef accretion (52). Such physiological trade-offs cast doubt on the ability of thermally tolerant clade D Symbiodinium to ameliorate the effects of climate change (22). The strong shifts in host immunologic and metabolic pathways described here indicate that to establish successful long-term symbioses, novel symbionts must overcome considerable challenges at the cellular and organismal levels. Furthermore, the ecological relevance of novel associations will depend on whether the symbiont provides any functional benefit to the coral’s physiology, beyond the capacity to simply form an association with a new host (53). Thus, the capability of novel symbionts to increase coral resilience to climate change will require each to run a gauntlet of host immune responses while balancing the exchange of multiple metabolite classes of functional significance, all while experiencing increasingly severe environmental stress. Confirming our observations in reef corals, not in the least to establish whether calcification has any impact on the pathways described, is critical to fully elucidate how coral reef communities may respond to a changing climate.
Materials and Methods
Aposymbiotic Aiptasia (23) (culture ID NZ1) were inoculated with either S. minutum (ITS2 type B1, originally isolated from NZ1) or S. trenchii (ITS2 type D1a, culture ID Ap2). Samples were collected at 5 wk after initial inoculation (n = 5). Total RNA was extracted, and cDNA libraries were prepared (54). The reference transcriptome was assembled and annotated, and differential expression was analyzed using RNA derived from the different symbiotic states. GC-MS–based metabolite profiling was conducted on polar and semipolar free metabolites extracted from the separated host tissues (55). A joint Wilcoxon pathway enrichment analysis of transcriptomic and metabolomic data was performed using the Integrated Molecular Pathway Level Analysis (IMPaLA) web interface (56). Methods are described in detail in SI Materials and Methods.
Supplementary Material
Acknowledgments
We acknowledge Siria Natera, Nirupama Samanmalie Jayasinghe, and Himasha Mendis for their contributions to the GC-MS analysis, which was performed at Metabolomics Australia (School of BioSciences, The University of Melbourne, Australia), a National Collaborative Research Infrastructure Strategy initiative under Bioplatforms Australia Pty Ltd. We thank the two anonymous reviewers for their helpful comments. This work was supported by the Royal Society of New Zealand Marsden Fund Grant 1202, to S.K.D., A.R.G., and V.M.W.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this study have been deposited in the Dryad repository, www.datadryad.org/.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1710733114/-/DCSupplemental.
References
- 1.Davy SK, Allemand D, Weis VM. Cell biology of cnidarian-dinoflagellate symbiosis. Microbiol Mol Biol Rev. 2012;76:229–261. doi: 10.1128/MMBR.05014-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Imbs AB, Yakovleva IM, Dautova TN, Bui LH, Jones P. Diversity of fatty acid composition of symbiotic dinoflagellates in corals: Evidence for the transfer of host PUFAs to the symbionts. Phytochemistry. 2014;101:76–82. doi: 10.1016/j.phytochem.2014.02.012. [DOI] [PubMed] [Google Scholar]
- 3.Burriesci MS, Raab TK, Pringle JR. Evidence that glucose is the major transferred metabolite in dinoflagellate-cnidarian symbiosis. J Exp Biol. 2012;215:3467–3477. doi: 10.1242/jeb.070946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kopp C, et al. Subcellular investigation of photosynthesis-driven carbon assimilation in the symbiotic reef coral Pocillopora damicornis. MBio. 2015;6:e02299-14. doi: 10.1128/mBio.02299-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoegh-Guldberg O, Ortiz JC, Dove S. The future of coral reefs. Science. 2011;334:1494–1495. author reply 1495–1496. doi: 10.1126/science.334.6062.1494-b. [DOI] [PubMed] [Google Scholar]
