Significance
Our work uncovers SUMOylation as a major regulator of α-synuclein fate and pathological aggregation. SUMOylation promotes α-synuclein accumulation by counteracting α-synuclein ubiquitination and inhibiting its degradation. SUMOylation also directly causes aggregation of α-synuclein in vitro and in cells, and the effects are much more prominent with the α-synuclein disease mutants. Most importantly, levels of SUMOylated α-synuclein and components of the SUMOylation machinery increase severalfold in Parkinson’s disease (PD) brains and are present in Lewy bodies. Our data indicate a major role of SUMOylation in the pathological aggregation of α-synuclein and Lewy body formation. Therefore, SUMOylation blockers provide a strategy to prevent intracellular α-synuclein accumulation, aggregation, and spreading in PD.
Keywords: Parkinson’s disease, α-synuclein, SUMOylation, ubiquitination, aggregation
Abstract
α-Synuclein accumulation is a pathological hallmark of Parkinson’s disease (PD). Ubiquitinated α-synuclein is targeted to proteasomal or lysosomal degradation. Here, we identify SUMOylation as a major mechanism that counteracts ubiquitination by different E3 ubiquitin ligases and regulates α-synuclein degradation. We report that PIAS2 promotes SUMOylation of α-synuclein, leading to a decrease in α-synuclein ubiquitination by SIAH and Nedd4 ubiquitin ligases, and causing its accumulation and aggregation into inclusions. This was associated with an increase in α-synuclein release from the cells. A SUMO E1 inhibitor, ginkgolic acid, decreases α-synuclein levels by relieving the inhibition exerted on α-synuclein proteasomal degradation. α-Synuclein disease mutants are more SUMOylated compared with the wild-type protein, and this is associated with increased aggregation and inclusion formation. We detected a marked increase in PIAS2 expression along with SUMOylated α-synuclein in PD brains, providing a causal mechanism underlying the up-regulation of α-synuclein SUMOylation in the disease. We also found a significant proportion of Lewy bodies in nigral neurons containing SUMO1 and PIAS2. Our observations suggest that SUMOylation of α-synuclein by PIAS2 promotes α-synuclein aggregation by two mutually reinforcing mechanisms. First, it has a direct proaggregatory effect on α-synuclein. Second, SUMOylation facilitates α-synuclein aggregation by blocking its ubiquitin-dependent degradation pathways and promoting its accumulation. Therefore, inhibitors of α-synuclein SUMOylation provide a strategy to reduce α-synuclein levels and possibly aggregation in PD.
Parkinson’s disease (PD) is caused by progressive degeneration of dopaminergic neurons in the substantia nigra (1). Mutations and multiplications of the α-synuclein gene cause autosomal-dominant PD (2). α-Synuclein is a major component of Lewy bodies in sporadic PD and its accumulation is also a feature of dementia with Lewy bodies (DLB) and multiple system atrophy (3, 4). It has been suggested that aggregated α-synuclein can spread to different brain regions (5). On the other hand, primary accumulation and aggregation of intracellular α-synuclein is widely regarded as a failure of degradation mechanisms (6, 7). Therefore, preventing α-synuclein accumulation is a major goal for novel therapies. Although α-synuclein is degraded by proteasomal, lysosomal, and autophagic pathways (8–10), the mechanisms that modulate its degradation remain obscure.
Lewy bodies are heavily ubiquitinated and α-synuclein purified from Lewy bodies is monoubiquitinated (11–13). Several ubiquitin ligases are capable of ubiquitinating α-synuclein (8, 14–17). The E3 ubiquitin-ligase seven in absentia homolog (SIAH) interacts with and monoubiquitinates α-synuclein (8, 17). SIAH is present in Lewy bodies (8) and monoubiquitinates α-synuclein at the same lysine residues that are monoubiquitinated in Lewy bodies (11, 17). Monoubiquitinated α-synuclein is degraded by the proteasome (18–20), while polyubiquitination by Nedd4 leads to its lysosomal degradation (15).
We previously found that the deubiquitinase USP9X dynamically deubiquitinates α-synuclein (18). Deubiquitination of α-synuclein by USP9X impairs SIAH-dependent α-synuclein proteasomal degradation and promotes degradation by autophagy (18). Another deubiquitinase, USP8, removes K63-linked ubiquitin chains of α-synuclein and prevents its lysosomal degradation (21). The regulation of α-synuclein ubiquitination processes and their role in α-synuclein homeostasis are still unclear.
α-Synuclein is also conjugated to small ubiquitin-like modifier (SUMO) at lysines, but its role in aggregation remains controversial (22–25). Some studies demonstrated increased α-synuclein aggregation by SUMOylation upon proteasomal inhibition (23, 24), but SUMOylation itself had limited or no effect on aggregation (22, 25). A lack of knowledge about the endogenous α-synuclein SUMO ligase hampered further progress in determining the physiological and pathological functions of α-synuclein SUMOylation. Therefore, we sought to identify the endogenous SUMO ligase for α-synuclein and to determine how SUMOylation affects α-synuclein ubiquitination, degradation, and aggregation. Our data highlight a role of SUMOylation in dynamically regulating α-synuclein levels and pathology.
Results
Interaction and SUMOylation of α-Synuclein by PIAS2.
