Significance
We explore the utility of expansion microscopy (ExM) in neuroscience and developmental biology using the zebrafish model. Regarding neuroscience studies, ExM enables the tracing of cellular processes in the zebrafish brain, as well as the imaging of synapses and their biomolecular content and organization. Regarding development, ExM enables the resolving of nuclear compartments, particularly nuclear invaginations and channels, and helps relate such cellular nanostructures to proteins of the cytoskeleton during embryogenesis.
Keywords: zebrafish, superresolution, microscopy, brain
Abstract
Expansion microscopy (ExM) allows scalable imaging of preserved 3D biological specimens with nanoscale resolution on fast diffraction-limited microscopes. Here, we explore the utility of ExM in the larval and embryonic zebrafish, an important model organism for the study of neuroscience and development. Regarding neuroscience, we found that ExM enabled the tracing of fine processes of radial glia, which are not resolvable with diffraction-limited microscopy. ExM further resolved putative synaptic connections, as well as molecular differences between densely packed synapses. Finally, ExM could resolve subsynaptic protein organization, such as ring-like structures composed of glycine receptors. Regarding development, we used ExM to characterize the shapes of nuclear invaginations and channels, and to visualize cytoskeletal proteins nearby. We detected nuclear invagination channels at late prophase and telophase, potentially suggesting roles for such channels in cell division. Thus, ExM of the larval and embryonic zebrafish may enable systematic studies of how molecular components are configured in multiple contexts of interest to neuroscience and developmental biology.
Expansion microscopy (ExM) enables 3D nanoscale resolution imaging of extended biological specimens by forming a dense, permeating, interconnected mesh of polyelectrolyte polymer throughout the specimen and then swelling the gel so as to move key biomolecules or labels apart from each other (1). In our recently developed protein retention ExM (proExM) protocol, an antibody-stained tissue is embedded in such a polyelectrolyte gel and proteins are covalently linked to the gel (2). The tissue is then proteolytically digested to mechanically homogenize the specimen in a fashion that spares the antibody stain, followed by addition of water to isotropically expand the gel-specimen composite. proExM linearly expands preserved cells and tissues by a factor of ∼4.5-fold, enabling a resolution improvement by this magnitude on standard, diffraction-limited microscopes (e.g., for a ∼300-nm diffraction limit lens, a final effective resolution of ∼300/4.5 or ∼60–70 nm is obtained). Since proExM-processed tissues can be imaged on fast, diffraction-limited microscopes, it enables scalable superresolution imaging, and thus may help open up the systematic exploration of how molecules are configured throughout cells and tissues with nanoscale precision.
Here, we explore the application of proExM to zebrafish, a genetically tractable vertebrate that is transparent throughout early life and, accordingly, has proven to be a useful model for neuroscience (3) and developmental biology (4). We explored topics in both fields. Regarding neuroscience, we show that ExM can enable the tracing of cellular processes too fine to trace in diffraction-limited images [a common problem in neuroscience (5, 6)], using radial glia of the tectum (7–10) as a testbed. ExM also enabled the detection and molecular analysis of synaptic connections in the intact brain, using circuitry responsible for the vestibulo-ocular reflex (11–13) and the escape response (14) as testbeds. Finally, ExM helped visualize the subsynaptic organization of neurotransmitter receptors, revealing the ring-like organization of glycine receptors on the Mauthner (M) cell of the zebrafish (15, 16). Toward answering questions in development, we examined nuclear architecture in gastrulating zebrafish embryos. In particular, we characterized nuclear envelope invaginations [previously described in other cell types and species (17–27)], as well as the configuration of nearby microtubules, and found that such structures could be easily visualized via ExM across the zebrafish embryo. The scalability of ExM allowed us to detect channels passing through the nucleus at specific, and potentially rare, points in the cycle of cell division, which may help illuminate the mechanics of this process. ExM helped with the visualization of chromatin with regard to molecular markers such as microtubules, which may help unravel how the structural and molecular organization of cells leads to successful mitosis in embryogenesis and development.
Methods
Fish Maintenance and Care.
All zebrafish (Danio rerio) larvae were raised in fish facility water at Harvard University according to protocols and procedures approved by the Harvard University/Faculty of Arts & Sciences Standing Committee on the Use of Animals in Research and Teaching (Institutional Animal Care and Use Committee), with the following exceptions: Larvae used in Fig. 1 and SI Appendix, Fig. S1 A, B, and D and one of two larvae used for the analysis shown in SI Appendix, Fig. S1C were raised in Danieau’s medium at the Max Planck Institute of Neurobiology. These animal procedures conformed to the institutional guidelines of the Max Planck Society and the local government (Regierung von Oberbayern). Experimental protocols were approved by Regierung von Oberbayern (55.2-1-54-2532-101-12 and 55.2-1-54-2532-31-2016). All larvae were raised on a standard 14-h light/10-h dark cycle at a temperature of 28 °C.
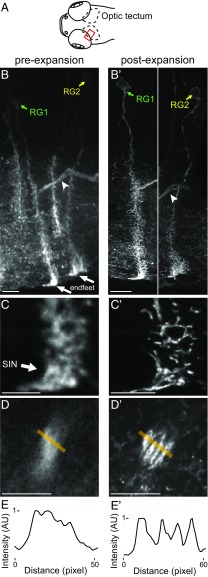
Fig. 1.
ExM helps resolve the morphology of fine cellular processes. (A) Schematic of the larval zebrafish brain showing the imaged area (red rectangle) within the left optic tectum. (B and B′) Maximum intensity projections of part of the tectum (highlighted in red in A) of a 6-d postfertilization larval zebrafish sparsely expressing membrane-bound EGFP and stained for GFP preexpansion (B) and postexpansion (B′), showing radial glial cells (two of which are labeled RG1 and RG2) and projection fiber bundles (arrowheads). (C and C′) Single confocal slices show projections of cell RG1, preexpansion (C) and postexpansion (C′). Endfeet processes of this cell wrap around the cell body of a superficial interneuron [SIN (113); arrow]. (D and D′) A bundle of extratectal fibers preexpansion (D) and postexpansion (D′) from the regions highlighted by arrowheads in B and B′, respectively, is shown. (E and E′) Intensity plots along the orange line in D and D′, respectively. AU, arbitrary units. [Scale bars: B, 10 μm; B′, 10 μm (physical size postexpansion, 35 μm); C and D, 5 μm; C′ and D′, 5 μm (17.5 μm).]
Transgenic Fish Lines.
The genotypes of the larvae and embryos used to generate each figure are detailed in SI Appendix, SI Methods. The transgenic fish lines that were crossed to produce these larvae and embryos were all previously described. All larval brain images are from 6-d postfertilization larvae, and all embryo images are from shield-stage (i.e., ∼6-h postfertilization) embryos.
Immunohistochemistry.
Immunohistochemistry was performed following standard, previously published procedures (28). The exact protocol, as well as a detailed list of antibodies used, is provided in SI Appendix, SI Methods.
Expansion.
Expansion was performed using the previously described proExM protocol (2) (SI Appendix, SI Methods).
Imaging.
Both pre- and postexpansion brains and embryos were imaged on an Andor spinning disk (CSU-X1 Yokogawa) confocal system with a 40×, 1.15 N.A. water immersion objective (Nikon), with the exception of some images in SI Appendix, Fig. S1: The first and third images in SI Appendix, Fig. S1A and all preexpansion images used to generate SI Appendix, Fig. S1C were acquired using a Deltavision OMX Blaze (GE Healthcare) structured-illumination microscope (SIM) with a 60×, 1.42 N.A. oil immersion objective (Olympus). These brains were immersed in SlowFade Diamond Antifade mounting medium (Invitrogen) for refractive index matching and suppression of bleaching. The first and third images in SI Appendix, Fig. S1B and all preexpansion images used to generate SI Appendix, Fig. S1D were acquired using a Leica TCS SP8 STED microscope, with a 100×, 1.4 N.A. oil immersion objective. These brains were immersed in SlowFade Gold Antifade mounting medium (Invitrogen) for refractive index matching and suppression of bleaching. The second and fourth images in SI Appendix, Fig. S1B and all postexpansion images used to generate SI Appendix, Fig. S1D were acquired using a Leica TCS SP8 confocal microscope, with a 40×, 1.1 N.A. water immersion objective. Images of the samples were also obtained with a 10×, 0.45 N.A. air objective and used to aid in the comparison of pre- and postexpansion data and computation of expansion factors. Details of excitation and emission collection are provided in SI Appendix, SI Methods. For embryos, postexpansion imaging was also performed via a Nikon Ti-E epifluorescence microscope with a 4×, 0.13 N.A. air objective to allow capturing of the entire sample for computation of expansion factors. For brains, expansion factors were computed by measuring the size of specific anatomical features (e.g., the axon cap) pre- vs. postexpansion, and taking the ratio of the respective sizes. For embryos, the diameter of the embryo pre- vs. postexpansion was compared. Scale bars on postexpansion images reflect these expansion factor computations. For the expanded embryos imaged in SI Appendix, Fig. S10, pre- and postexpansion images were taken from very different angles; thus, an exact expansion factor could not be computed. For SI Appendix, Fig. S10, an expansion factor of 4 [similar to the expansion factors computed for other embryos (3.8 and 4.1)] was estimated for the purpose of drawing scale bars.
Image Processing.
Each figure panel constitutes a single plane from a z-stack, where the area of interest was cropped out of the field of view using Fiji (29), or a maximal intensity projection, as indicated in the figure legends. The brightness and contrast of individual channels were adjusted in ImageJ (NIH) after cropping the area of interest. The STED preexpansion images shown in SI Appendix, Fig. S1B (first and third panels) and used in SI Appendix, Fig. S1D were deconvolved using Huygens (Scientific Volume Imaging). Tracing of cellular processes (shown in SI Appendix, Fig. S2) was performed using Imaris. This tracing algorithm is intensity-based. First, start and end points are detected, and then these points are connected with traces following the image intensity. Fig. 2B is a maximal intensity projection of four stacks acquired separately and stitched together using Fiji’s pairwise stitching plug-in (30). The data shown in Fig. 3 B–E and SI Appendix, Figs. S5 and S6 were cropped from stacks following illumination correction using CIDRE (31) and deconvolution using Huygens. Since the illumination model is dictated by the microscope optics, a single illumination model was learned using CIDRE by pooling the datasets together, and then this same model was used for the correction of both pre- and postexpansion datasets. After the application of illumination correction, a dataset-specific threshold was manually set according to the characteristic background noise level. Both pre- and postexpansion datasets were then deconvolved using the exact same procedure and parameters.
