Significance
Cyanobacteria contribute significantly to global primary production. Huge photosynthetic light-harvesting complexes, phycobilisomes, constitute up to 50% of the cellular protein. Their assembly, restructuring, and disassembly are major metabolic activities. Light is absorbed by chromophores related to animal bile pigments; they are attached to apoproteins by a complex set of enzymes. Two such lyases have previously been identified as β-barrel proteins. We report the structure of a third type that also catalyzes chromophore detachment and isomerization. CpcE/F belongs to a completely different protein family, α-solenoids, but also forms a cavity that transiently hosts the chromophore. Two other proteins were modeled on this structure: an isomerizing E/F-type lyase and the poorly characterized NblB for which we propose chromophore detaching function.
Keywords: photosynthesis, phycobilisome, chromophorylation, isomerase, phycobilin
Abstract
The light-harvesting phycobilisome in cyanobacteria and red algae requires the lyase-catalyzed chromophorylation of phycobiliproteins. There are three functionally distinct lyase families known. The heterodimeric E/F type is specific for attaching bilins covalently to α-subunits of phycocyanins and phycoerythrins. Unlike other lyases, the lyase also has chromophore-detaching activity. A subclass of the E/F-type lyases is, furthermore, capable of chemically modifying the chromophore. Although these enzymes were characterized >25 y ago, their structures remained unknown. We determined the crystal structure of the heterodimer of CpcE/F from Nostoc sp. PCC7120 at 1.89-Å resolution. Both subunits are twisted, crescent-shaped α-solenoid structures. CpcE has 15 and CpcF 10 helices. The inner (concave) layer of CpcE (helices h2, 4, 6, 8, 10, 12, and 14) and the outer (convex) layer of CpcF (h16, 18, 20, 22, and 24) form a cavity into which the phycocyanobilin chromophore can be modeled. This location of the chromophore is supported by mutations at the interface between the subunits and within the cavity. The structure of a structurally related, isomerizing lyase, PecE/F, that converts phycocyanobilin into phycoviolobilin, was modeled using the CpcE/F structure as template. A H87C88 motif critical for the isomerase activity of PecE/F is located at the loop between h20 and h21, supporting the proposal that the nucleophilic addition of Cys-88 to C10 of phycocyanobilin induces the isomerization of phycocyanobilin into phycoviolobilin. Also, the structure of NblB, involved in phycobilisome degradation could be modeled using CpcE as template. Combined with CpcF, NblB shows a low chromophore-detaching activity.
Many photosynthetic organisms have to cope with complex and strongly changing light environments. Cyanobacteria and red algae, which contribute significantly to global oxygen and biomass production, have met this challenge by evolution of the brilliantly colored phycobiliproteins (PBPs) that harvest light in spectral ranges where chlorophylls absorb poorly. PBPs are organized in supramolecular light-harvesting complexes, the phycobilisomes (PBSs), where their spatial arrangement and energetic tuning ensures directional energy transfer with high quantum efficiency to the photosynthetic reaction centers (1–3). Maturation of the constituent PBPs involves covalent attachment of bilin chromophores to the apoproteins (4–10). With a single exception (11, 12), these posttranslational chromophorylations are catalyzed by site- and chromophore-specific lyases.
So far, three functionally distinct lyase families have been characterized for catalyzing the chromophorylation, and there may be additional ones. E/F-type lyases are specific for attaching phycocyanobilin (PCB) or phycoerythrobilin (PEB) to the Cys-84 site of α-subunits of phycocyanins (PCs), phycoerythrins and phycoerythrocyanin (PEC) (9, 13–17). S/U-type lyases are specific for attaching bilins to the Cys-81 site of allophycocyanins and the Cys-84 site of β-subunits in PCs, phycoerythrins and PEC (5, 18–21). T-type lyases attach bilins to the Cys-155 site of β-subunits in PBPs (22–24).
PBP lyases are considered to act like chaperones or protective carriers that transiently bind the labile bilin chromophores (21, 25–28), but additional functions have been found (29, 30). Some E/F-type lyases are capable of isomerizing the chromophore concomitant with the attachment; an example is PecE/F. The protein complexes CpcE/F and PecE/F are homologs. Both bind PCB, but while CpcE/F transfers the bilin directly to Cys-84 of the apoprotein of the α-subunit (CpcA) of cyanobacterial PC (CPC) (9, 13, 27), PecE/F catalyzes its concomitant isomerization into phycoviolobilin (PVB) and attaches this chromophore to Cys-84 of the α-subunit of PEC (PecA) (15, 31–33). CpcE/F is also capable of detaching the chromophore from PCBT–CpcA. Another protein, NblB, that is distantly related to CpcE, is involved in the degradation of PBSs (29, 30).
The crystal structure of an S-type lyase, CpcS from Thermosynechococcus elongatus, has been determined in the absence of a chromophore (20). It adopts a β-barrel structure similar to those of fatty acid binding proteins, a subfamily of the calycin superfamily (Pfam0116) that binds a variety of small, mostly lipophilic molecules with high selectivity (34). The crystal structure of a T-type lyase, CpcT from Nostoc sp. PCC 7120 (Nostoc), has been resolved with and without the chromophore, PCB (24). This lyase and a homolog of CpcT from the cyanophage P-HM1, PhiCpeT (28), also adopt β-barrel structures. Upon monomerization of the CpcT dimer, the 3-ethylidene group of PCB becomes accessible for binding to the apoprotein of the β-subunit of CPC (CpcB). Asp-163 and Tyr-65 in CpcT near the ethylidene group of PCB have been identified to facilitate the acid-catalyzed nucleophilic addition of Cys-155 of CpcB to an N-acylimmonium intermediate of PCB (35).
Although E/F-types lyases have been characterized as early as 1992 (8, 13), their structure has withstood crystallization attempts. We now determine the crystal structure of the chromophore-free heterodimer of CpcE (Alr0532) and CpcF (Alr0533) from Nostoc at 1.89-Å resolution. Both subunits are twisted crescent-shaped α-solenoid structures. CpcE has 15 helices called h1–15, and CpcF has 10 helices called h16–25 from N- to C-terminal. The concave layer of CpcE (h2, 4, 6, 8, 10, 12, and 14) and the convex layer of CpcF (h16, 18, 20, 22, and 24) line a cavity into which the PCB chromophore could be modeled. Based on the CpcE/F structure, we modeled the lyase-isomerase, PecE/F, from Nostoc. Finally, we could also model NblB based on CpcE, thereby supporting its formerly proposed function in chromophore detachment.
