Significance
The pathological Duchenne muscular dystrophy (DMD) muscles show increased stretch-induced intracellular Ca2+ and nitrosative stress. Whether there is a link between the two, and how the former impacts the nitrosylated proteome, is unknown. Here, we report that transient receptor potential channel 6 (Trpc6) modulates increased nitrosative stress in dmdmdx:utrn+/− mice, as reflected by an increase in protein S-nitrosylation, and provide a broad high-throughput analysis of S-nitrosylation in this model. We found that S-nitrosothiol targets are conserved in dmdmdx:utrn+/− myocardium, but intensified in a Trpc6-dependent manner. Restoration of more normal S-nitrosylation profiles in dmdmdx:utrn+/− mouse hearts lacking Trpc6 corresponds to improved cardiac function and reduced fibrosis. These findings link Trpc6-mediated Ca2+ signaling and nitrosative stress in the redox pathobiology of DMD.
Keywords: Duchenne muscular dystrophy, protein S-nitrosylation, Trpc6, nitric oxide synthase signaling, mass spectrometry
Abstract
Duchenne muscular dystrophy (DMD) is an X-linked disorder with dystrophin loss that results in skeletal and cardiac muscle weakening and early death. Loss of the dystrophin–sarcoglycan complex delocalizes nitric oxide synthase (NOS) to alter its signaling, and augments mechanosensitive intracellular Ca2+ influx. The latter has been coupled to hyperactivation of the nonselective cation channel, transient receptor potential canonical channel 6 (Trpc6), in isolated myocytes. As Ca2+ also activates NOS, we hypothesized that Trpc6 would help to mediate nitric oxide (NO) dysregulation and that this would be manifest in increased myocardial S-nitrosylation, a posttranslational modification increasingly implicated in neurodegenerative, inflammatory, and muscle disease. Using a recently developed dual-labeling proteomic strategy, we identified 1,276 S-nitrosylated cysteine residues [S-nitrosothiol (SNO)] on 491 proteins in resting hearts from a mouse model of DMD (dmdmdx:utrn+/−). These largely consisted of mitochondrial proteins, metabolic regulators, and sarcomeric proteins, with 80% of them also modified in wild type (WT). S-nitrosylation levels, however, were increased in DMD. Genetic deletion of Trpc6 in this model (dmdmdx:utrn+/−:trpc6−/−) reversed ∼70% of these changes. Trpc6 deletion also ameliorated left ventricular dilation, improved cardiac function, and tended to reduce fibrosis. Furthermore, under catecholamine stimulation, which also increases NO synthesis and intracellular Ca2+ along with cardiac workload, the hypernitrosylated state remained as it did at baseline. However, the impact of Trpc6 deletion on the SNO proteome became less marked. These findings reveal a role for Trpc6-mediated hypernitrosylation in dmdmdx:utrn+/− mice and support accumulating evidence that implicates nitrosative stress in cardiac and muscle disease.
Duchenne muscular dystrophy (DMD) is a muscle disease affecting approximately one in 3,500 live-birth males. Patients with DMD develop severe skeletal and cardiac muscle weakness leading to immobility and early-onset mortality (1, 2). Heart failure with dilated ventricular chambers follows in a majority of patients, and accounts for an increasing proportion of deaths attributable to DMD (3). Treatment strategies have included immune suppression, gene replacement, and gene-editing methods targeting muscle weakness (4, 5). Heart failure therapy has followed standard-of-care guidelines developed for adult dilated cardiomyopathy, with none yet available that are tailored to DMD cardiomyopathy (4).
An exaggerated mechanosensitive response to stress is a primary hallmark of DMD muscle. The dystrophin–sarcoglycan complex (DSC) resides in the plasma membrane and links intracellular actin with extracellular laminin to couple mechanical signaling (6). Loss of dystrophin dissolves the entire complex, and increases muscle cell susceptibility to stretch-induced injury (7–9) coupled to increased intracellular Ca2+ entry (9, 10). The exact source of excess intracellular Ca2+ remains subject to debate, and has been attributed to membrane leaks; promiscuous release from internal Ca2+ stores; and enhanced activity of plasma membrane stretch-activated nonselective cation channels, including the transient receptor potential channels Trpc1, Trpc6, and Trpv2 (11–17). Of these, Trpc6 has been identified as particularly important in mechanosensing in DMD myocardium and isolated myocytes, with selective pharmacological suppression or Trpc6 gene deletion normalizing mechanosensitive Ca2+ and force responses in juvenile mice aged 2–3 months of age (12).
DMD muscle pathophysiology also involves changes in intracellular nitrosative stress (18). Neuronal NOS (NOS1) binds to α-syntrophin, an intracellular member of the DSC; with loss of the complex in DMD, NOS1 disappears from the plasma membrane, and its delocalization is proposed to disrupt nitric oxide (NO) signaling (19). In the heart, disruption also involves endothelial NOS (NOS3), the dominant isoform localized to the plasma membrane (20). Enhanced nitrosative stress (nitrotyrosine) in DMD skeletal muscle has been linked to reduced NOS1-plasma membrane localization (18). Other studies found increased S-nitrosylation of both the skeletal and cardiac muscle sarcoplasmic reticulum (SR) Ca2+ release channel in murine DMD (21, 22), which was proposed to cause promiscuous Ca2+ leaks. The specific cysteine residues modified are unknown, and broad proteomic-based analysis of S-nitrosothiol (SNO) in DMD has not yet been reported.
Abnormal Ca2+ regulation in DMD may itself contribute to disruption of NO signaling and downstream S-nitrosylation, as Ca2+ is required for both NOS1 and NOS3 activation. Both NOS isoforms are expressed in the heart, where they play important roles in regulating Ca2+ homeostasis by direct NO-protein modification and by activation of cyclic guanosine monophosphate and, in turn, protein kinase G (23). Interactions between these pathways are known, but how abnormal Ca2+ sources and/or handling is relevant to the loss of dystrophin in DMD and if and how such Ca2+ broadly impacts protein S-nitrosylation are not.
Here, we employed a robust proteomic approach to identify site-specific S-nitrosylation in cardiac proteins, and found that this modification is augmented in a DMD mouse model (dmdmdx:utrn+/−) at similar cysteine residues as those observed in healthy controls. Trpc6 deletion improved this cardiac dysfunction and histopathology, and partially reversed S-nitrosylation. We further examined the impact of adrenergic stimulation on S-nitrosylation and the role of Trpc6.
Results
Myocardial S-Nitrosylation Targets Are Shared with Wild Type but Increased in Level in dmdmdx:utrn+/− Mice.
Using a dual biotin switch protocol (24) (Fig. 1), we identified 1,166 S-nitrosylated amino acid residues that corresponded to 471 unambiguously identified proteins in control hearts (n = 4; Dataset S1). These proteins were predominantly in the sarcomeres or part of either mitochondrial or metabolic pathways. In a parallel analysis of dmdmdx:utrn+/− myocardium (n = 5), we found 1,276 S-nitrosylated cysteine residues representing 491 proteins. These included cysteine residues such as ryanodine receptor 2 (RyR2) at Cys158, Cys1282, Cys2520, Cys3836, and Cys4956.
