Significance
Following envelope-mediated fusion, the HIV-1 viral core, which houses the viral RNA and proteins required for virus reverse transcription and integration, must traffic toward the nucleus for subsequent nuclear import of the viral genome. In this study we examined the role of BICD2, a known dynein adaptor protein, for its role during the postentry trafficking of HIV-1 virions. We show that BICD2 binds viral capsid and mediates the postentry trafficking of HIV-1. Moreover, we also show that depletion of BICD2 sensitizes the virus to detection by innate immune sensing, revealing this BICD2 is necessary for the virus to avoid detection by innate sensing mechanisms in macrophages.
Keywords: BICD2, dynein, HIV-1, microtubules, trafficking
Abstract
Numerous viruses, including HIV-1, exploit the microtubule network to traffic toward the nucleus during infection. Although numerous studies have observed a role for the minus-end microtubule motor dynein in HIV-1 infection, the mechanism by which the viral core containing the viral genome associates with dynein and induces its perinuclear trafficking has remained unclear. Here, we report that the dynein adapter protein bicaudal D2 (BICD2) is able to interact with HIV-1 viral cores in target cells. We also observe that BICD2 can bind in vitro-assembled capsid tubes through its CC3 domain. We observe that BICD2 facilitates infection by promoting the trafficking of viral cores to the nucleus, thereby promoting nuclear entry of the viral genome and infection. Finally, we observe that depletion of BICD2 in the monocytic cell line THP-1 results in an induction of IFN-stimulated genes in these cells. Collectively, these results identify a microtubule adapter protein critical for trafficking of HIV-1 in the cytoplasm of target cells and evasion of innate sensing mechanisms in macrophages.
Following envelope-mediated fusion many viruses rely on microtubule transport to traffic their genome toward the nucleus during infection (1–3). This transport is necessitated by the density of the cytoplasmic environment, which precludes diffusion of protein complexes the size of most viral ribonucleoprotein complexes (RNPs) (4). Cytoplasmic dynein along with the dynactin complex is involved in the retrograde transport of a range of cellular cargoes toward the microtubule organizing center, which resides next to the nucleus. Numerous members of the retroviridae family have been shown to utilize microtubules and microtubular motor dynein for retrograde trafficking (reviewed in ref. 5) In the case of HIV-1, the minus-end motor dynein has been implicated in the retrograde trafficking of viral RNPs (6, 7). Additionally, we and others have observed that inhibiting dynein pharmacologically or genetically perturbs the normal uncoating process, which is defined as the process by which the viral capsid core, which houses the viral genome, disassembles during infection (6, 8). Although a number of reports have identified viral proteins, including Vpr and integrase, capable of undergoing dynein-mediated trafficking (9–12), it remains unclear how the viral capsid core, which houses the viral genome during infection, engages the microtubule machinery to induce the trafficking of the core and genome toward the nucleus.
Numerous studies have revealed a range of adaptor proteins that interact with various dynein subunits to regulate dynein function and enable cargo binding onto the dynein–dynactin motor complex (13–15). One such adapter, bicaudal D2 (BICD2), was identified in a genome-wide screen for host factors essential for HIV-1 replication (16), although it was not identified in two similar genome-wide screens (17, 18). BICD2 is a highly conserved and well-studied dynein motor adaptor consisting of three coiled-coil segments separated by highly flexible regions (14). The N-terminal CC1 region of BICD2 has been shown to bind to the dynein–dynactin complex, thus improving dynein processivity along microtubules (19, 20). The C-terminal CC3 region acts as the cargo-binding domain of BICD2 and has been shown to directly interact with various dynein cargoes (21, 22). Binding of BICD2 to the dynein–dynactin complex activates dynein processivity (23–25). As such, BICD2 acts to couple cargo binding and dynein activation to coordinate trafficking of cellular cargoes.
In this study we examined the function of BICD2 during HIV-1 infection. We utilized cells depleted of BICD2 using CRISPR-Cas9, which confirmed a previous finding that BICD2 is an essential host factor required for infection. Our study further shows that BICD2 interacts with incoming viral particles. Depletion of BICD2 reduces cytoplasmic trafficking of fused viral particles and prevents the nuclear import of the viral genome, as measured by 2-LTR formation. We also demonstrate that BICD2 binds to the in vitro-assembled viral capsid–nucleocapsid (CA-NC) cores through its CC3 domain. Finally, we show that BICD2 depletion in monocytic cells leads to an increased expression of IFN-stimulated genes. This suggests that HIV-1 utilizes this microtubule adaptor protein for trafficking in the cytoplasm of target cell and to evade host innate sensing machinery.
Results
HIV-1 Infection Requires BICD2.
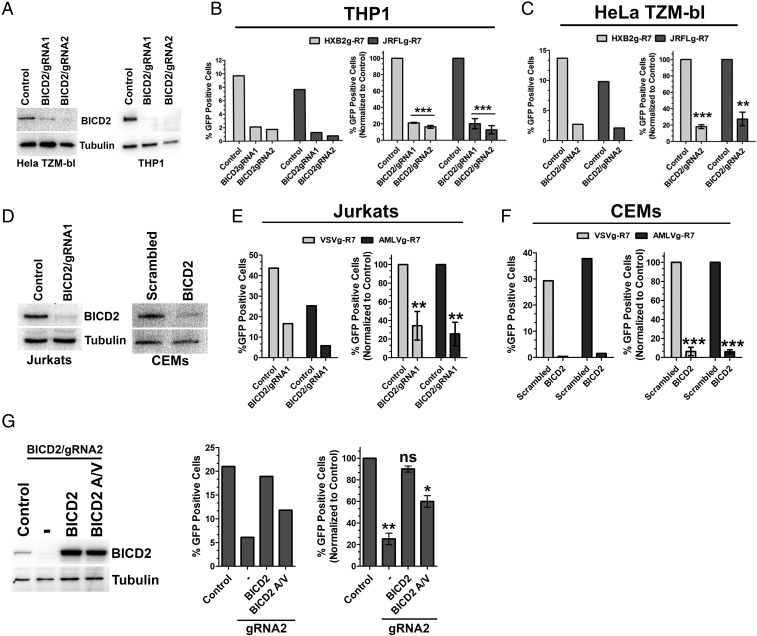
BICD2 was previously identified in a genome-wide screen for host cell dependency factors required for optimal HIV-1 infection (16). To confirm and extend this observation, we generated THP1 and HeLa TZM-bl cell lines in which the BICD2 gene was disrupted using CRSIPR/Cas9 technology. THP1 cells are a monocytic cell line which can be terminally differentiated into a macrophage-like cell upon phorbol myristate acetate (PMA) treatment, and HeLa TZM-bl cells are HeLa cells which express high levels of CD4 and coreceptors, similar in nature to the cells in which BICD2 was originally identified as a host factor supporting HIV-1 replication (16). Two guide RNAs (gRNAs) were designed targeting either exon 1 or exon 3 of BICD2 and cloned into a LentiCRISPR plasmid allowing for selection of cells depleted of BICD2. Both gRNAs were able to deplete BICD2 in THP1 cells, whereas only one of the gRNAs (BICD2/gRNA2) was successful in HeLa TZM-bl cells (Fig. 1A). Next, we looked at the level of viral infections in these knockout cells by infecting with HIV-1 pseudotyped with CCR5 tropic (JRFL) or CXCR4 tropic (HXB2) HIV-1 glycoproteins. BICD2 depletion resulted in a significant reduction in HIV-1 infectivity in both the cell types tested (Fig. 1 B and C). To understand if these results were specific to entry mediated by the HIV-1 envelope, we next infected cells with HIV-1 pseudotyped with the pH-dependent envelope protein from vesicular stomatitis virus (VSV-g) or the pH-independent amphotropic murine leukemia virus envelope protein (A-MLV). As seen in Fig. S1E, BICD2-deficient THP1 cells showed reduced viral infectivity when infected with VSV-g pseudotyped HIV-1, while HeLa TZM-bl cells displayed no change in viral infectivity in the BICD2-depleted cells (Fig. S1C). In contrast, infection with A-MLV pseudotyped HIV-1 resulted in similar infectivity defects in TZM-bl and THP-1 cell lines depleted of BICD2 (Fig. S1 C and E). Next, we asked whether BICD2 is important for infection in T cells. We used Jurkat T cells depleted of BICD2 by CRISPR technology and CEM cells where BICD2 depletion was achieved by siRNA as we were unable to select stable CRISPR-depleted CEM cells (Fig. 1D). Depletion of BICD2 in both the T-cell lines resulted in a significant reduction in virus infection (Fig. 1 E and F). Since BICD2 is well-studied as a dynein adaptor, we tested whether knockdown of other known dynein adaptors influenced HIV-1 infection. As seen in Fig. S1 A and B, knockdown of two dynein adaptors, Hook3 (26) and ZW10 (27, 28), had no effect on HIV-1 infection. To rule out off-target effects resulting from BICD2 depletion we transduced the depleted TZM-bl cells with a CRISPR-resistant full-length BICD2 construct (Fig. 1G) and infected these cells with a JRFL pseudotyped virus. As seen in Fig. 1G, complementing the depleted cells with full-length BICD2 restores viral infection to levels similar to those in control cells. Then, we asked whether the interaction between dynein and BICD2 is important for HIV-1 infection. To address this, we transduced the control and knockout TZM-bl cells with a CRISPR-resistant full-length BICD2 construct harboring mutations at the N terminus at amino acid positions 43 and 44 (Fig. 1G, BICD2 A/V). These mutations, as described before, resulted in a weaker interaction between BICD2 and dynein (23). Upon infection, a modest but significant reduction in viral infectivity was still observed in the depleted cells transduced with BICD2 A/V construct (Fig. 1G). This suggests that the interaction between BICD2 and dynein is key in establishing a productive infection. To test whether the requirement of BICD2 was specific for HIV-1 infection we also infected the depleted TZM-bl cells with a B tropic murine leukemia virus (B-MLV) pseudotyped with A-MLV or VSV envelope proteins. As seen in Fig. S1D, no defect in B-MLV infection was observed. Taken together, these results show that BICD2 is required for efficient HIV-1 infection but not for MLV infection.
