Significance
Synthetic sick or lethal (SS/L) interactions occur when disruption of two genes reduces cell viability to a greater extent than expected based on the individual gene disruptions. SS/L interactions involving tumor suppressors represent candidate drug targets for cancers because treatment is expected to kill tumor cells carrying the tumor suppressor mutation but leave healthy cells unaffected. Identification of SS/L interactions is of vital importance to develop new therapies for tumorigenic disease. We have developed an RNAi-based approach called variable dose analysis, which improves both sensitivity and robustness to noise compared with dsRNA-based methods for screening in Drosophila. Using this method, we identified four Food and Drug Administration-approved drugs with specific effects on cells deficient for the TSC1 and TSC2 tumor suppressor genes.
Keywords: synthetic lethality, Drosophila, RNAi, high-throughput screening, drug target discovery
Abstract
Synthetic sick or synthetic lethal (SS/L) screens are a powerful way to identify candidate drug targets to specifically kill tumor cells, but this approach generally suffers from low consistency between screens. We found that many SS/L interactions involve essential genes and are therefore detectable within a limited range of knockdown efficiency. Such interactions are often missed by overly efficient RNAi reagents. We therefore developed an assay that measures viability over a range of knockdown efficiency within a cell population. This method, called Variable Dose Analysis (VDA), is highly sensitive to viability phenotypes and reproducibly detects SS/L interactions. We applied the VDA method to search for SS/L interactions with TSC1 and TSC2, the two tumor suppressors underlying tuberous sclerosis complex (TSC), and generated a SS/L network for TSC. Using this network, we identified four Food and Drug Administration-approved drugs that selectively affect viability of TSC-deficient cells, representing promising candidates for repurposing to treat TSC-related tumors.
A genetic interaction occurs when the combined disruption of two genes produces a phenotype that differs from that expected based on the effects of each individual gene disruption. One type of genetic interaction in which cell viability is reduced only following combined disruption of two genes, but not following disruption of either gene alone, is called a synthetic sick or synthetic lethal (SS/L) interaction depending on the severity of the viability effect. SS/L interactions have received considerable interest for the development of drug targets for cancers because targeting of a gene that has a SS/L interaction with a tumor suppressor is expected to specifically reduce viability of tumor cells but leave wild-type cells unaffected (1–3). In addition, large-scale knowledge of SS/L interactions can be used to gain functional insight into individual genes and network structures (4–6).
SS/L screens have been performed covering most of the possible pairwise gene combinations in the yeast Saccharomyces cerevisiae, leading to insight into the global molecular wiring of a cell (6). Furthermore, SS/L screens have been performed with the aim of identifying drug targets in cultured mammalian cells, including tumor-derived lines (e.g., refs. 7–10). However, SS/L screens in general have suffered from limited consistency between screens and have resulted in the identification of relatively few effective drug targets (2, 11–13). This may in part be due to the noisy nature of high-throughput screens. However, a major contributor to this lack of concordance between studies is likely to be context dependence of cancer cell line dependencies, as illustrated by widely varying responses to existing therapeutic agents (14, 15). Thus, SS/L interactions can be classified either as “hard” interactions, which function independently of genetic or cellular context, or “soft” interactions, which may reflect cell context (i.e., the genes expressed in one cell line versus others or growth conditions) (10, 16). Consistent with this, a recent study in which SS/L interaction screens were performed in multiple Ras-dependent and Ras-independent acute myeloid leukemia cell lines found relatively few dependencies common specifically to Ras-dependent lines, suggesting that the majority of Ras-induced dependencies are specific to an individual context (10). These issues highlight the need to develop robust SS/L screening methods and assess SS/L interactions across diverse genetic backgrounds to find effective drug targets that will function in many contexts.
Toward this goal, we reported previously the use of a cross-species screening approach to identify SS/L interactions with TSC1 and TSC2, the two tumor suppressor genes underlying tuberous sclerosis complex (TSC) (17, 18). We performed focused dsRNA screens targeting all kinases and phosphatases in wild-type, TSC1, and TSC2 mutant Drosophila cells. By comparing screens, we identified three genes that specifically reduce viability of TSC1 and TSC2 mutant cells when knocked down. All three had conserved SS/L interactions with TSC2 in mouse and human cell lines, illustrating the potential of this approach to identify candidate drug targets relevant to humans. By performing these screens in Drosophila and then validating hits in diverse mammalian systems, we hoped to identify core SS/L interactions that were not specific to a single human cell type.
Here, we report an additional advance toward improved SS/L screening methods. Using a similar cross-species screening approach, we performed genome-wide screens to identify SS/L interactions with the TSC1 and TSC2 genes. Analysis of screen results and comparison between screens revealed that many SS/L interactions were missed in each screen, which accords with the low rate of consistency between previous screens. Further investigation revealed that many SS/L interactions involve essential genes that were likely missed due to overly efficient knockdown, which reduced the viability of both wild-type and TSC1/2 mutant cells. We therefore developed an RNAi-based screening assay that measures phenotypes over a range of knockdown efficiencies in a single sample. This method improved signal-to-noise ratio in viability assays by ∼2.5-fold compared with dsRNA assays and detected 86% of positive control SS/L interactions. Using this method in combination with our previous screen results and other preexisting datasets, we generated a high-confidence SS/L interaction network surrounding the TSC complex. Finally, using this network, we identified four Food and Drug Administration (FDA)-approved drugs showing selective effects on the viability of TSC-deficient cells that may represent promising candidates for drug repurposing to treat TSC tumors. Importantly, all four of these drugs showed conserved effects in three mammalian cell culture models of TSC, including two diverse tumor-derived cell lines, illustrating that this screening approach improves the identification of context-independent candidate therapeutic drugs.
Results
SS/L Interactions Are Enriched for Essential Genes.