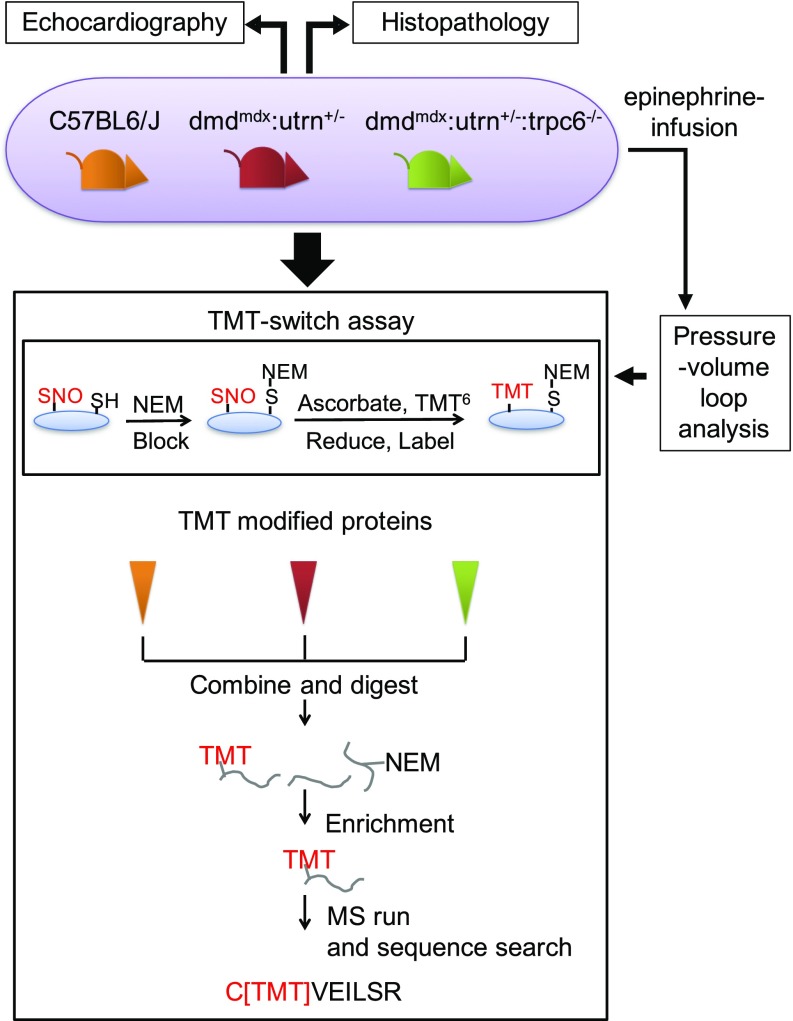
Fig. 1.
Multiple experimental approaches were employed to search for a linkage between Trpc6, nitrosative stress, and the pathobiology of DMD in the dmdmdx:utrn+/− mice. To capture protein S-nitrosylation, by an MS-coupled dual-labeling strategy, protein mouse heart lysates were labeled with multiplex cys- or iodoTMT6. Samples processed in the absence of ascorbate served as negative controls. Modified proteins were digested, enriched, desalted, and analyzed using MS. C[TMT]VEILSR, an example of TMT-labeled peptides; SH, free cysteine; S-NEM, cysteine blocked with N-ethylmaleimide (NEM).
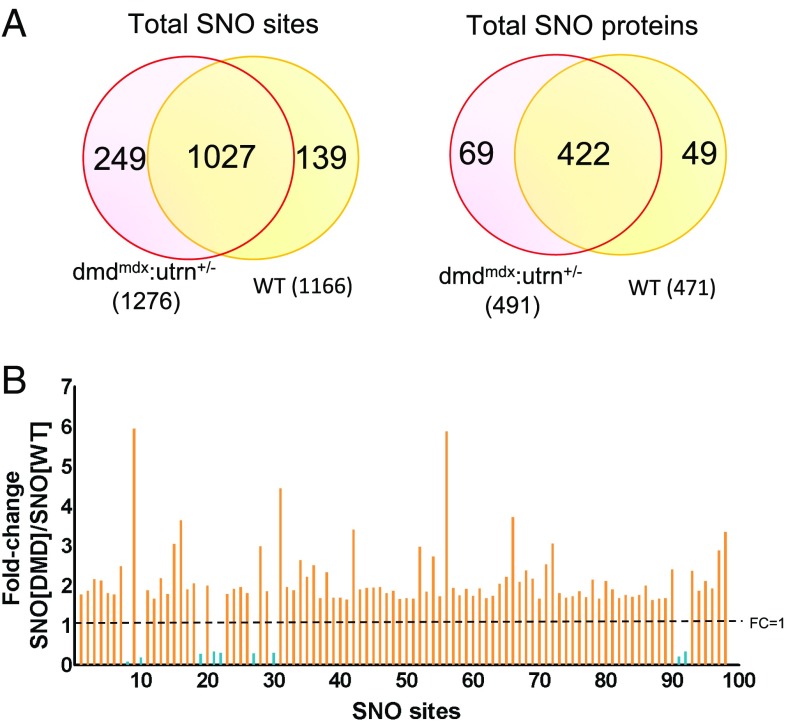
As shown by a Venn diagram (Fig. 2A), 1,027 (80% of total) dmdmdx:utrn+/− modification sites were commonly identified in control (Fig. 2A). At the protein level, 69 and 49 proteins were exclusively measured in dmdmdx:utrn+/− and control, respectively, while 422 proteins were in common (Fig. 2A). Gene ontology analysis of the 422 common proteins revealed that they were highly enriched for oxidoreductase activity (P value: 4.60E-31), whereas this was less strongly represented in the 69 proteins uniquely modified in dmdmdx:utrn+/− myocardium (P value: 2.30E-02; Fig. S1 and Dataset S2).
Fig. 2.
Hypernitrosylation of sites found in dmdmdx:utrn+/− cardiac proteins. (A) Bioinformatics identified 1,276 S-nitrosylated residues in dmdmdx:utrn+/− cardiac proteins, and these individual sites (Left) and related proteins (Right) displayed marked commonality between the WT and dmdmdx:utrn+/−. (B) Fold change (FC) in S-nitrosylation level at shared sites between WT and dmdmdx:utrn+/− that exceeded the coefficient of variation (either higher than 1.64 or lower than 0.36) and was detected in at least two biological replicates. Of these 98 sites, the clear majority were more frequently detected in dmdmdx:utrn+/− than in WT [hypernitrosylated (orange), hyponitrosylated (blue)].
We next probed whether the level of S-nitrosylation differed among the commonly identified cysteine residues. To determine this, we compared the fold change between the diseased versus healthy state by comparing the intensity of tandem mass tag (TMT) reporter ions generated by isobaric TMT tags. We further restricted our analysis to those sites that were detected in at least two biological replicates, and for which the difference between dmdmdx:utrn+/− and control exceeded the average coefficient of variation derived from repeated measures for that site. These criteria yielded ∼100 cysteine residues (Dataset S3A). Strikingly, we found that ∼90% of these residues showed intensified S-nitrosylation in dmdmdx:utrn+/− versus control wild type (WT) (Fig. 2B). This list included Cys173 of peroxiredoxin-1 (Prdx1) and Cys190 of tropomyosin α1 (Tpm1). Only nine residues showed a decline in S-nitrosylation in dmdmdx:utrn+/−, including two cysteines in sarco/endoplasmic reticulum Ca2+-ATPase (SERCA2; Atp2a2). Pathway analysis was next performed to better understand these two modification groups (hyper-SNO versus hypo-SNO), for which we loosened the inclusion criteria to accept detection in one biological sample to enhance the cluster analysis (Dataset S3B). The two groups similarly included proteins involved in mitochondrial function (e.g., glycolysis, oxidative phosphorylation) and actin cytoskeletal signaling (e.g., actin, myosin-6, myosin-binding protein C) (Fig. S2 and Dataset S3B).
Gene Deletion of Trpc6 Reverses Most S-Nitrosylation Found in dmdmdx:utrn+/−.
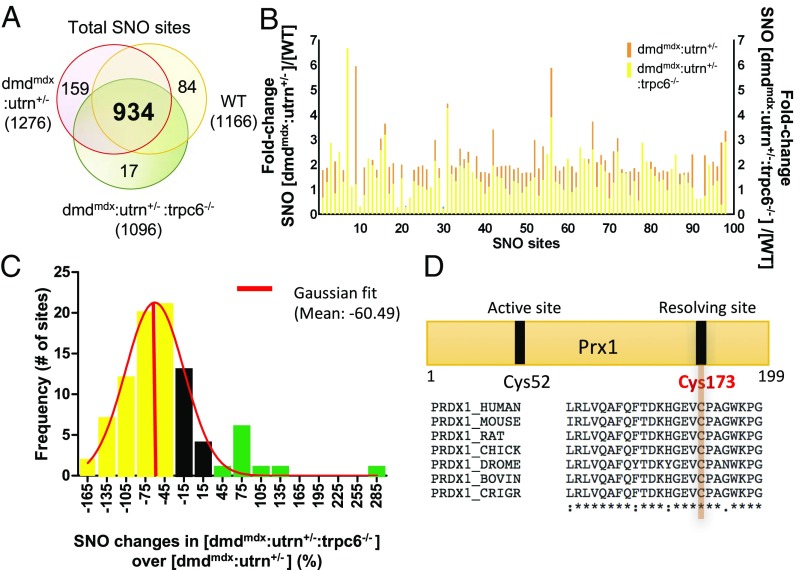
We next tested whether deletion of Trpc6 modified the S-nitrosylation profile in the dmdmdx:utrn+/− heart. Myocardium from Trpc6-deficient dystrophic mice (n = 5) yielded 1,096 S-nitrosylated residues, most of which were again overlapping (Fig. 3A) but different in magnitude of modification (Fig. 3B) compared with the dmdmdx:utrn+/− and healthy myocardium according to a three-way comparison. Eighty-five percent of these S-nitrosylated cysteine residues were shared among all groups, with <2% (17 sites) modified solely in the dmdmdx:utrn+/−:trpc6−/− (Fig. 3A). Interestingly, only seven proteins were unique to dmdmdx:utrn+/−:trpc6−/− hearts (Fig. S3), and these included calpain-2, talin-1, dead box protein 6, histone h3.2, cellular nucleic acid-binding protein, α2-macroglobulin, peroxin-19, and syndapin-2 (Dataset S1). We noted that these proteins are mostly located in the plasma membrane and involved in cell adhesion.