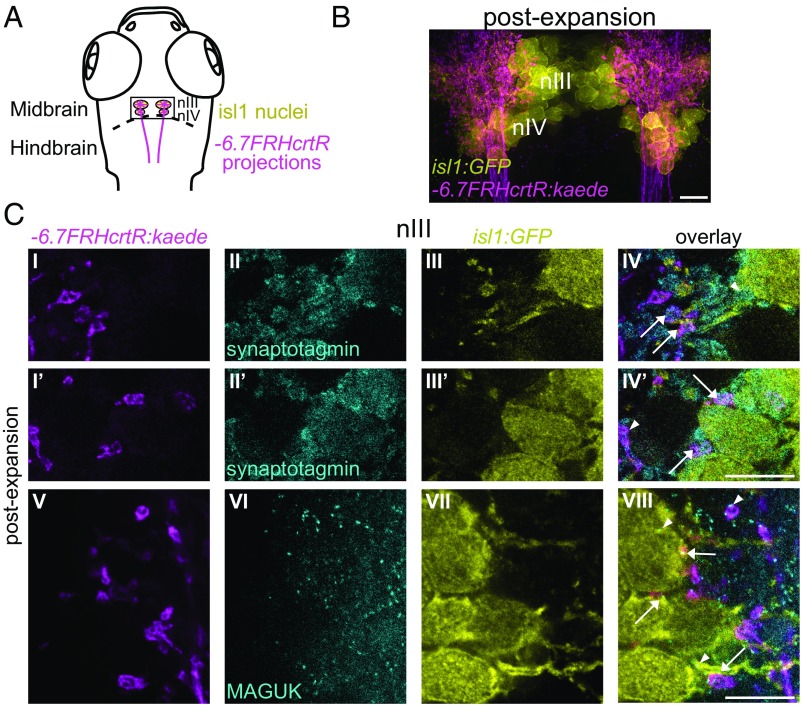
Fig. 2.
ExM analysis of synaptic connections. (A) Schematic of larval zebrafish brain showing nIII and nIV nuclei, labeled by Tg(isl1:GFP)rw0 (GFP, yellow) and neural projections labeled by Tg(−6.7FRhcrtR:Gal4VP16);Tg(UAS:Kaede) (Kaede, magenta). The rectangular area is imaged in B. (B) Maximal intensity projection of an ∼33-μm-thick volume corresponding to the rectangular area shown in A. The fish is 6 d postfertilization (dpf), and is stained with anti-GFP (yellow), anti-Kaede (magenta), and anti–pan-MAGUK (not shown). (C) GFP-labeled cells (yellow) and Kaede-labeled projections (magenta) in the nIII nucleus. (C, I–IV and I′–IV′) Two nearby planes (one in each row) from an expanded 6-dpf brain stained with anti-GFP (yellow), anti-Kaede (magenta), and anti-synaptotagmin2b (cyan). Arrows point to Kaede-expressing and synaptotagmin2b-stained varicosities and terminals next to GFP-labeled neuropil (IV) and cell bodies (IV′). Arrowheads point to a cluster of synaptotagmin2b, unlabeled by Kaede, next to a GFP-labeled cell (IV) and a Kaede-labeled synaptotagmin2b-stained varicosity next to a GFP-negative cell (IV′). (C, V–VIII) Single plane from a brain stained with anti–pan-MAGUK (cyan). Arrows point to Kaede-labeled varicosities and terminals next to GFP-labeled cells and neuropil, exhibiting colocalized MAGUK puncta. Arrowheads point to a MAGUK punctum on a GFP-negative cell opposed to a Kaede-labeled terminal (Top arrowhead) and to MAGUK puncta on GFP-labeled cell bodies and neuropil in the absence of nearby Kaede-labeled projections (Bottom two arrowheads). [Scale bars: B, 10 μm (38 μm); C, I–IV and I′–IV′, 5 μm (23 μm); C, V–VIII, 5 μm (19 μm).]
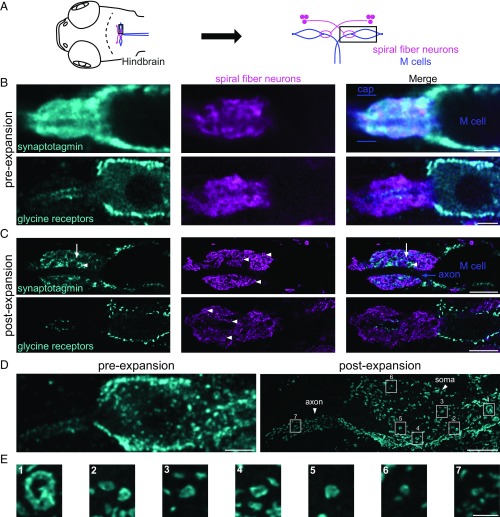
Fig. 3.
Expansion enables the resolving of synaptic heterogeneity and structure in intrasynaptic protein distributions. (A) Schematic of a larval zebrafish brain showing the M cells (blue) and spiral fiber neurons (magenta). The rectangle illustrates the region focused in on in B–D, consisting of the axon cap and a part of the M cell body. (B) Preexpansion images of the axon-cap area showing spiral fiber neurons (magenta) wrapping around the M cell axon initial segment (the unlabeled “tube” passing through these fibers, better visualized as a black stripe in C), as well as synaptotagmin2b (Top, cyan) and glycine receptors (Bottom, cyan). (C) Same as in B, but postexpansion. (Note: The synaptotagmin2b axon cap shown (Top) is not from the same brain as in B, Top.) (Top Left and Right) Arrows point to a Kaede-labeled varicosity bearing synaptotagmin2b at a low density. (Top Left and Right) Arrowheads point to a Kaede-negative varicosity bearing dense synaptotagmin2b staining. (Center) Arrowheads point to varicosities in spiral fiber neuron projections forming the M cell axon cap. (D) Maximal intensity projection of the M cell body and axon initial segment area showing the distribution of glycine receptors (cyan) preexpansion (Left) vs. postexpansion (Right). (Note: This is the same axon cap as shown in B and C, Bottom.) Boxes highlight seven examples of ring-shaped clusters zoomed in on in E. (E) Seven examples of ring-shaped clusters of various sizes present on the M cell body (1–6), and axon (7). [Scale bars: B, Top and Bottom, 5 μm; C, Top, 5 μm (23 μm); C, Bottom, 5 μm (20 μm); D, Left, 5 μm; D, Right, 5 μm (20 μm); E, 1 μm (4 μm).]
Measurement Error Quantification.
Errors were quantified using the same procedures as previously described (2, 32), with a few exceptions (SI Appendix, SI Methods).
Results
Validation of ExM of Zebrafish.
The proExM protocol has been previously successfully used in a variety of applications and tissue types, including cell culture as well as mouse brain slices (2, 33); mouse lung, spleen, and pancreas slices (2); isolated mouse mitochondria (34); planaria (35); the central nervous system and the germarium tip of ovaries in Drosophila (36, 37) and human tissue specimens prepared in a variety of different manners (32, 38). In many of these tissue types, the isotropy of expansion at the nanoscale was validated by comparison of postexpansion confocal images with superresolution structured-illumination microscopy (SIM) images of the same samples preexpansion [cell culture, mouse brain, and pancreas slices (2) and human breast biopsy tissue slices (32)]. Interestingly, ExM has rapidly become trusted enough that it has also been used without such validation, sometimes even in novel species or tissue types (33, 36, 38). While the widespread validation and trust in ExM bodes well for the use of the technology, we here sought to pursue validation nonetheless.
We first validated the isotropy of expansion of larval zebrafish brains at the nanoscale, using the previously developed methodology for validation by comparison with a classical superresolution method over small regions that could be imaged by both methods. Thus, we imaged larval zebrafish brains preexpansion with two different types of superresolution microscopes, SIM and a stimulated emission depletion (STED) microscope, and found these images to be nearly identical to postexpansion images of the same regions in the same brains (SI Appendix, Fig. S1 A and B). We do note, however, that depths of field vary in pre- and postexpansion images [due to expansion occurring in the axial dimension (1) as well as in lateral directions and, for the images shown here, also due to the use of different microscopes and objectives]; thus, sample features sometimes appear in one image but not the other. This difference in appearance is further aggravated by higher tissue scattering preexpansion compared with the optically clear (1) postexpansion samples, causing reduced signal-to-noise ratios that may obscure some features in preexpansion samples. Finally, orientation differences (even ones remaining after image registration) result in pre- and postexpansion images representing overlapping, yet distinct, optical sections in the samples.
Nevertheless, a quantitative comparison of the data shows that, in length measurements performed using postexpansion images, the root mean square (rms) of measurement errors relative to the lengths measured was comparable to the results obtained in previously published ExM papers. In particular, the rms of measurement errors was no larger than 5% for preexpansion images acquired with SIM and 2% for preexpansion images acquired with STED microscopy, on average (SI Appendix, Fig. S1 C and D). We note that these estimates constitute higher bounds on actual deformations in the samples, as they are affected by imperfections in the preliminary registration of datasets in three dimensions using rigid transformation and scaling (Methods).
Expansion Facilitates Tracing Cellular Processes in the Zebrafish Tectum.
We next set out to assess whether ExM could enable the tracing of fine cellular processes. We used a larva (strain details are provided in Methods) that strongly expresses membrane-bound GFP in a limited number of cells in the tectum (Fig. 1A), and particularly in radial glial cells (Fig. 1B). Expansion made their processes more visible vs. in preexpansion images (compare maximal intensity projections in Fig. 1B′ vs. Fig. 1B and single-plane images of specific regions in Fig. 1C′ vs. Fig. 1C). When we applied commercially available automated tracing software to quantitatively analyze these processes, the number of automatically identified branch points in the traced cells increased approximately sixfold postexpansion vs. when tracing was performed preexpansion (compare SI Appendix, Fig. S2A′ vs. SI Appendix, Fig. S2A and zoomed-in regions in SI Appendix, Fig. S2B′ vs. SI Appendix, Fig. S2B; numbers in the images reflect branch points of the cells indicated; also Movie S1). We were also able to resolve multiple processes within bundles that were not resolvable preexpansion (Fig. 1B′ vs. Fig. 1B, arrowheads and Fig. 1 D′ and E′ vs. Fig. 1 D and E). Thus, ExM may help clarify fine processes in zebrafish cells, as we had earlier found for cellular processes in mouse hippocampus (2).