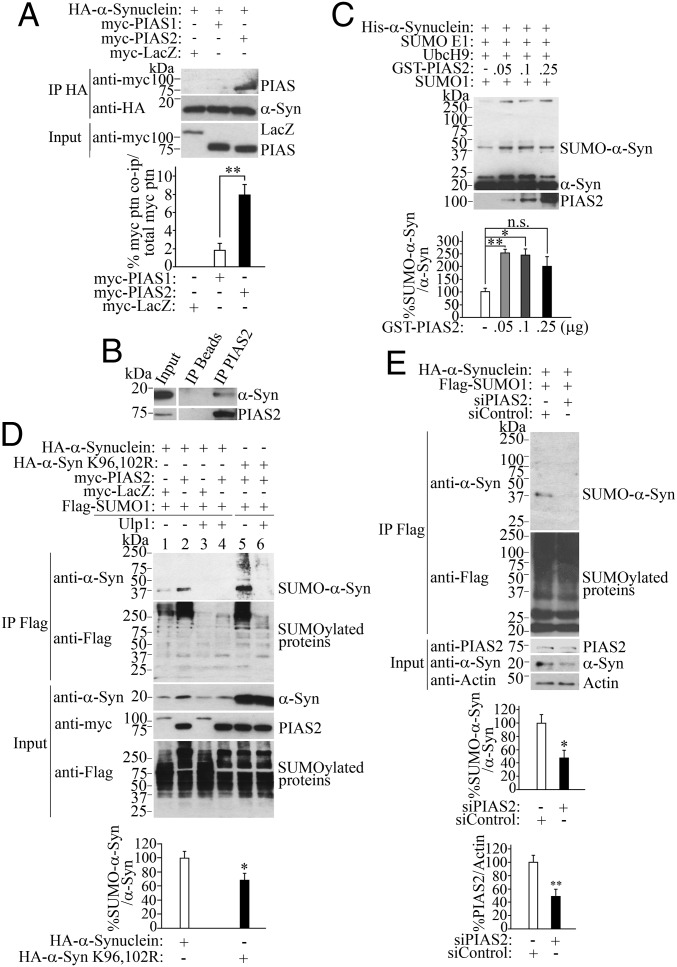
We found that PIAS2 coimmunoprecipitated with α-synuclein from transfected cells (Fig. 1A), rat brain tissues (Fig. 1B), and in in vitro pull-down assays (Fig. S1A). The close homolog PIAS1 also interacted with α-synuclein in transfected cells (Fig. 1A), in vitro pull-down assays (Fig. S1A), and rat brain tissues (Fig. S1B). However, a direct comparison with PIAS2 in cells and in in vitro binding assays indicates that the interaction of α-synuclein and PIAS1 is much weaker than PIAS2 (Fig. 1A and Fig. S1A). Thus, in subsequent experiments, we studied in more detail the interaction of PIAS2 and α-synuclein.
Fig. 1.
PIAS2 interacts with and SUMOylates α-synuclein. (A) Hemagglutinin (HA)–α-synuclein from transfected HEK293 cells was immunoprecipitated with anti-HA antibody (Middle), and coimmunoprecipitation with PIAS1 or PIAS2 was detected with anti-myc (Upper). Input levels were determined using anti-myc (Lower). Graph depicts the percentage of myc-PIAS1 and myc-PIAS2 coimmunoprecipitated with HA–α-synuclein relative to the total amount of each myc protein added per immunoprecipitation. (B) PIAS2 was immunoprecipitated from rat brain using anti-PIAS2 antibody (Lower), and coimmunoprecipitation was determined with anti–α-synuclein antibody (Upper). (C) In vitro SUMOylation of α-synuclein by PIAS2. His–α-synuclein was incubated with SUMOylation components and PIAS2. Levels of SUMOylated α-synuclein in relation to non-SUMOylated α-synuclein were quantified with anti–α-synuclein (graph). (D) SUMOylated proteins from transfected HEK293 cells were immunoprecipitated with anti-Flag antibody, and immunoprecipitates were further incubated with the purified deSUMOylase Ulp1 (lanes 3, 4, and 6). SUMOylated α-synuclein was detected with anti–α-synuclein (first panel). Immunoprecipitated SUMOylated proteins were detected with anti-Flag antibody (second panel). Graph represents the percentage of SUMO–α-synuclein in cells (lanes 2 and 5) relative to non-SUMOylated α-synuclein. (E) PIAS2 knockdown decreases α-synuclein SUMOylation. HEK293 cells were transfected in the presence of siRNA control or a PIAS2 siRNA. Graphs represent the percentage of SUMO–α-synuclein in cells relative to non-SUMOylated α-synuclein and the percentage of PIAS2 relative to β-actin. Values represent the average ± SEM of three experiments. Different from control at *P < 0.05 and **P < 0.01, respectively (Student’s t test). n.s., not significant.
PIAS2 increased α-synuclein SUMOylation in vitro (Fig. 1C). Basal levels of SUMOylated α-synuclein were observed in the absence of PIAS2, which is consistent with the ability of the E2-conjugating enzyme UbcH9 to promote limited SUMO transfer to substrates in the absence of E3 SUMO ligases (26). Nevertheless, very small amounts of PIAS2, from 50 ng per reaction, significantly increased SUMOylated α-synuclein levels by almost threefold (Fig. 1C, graph). The size of the SUMOylated α-synuclein product by PIAS2 was consistent with modifications by two individual SUMO1 molecules at two different sites, since SUMO1 alone cannot form polySUMO chains (27, 28). In agreement, methylated SUMO, which does not allow the formation of chains (29), did not affect the SUMOylation profile of α-synuclein (Fig. S2A). In addition, we observed the accumulation of high–molecular-weight α-synuclein species (above 250 kDa) in the presence of PIAS2 with either SUMO1 or methylated SUMO1 (Fig. 1C and Fig. S2A), suggesting the formation of α-synuclein aggregates, a phenomenon characterized in detail in Fig. 5.
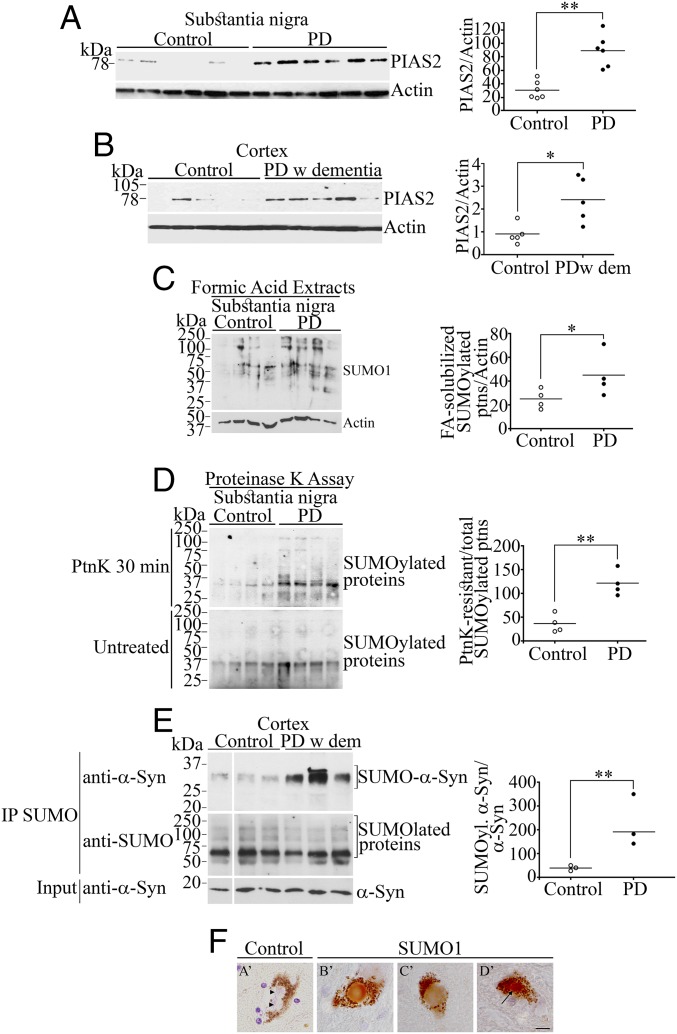
Fig. 5.