Fig. 3.
Trpc6 deletion reverses many SNO changes in dmdmdx:utrn+/−. (A) Three-way comparison of S-nitrosylation between WT, dmdmdx:utrn+/−, and dmdmdx:utrn+/−:trpc6−/−. More than 78% of sites were detected in all three models. (B) Fold change in S-nitrosylation level detected at a given site comparing dmdmdx:utrn+/− with dmdmdx:utrn+/−:trpc6−/−. Most the sites had less S-nitrosylation detected if Trpc6 was genetically deleted. (C) Histogram of the frequency of percent change in S-nitrosylation between dmdmdx:utrn+/− and dmdmdx:utrn+/−:trpc6−/−. The average of SNO changes in dmdmdx:utrn+/−:trpc6−/− over dmdmdx:utrn+/− was −46.95%; 70% were reversed by 30% or more with Trpc6 deletion (yellow), and 10 sites were increased further by 30% or more (green). The mean of best-fit Gaussian distribution was −60.49%. (D) peroxiredoxin-1 was selected as an example of a potential therapeutic target of Trpc6-dependent SNO proteins. The Cys173 of peroxiredoxin-1 is highly conserved across species, and this residue reacts with Cys52 to form a dimer, which affects the enzymatic activity of peroxiredoxin-1. The Cys173 residue was S-nitrosylated 1.9-fold more in dmdmdx:utrn+/− than control and was restored to normal by >43% in dmdmdx:utrn+/−:trpc6−/−.
Deletion of Trpc6 quantitatively reversed a majority of hyper-SNO residues found in dmdmdx:utrn+/−, restoring modification levels closer to the healthy myocardium (Fig. 3B and Table 1). A histogram of percent change in S-nitrosylation between dmdmdx:utrn+/− and dmdmdx:utrn+/−:trpc6−/− shows that more than two-thirds was reversed by 30% or more in dmdmdx:utrn+/−:trpc6−/− myocardium (Fig. 3C and Table 1). Nearly 80% of sites that were hyponitrosylated in dmdmdx:utrn+/− were also reversed (P = 0.07; Table 1). Thus, removal of Trpc6, a source of excess Ca2+ in dmdmdx:utrn+/−, led to amelioration of broad-based SNO abnormalities in the myocardium (Dataset S3A). One of the modifications involved the antioxidant protein peroxiredoxin-1 at Cys173 (Fig. 3D). This residue is highly conserved and is a critical cysteine residue in forming oxidized dimeric peroxiredoxin-1 that impacts its enzymatic activity (25–27). S-nitrosylation at this site was increased twofold in the dmdmdx:utrn+/− compared with the healthy control, and nearly restored to normal in dmdmdx:utrn+/−:trpc6−/−; percent changes compared with control were 91% and 52% in dmdmdx:utrn+/− and dmdmdx:utrn+/−:trpc6−/−, respectively.
Table 1.
Number of SNO sites different at baseline
| Category | No. SNO sites | Change in dmdmdx:utrn+/− vs. WT | No. SNO sites (%) | Change in dmdmdx:utrn+/−:trpc6−/− vs. dmdmdx:utrn+/− | No. SNO sites (%) |
| SNO sites selected with stringent criteria | 98 | Hyper-SNO | 89 (91%) | Reversed by ≥30% with Trpc6 deletion | 62 (70%) |
| Increased ≥30% with Trpc6 deletion | 10 (11%) | ||||
| Unchanged | 17 | ||||
| Hypo-SNO | 9 (9%) | SNO increased with Trpc6 deletion | 7 | ||
| SNO further decreased with Trpc6 deletion | 2 |
Trpc6 Deletion Improves dmdmdx:utrn+/− Cardiac Dysfunction and Remodeling.
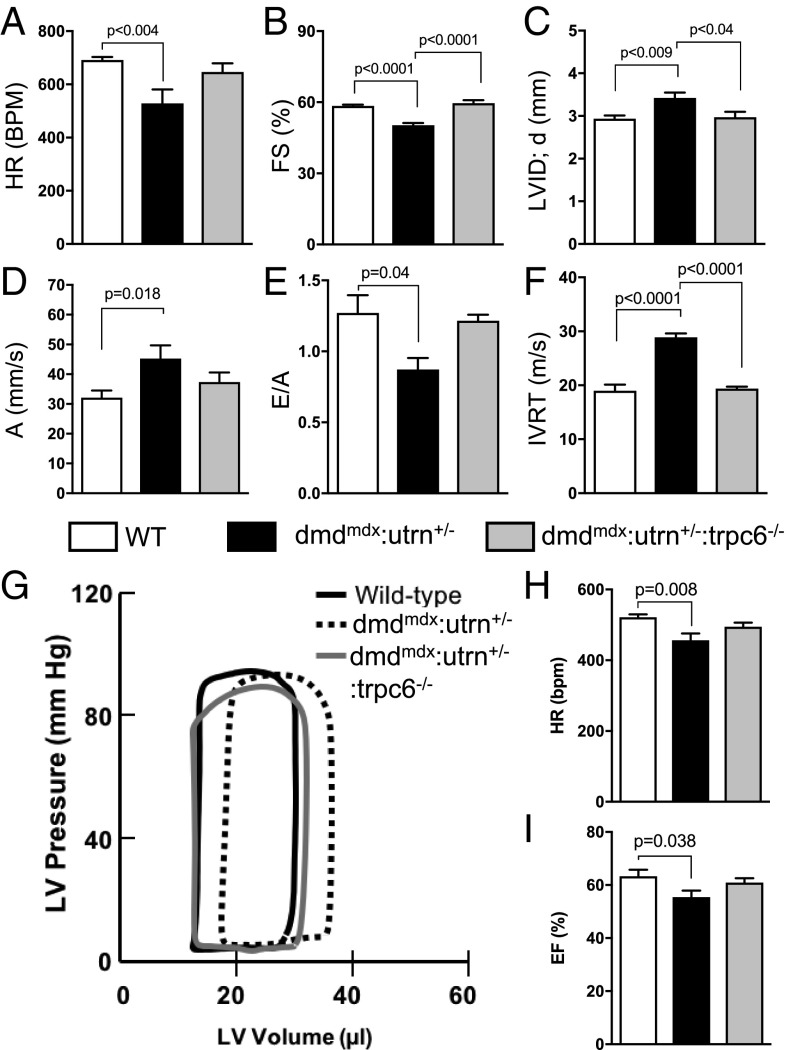
To determine the functional ramification of the reversal in the SNO proteome observed with Trpc6 deletion, we next examined cardiac function, morphology, and selected molecular signaling in the three groups (Fig. 1). Consistent with prior reports (12, 28, 29), we found that dmdmdx:utrn+/− hearts have a slowed heart rate, and that the left ventricle (LV) is dilated with a reduction in fractional shortening (Fig. 4 A–C). These changes were reversed by genetic deletion of Trpc6. The dmdmdx:utrn+/− hearts also displayed abnormal diastolic dysfunction, all of which were again reversed with Trpc6 deletion (Fig. 4 D–F). Invasive pressure-volume analysis (Dataset S4) of the dmdmdx:utrn+/− confirmed bradycardia and reduction in ejection fraction (Fig. 4 G–I) with greater end-diastolic volume.