Resolving Synapses Mediating Sensory/Motor Transformations.
We next used ExM to resolve synaptic connections, using larvae (Methods) in which the labeled neurons include some of the key neurons involved in the vestibulo-ocular reflex. In particular, the fluorophore Kaede (39) was expressed in projections into the oculomotor (nIII) and trochlear (nIV) nuclei, including putative tangential neuron projections (12) important for this reflex; furthermore, GFP was expressed in neurons in the nIII and nIV nuclei (highlighted in Fig. 2 A and B and Movie S2), which consist of extraocular motoneurons (40). We found that ExM revealed fine details of Kaede-labeled projections (compare Fig. 2C and SI Appendix, Figs. S3A and S4, examples 1–12, Bottom with SI Appendix, Figs. S3B and S4, examples 1–12, Top), enabling their terminations, which had the appearance of synaptic varicosities, to be visualized. The presynaptic protein synaptotagmin2b (Fig. 2C, I–IV and I′–IV′ and SI Appendix, Fig. S3A, I–IV) was localized to such varicosities, and particularly to their boundaries, consistent with these varicosities being presynaptic terminals (arrows in Fig. 2C, IV and IV′ and in SI Appendix, Fig. S3A, IV point to such varicosities abutting right next to GFP-positive cell bodies and neuropil). Similarly, when we immunostained with antibodies against postsynaptic proteins of the MAGUK class, with a pan-MAGUK antibody (41) (Fig. 2C, V–VIII and SI Appendix, Fig. S3A, V–VIII), postexpansion data clearly revealed MAGUK-positive puncta in GFP-labeled cells (arrows in Fig. 2C, VIII and SI Appendix, Fig. S3A, VIII), which were located in apposition to varicosities of Kaede-labeled projections, and thus were putative postsynaptic targets. The ability to characterize the location of synaptic proteins in regard to cell–cell contacts may help pinpoint fine details of synaptic wiring. In particular, examining the Kaede-labeled terminals abutting next to GFP-positive cells in preexpansion data from this pan-MAGUK–stained brain, we identified 12 example terminals where the data suggested more than one GFP-positive cell as a putative postsynaptic target (SI Appendix, Fig. S4, Top). However, examining the same regions in the same brain postexpansion, we found that MAGUK puncta were often present only on a subset of these putative postsynaptic targets, perhaps indicating a selectivity of functional contact not visible in preexpansion data (SI Appendix, Fig. S4, Bottom). The number of GFP-positive targets identified pre- vs. postexpansion for each terminal is indicated in SI Appendix, Table S1. This number is lower in six of the 12 examples in postexpansion compared with preexpansion data. Thus, this analysis suggests that ExM enables more specific assignment of putative postsynaptic partners in the larval zebrafish nervous system.
Expansion Allows Resolving of Subsynaptic Structures.
We next explored how ExM could reveal heterogeneity of synaptic composition in dense neuropil, focusing on synapses onto the M cell, which triggers a fast-escape response by firing a single action potential (42, 43). The axon cap is a dense area of neuropil that surrounds the axon initial segment and hillock of the M cell (44) (Fig. 3 A and B, Top Right). To visualize this area, we used larvae (Methods) in which the spiral fiber neurons whose projections form the axon cap (15, 45) express the fluorophore Kaede (46). Putative presynaptic terminals (as indicated with anti-synaptotagmin2b staining) were visible throughout the cap in both preexpansion (Fig. 3B, Top) and postexpansion (Fig. 3C, Top) data, consistent with electron microscopy data indicating the existence of synapses between spiral fiber neurons and themselves, as well as from these neurons onto the M cell axon (15, 47, 48) in the cap. However, individual putative synaptic varicosities were only resolvable postexpansion (Fig. 3C, Center, arrowheads and Movie S3). Furthermore, different synaptotagmin2b-positive varicosities exhibited different amounts of synaptotagmin2b staining (compare arrow vs. arrowhead in Fig. 3C, Top Right and Left), reflecting either synapse-to-synapse variability within a single type of synapse or the presence of multiple molecularly distinct synapse types in the axon cap (15, 47, 48). This heterogeneity also held for the synaptic vesicle glycoprotein 2a (SV2; arrows and top arrowhead vs. bottom arrowhead in SI Appendix, Fig. S5A, Bottom) and the presynaptic proteins synapsin1/2 (SI Appendix, Fig. S5B, Right, arrow vs. arrowhead). To examine whether proExM could go beyond the analysis of synaptic composition and help with the analysis of synaptic structure, we examined the distribution of glycine receptors on the M cell (Fig. 3 B and C, Bottom). These synapses were present on the M cell body and axon, but absent from the rest of the cap. Using a larva that additionally expressed GFP under the control of the glycine transporter-2 (glyt2) promoter (49), we found that postexpansion, “leg”-like protrusions from glycinergic neurons that approached glycine receptor patches on the M cell were visible (SI Appendix, Fig. S5C, Bottom Right, arrows). Furthermore, glycine receptors formed ring-like patches that were not resolvable preexpansion (Fig. 3D, Right and Movie S4 vs. Fig. 3D, Left). The sizes of these rings varied, with a gradient of decreasing size along the soma toward the axon (Fig. 3E). These findings are consistent with earlier findings in goldfish (16): With ExM, these rings, which are much smaller in the genetically tractable zebrafish, become visible.
Finally, these data also suggested that ExM may enable the better identification of glycine receptor clusters on such putative synapses (SI Appendix, SI Methods). To examine this, we segmented images of four M cell axons in two brains with labeled glycine receptors (SI Appendix, Fig. S6A) and counted the number of distinct receptor clusters as a function of the intensity threshold used in this segmentation. When segmentation was performed with low-intensity thresholds, clusters that visually seemed distinct were merged together, while at high thresholds, some clusters were eliminated. Thus, a peak in the identified number of clusters occurred at an intermediate threshold, representing a trade-off between these two alternatives. Importantly, this analysis revealed that at the peak and for a broad range of thresholds around it, the number of distinct clusters identified in postexpansion images was at least sixfold higher than in preexpansion images (SI Appendix, Fig. S6B). In preexpansion images, a low signal-to-noise ratio and limited resolution caused distinct clusters to merge together and small clusters to blend with the background. Thus, this analysis demonstrates that ExM data may be beneficial in counting distinct putative synapses.
Expansion Reveals the Shape of Intranuclear Invaginations in Embryos.
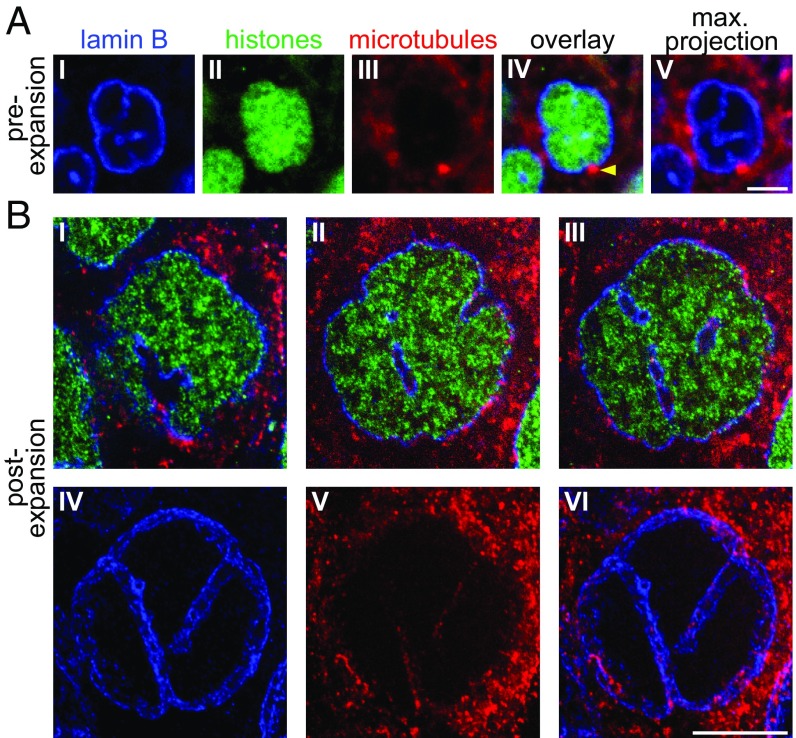
To explore ExM in the context of zebrafish developmental biology, we investigated the morphology of nuclei at an early embryonic stage. We used embryos from a fish line where EGFP is fused to histone-2B (50) (details are provided in Methods), and we used anti-lamin B and anti-tubulin to visualize the nuclear boundary and microtubules, respectively. Of ∼350 nuclei across two ∼6-h-old zebrafish embryos (∼250 and ∼100 nuclei in the two embryos, respectively), we found that ∼35 nuclei were between the prometaphase and telophase stages, whereas the other ∼315 nuclei were in interphase or prophase (i.e., had intact nuclear envelopes). Preexpansion data revealed lamin B within the nucleus, overlapping with labeled histones (Fig. 4A). Postexpansion, such lamin B staining could be resolved as nuclear envelope invaginations containing a cytoplasmic core (i.e., void of histone staining), as had been previously reported in interphase cells (20, 26) (Fig. 4B, Top and Movie S5). Indeed, with ExM, we could see highly variable organizations of such invaginations, ranging from just one or two channels passing through the nucleus (Fig. 4B and SI Appendix, Fig. S7, A and C, I–IV and I′) to complex networks of intranuclear channels (SI Appendix, Fig. S7 B and C, V–VII, VI′, and VII′).