Fig. 1.
BICD2 is required for productive HIV-1 infection. HeLa TZM-bl and THP1 cells depleted of BICD2 by using CRISPR-Cas9 genome editing. (A) Protein expression in the depleted cells confirmed by Western blotting. (B) Differentiated THP1 cells lacking BICD2 were infected with R7ΔEnvGFP virus pseudotyped with CCR5 tropic JRFL or the CXCR4 tropic HXB2 envelope glycoproteins. Cells were harvested 48 h after infection and GFP expression was measured by flow cytometry. Data from single independent experiments (Left) and normalized and average data (±SD) from three or more independent experiments (Right) are shown. (C) Similar infectivity experiments as in B in HeLa TZM-bl cells. (D) Protein expression in Jurkats and CEM BICD2-depleted cells confirmed by Western blotting. (E and F) Infectivity experiments in BICD2-depleted Jurkats and CEM cells. (G) Protein expression in BICD2-depleted HeLa TZM-bl cells transduced with either CRISPR-resistant full-length BICD2 or a mutant (BICD2 A/V). Control and BICD2-depleted TZM-bl cells stably expressing BICD2 or BICD2 A/V were infected with R7ΔEnvGFP virus pseudotyped with JRFL envelope glycoproteins. Infectivity measured 48 h after infection. ***P < 0.001, **P < 0.01, *P < 0.05; ns, not significant.
BICD2 Depletion Does Not Affect Viral Fusion or Reverse Transcription but Reduces HIV-1 Nuclear Import.
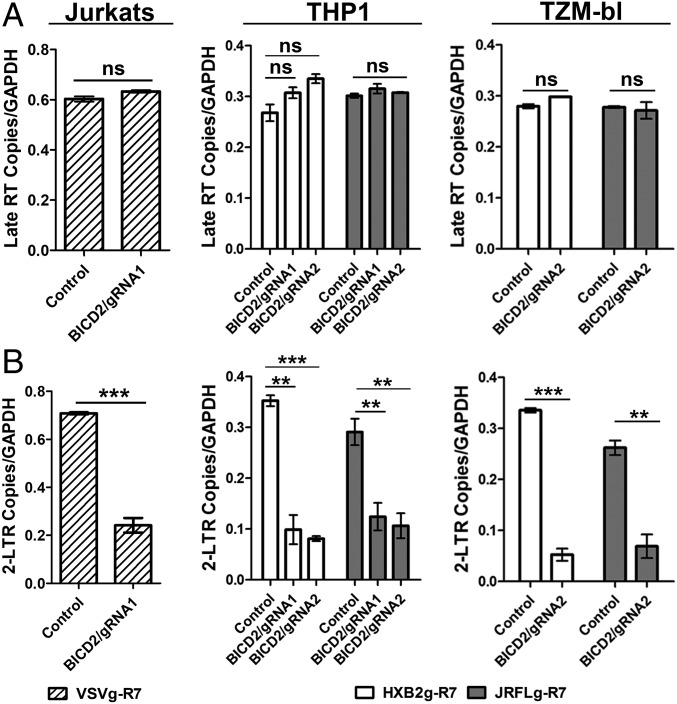
To determine at what step during the viral life cycle that BICD2 exerts its effect we measured individual steps during the early phase of virus infection in BICD2-depleted cells. We first measured fusion using a Vpr-β-lactamase fusion assay, which measures the cleavage of cellular substrate by the β-lactamase enzyme loaded into virions by fusion to Vpr (29). Infection with JRFL-R7 showed no significant differences in viral fusion in the knockout cells compared with control TZM (Fig. S2). We next measured viral reverse transcription and nuclear import (as measured by 2-LTR formation) in the BICD2-depleted cells using quantitative PCR. As seen in Fig. 2A, we observed no change in the amount of late reverse transcription products in either Jurkats, THP1 differentiated macrophages, or TZM-bl cells lacking BICD2. Next, we assessed nuclear import of the virus by measuring the amount of 2-LTR formation in cells. Depletion of BICD2 showed a significant reduction in 2-LTR formation in all cell lines tested, indicating a strong defect in viral nuclear entry (Fig. 2B). Collectively, these data demonstrate that BICD2 facilitates a step in HIV-1 infection between fusion and the nuclear import of the viral genome.
Fig. 2.
BICD2 depletion prevents HIV-1 nuclear import. Control and BICD2-depleted Jurkats, THP1 differentiated macrophages, and HeLa TZM-bl cells were subjected to synchronized infection with R7ΔEnvGFP virus pseudotyped with envelope proteins from either the JRFL (MOI 0.2), HXB2 strain (MOI 0.2), or VSV (MOI 0.4). Cells were collected 24 h postinfection and qPCR was performed using specific primers to quantify Late RT (A) and 2-LTR circles (B). Error bar represents SD of samples run in triplicate. The data shown here are representative of three or more experiments. ***P < 0.001, **P < 0.01; ns, not significant.
BICD2 Depletion Perturbs HIV-1 Uncoating.