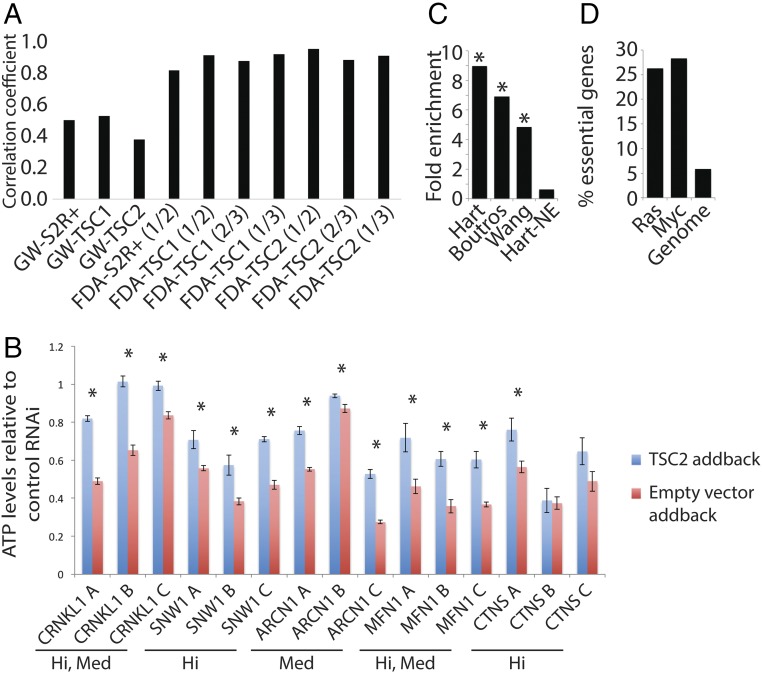
To identify SS/L interactions with the TSC complex, we performed SS/L interaction screens using two dsRNA libraries. The first library targeted 13,099 genes, representing the majority of the Drosophila genome. The second library targeted 466 Drosophila orthologs of putative targets of FDA-approved drugs. By screening this group of genes with high coverage, we improved the chances of identifying SS/L interactions with genes for which clinically approved drugs already exist, which therefore may be rapidly repurposed for clinical use to treat TSC tumors. From these two screens, 288 genes that had SS/L interactions with TSC1 and/or TSC2 were identified (Datasets S1 and S2). Replicate screens showed good correlation in general, indicating that the results are sound (Fig. 1A). We note that the genome-wide screen results had lower correlation coefficients than the FDA screens. This is likely due to edge effects, which are avoided in the FDA library by leaving edge wells empty. To confirm the validity of these hits, we selected five genes from the genome-wide screen with varying confidence levels and tested whether their human orthologs also showed SS/L interactions with TSC2 in a tumor-derived human cell line. Similar to the high validation rate in mammalian systems observed in our previous studies (18), all five SS/L interactions were validated in this system (Fig. 1B). Furthermore, two additional genes not expected to show SS/L interactions with TSC1 or TSC2 showed no selective viability effect in similar assays performed previously (18), indicating that the TSC2 addback cells are not simply less sensitive to viability changes.
Fig. 1.
SS/L interactions are enriched with essential genes. (A) Bar graph illustrating the correlation coefficients between pairs of replicates from the genome-wide (GW) or FDA screens as indicated. For the FDA screens, the numbers in parentheses indicate which replicates are compared in each case. (B) Bar graphs showing relative viability of TSC2-deficient RA-derived cells with either TSC2 cDNA (blue bars) or empty vector (red bars) addback transfected with the indicated shRNA constructs relative to control shRNA transfection, measured using CellTiter-glo assays. Bars represent average values from nine replicates in each case, and error bars indicate SEM. Asterisks indicate cases where viability of empty vector addback samples is significantly lower than TSC2 cDNA addback samples (P < 0.05) as determined using t tests. The confidence class with which each gene was identified in the genome-wide screen is indicated below the gene names (Hi, high confidence; Med, medium confidence). (C) Bar graph showing fold enrichment of essential genes among SS/L genes identified from the genome-wide screen. Three independent datasets were used to define essential (19, 20, 21) or nonessential genes [Hart-NE (20)]. Asterisks indicate statistically significant enrichment (P < 0.05) based on Z-tests to compare with 1,000 permutations of randomly selected genes. (D) Bar graph illustrating the percentage of genes identified as SS/L with Ras or Myc overexpression or activation in previous studies. “Genome” indicates the percentage of essential genes in the whole genome assessed using the same datasets as in C combined to define essential genes (19, 20, 21).
Despite the apparent high quality of the SS/L interactions identified, relatively little overlap was observed between the independent libraries screened. For example, a total of 50 SS/L genes were screened in the combined kinase, phosphatase (KP)/FDA libraries and the genome-wide screen, yet only two were identified as SS/L in both datasets. In addition, only 4.6% of the identified SS/L interactions in these screens were observed with multiple independent dsRNA reagents despite the fact that 67% of SS/L genes were represented by more than one dsRNA in the screens. This observation is consistent with the limited consistency between SS/L screens previously performed in mammalian systems, which is likely due to the use of different RNAi reagents. To investigate the reasons underlying the inconsistency between dsRNA reagents and screens, we assessed the effects of dsRNA reagents that did not identify SS/L interactions despite targeting genes that were identified as SS/L using independent dsRNA reagents. We found that ∼72.5% of reagents had no detectable effect on viability in wild-type, TSC1, or TSC2 mutant cells and likely represent ineffective reagents. By contrast, 27.5% of reagents failed to identify SS/L interactions because they reduced viability of all cell types, suggesting that their targets are essential genes or that the viability effects are due to disruption of off-target genes (Dataset S3). In addition, the 288 SS/L genes identified from the screens were highly enriched for known essential genes identified in multiple previous studies (Fig. 1C). These results indicate that genes with SS/L interactions are enriched for essential genes, consistent with previous observations in yeast (6). Furthermore, we found that genes identified from SS/L screens in mammalian systems with activated Ras or Myc also showed enrichment of essential genes (Fig. 1D), suggesting that this is a general property of SS/L interactions.
Variable Dose Analysis Allows Sensitive Detection of Viability Phenotypes.
