Significance
Under environmental stress, animals can adopt different forms and behaviors through phenotypic plasticity. The roundworm Caenorhabditis elegans can exit reproductive growth and enter the stress-resistant dauer larval stage. We investigated phenotypic plasticity in a whole organism by comparing gene expression during dauer and reproductive development using RNA sequencing. As animals entered dauer, we observed striking up-regulation of neuronal signaling peptides, which promote the dauer-entry decision instead of reproductive growth. These neuropeptides also enable new behaviors in dauers for exploiting carrier animals for dispersal. Neuropeptides are similarly up-regulated in the infective, dauer-like stages of parasitic roundworms, indicating dauer as a strong model for studying parasitic behaviors. Our investigation reveals that neuropeptides can alter developmental decision-making and behavior in stressed C. elegans.
Keywords: phenotypic plasticity, RNA-seq, dauer, neuropeptide, FMRFamide
Abstract
Animals, including humans, can adapt to environmental stress through phenotypic plasticity. The free-living nematode Caenorhabditis elegans can adapt to harsh environments by undergoing a whole-animal change, involving exiting reproductive development and entering the stress-resistant dauer larval stage. The dauer is a dispersal stage with dauer-specific behaviors for finding and stowing onto carrier animals, but how dauers acquire these behaviors, despite having a physically limited nervous system of 302 neurons, is poorly understood. We compared dauer and reproductive development using whole-animal RNA sequencing at fine time points and at sufficient depth to measure transcriptional changes within single cells. We detected 8,042 genes differentially expressed during dauer and reproductive development and observed striking up-regulation of neuropeptide genes during dauer entry. We knocked down neuropeptide processing using sbt-1 mutants and demonstrate that neuropeptide signaling promotes the decision to enter dauer rather than reproductive development. We also demonstrate that during dauer neuropeptides modulate the dauer-specific nictation behavior (carrier animal-hitchhiking) and are necessary for switching from repulsion to CO2 (a carrier animal cue) in nondauers to CO2 attraction in dauers. We tested individual neuropeptides using CRISPR knockouts and existing strains and demonstrate that the combined effects of flp-10 and flp-17 mimic the effects of sbt-1 on nictation and CO2 attraction. Through meta-analysis, we discovered similar up-regulation of neuropeptides in the dauer-like infective juveniles of diverse parasitic nematodes, suggesting the antiparasitic target potential of SBT-1. Our findings reveal that, under stress, increased neuropeptide signaling in C. elegans enhances their decision-making accuracy and expands their behavioral repertoire.
Stress during development can have long-lasting effects on animal physiology and behavior. For instance, trauma during early human childhood can lead to difficulties with coping against depression and anxiety in adults (1–4). Phenotypic plasticity allows animals to adapt to stresses from the environment (5). Examples of phenotypic plasticity include the production of new antibodies by the mammalian immune system (6), temperature-dependent sex determination in reptiles (7, 8), and crowding-induced cannibalism in the spadefoot toad (5, 9).
The free-living bacterivore Caenorhabditis elegans can adapt to stressful conditions by exiting reproductive development and entering the stress-resistant dauer larval stage (10–12). In reproductive development, C. elegans develops through four larval stages, L1, L2, L3, and L4, to become a reproductive adult. Declining food and temperature and crowding conditions, however, promote L1 entry into the predauer L2d stage. If conditions improve, L2d commit to reproductive development through amplification of the dafachronic acid (DA) growth hormone across the animal body (13). Unimproved conditions cause L2d larvae to commit to dauer development through a process that is not well understood. Following this event, L2d larvae molt into dauer larvae and halt their feeding.
Dauers are less metabolically active than nondauers and can survive long periods of starvation by utilizing stored lipids in lieu of aerobic respiration (14, 15). Morphologically, they have a highly impermeable cuticle that allows them to resist environmental assaults (10). In addition, they have a rewired nervous system (16, 17) and specialized dispersal behaviors for finding and stowing onto carrier animals (18, 19). Dauer lifespans are 10 times longer than those of nondauers (20), and dauers can resume reproductive development with an unaffected adult lifespan once they recover under favorable conditions (21). Thus, dauers have much to reveal about the biological control of longevity, stress-resistance, and neural state.
The genetic and anatomical tractability of C. elegans make it an advantageous model for studying phenotypic plasticity in a whole organism. Previous systems-level studies have analyzed the C. elegans transcriptome during molt into dauer, during dauer, and during recovery from dauer (22–24). However, the transcriptome of L2d larvae during the dauer-entry decision has not been characterized, likely because L2d larvae lack strong distinguishing traits that can be used to isolate them (25). Furthermore, dauer and reproductive development have not been compared under parallel growth conditions, a lack that has limited the discovery of genes that are differentially expressed in the two developments.
Therefore, we have used techniques from our previous analysis of daf-9(dh6) loss-of-function animals (13) to add crucial details to dauer development. Using DA, we controlled the developmental decisions of daf-9(dh6) L2d animals under parallel conditions. We coupled this with our previous timing of daf-9(dh6) development to collect pure populations of uncommitted L2d, dauer-committed L2d, L3-committing L2d, dauers, and L4 animals. We performed whole-animal RNA sequencing (RNA-seq) on these populations, revealing 8,042 genes differentially expressed during dauer and reproductive development. Through enrichment analysis, we observed striking up-regulation of neuropeptide genes during dauer commitment. Using the sbt-1(ok901)-null mutant to knock down neuropeptide processing (26), we demonstrate that peptidergic signaling promotes the dauer-entry decision, promotes coordination during nictation (a hitchhiking behavior), and is necessary for switching from CO2 repulsion in nondauers to CO2 attraction in dauers.
Testing individual neuropeptides using CRISPR knockouts and existing strains, we demonstrate uncoordinated nictation and CO2 avoidance phenotypes in flp-10(n4543) flp-17(n4894) double mutants, similar to those in sbt-1(ok901). Through a meta-analysis, we discovered similar up-regulation of neuropeptides in the dauer-like infective juveniles (IJs) of diverse parasitic nematodes, suggesting the antiparasitic target potential of SBT-1. Our results reveal that the C. elegans nervous system responds to environmental stress by increasing neuropeptide signaling to enhance decision-making and to enable specialized dispersal behaviors. The expansion of the neuropeptide genes, especially the FMRFamide-like neuropeptide (flp) genes, in nematodes has been a puzzle (27, 28), and our results provide one reasonable explanation.
Results
RNA-Seq Was Used to Investigate Dauer and Reproductive Development.