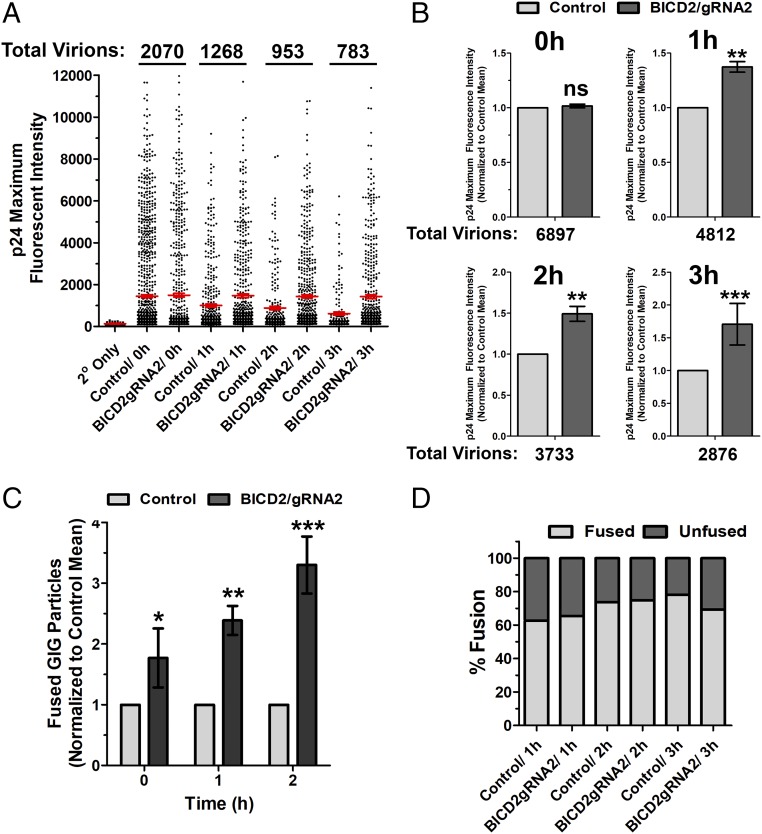
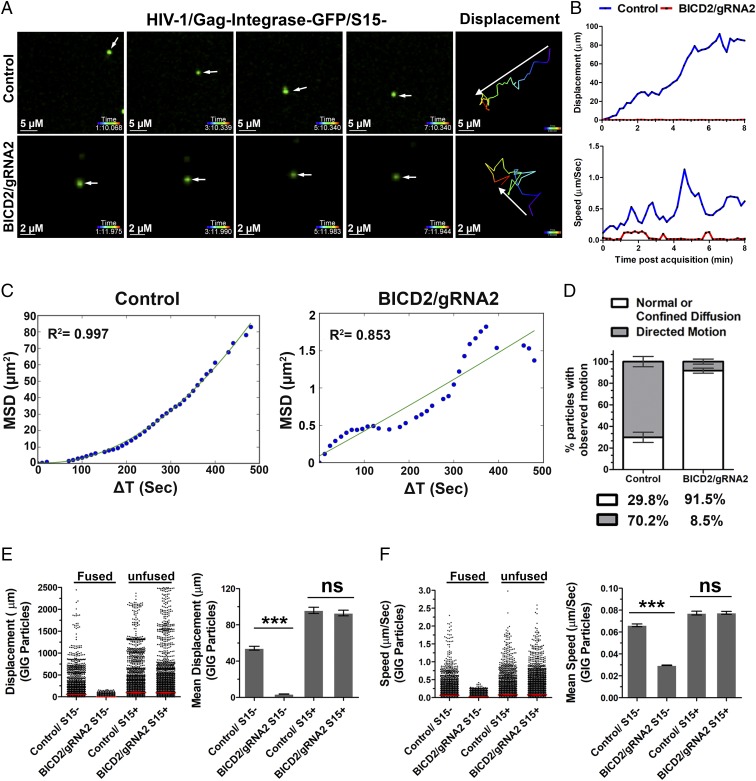
We and others have observed that microtubule motor dynein is required for HIV-1 uncoating (6, 8). To determine if the dynein adaptor BICD2 is required for HIV-1 uncoating we performed an in situ uncoating assay (6) in control and BICD2-depleted TZM-bl cells. This assay measures the amount of CA which remains associated with individual viral particles during infection. For this assay, we labeled HIV-1 virions with a Gag-integrase GFP construct (GIG) (30). We also labeled viral particles with the S15-mCherry protein (S15mCh). This protein contains the 15 N-terminal residues of the Src protein, which facilitates membrane association and incorporation into HIV-1 virions through a myristoylation sequence present in these residues (31). This label becomes lost from viral particles upon fusion, allowing particles that have productively entered the cytoplasm via fusion to be distinguished from particles that have not. Double-labeled virus pseudotyped with JRFL envelope was used to synchronously infect TZM-bL cells and fixed at various times postinfection. The amount of p24 associated with individual virions that have productively entered the cells (S15-negative) was determined by staining p24 with a monoclonal antibody and measuring p24 intensity using wide-field deconvolution microscopy. We observed that depletion of BICD2 delayed uncoating, as measured by p24 staining of individual GIG puncta (Fig. 3 A and B). This delay in uncoating was not due to a defect in viral fusion, as a similar percentage of virions had lost their S15 membrane label at the time points examined (Fig. 3D), supporting our previous observations using the Vpr-β-lactamase assay (Fig. S2). Also in these imaging experiments we observed a higher number of fused GIG particles in the cytoplasm of BICD2-depleted cells compared with control cells (Fig. 3C). This accumulation of viral particles in BICD2-depleted cells is also consistent with the hypothesis that BICD2 depletion delays the normal uncoating of HIV-1.
Fig. 3.
BICD2 depletion delays HIV-1 uncoating as measured using an in situ uncoating assay. Control and BICD2-depleted HeLa TZM-bl cells were synchronously infected with S15-mCherry/GIG JRFLg-HIV-1. (A) Cells were fixed at the indicated time points, and the p24 intensities associated with individual virions lacking the S15 membrane label (1–3 h postinfection) or all virions (0-h point) are shown. Red line represents average p24 intensity measured for all fused viruses at the indicated time points; 20–25 cells were imaged at each time point. Error bar represents SEM. (B) Data from three independent experiments, as shown in A, were normalized to the mean p24 intensity observed in control cells and averaged. (C) Fused GIG particles in control and BICD2-depleted cells from three independent experiments. Normalized to the number of cells observed in control cells at the indicated time points. (D) Percent fusion was calculated from the experiment described in A. Data are representative of three independent experiments. ***P < 0.001, **P < 0.01, *P < 0.05; ns, not significant.
BICD2 Interacts with Viral Capsid in Vivo and Binds to in Vitro-Assembled HIV-1 CA-NC Complexes Through the CC3 Domain.
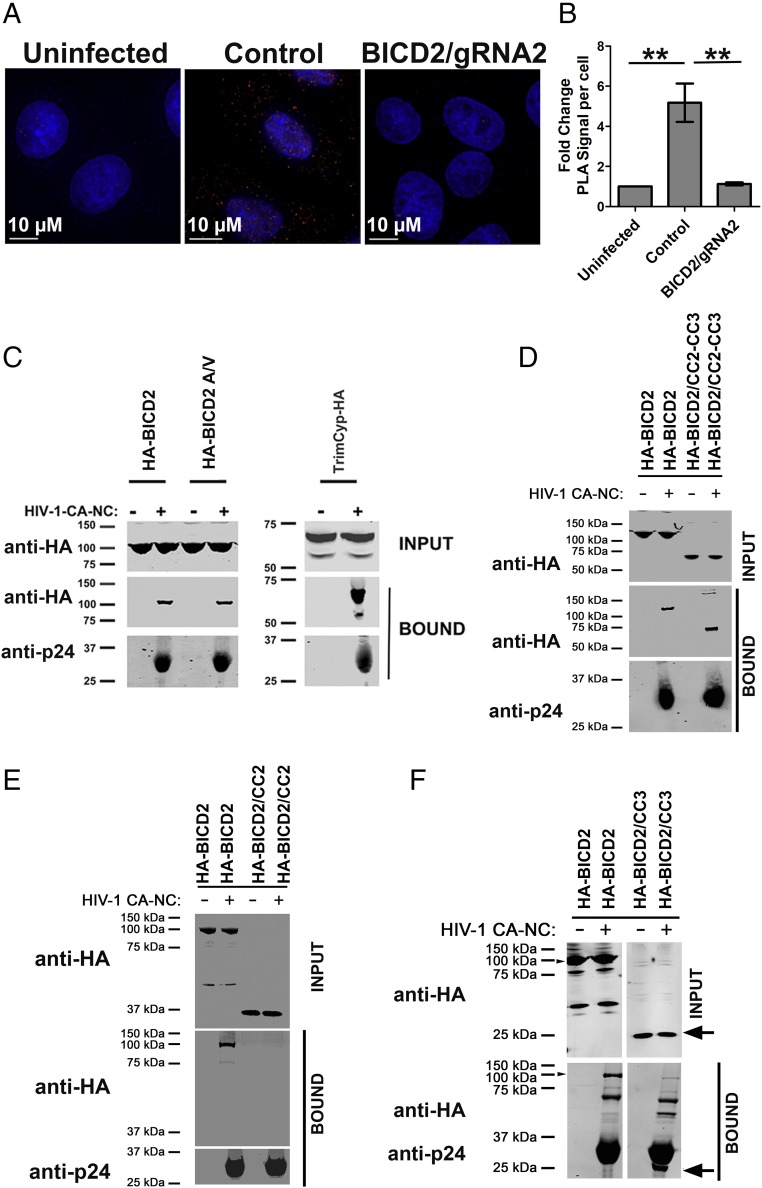
We next sought to determine if BICD2 could interact with determinants present in the mature capsid core of HIV-1 in vivo and in vitro. First, we examined the ability of BICD2 to associate with HIV-1 cores during infection by employing the proximity ligation assay (PLA). This assay detects close proximity between two antibodies (<30–40 nm) as bright fluorescent puncta, thereby measuring protein–protein interaction with high specificity and sensitivity (32). In a PLA using primary antibodies to CA and BICD2, PLA puncta were readily detected in the cytoplasm of TZM-bl cells upon infection with JRFL pseudotyped virus (Fig. 4A). The number of puncta quantified following infection showed a significant fold increase compared with uninfected control cells (Fig. 4B). The BICD2 knockout cells failed to induce PLA puncta, demonstrating the specificity of the assay (Fig. 4 A and B). This implies a specific association between HIV-1 viral cores and BICD2 in the cytoplasm of target cells. Next, we measured the ability of BICD2 to bind in vitro-assembled CA-NC tubes (33); 293T cells were transfected with a construct expressing HA-BICD2 or a mutant in which the dynein-binding domain of the protein was disrupted (HA-BICD2 A/V) (23) and these lysates were incubated with CA-NC tubes. CA-NC tubes were then pelleted by ultracentrifugation and BICD2 binding was measured using an anti-HA antibody. We observed that BICD2 binds to in vitro-assembled capsid in a similar fashion compared with the HIV-1 capsid-interacting protein TRIM-Cyp. This interaction was present in the pelleted material following incubation with CA-NC tubes but not in the material pelleted in the absence of assembled CA-NC tubes (Fig. 4C). The BICD2 mutant (A/V), which is unable to bind dynein, also pelleted in the presence of CA-NC but did not pellet in the absence of CA-NC tubes (Fig. 4C). To further define the region of BICD2 capable of binding CA-NC we examined the ability of the CC2 and CC3 domain to bind CA-NC tubes in tandem and individually. As seen in Fig. 4D, a construct containing both the CC2 and CC3 domains pelleted with CA-NC tubes. Analysis of individual domains revealed that the CC3 domain, but not the CC2 domain, was able to pellet with CA-NC tubes (Fig. 4 E and F). These results show that BICD2 through the CC3 domain can bind CA-NC, in a manner that is not dependent on dynein, suggesting that BICD2 is the adapter protein which tethers dynein to the viral capsid during infection.