Results and Discussion
α-Solenoid Structure of CpcE and CpcF.
Neither subunit E nor F could be crystallized individually, so we tested to crystallize the CpcE/F complex that was reported to be stable (9). A His6 tag was added to the C terminus of subunit F, whereas no tag was added to CpcE. This allowed CpcE to be copurified with CpcF, yielding a pure CpcE/F complex that was subjected to crystallization. In addition, the Se-methionine variant was prepared for phase determination, and the crystal structure was determined at 1.89 Å (Table 1).
Table 1.
Data collection and refinement statistics of X-ray crystallography
| Parameter | CpcE/F | CpcE/F-SeMet |
| Data collection | ||
| Beamline | DESY, EMBL, P11 | DESY, EMBL, P13 |
| Space group | I121 | C2 |
| Cell parameters, Å | a = 74.60, b = 59.83, c = 110.63 | a = 123.72, b = 60.80, c = 74.40 |
| α = 90.00°, β = 99.08°, γ = 90° | α = 90.00°, β = 117.62°, γ = 90° | |
| Resolution, Å | 46.44–1.89 (1.96–1.89) | 65.94–2.99 (3.18–2.99) |
| Observed reflections | 73,291 (5,464) | 51,830 (10,279) |
| Unique reflections | 36,870 (2,831) | 15,550 (2,984) |
| Rmerge | 0.019 (0.215) | 0.035 (0.068) |
| <I/σ, I> | 19.34 (3.97) | 26.1 (14.7) |
| Completeness, % | 95.3 (82.2) | 80.5 (96.9) |
| Redundancy | 2.0 (1.9) | 3.3 (3.4) |
| Structure refinement | ||
| Resolution, Å | 46.44–1.89 (1.96–1.89) | |
| No. of unique reflections | 36,868 (2,831) | |
| Rwork, % | 18.2 | |
| Rfree, % | 21.6 | |
| Total no. of atoms | 3,458 | |
| No. of molecules/ASU | 2 monomers | |
| Waters and ligands | 148 H2O | |
| Solvent content, % | 47.3 | |
| Wilson B-factor, Å2 | 26.9 | |
| rms, bonds | 0.022 | |
| rms, angles | 1.96 | |
| Ramachandran | ||
| Preferred regions, no. (%) | 425 (99.3) | |
| Allowed, no. (%) | 3 (0.7) | |
| Outliers, no. (%) | 0 (0) | |
| PDB ID code | 5N3U |
Values in parentheses are for the highest-resolution shell.
The asymmetric unit contains one heterodimer (i.e., subunits E and F) consisting mainly of 25 helices (Fig. 1). The electron density is clearly defined for most of the sequence of CpcE (96%) and CpcF (82%). In CpcE, the first three N-terminal residues and residues 253–259 from the loop connecting helices h14 and h15 could not be traced, and in CpcF, residues 1–29 from the N terminus and residues 200–208 from the C terminus (including the His6 tag) were not resolved in the electron density, reflecting flexible regions of the protein chains (Fig. S1). Within the resolved complex, the side chains of Ile-261 of CpcE and Gln-58 of CpcF are poorly defined by the electron density. In the Ramachandran diagram (36), all resolved residues of both CpcE and CpcF of the heterodimer are found in the energetically allowed areas, with no outliers (Table 1).
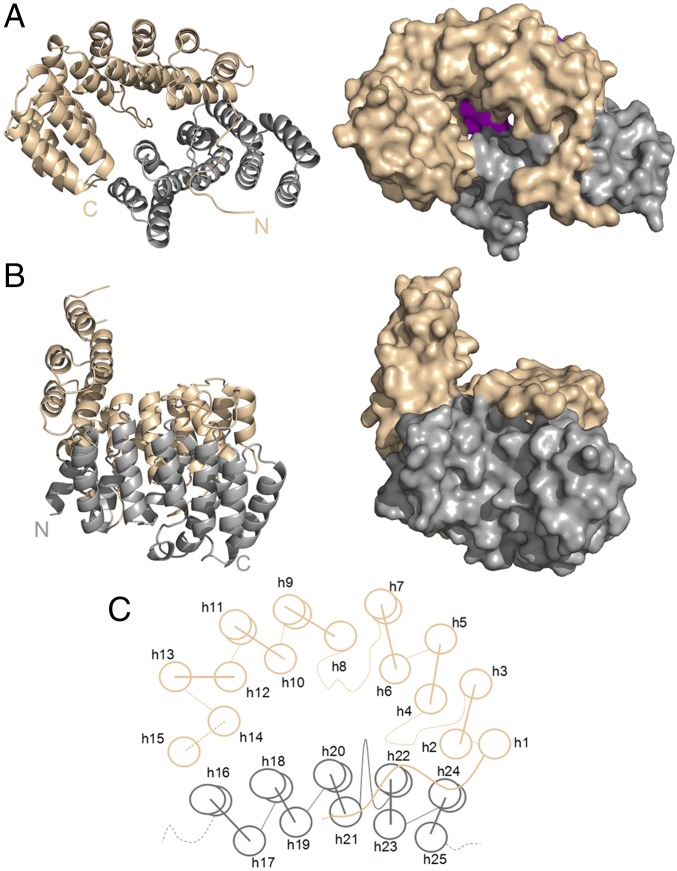
Fig. 1.
Structure and overview of CpcE/F heterodimer. Subunits E (brown) and F (gray) correspond to chains A and B in PDB ID code 5N3U. In the asymmetric unit, there is one heterodimer consisting of two chains containing 15 (CpcE) and 10 (CpcF) helices. Both subunits appear as α-solenoid proteins in the shape of twisted crescents. Each is composed of two layers of α-helices, and the two subunits are arranged in a spooning fashion (see main text for details). (A) Top view into the cavity formed between subunits E and F that fits a PCB chromophore. The cavity has a wide opening at the top and a smaller one at the bottom that is closed by three loops and two short helices that are in part visible (magenta) looking down the cavity (upper right). (B) Side view with subunit F in front, and the handle formed by the C-terminal helices of subunit E on Left. (C) Schematic structure, seen from the top into the cavity as in A. Dashed lines indicate unresolved stretches; curved lines indicate extended structures.