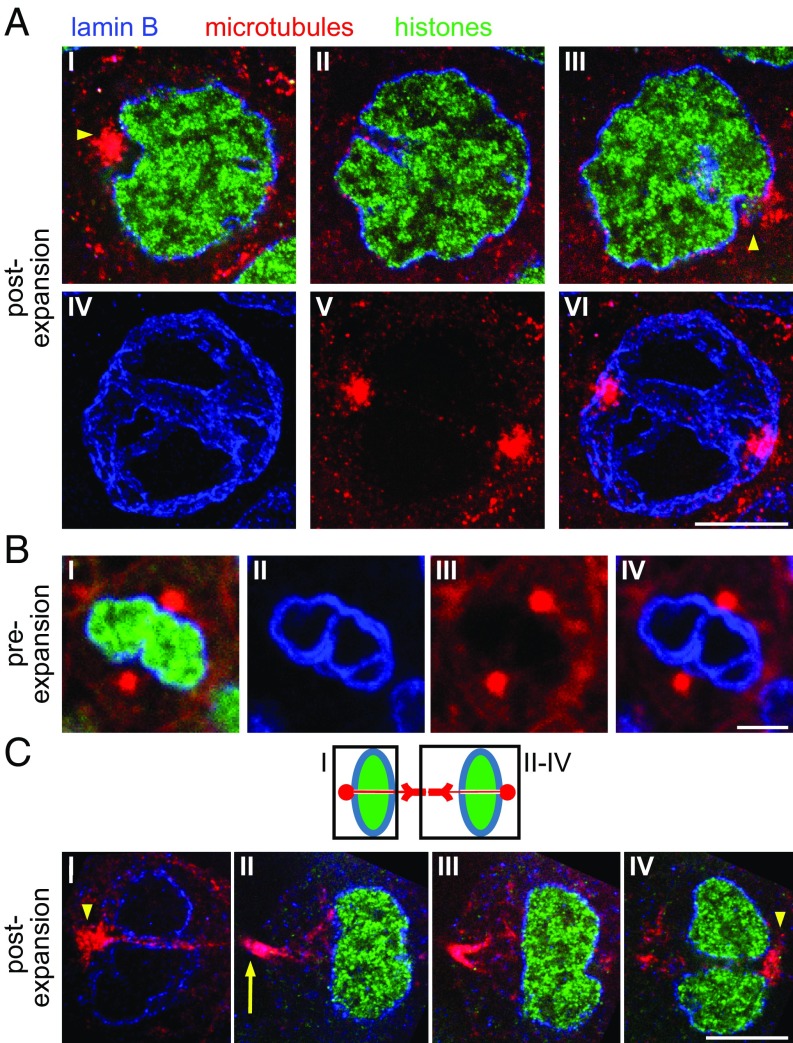
Fig. 4.
ExM examination of intranuclear invaginations. In all panels in this figure, in Figs. 5 and 6, and in Movies S5–S8: blue, anti-lamin B; green, histone 2B [EGFP fused to histone 2B in Tg(actb2:h2b-egfp/actb2:mem-mCherry2) and then stained with anti-GFP]; red, anti–α-tubulin (microtubules). All nuclei in this figure are from the same shield-stage embryo, either preexpansion (A) or postexpansion (B). (A, I–IV) Plane within a nucleus from this embryo. (A, IV) Arrowhead points to a centrosome positioned at the exit point of an indentation ending within the nucleus. (A, V) Maximal intensity projection of the invagination-containing area of this nucleus. (B, I–III) Three planes within a nucleus, postexpansion. (B, IV–VI) Maximal intensity projection of the invagination-containing area of this nucleus (shown is the same projection in all three panels, with stains as defined above). [Scale bars: A, 5 μm; B, 5 μm (20.5 μm).]
To compare the extent to which the structure of such intranuclear channels could be quantitatively captured using pre- vs. postexpansion images, we sampled 30 nuclei from each dataset (SI Appendix, SI Methods and Figs. S8 and S9). For each nucleus in these samples, the number of disconnected channel structures identified within the nucleus, the number of exit points from the nucleus, and the number of internal end points within the nucleus are enumerated in SI Appendix, Tables S2 and S3 for pre- and postexpansion samples, respectively. In preexpansion data, it was sometimes difficult to establish whether structures were connected or disconnected, and whether a channel reached all of the way to the nuclear boundary or ended within the nucleus, close to its boundary. Consequently, to capture multiple possible interpretations of the data, we estimated both minimal and maximal values for the above-described features, given the possible image interpretations (SI Appendix, Table S2). We found that, due to the enhanced resolution, complex structures were better captured in postexpansion data, leading to higher estimates for the mean number of disconnected structures and the mean number of exit points identified in nuclei when comparing postexpansion estimates with the minimal preexpansion estimates. Higher exit point counts were found in postexpansion data as well, compared with maximal preexpansion estimates. The number of end points within the nucleus was similar pre- and postexpansion. Put together, this analysis shows that ExM improved the ability to capture the complexity of intranuclear invagination structures.
We were further able to map out microtubule organization in and around nuclear invaginations, which is of interest since many kinds of cytoskeletal filaments have been associated with such invaginations (20, 22, 24, 26, 27, 51). For example, many centrosomes (clusters of microtubules highlighted with arrowheads in Fig. 4 and SI Appendix, Fig. S7) were localized at invagination openings, although some were offset from the openings (e.g., SI Appendix, Fig. S7, VII, arrow). We also observed microtubule staining at the boundaries of invaginations (Fig. 4B, Bottom and SI Appendix, Fig. S10), although such staining was only visible in a small number of nuclei and the microtubules did not appear continuous. One possibility is that these samples had poor antibody staining, since the dye we used (Methods) was one known to be poorly retained in proExM (2). An additional experiment performed supported this possibility: In two supplementary specimens labeled with dyes known to persist well in proExM (SI Appendix, Fig. S10 and Movie S6), anti-tubulin staining was of higher quality postexpansion. In ∼650 nuclei from these two embryos (∼400 and ∼250 nuclei, respectively), approximately half exhibited microtubules throughout the lengths of nuclear channels (which may still be an underestimate, as the tubulin antibody did not stain samples evenly; Methods). In summary, microtubule staining along the boundaries of intranuclear channels appears to be a widespread phenomenon in shield-stage embryonic nuclei.
Expansion Reveals Intranuclear Channels at Late Prophase and Telophase.
Of the ∼350 nuclei we analyzed in our main dataset, we found 16 cells (Fig. 5 and SI Appendix, Figs. S7 and S11) in late prophase, with centrosomes on opposing sides of the cell. At this stage, previous studies in cultured mammalian cells suggested that microtubule-containing nuclear envelope indentations form next to centrosomes as the nuclear envelope begins to disassemble (21, 52–56; reviewed in ref. 57.) Indeed, four of the 16 nuclei exhibited indentations ending within the nucleus and starting next to centrosomes (SI Appendix, Fig. S11B, V–VIII). However, in 12 of the 16 nuclei, a channel passed through the nucleus, connecting the two centrosomes (one example is shown in Fig. 5A and Movie S7; further examples are shown in SI Appendix, Figs. S7C, VI and VII and S11; arrowheads in all figure panels point to centrosomes). Microtubule staining was sometimes visible within the channel (Fig. 5A, V). Such channels were not resolved preexpansion (although hints were visible; Fig. 5B). We further observed that such nuclear channels might link cytoskeletal components in unanticipated ways in cells in late telophase. In particular, in late-telophase nuclei (nine of the ∼350 total nuclei; Fig. 5C and SI Appendix, Fig. S12 and Movie S8), we identified intranuclear channels running through daughter nuclei. A centrosome was positioned at one opening of each of these channels (Fig. 5C, I and IV, arrowheads), and microtubules passed through the channels (Fig. 5C, I and IV), converging at the midbody (Fig. 5C, II, arrow); microtubule staining is absent from the midbody center, likely due to the density of proteins causing epitope masking (58). Thus, these nuclear channels may permit microtubule connections to be made between the now-distant centrosomes. Heterogeneity in the shape and appearance of these structures was apparent from cell to cell; for example, the midbodies of SI Appendix, Fig. S12, I, IV, VI, and VIII seem to exhibit only one microtubule bundle emerging from each side of the midbody rather than two. Overall, however, this configuration (schematized in Fig. 5C, Top) suggests that the nuclear channels might mediate a cross-daughter cell mechanical connection at a time during which the reassembled nuclear envelope would otherwise topologically prohibit such a connection. Another possibility is that the daughter cells have simply advanced beyond telophase, and the phenomena of Fig. 5C are similar to the other phenomena we saw in Fig. 5A (as discussed above); this possibility has been commented on in earlier studies of cells immediately after division (19, 21). Future studies can take advantage of the extended scale imaging capacity of ExM to enable nanoscale imaging of dividing cells, hopefully unraveling structural relations between a variety of proteins and cellular compartments participating in this process (59, 60).
Fig. 5.
ExM reveals intranuclear channels in late-prophase and telophase nuclei. (A) Nucleus at late prophase, postexpansion. (A, I–III) Three planes from the nucleus. (A, I) Arrowhead points to a centrosome at one opening of an intranuclear channel. (A, III) Arrowhead points to a centrosome at the other opening of the intranuclear channel. (A, IV–VI) Maximal intensity projection of the intranuclear channel-containing area, showing that the channel runs between the two centrosomes located at opposing edges of this nucleus. (B) Nucleus at late prophase, preexpansion, containing two centrosomes at opposite ends of a lamin B-stained channel. (B, I) Representative plane from the nucleus. (B, II–IV) Maximal intensity projection of the invagination-containing region of the nucleus. (C) Structure of a late-telophase nucleus, postexpansion. (Top) Schematic of the structure observed in 10 late-telophase nuclei (eight more nuclei are shown in SI Appendix, Fig. S12). Chromatin (green) is unpacked and fills the daughter nuclei that are surrounded by continuous lamin B staining (blue). Two microtubule bundles (red) diverge from their convergence point at the midbody toward each daughter nucleus and disperse when they approach the nucleus. Unbundled microtubules pass through a channel through each daughter nucleus to connect with the centrosome at the other end. Black boxes outline the areas indicated in I–IV. (C, I) Maximal intensity projection of the invagination-containing area in one of the two daughter nuclei, showing a channel traversing through this nucleus. The arrowhead points to a centrosome at the channel opening. (C, II–IV) Different planes showing the second daughter nucleus. (C, II) Arrow points to a dip in the intensity of microtubule staining at the midbody. (C, IV) Arrowhead points to a centrosome at the channel opening within the second daughter nucleus. [Scale bars: A, 5 μm (19 μm); B, 5 μm; C, 5 μm (20.5 μm).]