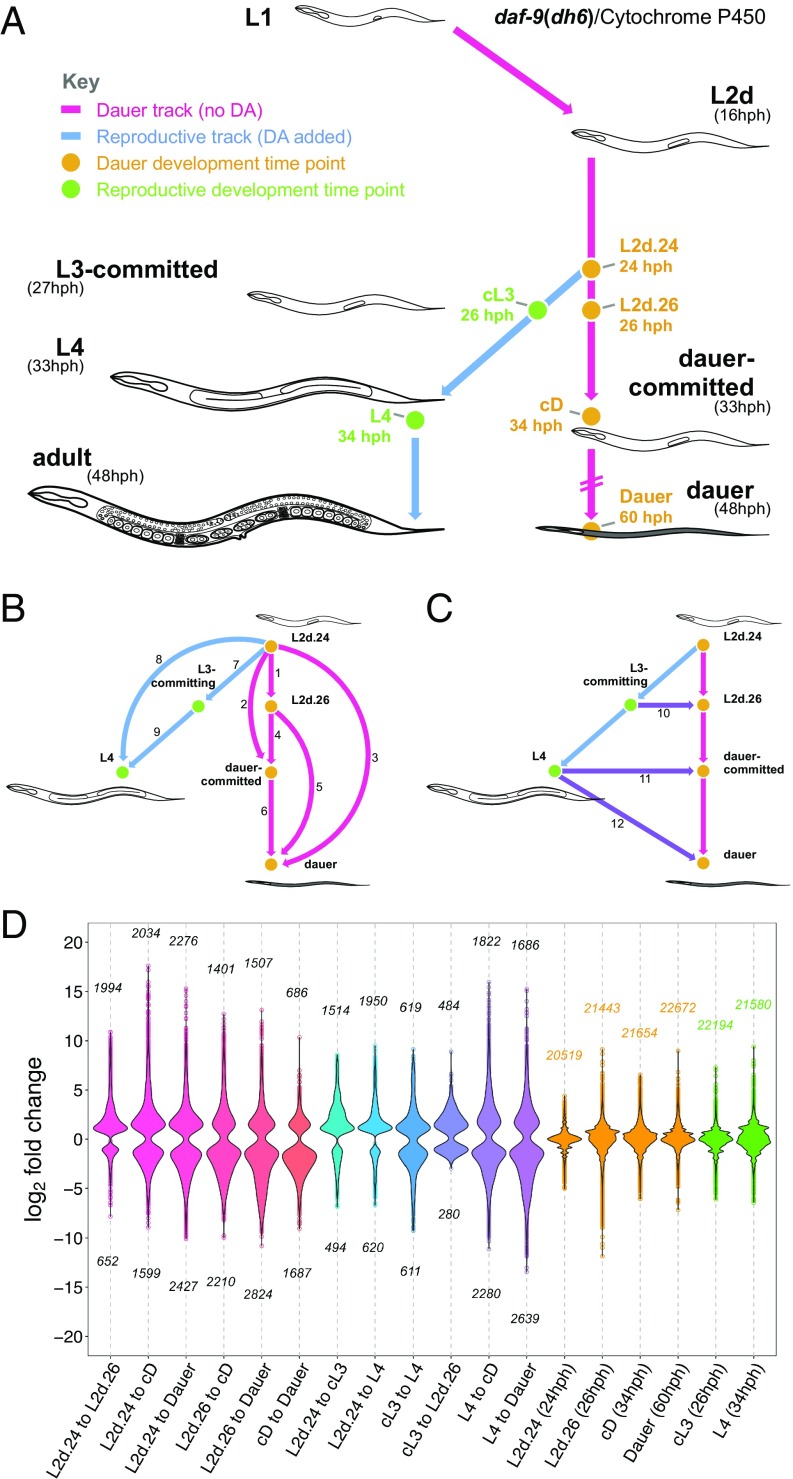
Our previous analysis of the daf-9(dh6)-null mutant (13) allowed us to sequence cDNA from large quantities of dauer- and reproductive-developing animals. Briefly, C. elegans DAF-9 is a cytochrome P450 enzyme that synthesizes the growth hormone DA (29, 30). Commitment to reproductive development occurs only when the level of DA in the animal body is high enough to trigger feedback amplification of DA across the entire organism, thus locking the developmental decision (13). daf-9(dh6)-null mutants cannot produce their own DA and therefore constitutively form dauers unless synthetic DA is added to induce reproductive development (13, 30, 31). We previously characterized the timing of development in daf-9(dh6) animals: Dauer commitment occurs at 33 h post hatch (hph), followed by molt into dauer at 48 hph (13). However, if DA is added at 24 hph, daf-9(dh6) L2d animals commit to reproductive development by 27 hph and molt into L4 animals at 34 hph.
Using these conditions, we grew synchronized populations of dauer-developing daf-9(dh6) animals by withholding synthetic DA and collected L2d animals at 24 hph and 26 hph, dauer-committed L2d animals at 34 hph, and fully developed dauers at 60 hph for RNA-seq (Fig. 1A). In parallel, we added synthetic DA to a subculture, forcing it into reproductive development, from which we collected L3-committing larvae at 26 hph and L4s at 34 hph for sequencing. We sequenced each sample using two biological replicates, at an average depth of 100 million reads (the sum of the replicates) (Dataset S1). Since C. elegans animals contain 959 cells (32), and each cell likely expresses 100,000 mRNA transcripts (33, 34), we estimated that we had sequenced one read for every transcript in the whole animal at each time point. Therefore, we expected to detect signals from transcripts as long as they were not expressed at low abundance in only a few cells. For each sample, we detected between 20,519 and 22,672 expressed genes of the total 20,362 coding and 24,719 noncoding genes of the C. elegans genome (Fig. S1A and Dataset S2) (35, 36).
Fig. 1.
Thirty-nine percent of the C. elegans genome is differentially expressed during dauer and reproductive development. (A) Experimental design for collecting dauer and reproductively developing daf-9(dh6) animals. The timing of molt events is indicated in parentheses (13). (B and C) Twelve comparisons between the six time points. Arrows are directed from the reference point to the end point of each comparison. (D) Violin plots of the significantly up- and down-regulated genes in each comparison. The numbers of up- and down-regulated genes in each comparison are indicated above and below the violin plot. The fold changes between the replicates of each sequenced time point are plotted for reference (orange and green plots). cD, dauer-committed; cL3, L3-committing; DA, dafachronic acid; hph, hours post hatch.
PCA Analysis Revealed the Extent of Variation Between Developmental Stages.
We analyzed the variation between our transcriptomes using principal component analysis (PCA) (Fig. S1 B and C). Replicate samples had similar principal component (PC) scores, indicating that our biological replicates were well correlated in their gene expression. Our analysis also revealed that the largest sources of variation between our transcriptomes were the differences between dauer commitment/differentiation versus the remaining time points (PC1, 65% of overall variation), and the difference between 24-hph L2d and 26-hph L2d (PC2, 17% of overall variation). Along the first two PCs, 24-hph L2d and L4 demonstrated close similarity, as did 26-hph L2d and L3-committing larvae and dauer and dauer-committed larvae.
8,042 Genes Are Differentially Expressed During Dauer and Reproductive Development.
Using the differential gene-expression analysis program DESeq (37, 38), we performed pairwise comparisons between 24-hph L2d and 26-hph L2d animals to identify gene-expression changes during L2d sensory integration; between L2d and dauer-committed larvae for changes during dauer commitment; and between L2d and dauer larvae for changes during differentiation and maintenance of dauer (Fig. 1B). With our reproductive development samples, we performed pairwise comparisons between L2d and L3-committing larvae for changes during commitment to reproductive development and between L2d and L4 animals for changes during reproductive growth. In addition, our design allowed us to perform pairwise comparisons between age-matched dauer and reproductive-developing animals at 26 hph (L2d versus L3-committing larvae) and 34 hph (dauer-committed larvae versus L4 animals) to identify gene-expression changes specific to one developmental track (Fig. 1C). We avoided sequencing an age-matched reproductive sample for 60-hph dauers since reproductive animals at 60 hph are gravid and not suitable for studying a stage-specific transcriptome. In total, we performed 12 pairwise comparisons between our dauer and reproductive time points. In each comparison, we detected between 484 and 2,276 up-regulated genes and 280–2,824 down-regulated genes (Fig. 1D and Dataset S3). Overall, we observed 8,042 differentially expressed genes from the 12 comparisons. These genes corresponded to 7,866 coding genes and 77 ncRNA genes, indicating that 39% of the C. elegans protein-coding genome is differentially expressed during dauer and reproductive development.