Both subunits have the shape of twisted crescents, each formed by an inner (concave) and outer (convex) layer of α-helices. Their structures are of the α-solenoid type, similar to that of importin β1 or to artificial α-helicoidal repeat proteins that are designed based on thermostable HEAT-like repeats (37). Subunit E has 15 helices (h1–15), and subunit F has 10 helices (h16–25) counted from N to C terminus (Fig. 1). The two subunits are in a “spooning” geometry, with a long N-terminal “arm” lacking secondary structure (Pro-4–19) of CpcE reaching “over the top” of CpcF (see below). The total contact interface between E/F subunits buries an area of 2,397 Å2 (38). As CpcE is more strongly curved than CpcF, a central cavity is formed that is lined by part of the concave helix layer of CpcE (h2, 4, 6, 8, 10, and 12) and of the convex layer of CpcF (h16, 18, 20, 22, and 24). Part of the convex helix layer of CpcE (h1, 3, 5, 7, and 9) and of the concave layer of CpcF (h17, 19, 21, 23, and 25) form the outside of the cavity. While the concave layers of both subunits are well-formed helices, many helices of the convex layers are relatively short (e.g., h1) or presented as kinked by the program used (Biovia Discovery Suite; Version 16) (mainly due to insertion of prolines), indicating less order or strain (Fig. 1). Two of the loops of CpcE (h3–4 and h7–8) and one of CpcF (h21–22) are long and contain single-turn helical structures; together, they form the bottom of the cavity (Fig. 1; see below). Neighboring helices within each layer are in an approximately parallel arrangement, whereas neighboring helices between the layers are tilted by up to 30°, thereby generating an overall twist. This twist is relatively small in CpcF (22° between the N- and C-terminal helices). In CpcE, it is much bigger (up to 90°), but largely confined to the central part. Thereby, the N termini of h10 and h12 of CpcE locate sideways on h16 of CpcF in a perpendicular arrangement. The C-terminal h13–15 of CpcE are stacked nearly parallel to h12 and form a “handle” protruding from the bowl-shaped main body of the heterodimer (Fig. 1).
The overall shape of the cavity is calyx-like and probably acts, like the similarly shaped insides of the β-barrels in CpcS and CpcT lyases (20, 24, 28), as a container for the bilin chromophore (Fig. 2). This arrangement protects the chromophore and allows its delivery to the apoprotein (27). The unresolved residues at the C terminus of CpcF, including the C-terminal His6 tag, are located on the outside of the “bowl” and distant from the handle. Their location makes contact with the E subunit unlikely, as well as an interference with the chromophore binding of the lyase (see below). However, the 29 unresolved N-terminal residues of CpcF might be contacting the E subunit. The N terminus of CpcE (i.e., the arm) is near to the handle, which interferes with the C-terminal His6 tag of the CpcA–His6 tag contacting the opening of the cavity (Fig. 2 A and B), so that the CpcA–His6 tag could not accept the PCB chromophore from chromophore carrying the CpcE/CpcF–His6 tag (Table S1). The N terminus of CpcE, up to Pro-19, is in an extended arrangement that reaches over the ends of h20, 21, and 23 of CpcF. This arm forms part of the rim of the large opening of the cavity. At the same time, it contributes substantially to the contacts between both subunits.
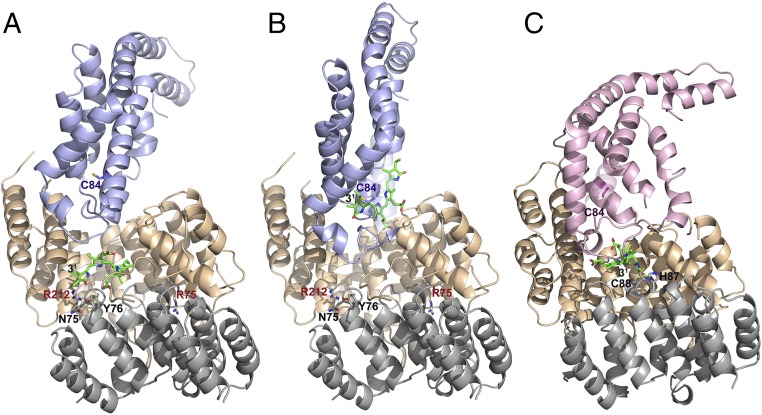
Fig. 2.
Docking of E/F-type lyase with PBPs. (A) Energy-minimized arrangement of apo-CpcA docked to PCB-loaded CpcE/F. This arrangement mimics the chromophorylation process. A zoom into the active site is shown in Fig. S4A. (B) PCBT–CpcA docked to empty CpcE/F. This situation corresponds to the beginning of detachment of the PCB chromophore from PCB–CpcA. A zoom into the active site is shown in Fig. S4B. (C) Docking of PecE/F loaded with PCB to apo-PecA. In the PecE/F complex, H87C88 are located at the loop between h20 and h21; as these residues are necessary for the isomerase activity, the complexed PCB chromophore is placed in the vicinity of H87C88 of PecF. The zoomed active site is shown in Fig. S4D. The structure of PCBT–CpcA is taken from the reported structure of CPC from Synechococcus elongatus (PDB ID code 4ZIZ), and that of the apoprotein is modeled from CPC holoprotein (PDB ID code 4ZIZ). The structure of CpcE/F loaded with PCB is obtained by fitting PCB from the CpcT–PCB crystal structure (PDB ID code 4O4S) into the empty CpcE/F heterodimer (this work). The structure of PecA is modeled from the reported structure of α-PEC (PDB ID code 2J96). The structure of PecE/F is simulated based on the present structure of CpcE/F (Fig. S2), followed by fitting the chromophore into the cavity and energy-minimization. CpcE and PecE are shown in brown, CpcF and PecF in gray, CpcA in blue, and PecA in violet.
Simulated Structure of PecE and PecF.
In Nostoc, there is, besides CpcE/F, a second E/F-type lyase, PecE/F. It delivers PCB to the α-subunit of PEC (α-PEC) and, in addition, isomerizes it to PVB concomitant with the attachment. Neither the individual subunits, PecE or PecF, nor the complex of PecE/F could be crystallized. Because of the high degree of homology between the two lyases (Fig. S2A), we simulate the structure of PecE/F based on the here-presented structure of CpcE/F. According to the resulting model, the organization of the two lyases is very similar, and their overall structures overlap well (Fig. S2 B and C). Distinct differences include the cavity between the subunits, the extended structure at the N terminus of PecE, and the extended loops. PecE lacks the long loop connecting h7 and h8. Accordingly, the bottom opening of the cavity is larger in PecE/F than in CpcE/F. By contrast, the top opening is narrowed by an additional extended loop of the F subunit; thereby, the cavity of PecE/F assumes a more cylindrical shape. The loop connecting h20 and h21 is extended by insertion of the NHCQ motif that is characteristic for isomerizing lyases (32), leading to the formation of a short helix (Fig. 2C and Fig. S2B): Its His and Cys are required for the isomerase activity of PecE/F (Fig. 3 and Table S1). The differences in cavity topology may come along with the different geometries of PVB and PCB in the ring A region and/or the different functions: PecE/F has both isomerase and lyase activity, and these reactions are irreversible (15, 31). CpcE/F has only lyase activity, but the reaction is reversible: It cannot only ligate PCB to apo-CpcA, but also detach it from the holoprotein, PCBT–CpcA (9, 39). The N-terminal arm in CpcE forms part of the upper rim and is, therefore, relevant both to the entry and the exit of the chromophore and to the interaction with the target protein. Deletion of this arm in CpcE enhances chromophore detachment (Fig. S3; see below).