Fig. 4.
BICD2 interacts with HIV-1 capsid in vivo and in vitro. (A) Control and BICD2 knockout HeLa TZM-bl cells were synchronously infected with JRFLg pseudotyped HIV-1 reporter virus (MOI of 0.4). Cells fixed 1 h post infection and PLA assay performed with antibodies to BICD2 and HIV-1 capsid protein p24. (B) Quantification of average fold increase in PLA puncta; 20 or more cells were analyzed in each experiment. Error bar represent the SEM (**P < 0.01). (C) The 293T cells were transfected with HA-tagged WT BICD2 and mutant (BICD2A/V). Cells were lysed 48 h posttransfection and the lysates incubated at room temperature for 1 h with in vitro-assembled HIV-1 CA-NC complexes. Samples were taken either before (INPUT) or after sedimentation through a sucrose cushion (BOUND) and analyzed by WB using anti-HA and anti-p24 antibodies. (D) Binding of BICD2 CC2-CC3 domain. (E) Binding of BICD2 CC2 domain. (F) Binding of BICD2 CC3 domain. A representative experiment is shown.
BICD2 Depletion Perturbs the Cytoplasmic Trafficking of HIV-1 During Infection.
The defect in nuclear import observed in BICD2-depleted cells and the ability of BICD2 to associate and traffic with incoming viral particles is consistent with the hypothesis that BICD2 mediates the cytoplasmic trafficking of HIV-1 toward the nucleus. To determine if BICD2 depletion affected viral trafficking we generated HIV-1 pseudotyped with the JRFL envelope and labeled with S15mCh and GIG. Double-labeled virus was used to synchronously infect TZM-bl cells, as these cells allowed for efficient fusion mediated by the JRFL envelope, owing to the overexpression of CD4 and CCR5 on these cells. Live-cell movies were acquired with cells imaged at 10-s intervals for 1 h immediately following infection. We employed particle tracking to map the trajectories of unfused (S15+) and cytoplasmic (S15−) viral populations and analyzed the behavior of each population of virions in both control and BICD2-depleted cells. Tracks of individual particles revealed that cytoplasmic particles (S15mCh-negative) typically exhibited greater displacement during the acquisition period in control cells compared with BICD2-depleted cells (Fig. 5 A, B, and E). This was evident when the trajectories of individual particles were examined (Fig. 5 A and B) and when viewed as a population (Fig. 5E). Similarly, peak particle speed was lower in BICD2-depleted cells compared with control cells (Fig. 5 B and F). In contrast, no difference in displacement and particle speed was observed in the unfused (S15mCh-positive) viral particles (Fig. 5 E and F). Mean square displacement (MSD) analysis was then employed to characterize the motion of the S15-/GIG–labeled viral particles. MSD analysis determines three types of particle motion: normal diffusion, confined diffusion, and directed motion. Active transport along microtubules is directed motion which can be described by a quadratic dependence of MSD on time interval ΔT. The MSD analysis shows almost 70% of the viral particles in the control cells and the one depicted in Fig. 5A can be fitted into a quadratic curve (Fig. 5 C and D), reflecting directed motion, whereas in the BICD2-deficient cells ∼91% of the particles analyzed showed a linear dependence or asymptotic behavior in the MSD ΔT plots (Fig. 5 C and D), consistent with normal and confined motion, respectively. These data collectively demonstrate that HIV-1 microtubule trafficking is dependent on BICD2.
Fig. 5.
BICD2 is required for efficient cytoplasmic trafficking of HIV-1 virions. Control and BICD2 knockout HeLa TZM-bl cells infected with S15-mCherry and GIG labeled HIV-1 virions pseudotyped with JRFL glycoproteins. Live-cell imaging was performed immediately following a synchronized infection. (A) Snapshots of consecutive frames for an S15-negative virus particle in the control and BICD2 knockout cells. (B) Displacement from the starting position for the virus particles in A and speed of the virus particles in A over time. (C) The MSD of the virus particle (A) in the control cells and BICD2-depleted cells plotted against time interval (ΔT). (D) Proportion of viral particles with directed motion or diffusion in the control and BICD2 knockout cells. (E) Displacement of S15+ and S15− virus particles in the control and BICD2 knockout cells. Representative of three independent experiments (dot blot) and mean displacement (± SEM) of results from three independent experiments (bar graph) are shown. (F) Similar to blot in E showing speed of S15+ and S15− virus particles. ***P < 0.001; ns, not significant.
These data suggest that HIV-1 utilizes BICD2 to achieve dynein-dependent trafficking toward the nucleus during infection. To further validate this hypothesis, we determined the ability of HIV-1 to induce the cytoplasmic relocalization of NUP358 during infection in BICD2-depleted cells. We have previously shown that WT CA induces the kinesin-1–dependent removal of Nup358 from nuclear pore complexes (34). As we have previously demonstrated, infection with HIV-1 induced substantial relocalization of NUP358 into the cytoplasm of these cells (Fig. S3 A and B). This relocalization was largely abrogated in BICD2-depleted cells (Fig. S3 A and B). Similarly, the association of NUP358 and HIV-1 capsids in the cytoplasm was reduced in BICD2, as measured by colocalization and PLA using antibodies to HIV-1 CA and Nup358 (Fig. S3 C–E).
BICD2 Depletion Increases Innate Sensing of HIV-1 Infection.