Fig. 4.
LV dilatation increases preload to depress dmdmdx:utrn+/− cardiac function. (A–F) In vivo cardiac function assessed by echocardiography. A, atrial contraction tissue velocity; BPM, beats per minute; E/A, early-to-atrial filling ratio; FS, fractional shortening; HR, heart rate; IVRT, isovolumic ventricular relaxation time; LVID;d, LV diastolic dimension. Pathological changes in all parameters were restored with gene deletion of Trpc6. (WT, n = 8; dmdmdx:utrn+/− dystrophy model, n = 6; and dmdmdx:utrn+/−:trpc6−/− dystrophy model lacking Trpc6, n = 4.) (G–I) Representative pressure-volume loop traces and summary data at rest. The HR and ejection fraction (EF) were reduced in dmdmdx:utrn+/− and restored by Trpc6 deletion. (n = 13, n = 19, and n = 17 for the three groups, respectively.)
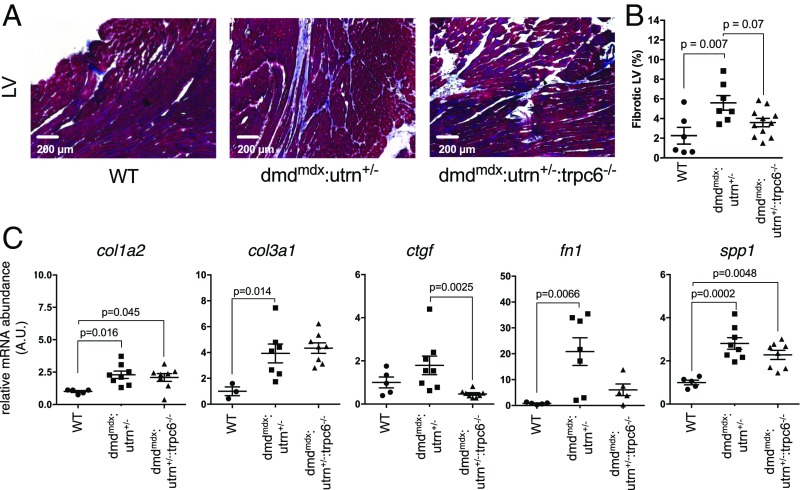
Histopathological study revealed a severe myocardial fibrosis in dmdmdx:utrn+/− that was partially reversed by deleting Trpc6 (Fig. 5 A and B). Gene expression of collagen 1a and 3a (col1a2 and col3a1) and profibrotic proteins [fibronectin1 (fn1) and osteopontin1 (spp1)] was increased in dmdmdx:utrn+/− myocardium (Fig. 5C). Although connective tissue growth factor (ctgf) was not significantly increased in the dystrophic heart, Trpc6 gene deletion reduced its expression but did not significantly alter the other transcripts. Taken together, these data show Trpc6 gene deletion improved contractile dysfunction, chamber remodeling, and fibrosis, all in association with its restoration of a more normal broad S-nitrosylated proteome.
Fig. 5.
Trpc6 deletion reverses fibrosis in dmdmdx:utrn+/− hearts. (A) LV myocardium stained with Masson’s trichrome shows increased fibrosis in dmdmdx:utrn+/− mice. This tended to decline with Trpc6 deletion. (B) Summary data (one-way ANOVA, with Tukey’s post hoc test; n = 5–7 per group.) (C) Gene expression for collagen types 1A and 3A (col1a2 and col3a1), ctgf, fn1, and spp1. Most were elevated in dmdmdx:utrn+/−, with Ctgf declining in dmdmdx:utrn+/−:trpc6−/− (one-way ANOVA, with Tukey’s post hoc test, for col1a2 and fn1; Kruskal–Wallis test for col3a1, ctgf, and spp1; n = 5–7 per group.) A.U., arbitrary unit.
Impact of Adrenergic Stimulation/Stress on Dystrophic SNO Modulation.
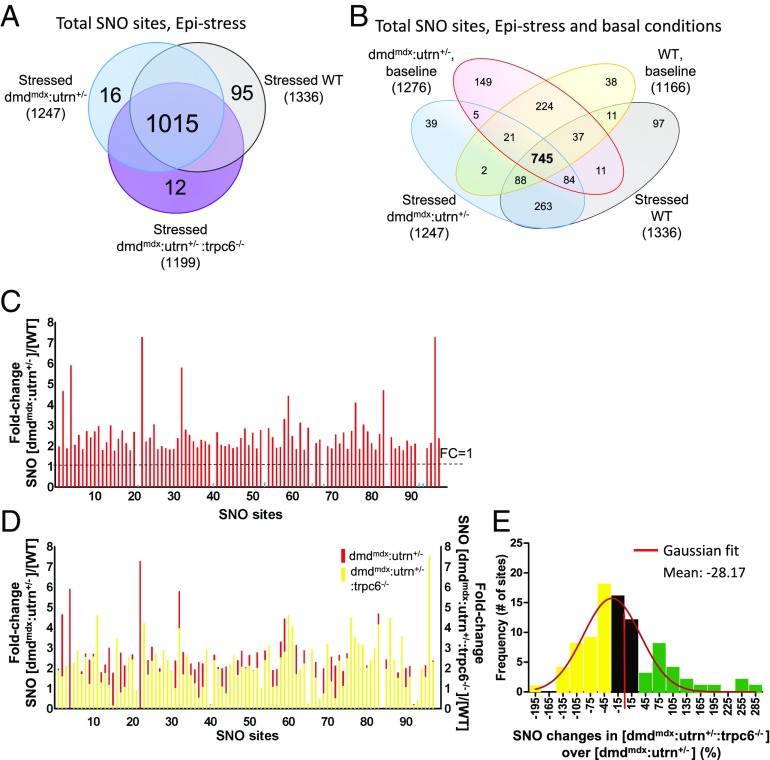
The preceding results were all obtained in hearts at rest. However, myocardial Ca2+, NO, and workload are enhanced with combined α- and β-adrenergic stimulation as with exercise (30, 31), and we speculated that such stress might further amplify SNO disparities between control and dmdmdx:utrn+/− (Fig. 1). To test this, we exposed mice to 10 min of epinephrine (2 μg⋅kg−1⋅min−1, i.v.) to engage α- and β-adrenergic pathways as a simulation of exercise. Hearts were again harvested in the dark, and proteomic analysis was performed. All three models displayed substantial increases in minute work (stroke work × heart rate) compared with baseline (WT: 76 ± 36%; dmdmdx:utrn+/−: 75 ± 39%; dmdmdx:utrn+/−:trpc6−/−: 107 ± 24%; P = 0.8, one-way ANOVA). In this state, 1,247, 1,199, and 1,336 S-nitrosylated residues, respectively in dmdmdx:utrn+/−, dmdmdx:utrn+/−:trpc6−/−, and control hearts were identified (Fig. 6A and Dataset S5). The majority of the modified cysteine residues were detected in all models, with 95% of sites in dmdmdx:utrn+/− being identical to those identified in WT and 99% shared with dmdmdx:utrn+/−:trpc6−/− or WT (Fig. 6 A and B). Seventy-two percent of a pool of these residues in WT and dmdmdx:utrn+/− were also found in baseline (Fig. 6B). As under rest conditions, most residues found in dmdmdx:utrn+/− were hypernitrosylated over control in the presence of epinephrine (Fig. 6C and Dataset S6). However, in this setting, Trpc6 gene deletion reversed fewer of these changes in dmdmdx:utrn+/− (Fig. 6D and Table 2) than it had done under rest conditions (44% versus >70%; P < 0.005, Fisher’s exact χ2 test; Figs. 3C and 6E). The average magnitude of decline was −4% under stress versus −47% at rest (P < 0.001; Figs. 3C and 6E). These data indicate that alternative Ca2+- and G protein-coupled NOS stimulation and/or altered NO chemistry occurs under sympathetic stimulation that, while still resulting in greater S-nitrosylation in dmdmdx:utrn+/−, reduces the relative contribution of Trpc6.
Fig. 6.