Differential Expression Was Detected with High Accuracy and at Single-Cell Resolution.
To analyze the resolution of our RNA-seq dataset, we used WormBase anatomical-level gene-expression data to search our 8,042 differentially expressed genes for genes previously reported to have tissue-specific expression. We detected transcriptional changes in 47 epithelial system genes (out of 74 total epithelium-specific genes in WormBase), 56 muscular system genes (out of 89), 181 alimentary system genes (out of of 310 total), 108 reproductive system genes (out of 233), 139 nervous system genes (out of 599 total), 9 amphid sensillum genes (of 62 total), and one XXX cell gene (of four total) (Fig. S2A). This analysis suggests that we could detect differential expression from within tissues and single cells. Indeed, we constructed fluorescent transcriptional reporters for col-40 and srt-41, which were up-regulated in our data during dauer development, and we observed previously unreported, specific expression for the two genes in the hypodermis and the AWC neuron, respectively (Fig. S2 B–O).
We examined the accuracy of our dataset by comparing it with SAGE data (39) and microarray data (24), which have identified genes that are enriched in wild-type dauers versus mixed populations or postdauer adults, respectively (24, 39). When tested for differential expression in our data, 141 (45%) of the dauer-enriched genes from ref. 39 were significantly up-regulated at dauer commitment and dauer relative to L2d and L4, respectively (Fig. S2P). Similarly, 312 (69%) of the dauer-enriched genes from ref. 24 were significantly up-regulated at dauer commitment and dauer. Thus, our calculations for differential expression (negative binomial testing at a Benjamini–Hochberg controlled false discovery rate <0.01) may have been more stringent than the calculations in refs. 24 and 39. Other differences may be explained by gene-expression changes that are observable only between dauer and postdauer and by differences in the daf-9(dh6) strain we used versus the wild-type strain. Nonetheless, we conclude from this analysis that daf-9(dh6) and wild-type transcriptomes demonstrate high comparability and that our differential expression testing is conservative.
Clustering Revealed Six Common Expression Profiles During Dauer and Reproductive Development.
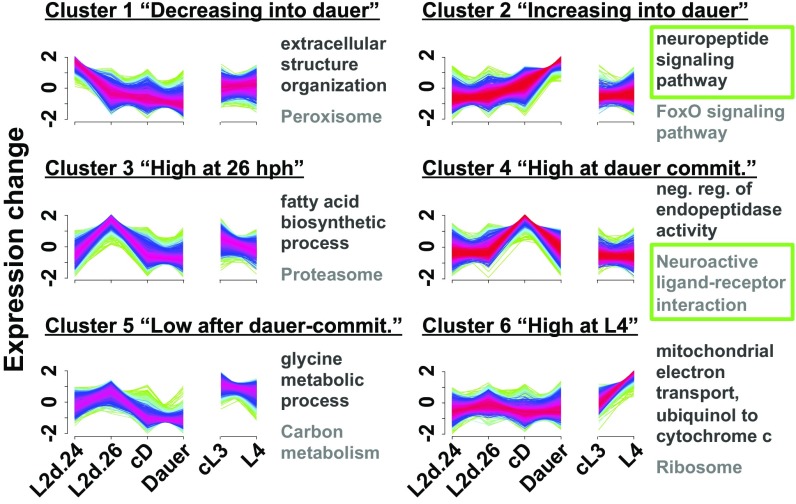
Soft clustering is a sensitive method for identifying common expression profiles within a dataset (40, 41). We performed soft clustering on our 8,042 differentially expressed genes to group the genes by similarities in their expression profiles. This revealed six clusters, indicating that differential gene expression through dauer and reproductive development can be described by six common expression profiles (Fig. 2): The expression levels of 1,102 genes gradually decreased into dauer (cluster 1); 1,921 genes gradually increased into dauer (cluster 2); 1,025 genes increased transiently at 26 hph (cluster 3); 1,497 genes increased transiently during dauer commitment (cluster 4); 1,332 genes decreased after dauer commitment (cluster 5); and 1,165 genes increased in L4 (cluster 6) (Dataset S4).
Fig. 2.
Clustering revealed six common expression profiles during dauer and reproductive development. Soft clustering of the 8,042 differentially expressed genes into six common expression profiles. Yellow-green lines indicate genes with low cluster membership scores, and purple-red lines indicate genes with high membership scores. The top enriched GO and KEGG terms for each cluster are listed in black and gray, respectively. Green boxes indicate terms that are emphasized in the text. commit, commitment.
The genes from clusters 2 and 4 encompass 3,418 genes likely involved in commitment, differentiation, and maintenance of dauer. These highly dauer-specific genes represent a 7.6-fold expansion over the 449 dauer-enriched genes described previously (24). In addition, the genes from clusters 1, 3, 5, and 6 encompass 4,624 genes whose expressions are largely excluded during dauer commitment and dauer.
We examined the six clusters for enriched biochemical pathways and gene ontology (GO) terms using Kyoto Encyclopedia of Genes and Genomes (KEGG) biochemical pathway data (42–44), and the Protein Analysis Through Evolutionary Relationships (PANTHER) classification system (45). We observed that cluster 1 (genes with decreasing expression into dauer) was enriched for the peroxisome KEGG pathway, suggesting that peroxisomal activity is reduced in dauers (Fig. 2 and Dataset S4). This is consistent with the reduction of ascaroside pheromone production in dauers (46), since a key step of ascaroside biosynthesis occurs via peroxisomal β-oxidation (47). In fact, cluster 1 contains genes for the β-oxidation enzymes ACOX-1, MAOC-1, and DHS-28, which perform three of the four steps of ascaroside side-chain biosynthesis (47).
Cluster 2, genes with increasing expression into dauer, was enriched for the FoxO signaling pathway and longevity-regulating pathway–worm KEGG pathways, as would be expected from the extended longevity of dauers (20) and the role of FOXO signaling in modulating longevity and stress resistance (Fig. 2 and Dataset S4) (11). We observed 13 members (18%) of the longevity-regulating pathway–worm in cluster 2, including members in the branches of the pathway that respond to environmental cues, dietary restriction, oxidative stress, germline state, and the mitochondrial unfolded protein response to affect lifespan (42–44). The remaining input into the longevity-regulating pathway, the hypoxia branch, did not have any members in cluster 2. This suggests the intriguing possibility that the extreme longevity of dauers arises from the simultaneously enhanced activity of five of the six branches of the longevity pathway.
We also observed enrichment of the neuropeptide signaling pathway GO term in cluster 2 (Fig. 2 and Dataset S4), suggesting that neuropeptides modulate the dauer-commitment decision and/or neural functions downstream of the decision. Indeed, some neuropeptides have been shown to affect dauer biology: Insulin signaling via DAF-28, INS-4, and INS-6 promotes reproductive growth over dauer development, and INS-1 and INS-18 antagonize this pathway (reviewed in ref. 48). In addition, FMRFamide signaling via FLP-18 acts in parallel with the TGF-β signaling pathway to inhibit dauer development (49). Furthermore, ins-3, -6, -18, -20, -27, -28, -31, and -34 and daf-28 have been shown to affect the fraction of time that is spent nictating in dauer (50).