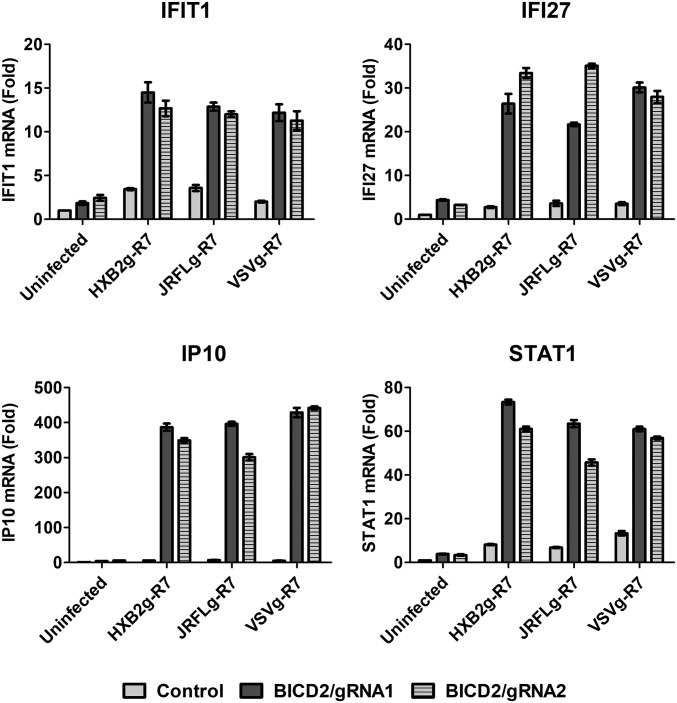
Given that BICD2 depletion does not affect the formation of RT products but reduces the ability of the viral genome to enter the nucleus, we reasoned that accumulation of viral particles in the cell cytoplasm might trigger innate immune activation by sensors that recognize cytoplasmic DNA or other viral pathogen-associated molecular patterns. To test this hypothesis, we measured the expression of genes which are induced following innate sensing of HIV-1 (35–37). Control and BICD2-depleted THP-1 cells were infected with HIV-1 pseudotyped with HIV or VSV-g envelope and the mRNA levels of several IFN-stimulated genes (ISGs) were measured by qRT-PCR. As seen in Fig. 6, expression of antiviral genes was significantly increased in BICD2-depleted cells compared with control cells. These results demonstrate that depletion of BICD2 sensitizes the virus to detection by innate sensing in THP1 differentiated macrophages.
Fig. 6.
Increased antiviral gene expression in response to HIV-1 infection in BICD2 knockout cells. Control and BICD2 knockout THP1 differentiated macrophages infected with R7ΔEnvGFP pseudotyped with either JRFL, HXB2, or VSV glycoproteins. Cells were harvested 24 h postinfection and expression of IFIT1, IFI27, IP10, and STAT1 was measured using qRT-PCR with gene specific primers. Fold increase in mRNA expression compared with uninfected control. Error bar represents SD of samples run in triplicate. The data shown here are representative of three or more experiments.
Discussion
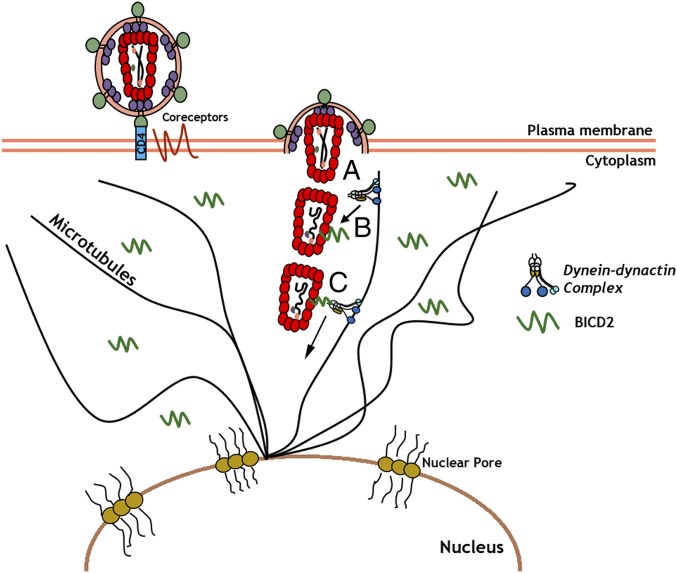
In this study we found that the microtubule adapter protein BICD2 mediates the dynein-dependent trafficking of HIV-1 particles toward the nucleus during infection (Fig. 7). We observed that BICD2 depletion inhibits HIV-1 infection in two T-cell lines, differentiated THP-1 cells as well as HeLa TZM-bl cells (Fig. 1). We show that this inhibition of infection is manifested at a postfusion stage of the viral life cycle, as fusion and reverse transcription are not perturbed by BICD2 depletion (Fig. 2A and Fig. S2). However, viral uncoating and nuclear import of the viral genome is reduced in BICD2-depleted cells (Figs. 2B and 3), consistent with the hypothesis that BICD2 facilitates the cytoplasmic trafficking of the viral core toward the nucleus during infection. This was further supported by the observation that BICD2 interacts with incoming viral particles as measured using a PLA (Fig. 4). We also observe that in vitro-assembled CA can bind BICD2 present in target cell lysates (Fig. 4). This interaction was mapped to the CC3 domain in BICD2 (Fig. 4F), which is thought to function as a cargo binding domain. Future studies are required to determine if this interaction is direct or dependent on other host-cell factors. However, the defects in trafficking and infection observed following BICD2 knockout demonstrate BICD2 as a critical mediator of viral trafficking.
Fig. 7.
Model of BICD2-mediated HIV-1 trafficking toward the nucleus. HIV-1 infection initiates upon binding of the viral envelope with CD4 and coreceptors on the cell surface. (A) Viral fusion occurs and viral capsid core released into the cytoplasm. (B) Viral core recruits the dynein adaptor protein BICD2 present in the cytoplasm, which in turn enables dynein recruitment to the viral capsid. (C) BICD2-mediated dynein recruitment enables the viral capsid to traffic along microtubules toward the nuclear import sites for delivery of the viral genome to the nucleus.
In addition, cytoplasmic HIV-1 virions exhibit altered trafficking in the cytoplasm in cells depleted of BICD2, as evidenced by the reduced average speed and displacement of viral particles in BICD2-depleted cells compared with control (Fig. 5). This defect was specific for virions which had productively entered the target cell (S15−), as a similar defect in trafficking speed and displacement was not observed in virions which retained their S15mCh membrane label (Fig. 5 E and F). The MSD ΔT plots (Fig. 5 C and D) support previous findings that HIV-1 undergoes directed motion along microtubules consistent with dynein-mediated retrograde trafficking. This type of directed motion was absent in BICD2-deficient cells (Fig. 5 C and D), highlighting the importance of BICD2 for HIV-1 trafficking toward the nucleus.
A number of studies have demonstrated that innate immune sensors can recognize the viral genome, especially in cases where interactions with host factors that facilitate infection are perturbed (35, 38, 39). In keeping with this finding, we observe that BICD2 knockout sensitizes the virus to innate immune sensors, leading to the up-regulation of IFN-stimulated genes following HIV-1 infection (Fig. 6). The potential for IFN induction to influence viral replication is supported by studies finding that IFN treatment can influence viral rebound in patients following antiretroviral therapy cessation (40), prevent infection in a simian immunodeficiency virus model of infection in macaques (41) and viral replication in culture (35, 42, 43). Although the ability of IFN to induce sustained control of HIV-1 replication is uncertain (44, 45), our observation that depletion of BICD2 exposes HIV-1 to innate sensing that induces the expression of ISGs reveals this interaction as a potential target that might be utilized to increase the innate immune response to HIV-1 infection. Future studies are necessary to identify the mechanism by which HIV-1 is sensed in the context of BICD2 depletion and how this sensing may influence viral replication in vivo.
Materials and Methods
Cell Lines and Constructs.
HeLa TZM-bl cells stably expressing CD4 and CCR5 were obtained through the NIH AIDS Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases (NIAID), NIH (TZM-bl from John C. Kappes, Xiaoyun Wu, and Tranzyme Inc.); 293T and THP1 cells were obtained from ATCC. HeLa TZM-bl and 293T cells were cultured in DMEM and THP1 cells in RPMI medium (Cellgrow) supplemented with 10% characterized FBS, 1,000 U/mL penicillin, 1,000 U/mL streptomycin, and 10 µg/mL ciprofloxacin hydrochloride. THP1 cells were differentiated by treating the cells with 100 ng/mL PMA for 48 h. HA-tagged BICD2 constructs (23) were kind gift from Casper C. Hoogenraad, Erasmus Medical Center, Rotterdam. Plasmids encoding BICD2 coiled-coil (CC) domains were generated by PCR-based strategy using the HA-BICD2 construct. The CC2 (amino acids 340–539), CC2-CC3 (amino acids 340–804) and CC3 (amino acids 662–804) domain were subcloned into Flag-HA-pcDNA3.1, a gift from Adam Antebi, Max Planck Institute for Biology of Ageing, Cologne, Germany (Addgene plasmid no. 52535).
Virus Production and Synchronized Infection.