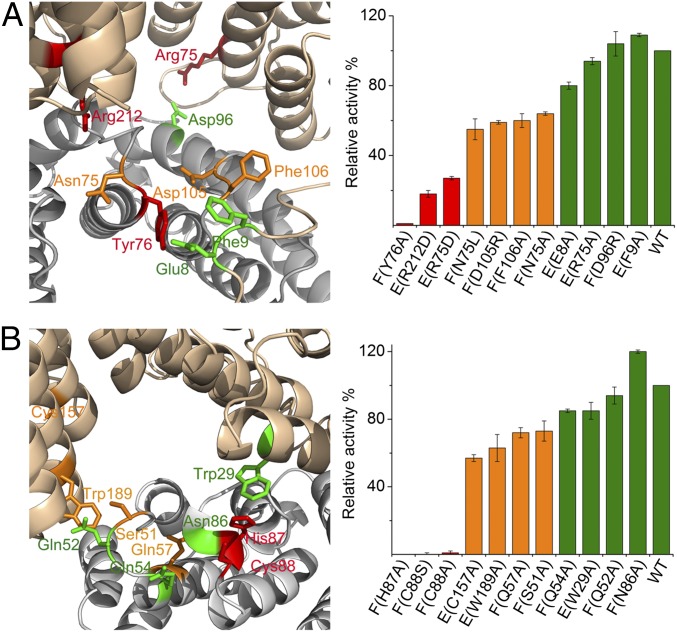
Fig. 3.
Selected mutations of the lyase, CpcE/F, and the lyase-isomerase, PecE/F. (A) Selected variants of CpcE/F (Left; color coded as the bars on the Right) and relative activities for chromophorylation of CpcA with PCB (Right). Arg-75 at h4 and Arg-212 at h12 of CpcE and Asn-75 and Tyr-76 at the loop between h18 and h19 of CpcF are important for the lyase activity. (B) Selected variants of PecE/F (Left) and relative activities for chromophorylation of PecA with PCB and isomerization to PVB. His-87 and Cys-88 of PecF are essential amino acids for activity. More details on relative enzymatic and PCB-binding activities of the two lyases are given in Table S1.
The PCB Chromophore May Be (Partly) Complexed in the Cavity Formed by E and F Subunits.
CpcE/F (27), PecE/F (32), CpcS (20, 21, 25), and CpcT (24) can reversibly bind the PCB chromophore in a noncovalent manner, which is important for the lyase activity. CpcT is the only lyase for which the chromophore-bearing structure is known: The PCB molecule is located in the cavity formed by the β-barrel protein (24). This calyx-like cavity is open at the top, but due to the homodimeric structure, the chromophore is protected during transport. A similar function is likely for CpcS that adopts a β-barrel conformation, too, and also forms homodimers or heterodimers with CpcU in one subclass (19, 20). The E/F-type lyases belong to a completely different class of proteins. Here, the cavity is not formed by a single protein, but is generated upon heterodimerization of the two subunits. It is tempting to speculate that in these lyases, the chromophore is likewise located in the cavity. This is supported by mutations (Fig. 3 and Table S1): Functionally relevant amino acids either line the cavity or are involved in subunit interactions.
Three different functions have been studied independently for the variants: chromophore binding, chromophore attachment to the target apoprotein, and chromophore detachment from the holoprotein. Few mutations—in particular, Tyr-76, Asp-105, and Phe-106 of CpcF lining the rim of the cavity—affect primarily the attachment reaction, but not PCB binding; they are likely involved in the interaction with the target protein (see below). Others—in particular, Arg-75 and -212 of CpcE or Asn-75 of CpcF—affect both PCB binding and the attachment reaction; they are located deeper in the cavity and/or at the interface of the two subunits. This would be compatible with a situation where the chromophore is at least partly within the cavity during transport. In the S- and T-type lyases, dimerization of the calycin-type β-barrels has been suggested to protect the chromophore during transport (20, 24). Interestingly, the CpcE/F heterodimer does not aggregate further to heterotetramers (E2F2), which would leave the chromophore solvent-exposed during transport. The partial protection in the cavity may suffice for the function. However, the opening could also be protected by interaction with another protein, including the target apoprotein (Fig. S4), but also the reductase, PcyA, that generates PCB from biliverdin and has been implied in substrate channeling (40, 41). It is, furthermore, conceivable that the N-terminal arm of the E subunit swings over the opening of the cavity upon chromophore binding (see below).
PecE/F is characterized by its isomerase activity. Isomerization of PCB to PVB has been related in both PBPs and phytochrome-type photoreceptors to cysteines in preserved motifs (HC and DXCF, respectively) (32, 42). Nucleophilic addition to C10 of the chromophore, between ring B and C, is discussed as initiating the isomerization (42–45). For PecE/F from Mastigocladus laminosus, amino acids H121C122 of PecF have been identified as necessary for the isomerization (32), which correspond to H87C88 of PecF in Nostoc. In fact, this function is confirmed by the respective site-directed mutations for the Nostoc proteins (Fig. 3 and Table S1). From the simulated structure of PecE/F, H87C88 of PecF from Nostoc are located in the extra loop between h20 and h21 that distinguishes it from its nonisomerizing homolog, CpcF (Fig. S2). Therefore, during model building, we position C10 of the PCB chromophore near to this loop (Fig. 2C and Fig. S4). However, it may also be buried more deeply and only move there after encountering the target protein. Since Asp-105 and Phe-106 at the corresponding (shorter) loop between h20 and h21 of CpcF are important to its lyase activity (Table S1), we similarly place PCB near to this loop in CpcE/F (Fig. 2 and Fig. S4). The N75Y76 motif at the loop between h18 and h19 of CpcF is important to the lyase activity (Table S1); Tyr-76 is presumed to be the proton-donating residue that facilitates nucleophilic addition of the chromophore-binding Cys-84 (Fig. 2 and Fig. S4; see below), and, accordingly, ring A and/or B of PCB would be near to this motif. Taking these constraints into consideration, energy minimization results in the models of PCB:CpcE/F and PCB:PecE/F that are shown in Fig. 2, Fig. S4, and Table S2.