In cluster 3, genes with increased expression at 26 hph, we observed enrichment for biomolecule synthesis and turnover pathways, including the proteasome, lysosome, fatty acid degradation, and fatty acid elongation KEGG pathways (Fig. 2, Fig. S3, and Dataset S4). This observation may reflect the developmental uncertainty in L2d larvae and perhaps represents a bet-hedging strategy of cycling biomolecules in preparation for either commitment decision. Consistent with the prediction of developmental uncertainty, genes with the molting cycle GO term were enriched (2.9-fold enrichment, P = 2.54 × 10−14) among the genes up-regulated in 26-hph L2d versus 24-hph L2d larvae (Dataset S3) although the molt into dauer, one of the two possible molts from L2d, occurs 22 h later, at 48 hph.
In cluster 4, genes with increased expression at dauer commitment, we observed enrichment of the neuroactive ligand–receptor interaction and calcium signaling KEGG pathways (Fig. 2, Fig. S3, and Dataset S4). Together with cluster 2, this indicates that several genes with neuronal functions have increased expression during dauer commitment and dauer.
Genes that are down-regulated during dauer commitment are likely repressed to exclude nondauer physiologies, and, indeed, our enrichment data for cluster 5, genes with decreased expression after dauer commitment, are consistent with the reduction of TCA cycle activity in favor of long-term lipid metabolism in dauer (51), as we observed enrichment of the fatty acid degradation and citrate cycle (TCA cycle) KEGG pathways (Fig. S3 and Dataset S4).
Cluster 6, genes with increased expression at L4, was enriched for terms related to translation and respiration, including the mitochondrial electron transport, ubiquinol to cytochrome c GO term and the ribosome KEGG pathway (Fig. 2 and Dataset S4), which likely reflects growth during reproductive development and gametogenesis in the L4 animals (52–54).
Differential Expression of the Neuronal Genome During Dauer Development.
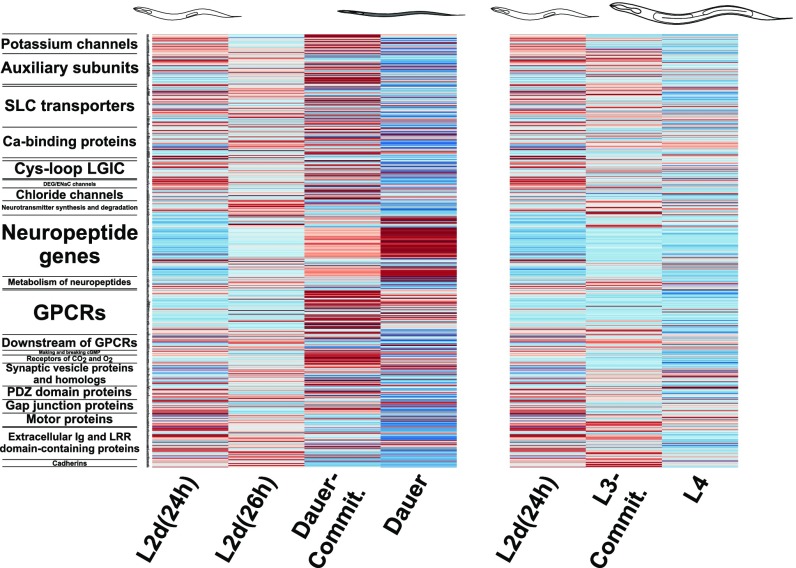
Our KEGG and GO enrichment analyses indicated the strong involvement of neuronal effector genes during dauer commitment and dauer. To investigate this further, we examined the expression of the neuronal genome of C. elegans during dauer and reproductive development. The neuronal genome of C. elegans encodes 3,114 genes from 30 gene classes, including the calcium channels, neurotransmitters, G protein-coupled receptors (GPCRs), and CO2 receptors (55). We detected the differential expression of 606 neuronal genes during dauer and reproductive development, corresponding to 19% of the total neuronal genome, with members from all of the 30 gene classes (Fig. 3).
Fig. 3.
Differential expression of the neuronal effector genome of C. elegans during dauer and reproductive development. In the heatmap of the expression of 606 differentially expressed neuronal effector genes, each row represents a single gene; the class to which the gene belongs is indicated on the left. Red and blue indicate high and low expression scores, respectively. Commit, commitment.
Five gene classes were enriched in one of the soft clusters 1–6, indicating that for these classes a high proportion of their members followed a certain expression profile during dauer and reproductive development (Dataset S5). The extracellular Ig and leucine-rich repeat domain gene class was overrepresented in cluster 1 (decreasing expression into dauer), with 2.8-fold enrichment and P = 6.42 × 10−3. The neuropeptide gene class was overrepresented in cluster 2 (increasing expression into dauer), with 3.1-fold enrichment and P = 6.32 × 10−21. Finally, the GPCR (2.9-fold enrichment, P = 2.41 × 10−8), CO2 and O2 receptor (4.9-fold enrichment, P = 7.86 × 10−5), and potassium channel (3.0-fold enrichment, P = 2.84 × 10−3) gene classes were overrepresented in cluster 4 (increased expression at dauer commitment).
GPCR gene expression increases sharply during dauer commitment, before neuropeptide gene expression reaches its peak during dauer (Fig. 3). In addition, the 34 GPCRs in cluster 4 include one biochemically deorphanized neuropeptide GPCR (npr-11) and nine putative neuropeptide GPCRs (ckr-1, frpr-7, -19, npr-17, -31, C01F1.4, F13H6.5, Y37E11AL.1, and Y70D2A.1) (Dataset S5) (27, 55). This suggests that neuropeptide receptors are up-regulated during dauer commitment in anticipation of increasing neuropeptide gene expression during dauer commitment and dauer.
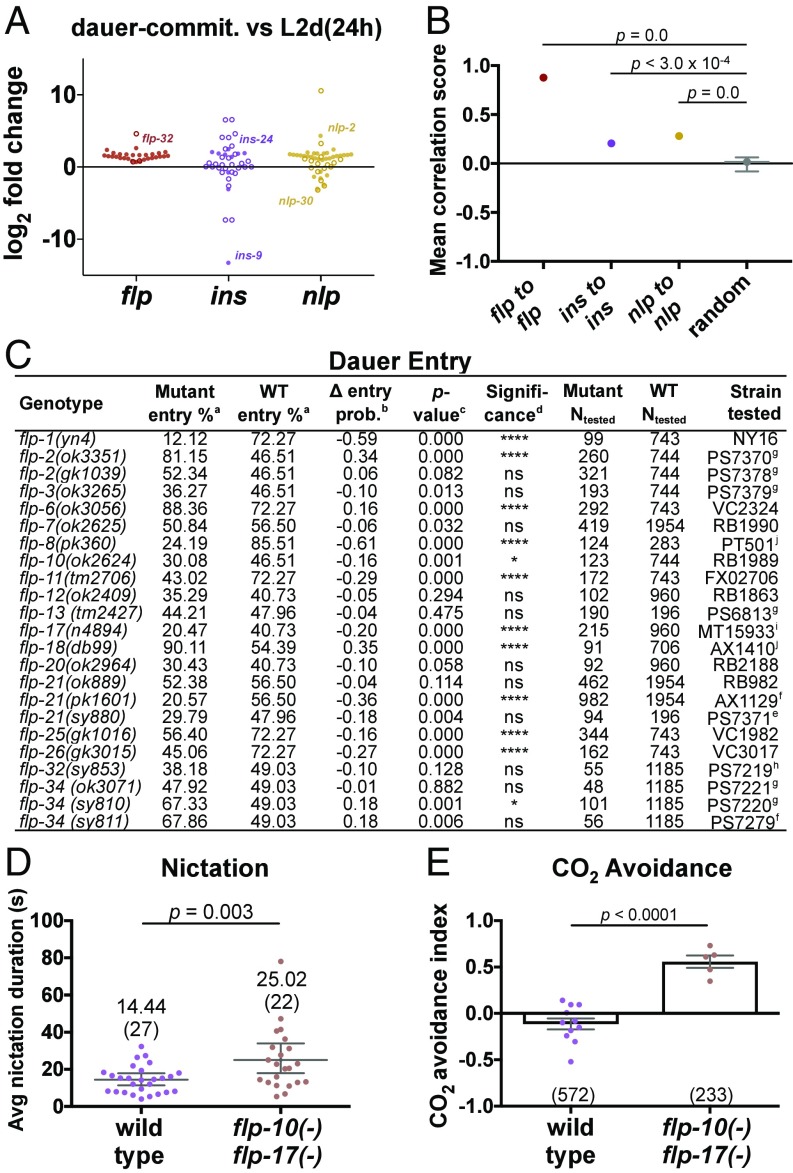
Notably, the neuropeptide gene class was the only class that was enriched for increasing expression into dauer. We observed extensive up-regulation of the neuropeptides during dauer development: In dauer commitment (34 hph) versus L2d (24 hph), 60 of the 118 total neuropeptide genes were up-regulated, while nine were down-regulated (see Fig. 5A). Similarly, at dauer commitment (34 hph) versus L4 (34 hph), 43 neuropeptide genes were up-regulated, while 10 were down-regulated (Fig. S5A), and in dauer (60 hph) versus L2d (24 hph), 64 neuropeptide genes were up-regulated, while nine were down-regulated (Fig. S5B). The up-regulation of 64 neuropeptide genes during dauer versus L2d is remarkable, as it corresponds to the majority of all neuropeptide genes in the C. elegans genome. Furthermore, the 64 genes encode for 215 putative or biochemically isolated peptides (48). By comparison, the human genome contains 97 neuropeptide genes that encode for 270 peptides (56).