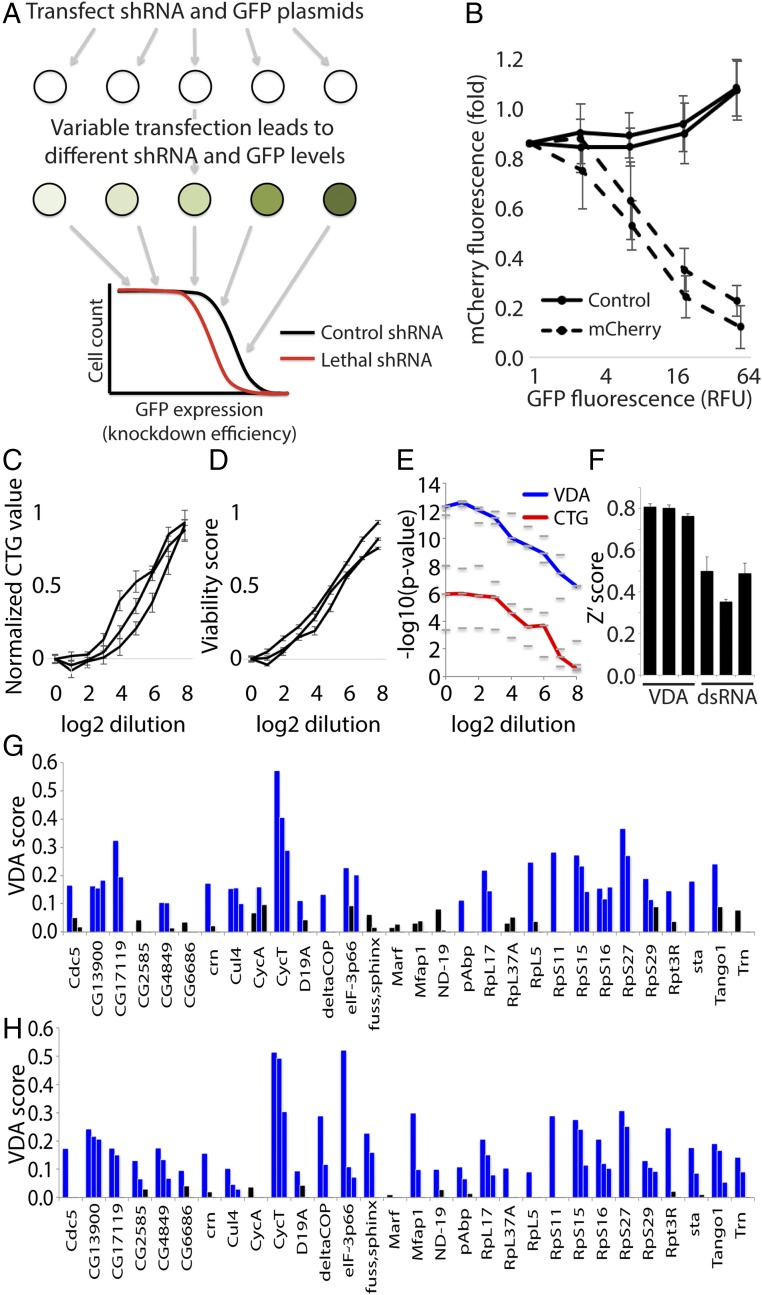
SS/L interactions involving essential genes are detectable within a limited range of target-gene knockdown efficiency because weak knockdown is ineffective and strong knockdown is lethal to both wild-type and mutant cells. Therefore, to improve detection of this type of SS/L interaction, an assay is required that allows assessment of viability effects over a range of target-gene knockdown efficiencies. To achieve this, we took advantage of the variable efficiency of plasmid transfections between individual cells in a population, resulting in variable plasmid copy number in each cell. By cotransfecting a GFP-expressing plasmid and a shRNA-expressing plasmid, the GFP intensity can be used as an indirect readout of shRNA expression and therefore target-gene knockdown efficiency (Fig. 2A). We named this method variable dose analysis (VDA). To test the correlation between GFP expression and knockdown efficiency using this approach, we used S2 cells that express mCherry from a genomic transgene insertion into the CLIC locus (22). VDA assays were performed in this cell line targeting either a control gene (white) or the mCherry transgene. mCherry fluorescence was then compared with GFP fluorescence, and mCherry fluorescence was found to decrease as GFP intensity increased with a nonlinear relationship (Fig. 2B). Therefore, GFP fluorescence can be used as a measure of relative target-gene knockdown efficiency.
Fig. 2.
VDA is an effective method to measure viability phenotypes and detect SS/L interactions. (A) Schematic illustrating the experimental setup of VDA assays. (B) Graph illustrating the relationship between GFP fluorescence and target-gene knockdown efficiency. mCherry fluorescence was used as a measure of knockdown efficiency and is displayed as a fold change to cells with no detectable GFP expression. Each line represents the average of three biological replicate experiments using an independent shRNA reagent targeting mCherry (dashed lines) or white (solid lines). Error bars indicate SEM. (C and D) Graphs showing cell viability measured using either CTG (C) or VDA (D) assays over a range of thread RNAi dilutions. Measurements are normalized to both positive (thread RNAi) and negative (white RNAi) controls to allow simple comparison between the two methods. Lines show average readings from six replicates in each case. Multiple groups of replicates are shown to better illustrate consistency between experiments. Error bars indicate SEM. (E) Graph illustrating the improved ability of VDA assays (blue line) to detect viability phenotypes compared with CellTiter-glo assays (red line) performed on the same populations of cells. Dashes represent −log10 P values calculated from three independent groups of six replicate experiments. The lines represent the median value of the three dashes. These data were calculated from the same experiments represented in C and D. (F) Bar graph illustrating average Z′scores from three independent replicate experiments, each consisting of 30 biological positive control replicates and 30 biological negative control replicate measurements. Each bar represents a different pair of positive and negative control reagents measured using dsRNA/CellTiter-glo or VDA assays as indicated. Error bars indicate SEM. (G) Results from VDA assays targeting 30 different genes as indicated. Each bar represents a different shRNA reagent (three per gene). Blue bars indicate samples with a VDA score greater than 0.1, and black bars represent VDA scores less than 0.1. (H) Graph displaying VDA results as in G but with VDA analysis performed including cell size correction.
Next, we performed experiments to assess the sensitivity of this method relative to an established viability assay. We cotransfected the GFP reporter plasmid with shRNA plasmids targeting thread, an apoptosis inhibitor that robustly induces cell death when inhibited, or a control gene, white. Signal strength was varied by serially diluting the thread shRNA plasmid with the white shRNA plasmid. In addition, the same samples were analyzed using CellTiter-Glo (CTG) assays, a standard readout that has been widely used in high-throughput viability screens. We found that VDA outperformed CTG assays for detection of weak phenotypes and P values remained highly significant even when the thread shRNA was diluted 256-fold compared with the standard dose (Fig. 2 C–E).