- 6.Berkelmans R, van Oppen MJ. The role of zooxanthellae in the thermal tolerance of corals: A “nugget of hope” for coral reefs in an era of climate change. Proc Biol Sci. 2006;273:2305–2312. doi: 10.1098/rspb.2006.3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boulotte NM, et al. Exploring the Symbiodinium rare biosphere provides evidence for symbiont switching in reef-building corals. ISME J. 2016;10:2693–2701. doi: 10.1038/ismej.2016.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loram JE, Trapido-Rosenthal HG, Douglas AE. Functional significance of genetically different symbiotic algae Symbiodinium in a coral reef symbiosis. Mol Ecol. 2007;16:4849–4857. doi: 10.1111/j.1365-294X.2007.03491.x. [DOI] [PubMed] [Google Scholar]
- 9.Voolstra CR, et al. The host transcriptome remains unaltered during the establishment of coral-algal symbioses. Mol Ecol. 2009;18:1823–1833. doi: 10.1111/j.1365-294X.2009.04167.x. [DOI] [PubMed] [Google Scholar]
- 10.Schnitzler CE, Weis VM. Coral larvae exhibit few measurable transcriptional changes during the onset of coral-dinoflagellate endosymbiosis. Mar Genomics. 2010;3:107–116. doi: 10.1016/j.margen.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 11.Mohamed AR, et al. The transcriptomic response of the coral Acropora digitifera to a competent Symbiodinium strain: The symbiosome as an arrested early phagosome. Mol Ecol. 2016;25:3127–3141. doi: 10.1111/mec.13659. [DOI] [PubMed] [Google Scholar]
- 12.Leal MC, et al. Symbiont type influences trophic plasticity of a model cnidarian-dinoflagellate symbiosis. J Exp Biol. 2015;218:858–863. doi: 10.1242/jeb.115519. [DOI] [PubMed] [Google Scholar]
- 13.Starzak DE, Quinnell RG, Nitschke MR, Davy SK. The influence of symbiont type on photosynthetic carbon flux in a model cnidarian–dinoflagellate symbiosis. Mar Biol. 2014;161:711–724. [Google Scholar]
- 14.Grajales A, Rodriguez E. Morphological revision of the genus Aiptasia and the family Aiptasiidae (Cnidaria, Actiniaria, Metridioidea) Zootaxa. 2014;3826:55–100. doi: 10.11646/zootaxa.3826.1.2. [DOI] [PubMed] [Google Scholar]
- 15.Weis VM, Davy SK, Hoegh-Guldberg O, Rodriguez-Lanetty M, Pringle JR. Cell biology in model systems as the key to understanding corals. Trends Ecol Evol. 2008;23:369–376. doi: 10.1016/j.tree.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 16.Baumgarten S, et al. The genome of Aiptasia, a sea anemone model for coral symbiosis. Proc Natl Acad Sci USA. 2015;112:11893–11898. doi: 10.1073/pnas.1513318112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lajeunesse TC, Parkinson JE, Reimer JD. A genetics-based description of Symbiodinium minutum sp. nov. and S. psygmophilum sp. nov. (Dinophyceae), two dinoflagellates symbiotic with cnidaria. J Phycol. 2012;48:1380–1391. doi: 10.1111/j.1529-8817.2012.01217.x. [DOI] [PubMed] [Google Scholar]
- 18.Thornhill DJ, Xiang Y, Pettay DT, Zhong M, Santos SR. Population genetic data of a model symbiotic cnidarian system reveal remarkable symbiotic specificity and vectored introductions across ocean basins. Mol Ecol. 2013;22:4499–4515. doi: 10.1111/mec.12416. [DOI] [PubMed] [Google Scholar]
- 19.LaJeunesse TC, et al. Ecologically differentiated stress-tolerant endosymbionts in the dinoflagellate genus Symbiodinium (Dinophyceae) clade D are different species. Phycologia. 2014;53:305–319. [Google Scholar]
- 20.Stat M, Gates RD. Clade D Symbiodinium in scleractinian corals: A “nugget” of hope, a selfish opportunist, an ominous sign, or all of the above? J Mar Biol. 2011;2011:1–9. [Google Scholar]
- 21.Cunning R, Gillette P, Capo T, Galvez K, Baker AC. Growth tradeoffs associated with thermotolerant symbionts in the coral Pocillopora damicornis are lost in warmer oceans. Coral Reefs. 2015;34:155–160. [Google Scholar]