Elevated SUMOylated α-synuclein and SUMOylation components in PD brains. (A) Homogenates prepared from substantia nigra of PD and control patients were probed with anti-PIAS2 antibody (Upper). Graph depicts PIAS2 levels normalized to β-actin. (B) Homogenates from frontal cortex tissue of PD with dementia and matched controls were analyzed and quantified as in A. (C) Substantia nigra tissues of PD and control patients were homogenized in formic acid and probed with anti-SUMO1 antibody (Upper). Graph depicts SUMO1 levels normalized to β-actin. (D) Homogenates from substantia nigra tissue of PD and controls were incubated with proteinase K for 30 min and probed with anti-SUMO1 antibody (Upper). A loading control was included by immediately adding 2% SDS to the samples (Lower). Graph depicts SUMOylated protein levels of PD after 30-min proteinase K normalized to SUMOylated protein levels in untreated substantia nigra samples. (E) Homogenates from frontal cortex tissues of PD with dementia and matched controls were immunoprecipitated with anti-SUMO1 antibody (Middle). Levels of SUMOylated α-synuclein were determined using anti–α-synuclein antibody (Upper). Lower shows the input levels of α-synuclein. Graph depicts SUMOylated α-synuclein levels normalized to non-SUMOylated α-synuclein. (F) Immunohistochemistry of substantia nigra from PD patients using anti-SUMO1 antibody (B′–D′). Control lacks the primary antibody (A′). Arrow in D′ points to a SUMO1-positive Lewy bodies. Arrowheads in A′ point to control unstained Lewy bodies. Sections were counterstained with hematoxylin. (Scale bar, 10 μm.) Significantly different from control at *P < 0.05 and **P < 0.01 (Student’s t test).
PIAS2 significantly increased α-synuclein SUMOylation in cultured cells (Fig. 1D). Treatment of SUMO1 immunoprecipitates with the purified deSUMOylase Ulp1 (30) removed all SUMO conjugates and abolished the SUMOylated α-synuclein signal (Fig. 1D, lanes 3, 4, and 6), confirming the specificity of SUMO1–α-synuclein conjugates. Longer exposure of the blots also demonstrates low levels of α-synuclein SUMOylation in the absence of PIAS2 (Fig. S2B).
In agreement with its weaker binding to α-synuclein (Fig. 1A), PIAS1 promoted much less α-synuclein SUMOylation in cells compared with PIAS2 (Fig. S2C). On the other hand, addition of the catalytic region of the E3 SUMO-ligase RanBP2 significantly increased the in vitro SUMOylation of α-synuclein, but without the formation of high–molecular-weight α-synuclein species (Fig. S2D).
α-Synuclein has two lysine consensus sites for SUMOylation, K96 and K102 (22). Nevertheless, mass spectrometry analysis revealed that 11 of 15 α-synuclein lysines are SUMOylated (25). In support of widespread SUMOylation of α-synuclein lysines by PIAS2, we found that mutation of the α-synuclein lysines K96 and 102R decreased SUMOylation in cells by only 30% (Fig. 1D, graph). Most importantly, partial knockdown of PIAS2 by short interfering RNA (siRNA) significantly decreased the levels of SUMOylated α-synuclein (Fig. 1E). siPIAS2 also decreased the steady-state levels of α-synuclein (Fig. 1E, fourth panel; characterized in detail in Fig. 3). However, siPIAS2 still decreased the levels of SUMO–α-synuclein by 50% after correcting for the lower steady-state levels of α-synuclein (Fig. 1E, graph).
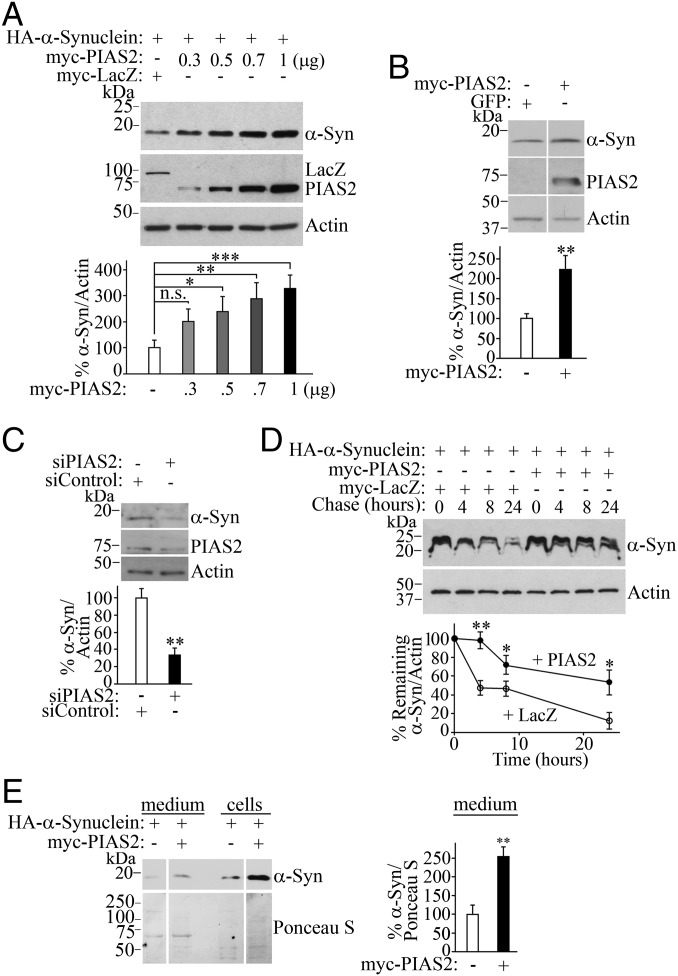
Fig. 3.