Interaction of Lyases and Target Proteins.
To obtain more information on the mechanism, docking complexes are modeled between the two lyases, CpcE/F and PecE/F, and their substrates. This is a fairly roundabout process detailed in SI Materials and Methods. The results of the chromophore dockings to the lyases are summarized in the first two columns of Table S2. In case of the CpcA:PCB:CpcE/F complex, the C-terminal helices of CpcA [G and H in the nomenclature of Schirmer et al. (46)] contact the large hydrophobic patch of the handle of CpcE (Fig. 2A and Fig. S4A). This region of CpcA is neither involved in the heterodimer formation with holo-CpcB, nor in the subsequent trimerization of the αβ-protomer. In the hexamer, it is engaged in intertrimer interactions with one CpcB and two CpcA subunits, but, in particular, helix G is still partly accessible. This indicates that interactions of the lyase CpcE/F with the respective regions of CpcA may still be possible in PBS rods. Partial disassembly of the PBSs resulting from certain linker dephosphorylation would also facilitate access of the lyase by destabilizing the hexamers (47). In this model, the PCB chromophore complexes mainly with CpcF and becomes quite twisted in the docked complex: The adjacent pyrrolic rings of PCB are nearly perpendicular to each other. Otherwise, the chromophore remains at nearly the same position in the lyase as in the starting complex, near to Asn-75 (∼10 Å) and Tyr-76 (∼10 Å), that have been implied as proton donors. Its distance to Cys-84 of CpcA is ∼21 Å, and binding would require transient loosening of the helix–helix interactions of CpcA to allow transfer of the chromophore to the binding site. In the active site, one important residue, Arg-75 of CpcE, contacts the loop between h18 and h19 of CpcF. Mutation of Tyr-76 in the same loop of CpcF fully inactivates the lyase (Fig. 3 and Table S1). Moreover, Tyr-76 contacts Ala-107 of the neighboring loop (h20–21) and also the N-terminal arm of CpcE, where its phenolic hydroxyl group is in H-bonding distance to the carboxyl-group of Glu-8 (Fig. 2A).
We also simulated the structure of PCB:CpcE/F docked onto CpcB, which is not a substrate of the E/F lyase (Fig. S4C). In this model, Cys-155 is too distant to be attached at the PCB chromophore, coinciding with the fact that this site is the target of CpcT, a β-barrel–type lyase (22–24). However, Cys-84 is accessible according to these simulations, although this site is not targeted by E/F type lyases (4, 6, 7). The amino acid sequences around the PCB84-binding site between CpcA and CpcB are distinctly different (5), but at the current state of docking simulations, these differences are not indicative enough (Fig. S4 and Table S2).
The aforementioned complex of CpcA and PCB:CpcE/F can be regarded as simulating the encounter preceding the transfer and attachment of PCB to CpcA. The process starting the detachment reaction of the lyase, simulated by docking complex PCBT–CpcA to CpcE/F, has a distinctly different arrangement (Fig. 2B and Fig. S4B). The PBP is tilted by ∼60° compared with the attachment encounter, thereby reducing the interactions of helices G and H with the hydrophobic surface of the handle of CpcE and increasing contact with the N-terminal arm. This brings the chromophore closer to the opening of the cavity of the lyase, where it interacts directly with the N-terminal arm of CpcE with only little hindrance for a movement to the lyase. Tyr-76 is ∼22 Å away from the chromophore binding Cys-84, but it is protruding from the loop between helices h18 and h19, which may allow a closer approach. PCBT–CpcA contacts mainly CpcE, so C31 of the bound PCB chromophore is slightly distant from Tyr-76 of CpcF. One can imagine that the PCB chromophore (interacting mainly with CpcF) moves during chromophorylation more in the direction of CpcE, as CpcA contacts mainly with CpcE, and thus this movement will facilitate PCB attachment to CpcA. Conversely, during the detachment of PCB from PCBT–CpcA, the bound PCB chromophore might be attracted to CpcF and finally complexed with CpcF, thus facilitating PCB detachment. Hindrance of this movement by the unstructured N-terminal arm of CpcE located in front of CpcF would explain the enhanced detachment by truncation of this arm (Table S1).
The docking structure of the loaded isomerizing lyase, PCB:PecE/F, with PecA is very similar to that of the respective nonisomerizing lyase with its target protein (Fig. 2C, Fig. S4D, and Table S2). The essential HC motif is close to the chromophore, with the sulfhydryl group positioned at a distance of ∼9 Å from C-10 and -5 and of ∼16 Å from C31. Taking into account the flexibility of this region, it is conceivable that protonation by this moiety and/or transient binding to Cys-88 initiates the isomerization reaction of the chromophore. However, the distance to the binding site of PecA is still ∼16 Å away, and the chromophore would have to slip in again between helices B and G to approach the binding Cys-84. This sequence of events would require no large rearrangements of the proteins, but it would imply that the chromophore is isomerized before or at least during transfer to PecA and binding.
Is NblB a Lyase Subunit?
NblB is involved in the degradation of PBPs. Its homology to CpcE has been noted early on, but its precise function has remained unclear (29). Based on the now-available crystal structure of CpcE/F, the structure of NblB from Nostoc could be simulated (Fig. S5). It is, indeed, very similar to that of CpcE (brown), but lacks the N-terminal extended stretch (arm), the C-terminal h15, and the long loop (Val-138–Pro-147) between h7 and h8. This opens the possibility that NblB, like the structurally unrelated NblA, interacts with CpcA. Furthermore, it may be directly involved in PBS degradation by detaching the PCB from the Cys-84 site (47–50). This possibility is supported by the following results: NblB is dimeric in solution and binds PCB. It is inactive, however, in both the attachment and detachment assays, but so is CpcE. To further explore this line, the activity of NblB has been tested in combination with CpcF, and CpcE variants were constructed that lack some of the distinctive features (Table S1). Removal of the N-terminal arm of CpcE [CpcE(Δ2–10)/F] increases the chromophore detachment activity of CpcE/F. The affinity of CpcE(Δ2–10)/F for PCBT–CpcA decreases ∼8.7-fold, but the catalytic activity, kcat, for detaching PCB increases by ∼7.9-fold, and chromophore transfer to apo-Cph1 (the apoprotein of cyanobacterial phytochrome from Synechocystis sp. PCC 6803) is accelerated (Fig. S3 and Table S3).