- 22.Ortiz JC, González-Rivero M, Mumby PJ. Can a thermally tolerant symbiont improve the future of Caribbean coral reefs? Glob Change Biol. 2013;19:273–281. doi: 10.1111/gcb.12027. [DOI] [PubMed] [Google Scholar]
- 23.Matthews JL, et al. Menthol-induced bleaching rapidly and effectively provides experimental aposymbiotic sea anemones (Aiptasia sp.) for symbiosis investigations. J Exp Biol. 2016;219:306–310. doi: 10.1242/jeb.128934. [DOI] [PubMed] [Google Scholar]
- 24.Janeway CA, Jr, Medzhitov R. Innate immune recognition. Annu Rev Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 25.Meyer E, Weis VM. Study of cnidarian-algal symbiosis in the “omics” age. Biol Bull. 2012;223:44–65. doi: 10.1086/BBLv223n1p44. [DOI] [PubMed] [Google Scholar]
- 26.Lehnert EM, et al. Extensive differences in gene expression between symbiotic and aposymbiotic cnidarians. G3 (Bethesda) 2014;4:277–295. doi: 10.1534/g3.113.009084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DeSalvo MK, et al. Coral host transcriptomic states are correlated with Symbiodinium genotypes. Mol Ecol. 2010;19:1174–1186. doi: 10.1111/j.1365-294X.2010.04534.x. [DOI] [PubMed] [Google Scholar]
- 28.Schoenberg D, Trench R. Genetic variation in Symbiodinium (= Gymnodinium) microadriaticum Freudenthal, and specificity in its symbiosis with marine invertebrates, III: Specificity and infectivity of Symbiodinium microadriaticum. Proc R Soc Lond B Biol Sci. 1980;207:445–460. [Google Scholar]
- 29.Beyer RE. An analysis of the role of coenzyme Q in free radical generation and as an antioxidant. Biochem Cell Biol. 1992;70:390–403. doi: 10.1139/o92-061. [DOI] [PubMed] [Google Scholar]
- 30.Berking S, et al. A newly discovered oxidant defence system and its involvement in the development of Aurelia aurita (Scyphozoa, Cnidaria): Reactive oxygen species and elemental iodine control medusa formation. Int J Dev Biol. 2005;49:969–976. doi: 10.1387/ijdb.052024sb. [DOI] [PubMed] [Google Scholar]
- 31.Dringen R. Metabolism and functions of glutathione in brain. Prog Neurobiol. 2000;62:649–671. doi: 10.1016/s0301-0082(99)00060-x. [DOI] [PubMed] [Google Scholar]
- 32.Moné Y, Monnin D, Kremer N. The oxidative environment: A mediator of interspecies communication that drives symbiosis evolution. Proc Biol Sci. 2014;281:20133112. doi: 10.1098/rspb.2013.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lesser MP. Coral Reefs: An Ecosystem in Transition. Springer; Berlin: 2011. Coral bleaching: Causes and mechanisms; pp. 405–419. [Google Scholar]
- 34.Detournay O, Schnitzler CE, Poole A, Weis VM. Regulation of cnidarian-dinoflagellate mutualisms: Evidence that activation of a host TGFβ innate immune pathway promotes tolerance of the symbiont. Dev Comp Immunol. 2012;38:525–537. doi: 10.1016/j.dci.2012.08.008. [DOI] [PubMed] [Google Scholar]
- 35.Hawkins TD. 2013. Bleaching of the cnidarian-dinoflagellate symbiosis: Aspects of innate immunity and the role of nitric oxide. PhD thesis (Victoria University of Wellington, Wellington, New Zealand)
- 36.Tuteja N. Signaling through G protein coupled receptors. Plant Signal Behav. 2009;4:942–947. doi: 10.4161/psb.4.10.9530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peng SE, et al. Proteomic analysis of symbiosome membranes in cnidaria-dinoflagellate endosymbiosis. Proteomics. 2010;10:1002–1016. doi: 10.1002/pmic.200900595. [DOI] [PubMed] [Google Scholar]
- 38.Dunn SR, Weis VM. Apoptosis as a post-phagocytic winnowing mechanism in a coral-dinoflagellate mutualism. Environ Microbiol. 2009;11:268–276. doi: 10.1111/j.1462-2920.2008.01774.x. [DOI] [PubMed] [Google Scholar]
- 39.Capdevila JH, Falck JR, Harris RC. Cytochrome P450 and arachidonic acid bioactivation: Molecular and functional properties of the arachidonate monooxygenase. J Lipid Res. 2000;41:163–181. [PubMed] [Google Scholar]