To generate HIV-1 particles harboring HXB2 or JRFL envelope proteins, 293T cells seeded in a 10-cm dish at 70% confluency were transfected with 7 µg pCMV-HXB2g or pCMV-JRFLg and 3 µg of R7ΔEnvGFP using polyethylenimine (PEI) (molecular weight 25,000; Polysciences). HIV-1 particles pseudotyped with VSVg or AMLVg were generated by transfecting 293T cells in 10 cm with 7 µg of R7ΔEnvGFP and 3 µg of pCMV-VSVg or AMLVg. B tropic MLV (B-MLV) particles were generated by transfecting 293T in 10 cm with 5 µg of GFP reporter vector, 3 µg of pCigB, and 2 µg of pCMV-VSVg or AMLVg. Fluorescently labeled HIV-1 particles were generated by transfecting 293T in a 10-cm dish with 4.5 µg pCMV-JRFLg, 3.5 µg R7ΔEnvGFP, and 2 µg Gag-integrase-Ruby. S15-labeled HIV-1 particles were generated by transfecting 293T cells in a 10-cm dish with 4 µg pCMV-JRFLg, 3 µg R7ΔEnvGFP, 1.2 µg GIG, and 1.8 µg S15-mCherry. Virus with S15 labeling of GIG+ particles >90% was used for experiments. Vpx-containing virus-like particles (VLPs) were produced by the transfection of 293T cells in a 10-cm dish with 7 µg pSIV3+ and 3 µg of pCMV-VSVg. Viruses were harvested 48 h after transfection, spun for 5 min at 1,200 rpm, and filtered through a 0.45-μm filter (Milipore). THP1 cells were pretreated with VPX containing VLPs for 4 h before synchronized infection. Synchronized infection was performed by spinoculation at 13 °C for 2 h at 1,200 × g, after which virus-containing medium was removed and replaced with 37 °C media. Infectivity was measured 48 h after synchronized infection and percentage of GFP-positive cells was determined using a BD FACSCanto II cytometer (BD Bioscience). Multiplicity of infection (MOI) was calculated based on the equation MOI = −lnP(0), where P(0) is the proportion of uninfected cells.
Blam-Vpr Fusion Assay.
Blam-Vpr–labeled HIV-1 particles were generated by transfecting 293T cells in a 60-mm dish with 1.5 µg of pCMV-JRFLg, 1 µg of R7ΔEnvGFP, and 0.5 µg of Blam-Vpr. HeLa TZM-bl cells were infected with Blam-Vpr–labeled virus by spinoculation. Following spinoculation, medium was replaced with warm medium and incubated at 37 °C for 2 h, followed by CCF2-AM (Invitrogen) loading of the cells according to the manufacturer’s protocol. The reaction was allowed to proceed overnight at room temperature and the cells were subsequently collected for flow cytometery analysis to measure CCF2 cleavage.
Antibodies and Chemicals.
HIV-1 capsid protein p24 was stained using anti-p24 183-H12-5C (HIV-1 p24 hybridoma, from Bruce Chesebro, NIAID, Bethesda) and were obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH. Rabbit polyclonal antibodies against BICD2 (ab117818) and Nup358 (ab64276) were purchased from Abcam. Mouse monoclonal antibodies against Hook3 (sc-398924) and ZW10 (sc-81430) were purchased from Santa Cruz Biotechnology, Inc. Secondary antibodies conjugated to fluorophore for immunofluorescence studies were purchased from Jackson Immunoresearch Laboratories. DAPI to stain the nucleus was obtained from Sigma-Aldrich.
Western Blotting.
Cell lysates were prepared by lysing cells with Nonidet P-40 lysis buffer (100 mM Tris, pH 8.0, 1% Nonidet P-40, and 150 mM NaCl) containing protease inhibitor mixture (Roche) for 10 min on ice. Following incubation, lysates were spun down at 13,000 rpm for 10 min and supernatant collected for Western blot analysis. In brief, 2× Laemmli sample buffer was added to the lysed sample and incubated at 100 °C for 5 min. Protein concentration was measured using Pierce BCA protein assay kit (Thermo Scientific) and an equal amount of protein was loaded into an 8% polyacrylamide gel for SDS/PAGE. Upon separation, the proteins were transferred to a nitrocellouse membrane (Bio-Rad). Membranes were probed using specific primary antibodies and then probing with secondary antibodies conjugated to horseradish peroxidase (Thermo Scientific). Antibody complexes were detected using SuperSignal West Femto Chemiluminescent Substrate (Thermo Scientific). Chemiluminescence was detected using the ChemiDoc Imaging System (Bio-Rad).
Generation of Knockout Cell Lines and RNA Interference.
BICD2 knockout THP1 and HeLa TZM-bl cells were generated by using LentiCRISPRv2 (Addgene plasmid no. 52961), a gift from Feng Zhang, Massachusetts Institute of Technology, Cambridge, MA (46). gRNA targeting BICD2 exon 1 gRNA1: 5′- GATCTTCTCACGCGTGGTCT-3′ and exon 3 gRNA2: 5′- GCTGCAGGACTACTCGGAAC-3′ were generated using the online tool (crispr.mit.edu/). The gRNAs were annealed and cloned into LentiCRISPRv2. Lentivirus was prepared by transfecting 1 µg LentiCRISPR vector, 1 µg pCMV-VSVg, and 1 µg of psPAX2 into 293T in a 60-mm dish using PEI. Viruses were harvested 48 h posttransfection and were used to transduce HeLa TZM-bl and THP1 cells. The medium was supplemented with 5 μg/mL of puromycin 2 d posttransduction for HeLa TZM-bl and THP1 cells and 1 μg/mL for Jurkat cells. Following selection, cells were collected for Western blot to analyze knockout efficiency. siRNA targeting BICD2 (sc-92831), Hook3 (sc-60800), and ZW10 (sc-63259) were purchased from Santa Cruz Biotechnology, Inc. A control siRNA targeting luciferase was purchased from Fisher Scientific. siRNAs were transfected in to HeLa TZM-bl cells using Lipofectamine 2000 (Invitrogen) as per the manufacturer’s protocol. siRNAs were introduced into CEM cells by electroporation. In brief, 4 × 105 cells in OptiMEM were electroporated with 50 nM siRNA with settings 0.260 kV and 800 µF. After electroporation, cells were resuspended in media with 10% FBS. Three days after electroporation cells were collected for Western blot and to perform infectivity experiments.
Binding of BICD2 to in Vitro-Assembled CA-NC Complexes.
The binding of BICD2 protein to in vitro-assembled CA-NC complexes was carried out as described previously (47). In brief, 293T cells were transfected with plasmids expressing HA-tagged WT, mutant BICD2, or the BICD2 deletion proteins. Forty-eight hours after transfection, cell lysates were prepared as follows. Previously washed cells were resuspended in hypotonic lysis buffer (10 mM Tris, pH 7.4, 1.5 mM MgCl2, 10 mM KCl, and 0.5 mM DTT). The cell suspension was frozen and thawed and incubated on ice for 10 min. Afterward, the lysate was centrifuged at maximum speed in a refrigerated Eppendorf micro centrifuge (∼14,000 × g) for 5 min. The supernatant was supplemented with 1/10 volume of 10× PBS and then used in the binding assay. To test binding, 5 μL of CA-NC particles assembled in vitro were incubated with 200 μL of cell lysate at room temperature for 1 h. A fraction of this mixture was stored (input). The mixture was spun through a 70% sucrose cushion (70% sucrose, 1× PBS, and 0.5 mM DTT) at 100,000 × g in an SW55 rotor (Beckman) for 1 h at 4 °C. After centrifugation, the supernatant was carefully removed and the pellet resuspended in 1× SDS/PAGE loading buffer (pellet). The level of BICD2 proteins was determined by Western blotting with an anti-HA antibody and the level of HIV-1 CA-NC protein in the pellet was assessed by Western blotting with an anti-p24 CA antibody.
Microscopy and Image Acquisition.