Interestingly, a combination of NblB and CpcF also show detachment activity, albeit at a very low level (Table S1). The measured value is comparable to that of a CpcE variant lacking the N-terminal arm and the C-terminal h15 [CpcE(Δ2–10/h15)]; this construct was inactive in the chromophorylation. When h15 is truncated from CpcE(Δ2–10), the association with CpcF is damaged (Table S1), indicating that the C-terminal h15 is involved in regulation of the chromophore detachment activity. It is conceivable that, in vivo, NblB could realize the detaching activity together with other protein(s) and/or cofactor(s) (47–50), which might complement the function of h15. The lack of the long loop between h7 and h8 that in the CpcE/F complex covers the bottom of the cavity also raises the possibility that a complex like NblB/CpcF acts in a tunnel-like fashion for chromophore transfer.
Concluding Remarks
The present work completes the structural characterization of members of all three PBP lyase families. Although CpcE/F is not a β-barrel protein as the S/U- and T-type lyases, the E/F heterodimer provides a similar, calyx-shaped cavity fitting the chromophore. Its structural distinction probably reflects a functional distinction. First, E/F-type lyases have been shown to generate the chemically modified chromophores, PVB and phycourobilin bound to Cys-α84. It remains open if the lyases generating these chromophores at other binding sites will also turn out members of the α-solenoid type. Secondly, they serve the site implicated in PBS degradation (48, 51), which may relate to their distinguishing chromophore detachment activity. Such activities are relevant under several physiological conditions, including acclimation to varying light conditions and nutrient starvation (17, 29, 48, 52, 53). At high light, PBPs potentially induce photodamage by overexcitation of the reaction centers. The orange carotenoid protein is one of the components that prevent on a short time scale such damage by quenching excitons before transfer to the chlorophyll proteins (54). Degradation of the PBS is a longer-term mechanism coping with this problem. Partial degradation is also relevant in PBS restructuring during certain chromatic acclimations, where individual subunits are replaced by newly synthesized ones (17, 52), and also in coping with N or S deficits (29, 48, 53). If CpcE/F is involved in these processes, it is unclear how the two activities are differentially controlled. It is intriguing that NblB may be directly involved in the detachment reactions.
Materials and Methods
Crystallization and Data Collection.
Native and single-wavelength anomalous dispersion (SAD) X-ray diffraction data are collected from a single crystal in each case on beamline P11 or P13, Deutsches Elektronen-Synchrotron (DESY), European Molecular Biology Laboratory (EMBL) (Hamburg, Germany).
Data Processing and Structure Refinement.
All diffraction data are processed with the XDS program package and scaled by using XSCALE. The crystal structure is determined by the SAD method using the selenomethionine-substituted crystals. The initial model is obtained by using MRSAD on the Auto-Rickshaw server. The structure of native CpcE/F is determined by molecular replacement (Phaser in CCP4 using CpcE/F-SeMet structure as a starting model). The structures of native CpcE/F are refined by iterative cycles of manual refinement using Coot and Refmac5 from the CCP4 suite. Data refinement statistics and model content are summarized in Table 1. The structure of the complex of CpcE and CpcF from Nostoc is deposited at the Protein Data Bank (PDB ID code: 5N3U).
Materials and methods are detailed in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank the staff of beamline BM30-A (European Synchrotron Radiation Facility, Grenoble, France) for kind support during screening of the crystals. We also acknowledge the staff at beamline P11 and P13 (DESY, EMBL, Hamburg, Germany) for kind support during data collection. We thank Robert J. Porra (Canberra) for improving the English. This work was supported by National Natural Science Foundation of China Grants 21472055 and 31770822 (to K.-H.Z.) and 31370777 (to M.Z.). W.G. was supported by the Max Planck Society.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID code 5N3U).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1715495114/-/DCSupplemental.
References
- 1.Sidler WA. Phycobilisome and phycobiliprotein structures. In: Bryant DA, editor. The Molecular Biology of Cyanobacteria. Kluwer; Dordrecht, The Netherlands: 1994. pp. 139–216. [Google Scholar]
- 2.Gantt E, Grabowski B, Cunningham FX. Antenna systems of red algae: Phycobilisomes with photosystem II and chlorophyll complexes with photosystem I. In: Green B, Parson W, editors. Light-Harvesting Antennas in Photosynthesis. Kluwer; Dordrecht, The Netherlands: 2003. pp. 307–322. [Google Scholar]
- 3.Glazer AN. Phycobilisome:–A macromolecular complex optimized for light energy transfer. Biochim Biophys Acta. 1984;768:29–51. [Google Scholar]
- 4.Shen G, Schluchter WM, Bryant DA. Biogenesis of phycobiliproteins: I. cpcS-I and cpcU mutants of the cyanobacterium Synechococcus sp. PCC 7002 define a heterodimeric phyococyanobilin lyase specific for β-phycocyanin and allophycocyanin subunits. J Biol Chem. 2008;283:7503–7512. doi: 10.1074/jbc.M708164200. [DOI] [PubMed] [Google Scholar]
- 5.Zhao KH, et al. Phycobilin:cystein-84 biliprotein lyase, a near-universal lyase for cysteine-84-binding sites in cyanobacterial phycobiliproteins. Proc Natl Acad Sci USA. 2007;104:14300–14305. doi: 10.1073/pnas.0706209104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scheer H, Zhao KH. Biliprotein maturation: The chromophore attachment. Mol Microbiol. 2008;68:263–276. doi: 10.1111/j.1365-2958.2008.06160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schluchter WM, et al. Phycobiliprotein biosynthesis in cyanobacteria: Structure and function of enzymes involved in post-translational modification. Adv Exp Med Biol. 2010;675:211–228. doi: 10.1007/978-1-4419-1528-3_12. [DOI] [PubMed] [Google Scholar]