ExM Facilitates Analyses of Mitosis in Zebrafish Embryos.
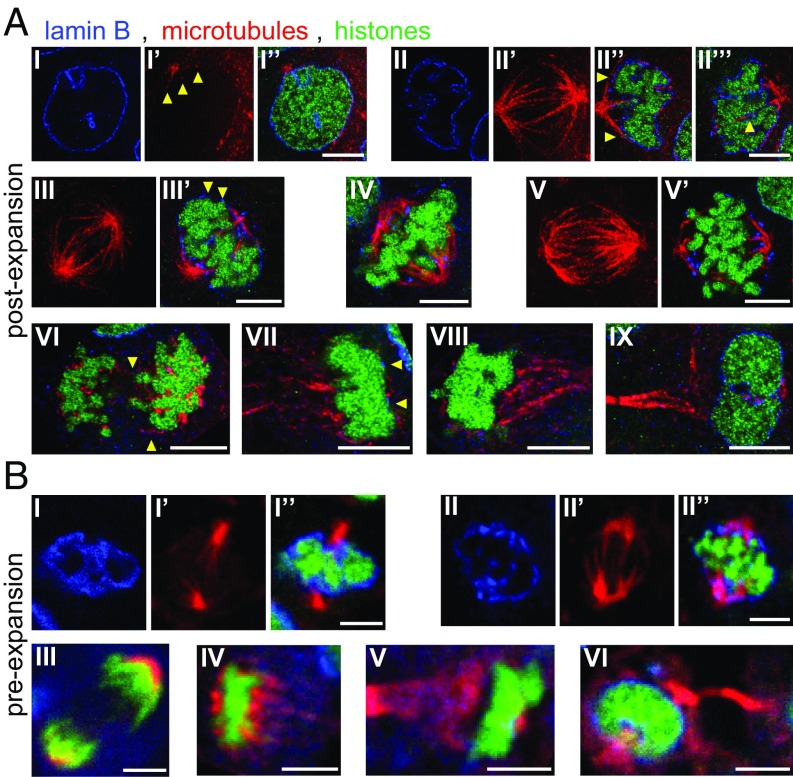
We examined cells in other stages of cell division as well. Post- and preexpansion data from mitotic cells were highly consistent (e.g., compare Fig. 6A vs. Fig. 6B), with some stages nearly identical in appearance (e.g., metaphase images in Fig. 6A, IV vs. Fig. 6B, II′′; late anaphase images in Fig. 6A, VII and VIII vs. Fig. 6B, IV and V). However, the structure of microtubules was more accurately captured in postexpansion data vs. preexpansion data, with putative kinetochore-attached microtubule bundles (61) observed to traverse longer distances within nuclei postexpansion [e.g., compare before expansion (Fig. 6B, I′ and II′) vs. after expansion (Fig. 6A, II′ and III)], and even the fine and short microtubules of prometaphase were visible postexpansion (Fig. 6A, I′, arrowheads). Furthermore, microtubules near the nucleus boundary (as demarcated with lamin B staining) were easily identified postexpansion in four of five prometaphase nuclei identified in our dataset (Fig. 6A, II′′, arrowheads and SI Appendix, Fig. S13, yellow arrowheads; the fifth nucleus was excluded since it was oriented along the optical axis of the objective lens, with its poorer resolution in this direction). Even fine microtubules at early anaphase (Fig. 6A, VI, arrowheads) were observable postexpansion. These various structures are consistent with earlier electron microscopy studies (62, 63), with the exception of the observation of microtubules at nuclear boundaries, which were not described in these earlier studies, perhaps since these studies focused on the internal part of the nucleus and did not explore the boundaries. Similar to the microtubules, lamin B patches were easier to resolve postexpansion (e.g., Fig. 6A, III′, arrowheads) than preexpansion (Fig. 6B, I and II). In particular, patches of lamin B were observable postexpansion to be associated with microtubules that protrude into the nucleus during prometaphase (Fig. 6A, II′′′ and SI Appendix, Fig. S13, white arrowheads). Finally, we examined chromatin postexpansion, using the common strategy of imaging EGFP fused to histones as a proxy for chromatin density (64, 65). Postexpansion data recapitulated discoveries made using diffraction-limited light microscopy (66–68). For example, chromatin appeared less densely packed at prophase and early prometaphase (Fig. 6A, I and II) than in late prometaphase and metaphase (Fig. 6A, III–V), and distinct chromosome arms are not apparent at late anaphase when chromatin is very densely packed (66) (Fig. 6A, VII and VIII). In postexpansion data, chromatin appears (at least in some cell cycle phases) as discrete spots, as in other superresolution microscopy studies (69–71), which may convey its degree of compaction more directly than intensity changes. This suggests that in combination with DNA FISH, it may be possible to use expansion to map out the location of specific genes in the context of higher level chromatin features.
Fig. 6.
ExM and the analysis of mitotic nuclei. (A) Mitotic nuclei, postexpansion. (A, I–I′′) Nucleus at the transition between prophase and prometaphase. (A, I) Lamin B staining. (A, I′) Microtubules. Arrowheads point to three short microtubules emerging from one of the centrosomes. (A, I′′) Overlay. (A, II–II′′′) Nucleus at prometaphase. (A, II) Lamin B staining. (A, II′) Maximal intensity projection of the microtubules in this nucleus. (A, II′′ and II′′′) Two distinct planes in this nucleus. (A, II′′) Arrowheads point to microtubules at the boundaries of this nucleus. (A, II′′′) Arrowhead points to lamin B staining next to a microtubule protruding into this nucleus. (A, III and III′) Nucleus at late prometaphase. (A, III) Maximal intensity projection of microtubules in this nucleus. (A, III′) Plane from this nucleus. Arrowheads point to discontinuous patches of lamin B surrounding the nucleus. (A, IV) Plane from a nucleus at metaphase. (A, V and V′) Nucleus at early anaphase. (A, V) Maximal intensity projection of microtubules in this nucleus. (A, V′) Plane from this nucleus. (A, VI) Plane from a nucleus at anaphase. Arrowheads point to fine microtubules located between chromosomes. (A, VII) Plane from a nucleus at late anaphase. Arrowheads point to lamin B patches at the boundary of nuclear chromatin. (A, VIII) Plane from a nucleus at telophase. (A, IX) Plane from a nucleus at late telophase. (B) Mitotic nuclei, preexpansion. (B, I–I′′) Plane from a nucleus at late prometaphase. (B, I) Lamin B staining. (B, I′) Microtubules. (B, I′′) Overlay. (B, II–II′′) Plane from a nucleus at metaphase. (B, II) Lamin B staining. (B, II′) Microtubules. (B, II′′) Overlay. (B, III) Plane from a nucleus at anaphase. (B, IV) Plane from a nucleus at late anaphase. (B, V) Plane from a nucleus at early telophase. (B, VI) Plane from a nucleus at late telophase. [Scale bars: A, I–IV, and IX, 5 μm (20.5 μm); A, V–VIII, 5 μm (19 μm); B, 5 μm.]
Discussion
In this work, we explore the utility of ExM in the zebrafish, a model system popular for the investigation of topics in developmental biology and neuroscience. We found that ExM helped along numerous neuroscience axes, assisting with the visualization and tracing of fine cellular processes, enabling the resolving and compositional mapping of synapses, and enabling subsynaptic protein distributions (i.e., glycine receptor rings) to be visualized. In the context of development, we found that ExM enabled scalable study of intranuclear invaginations, as well as the analysis of how these are configured in specific stages of mitosis. Such static snapshots, while spatially precise, must of course be coupled to functional studies in the future; the transparent and genetically tractable nature of the zebrafish will undoubtedly facilitate this process.
In this study, we primarily compare our data with published electron microscopy studies of larval zebrafish brains and embryos (or cells from other species) rather than with superresolution microscopy imaging studies. This is because, presumably due to the limited z-depth accessible to classical superresolution methods, only a few studies have performed superresolution imaging in zebrafish to date. Techniques used included SIM (72–74), providing a resolution of ∼140 nm and depth penetration of 50–100 μm, and STED microscopy, providing ∼70-nm resolution but limited depth penetration [14-μm-thick sections of larval zebrafish retina were imaged in ref. 75; a commercial system (Leica TCS SP5 STED) with a reported 90-nm full-width at half-maximum lateral resolution and an objective with a working distance of 130 μm was used in ref. 76]. Thus, the successful application of ExM to whole-zebrafish brains and embryos described here may enable the systematic characterization of fine structures in this animal model.
Due to its transparency throughout development, the larval zebrafish is a well-established model for imaging- and optogenetics-based studies of nervous system function (77–79). Synaptic and morphological information provided by ExM would complement such data, and allow linkages to be made between synaptic connectivity and function. In particular, expanding brains after monitoring or manipulating activity in specific cell groups defined by molecular type or connectivity pattern could provide anatomical insights into how information flows or is transformed in the context of behavior. In the future, ExM may be combined with other markers of cellular membranes or cytosol designed to facilitate tracing (e.g., the “spaghetti monster” fluorescent proteins equipped with multiple immunoepitope tags described in ref. 80). Using multicolor labeling (e.g., with Brainbow constructs, as in refs. 81 and 82) could help disambiguate nearby or touching cells that express different combinations of fluorophores or immunoepitopes, reducing the effort associated with neuron tracing (5, 83, 84). Because ExM fills the sample with water, resulting in a transparent, refractive index homogenized specimen (1), such expanded samples are easily scanned with light-sheet microscopes, as we have demonstrated in the context of RNA imaging in expanded mouse brain specimens (85). Embedding preserved biological specimens in polymer hydrogels for imaging purposes has a long history (86), but the use of evenly synthesized swellable polymers to isotropically expand biomolecules away from each other is particularly useful because it can enhance the power of a diversity of rapidly evolving techniques, as described here. In particular, multicolor labeling facilitates antibody staining of multiple proteins at nanoscale resolution with ExM, which is difficult with electron microscopy (87). We regard it as promising that, in parallel to our development of the proExM variant of ExM, two other groups independently developed protocols related to proExM (33, 88), suggesting that proExM is a robust protocol that is easily implemented and deployed.