Finally, to directly compare VDA with established dsRNA-based methods, we generated three pairs of positive and negative control shRNAs for cell viability targeting thread and white, respectively. We then performed VDA assays in S2R+ cells and calculated Z′ scores for each pair of reagents. In addition, we used three pairs of positive and negative control reagents from the Drosophila RNAi Screening Center dsRNA collection (19, 23) and performed similar assays using a CTG readout. Comparison between these results showed that the Z′ scores for VDA assays were consistently higher than for dsRNA assays, corresponding to an increase in signal-to-noise ratio of ∼2.5-fold (Fig. 2F). In addition, VDA assays had reduced variation between control reagent pairs, indicating that these assays may be more robust to differences in reagent efficiency.
Overall, these experiments demonstrate that VDA is a highly sensitive and robust method for the detection of viability phenotypes.
VDA Assays Efficiently Identify SS/L Interactions with Essential and Nonessential Genes.
To test whether VDA assays are able to robustly identify known SS/L interactions with both essential and nonessential genes, we generated three shRNA reagents per gene targeting 27 genes identified as SS/L in the dsRNA screens. In addition, to assess the ability of VDA assays to identify SS/L interactions with essential genes, we included three genes that were identified as essential (lethal to all cell types) but that had Z-scores at least 1.5-fold lower in both TSC1 and TSC2 mutant cells compared with wild type. Of these 30 genes, 18 were identified as essential genes in previous screens (19, 20, 21).
SS/L interactions were identified as shRNA reagents that cause a significantly greater viability reduction in TSC1 cells compared with wild type. Positive control genes (70.4%, or 19/27) were identified as having significant SS/L interactions with TSC1, illustrating the high sensitivity of this assay (Fig. 2G and Dataset S4). In addition, the three genes that failed to score as SS/L in dsRNA assays due to viability effects in wild-type cells were all identified as SS/L using this assay. Finally, 33% of the genes assessed were identified as SS/L with multiple shRNA reagents (Fig. 2G and Dataset S4), indicating a higher rate of consistency between reagents compared with dsRNA assays.
Notably, SS/L interactions were not identified for eight positive control genes. Viability phenotypes were detected for all but one of these genes, indicating that the failure of validation was not due to ineffective reagents. Another possible explanation is that, in addition to affecting cell viability, these genes alter cell size specifically in TSC1/2-deficient cells and are therefore detected as SS/L in the ATP-based assays used in the dsRNA screens but not in cell count-based VDA assays. To assess this possibility, VDA data were reanalyzed as before but GFP measurements were normalized to cell sizes based on forward-scatter (FSC) readings collected in parallel with GFP fluorescence data. This allows for detection of cell size phenotypes as well as viability effects. Using this analysis approach, 28/30 genes tested were identified as SS/L, and 19 were identified with multiple reagents (Fig. 2H and Dataset S4). This demonstrates the ability of VDA assays to detect multiple different phenotypes in a single assay and to characterize hits in more detail than simple ATP-based assays.
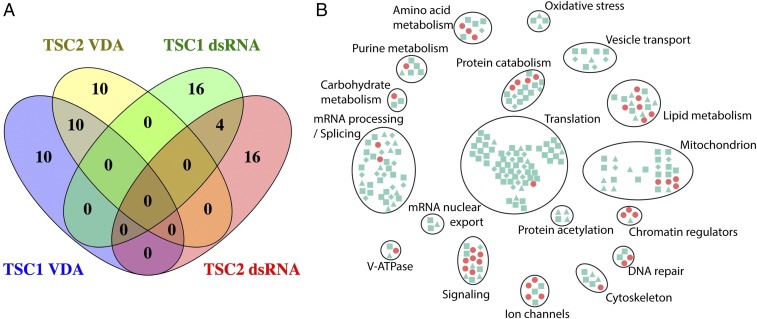
Given the improved ability of VDA assays to identify known SS/L interactions compared with the dsRNA/CellTiter-Glo assays used in our previous screens, we used this method to screen 154 genes that can be targeted with well-characterized FDA-approved drugs in wild-type, TSC1, and TSC2 cells. Forty-four genes were identified as having SS/L relationships with TSC1 and/or TSC2 (Dataset S5). Surprisingly, only one gene (porin) was identified by both VDA and dsRNA assays. TSC1 and TSC2 act together in a protein complex and are thought to share the majority of their functions. Therefore, as SS/L interactions are related to gene function, these genes are expected to have similar SS/L interaction profiles. To assess the relative quality of dsRNA and VDA screens, we therefore compared SS/L interactions identified with TSC1 or TSC2 for each method. We selected the top 20 genes from dsRNA and VDA screens in TSC1 and TSC2 cells based on either VDA scores or differences in Z-score compared with wild-type cells. For dsRNA assays, 11% (4/36) of the top-ranked SS/L genes were identified with both TSC1 and TSC2. By contrast, for VDA assays, 33% (10/30) of the top-ranked SS/L genes were shared, suggesting that VDA is a much more robust method for identification of SS/L interactions (Fig. 3A).
Fig. 3.
An integrated network analysis of TSC dependencies. (A) Venn diagram showing overlap of genes between the top 20 ranked hits from the VDA or dsRNA screens performed in TSC1 or TSC2 cells. (B) Network diagram showing genes identified as SS/L with TSC1 and/or TSC2. Symbols represent individual genes identified from dsRNA screens (light green) or VDA screens (red). Symbol shapes indicate confidence with which hits from dsRNA screens were identified (diamond, high confidence; square, medium confidence, and triangle, low confidence). Gene cluster functions were defined based on manual curation of component functions.
Integrated Analysis of SS/L Screen Data Results in a High-Quality SS/L Network That Is Predictive of Selective Drug Effects.
Previous studies have shown that genes that physically interact and have related functions share SS/L interaction partners (5, 6). Therefore, to identify the most robust hits from the screens and remove false positives, the hits from the dsRNA and VDA screens were pooled (331 genes) and mapped onto a high-confidence (score > 0.9) protein–protein interaction (PPI) network using the STRING database (24), and genes were clustered based on their PPIs to identify those most likely to have related functions. Due to the relatively low coverage of PPI data, we also manually assigned genes to clusters in cases where gene functions were known to be similar based on literature annotation (Fig. 3B and Dataset S6). Using this approach, we defined 18 biological processes, each of which were identified as SS/L with the TSC complex by multiple genes. Fourteen of these processes were supported by both dsRNA and VDA screen results. Furthermore, three of the processes have been identified previously as dependencies of TSC-deficient cells [e.g., ROS (oxidative stress), proteasome (protein catabolism), and lipid metabolism (25–31)], indicating that the network is a robust representation of the functional interactions of the TSC complex. Finally, 86.4% (38/44) of the genes identified using VDA assays were assigned to clusters, compared with 68.4% (197/288) of the genes identified using dsRNAs, suggesting that VDA is a more reliable approach for detection of genuine SS/L interactions.