- 8.Zhou J, Gasparich GE, Stirewalt VL, de Lorimier R, Bryant DA. The cpcE and cpcF genes of Synechococcus sp. PCC 7002. Construction and phenotypic characterization of interposon mutants. J Biol Chem. 1992;267:16138–16145. [PubMed] [Google Scholar]
- 9.Fairchild CD, Glazer AN. Oligomeric structure, enzyme kinetics, and substrate specificity of the phycocyanin alpha subunit phycocyanobilin lyase. J Biol Chem. 1994;269:8686–8694. [PubMed] [Google Scholar]
- 10.Jung LJ, Chan CF, Glazer AN. Candidate genes for the phycoerythrocyanin alpha subunit lyase. Biochemical analysis of pecE and pecF interposon mutants. J Biol Chem. 1995;270:12877–12884. doi: 10.1074/jbc.270.21.12877. [DOI] [PubMed] [Google Scholar]
- 11.Tang K, et al. The terminal phycobilisome emitter, LCM: A light-harvesting pigment with a phytochrome chromophore. Proc Natl Acad Sci USA. 2015;112:15880–15885. doi: 10.1073/pnas.1519177113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao KH, et al. Reconstitution of phycobilisome core-membrane linker, LCM, by autocatalytic chromophore binding to ApcE. Biochim Biophys Acta. 2005;1706:81–87. doi: 10.1016/j.bbabio.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 13.Fairchild CD, et al. Phycocyanin alpha-subunit phycocyanobilin lyase. Proc Natl Acad Sci USA. 1992;89:7017–7021. doi: 10.1073/pnas.89.15.7017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kahn K, Mazel D, Houmard J, Tandeau de Marsac N, Schaefer MR. A role for cpeYZ in cyanobacterial phycoerythrin biosynthesis. J Bacteriol. 1997;179:998–1006. doi: 10.1128/jb.179.4.998-1006.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao KH, et al. Novel activity of a phycobiliprotein lyase: Both the attachment of phycocyanobilin and the isomerization to phycoviolobilin are catalyzed by the proteins PecE and PecF encoded by the phycoerythrocyanin operon. FEBS Lett. 2000;469:9–13. doi: 10.1016/s0014-5793(00)01245-x. [DOI] [PubMed] [Google Scholar]
- 16.Blot N, et al. Phycourobilin in trichromatic phycocyanin from oceanic cyanobacteria is formed post-translationally by a phycoerythrobilin lyase-isomerase. J Biol Chem. 2009;284:9290–9298. doi: 10.1074/jbc.M809784200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shukla A, et al. Phycoerythrin-specific bilin lyase-isomerase controls blue-green chromatic acclimation in marine Synechococcus. Proc Natl Acad Sci USA. 2012;109:20136–20141. doi: 10.1073/pnas.1211777109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao KH, et al. Chromophore attachment to phycobiliprotein β-subunits: Phycocyanobilin:cysteine-β84 phycobiliprotein lyase activity of CpeS-like protein from Anabaena Sp. PCC7120. J Biol Chem. 2006;281:8573–8581. doi: 10.1074/jbc.M513796200. [DOI] [PubMed] [Google Scholar]
- 19.Saunée NA, Williams SR, Bryant DA, Schluchter WM. Biogenesis of phycobiliproteins: II. CpcS-I and CpcU comprise the heterodimeric bilin lyase that attaches phycocyanobilin to CYS-82 OF β-phycocyanin and CYS-81 of allophycocyanin subunits in Synechococcus sp. PCC 7002. J Biol Chem. 2008;283:7513–7522. doi: 10.1074/jbc.M708165200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kronfel CM, et al. Structural and biochemical characterization of the bilin lyase CpcS from Thermosynechococcus elongatus. Biochemistry. 2013;52:8663–8676. doi: 10.1021/bi401192z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Overkamp KE, et al. Insights into the biosynthesis and assembly of cryptophycean phycobiliproteins. J Biol Chem. 2014;289:26691–26707. doi: 10.1074/jbc.M114.591131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen G, et al. Identification and characterization of a new class of bilin lyase: The cpcT gene encodes a bilin lyase responsible for attachment of phycocyanobilin to Cys-153 on the β-subunit of phycocyanin in Synechococcus sp. PCC 7002. J Biol Chem. 2006;281:17768–17778. doi: 10.1074/jbc.M602563200. [DOI] [PubMed] [Google Scholar]
- 23.Zhao KH, et al. Lyase activities of CpcS- and CpcT-like proteins from Nostoc PCC7120 and sequential reconstitution of binding sites of phycoerythrocyanin and phycocyanin β-subunits. J Biol Chem. 2007;282:34093–34103. doi: 10.1074/jbc.M703038200. [DOI] [PubMed] [Google Scholar]
- 24.Zhou W, et al. Structure and mechanism of the phycobiliprotein lyase CpcT. J Biol Chem. 2014;289:26677–26689. doi: 10.1074/jbc.M114.586743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kupka M, et al. Catalytic mechanism of S-type phycobiliprotein lyase: Chaperone-like action and functional amino acid residues. J Biol Chem. 2009;284:36405–36414. doi: 10.1074/jbc.M109.056382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schluchter WM, Glazer AN. Biosynthesis of phycobiliproteins in cyanobacteria. In: Peschek GA, Löffelhardt W, Schmetterer G, editors. The Phototrophic Prokaryotes. Kluwer/Plenum; New York: 1999. pp. 83–95. [Google Scholar]
- 27.Zhao KH, et al. Chromophore attachment in phycocyanin. Functional amino acids of phycocyanobilin–α-phycocyanin lyase and evidence for chromophore binding. FEBS J. 2006;273:1262–1274. doi: 10.1111/j.1742-4658.2006.05149.x. [DOI] [PubMed] [Google Scholar]
- 28.Gasper R, et al. Distinct features of cyanophage-encoded T-type phycobiliprotein lyase ΦCpeT: The role of auxiliary metabolic genes. J Biol Chem. 2017;292:3089–3098. doi: 10.1074/jbc.M116.769703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dolganov N, Grossman AR. A polypeptide with similarity to phycocyanin alpha-subunit phycocyanobilin lyase involved in degradation of phycobilisomes. J Bacteriol. 1999;181:610–617. doi: 10.1128/jb.181.2.610-617.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H, Sherman LA. Characterization of Synechocystis sp. strain PCC 6803 and deltanbl mutants under nitrogen-deficient conditions. Arch Microbiol. 2002;178:256–266. doi: 10.1007/s00203-002-0446-y. [DOI] [PubMed] [Google Scholar]