Influence of epinephrine (Epi) stimulation on SNO proteome in three experimental groups. (A) Venn diagram shows total number of SNO sites identified in Epi-stressed control, dmdmdx:utrn+/− and dmdmdx:utrn+/−:trpc6−/−. (B) Four-way comparison of SNO sites observed in WT and dmdmdx:utrn+/− myocardium ± Epi. (C) Fold change in S-nitrosylation level comparing dmdmdx:utrn+/− with WT. Overall, SNO changes were increased at similar sites in dmdmdx:utrn+/− in the presence of Epi. (D) Fold change in S-nitrosylation level comparing dmdmdx:utrn+/− (red) with dmdmdx:utrn+/−:trpc6−/− (yellow). While some S-nitrosylation levels declined in individual sites, the overall effect was more modest than observed without Epi. (E) Histogram of the relative percent change in S-nitrosylation between dmdmdx:utrn+/− and dmdmdx:utrn+/−:trpc6−/−. Among 90 hyper-SNO sites, 44% were reversed by 30% or more with Trpc6 deletion (yellow), and 22 sites were increased further by 30% or more (green).
Table 2.
Number of SNO sites different with epinephrine-stimulation
| Category | No. SNO sites | Change in dmdmdx:utrn+/− vs. WT | No. SNO sites (%) | Change in dmdmdx:utrn+/−:trpc6−/− vs. dmdmdx:utrn+/− | No. SNO sites (%) |
| SNO sites selected with stringent criteria | 97 | Hyper-SNO | 90 (93%) | Reversed by ≥30% with Trpc6 deletion | 40 (44%) |
| Decreased ≥30% with Trpc6 deletion | 22 (24%) | ||||
| Unchanged | 28 | ||||
| Hypo-SNO | 7 (7%) | SNO level increased with Trpc6 deletion | 5 | ||
| SNO level further decreased with Trpc6 deletion | 2 |
Bioinformatic Synthesis of SNO Networks.
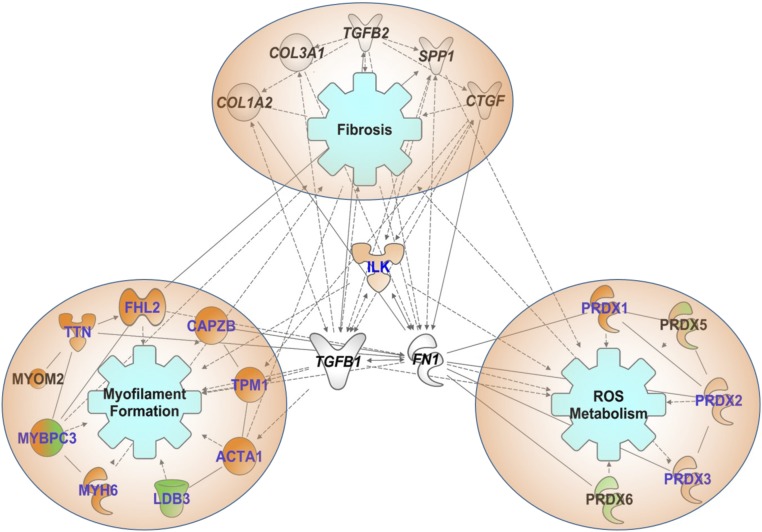
To better understand broad SNO changes at a systems level, we compared the 1,000+ SNO sites identified in dmdmdx:utrn+/−, dmdmdx:utrn+/−:trpc6−/−, and WT using protein network analysis. This analysis also included gene expression data related to tissue fibrosis, and identified relationships between S-nitrosylated proteins in the dmdmdx:utrn+/− and dmdmdx:utrn+/−:trpc6−/− myocardium with/without epinephrine stimulation. Under rest conditions, reactive oxygen species (ROS) metabolism, fibrosis, and myofilament proteins emerged as interrelated networks (Fig. 7). Similar networks were identified under epinephrine stimulation, although the specific sites modified in each protein were different between rest and stressed conditions (Dataset S7).
Fig. 7.
Bioinformatics analysis provided plausible associations between selected SNO proteins and fibrosis pathways. Proteins that showed SNO change in dmdmdx:utrn+/− were analyzed. Filled shapes indicated the following: proteins where a (or most of the) site(s) was/were hyper-SNO in dmdmdx:utrn+/− (dark orange); proteins where a (or most of the) site(s) was/were hypo-SNO in dmdmdx:utrn+/− (dark green); hyper-SNO or hypo-SNO, but the change did not exceed the average coefficient of variation (light orange or light green); some sites were hyper-SNO and others were hypo-SNO on that protein (mixed orange/green); fibrosis markers confirmed in the level of transcriptome (no filling). Letters in blue indicate SNO on (a) site(s) of the protein was/were reversed with Trpc6-deletion. Of the enriched functional categories, networks of myofilament proteins, multiple isoforms of Prdxs and ILK were selected as being of particular interest. In addition, based on our study on fibrosis, fibrotic markers were included for internetwork generation. Proteins where this network analysis provided the associations between the SNO genes that might affect fibrosis pathways directly/indirectly in DMD pathophysiology are shown.
Discussion
The current study provides insight into the cardiac pathophysiology of DMD using aged (∼1 y) dmdmdx:utrn+/− mice, finding evidence for Trpc6-dependent, disease-associated hypernitrosylation of myocardial proteins. S-nitrosylated residues were mostly shared between normal and dmdmdx:utrn+/− hearts, but with important exceptions (e.g., RyR, where multiple sites were identified). This was also true in the presence of epinephrine stimulation, where Trpc6 deletion played a less prominent role. These findings appear at odds with the view that NOS delocalization in the DMD skeletal muscle would result in promiscuous S-nitrosylation (18). There are a number of possible explanations for our finding. One is that the set of targetable cysteine residues is selective, related to the chemistry around the cysteine and its probability for S-nitrosylation. Our previous study has shown that there is a range of sensitivity for different cysteines for S-nitrosylation, such that different amounts of NO are required to induce this modification at each cysteine residue (32).
Amplification of S-nitrosylation in dmdmdx:utrn+/− myocardium was associated with abnormalities in cardiac function and structure, suggesting a deleterious role for increased S-nitrosylation in dystrophic cardiomyopathy. S-nitrosylation has been previously suggested to be protective, particularly in the setting of ischemic injury or other oxidative damage, shielding specific cysteine residues from irreversible oxidation (33). Nevertheless, excessive S-nitrosylation is thought to contribute to disease pathogenesis, as in neurodegenerative diseases, including Alzheimer’s and Parkinson’s diseases (34, 35). In a similar fashion, increased S-nitrosylation has been linked to arrhythmias in focused studies examining RyRs in DMD muscles (21, 22). Our results provide evidence in further support of this prediction at the systems level, indicating that S-nitrosylation may be deleterious in DMD.
SNO targets have scarcely been studied for changes in the DMD setting in particular, but RyRs have been a notable exception (21, 22). Direct S-nitrosylation of the RyR2 results in increased channel open probability in vitro (36), and this has been suggested to cause Ca2+ leakiness and trigger arrhythmias in the dystrophic heart (22). In this previous report (22), no modified cysteine residues were determined in the cardiac isoform RyR2. In the skeletal muscle RyR1, Cys3635 is the only site reported to be functionally relevant, as S-nitrosylation on the cysteine residue contributed to the calmodulin-dependent effect of NO on the channel in vitro (37). Our study detected 11 SNO sites on RyR2, including Cys158, Cys1282, Cys2520, and Cys4956. Cys4956 is in the C terminus, which includes the 12.6-kDa FK506-binding protein (FKBP12.6)-binding site (38). Nine of the 11 residues were not detected in controls but only in dmdmdx:utrn+/−, while Cys36 and Cys3836 were detected in both dmdmdx:utrn+/− and control hearts and declined in dmdmdx:utrn+/− compared with WT. Functional ramifications of these alterations warrant further study, and may reveal important ramifications for Ca2+ signaling. Such findings would be intriguing for a possible interaction with Trpc6 hyperactivity in DMD (12) to bring about disease pathology.