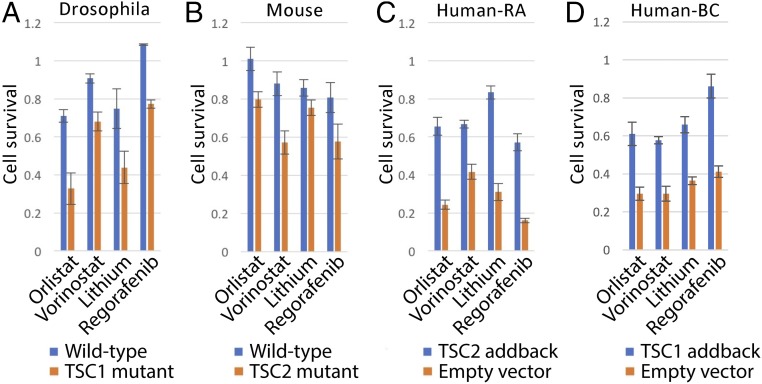
Genes within the SS/L interaction network represent candidate drug targets to specifically reduce viability of TSC-deficient tumor cells. We therefore selected nine FDA-approved drugs that target high-scoring components of the SS/L network and tested for specific viability effects on TSC2-deficient Drosophila cells. Of these, four had a greater effect on the viability of TSC2 cells compared with wild type (Fig. 4A). In addition, all four of these drugs had conserved effects on TSC2-deficient mouse embryonic fibroblasts, TSC2-deficient renal angiomyolipoma (RA)-derived human cells, and TSC1-deficient bladder cancer-derived human cells (32) (Fig. 4 B–D), although the quantitative difference in viability effect varied between cell types. Thus, the combined use of improved screening methods with a network-based analysis approach is a powerful method to identify drugs with reproducible viability effects specific to a given genetic background.
Fig. 4.
Identification of drugs with selective effects on TSC-deficient cells. (A–D) Bar graphs illustrating cell viability as measured using CellTiter-glo assays in the presence of the indicated drugs, normalized to vector alone. Bars represent the average of at least six replicate measurements either in wild-type (blue bars) or TSC1/2-deficient (orange bars) (A and B) or in TSC1/2-deficient cells with empty vector (orange bars) or TSC1/2 cDNA (blue bars) addback (C and D) cells as indicated. Error bars indicate SEM. “Human-RA” indicates human RA-derived cells and “Human-BC” indicates human bladder cancer-derived cells.
Discussion
SS/L interaction screens have long been considered a powerful approach for drug target discovery, yet have resulted in the identification of relatively few effective drugs. This appears to be due at least in part to a lack of consistency between screens despite apparently robust results within studies (13). Two technical factors likely contribute to this lack of consistency. First, high-throughput screens are inherently noisy, resulting in both false-positive and false-negative results. Second, as we and others have shown (6), SS/L interactions are enriched for essential genes, which are often missed due to overly efficient gene disruption, resulting in general toxicity to all cell types. In particular, this is likely to be an issue for CRISPR screens, which often result in null mutations of the target genes.
In this study, we have addressed both of these issues. First, we developed an assay for synthetic lethality called VDA. This approach enables differences in viability between genetic backgrounds to be measured over a range of knockdown efficiencies and can therefore detect SS/L interactions with essential genes at sublethal efficiency. In addition, the VDA method is more sensitive and robust to noise than other well-established methods to measure cell viability in Drosophila high-throughput screens. Finally, by combining results from two independent screening technologies using a network-based analysis method, we have generated a high-confidence SS/L interaction network for the TSC complex. The quality of this network is illustrated by the identification of four effective FDA-approved drugs that represent promising candidates for therapeutic strategies to treat TSC tumors.
Of the four drugs identified, all have previously shown promise as therapeutic targets for cancers. For example, vorinostat and regorafenib are FDA-approved for the treatment of a number of different cancers and are under investigation for multiple others. In addition, lithium has shown antiproliferative, apoptotic, or antimetastatic effects in multiple cancer models (33–36), and orlistat has been shown to inhibit breast cancer cell proliferation (37). Orlistat likely functions in this context by reducing the availability of lipids upon which TSC-deficient cells are known to be dependent (29). The mechanism by which lithium functions in this context is less obvious. Lithium has been used for over five decades for the treatment of bipolar disorder and is efficacious in this role (38, 39). However, despite the long history of clinical use, the mechanism of lithium’s action remains poorly understood (40). Multiple proteins have been identified as direct targets of lithium (e.g., IMPA1, IMPA2, GSK3, and GRIA3; https://www.drugbank.ca), but the effect of this drug in treating bipolar disorder has not been robustly linked with any of these proteins. In addition, lithium can disrupt protein function by displacing magnesium ions (41). Given that many hundreds of proteins rely on magnesium ions as a cofactor for their function (including all enzymes that utilize ATP), the mechanism of lithium’s action may involve complex effects on many different proteins, thus explaining the difficulties encountered in determining a single therapeutic target. Thus, while this may represent a promising candidate for the treatment of TSC, determining the mechanism by which the selective effect is achieved may be challenging.
In addition to the technical issues associated with SS/L screens, context dependence of SS/L interactions is also likely to reduce consistency between screens in different experimental systems. A common approach to limit this effect is to perform screens in systems that are as closely related to the target tumors as possible, often using tumor-derived cells. More recently, efforts to identify SS/L interactions in panels of divergent cell lines sharing a common tumorigenic driver mutation have been used to identify shared dependencies (9, 10, 42). However, this approach requires extensive screening and is limited by high costs and available well-characterized cell lines. Instead, we chose an experimental paradigm in which screens are performed in Drosophila cells, which represent a genetic background highly divergent from the target human tumors. Given that many SS/L interactions are highly conserved (43), our expectation was that SS/L interactions identified in Drosophila cells that could be validated in mouse or human cell lines would represent fundamental context-independent interactions (hard interactions), which may therefore have a higher success rate when transferred to mammalian systems and to clinical use. Strikingly, we found that, of eight SS/L interactions identified in our Drosophila screens, all could be validated in divergent mammalian cell lines (Fig. 1B) (18). One possible explanation of this is that the increased complexity of mammalian genomes compared with Drosophila results in greater network plasticity and therefore more soft interactions. In this case, SS/L interactions identified in Drosophila have a greater chance of being context-independent and therefore more likely to be conserved between divergent systems. This possibility is supported by the similar effects observed for the four identified drugs in diverse backgrounds including mouse and human tumor-derived cell lines. In particular, similar effects were observed in bladder cancer-derived cells, which have one of the highest mutation rates of any cancer type (44), likely resulting in a highly divergent genetic background compared with the mouse and RA-derived cells.