- 40.Lõhelaid H, Teder T, Samel N. Lipoxygenase-allene oxide synthase pathway in octocoral thermal stress response. Coral Reefs. 2015;34:143–154. [Google Scholar]
- 41.Magtanong L, Ko PJ, Dixon SJ. Emerging roles for lipids in non-apoptotic cell death. Cell Death Differ. 2016;23:1099–1109. doi: 10.1038/cdd.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Levin G, et al. Differential metabolism of dihomo-γ-linolenic acid and arachidonic acid by cyclo-oxygenase-1 and cyclo-oxygenase-2: Implications for cellular synthesis of prostaglandin E1 and prostaglandin E2. Biochem J. 2002;365:489–496. doi: 10.1042/BJ20011798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.DeSalvo MK, Sunagawa S, Voolstra CR, Medina M. Transcriptomic responses to heat stress and bleaching in the elkhorn coral Acropora palmata. Mar Ecol Prog Ser. 2010;402:97–113. [Google Scholar]
- 44.Wright EM, Turk E. The sodium/glucose cotransport family SLC5. Pflugers Arch. 2004;447:510–518. doi: 10.1007/s00424-003-1063-6. [DOI] [PubMed] [Google Scholar]
- 45.Pernice M, et al. A nanoscale secondary ion mass spectrometry study of dinoflagellate functional diversity in reef-building corals. Environ Microbiol. 2015;17:3570–3580. doi: 10.1111/1462-2920.12518. [DOI] [PubMed] [Google Scholar]
- 46.Cantin NE, van Oppen MJ, Willis BL, Mieog JC, Negri AP. Juvenile corals can acquire more carbon from high-performance algal symbionts. Coral Reefs. 2009;28:405–414. [Google Scholar]
- 47.Jones AM, Berkelmans R. Tradeoffs to thermal acclimation: Energetics and reproduction of a reef coral with heat tolerant Symbiodinium type-D. J Mar Biol. 2011;2011:1–12. [Google Scholar]
- 48.Shigesada K, Tatibana M. Role of acetylglutamate in ureotelism, I: Occurrence and biosynthesis of acetylglutamate in mouse and rat tissues. J Biol Chem. 1971;246:5588–5595. [PubMed] [Google Scholar]
- 49.Cooper TF, et al. Symbiodinium genotypic and environmental controls on lipids in reef-building corals. PLoS One. 2011;6:e20434. doi: 10.1371/journal.pone.0020434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oku H, Yamashiro H, Onaga K, Sakai K, Iwasaki H. Seasonal changes in the content and composition of lipids in the coral Goniastrea aspera. Coral Reefs. 2003;22:83–85. [Google Scholar]
- 51.Coffroth MA, Poland DM, Petrou EL, Brazeau DA, Holmberg JC. Environmental symbiont acquisition may not be the solution to warming seas for reef-building corals. PLoS One. 2010;5:e13258. doi: 10.1371/journal.pone.0013258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pettay DT, Wham DC, Smith RT, Iglesias-Prieto R, LaJeunesse TC. Microbial invasion of the Caribbean by an Indo-Pacific coral zooxanthella. Proc Natl Acad Sci USA. 2015;112:7513–7518. doi: 10.1073/pnas.1502283112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee MJ, et al. Most low-abundance “background” Symbiodinium spp. are transitory and have minimal functional significance for symbiotic corals. Microb Ecol. 2016;71:771–783. doi: 10.1007/s00248-015-0724-2. [DOI] [PubMed] [Google Scholar]
- 54.Meyer E, Aglyamova GV, Matz MV. Profiling gene expression responses of coral larvae (Acropora millepora) to elevated temperature and settlement inducers using a novel RNA-seq procedure. Mol Ecol. 2011;20:3599–3616. doi: 10.1111/j.1365-294X.2011.05205.x. [DOI] [PubMed] [Google Scholar]
- 55.Roessner U, Wagner C, Kopka J, Trethewey RN, Willmitzer L. Technical advance: Simultaneous analysis of metabolites in potato tuber by gas chromatography-mass spectrometry. Plant J. 2000;23:131–142. doi: 10.1046/j.1365-313x.2000.00774.x. [DOI] [PubMed] [Google Scholar]
- 56.Kamburov A, Cavill R, Ebbels TM, Herwig R, Keun HC. Integrated pathway-level analysis of transcriptomics and metabolomics data with IMPaLA. Bioinformatics. 2011;27:2917–2918. doi: 10.1093/bioinformatics/btr499. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.