The decline in SNO magnitude found in dmdmdx:utrn+/− hearts lacking Trpc6 supports an important link between this channel and dysregulated S-nitrosative chemistry. Trpc6 conducts calcium sixfold more than sodium, and as only Ca2+ activates NOS, this is a likely mechanism linking these proteins. By deleting Trpc6, a pathological source of Ca2+ is removed (12), which could rebalance the NO-ROS axis, reducing SNO levels. The current data support a potential therapeutic value of such agents in DMD, and would warrant the development of specific Trpc6 inhibitors. There is also potential negative feedback on TRPC6 due to enhanced NOS activity generating NO, as cGMP-stimulated protein kinase G (PKG) phosphorylation of Trpc6 reduces channel activity, which is particularly notable in the same dystrophic model (12). Trpc6 is hyperactivated at baseline, however, and blockade by PKG required exogenous stimulation of the kinase, so the net balance does not favor the negative feedback. Lastly, it is feasible that S-nitrosylation of Trpc6 occurs, although this was not detected in the proteomic analysis, perhaps due to limitations in the dynamic range of the assay.
The majority of proteins with increased SNO in dmdmdx:utrn+/− myocardium were primarily mitochondrial or sarcomeric proteins. This is not surprising given that the heart tissue largely consists of myocytes, but the specifics are worth considering. Mitochondrial proteins included members of the oxidoreductase family (dehydrogenases, oxidases, and oxygenases), and may reflect metabolic alterations that are early features of dystrophic myocardium (39–41). Increased fibrosis is also a hallmark of DMD (42), and in general oxidative stress is associated with increased fibrogenic gene expression and fibrosis. This signaling was abnormal in dmdmdx:utrn+/− and was partially reversed by deleting Trpc6, so we further examined Trpc6-dependent SNO antioxidant proteins. We detected substantial S-nitrosylation at Cys173 of peroxiredoxin-1 in DMD that was reduced in hearts lacking Trpc6. This residue plays key antioxidant roles that determine the enzyme’s peroxide activity. The N-terminal Cys52 is oxidized to form a disulfide bond with the Cys173 in adjacent peroxiredoxin-1 to form a dimer (27). This is a requisite step for protein enzymatic activity and localization (25–27). A prior study found S-nitrosylation at Cys52 promotes structural and functional changes of peroxiredoxin-1 (26). The study also found S-nitrosylation occurs at Cys173, but at low SNO occupancy (26). Another intriguing target is integrin-linked protein kinase (ILK), which also plays a role in pathological fibrosis and mediates TGF-β1–initiated fibronectin expression (43). ILK also links the actin-based cytoskeleton and can phosphorylate myosin (44), suggesting it may play a role in muscle contraction (44) and affect sarcomere proteins (45). S-nitrosylation on cysteines in actin, myosin-6, myosin-binding protein C, four-and-a-half LIM domains protein 2, Tpm1, and myomesin-2 were elevated in dmdmdx:utrn+/− and reversed by Trpc6 deletion, although the functional role of these changes remains to be determined.
Epinephrine-stimulated hearts also displayed amplified S-nitrosylation in the dmdmdx:utrn+/− (compared with stimulated WT) as observed in resting hearts. Interestingly, unlike the resting condition where removing Trpc6 had a major impact, this became less notable in the presence of adrenergic stimulation. This may be because enhanced Ca2+ influx via L-type Ca2+ channels and SR release (46) are themselves coupled to NOS activation (47), and these alternative sources overwhelm changes deriving from Trpc6. In addition, these results suggest that loss of dystrophin may disrupt β-adrenergic receptor localization or sensitize the β-adrenergic signaling so as to increase S-nitrosylation in response to an endogenous stimulation of this pathway. The same outcome may be expected if denitrosylation was regulated in the dystrophic hearts (48). Although the mechanism remains elusive, our results may explain why the use of β-blockers exerts a preventative effect in patients who have DMD (49).
This study provides the largest unbiased proteomics detection of S-nitrosylation in the dystrophic myocardium to date, and also specifically links a Ca2+ channel to abnormal mechano- and hormone-stimulated cation entry in a dmdmdx:utrn+/− model of DMD with S-nitrosylation. Unlike prior work, where S-nitrosylation was manipulated artificially using S-nitrosoglutathione (GSNO) or other direct S-nitrosylation modifiers, the changes in the present study are intrinsic to the dystrophin/utrophin genetic modulation and consequent disease. Our study is nevertheless limited in that the specific functional impact of individual S-nitrosylation remains to be determined. To limit the false-positive detection of disulfides that may be observed in a classical biotin-switch assay (50, 51), low concentrations of ascorbate were used in our study. Forrester et al. (52) suggested that ascorbate-dependent reduction of disulfides is thermodynamically unfavorable and does not contribute to the SNO signal. We have previously monitored this in a prior study and found that less than 3% of the SNO proteome can be attributed to disulfide bonds (32). Taking titin as an example in our study, in which we found the largest number of S-nitrosylated cysteine residues (163 sites), we saw that less than 10% (16 sites) were known to be potential disulfide pairs according to the UniProt annotation database. We also included controls that were not treated with ascorbate, and thus not chemically reduced. This was done for all samples in parallel to ensure that we would exclude false-positive results.
In conclusion, myocardium in healthy animals displays extensive S-nitrosylation, and this is broadly enhanced in dmdmdx:utrn+/−. Such hyper-SNO is impacted by the nonselective cation channel Trpc6, and deleting this channel in dmdmdx:utrn+/− reverses many of the S-nitrosylation changes and improves cardiac pathobiology. As newer methods are being explored to replace or edit the mutant dystrophin gene, S-nitrosylation profiling could be a relatively easy way to detect if the underlying molecular signaling has been restored toward normal. Our studies also broaden the impact of Trpc6 on DMD pathophysiology and support ongoing efforts to develop specific inhibitors that can be used for chronic studies in vivo.
Materials and Methods
Animals.
Male mice hemizygous for the dystrophin-truncating mutation mdx and heterozygous for utrophin deletion (dmdmdx:utrn+/−), or those homozygously deficient also in Trpc6 (dmdmdx:utrn+/−:trpc6−/−) (12), and WT controls were used. Mice were bred as reported (12); all mice were on a C57BL6/J background. The average age of all mice included in the study was 50.2 ± 1.4 wk. All studies were conducted in accordance with the NIH Guidelines for the Care and Use of Laboratory Animals (53) and were reviewed by the Institutional Animal Care and Use Committee at Johns Hopkins University.
TMT-Switch Assay and Proteomics Analysis.
The proteomic assay workflow is highlighted in Fig. 1. Data were obtained in mice under resting conditions [n = 5 per cohort, except WT (n = 4)], as well as in animals stimulated with epinephrine (0.5–2.0 μg⋅kg−1⋅min−1, n = 5 per cohort) to increase cardiac minute work. A separate set of sample isolations was then performed under these work conditions. To assess SNO proteins, LV tissue is extracted, and homogenates are then generated in the dark and subjected to a dual-labeling TMT-switch assay (24). Briefly, ice-cold homogenization buffer containing sucrose (300 mM), PBS (pH 8.0), EDTA (1 mM), neocuproine (0.1 mM), N-ethylmaleimide (NEM; 20 mM), and an EDTA-free protease inhibitor tablet (Roche) is used. Lysates are centrifuged for 2 min at 1,500 relative centrifugal force (rct), and samples are blocked with 20 mM NEM in the presence of 2.5% (wt/vol) SDS and incubated for 20 min at 50 °C. Excess NEM is removed by a Zeba desalt spin column (Thermo Fisher) equilibrated with a buffer of PBS (pH 8.0), EDTA (1 mM), and neocuproine (0.1 mM). The resulting samples are divided and incubated with either cysTMT (Thermo Fisher) or iodoTMT (Thermo Fisher) to enhance SNO proteome coverage. Samples are prepared with or without ascorbate to yield dual-labeled samples and controls, respectively, minimizing false-positive results (24). Following labeling of SNO sites, proteins are digested, enriched, cleaned, and analyzed by mass spectrometry. SNO site identification and quantification of the modification are as previously described (24) and the Ingenuity Pathway Analysis (QIAGEN) tool (www.qiagen.com/ingenuity) and DAVID bioinformatics resources (ver. 6.8) (54, 55) are used for bioinformatics analysis. An extended description of methods used is available in SI Materials and Methods.
Echocardiography.
In vivo cardiac geometry and function were assessed by transthoracic echocardiography in both M-mode and Doppler imaging modalities (Acuson Sequoia C256, 13-MHz transducer; Siemens). Heart rate, LV fractional shortening, and LV end-diastolic inner diameter were calculated or determined from an average of three to five cardiac cycles from the M-mode (12). Diastolic function was assessed by A-wave tissue Doppler velocity, early/atrial filling ratio, and isovolumic relaxation time. All parameters were determined by an observer blinded as to condition.