Previous screening approaches have generally focused on developing the most efficient RNAi reagents possible to maximize resulting phenotypes (45–47). More recently, CRISPR has emerged as a powerful screening technology to generate primarily null mutations in target genes and therefore further increases phenotype strength (48, 49). However, these screening paradigms, based on maximizing target-gene disruption efficiency, may often be less representative of effects that can be achieved with small-molecule inhibitors, which generally incompletely inhibit their targets (47). It is therefore possible that approaches that are optimized for efficient gene disruption may result in a lower rate of consistency with pharmacological assays. In this case, genes identified using VDA may be expected to have more consistent effects using pharmacological assays because phenotypes can be detected at a relatively low gene disruption efficiency. Consistent with this, three of the four drugs identified in this study were detected only using VDA assays and only one (vorinostat) was identified by both VDA and dsRNA assays. However, neither approach was able to quantitatively predict specific drug effects with no correlation detected between screen score and pharmacological selectivity (Fig. S1), which is likely due to differences in the mechanism of disruption between RNAi and small-molecule inhibitors (47). Thus, identification of the most promising targets for pharmacological targeting remains a complex and unresolved issue.
Despite the clear advantages of VDA over the previous dsRNA-based methods available for screening in Drosophila cells, there are some limitations, which must be overcome to enable the use of VDA in other systems and for other applications. In particular, VDA requires the use of transfection, which may limit its use in mammalian cell lines to those that are readily transfected without the use of viruses. In addition, the use of GFP as a reporter of transfection efficiency may result in nonspecific toxicity when expressed at high levels in some cell lines. Furthermore, VDA requires higher cell numbers per sample compared with viability assays performed using a CellTiter-Glo readout. Therefore, screens must be performed in a 96-well format, reducing the throughput. In addition, the cytometry readout requires ∼25 min to analyze each 96-well plate compared with 1–2 min per 384-well plate in CellTiter-Glo assays. Finally, while we have delivered shRNA plasmids and the GFP reporter as separate plasmids, it is likely that combining these elements into a single construct will improve results of future VDA applications especially in systems where plasmids are not necessarily delivered with a constant ratio. Thus, further methodological developments will be needed to allow VDA screens to be fully exploited for a wide range of screening applications.
Materials and Methods
Construction of the FDA RNAi Libraries.
We retrieved the FDA drug list from DrugBank (Version 4.0, https://www.drugbank.ca) and extracted drug gene targets using the DrugBank.xml file. Human drug target genes were mapped to Drosophila genes using DIOPT vs.4.0 (50), and only the high-confidence orthologous relationships supported by at least seven different algorithms were selected for Drosophila FDA target libraries. The final FDA library contains 458 Drosophila genes. Two quality amplicons were selected to make the dsRNA library (23) while three different shRNAs were designed based on the DSIR tool (51) and were cloned into the Valium20 vector (52) to make the shRNA library for VDA assays.
The dsRNA Screens.
The dsRNA screens were performed using the genome-wide and FDA libraries available from the Drosophila RNAi Screening Center (https://fgr.hms.harvard.edu/), following the bathing protocol (https://fgr.hms.harvard.edu/fly-cell-rnai-384-well-format). Hits from each replicate screen were defined using a Z-score cutoff of −1.5. The SS/L hits from the genome-wide screen were identified as dsRNA reagents that did not score in either replicate screen in S2R+ cells but scored in both replicate screens in TSC1 cells and in both replicates in TSC2 cells (high confidence), both replicates of either the TSC1 or TCS2 screen (medium confidence), or one replicate from each of the TSC1 and TSC2 screens (low confidence).
VDA Assays.
VDA assays were performed by first transfecting a mixture of 10 ng actin–GFP, 45 ng actin–Gal4, and 45 ng shRNA plasmids into wild-type, TSC1, or TSC2 cells seeded into 96-well plates with 30,000 cells per well, following the standard Effectene transfection reagent protocol (Qiagen, 301427). Following 5 d of culture at 25 °C, culture plates were analyzed using a BD LSR II flow cytometer. The 20,000 events were measured per sample, and GFP intensities and FSC measurements were exported for all GFP-expressing cells as .csv files for further analysis.
Cytometry data were analyzed using custom Matlab scripts by first normalizing all GFP intensities to cell size measurements (FSC). Next, events were divided into 500 bins based on GFP fluorescence and GFP distributions normalized between all samples. Finally, the area under the cumulative GFP distribution plots was calculated and compared with negative and positive control samples to calculate viability scores.
Viability scores were calculated as the area between cumulative distribution plots for each sample and the median negative control cumulative distribution plot for each plate (based on nine negative controls targeting the white gene per plate) divided by the area between the sample curve and the median positive control curve (based on nine positive controls targeting the thread gene per plate). VDA scores were calculated as the viability score in TSC1 or TSC2 cells minus the viability score in wild-type cells.
The VDA-score cutoff of 0.1 used to identify SS/L interactions was determined by performing 12 replicate VDA assays on each of six shRNA reagents determined previously to have no selective effect on TSC-deficient cells (18). We then determined the VDA score corresponding to a Z-score of 1.5 based on the distribution of VDA scores from this negative control dataset.
Hits were selected as genes that scored with a VDA score ≥0.1 in at least two replicates of the relevant screen and scored with either multiple shRNA reagents or a single reagent in both TSC1 and TSC2 screens.
shRNA Assays in Human RA Tumor-Derived Cells.