ExM could be particularly powerful in conjunction with reverse genetic technologies that have been successfully applied to zebrafish (89–92). For example, the structure of radial glial processes and neural projections could be characterized in mutants with compromised development (93–95) to reveal the roles different genes play in wiring the brain. As another example, the scaffolding protein gephyrin localizes glycine (and GABAA) receptors to inhibitory synapses (96). Using ExM in zebrafish, it would be possible to examine the shapes of glycine receptor clusters in mutants where the functions of genes encoding synapse scaffolding components (97, 98) or of other proteins involved in the regulation of such clustering are compromised (99–101).
In the context of development, expanding zebrafish embryos allowed us to identify intranuclear invaginations previously described in other models (20, 26) and to characterize their relationship with microtubules. Previous studies have identified various cytoskeletal filaments in relation to these compartments (20, 22, 24, 26, 27, 51); ExM may enable such analyses to be done rapidly and systematically. Expanding zebrafish embryos could also help link these structures to specific functions. For example, such invaginations (in cells from other animals) have been proposed to allow regulation of signaling molecule (e.g., Ca2+) concentration within parts of the nucleus (102, 103), which, in turn, could support spatial regulation of gene expression (23, 104, 105). ExM could allow for the mapping of epigenetic markers with respect to these invaginations and associated proteins, as well as the relationship between all of these variables and emergent cell phenotypes (22, 106). Our observation that internuclear invagination channels exist at late prophase and telophase demonstrates the value of a multiscale 3D nanoscopy method such as ExM. Such channels might serve a role in mechanically augmenting the spindle, by allowing microtubules to pass through them and to connect centrosomes at a point in time when the nuclear envelope may otherwise interfere with such direct connections. Of course, from the observation of structure alone, it is impossible to deduce the functional role of these channels, but such hypotheses could be tested in future studies. Studies in zebrafish have revealed a variety of mutants with cell cycle defects [e.g., ones caused by mutations in early-arrest genes (107)] or deficiencies in chromosome condensation and organization in both interphase and mitosis (108, 109), which could be investigated with ExM. In addition, since incorrect chromosome segregation in mitosis is a potential cause of genomic instability and cancer (110), ExM in zebrafish could be utilized to relate division errors to the development of malignancies (111, 112).
Supplementary Material
Acknowledgments
We thank Alex Schier for his support of A.R., J.A.G., and O.R., and for helpful discussions of the paper. We also thank Iain Cheeseman for useful comments on the paper. We thank Dr. Avi Jacobs for his help with STED imaging, Dr. Eliza Vasile for her help with SIM imaging, and Dr. Alexandra Lichtenstein and Dr. Miriam Shaharabani for their help with confocal microscopy. We thank Daniel M. Estandian and Giovanni Talei Franzesi for their help in larvae preparation for validation experiments. We thank Dr. Natalie Elia and Dr. Dikla Nachmias for their help with SIM imaging in preliminary validation experiments. We thank Yongxin Zhao and Fei Chen for their advice and code for validation data analysis. We thank Dr. Pablo Blinder for use of his laboratory facilities for validation experiments described in SI Appendix, Fig. S1 B and D. L.F. was supported by a postdoctoral fellowship from the Simons Center for the Social Brain at the Massachusetts Institute of Technology (MIT). D.F. was supported by a European Molecular Biology Organization fellowship (ALTF 104-2013). A.R. is a Howard Hughes Medical Institute Gilliam Fellow. E.K.C. was funded by a MIT Peter J. Eloranta Summer Undergraduate Research Fellowship. D.A.M.-A. was funded by a McGovern Institute Shelly Razin Fund Fellowship. This work was funded, in part, by the Max Planck Society. E.S.B. was funded by the Howard Hughes Medical Institute-Simons Faculty Scholars Program, the Open Philanthropy Project, the MIT Media Laboratory, the New York Stem Cell Foundation, NIH Grant 1R01EB024261, NIH Grant 1R01MH110932, NIH Grant 2R01DA029639, NIH Grant 1R01NS087950, NIH Grant 1U01MH106011, John Doerr, and Jeremy and Joyce Wertheimer.
Footnotes
Conflict of interest statement: E.S.B. and R.G. are inventors on one or more patent applications related to expansion microscopy (ExM). E.S.B. is co-founder of a company, Expansion Technologies, that aims to provide ExM kits and services to the community.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1706281114/-/DCSupplemental.
References
- 1.Chen F, Tillberg PW, Boyden ES. Expansion microscopy. Science. 2015;347:543–548. doi: 10.1126/science.1260088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tillberg PW, et al. Protein-retention expansion microscopy of cells and tissues labeled using standard fluorescent proteins and antibodies. Nat Biotechnol. 2016;34:987–992. doi: 10.1038/nbt.3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Friedrich RW, Genoud C, Wanner AA. Analyzing the structure and function of neuronal circuits in zebrafish. Front Neural Circuits. 2013;7:71. doi: 10.3389/fncir.2013.00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haffter P, et al. The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development. 1996;123:1–36. doi: 10.1242/dev.123.1.1. [DOI] [PubMed] [Google Scholar]
- 5.Helmstaedter M. Cellular-resolution connectomics: Challenges of dense neural circuit reconstruction. Nat Methods. 2013;10:501–507. doi: 10.1038/nmeth.2476. [DOI] [PubMed] [Google Scholar]
- 6.Peng H, et al. BigNeuron: Large-scale 3D neuron reconstruction from optical microscopy images. Neuron. 2015;87:252–256. doi: 10.1016/j.neuron.2015.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiao T, Baier H. Lamina-specific axonal projections in the zebrafish tectum require the type IV collagen Dragnet. Nat Neurosci. 2007;10:1529–1537. doi: 10.1038/nn2002. [DOI] [PubMed] [Google Scholar]
- 8.Scott EK, Baier H. The cellular architecture of the larval zebrafish tectum, as revealed by gal4 enhancer trap lines. Front Neural Circuits. 2009;3:13. doi: 10.3389/neuro.04.013.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nevin LM, Robles E, Baier H, Scott EK. Focusing on optic tectum circuitry through the lens of genetics. BMC Biol. 2010;8:126. doi: 10.1186/1741-7007-8-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiao T, et al. Assembly of lamina-specific neuronal connections by slit bound to type IV collagen. Cell. 2011;146:164–176. doi: 10.1016/j.cell.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boyden ES, Katoh A, Raymond JL. Cerebellum-dependent learning: The role of multiple plasticity mechanisms. Annu Rev Neurosci. 2004;27:581–609. doi: 10.1146/annurev.neuro.27.070203.144238. [DOI] [PubMed] [Google Scholar]
- 12.Bianco IH, et al. The tangential nucleus controls a gravito-inertial vestibulo-ocular reflex. Curr Biol. 2012;22:1285–1295. doi: 10.1016/j.cub.2012.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cullen KE. The vestibular system: Multimodal integration and encoding of self-motion for motor control. Trends Neurosci. 2012;35:185–196. doi: 10.1016/j.tins.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Korn H, Faber DS. The Mauthner cell half a century later: A neurobiological model for decision-making? Neuron. 2005;47:13–28. doi: 10.1016/j.neuron.2005.05.019. [DOI] [PubMed] [Google Scholar]
- 15.Koyama M, Kinkhabwala A, Satou C, Higashijima S, Fetcho J. Mapping a sensory-motor network onto a structural and functional ground plan in the hindbrain. Proc Natl Acad Sci USA. 2011;108:1170–1175. doi: 10.1073/pnas.1012189108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seitanidou T, Triller A, Korn H. Distribution of glycine receptors on the membrane of a central neuron: An immunoelectron microscopy study. J Neurosci. 1988;8:4319–4333. doi: 10.1523/JNEUROSCI.08-11-04319.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bourgeois CA, Hemon D, Bouteille M. Structural relationship between the nucleolus and the nuclear envelope. J Ultrastruct Res. 1979;68:328–340. doi: 10.1016/s0022-5320(79)90165-5. [DOI] [PubMed] [Google Scholar]
- 18.Broers JL, et al. Dynamics of the nuclear lamina as monitored by GFP-tagged A-type lamins. J Cell Sci. 1999;112:3463–3475. doi: 10.1242/jcs.112.20.3463. [DOI] [PubMed] [Google Scholar]
- 19.Ellenberg J, et al. Nuclear membrane dynamics and reassembly in living cells: Targeting of an inner nuclear membrane protein in interphase and mitosis. J Cell Biol. 1997;138:1193–1206. doi: 10.1083/jcb.138.6.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fricker M, Hollinshead M, White N, Vaux D. Interphase nuclei of many mammalian cell types contain deep, dynamic, tubular membrane-bound invaginations of the nuclear envelope. J Cell Biol. 1997;136:531–544. doi: 10.1083/jcb.136.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manilal S, Nguyen TM, Morris GE. Colocalization of emerin and lamins in interphase nuclei and changes during mitosis. Biochem Biophys Res Commun. 1998;249:643–647. doi: 10.1006/bbrc.1998.9209. [DOI] [PubMed] [Google Scholar]
- 22.Johnson N, et al. Actin-filled nuclear invaginations indicate degree of cell de-differentiation. Differentiation. 2003;71:414–424. doi: 10.1046/j.1432-0436.2003.7107003.x. [DOI] [PubMed] [Google Scholar]