Pressure-Volume Loop Analysis.
Hemodynamics were measured in anesthetized mice (1–2% isoflurane), using a pressure-volume catheter (PVR-1030; Millar, Inc.) (12). Data were recorded under resting steady-state conditions, and included brief periods in which the inferior vena cava was occluded to measure pressure-volume relations. Data were digitized at 2 kHz using PowerLab (AD Instruments), and analyzed with LabChart 8.0 (AD Instruments) and custom software (WinPVAN3.0).
Tissue Collection and Histology.
Ventricular tissue from the midchamber was harvested, embedded in optimal cutting temperature (OCT), and frozen in liquid nitrogen. Sections were then stained with Masson’s trichrome, and each cross-section was divided into four quadrants and imaged under a magnification of 2.5×, with fibrosis quantified by the percentage of blue stain per total area of tissue (ImageJ; NIH). Two cross-sectional micrographs were imaged per animal.
RNA Isolation and RT-qPCR.
TRIzol (Ambion) was used to extract total RNA, which was subsequently transcribed into cDNA using a High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher). Transcript expression level was measured using Taqman probes: GAPDH (Mm99999915_g1), Col1a2 (Mm00483888_m1), Col3a1 (Mm01254476_m1), Ctgf (Mm01192932_g1), Fn1 (Mm01256744_m1), and Spp1 (Mm00436767_m1) (all probes were from Applied Biosystems). Relative change in expression level was normalized to GAPDH by using the relative change in cycle threshold (ΔΔCT) method.
Statistical Analysis.
Where indicated, an unpaired t test with two tails or a one-way ANOVA test was performed for significance between data showing a normal distribution and equal variance among groups. Otherwise, a Kruskal–Wallis nonparametric test was used. For post hoc analysis, we used a Tukey–Kramer test or Mann–Whitney U test as appropriate. The Fisher’s exact test was used for two-by-two comparisons. Analysis was performed with SigmaStat (version 13). Data are plotted as mean ± SEM using GraphPad Prism. A P value of ≤0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
This work was supported by an American Heart Association MidAtlantic Fellowship Grant (to H.S.C.); T32 Grant T32-HL-7227 (to G.E.K.); American Heart Association Go Red for Women Network Grant 16SFRN27870000 (to D.A.K.); National Institute of Health (NIH) Grants HL131358 and R35-HL135827 (to D.A.K.); Muscular Dystrophy Association Grant 186454 (to D.A.K.); National Heart, Lung, and Blood Institute (NHLBI) Grants R01HL119012, P01HL10026, and 5P01HL112730-03 (to J.E.V.E.); the Erika J. Glazer chair in Women's Heart Health (J.E.V.E.); and the Barbra Streisand Women's Heart Center (J.E.V.E.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.S.S. is a guest editor invited by the Editorial Board.
Data deposition: The MS data have been deposited in the PeptideAtlas repository, www.peptideatlas.org (identifier no. PASS01074).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1712623114/-/DCSupplemental.
References
- 1.Davies KE, Nowak KJ. Molecular mechanisms of muscular dystrophies: Old and new players. Nat Rev Mol Cell Biol. 2006;7:762–773. doi: 10.1038/nrm2024. [DOI] [PubMed] [Google Scholar]
- 2.Hoffman EP, Brown RH, Jr, Kunkel LM. Dystrophin: The protein product of the Duchenne muscular dystrophy locus. Cell. 1987;51:919–928. doi: 10.1016/0092-8674(87)90579-4. [DOI] [PubMed] [Google Scholar]
- 3.Hermans MCE, et al. Hereditary muscular dystrophies and the heart. Neuromuscul Disord. 2010;20:479–492. doi: 10.1016/j.nmd.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 4.Nelson CE, et al. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science. 2016;351:403–407. doi: 10.1126/science.aad5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tabebordbar M, et al. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science. 2016;351:407–411. doi: 10.1126/science.aad5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ehmsen J, Poon E, Davies K. The dystrophin-associated protein complex. J Cell Sci. 2002;115:2801–2803. doi: 10.1242/jcs.115.14.2801. [DOI] [PubMed] [Google Scholar]
- 7.Head SI, Williams DA, Stephenson DG. Abnormalities in structure and function of limb skeletal muscle fibres of dystrophic mdx mice. Proc Biol Sci. 1992;248:163–169. doi: 10.1098/rspb.1992.0058. [DOI] [PubMed] [Google Scholar]
- 8.Whitehead NP, Streamer M, Lusambili LI, Sachs F, Allen DG. Streptomycin reduces stretch-induced membrane permeability in muscles from mdx mice. Neuromuscul Disord. 2006;16:845–854. doi: 10.1016/j.nmd.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 9.Allen DG, Gervasio OL, Yeung EW, Whitehead NP. Calcium and the damage pathways in muscular dystrophy. Can J Physiol Pharmacol. 2010;88:83–91. doi: 10.1139/Y09-058. [DOI] [PubMed] [Google Scholar]
- 10.Allen DG, Zhang BT, Whitehead NP. Stretch-induced membrane damage in muscle: Comparison of wild-type and mdx mice. Adv Exp Med Biol. 2010;682:297–313. doi: 10.1007/978-1-4419-6366-6_17. [DOI] [PubMed] [Google Scholar]
- 11.Vandebrouck C, Martin D, Colson-Van Schoor M, Debaix H, Gailly P. Involvement of TRPC in the abnormal calcium influx observed in dystrophic (mdx) mouse skeletal muscle fibers. J Cell Biol. 2002;158:1089–1096. doi: 10.1083/jcb.200203091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seo K, et al. Hyperactive adverse mechanical stress responses in dystrophic heart are coupled to transient receptor potential canonical 6 and blocked by cGMP-protein kinase G modulation. Circ Res. 2014;114:823–832. doi: 10.1161/CIRCRESAHA.114.302614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gervásio OL, Whitehead NP, Yeung EW, Phillips WD, Allen DG. TRPC1 binds to caveolin-3 and is regulated by Src kinase–Role in Duchenne muscular dystrophy. J Cell Sci. 2008;121:2246–2255. doi: 10.1242/jcs.032003. [DOI] [PubMed] [Google Scholar]
- 14.Spassova MA, Hewavitharana T, Xu W, Soboloff J, Gill DL. A common mechanism underlies stretch activation and receptor activation of TRPC6 channels. Proc Natl Acad Sci USA. 2006;103:16586–16591. doi: 10.1073/pnas.0606894103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muraki K, et al. TRPV2 is a component of osmotically sensitive cation channels in murine aortic myocytes. Circ Res. 2003;93:829–838. doi: 10.1161/01.RES.0000097263.10220.0C. [DOI] [PubMed] [Google Scholar]
- 16.Iwata Y, Katanosaka Y, Arai Y, Shigekawa M, Wakabayashi S. Dominant-negative inhibition of Ca2+ influx via TRPV2 ameliorates muscular dystrophy in animal models. Hum Mol Genet. 2009;18:824–834. doi: 10.1093/hmg/ddn408. [DOI] [PubMed] [Google Scholar]
- 17.Zanou N, et al. Essential role of TRPV2 ion channel in the sensitivity of dystrophic muscle to eccentric contractions. FEBS Lett. 2009;583:3600–3604. doi: 10.1016/j.febslet.2009.10.033. [DOI] [PubMed] [Google Scholar]
- 18.Li D, Yue Y, Lai Y, Hakim CH, Duan D. Nitrosative stress elicited by nNOSµ delocalization inhibits muscle force in dystrophin-null mice. J Pathol. 2011;223:88–98. doi: 10.1002/path.2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Percival JM, Adamo CM, Beavo JA, Froehner SC. Evaluation of the therapeutic utility of phosphodiesterase 5A inhibition in the mdx mouse model of duchenne muscular dystrophy. Handb Exp Pharmacol. 2011:323–344. doi: 10.1007/978-3-642-17969-3_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Su JB, et al. Bradykinin restores left ventricular function, sarcomeric protein phosphorylation, and e/nNOS levels in dogs with Duchenne muscular dystrophy cardiomyopathy. Cardiovasc Res. 2012;95:86–96. doi: 10.1093/cvr/cvs161. [DOI] [PubMed] [Google Scholar]