RA cells with stable vector (621-102) or TSC2 (621-103) addback (53) were cultured in DMEM (#45000–312; VWR) +10% heat-inactivated FBS (#10437–028; ThermoFisher Scientific) +1× penicillin/streptomycin (30–002-CI; CellGRO). Cells were transfected with shRNA targeting ARCN1, CRNKL1, SNW1, CTNS, or MFN1 (three nonoverlapping shRNAs per target) in pLKO.1 vector using Lipofectamine 3000 transfection reagent (#L3000015; ThermoFisher Scientific) according to the manufacturer’s instructions. A scrambled shRNA in pLKO.1 vector was transfected as a control. Six hours after transfection, cells were washed with 1× PBS, and fresh growth media was added. Cell viability was measured 48 h later using the CellTiter-Glo Luminescent Cell Viability Assay (#G7573; Promega) according to the manufacturer’s instructions. The following shRNA constructs were purchased from the Dana Farber/Harvard Cancer Center Plasmid Information Database (PlasmID): HsSH00157011, HsSH00157016, HsSH00157036, HsSH00167269, HsSH00167262, HsSH00167285, HsSH00152131, HsSH00152139, HsSH00152225, HsSH00116873, HsSH00116854, HsSH00116860, HsSH00157364, HsSH00157314, and HsSH00157319.
Network Analysis.
SS/L genes identified from dsRNA and VDA screens were pooled, and the online STRING database tool (24) was used to identify high-confidence (score ≥ 0.9) PPIs and cluster the genes. In addition, gene functions were annotated manually based on Drosophila or human data in cases where clear orthologs could be identified. Additional SS/L genes were added to network clusters, and new clusters were created in cases where several genes shared similar functions. The network map was generated using Cytoscape (54).
Supplementary Material
Acknowledgments
We thank D. Kwiatkowski for the RT4 bladder cancer-derived cell line and E. Henske for the 621-102 and 621-103 RA-derived cell lines. This work was supported by NIH Grant P01CA120964; University of Pennsylvania Orphan Disease Program Grant MDBR-15-103-LAM; and Department of Defense Grant W81XWH-16-1-0127. N.P. is a Howard Hughes Medical Institute Investigator.
Footnotes
The authors declare no conflict of interest.
Data deposition: RNAi screen data are provided in Datasets S1–S6 and in the Drosophila RNAi Screening Center (DRSC) database and are publicly available at http://www.flyrnai.org/cgi-bin/RNAi_public_screen.pl?project_id=197 and http://www.flyrnai.org/cgi-bin/RNAi_public_screen.pl?project_id=210.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1713362114/-/DCSupplemental.
References
- 1.Nijman SMB. Synthetic lethality: General principles, utility and detection using genetic screens in human cells. FEBS Lett. 2011;585:1–6. doi: 10.1016/j.febslet.2010.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaelin WG., Jr The concept of synthetic lethality in the context of anticancer therapy. Nat Rev Cancer. 2005;5:689–698. doi: 10.1038/nrc1691. [DOI] [PubMed] [Google Scholar]
- 3.Thompson JM, Nguyen QH, Singh M, Razorenova OV. Approaches to identifying synthetic lethal interactions in cancer. Yale J Biol Med. 2015;88:145–155. [PMC free article] [PubMed] [Google Scholar]
- 4.Boone C, Bussey H, Andrews BJ. Exploring genetic interactions and networks with yeast. Nat Rev Genet. 2007;8:437–449. doi: 10.1038/nrg2085. [DOI] [PubMed] [Google Scholar]
- 5.Tong AH, et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- 6.Costanzo M, et al. A global genetic interaction network maps a wiring diagram of cellular function. Science. 2016;353:aaf1420. doi: 10.1126/science.aaf1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luo J, et al. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell. 2009;137:835–848. doi: 10.1016/j.cell.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hart T, et al. High-resolution CRISPR screens reveal fitness genes and genotype-specific cancer liabilities. Cell. 2015;163:1515–1526. doi: 10.1016/j.cell.2015.11.015. [DOI] [PubMed] [Google Scholar]
- 9.Cowley GS, et al. Parallel genome-scale loss of function screens in 216 cancer cell lines for the identification of context-specific genetic dependencies. Sci Data. 2014;1:140035, and erratum (2014) 1:140044. doi: 10.1038/sdata.2014.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang T, et al. Gene essentiality profiling reveals gene networks and synthetic lethal interactions with oncogenic Ras. Cell. 2017;168:890–903.e15. doi: 10.1016/j.cell.2017.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cox AD, Fesik SW, Kimmelman AC, Luo J, Der CJ. Drugging the undruggable RAS: Mission possible? Nat Rev Drug Discov. 2014;13:828–851. doi: 10.1038/nrd4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vyse S, Howitt A, Huang PH. Exploiting synthetic lethality and network biology to overcome EGFR inhibitor resistance in lung cancer. J Mol Biol. 2017;429:1767–1786. doi: 10.1016/j.jmb.2017.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Downward J. RAS synthetic lethal screens revisited: Still seeking the elusive prize? Clin Cancer Res. 2015;21:1802–1809. doi: 10.1158/1078-0432.CCR-14-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barretina J, et al. The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garnett MJ, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483:570–575. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ashworth A, Lord CJ, Reis-Filho JS. Genetic interactions in cancer progression and treatment. Cell. 2011;145:30–38. doi: 10.1016/j.cell.2011.03.020. [DOI] [PubMed] [Google Scholar]
- 17.Housden BE, Nicholson HE, Perrimon N. Synthetic lethality screens using RNAi in combination with CRISPR-based knockout in Drosophila cells. Bio Protoc. 2017;7:e2119. doi: 10.21769/BioProtoc.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Housden BE, et al. Identification of potential drug targets for tuberous sclerosis complex by synthetic screens combining CRISPR-based knockouts with RNAi. Sci Signal. 2015;8:rs9. doi: 10.1126/scisignal.aab3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boutros M, et al. Heidelberg Fly Array Consortium Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science. 2004;303:832–835. doi: 10.1126/science.1091266. [DOI] [PubMed] [Google Scholar]