SUMOylation by PIAS2 regulates α-synuclein levels and degradation. (A) PIAS2 increases the steady-state levels of α-synuclein in transfected HEK293 cells. Graph depicts the percentage of α-synuclein steady-state levels relative to β-actin at increasing amounts of myc-PIAS2. (B) Endogenous levels of α-synuclein increase upon myc-PIAS2 transfection. (C) PIAS2 knockdown decreases endogenous α-synuclein levels. HEK293 cells were transfected with siRNA control or siRNA to PIAS2. Endogenous levels of α-synuclein were determined using anti–α-synuclein antibody. (D) PIAS2 prolongs α-synuclein half-life in transfected HEK293 cells treated with 50 μM cycloheximide. Graph depicts α-synuclein levels relative to β-actin in the presence of PIAS2 (●) or control LacZ (○). (E) PIAS2 elevates α-synuclein in extracellular medium. Levels of α-synuclein in intracellular and extracellular HEK293 cell compartments were monitored with anti-HA (Upper). Total protein loading was determined by Ponceau S staining (Lower). Graph depicts the percentage of α-synuclein in the medium relative to total protein loading monitored by Ponceau S staining. Values represent the average ± SEM of three to four experiments. Different from control at *P < 0.05, **P < 0.01, and ***P < 0.001. Repeated-measures one-way ANOVA with Bonferroni post hoc test (A) or Student’s t test (B–E). n.s., not significant.
Inhibitory Role of SUMOylation on α-Synuclein Ubiquitination.
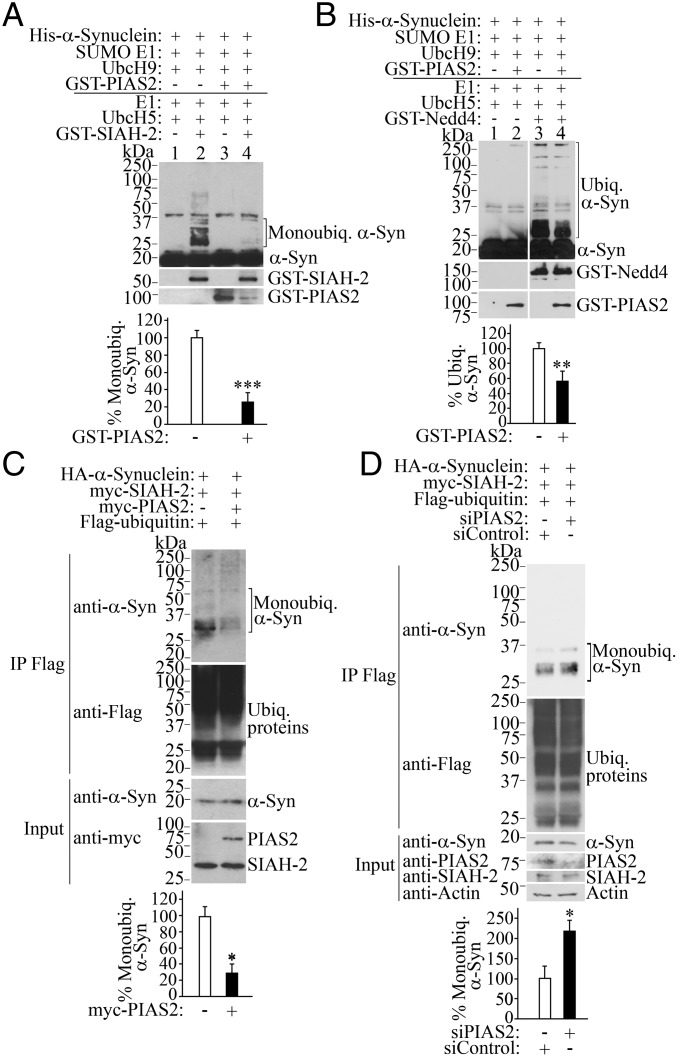
SUMOylation and ubiquitination target lysine residues, but the relationship between these modifications has not been investigated for α-synuclein. We observed that in vitro SUMOylation by PIAS2 decreased by 80% the monoubiquitination of α-synuclein promoted by SIAH-2 (Fig. 2A, graph). In addition, we noticed a decrease in PIAS2 levels in the presence of SIAH-2 (Fig. 2A, compare lanes 3 and 4). This was due to the formation of high–molecular-weight glutathione S-transferase (GST)–PIAS2 species that were retained in the stacking gel following the prolonged SUMOylation/monoubiquitination assays, so that less monomeric GST-PIAS2 remained in the presence of SIAH-2 (Fig. S3). As control, the addition of PIAS2 without the SUMOylation components (SUMO E1 and UbcH9) had no effect on in vitro α-synuclein monoubiquitination (Fig. S4), indicating that SUMOylation is responsible for the decrease in α-synuclein monoubiquitination.
Fig. 2.
SUMOylation by PIAS2 prevents α-synuclein monoubiquitination. (A) Inhibition of SIAH-2–mediated monoubiquitination by PIAS2. In vitro SUMOylation/ubiquitination reactions were carried out by incubating His–α-synuclein with purified SUMOylation components for 60 min, followed by the addition of ubiquitination components for another 60 min. Levels of His–α-synuclein monoubiquitination were determined with anti–α-synuclein. Graph shows the percentage of SIAH-2–dependent α-synuclein monoubiquitination, with and without GST-PIAS2 by comparing lanes 2 and 4. (B) PIAS2 inhibits Nedd4-mediated α-synuclein ubiquitination. Conditions were the same as in A. Graph depicts the percentage of Nedd4-dependent α-synuclein ubiquitination in the absence and in the presence of GST-PIAS2. (C) PIAS2 decreases α-synuclein monoubiquitination in cells. Ubiquitinated proteins from transfected HEK293 cells were immunoprecipitated with anti-Flag antibody, and monoubiquitinated α-synuclein was detected with anti–α-synuclein (first panel). Graph depicts the percentage of monoubiquitinated α-synuclein in cells relative to unmodified α-synuclein. (D) HEK293 cells were transfected as indicated, and in the presence of siRNA control or PIAS2 siRNA. Immunoprecipitation was carried out and analyzed as in C. Graph represents the percentage of monoubiquitinated α-synuclein in cells relative to unmodified α-synuclein. Values represent the average ± SEM of three experiments. Different from control at *P < 0.05, **P < 0.01, and ***P < 0.001 (Student’s t test).
SUMOylation by PIAS2 significantly decreased the in vitro ubiquitination of α-synuclein promoted by Nedd4 (Fig. 2B, compare lanes 3 and 4, graph). Therefore, PIAS2-mediated SUMOylation negatively modulates different forms of α-synuclein ubiquitination.