- 23.Lee RKY, et al. The nuclear tubular invaginations are dynamic structures inside the nucleus of HeLa cells. Can J Physiol Pharmacol. 2006;84:477–486. doi: 10.1139/y05-110. [DOI] [PubMed] [Google Scholar]
- 24.Storch KN, et al. Alpha smooth muscle actin distribution in cytoplasm and nuclear invaginations of connective tissue fibroblasts. Histochem Cell Biol. 2007;127:523–530. doi: 10.1007/s00418-007-0275-9. [DOI] [PubMed] [Google Scholar]
- 25.Wittmann M, et al. Synaptic activity induces dramatic changes in the geometry of the cell nucleus: Interplay between nuclear structure, histone H3 phosphorylation, and nuclear calcium signaling. J Neurosci. 2009;29:14687–14700. doi: 10.1523/JNEUROSCI.1160-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malhas A, Goulbourne C, Vaux DJ. The nucleoplasmic reticulum: Form and function. Trends Cell Biol. 2011;21:362–373. doi: 10.1016/j.tcb.2011.03.008. [DOI] [PubMed] [Google Scholar]
- 27.Jorgens DM, et al. Deep nuclear invaginations are linked to cytoskeletal filaments–Integrated bioimaging of epithelial cells in 3D culture. J Cell Sci. 2017;130:177–189. doi: 10.1242/jcs.190967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Randlett O, et al. Whole-brain activity mapping onto a zebrafish brain atlas. Nat Methods. 2015;12:1039–1046. doi: 10.1038/nmeth.3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schindelin J, et al. Fiji: An open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Preibisch S, Saalfeld S, Tomancak P. Globally optimal stitching of tiled 3D microscopic image acquisitions. Bioinformatics. 2009;25:1463–1465. doi: 10.1093/bioinformatics/btp184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith K, et al. CIDRE: An illumination-correction method for optical microscopy. Nat Methods. 2015;12:404–406. doi: 10.1038/nmeth.3323. [DOI] [PubMed] [Google Scholar]
- 32.Zhao Y, et al. Nanoscale imaging of clinical specimens using pathology-optimized expansion microscopy. Nat Biotechnol. 2017;35:757–764. doi: 10.1038/nbt.3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chozinski TJ, et al. Expansion microscopy with conventional antibodies and fluorescent proteins. Nat Methods. 2016;13:485–488. doi: 10.1038/nmeth.3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suofu Y, et al. Dual role of mitochondria in producing melatonin and driving GPCR signaling to block cytochrome c release. Proc Natl Acad Sci USA. 2017;114:E7997–E8006. doi: 10.1073/pnas.1705768114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang IE, Lapan SW, Scimone ML, Clandinin TR, Reddien PW. Hedgehog signaling regulates gene expression in planarian glia. Elife. 2016;5:e16996. doi: 10.7554/eLife.16996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cahoon CK, et al. Superresolution expansion microscopy reveals the three-dimensional organization of the Drosophila synaptonemal complex. Proc Natl Acad Sci USA. 2017;114:E6857–E6866. doi: 10.1073/pnas.1705623114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mosca TJ, Luginbuhl DJ, Wang IE, Luo L. Presynaptic LRP4 promotes synapse number and function of excitatory CNS neurons. Elife. 2017;6:e27347. doi: 10.7554/eLife.27347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deshpande T, et al. Subcellular reorganization and altered phosphorylation of the astrocytic gap junction protein connexin43 in human and experimental temporal lobe epilepsy. Glia. 2017;65:1809–1820. doi: 10.1002/glia.23196. [DOI] [PubMed] [Google Scholar]
- 39.Hatta K, Tsujii H, Omura T. Cell tracking using a photoconvertible fluorescent protein. Nat Protoc. 2006;1:960–967. doi: 10.1038/nprot.2006.96. [DOI] [PubMed] [Google Scholar]
- 40.Greaney MR, Privorotskiy AE, D’Elia KP, Schoppik D. Extraocular motoneuron pools develop along a dorsoventral axis in zebrafish, Danio rerio. J Comp Neurol. 2017;525:65–78. doi: 10.1002/cne.24042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meyer MP, Trimmer JS, Gilthorpe JD, Smith SJ. Characterization of zebrafish PSD-95 gene family members. J Neurobiol. 2005;63:91–105. doi: 10.1002/neu.20118. [DOI] [PubMed] [Google Scholar]
- 42.Eaton RC, Farley RD. Mauthner neuron field potential in newly hatched larvae of the zebra fish. J Neurophysiol. 1975;38:502–512. doi: 10.1152/jn.1975.38.3.502. [DOI] [PubMed] [Google Scholar]
- 43.Watanabe T, et al. Coexpression of auxiliary Kvβ2 subunits with Kv1.1 channels is required for developmental acquisition of unique firing properties of zebrafish Mauthner cells. J Neurophysiol. 2014;111:1153–1164. doi: 10.1152/jn.00596.2013. [DOI] [PubMed] [Google Scholar]
- 44.Eaton RC, Farley RD, Kimmel CB, Schabtach E. Functional development in the Mauthner cell system of embryos and larvae of the zebra fish. J Neurobiol. 1977;8:151–172. doi: 10.1002/neu.480080207. [DOI] [PubMed] [Google Scholar]
- 45.Lorent K, Liu KS, Fetcho JR, Granato M. The zebrafish space cadet gene controls axonal pathfinding of neurons that modulate fast turning movements. Development. 2001;128:2131–2142. doi: 10.1242/dev.128.11.2131. [DOI] [PubMed] [Google Scholar]
- 46.Lacoste AMB, et al. A convergent and essential interneuron pathway for Mauthner-cell-mediated escapes. Curr Biol. 2015;25:1526–1534. doi: 10.1016/j.cub.2015.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kimmel CB, Sessions SK, Kimmel RJ. Morphogenesis and synaptogenesis of the zebrafish Mauthner neuron. J Comp Neurol. 1981;198:101–120. doi: 10.1002/cne.901980110. [DOI] [PubMed] [Google Scholar]
- 48.Ito R, Kohno K. Development of the axon cap neuropil of the Mauthner cell in the goldfish. Cell Tissue Res. 1984;237:49–55. doi: 10.1007/BF00229199. [DOI] [PubMed] [Google Scholar]
- 49.McLean DL, Fan J, Higashijima S, Hale ME, Fetcho JR. A topographic map of recruitment in spinal cord. Nature. 2007;446:71–75. doi: 10.1038/nature05588. [DOI] [PubMed] [Google Scholar]
- 50.Xiong F, et al. Interplay of cell shape and division orientation promotes robust morphogenesis of developing epithelia. Cell. 2014;159:415–427. doi: 10.1016/j.cell.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kamei H. Relationship of nuclear invaginations to perinuclear rings composed of intermediate filaments in MIA PaCa-2 and some other cells. Cell Struct Funct. 1994;19:123–132. doi: 10.1247/csf.19.123. [DOI] [PubMed] [Google Scholar]
- 52.Robbins E, Gonatas NK. The ultrastructure of a mammalian cell during the mitotic cycle. J Cell Biol. 1964;21:429–463. doi: 10.1083/jcb.21.3.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Paweletz N, Lang U. Fine structural studies of early mitotic stages in untreated and nocodazole-treated HeLa cells. Eur J Cell Biol. 1988;47:334–345. [PubMed] [Google Scholar]
- 54.Georgatos SD, Pyrpasopoulou A, Theodoropoulos PA. Nuclear envelope breakdown in mammalian cells involves stepwise lamina disassembly and microtubule-drive deformation of the nuclear membrane. J Cell Sci. 1997;110:2129–2140. doi: 10.1242/jcs.110.17.2129. [DOI] [PubMed] [Google Scholar]
- 55.Salina D, et al. Cytoplasmic dynein as a facilitator of nuclear envelope breakdown. Cell. 2002;108:97–107. doi: 10.1016/s0092-8674(01)00628-6. [DOI] [PubMed] [Google Scholar]
- 56.Beaudouin J, Gerlich D, Daigle N, Eils R, Ellenberg J. Nuclear envelope breakdown proceeds by microtubule-induced tearing of the lamina. Cell. 2002;108:83–96. doi: 10.1016/s0092-8674(01)00627-4. [DOI] [PubMed] [Google Scholar]
- 57.Güttinger S, Laurell E, Kutay U. Orchestrating nuclear envelope disassembly and reassembly during mitosis. Nat Rev Mol Cell Biol. 2009;10:178–191. doi: 10.1038/nrm2641. [DOI] [PubMed] [Google Scholar]
- 58.D’Avino PP, Capalbo L. Regulation of midbody formation and function by mitotic kinases. Semin Cell Dev Biol. 2016;53:57–63. doi: 10.1016/j.semcdb.2016.01.018. [DOI] [PubMed] [Google Scholar]
- 59.Glotzer M. The 3Ms of central spindle assembly: Microtubules, motors and MAPs. Nat Rev Mol Cell Biol. 2009;10:9–20. doi: 10.1038/nrm2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Green RA, Paluch E, Oegema K. Cytokinesis in animal cells. Annu Rev Cell Dev Biol. 2012;28:29–58. doi: 10.1146/annurev-cellbio-101011-155718. [DOI] [PubMed] [Google Scholar]
- 61.Meunier S, Vernos I. Microtubule assembly during mitosis–From distinct origins to distinct functions? J Cell Sci. 2012;125:2805–2814. doi: 10.1242/jcs.092429. [DOI] [PubMed] [Google Scholar]
- 62.Mastronarde DN, McDonald KL, Ding R, McIntosh JR. Interpolar spindle microtubules in PTK cells. J Cell Biol. 1993;123:1475–1489. doi: 10.1083/jcb.123.6.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McDonald KL, O’Toole ET, Mastronarde DN, McIntosh JR. Kinetochore microtubules in PTK cells. J Cell Biol. 1992;118:369–383. doi: 10.1083/jcb.118.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kanda T, Sullivan KF, Wahl GM. Histone-GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Curr Biol. 1998;8:377–385. doi: 10.1016/s0960-9822(98)70156-3. [DOI] [PubMed] [Google Scholar]
- 65.Pauls S, Geldmacher-Voss B, Campos-Ortega JA. A zebrafish histone variant H2A.F/Z and a transgenic H2A.F/Z:GFP fusion protein for in vivo studies of embryonic development. Dev Genes Evol. 2001;211:603–610. doi: 10.1007/s00427-001-0196-x. [DOI] [PubMed] [Google Scholar]
- 66.Mora-Bermúdez F, Gerlich D, Ellenberg J. Maximal chromosome compaction occurs by axial shortening in anaphase and depends on Aurora kinase. Nat Cell Biol. 2007;9:822–831. doi: 10.1038/ncb1606. [DOI] [PubMed] [Google Scholar]
- 67.Liang Z, et al. Chromosomes progress to metaphase in multiple discrete steps via global compaction/expansion cycles. Cell. 2015;161:1124–1137. doi: 10.1016/j.cell.2015.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gerlich D, Hirota T, Koch B, Peters J-M, Ellenberg J. Condensin I stabilizes chromosomes mechanically through a dynamic interaction in live cells. Curr Biol. 2006;16:333–344. doi: 10.1016/j.cub.2005.12.040. [DOI] [PubMed] [Google Scholar]