Z-stack images were collected with a DeltaVision wide-field fluorescent microscope (Applied Precision, GE) equipped with a digital camera (CoolSNAP HQ; Photometrics), using a 1.4-numerical aperture 100× objective lens. Excitation light was generated with an Insight SSI solid-state illumination module (Applied Precision, GE) and was deconvolved with SoftWoRx deconvolution software (Applied Precision, GE). For live cell experiments, cells were plated on delta DPG dishes (Thermo Fisher Scientific) and maintained at 5% CO2 and 37 °C in an environmental chamber on a DeltaVision microscope. Images were captured in a z series on an electron multiplied charge coupled device digital camera (EMCCDCascade 2; Photometrics) and deconvolved using SoftWoRx deconvolution software. In any experiment, identical acquisition conditions were used to acquire all images. Following deconvolution, images were analyzed by Imaris 8.3.1 (Bitplane). An algorithm was designed using the spot or surface function in Imaris to generate surfaces around signal of interest and the maximum fluorescence intensity associated within these surfaces was measured. The algorithm was applied to all images within the same experiment. The trajectories of viral particles were generated using the track spots over time function in Imaris. The parameters (speed, displacement, and position) of the viral trajectories were directly extracted from the Imaris program. The displacement is defined as the changes in the position and given by , where t is the time index. MSD was calculated by using Python script based on the following equation:
where N is the total number of frames collected, ∆t is the time interval between two successive frames, and n and i are positive integers.
In Situ Uncoating Assay.
Fluorescently labeled HIV-1 was generated by transfecting 293T cells on a 10-cm dish with 2 μg S15-mCherry, 1 μg GIG, 3 μg R7ΔEnvGFP, and 4 μg pCMV-JRFLg using PEI. Following fusion, S15-mCherry–labeled viral membrane is lost, which allows one to effectively discriminate between viruses that have not productively endocytosed by the target cells (S15-mCherry+, GIG+) from those that have productively fused into the cytoplasm (S15-mCherry−, GIG+). A synchronized infection performed on HeLa TZM-bl cells with these fluorescently labeled virions (labeling efficiency >90%). Following spinoculation, media was aspirated and changed to 37 °C warm media. Cells were incubated at 37 °C and fixed at various time points postinfection. Coverslips were then subjected to indirect immunofluorescence to stain for viral capsid protein p24 using the anti-p24 mAb 183-H12-5C and a Cy5 conjugated secondary antibody (Jackson Immunoresearch) and mounted on glass slides. Images were acquired at 100× magnification using DeltaVision wide-field fluorescent microscope; 20–25 fields were acquired per coverslip. After deconvolution, GIG viral complexes were identified using the surface function in Imaris (Bitplane) software and the maximal S15-mCherry and p24 signal present within these individual GIG generated surfaces was determined. From the large sets of data acquired the average maximal p24 intensity of fused (S15-mCherry–negative) populations of virions was determined.
qPCR.
Cells were infected with equal MOI and RT-PCR was performed to determine the late reverse transcription (Late RT), 2-LTR products, and ISG expression. To measure ISG expression, total RNA was purified from cell lysate 24 h after synchronized infection using NucleoSpin RNA extraction kit (Macherey-Nagel). cDNA was synthesized using the GoScript RT System (Promega) and quantitative PCR was performed using gene-specific primers and Syber green master Mix (Roche). The following primer pairs were used in this study: STAT1 fwd, 5′-CCGTTTTCATGACCTCCTGT-3′; STAT1 rev, 5′-TGAATATTCCCCGACTGAGC-3′; IFI27 fwd, 5′-TCTGGCTCTGCCGTAGTTTT-3′; IFI27 rev, 5′- GAACTTGGTCAATCCGGAGA-3′; IFIT1 fwd, 5′- CAACCATGAGTACAAATGGTG-3′; IFIT1 rev, 5′- CTCACATTTGCTTGGTTGTC-3′; IP10 fwd, 5′-TGAAATTATTCCTGCAAGCCAATT-3′; and IP10 rev, 5′-CAGACATCTCTTCTCACCCTTCTTT-3′. Late RT was determined using GFP-specific primers and 2-LTR primers as described previously (48). GAPDH primers were used as a housekeeping gene for normalization. In brief, genomic DNA from cells was extracted following the DNeasy blood and tissue kit protocol (Qiagen). DNA concentration was determined and an equal amount of DNA was digested with Dpn1 (New England BioLabs) before performing RT-PCR.
PLA.
A Duolink PLA kit was purchased from Sigma and assay performed as described by the manufacturer (Olink Bioscience). Cells grown on coverslips were fixed with 3.7% paraformaldehyde 3 h after synchronized infection. To detect interaction between viral capsid protein p24 and Nup358 or BICD2, cells were permeabilized and blocked in 3% BSA followed by incubation with primary antibodies targeting viral protein p24 (mouse monoclonal) and Nup358 or BICD2 (rabbit polyclonal). After primary staining, coverslips containing cells were washed and incubated (1 h at 37 °C) with secondary anti-mouse conjugated with minus and anti-rabbit conjugated with plus Duolink II PLA probes. Coverslips were washed again and incubated with ligation-ligase solution (30 min at 37 °C) followed by washing and subsequent incubation with amplification-polymerase solution (100 min at 37 °C) containing Duolink II in Situ Detection Reagents Red. Finally, coverslips were washed and mounted with Duolink II mounting medium containing DAPI. Interactions were detected as fluorescent spots (λ excitation/emission 598/634 nm) under a fluorescence microscope.
Quantification of Perinuclear and Cytoplasmic Protein Signal.
The amount of Nup358 signal present in the cytoplasm and around the nucleus was determined as described before (34). In brief, an algorithm was designed using the DAPI channel and surface function to detect the cell nuclei. To detect nuclear or perinuclear signal, all signal outside the nuclei surface generated was masked using the masking tool in Imaris and saved as a separate channel. Then, a new algorithm was designed utilizing this new channel and the surface function to detect the p24 or Nup358 signal present around the nucleus. The total sum intensity of these surface masks was calculated to determine the amount of protein present around the nucleus. Similarly, to detect cytoplasmic signal, all signal outside of the nuclear surface mask was determined. The relative fraction of the perinuclear signal was calculated as a percentage of the total signal (perinuclear + cytoplasmic).
Statistical Analysis.
GraphPad Prism version 5.00 (GraphPad Software, Inc.) was employed for statistical analysis and to make graphs. Statistical significance was assessed using one-way or two-way ANOVA and Bonferroni posttest. P < 0.05 was considered significant in our experiments. Data are represented as mean ± SEM depending on the graph.
Supplementary Material
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1712033114/-/DCSupplemental.