- 31.Storf M, et al. Chromophore attachment to biliproteins: Specificity of PecE/PecF, a lyase-isomerase for the photoactive 3(1)-cys-α 84-phycoviolobilin chromophore of phycoerythrocyanin. Biochemistry. 2001;40:12444–12456. doi: 10.1021/bi010776s. [DOI] [PubMed] [Google Scholar]
- 32.Zhao KH, et al. Amino acid residues associated with enzymatic activities of the isomerizing phycoviolobilin-lyase PecE/F. Biochemistry. 2005;44:8126–8137. doi: 10.1021/bi0500168. [DOI] [PubMed] [Google Scholar]
- 33.Böhm S, Endres S, Scheer H, Zhao KH. Biliprotein chromophore attachment: Chaperone-like function of the PecE subunit of alpha-phycoerythrocyanin lyase. J Biol Chem. 2007;282:25357–25366. doi: 10.1074/jbc.M702669200. [DOI] [PubMed] [Google Scholar]
- 34.Flower DR, North AC, Sansom CE. The lipocalin protein family: Structural and sequence overview. Biochim Biophys Acta. 2000;1482:9–24. doi: 10.1016/s0167-4838(00)00148-5. [DOI] [PubMed] [Google Scholar]
- 35.Grubmayr K, Wagner UG. On the chemistry of the addition of thiols to 2,3-dihydro-3-ethylidene-dihydrodipyrrin-1(1OH)-ones - A model study on the covalent chromophore-protein-linkage in biliproteins. Monatsh Chem. 1988;119:965–983. [Google Scholar]
- 36.Ramachandran GN, Sasisekharan V. Conformation of polypeptides and proteins. Adv Protein Chem. 1968;23:283–438. doi: 10.1016/s0065-3233(08)60402-7. [DOI] [PubMed] [Google Scholar]
- 37.Urvoas A, et al. Design, production and molecular structure of a new family of artificial alpha-helicoidal repeat proteins (αRep) based on thermostable HEAT-like repeats. J Mol Biol. 2010;404:307–327. doi: 10.1016/j.jmb.2010.09.048. [DOI] [PubMed] [Google Scholar]
- 38.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 39.Zhao KH, et al. Photochromic biliproteins from the cyanobacterium Anabaena sp. PCC 7120: Lyase activities, chromophore exchange, and photochromism in phytochrome AphA. Biochemistry. 2004;43:11576–11588. doi: 10.1021/bi0491548. [DOI] [PubMed] [Google Scholar]
- 40.Okada K. HO1 and PcyA proteins involved in phycobilin biosynthesis form a 1:2 complex with ferredoxin-1 required for photosynthesis. FEBS Lett. 2009;583:1251–1256. doi: 10.1016/j.febslet.2009.03.052. [DOI] [PubMed] [Google Scholar]
- 41.Hagiwara Y, Sugishima M, Takahashi Y, Fukuyama K. Crystal structure of phycocyanobilin:ferredoxin oxidoreductase in complex with biliverdin IXalpha, a key enzyme in the biosynthesis of phycocyanobilin. Proc Natl Acad Sci USA. 2006;103:27–32. doi: 10.1073/pnas.0507266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rockwell NC, Martin SS, Gulevich AG, Lagarias JC. Phycoviolobilin formation and spectral tuning in the DXCF cyanobacteriochrome subfamily. Biochemistry. 2012;51:1449–1463. doi: 10.1021/bi201783j. [DOI] [PubMed] [Google Scholar]
- 43.Rockwell NC, Martin SS, Lagarias JC. Mechanistic insight into the photosensory versatility of DXCF cyanobacteriochromes. Biochemistry. 2012;51:3576–3585. doi: 10.1021/bi300171s. [DOI] [PubMed] [Google Scholar]
- 44.Ma Q, et al. A rising tide of blue-absorbing biliprotein photoreceptors: Characterization of seven such bilin-binding GAF domains in Nostoc sp. PCC7120. FEBS J. 2012;279:4095–4108. doi: 10.1111/febs.12003. [DOI] [PubMed] [Google Scholar]
- 45.Tu JM, et al. Toward a mechanism for biliprotein lyases: Revisiting nucleophilic addition to phycocyanobilin. J Am Chem Soc. 2009;131:5399–5401. doi: 10.1021/ja9002348. [DOI] [PubMed] [Google Scholar]
- 46.Schirmer T, et al. Crystal structure analysis and refinement at 2.5 A of hexameric C-phycocyanin from the cyanobacterium Agmenellum quadruplicatum. The molecular model and its implications for light-harvesting. J Mol Biol. 1986;188:651–676. doi: 10.1016/s0022-2836(86)80013-4. [DOI] [PubMed] [Google Scholar]
- 47.Piven I, Ajlani G, Sokolenko A. Phycobilisome linker proteins are phosphorylated in Synechocystis sp. PCC 6803. J Biol Chem. 2005;280:21667–21672. doi: 10.1074/jbc.M412967200. [DOI] [PubMed] [Google Scholar]
- 48.Nguyen AY, et al. The proteolysis adaptor, NblA, binds to the N-terminus of β-phycocyanin: Implications for the mechanism of phycobilisome degradation. Photosynth Res. 2017;132:95–106. doi: 10.1007/s11120-016-0334-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baier A, Winkler W, Korte T, Lockau W, Karradt A. Degradation of phycobilisomes in Synechocystis sp. PCC6803: Evidence for essential formation of an NblA1/NblA2 heterodimer and its codegradation by A Clp protease complex. J Biol Chem. 2014;289:11755–11766. doi: 10.1074/jbc.M113.520601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dines M, Sendersky E, David L, Schwarz R, Adir N. Structural, functional, and mutational analysis of the NblA protein provides insight into possible modes of interaction with the phycobilisome. J Biol Chem. 2008;283:30330–30340. doi: 10.1074/jbc.M804241200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bienert R, Baier K, Volkmer R, Lockau W, Heinemann U. Crystal structure of NblA from Anabaena sp. PCC 7120, a small protein playing a key role in phycobilisome degradation. J Biol Chem. 2006;281:5216–5223. doi: 10.1074/jbc.M507243200. [DOI] [PubMed] [Google Scholar]
- 52.Gan F, et al. Extensive remodeling of a cyanobacterial photosynthetic apparatus in far-red light. Science. 2014;345:1312–1317. doi: 10.1126/science.1256963. [DOI] [PubMed] [Google Scholar]
- 53.Schwarz R, Forchhammer K. Acclimation of unicellular cyanobacteria to macronutrient deficiency: Emergence of a complex network of cellular responses. Microbiology. 2005;151:2503–2514. doi: 10.1099/mic.0.27883-0. [DOI] [PubMed] [Google Scholar]
- 54.Kirilovsky D, Kerfeld CA. Cyanobacterial photoprotection by the orange carotenoid protein. Nat Plants. 2016;2:16180. doi: 10.1038/nplants.2016.180. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.