Fig. 5.
FMRFamide-like peptides are coordinately up-regulated during dauer development. (A) Fold changes in gene expression for all 118 C. elegans neuropeptide genes. Closed and open circles indicate significant and nonsignificant differential expression, respectively. The most up- and down-regulated genes of each family are labeled for reference. (B) Average Spearman correlation scores of genes to other genes in the same neuropeptide family, calculated across our RNA-seq dataset. The bootstrapped mean and 99% CI are indicated. (C) Survey for flp genes involved in the dauer-entry decision. Footnotes: a, mean percentage calculated by nonparametric bootstrapping; b, the mean difference in dauer-entry probability between wild-type and mutant animals, calculated using Bayesian probability; c, calculated via permutation test; d, determined using a cutoff of Bonferroni-corrected P value < 0.05: ****P < 0.0001, *P < 0.05, ns, not significant; e–j, strains outcrossed 1, 2, 3, 4, >4, and 6 times, respectively. (D and E) Nictation duration (D) and CO2 avoidance (E) assays. Avg, average; commit, commitment; prob, probability.
Peptidergic Signaling Downstream of SBT-1 Promotes Dauer Entry and Nictation Coordination and Is Necessary for CO2 Chemoattraction in Dauer.
Neuropeptides become functional transmitters and neuromodulators only after they are cleaved from longer proneuropeptide chains (Fig. S4A) (48). SBT-1/7B2 is a chaperone for the proprotein convertase EGL-3/PC2, which cleaves proneuropeptides (57), and as a result, sbt-1(ok901)-null mutants have reduced levels of mature neuropeptides compared with wild-type animals (Fig. S4B) (26). Previously, sbt-1(ok901) mutants were reported to possess aldicarb resistance and extended lifespans (58, 59), but to our knowledge no functions for sbt-1 in dauer biology have been reported.
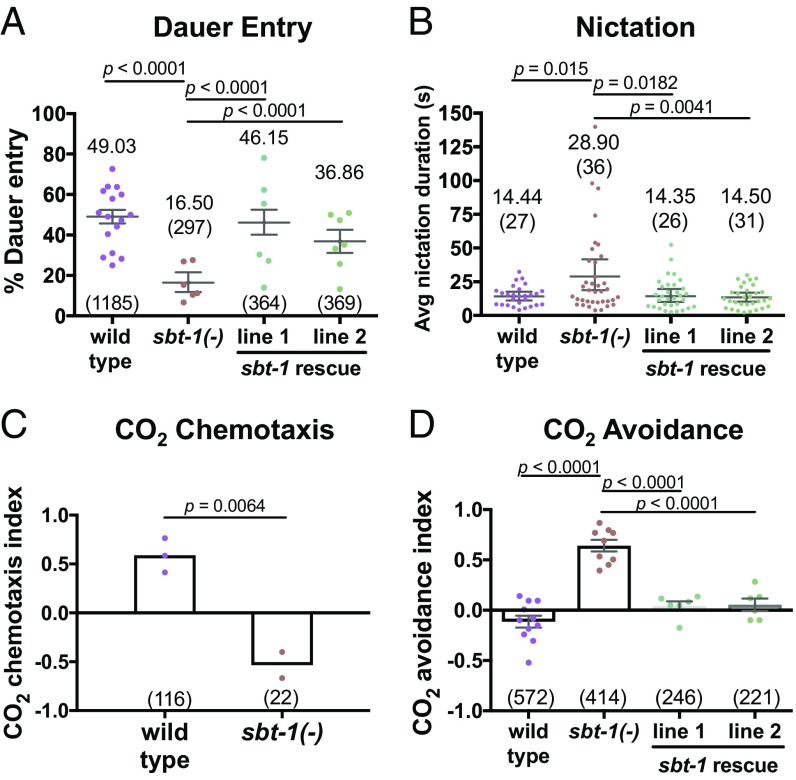
Because we observed up-regulation of neuropeptides starting from dauer commitment, we tested the ability of sbt-1(ok901) mutants to enter dauer, using crude pheromone to induce dauer entry. Under the same dauer-inducing conditions, wild-type animals entered dauer 49% of the time, while sbt-1(ok901) mutants entered dauer 16.5% of the time (Fig. 4A). We also observed that expressing sbt-1 genomic DNA in sbt-1(ok901) mutants (under the control of the endogenous promoter) rescued the dauer-entry phenotype in two independent lines: Rescue line 1 entered dauer 46% of the time, and rescue line 2 entered dauer 37% of the time (Fig. 4A). These results suggest that the net effect of peptidergic signaling downstream of SBT-1 promotes dauer entry over reproductive development.
Fig. 4.
Neuropeptide signaling promotes dauer entry and dispersal behaviors. (A and B) Dauer-entry (A) and nictation duration (B) assays. Bootstrapped means and 99% CIs are indicated. (C and D) CO2 chemotaxis (C) and avoidance (D) assays. Means and SEM are indicated. In A–D, each dot is one trial, and the number tested is in parentheses. Statistics: permutation test (A and B); two-tailed t (C); one-way ANOVA (D). Avg, average.