Ectopic expression of PIAS2 led to a 70% decrease in α-synuclein monoubiquitination levels in cells (Fig. 2C, graph). Conversely, PIAS2 knockdown by siRNA increased the levels of monoubiquitinated α-synuclein by SIAH-2 by more than twofold (Fig. 2D), supporting the role of PIAS2-mediated SUMOylation in counteracting α-synuclein monoubiquitination in cells.
Role of SUMOylation in α-Synuclein Degradation.
We found that PIAS2 increased the steady-state levels of α-synuclein by up to threefold (Fig. 3A), but it did not modify the steady-state levels of the control protein LacZ (Fig. S5). Endogenous α-synuclein levels were increased twofold by PIAS2 overexpression (Fig. 3B), whereas PIAS2 knockdown down-regulated endogenous α-synuclein (Fig. 3C). Moreover, PIAS2 prolonged the half-life of α-synuclein in the presence of the protein synthesis inhibitor cycloheximide (Fig. 3D), suggesting that PIAS2-dependent SUMOylation leads to α-synuclein accumulation by reducing its degradation.
Since α-synuclein export has been proposed to spread PD, we monitored extracellular α-synuclein levels. We found that overexpression of PIAS2 increased the levels of extracellular α-synuclein as well (Fig. 3E).
To explore the physiologic relevance of α-synuclein SUMOylation, we used ginkgolic acid, an E1 SUMO inhibitor (31). Ginkgolic acid decreased the steady-state levels of α-synuclein in cells by up to 90% (Fig. S6A), and this was preventable by the proteasome inhibitor lactacystin, suggesting that SUMOylation inhibition accelerates α-synuclein degradation by the proteasome. Ginkgolic acid also down-regulated the endogenous levels of α-synuclein in primary neuronal cultures (Fig. S6B), confirming the role of SUMOylation in regulating endogenous α-synuclein levels.
We next investigated the presence of α-synuclein SUMOylation in normal human brain tissue. We immunoprecipitated SUMOylated proteins from human frontal cortex and identified a SUMOylated α-synuclein signal in the human brain (Fig. S6C). Treatment of the immunoprecipitate with the de-SUMOylase Ulp1 abolished the SUMOylated–α-synuclein signal (Fig. S6C, lane 2), indicating that SUMOylation of α-synuclein occurs in vivo.
SUMOylation of Mutant α-Synuclein Species and Aggregation.
SUMOylation of the α-synuclein disease mutants A30P, A53T, and E46K was severalfold higher than the wild-type α-synuclein in vitro (Fig. 4A, graph). Addition of PIAS2 further increased SUMOylation of all α-synuclein types, although the percentage of SUMOylation increase promoted by PIAS2 was similar among wild type and disease mutants (Fig. 4A). Moreover, SUMOylation of the disease mutants by PIAS2 led to the formation of high–molecular-weight α-synuclein species suggestive of aggregates, which were much more prominent in all disease mutants than wild-type α-synuclein (Fig. 4A, Middle).
Fig. 4.
α-Synuclein disease mutants are more susceptible to SUMOylation and aggregation. (A) In vitro SUMOylation was carried out by incubating His–α-synuclein WT and disease variants with SUMOylation components, with and without GST-PIAS2. Levels of His–α-synuclein SUMOylation were determined with anti–α-synuclein antibody (first and second panels). Third panel shows a short exposure to reveal α-synuclein levels. Graph depicts the percent of in vitro SUMOylated α-synuclein normalized to non-SUMOylated α-synuclein. (B) SUMOylated proteins from transfected HEK293 cells were immunoprecipitated with anti-Flag antibody (second panel), and SUMOylated α-synuclein was detected with anti–α-synuclein (first panel). IgG heavy chain is indicated. Graph depicts the percentage of SUMOylated α-synuclein in cells normalized to non-SUMOylated α-synuclein. (C) SH-SY5Y cells were transfected with HA–α-synuclein (wild type, A30P, A53T, and E46K disease mutations) and Flag-SUMO1, in the presence of myc-FKBP12 or myc-PIAS2. Cells were treated with proteinase K before fixation and processed for immunocytochemistry with anti-HA antibody. (Scale bar, 25 μm.) Arrows in B′ indicate cells with aggregates. (D) SH-SY5Y cells were transfected, processed, and analyzed as in C. Graph represents the percentage of cells with HA–α-synuclein aggregates. Values represent the average ± SEM of three independent experiments. Different from WT at *P < 0.05, **P < 0.01, and ***P < 0.001 (repeated-measures one-way ANOVA with Bonferroni post hoc test).
α-Synuclein disease mutants were two to three times more SUMOylated in cells in the presence of PIAS2 compared with the wild-type protein (Fig. 4B). In agreement, the steady-state levels of α-synuclein disease mutants were more pronouncedly increased by PIAS2 compared with the wild-type protein (Fig. S7A). Furthermore, α-synuclein becomes partially resistant to proteinase K treatment in the presence of PIAS2 (Fig. S7B), suggesting that SUMOylation increases the aggregation state of α-synuclein in cells. Resistance to proteinase K treatment was more pronounced with α-synuclein disease mutants (Fig. S7B), which is compatible with their higher aggregation state following PIAS2-mediated SUMOylation.
We then investigated the aggregation and inclusion formation of α-synuclein by immunocytochemistry. To improve the sensitivity of our assay, we treated cells with proteinase K before fixation for immunocytochemistry (32). Levels of proteinase K-resistant α-synuclein aggregates were higher in disease mutants than wild type in the presence of PIAS2 (Fig. 4 C and D). No significant inclusion formation was observed when a control protein (myc-FKBP12) was transfected instead of myc-PIAS2 (Fig. 4 C and D). Moreover, SUMO1 was present in α-synuclein inclusions (Fig. S8).
Levels of SUMO–α-Synuclein and SUMOylation Machinery in PD Patients.
Since α-synuclein SUMOylation takes place in normal human brain tissues (Fig. S6C), we investigated the levels of PIAS2 in PD brains. We observed that the levels of PIAS2 are about threefold higher in the substantia nigra of PD patients (Fig. 5A) and in the cerebral cortex of PD subjects that progressed to dementia (Fig. 5B). Upon solubilization of brain tissues with formic acid (33), the total levels of SUMOylated proteins are twofold higher in the substantia nigra of PD brains compared with that of controls (Fig. 5C). In addition, we treated with proteinase K to determine the levels of aggregated proteins (32, 34). The substantia nigra of PD subjects contained significantly more proteinase K-resistant SUMOylated proteins than controls (Fig. 5D). Most importantly, the levels of SUMOylated α-synuclein isolated by immunoprecipitation are strikingly increased in the cerebral cortex of PD patients with dementia (Fig. 5E). In addition, we found strong immunoreactivity of SUMO1 in ∼25% (10 out of 41 α-synuclein–positive) Lewy bodies of three different PD cases (Fig. 5F), while PIAS2 was detected in ∼50% (18 out of 37 α-synuclein–positive) Lewy bodies in three different PD cases (Fig. S9).