- 21.Bellinger AM, et al. Hypernitrosylated ryanodine receptor calcium release channels are leaky in dystrophic muscle. Nat Med. 2009;15:325–330. doi: 10.1038/nm.1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fauconnier J, et al. Leaky RyR2 trigger ventricular arrhythmias in Duchenne muscular dystrophy. Proc Natl Acad Sci USA. 2010;107:1559–1564. doi: 10.1073/pnas.0908540107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carnicer R, Crabtree MJ, Sivakumaran V, Casadei B, Kass DA. Nitric oxide synthases in heart failure. Antioxid Redox Signal. 2013;18:1078–1099. doi: 10.1089/ars.2012.4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chung HS, et al. Dual labeling biotin switch assay to reduce bias derived from different cysteine subpopulations: A method to maximize S-nitrosylation detection. Circ Res. 2015;117:846–857. doi: 10.1161/CIRCRESAHA.115.307336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu C, et al. Redox regulatory mechanism of transnitrosylation by thioredoxin. Mol Cell Proteomics. 2010;9:2262–2275. doi: 10.1074/mcp.M110.000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Engelman R, et al. Multilevel regulation of 2-Cys peroxiredoxin reaction cycle by S-nitrosylation. J Biol Chem. 2013;288:11312–11324. doi: 10.1074/jbc.M112.433755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mullen L, Hanschmann E-M, Lillig CH, Herzenberg LA, Ghezzi P. Cysteine oxidation targets peroxiredoxins 1 and 2 for exosomal release through a novel mechanism of redox-dependent secretion. Mol Med. 2015;21:98–108. doi: 10.2119/molmed.2015.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chun JL, O’Brien R, Berry SE. Cardiac dysfunction and pathology in the dystrophin and utrophin-deficient mouse during development of dilated cardiomyopathy. Neuromuscul Disord. 2012;22:368–379. doi: 10.1016/j.nmd.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 29.Chu V, et al. Electrocardiographic findings in mdx mice: A cardiac phenotype of Duchenne muscular dystrophy. Muscle Nerve. 2002;26:513–519. doi: 10.1002/mus.10223. [DOI] [PubMed] [Google Scholar]
- 30.Petroff MGV, et al. Endogenous nitric oxide mechanisms mediate the stretch dependence of Ca2+ release in cardiomyocytes. Nat Cell Biol. 2001;3:867–873. doi: 10.1038/ncb1001-867. [DOI] [PubMed] [Google Scholar]
- 31.Balligand JL. Regulation of cardiac beta-adrenergic response by nitric oxide. Cardiovasc Res. 1999;43:607–620. doi: 10.1016/s0008-6363(99)00163-7. [DOI] [PubMed] [Google Scholar]
- 32.Murray CI, Uhrigshardt H, O’Meally RN, Cole RN, Van Eyk JE. Identification and quantification of S-nitrosylation by cysteine reactive tandem mass tag switch assay. Mol Cell Proteomics. 2012;11:M111.013441. doi: 10.1074/mcp.M111.013441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murphy E, Kohr M, Sun J, Nguyen T, Steenbergen C. S-nitrosylation: A radical way to protect the heart. J Mol Cell Cardiol. 2012;52:568–577. doi: 10.1016/j.yjmcc.2011.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shao Q, et al. Adenosine A1 receptor activation increases myocardial protein S-nitrosothiols and elicits protection from ischemia-reperfusion injury in male and female hearts. PLoS One. 2017;12:e0177315. doi: 10.1371/journal.pone.0177315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakamura T, et al. Aberrant protein S-nitrosylation contributes to the pathophysiology of neurodegenerative diseases. Neurobiol Dis. 2015;84:99–108. doi: 10.1016/j.nbd.2015.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu L, Eu JP, Meissner G, Stamler JS. Activation of the cardiac calcium release channel (ryanodine receptor) by poly-S-nitrosylation. Science. 1998;279:234–237. doi: 10.1126/science.279.5348.234. [DOI] [PubMed] [Google Scholar]
- 37.Sun J, Xin C, Eu JP, Stamler JS, Meissner G. Cysteine-3635 is responsible for skeletal muscle ryanodine receptor modulation by NO. Proc Natl Acad Sci USA. 2001;98:11158–11162. doi: 10.1073/pnas.201289098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zissimopoulos S, Lai FA. Interaction of FKBP12.6 with the cardiac ryanodine receptor C-terminal domain. J Biol Chem. 2005;280:5475–5485. doi: 10.1074/jbc.M412954200. [DOI] [PubMed] [Google Scholar]
- 39.Khairallah M, et al. Metabolic and signaling alterations in dystrophin-deficient hearts precede overt cardiomyopathy. J Mol Cell Cardiol. 2007;43:119–129. doi: 10.1016/j.yjmcc.2007.05.015. [DOI] [PubMed] [Google Scholar]
- 40.Gulston MK, et al. A combined metabolomic and proteomic investigation of the effects of a failure to express dystrophin in the mouse heart. J Proteome Res. 2008;7:2069–2077. doi: 10.1021/pr800070p. [DOI] [PubMed] [Google Scholar]
- 41.Lewis C, Jockusch H, Ohlendieck K. Proteomic profiling of the dystrophin-deficient MDX heart reveals drastically altered levels of key metabolic and contractile proteins. J Biomed Biotechnol. 2010;2010:648501. doi: 10.1155/2010/648501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou L, Lu H. Targeting fibrosis in Duchenne muscular dystrophy. J Neuropathol Exp Neurol. 2010;69:771–776. doi: 10.1097/NEN.0b013e3181e9a34b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li Y, et al. Inhibition of integrin-linked kinase attenuates renal interstitial fibrosis. J Am Soc Nephrol. 2009;20:1907–1918. doi: 10.1681/ASN.2008090930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu C, Dedhar S. Integrin-linked kinase (ILK) and its interactors: A new paradigm for the coupling of extracellular matrix to actin cytoskeleton and signaling complexes. J Cell Biol. 2001;155:505–510. doi: 10.1083/jcb.200108077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chaillou T, Lee JD, England JH, Esser KA, McCarthy JJ. Time course of gene expression during mouse skeletal muscle hypertrophy. J Appl Physiol (1985) 2013;115:1065–1074. doi: 10.1152/japplphysiol.00611.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kamp TJ, Hell JW. Regulation of cardiac L-type calcium channels by protein kinase A and protein kinase C. Circ Res. 2000;87:1095–1102. doi: 10.1161/01.res.87.12.1095. [DOI] [PubMed] [Google Scholar]
- 47.Campbell DL, Stamler JS, Strauss HC. Redox modulation of L-type calcium channels in ferret ventricular myocytes. Dual mechanism regulation by nitric oxide and S-nitrosothiols. J Gen Physiol. 1996;108:277–293. doi: 10.1085/jgp.108.4.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Benhar M, Forrester MT, Stamler JS. Protein denitrosylation: Enzymatic mechanisms and cellular functions. Nat Rev Mol Cell Biol. 2009;10:721–732. doi: 10.1038/nrm2764. [DOI] [PubMed] [Google Scholar]
- 49.Ogata H, Ishikawa Y, Ishikawa Y, Minami R. Beneficial effects of beta-blockers and angiotensin-converting enzyme inhibitors in Duchenne muscular dystrophy. J Cardiol. 2009;53:72–78. doi: 10.1016/j.jjcc.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 50.Huang B, Chen C. An ascorbate-dependent artifact that interferes with the interpretation of the biotin switch assay. Free Radic Biol Med. 2006;41:562–567. doi: 10.1016/j.freeradbiomed.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 51.Wolhuter K, Eaton P. How widespread is stable protein S-nitrosylation as an end-effector of protein regulation? Free Radic Biol Med. 2017;109:156–166. doi: 10.1016/j.freeradbiomed.2017.02.013. [DOI] [PubMed] [Google Scholar]
- 52.Forrester MT, Foster MW, Stamler JS. Assessment and application of the biotin switch technique for examining protein S-nitrosylation under conditions of pharmacologically induced oxidative stress. J Biol Chem. 2007;282:13977–13983. doi: 10.1074/jbc.M609684200. [DOI] [PubMed] [Google Scholar]
- 53.National Research Council . Guide for the Care and Use of Laboratory Animals. 8th Ed National Academies Press; Washington, DC: 2011. [Google Scholar]
- 54.Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.