We examined if neuropeptides play a role during dauer, when the majority of the C. elegans neuropeptide genes in our dataset were expressed at the highest level. C. elegans dauers have been found to associate with invertebrate carriers, likely for transportation to new niches (60). Nictation, during which dauers stand on their tails and wave their bodies, and CO2 chemoattraction are two dauer-specific behaviors that likely enable dauers to migrate toward and attach onto carriers (18, 61). We tested sbt-1(ok901) nictation on microdirt chips, which provide substrates on which dauers can nictate, and observed that the average nictation duration doubled in sbt-1(ok901) mutants (mean = 28.90 s, maximum = 139.63 s) compared with wild-type animals (mean = 14.44 s, maximum = 32.32 s). Moreover, the phenotype was rescued by sbt-1 genomic DNA expression under the endogenous promoter (line 1: mean = 14.35 s and maximum = 78.00 s, line 2: mean = 14.50 s and maximum = 52.27 s) (Fig. 4B). We observed a difference in the degree of 3D movement during nictation in wild-type and sbt-1(ok901) mutant animals that may explain the increased duration of nictation in sbt-1(ok901) mutants: While wild-type animals displayed a wide range of motion and fell back to the chip easily, sbt-1(ok901) animals displayed a limited range of motion and slow, uncoordinated waving that likely increased stability during nictation (Movies S1 and S2). We did not observe significant differences between wild-type animals and sbt-1(ok901) mutants in other components of the nictation behavior, such as initiation frequency and the proportion of time spent nictating (Fig. S4 C and D) (19).
CO2 has been shown to be attractive to dauers and repulsive to nondauers (18, 62). Using chemotaxis assays, we observed attraction to CO2 in wild-type dauers (chemotaxis index = 0.59) and repulsion to CO2 in sbt-1(ok901) dauers (chemotaxis index = −0.53) (Fig. 4C). We further performed CO2 acute avoidance assays by delivering CO2 directly to the nose of forward-moving dauers and scoring reversal. While wild-type dauers did not avoid CO2 (avoidance index = −0.11), we observed rapid reversal in sbt-1(ok901) mutants in response to CO2 (avoidance index = 0.64). In addition, the CO2 repulsion phenotype of sbt-1(ok901) mutants was rescued by sbt-1 genomic DNA (line 1: avoidance index = 0.04; line 2: avoidance index = 0.06) (Fig. 4D and Movies S3–S6). Together, our results indicate that the neuropeptides downstream of SBT-1 modulate proper nictation coordination and are necessary for the correct CO2 preference switch from aversion in nondauers to attraction in dauers.
The 31-Gene flp Family Is Coordinately Up-Regulated During Dauer Development.
The C. elegans genome encodes for three families of neuropeptides: the FMRFamide-like peptides (31 flp genes), the insulin-related peptides (40 ins genes), and the neuropeptide-like proteins (47 nlp genes) (48). In dauer commitment versus L2d, we observed the up-regulation of almost all the flp genes, with significant increases in expression for flp-1, -2, -4–9, -11–22, -24–28, and -32–34 (28 of the total 31) (Fig. 5A and Dataset S3). In contrast, smaller proportions of ins and nlp genes were up-regulated during dauer commitment versus L2d: ins-1, -17–18, -24, -28, and -30 and daf-28 (7 of the total 40) and nlp-1–3, -6, -8–15, -17–18, -21, -35, -37, -38, -40–42, and -47, ntc-1, pdf-1, and snet-1 (25 of the total 47). Similar results were observed during dauer commitment versus L4 (Fig. S5A) and dauer versus L2d (Fig. S5B). Therefore, the flp genes, more than the ins or nlp genes, are coordinately up-regulated during dauer development.
We quantified this coordination in flp gene expression by pairing every combination of the 31 flp genes and scoring the correlation between the expression of each pair of genes across our RNA-seq dataset. The average correlation score between the flp genes was 0.88, with possible scores ranging from −1 (perfectly anticorrelated) to 1 (perfectly correlated) (Fig. 5B). By comparison, the average correlation scores for random sets of 31 genes (mimicking the size of the flp family) were distributed around a bootstrapped mean of 0.02. In addition, the ins and nlp genes had an average score of 0.21 and 0.28, respectively. Furthermore, we obtained similar results when we expanded our correlation analysis to include expression data from 246 publically available RNA-seq datasets describing a broad range of C. elegans life stages and experimental conditions (Fig. S5 C and D) (23).
Using dotplot analysis (63), we examined whether the 31 flp genes share regions of sequence similarity (Fig. S5E). We observed that there are no regions of shared sequence among the flp genes that extend beyond 20 nt, with only two regions sharing a 20-nt match (between flp-2 and flp-22, and between flp-27 and flp-28). Therefore, it is unlikely that the high correlation in flp gene expression was caused by any cross-mapping of RNA-seq reads among the 31 flp genes, since our sequenced reads were 50–100 bp in length. Together, these results strongly suggest that the flp genes are coregulated and are coordinately up-regulated during dauer development.
FLPs Modulate the Dauer-Entry Decision, Nictation, and CO2 Chemoattraction.
We investigated whether FLP neuropeptides modulate the dauer-entry decision by assaying four CRISPR-generated knockout alleles and 19 available flp alleles corresponding to mutations in 18 flp genes. We induced dauer entry using crude pheromone and compared the dauer-entry percentage of each genotype with the wild-type control. We recapitulated the previously reported increased dauer-entry phenotype of flp-18(db99) as a positive control (49). We observed increased dauer entry in three additional flp mutants: flp-2(ok3351), flp-6(ok3056), and flp-34(sy810). Furthermore, we detected decreased dauer entry in eight mutants: flp-1(yn4), flp-8(pk360), flp-10(ok2624), flp-11(tm2706), flp-17(n4894), flp-21(pk1601), flp-25(gk1016), and flp-26(gk3015) (Fig. 5C). These results suggest that FLPs can act redundantly and with opposed effects on dauer entry.
flp-10 and flp-17 are expressed in the CO2-sensing BAG neuron (64, 65) and act synergistically with the neurotransmitter acetylcholine, which promotes the nictation behavior (19), to inhibit egg-laying (66). Because of these connections to nictation and CO2-sensing, we examined nictation and CO2 chemoattraction in flp-10(n4543) flp-17(n4894) double-mutant animals. Using the microdirt chip, we observed an average nictation duration of 14.44 s for wild-type animals and an increased average duration of 25.02 s in flp-10(n4543) flp-17(n4894) mutants (Fig. 5D). For CO2 avoidance, we observed that flp-10(n4543) flp-17(n4894) mutants displayed increased reversal behavior in response to CO2 (avoidance index = 0.56) compared with wild-type animals (avoidance index = −0.11) (Fig. 5E). These data suggest that flp-10 and flp-17 contribute to SBT-1 functions in mediating nictation coordination and the switch to CO2 attraction in dauers.
flp Genes Are Coordinately Up-Regulated in Parasitic Nematode IJs.