- 20.Hart T, Brown KR, Sircoulomb F, Rottapel R, Moffat J. Measuring error rates in genomic perturbation screens: Gold standards for human functional genomics. Mol Syst Biol. 2014;10:733. doi: 10.15252/msb.20145216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang T, et al. Identification and characterization of essential genes in the human genome. Science. 2015;350:1096–1101. doi: 10.1126/science.aac7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neumüller RA, et al. Stringent analysis of gene function and protein-protein interactions using fluorescently tagged genes. Genetics. 2012;190:931–940. doi: 10.1534/genetics.111.136465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu Y, et al. FlyRNAi.org—The database of the Drosophila RNAi screening center and transgenic RNAi project: 2017 update. Nucleic Acids Res. 2017;45:D672–D678. doi: 10.1093/nar/gkw977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Szklarczyk D, et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y, Manning BD. mTORC1 signaling activates NRF1 to increase cellular proteasome levels. Cell Cycle. 2015;14:2011–2017. doi: 10.1080/15384101.2015.1044188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Medvetz D, et al. High-throughput drug screen identifies chelerythrine as a selective inducer of death in a TSC2-null setting. Mol Cancer Res. 2015;13:50–62. doi: 10.1158/1541-7786.MCR-14-0440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li J, et al. Synthetic lethality of combined glutaminase and Hsp90 inhibition in mTORC1-driven tumor cells. Proc Natl Acad Sci USA. 2015;112:E21–E29. doi: 10.1073/pnas.1417015112. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Kang YJ, Lu M-K, Guan K-L. The TSC1 and TSC2 tumor suppressors are required for proper ER stress response and protect cells from ER stress-induced apoptosis. Cell Death Differ. 2011;18:133–144. doi: 10.1038/cdd.2010.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Young RM, et al. Dysregulated mTORC1 renders cells critically dependent on desaturated lipids for survival under tumor-like stress. Genes Dev. 2013;27:1115–1131. doi: 10.1101/gad.198630.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Siroky BJ, et al. Human TSC-associated renal angiomyolipoma cells are hypersensitive to ER stress. Am J Physiol Renal Physiol. 2012;303:F831–F844. doi: 10.1152/ajprenal.00441.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, et al. Coordinated regulation of protein synthesis and degradation by mTORC1. Nature. 2014;513:440–443. doi: 10.1038/nature13492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo Y, et al. TSC1 involvement in bladder cancer: Diverse effects and therapeutic implications. J Pathol. 2013;230:17–27. doi: 10.1002/path.4176. [DOI] [PubMed] [Google Scholar]
- 33.Wang Y, Zhang Q, Wang B, Li P, Liu P. LiCl treatment induces programmed cell death of schwannoma cells through AKT- and MTOR-mediated necroptosis. Neurochem Res. 2017;42:2363–2371. doi: 10.1007/s11064-017-2256-2. [DOI] [PubMed] [Google Scholar]
- 34.Maeng Y-S, Lee R, Lee B, Choi S-I, Kim EK. Lithium inhibits tumor lymphangiogenesis and metastasis through the inhibition of TGFBIp expression in cancer cells. Sci Rep. 2016;6:20739. doi: 10.1038/srep20739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lan Y, et al. Lithium enhances TRAIL-induced apoptosis in human lung carcinoma A549 cells. Biometals. 2013;26:241–254. doi: 10.1007/s10534-012-9607-x. [DOI] [PubMed] [Google Scholar]
- 36.Han S, et al. Lithium enhances the antitumour effect of temozolomide against TP53 wild-type glioblastoma cells via NFAT1/FasL signalling. Br J Cancer. 2017;116:1302–1311. doi: 10.1038/bjc.2017.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Menendez JA, Vellon L, Lupu R. Antitumoral actions of the anti-obesity drug orlistat (XenicalTM) in breast cancer cells: Blockade of cell cycle progression, promotion of apoptotic cell death and PEA3-mediated transcriptional repression of Her2/neu (erbB-2) oncogene. Ann Oncol. 2005;16:1253–1267. doi: 10.1093/annonc/mdi239. [DOI] [PubMed] [Google Scholar]
- 38.Malhi GS, Gershon S. Ion men and their mettle. Aust N Z J Psychiatry. 2009;43:1091–1095. doi: 10.3109/00048670903279929. [DOI] [PubMed] [Google Scholar]
- 39.Grunze H, et al. WFSBP Task Force on Treatment Guidelines for Bipolar Disorders The World Federation of Societies of Biological Psychiatry (WFSBP) guidelines for the biological treatment of bipolar disorders: Update 2012 on the long-term treatment of bipolar disorder. World J Biol Psychiatry. 2013;14:154–219. doi: 10.3109/15622975.2013.770551. [DOI] [PubMed] [Google Scholar]
- 40.Malhi GS, Tanious M, Das P, Coulston CM, Berk M. Potential mechanisms of action of lithium in bipolar disorder. Current understanding. CNS Drugs. 2013;27:135–153. doi: 10.1007/s40263-013-0039-0. [DOI] [PubMed] [Google Scholar]
- 41.Can A, Schulze TG, Gould TD. Molecular actions and clinical pharmacogenetics of lithium therapy. Pharmacol Biochem Behav. 2014;123:3–16. doi: 10.1016/j.pbb.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luo B, et al. Highly parallel identification of essential genes in cancer cells. Proc Natl Acad Sci USA. 2008;105:20380–20385. doi: 10.1073/pnas.0810485105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Srivas R, et al. A network of conserved synthetic lethal interactions for exploration of precision cancer therapy. Mol Cell. 2016;63:514–525. doi: 10.1016/j.molcel.2016.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cancer Genome Atlas Research Network Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507:315–322. doi: 10.1038/nature12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kampmann M, et al. Next-generation libraries for robust RNA interference-based genome-wide screens. Proc Natl Acad Sci USA. 2015;112:E3384–E3391. doi: 10.1073/pnas.1508821112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fellmann C, et al. An optimized microRNA backbone for effective single-copy RNAi. Cell Rep. 2013;5:1704–1713. doi: 10.1016/j.celrep.2013.11.020. [DOI] [PubMed] [Google Scholar]
- 47.Housden BE, et al. Loss-of-function genetic tools for animal models: Cross-species and cross-platform differences. Nat Rev Genet. 2017;18:24–40. doi: 10.1038/nrg.2016.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shalem O, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hu Y, et al. An integrative approach to ortholog prediction for disease-focused and other functional studies. BMC Bioinformatics. 2011;12:357. doi: 10.1186/1471-2105-12-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vert J-P, Foveau N, Lajaunie C, Vandenbrouck Y. An accurate and interpretable model for siRNA efficacy prediction. BMC Bioinformatics. 2006;7:520. doi: 10.1186/1471-2105-7-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Perkins LA, et al. The transgenic RNAi project at Harvard Medical School: Resources and validation. Genetics. 2015;201:843–852. doi: 10.1534/genetics.115.180208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li C, et al. Estradiol and mTORC2 cooperate to enhance prostaglandin biosynthesis and tumorigenesis in TSC2-deficient LAM cells. J Exp Med. 2014;211:15–28. doi: 10.1084/jem.20131080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shannon P, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.