Discussion
Our observations indicate that increased SUMOylation may play a role in promoting α-synuclein accumulation and aggregation in PD. We demonstrated that α-synuclein is SUMOylated by PIAS2, and this is associated with α-synuclein accumulation, aggregation, and increased α-synuclein levels in the extracellular medium. SUMOylation by PIAS2 promotes the formation of α-synuclein inclusions in cells even in the absence of proteasome inhibitors. Furthermore, SUMOylation decreases the ubiquitination of α-synuclein by both SIAH and Nedd4, indicating a general inhibitory role of SUMOylation on α-synuclein ubiquitination. Most importantly, SUMOylated α-synuclein and PIAS2 are markedly elevated in the substantia nigra of PD brains, and Lewy bodies are positive for both SUMO1 and PIAS2.
We found that SUMOylation increases α-synuclein aggregation by two self-reinforcing mechanisms. First, SUMOylation by PIAS2 directly promotes the aggregation of α-synuclein. Second, SUMOylation impairs α-synuclein ubiquitination, which prevents α-synuclein degradation. This causes the accumulation of α-synuclein and facilitates its aggregation. In addition to blocking α-synuclein degradation, SUMOylation by PIAS2 increased the release of α-synuclein to the extracellular medium. The increase in extracellular α-synuclein may be due to the α-synuclein accumulation secondary to SUMOylation or indirectly by SUMOylation of the endosomal sorting complex, which is known to increase α-synuclein extracellular export (35).
α-Synuclein contains two lysine residues, K96 and K102, which follow the consensus for SUMOylation (22). A study using α-synuclein mutated at K96 and K102 suggested that SUMOylation of these residues decreases the aggregation of α-synuclein (25). A caveat of using K96/102 mutants is that at least nine additional α-synuclein lysines are SUMOylated in cells (25). In agreement, we found that mutations on K96 and K102 caused only a 30% decrease in the SUMOylation of α-synuclein by PIAS2. In support of our findings, SUMOylation of numerous proteins by SUMO ligases occurs in lysines that lack consensus sequence for SUMO conjugation (36). Thus, the effects of K96/102 mutants on aggregation observed in a previous study (25) may be related to a change in the specific amino acids rather than SUMOylation changes.
SUMOylation may have opposite effects on ubiquitination of target proteins. It can either prime SUMO-targeted ubiquitin ligases and increase ubiquitination or compete for the target lysines preventing the ubiquitin linkage (37). The latter appears to be the case for α-synuclein and PIAS2. In agreement, the E1 SUMO inhibitor ginkgolic acid accelerated the proteasomal degradation of α-synuclein by relieving the inhibition exerted on its ubiquitination. This effect was more pronounced in transfected cells than in neurons, suggesting that endogenous neuronal α-synuclein may have slower half-life or additional regulatory mechanisms that affect its proteasomal degradation, like glycation (38).
Previous studies reported that proteasomal inhibition promotes α-synuclein inclusions that are SUMOylated but did not ascribe a direct effect of SUMOylation on aggregation (23, 24). Proteasomal inhibition also favors the formation of monoubiquitinated α-synuclein aggregates (17, 18). We now show that PIAS2-mediated SUMOylation per se increases the aggregation of α-synuclein under intact proteasomal activity. It is possible that, akin to monoubiquitination (17), SUMOylation increases the hydrophobicity of α-synuclein and directly facilitates its aggregation.
α-Synuclein may be SUMOylated by additional SUMO ligases, albeit with different efficiencies. We found that PIAS1 also interacts with α-synuclein in the brain, but direct comparison by employing the same antibody for coimmunoprecipitations or recombinant α-synuclein in pull-down assays indicates weaker interaction of human PIAS1 with α-synuclein compared with human PIAS2. In agreement with a previous study (25), we found that the catalytic portion of RanBP2 SUMOylates α-synuclein in vitro. However, we did not detect formation of high–molecular-weight or aggregated α-synuclein species following RanBP2 SUMOylation. In this context, it is possible that the strong binding of PIAS2 to α-synuclein also contributes to the efficient aggregation of SUMOylated α-synuclein, an effect not shared by the catalytic part of RanBP2.
We found that the α-synuclein disease mutants are more prone to SUMOylation and SUMOylation-mediated aggregation than the wild-type protein. Further studies are necessary to better understand the mechanisms underlying the increased SUMOylation among α-synuclein disease mutants.
Abnormal SUMOylation has been implicated in the pathology of neurodegenerative diseases, including Huntington’s, Alzheimer’s, and amyotrophic lateral sclerosis (39–43). SUMO1 was also suggested to be present in some Lewy body preparations of PD and DLB brains (24). Unilateral rotenone-lesioned mouse models of PD have increased levels of SUMO1 (44). Our observation of a striking increase in SUMOylated proteins and SUMOylated α-synuclein in PD tissues provides compelling evidence supporting a general up-regulation of SUMOylation in PD and of the SUMOylated α-synuclein in particular. Our results demonstrating an increase in PIAS2 expression in PD tissues and the presence of SUMO1 and PIAS2 in Lewy bodies provides a causal mechanism underlying the accumulation of SUMOylated α-synuclein. Therefore, SUMOylation blockers may provide a strategy to prevent intracellular α-synuclein aggregation and the spread of α-synuclein.
Materials and Methods
In Vitro SUMOylation Assays.
Recombinant His–α-synuclein was incubated in medium containing 40 mM Tris⋅HCl (pH 7.6), 5 mM MgCl2, 2 mM DTT, 1 mM ATP-γ–S, 5 μg of SUMO1, 1 μM SUMO1 aldehyde, 100 ng of SUMO E1 (SAE1/UBA2; activating enzyme), and 200 ng of UbcH9 (SUMO E2; conjugating enzyme), in the presence or absence of 200 ng of PIAS2 or RanBP2ΔFG (E3 SUMO ligases), unless specified otherwise. Reactions were incubated at 37 °C for 1 h and resolved on PAGE/SDS gels. SUMOylated α-synuclein was determined by Western blot using anti–α-synuclein antibody.