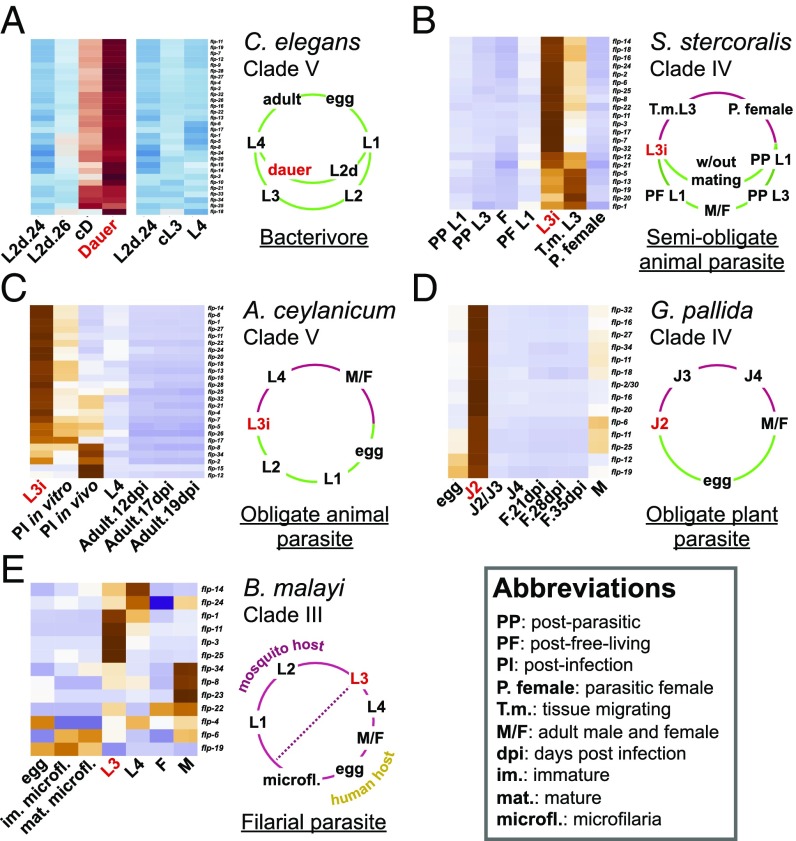
The IJ dispersal stage of parasitic nematodes is similar to dauer in several ways: Both are nonfeeding stages with a resistant cuticle (67), and both recognize and exploit carriers/hosts similarly (18, 68). One gene class that has been shown to affect dauers and IJs is the neuropeptide-encoding set of genes (27, 48). To investigate if the coordinated up-regulation of flp genes is a strategy shared by dauers and IJs, we performed a meta-analysis on flp gene expression in IJs of the semiobligate animal parasite Strongyloides stercoralis, the obligate animal parasite Ancylostoma ceylanicum, the obligate plant parasite Globodera pallida, and the filarial parasite Brugia malayi. We selected these distantly related parasitic species because orthologs/analogs of C. elegans flp genes have been identified in these nematodes (69). In addition, the transcriptomes of these species had been collected using RNA-seq from stages during, before, and after IJ (70–73).
There are 21 flp genes in S. stercoralis, 25 in A. ceylanicum, 14 in G. pallida, and 13 in B. malayi. We observed that each flp gene was expressed at its highest level during the IJ or postinfection IJ stage in S. stercoralis, A. ceylanicum, and G. pallida and was expressed at low levels in other stages, including the egg, the first larval, third larval, fourth larval, and adult stages (Fig. 6 B–D). Specifically, S. stercoralis expressed 16 flp genes at high levels in IJs and five flp genes at high levels in postinfection IJs (Fig. 6B). A. ceylanicum expressed 18 flp genes at high levels in IJs and five flp genes at high levels in postinfection IJs (Fig. 6C). G. pallida expressed 14 flp genes at high levels in IJs (Fig. 6D).
Fig. 6.
FMRFamide-like peptides are coordinately up-regulated in host-seeking IJs of parasitic nematodes. The life cycle and clade membership [in five major clades of Nematoda (107)] of each species are indicated to the right of each heatmap. Green life-cycle regions indicate stages that are free-living or external to a host. Red life-cycle regions indicate stages internal to a host. The dauer and IJ stages are indicated with red letters. (A) Expression of flp genes in the free-living C. elegans (Dataset S5). Red and blue indicate high and low expression scores, respectively. (B–E) Expression of flp orthologs/analogs in the transcriptomes of the semiobligate animal parasite S. stercoralis (B), the obligate animal parasite A. ceylanicum (C), the obligate plant parasite G. pallida (D), and the filarial parasite B. malayi (E). Orange and purple indicate high and low expression scores, respectively. Transcriptomic data from are from refs. 70–73.
By contrast, only 4 of the 13 flp genes in B. malayi were expressed at high levels in IJs. The expressions of the remaining flp genes were specialized to other stages, such as the microfilariae and the adult male (Fig. 6E). Unlike the other three parasitic nematodes, B. malayi spends its life cycle entirely within its hosts. Notably, the IJs of B. malayi infect humans through the aid of a mosquito vector (74). This differs from the IJs of S. stercoralis, A. ceylanicum, and G. pallida, which must find and infect their hosts (Fig. 6). We therefore observe that C. elegans dauers and the host-seeking IJs of S. stercoralis, A. ceylanicum, and G. pallida share a strategy of coordinately up-regulating the flp family.
Discussion
In the wild, C. elegans feeds on transient microbial communities that collapse approximately every three generations (75). To persist, C. elegans can enter the stress-resistant dauer larval stage, which can seek improved conditions by stowing onto carrier animals (60). We sequenced cDNA from dauer- and reproductively developing animals by culturing daf-9(dh6) animals under identical conditions apart from exposure to DA. This allowed us to collect transcriptomes of L2d animals during dauer commitment and commitment to reproductive development. Our design also allowed us to compare dauer and reproductive development to identify gene-expression changes along and between the two tracks. We have demonstrated that 8,042 genes are differentially expressed during dauer and reproductive development, including the up-regulation of 51% of the neuropeptide genes during dauer commitment.
Neuropeptides are short sequences of amino acids that are derived from longer proneuropeptide chains and can act as transmitters and neuromodulators. As neuromodulators, they can control the activity, polarity, sensitivity, and signaling repertoire of neurons (76, 77). Neuropeptides can also diffuse to facilitate signaling between synaptically unconnected neurons (78, 79). Through these modulatory functions, neuropeptides can shape which circuits are active in the nervous system, the membership of these circuits, and their functions (79).
C. elegans encodes three families of neuropeptides: the insulin-like peptides (INS), the neuropeptide-like proteins (NLP), and the FMRFamide-related peptides (FLP) (48). We have shown that the flp genes are coordinately up-regulated during dauer commitment. On the other hand, few of the ins genes and approximately half of the nlp genes are up-regulated during dauer commitment. The low level of ins up-regulation is not surprising, since insulins have conserved roles in growth and metabolism in Metazoa (80). In addition, signaling through the C. elegans insulin-like receptor DAF-2 promotes reproductive growth (48). Indeed, the only ins gene that was up-regulated between dauer commitment and L4 was ins-1, which is known to increase dauer entry, likely by antagonizing DAF-2 signaling (28). Likewise, the nlp genes would not be expected to be up-regulated as a family, since the NLPs are a miscellaneous group of non-INS, non-FLP neuropeptides (48) that likely function in several independent processes. On the other hand, FLPs have conserved roles in regulating feeding and reproduction in nematodes, arthropods, mollusks, and vertebrates (80–83). These roles correlate well with the inhibition of feeding and reproduction and the activation of specialized food-seeking behaviors in dauer. Therefore, the coordinated up-regulation of the flp family may function to generate a wide response to stress that is centered on feeding and reproduction control.
We took advantage of the knockdown of neuropeptide processing in sbt-1(ok901)-null mutants (26) to investigate the function of the neuropeptides during dauer development. We have shown that the net effect of peptidergic signaling downstream of sbt-1 is to promote dauer entry, perhaps by encoding prodauer information from the sensed environment or by modulating the food, temperature, and pheromone signaling pathways to affect the threshold (13) for dauer entry. We have also assayed 23 flp mutants and observed increased dauer entry in four mutants and decreased entry in eight mutants. These results suggest that FLPs can act redundantly and with opposed effects on dauer entry, perhaps to fine-tune the entry decision in response to environmental signals.