- 69.Schermelleh L, et al. Subdiffraction multicolor imaging of the nuclear periphery with 3D structured illumination microscopy. Science. 2008;320:1332–1336. doi: 10.1126/science.1156947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zessin PJM, Finan K, Heilemann M. Super-resolution fluorescence imaging of chromosomal DNA. J Struct Biol. 2012;177:344–348. doi: 10.1016/j.jsb.2011.12.015. [DOI] [PubMed] [Google Scholar]
- 71.Ricci MA, Manzo C, García-Parajo MF, Lakadamyali M, Cosma MP. Chromatin fibers are formed by heterogeneous groups of nucleosomes in vivo. Cell. 2015;160:1145–1158. doi: 10.1016/j.cell.2015.01.054. [DOI] [PubMed] [Google Scholar]
- 72.York AG, et al. Resolution doubling in live, multicellular organisms via multifocal structured illumination microscopy. Nat Methods. 2012;9:749–754. doi: 10.1038/nmeth.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Winter PW, et al. Two-photon instant structured illumination microscopy improves the depth penetration of super-resolution imaging in thick scattering samples. Optica. 2014;1:181–191. doi: 10.1364/OPTICA.1.000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.York AG, et al. Instant super-resolution imaging in live cells and embryos via analog image processing. Nat Methods. 2013;10:1122–1126. doi: 10.1038/nmeth.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lv C, Gould TJ, Bewersdorf J, Zenisek D. High-resolution optical imaging of zebrafish larval ribbon synapse protein RIBEYE, RIM2, and CaV 1.4 by stimulation emission depletion microscopy. Microsc Microanal. 2012;18:745–752. doi: 10.1017/S1431927612000268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Webb SE, Cheung CC, Chan CM, Love DR, Miller AL. Application of complementary luminescent and fluorescent imaging techniques to visualize nuclear and cytoplasmic Ca2+ signalling during the in vivo differentiation of slow muscle cells in zebrafish embryos under normal and dystrophic conditions. Clin Exp Pharmacol Physiol. 2012;39:78–86. doi: 10.1111/j.1440-1681.2011.05582.x. [DOI] [PubMed] [Google Scholar]
- 77.Wyart C, Del Bene F. Let there be light: Zebrafish neurobiology and the optogenetic revolution. Rev Neurosci. 2011;22:121–130. doi: 10.1515/RNS.2011.013. [DOI] [PubMed] [Google Scholar]
- 78.Del Bene F, Wyart C. Optogenetics: A new enlightenment age for zebrafish neurobiology. Dev Neurobiol. 2012;72:404–414. doi: 10.1002/dneu.20914. [DOI] [PubMed] [Google Scholar]
- 79.Portugues R, Severi KE, Wyart C, Ahrens MB. Optogenetics in a transparent animal: Circuit function in the larval zebrafish. Curr Opin Neurobiol. 2013;23:119–126. doi: 10.1016/j.conb.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 80.Viswanathan S, et al. High-performance probes for light and electron microscopy. Nat Methods. 2015;12:568–576. doi: 10.1038/nmeth.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pan YA, et al. Zebrabow: Multispectral cell labeling for cell tracing and lineage analysis in zebrafish. Development. 2013;140:2835–2846. doi: 10.1242/dev.094631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cai D, Cohen KB, Luo T, Lichtman JW, Sanes JR. Improved tools for the Brainbow toolbox. Nat Methods. 2013;10:540–547. [PubMed] [Google Scholar]
- 83.Maglione M, Sigrist SJ. Seeing the forest tree by tree: Super-resolution light microscopy meets the neurosciences. Nat Neurosci. 2013;16:790–797. doi: 10.1038/nn.3403. [DOI] [PubMed] [Google Scholar]
- 84.Lichtman JW, Pfister H, Shavit N. The big data challenges of connectomics. Nat Neurosci. 2014;17:1448–1454. doi: 10.1038/nn.3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chen F, et al. Nanoscale imaging of RNA with expansion microscopy. Nat Methods. 2016;13:679–684. doi: 10.1038/nmeth.3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hausen P, Dreyer C. The use of polyacrylamide as an embedding medium for immunohistochemical studies of embryonic tissues. Stain Technol. 1981;56:287–293. doi: 10.3109/10520298109067329. [DOI] [PubMed] [Google Scholar]
- 87.Maidorn M, Rizzoli SO, Opazo F. Tools and limitations to study the molecular composition of synapses by fluorescence microscopy. Biochem J. 2016;473:3385–3399. doi: 10.1042/BCJ20160366. [DOI] [PubMed] [Google Scholar]
- 88.Ku T, et al. Multiplexed and scalable super-resolution imaging of three-dimensional protein localization in size-adjustable tissues. Nat Biotechnol. 2016;34:973–981. doi: 10.1038/nbt.3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nasevicius A, Ekker SC. Effective targeted gene ‘knockdown’ in zebrafish. Nat Genet. 2000;26:216–220. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- 90.Bedell VM, et al. In vivo genome editing using a high-efficiency TALEN system. Nature. 2012;491:114–118. doi: 10.1038/nature11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hwang WY, et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gagnon JA, et al. Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PLoS One. 2014;9:e98186, and erratum (2014) 9:e106396. doi: 10.1371/journal.pone.0098186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Xiao T, Roeser T, Staub W, Baier H. A GFP-based genetic screen reveals mutations that disrupt the architecture of the zebrafish retinotectal projection. Development. 2005;132:2955–2967. doi: 10.1242/dev.01861. [DOI] [PubMed] [Google Scholar]
- 94.Muto A, et al. Forward genetic analysis of visual behavior in zebrafish. PLoS Genet. 2005;1:e66. doi: 10.1371/journal.pgen.0010066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Barresi MJF, et al. Essential genes for astroglial development and axon pathfinding during zebrafish embryogenesis. Dev Dyn. 2010;239:2603–2618. doi: 10.1002/dvdy.22393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tyagarajan SK, Fritschy J-M. Gephyrin: A master regulator of neuronal function? Nat Rev Neurosci. 2014;15:141–156. doi: 10.1038/nrn3670. [DOI] [PubMed] [Google Scholar]
- 97.Cui WW, et al. The zebrafish shocked gene encodes a glycine transporter and is essential for the function of early neural circuits in the CNS. J Neurosci. 2005;25:6610–6620. doi: 10.1523/JNEUROSCI.5009-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hirata H, et al. Zebrafish bandoneon mutants display behavioral defects due to a mutation in the glycine receptor β-subunit. Proc Natl Acad Sci USA. 2005;102:8345–8350. doi: 10.1073/pnas.0500862102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ogino K, et al. Duplicated gephyrin genes showing distinct tissue distribution and alternative splicing patterns mediate molybdenum cofactor biosynthesis, glycine receptor clustering, and escape behavior in zebrafish. J Biol Chem. 2011;286:806–817. doi: 10.1074/jbc.M110.125500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yamanaka I, et al. Glycinergic transmission and postsynaptic activation of CaMKII are required for glycine receptor clustering in vivo. Genes Cells. 2013;18:211–224. doi: 10.1111/gtc.12032. [DOI] [PubMed] [Google Scholar]
- 101.Ogino K, Hirata H. Defects of the glycinergic synapse in zebrafish. Front Mol Neurosci. 2016;9:50. doi: 10.3389/fnmol.2016.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Avedanian L, Jacques D, Bkaily G. Presence of tubular and reticular structures in the nucleus of human vascular smooth muscle cells. J Mol Cell Cardiol. 2011;50:175–186. doi: 10.1016/j.yjmcc.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 103.Lui PPY, Kong SK, Kwok TT, Lee CY. The nucleus of HeLa cell contains tubular structures for Ca2+ signalling. Biochem Biophys Res Commun. 1998;247:88–93. doi: 10.1006/bbrc.1998.8649. [DOI] [PubMed] [Google Scholar]
- 104.Bootman MD, Fearnley C, Smyrnias I, MacDonald F, Roderick HL. An update on nuclear calcium signalling. J Cell Sci. 2009;122:2337–2350. doi: 10.1242/jcs.028100. [DOI] [PubMed] [Google Scholar]
- 105.Echevarría W, Leite MF, Guerra MT, Zipfel WR, Nathanson MH. Regulation of calcium signals in the nucleus by a nucleoplasmic reticulum. Nat Cell Biol. 2003;5:440–446. doi: 10.1038/ncb980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Abe T, et al. Myocyte differentiation generates nuclear invaginations traversed by myofibrils associating with sarcomeric protein mRNAs. J Cell Sci. 2004;117:6523–6534. doi: 10.1242/jcs.01574. [DOI] [PubMed] [Google Scholar]
- 107.Kane DA, et al. The zebrafish early arrest mutants. Development. 1996;123:57–66. doi: 10.1242/dev.123.1.57. [DOI] [PubMed] [Google Scholar]
- 108.Seipold S, et al. Non-SMC condensin I complex proteins control chromosome segregation and survival of proliferating cells in the zebrafish neural retina. BMC Dev Biol. 2009;9:40. doi: 10.1186/1471-213X-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pfaff KL, et al. The zebra fish cassiopeia mutant reveals that SIL is required for mitotic spindle organization. Mol Cell Biol. 2007;27:5887–5897. doi: 10.1128/MCB.00175-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Shepard JL, Stern HM, Pfaff KL, Amatruda JF. Analysis of the cell cycle in zebrafish embryos. Methods Cell Biol. 2004;76:109–125. doi: 10.1016/s0091-679x(04)76007-0. [DOI] [PubMed] [Google Scholar]
- 111.Amatruda JF, Shepard JL, Stern HM, Zon LI. Zebrafish as a cancer model system. Cancer Cell. 2002;1:229–231. doi: 10.1016/s1535-6108(02)00052-1. [DOI] [PubMed] [Google Scholar]
- 112.Zhao X, et al. Interruption of cenph causes mitotic failure and embryonic death, and its haploinsufficiency suppresses cancer in zebrafish. J Biol Chem. 2010;285:27924–27934. doi: 10.1074/jbc.M110.136077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Del Bene F, et al. Filtering of visual information in the tectum by an identified neural circuit. Science. 2010;330:669–673. doi: 10.1126/science.1192949. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.