Brain Tissues.
Postmortem human brains were processed and analyzed as described in SI Materials and Methods. Informed written consent was obtained for all cases and the study was approved by the local research ethics committee (University College London Institute of Neurology, United Kingdom). For rat primary cortical cultures, animals were killed by decapitation after isoflurane anesthesia with the approval of the committee for the supervision of animal experiments (Technion–Israel Institute of Technology). A detailed description of the procedure was described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We are grateful to Prof. Daniela Rotin for Nedd4 cDNA. R.B. is funded by The Reta Lila Weston Foundation. This work was supported in part by Wellcome Trust/Medical Research Council (MRC) Joint Call in Neurodegeneration Award WT089698 to the UK Parkinson’s Disease Consortium whose members are from the University College London Institute of Neurology, University of Sheffield, and the MRC Protein Phosphorylation Unit of Dundee (R.B.). S.E. received funds from the Israel Academy of Sciences, Rappaport Family Institute for Research in the Medical Sciences, The Allen and Jewel Prince Center for Neurodegenerative Disorders of the Brain, and the Technion research funds.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1704351114/-/DCSupplemental.
References
- 1.Moore DJ, West AB, Dawson VL, Dawson TM. Molecular pathophysiology of Parkinson’s disease. Annu Rev Neurosci. 2005;28:57–87. doi: 10.1146/annurev.neuro.28.061604.135718. [DOI] [PubMed] [Google Scholar]
- 2.Trinh J, Farrer M. Advances in the genetics of Parkinson disease. Nat Rev Neurol. 2013;9:445–454. doi: 10.1038/nrneurol.2013.132. [DOI] [PubMed] [Google Scholar]
- 3.Takeda A, et al. Abnormal accumulation of NACP/alpha-synuclein in neurodegenerative disorders. Am J Pathol. 1998;152:367–372. [PMC free article] [PubMed] [Google Scholar]
- 4.Tu PH, et al. Glial cytoplasmic inclusions in white matter oligodendrocytes of multiple system atrophy brains contain insoluble alpha-synuclein. Ann Neurol. 1998;44:415–422. doi: 10.1002/ana.410440324. [DOI] [PubMed] [Google Scholar]
- 5.Luk KC, et al. Intracerebral inoculation of pathological α-synuclein initiates a rapidly progressive neurodegenerative α-synucleinopathy in mice. J Exp Med. 2012;209:975–986. doi: 10.1084/jem.20112457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Engelender S, Isacson O. The threshold theory for Parkinson’s disease. Trends Neurosci. 2017;40:4–14. doi: 10.1016/j.tins.2016.10.008. [DOI] [PubMed] [Google Scholar]
- 7.Walsh DM, Selkoe DJ. A critical appraisal of the pathogenic protein spread hypothesis of neurodegeneration. Nat Rev Neurosci. 2016;17:251–260. doi: 10.1038/nrn.2016.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liani E, et al. Ubiquitylation of synphilin-1 and alpha-synuclein by SIAH and its presence in cellular inclusions and Lewy bodies imply a role in Parkinson’s disease. Proc Natl Acad Sci USA. 2004;101:5500–5505. doi: 10.1073/pnas.0401081101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paxinou E, et al. Induction of alpha-synuclein aggregation by intracellular nitrative insult. J Neurosci. 2001;21:8053–8061. doi: 10.1523/JNEUROSCI.21-20-08053.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Webb JL, Ravikumar B, Atkins J, Skepper JN, Rubinsztein DC. Alpha-synuclein is degraded by both autophagy and the proteasome. J Biol Chem. 2003;278:25009–25013. doi: 10.1074/jbc.M300227200. [DOI] [PubMed] [Google Scholar]
- 11.Anderson JP, et al. Phosphorylation of Ser-129 is the dominant pathological modification of alpha-synuclein in familial and sporadic Lewy body disease. J Biol Chem. 2006;281:29739–29752. doi: 10.1074/jbc.M600933200. [DOI] [PubMed] [Google Scholar]
- 12.Hasegawa M, et al. Phosphorylated alpha-synuclein is ubiquitinated in alpha-synucleinopathy lesions. J Biol Chem. 2002;277:49071–49076. doi: 10.1074/jbc.M208046200. [DOI] [PubMed] [Google Scholar]
- 13.Tofaris GK, Razzaq A, Ghetti B, Lilley KS, Spillantini MG. Ubiquitination of alpha-synuclein in Lewy bodies is a pathological event not associated with impairment of proteasome function. J Biol Chem. 2003;278:44405–44411. doi: 10.1074/jbc.M308041200. [DOI] [PubMed] [Google Scholar]
- 14.Kalia LV, et al. Ubiquitinylation of α-synuclein by carboxyl terminus Hsp70-interacting protein (CHIP) is regulated by Bcl-2-associated athanogene 5 (BAG5) PLoS One. 2011;6:e14695. doi: 10.1371/journal.pone.0014695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tofaris GK, et al. Ubiquitin ligase Nedd4 promotes α-synuclein degradation by the endosomal–lysosomal pathway. Proc Natl Acad Sci USA. 2011;108:17004–17009. doi: 10.1073/pnas.1109356108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mulherkar SA, Sharma J, Jana NR. The ubiquitin ligase E6-AP promotes degradation of alpha-synuclein. J Neurochem. 2009;110:1955–1964. doi: 10.1111/j.1471-4159.2009.06293.x. [DOI] [PubMed] [Google Scholar]
- 17.Rott R, et al. Monoubiquitylation of alpha-synuclein by seven in absentia homolog (SIAH) promotes its aggregation in dopaminergic cells. J Biol Chem. 2008;283:3316–3328. doi: 10.1074/jbc.M704809200. [DOI] [PubMed] [Google Scholar]
- 18.Rott R, et al. α-Synuclein fate is determined by USP9X-regulated monoubiquitination. Proc Natl Acad Sci USA. 2011;108:18666–18671. doi: 10.1073/pnas.1105725108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abeywardana T, Lin YH, Rott R, Engelender S, Pratt MR. Site-specific differences in proteasome-dependent degradation of monoubiquitinated α-synuclein. Chem Biol. 2013;20:1207–1213. doi: 10.1016/j.chembiol.2013.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shabek N, et al. The size of the proteasomal substrate determines whether its degradation will be mediated by mono- or polyubiquitylation. Mol Cell. 2012;48:87–97. doi: 10.1016/j.molcel.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 21.Alexopoulou Z, et al. Deubiquitinase Usp8 regulates α-synuclein clearance and modifies its toxicity in Lewy body disease. Proc Natl Acad Sci USA. 2016;113:E4688–E4697. doi: 10.1073/pnas.1523597113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dorval V, Fraser PE. Small ubiquitin-like modifier (SUMO) modification of natively unfolded proteins tau and alpha-synuclein. J Biol Chem. 2006;281:9919–9924. doi: 10.1074/jbc.M510127200. [DOI] [PubMed] [Google Scholar]