References
- 1.Dodding MP, Way M. Coupling viruses to dynein and kinesin-1. EMBO J. 2011;30:3527–3539. doi: 10.1038/emboj.2011.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leopold PL, Pfister KK. Viral strategies for intracellular trafficking: Motors and microtubules. Traffic. 2006;7:516–523. doi: 10.1111/j.1600-0854.2006.00408.x. [DOI] [PubMed] [Google Scholar]
- 3.Merino-Gracia J, García-Mayoral MF, Rodríguez-Crespo I. The association of viral proteins with host cell dynein components during virus infection. FEBS J. 2011;278:2997–3011. doi: 10.1111/j.1742-4658.2011.08252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luby-Phelps K. Cytoarchitecture and physical properties of cytoplasm: Volume, viscosity, diffusion, intracellular surface area. Int Rev Cytol. 2000;192:189–221. doi: 10.1016/s0074-7696(08)60527-6. [DOI] [PubMed] [Google Scholar]
- 5.Arriagada G. Retroviruses and microtubule-associated motor proteins. Cell Microbiol. 2017;19:e12759. doi: 10.1111/cmi.12759. [DOI] [PubMed] [Google Scholar]
- 6.Lukic Z, Dharan A, Fricke T, Diaz-Griffero F, Campbell EM. HIV-1 uncoating is facilitated by dynein and kinesin 1. J Virol. 2014;88:13613–13625. doi: 10.1128/JVI.02219-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McDonald D, et al. Visualization of the intracellular behavior of HIV in living cells. J Cell Biol. 2002;159:441–452. doi: 10.1083/jcb.200203150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pawlica P, Berthoux L. Cytoplasmic dynein promotes HIV-1 uncoating. Viruses. 2014;6:4195–4211. doi: 10.3390/v6114195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Danappa Jayappa K, et al. Human immunodeficiency virus type 1 employs the cellular dynein light chain 1 protein for reverse transcription through interaction with its integrase protein. J Virol. 2015;89:3497–3511. doi: 10.1128/JVI.03347-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dumas A, et al. The HIV-1 protein Vpr impairs phagosome maturation by controlling microtubule-dependent trafficking. J Cell Biol. 2015;211:359–372. doi: 10.1083/jcb.201503124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caly L, et al. Fast track, dynein-dependent nuclear targeting of human immunodeficiency virus Vpr protein; impaired trafficking in a clinical isolate. Biochem Biophys Res Commun. 2016;470:735–740. doi: 10.1016/j.bbrc.2016.01.051. [DOI] [PubMed] [Google Scholar]
- 12.Desfarges S, et al. HIV-1 integrase trafficking in S. cerevisiae: A useful model to dissect the microtubule network involvement of viral protein nuclear import. Yeast. 2009;26:39–54. doi: 10.1002/yea.1651. [DOI] [PubMed] [Google Scholar]
- 13.Allan VJ. Cytoplasmic dynein. Biochem Soc Trans. 2011;39:1169–1178. doi: 10.1042/BST0391169. [DOI] [PubMed] [Google Scholar]
- 14.Cianfrocco MA, DeSantis ME, Leschziner AE, Reck-Peterson SL. Mechanism and regulation of cytoplasmic dynein. Annu Rev Cell Dev Biol. 2015;31:83–108. doi: 10.1146/annurev-cellbio-100814-125438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vallee RB, McKenney RJ, Ori-McKenney KM. Multiple modes of cytoplasmic dynein regulation. Nat Cell Biol. 2012;14:224–230. doi: 10.1038/ncb2420. [DOI] [PubMed] [Google Scholar]
- 16.Zhou H, et al. Genome-scale RNAi screen for host factors required for HIV replication. Cell Host Microbe. 2008;4:495–504. doi: 10.1016/j.chom.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 17.Brass AL, et al. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- 18.König R, et al. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell. 2008;135:49–60. doi: 10.1016/j.cell.2008.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoogenraad CC, et al. Bicaudal D induces selective dynein-mediated microtubule minus end-directed transport. EMBO J. 2003;22:6004–6015. doi: 10.1093/emboj/cdg592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoogenraad CC, et al. Mammalian Golgi-associated Bicaudal-D2 functions in the dynein-dynactin pathway by interacting with these complexes. EMBO J. 2001;20:4041–4054. doi: 10.1093/emboj/20.15.4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matanis T, et al. Bicaudal-D regulates COPI-independent Golgi-ER transport by recruiting the dynein-dynactin motor complex. Nat Cell Biol. 2002;4:986–992. doi: 10.1038/ncb891. [DOI] [PubMed] [Google Scholar]
- 22.Splinter D, et al. Bicaudal D2, dynein, and kinesin-1 associate with nuclear pore complexes and regulate centrosome and nuclear positioning during mitotic entry. PLoS Biol. 2010;8:e1000350. doi: 10.1371/journal.pbio.1000350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schlager MA, et al. Bicaudal D family adaptor proteins control the velocity of dynein-based movements. Cell Rep. 2014;8:1248–1256. doi: 10.1016/j.celrep.2014.07.052. [DOI] [PubMed] [Google Scholar]
- 24.Carter AP, Diamant AG, Urnavicius L. How dynein and dynactin transport cargos: A structural perspective. Curr Opin Struct Biol. 2016;37:62–70. doi: 10.1016/j.sbi.2015.12.003. [DOI] [PubMed] [Google Scholar]
- 25.Belyy V, et al. The mammalian dynein-dynactin complex is a strong opponent to kinesin in a tug-of-war competition. Nat Cell Biol. 2016;18:1018–1024. doi: 10.1038/ncb3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schroeder CM, Vale RD. Assembly and activation of dynein-dynactin by the cargo adaptor protein Hook3. J Cell Biol. 2016;214:309–318. doi: 10.1083/jcb.201604002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Starr DA, Williams BC, Hays TS, Goldberg ML. ZW10 helps recruit dynactin and dynein to the kinetochore. J Cell Biol. 1998;142:763–774. doi: 10.1083/jcb.142.3.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vallee RB, Varma D, Dujardin DL. ZW10 function in mitotic checkpoint control, dynein targeting and membrane trafficking: Is dynein the unifying theme? Cell Cycle. 2006;5:2447–2451. doi: 10.4161/cc.5.21.3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cavrois M, De Noronha C, Greene WC. A sensitive and specific enzyme-based assay detecting HIV-1 virion fusion in primary T lymphocytes. Nat Biotechnol. 2002;20:1151–1154. doi: 10.1038/nbt745. [DOI] [PubMed] [Google Scholar]
- 30.Hulme AE, Kelley Z, Foley D, Hope TJ. Complementary assays reveal a low level of CA associated with nuclear HIV-1 viral complexes. J Virol. 2015;89:5350–5361. doi: 10.1128/JVI.00476-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Campbell EM, Perez O, Melar M, Hope TJ. Labeling HIV-1 virions with two fluorescent proteins allows identification of virions that have productively entered the target cell. Virology. 2007;360:286–293. doi: 10.1016/j.virol.2006.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fredriksson S, et al. Protein detection using proximity-dependent DNA ligation assays. Nat Biotechnol. 2002;20:473–477. doi: 10.1038/nbt0502-473. [DOI] [PubMed] [Google Scholar]
- 33.Ganser BK, Li S, Klishko VY, Finch JT, Sundquist WI. Assembly and analysis of conical models for the HIV-1 core. Science. 1999;283:80–83. doi: 10.1126/science.283.5398.80. [DOI] [PubMed] [Google Scholar]
- 34.Dharan A, et al. KIF5B and Nup358 cooperatively mediate the nuclear import of HIV-1 during infection. PLoS Pathog. 2016;12:e1005700. doi: 10.1371/journal.ppat.1005700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rasaiyaah J, et al. HIV-1 evades innate immune recognition through specific cofactor recruitment. Nature. 2013;503:402–405. doi: 10.1038/nature12769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vermeire J, et al. HIV triggers a cGAS-dependent, Vpu- and Vpr-regulated type I interferon response in CD4(+) T cells. Cell Rep. 2016;17:413–424. doi: 10.1016/j.celrep.2016.09.023. [DOI] [PubMed] [Google Scholar]
- 37.Zahoor MA, et al. HIV-1 Vpr induces interferon-stimulated genes in human monocyte-derived macrophages. PLoS One. 2014;9:e106418. doi: 10.1371/journal.pone.0106418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao D, et al. Cyclic GMP-AMP synthase is an innate immune sensor of HIV and other retroviruses. Science. 2013;341:903–906. doi: 10.1126/science.1240933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lahaye X, et al. The capsids of HIV-1 and HIV-2 determine immune detection of the viral cDNA by the innate sensor cGAS in dendritic cells. Immunity. 2013;39:1132–1142. doi: 10.1016/j.immuni.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 40.Azzoni L, et al. Pegylated interferon alfa-2a monotherapy results in suppression of HIV type 1 replication and decreased cell-associated HIV DNA integration. J Infect Dis. 2013;207:213–222. doi: 10.1093/infdis/jis663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sandler NG, et al. Type I interferon responses in rhesus macaques prevent SIV infection and slow disease progression. Nature. 2014;511:601–605. doi: 10.1038/nature13554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goujon C, et al. Human MX2 is an interferon-induced post-entry inhibitor of HIV-1 infection. Nature. 2013;502:559–562. doi: 10.1038/nature12542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu Z, et al. The interferon-inducible MxB protein inhibits HIV-1 infection. Cell Host Microbe. 2013;14:398–410. doi: 10.1016/j.chom.2013.08.015. [DOI] [PubMed] [Google Scholar]
- 44.Bosinger SE, Utay NS. Type I interferon: Understanding its role in HIV pathogenesis and therapy. Curr HIV/AIDS Rep. 2015;12:41–53. doi: 10.1007/s11904-014-0244-6. [DOI] [PubMed] [Google Scholar]
- 45.Doyle T, Goujon C, Malim MH. HIV-1 and interferons: Who’s interfering with whom? Nat Rev Microbiol. 2015;13:403–413. doi: 10.1038/nrmicro3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fricke T, Buffone C, Opp S, Valle-Casuso J, Diaz-Griffero F. BI-2 destabilizes HIV-1 cores during infection and prevents binding of CPSF6 to the HIV-1 capsid. Retrovirology. 2014;11:120. doi: 10.1186/s12977-014-0120-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Anderson JL, et al. Proteasome inhibition reveals that a functional preintegration complex intermediate can be generated during restriction by diverse TRIM5 proteins. J Virol. 2006;80:9754–9760. doi: 10.1128/JVI.01052-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.