- 23.Oh Y, Kim YM, Mouradian MM, Chung KC. Human Polycomb protein 2 promotes α-synuclein aggregate formation through covalent SUMOylation. Brain Res. 2011;1381:78–89. doi: 10.1016/j.brainres.2011.01.039. [DOI] [PubMed] [Google Scholar]
- 24.Kim YM, et al. Proteasome inhibition induces α-synuclein SUMOylation and aggregate formation. J Neurol Sci. 2011;307:157–161. doi: 10.1016/j.jns.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krumova P, et al. Sumoylation inhibits alpha-synuclein aggregation and toxicity. J Cell Biol. 2011;194:49–60. doi: 10.1083/jcb.201010117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin JY, Ohshima T, Shimotohno K. Association of Ubc9, an E2 ligase for SUMO conjugation, with p53 is regulated by phosphorylation of p53. FEBS Lett. 2004;573:15–18. doi: 10.1016/j.febslet.2004.07.059. [DOI] [PubMed] [Google Scholar]
- 27.Knipscheer P, van Dijk WJ, Olsen JV, Mann M, Sixma TK. Noncovalent interaction between Ubc9 and SUMO promotes SUMO chain formation. EMBO J. 2007;26:2797–2807. doi: 10.1038/sj.emboj.7601711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tatham MH, et al. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J Biol Chem. 2001;276:35368–35374. doi: 10.1074/jbc.M104214200. [DOI] [PubMed] [Google Scholar]
- 29.Hershko A, Heller H. Occurrence of a polyubiquitin structure in ubiquitin-protein conjugates. Biochem Biophys Res Commun. 1985;128:1079–1086. doi: 10.1016/0006-291x(85)91050-2. [DOI] [PubMed] [Google Scholar]
- 30.Li SJ, Hochstrasser M. The Ulp1 SUMO isopeptidase: Distinct domains required for viability, nuclear envelope localization, and substrate specificity. J Cell Biol. 2003;160:1069–1081. doi: 10.1083/jcb.200212052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fukuda I, et al. Ginkgolic acid inhibits protein SUMOylation by blocking formation of the E1-SUMO intermediate. Chem Biol. 2009;16:133–140. doi: 10.1016/j.chembiol.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 32.Neumann M, et al. Misfolded proteinase K-resistant hyperphosphorylated alpha-synuclein in aged transgenic mice with locomotor deterioration and in human alpha-synucleinopathies. J Clin Invest. 2002;110:1429–1439. doi: 10.1172/JCI15777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iwatsubo T, et al. Purification and characterization of Lewy bodies from the brains of patients with diffuse Lewy body disease. Am J Pathol. 1996;148:1517–1529. [PMC free article] [PubMed] [Google Scholar]
- 34.Lopes da Fonseca T, Pinho R, Outeiro TF. A familial ATP13A2 mutation enhances alpha-synuclein aggregation and promotes cell death. Hum Mol Genet. 2016;25:2959–2971. doi: 10.1093/hmg/ddw147. [DOI] [PubMed] [Google Scholar]
- 35.Kunadt M, et al. Extracellular vesicle sorting of α-Synuclein is regulated by sumoylation. Acta Neuropathol. 2015;129:695–713. doi: 10.1007/s00401-015-1408-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hendriks IA, Vertegaal AC. A comprehensive compilation of SUMO proteomics. Nat Rev Mol Cell Biol. 2016;17:581–595. doi: 10.1038/nrm.2016.81. [DOI] [PubMed] [Google Scholar]
- 37.Denuc A, Marfany G. SUMO and ubiquitin paths converge. Biochem Soc Trans. 2010;38:34–39. doi: 10.1042/BST0380034. [DOI] [PubMed] [Google Scholar]
- 38.Vicente Miranda H, et al. Glycation potentiates alpha-synuclein-associated neurodegeneration in synucleinopathies. Brain. 2017;140:1399–1419. doi: 10.1093/brain/awx056. [DOI] [PubMed] [Google Scholar]
- 39.O’Rourke JG, et al. SUMO-2 and PIAS1 modulate insoluble mutant huntingtin protein accumulation. Cell Rep. 2013;4:362–375. doi: 10.1016/j.celrep.2013.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ochaba J, et al. PIAS1 regulates mutant huntingtin accumulation and Huntington’s disease-associated phenotypes in vivo. Neuron. 2016;90:507–520. doi: 10.1016/j.neuron.2016.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Subramaniam S, Sixt KM, Barrow R, Snyder SH. Rhes, a striatal specific protein, mediates mutant-huntingtin cytotoxicity. Science. 2009;324:1327–1330. doi: 10.1126/science.1172871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luo HB, et al. SUMOylation at K340 inhibits tau degradation through deregulating its phosphorylation and ubiquitination. Proc Natl Acad Sci USA. 2014;111:16586–16591. doi: 10.1073/pnas.1417548111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fei E, et al. SUMO-1 modification increases human SOD1 stability and aggregation. Biochem Biophys Res Commun. 2006;347:406–412. doi: 10.1016/j.bbrc.2006.06.092. [DOI] [PubMed] [Google Scholar]
- 44.Weetman J, et al. Increased SUMO-1 expression in the unilateral rotenone-lesioned mouse model of Parkinson’s disease. Neurosci Lett. 2013;544:119–124. doi: 10.1016/j.neulet.2013.03.057. [DOI] [PubMed] [Google Scholar]
- 45.Engelender S, et al. Synphilin-1 associates with alpha-synuclein and promotes the formation of cytosolic inclusions. Nat Genet. 1999;22:110–114. doi: 10.1038/8820. [DOI] [PubMed] [Google Scholar]
- 46.Eyal A, et al. Synphilin-1A: An aggregation-prone isoform of synphilin-1 that causes neuronal death and is present in aggregates from alpha-synucleinopathy patients. Proc Natl Acad Sci USA. 2006;103:5917–5922. doi: 10.1073/pnas.0509707103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.