Following dauer entry, dauer larvae demonstrate behaviors and preferences that are not observed in nondauers. For instance, dauers are the only stage that can nictate (10, 19) and are attracted to CO2, whereas nondauers are repelled (18, 84). These changes indicate that dauers possess a different neural state from nondauers, likely involving different or altered circuits in the nervous system. However, C. elegans possesses only 302 neurons that are densely interconnected (79, 85), with no synaptically compartmentalized circuits to switch between during dauer and nondauer. To overcome this constraint, dauers can rewire their neurons to access new behaviors (16, 19). We observed that, in addition to this strategy, peptidergic signaling downstream of sbt-1 promotes coordination during nictation and is necessary for the switch from CO2 repulsion to CO2 attraction in dauer. We have also shown that the combined effects of flp-10 and flp-17 strongly promote nictation coordination and the switch to CO2 attraction. Therefore, we have demonstrated that neuropeptides change the neural state of C. elegans during dauer, possibly by altering the composition and function of the active circuits in the nervous system (Fig. S6).
Considering these results, it is notable how many neuropeptides are up-regulated during dauer development. By dauer, 64 neuropeptide genes encoding 215 peptides are up-regulated; by comparison, the entire human genome contains only 97 neuropeptide genes encoding 270 peptides (56). Indeed, the neuropeptide gene families are expanded in C. elegans (48), and the FLP neuropeptides are especially expanded in the phylum Nematoda (27). We observed that FLPs are involved in establishing the C. elegans dauer neural state, and RNAi knockdown experiments have also shown that FLPs regulate the IJ dispersal behaviors of G. pallida (flp-12), Meloidogyne incognita (flp-18), and Steinernema carpocapsae (flp-21) (86, 87), indicating that FLPs are involved in establishing the IJ neural state as well. Because we observed coordinated up-regulation of the FLPs during dauer and in the IJs of the distantly related nematodes S. stercoralis, A. ceylanicum, and G. pallida, we speculate that the computational challenges of dauer and IJ were the driving force for flp expansion in Nematoda. This hypothesis is supported by the lack of such expansion in the nematodes Trichinella spiralis and Trichuris muris (69), which do not posses dauer or IJ stages (88, 89) and encode only four flp genes each. Therefore, flp expansion may have provided ancestral nematodes the means to overcome their constrained nervous systems (90) to adapt effectively to stress during dauer and IJ.
Our genetic data and meta-analysis also suggest that SBT-1 would be a potent target for anthelminthic control. Since sbt-1–null mutants are strongly defective in dauer entry and dispersal behaviors, we predict that targeting SBT-1 in parasitic nematodes will severely impair dispersal and host-seeking in their IJs. While RNAi against individual FLPs can affect IJ dispersal (86, 87), our meta-analysis indicates that multiple FLPs are up-regulated in several types of parasitic nematodes. We propose that inhibition of SBT-1 could be used efficiently to knock down multiple FLPs at once and in a wide range of parasitic nematodes. SBT-1 would also be an excellent target, since nematode SBT-1 is distinct in sequence from vertebrate 7B2 (57), reducing the risks of cross-species effects.
We have investigated phenotypic plasticity in a whole organism by studying C. elegans adaptation to stress during development. We uncovered the transcriptional dynamics of C. elegans during dauer development and discovered a strategy of massively up-regulating neuropeptide expression. This strategy functions to enhance the dauer-entry decision and expand the behavioral repertoire of dauers and appears to be evolutionarily shared by dauers and host-seeking IJs, suggesting SBT-1 as a potent anthelminthic target.
Materials and Methods
Animal Strains.
C. elegans strains were grown using standard protocols with the Escherichia coli strain OP50 (for plate cultures) or HB101 (for liquid cultures) as a food source (91). The wild-type strain was N2 (Bristol). Other animal strains are listed in SI Materials and Methods.
Transgenic Strains.
sbt-1 genomic DNA rescue strains were generated by injecting 15 ng/μL of sbt-1 genomic DNA (amplified by PCR with forward primer CTGTGAAGCGCTCATCTGAA and reverse primer TTCAGGCAAATCCATCATCA), 50 ng/μL coelomocyte-specific ofm-1p::rfp coinjection marker, and 135 ng/μL 1-kb DNA ladder (New England Biolabs) carrier DNA into the adult gonads of sbt-1(ok901) animals, followed by integration into the genome by X-ray (92, 93). Two independent integration lines were generated: PS7274 sbt-1(ok901); Is444[sbt-1p::sbt-1; ofm-1p::rfp] (line 1, outcrossed two times) and PS7275 sbt-1(ok901); Is445[sbt-1p::sbt-1; ofm-1p::rfp] (line 2, outcrossed three times). Transcriptional reporter strains for srt-41 (PS7128) and col-40 (PS6727) were built using methods described in SI Materials and Methods.
CRISPR-Generated Strains.
CRISPR mutagenesis with coconversion (94) was used to generate the PS7371 flp-21(sy880), PS7219 flp-32(sy853), PS7220 flp-34(sy810), and PS7279 flp-34(sy811) deletion strains. Detailed information about strain generation and about the deletion alleles is given in SI Materials and Methods.
daf-9(dh6) Culturing and Harvesting for RNA-Seq.
Synchronous, single-stage populations of daf-9(dh6) animals were grown using our previously described method for liquid culturing daf-9(dh6) (13). daf-9(dh6) animals were collected at six points over a branching time series from 24–60 hph. This period, as we have previously analyzed, includes L2d sensory integration, dauer commitment, dauer maintenance, reproductive commitment, and reproductive development (Fig. 1A) (13). The dauer-developing branch was obtained by withholding DA (Δ7-DA), and animals were collected at 24 hph (L2d), 26 hph (L2d), 34 hph (dauer commitment), and 60 hph (dauer). The reproductive-developing branch was obtained by adding 100 nM Δ7-DA at 24 hph, and animals were collected at 26 hph (L3 committing) and 34 hph (L4). Reproductive animals at 60 hph are gravid, making them inappropriate for single-stage transcriptome analysis, and were therefore not collected.
Cultures from the dauer and reproductive branches were grown in parallel, fed at the same time, and experienced the same batches of HB101 and Δ7-DA to minimize asynchronous development between the cultures. At each time point samples were collected from two independently cultured biological replicates. Each biological replicate was maintained separately for at least five generations. Harvested animals were spun three times in S basal medium (5.85 g NaCl, 1 g K2HPO4, 6 g KH2PO4 and 1 mL of 5 mg/mL cholesterol, all dissolved in 1 L water) to help clear the bacteria. The worm pellets (∼10,000 worms per pellet) were then treated with 1 mL TRIzol and 0.6 mg/mL linear polyacrylamide carrier before being flash frozen in liquid nitrogen and stored at −80 °C. RNA was purified as previously described (13).
RNA-Seq and Computational Analysis.
cDNA was prepared from the collected samples using the Illumina TruSeq RNA Sample Preparation kit or mRNA-Seq Sample Preparation kit. cDNA was sequenced using the Illumina HiSeq 2000 to generate paired-end or single-end libraries. Paired-end libraries were not multiplexed, were sequenced at a read length of 100 bp, and were sequenced to an average depth of 76 million reads. Single-end libraries were multiplexed at four libraries per lane, were sequenced at a read length of 50 bp, and were sequenced to an average depth of 33 million reads. All raw sequences have been deposited in the National Center for Biotechnology Information Sequence Read Archive (SRA) database (accession no. SRP116980). Dataset S1 contains the detailed metadata for the sequenced libraries. Codes used for data analysis have been deposited in GitHub at https://github.com/WormLabCaltech/dauerRNAseq.
Read mapping and differential expression testing.
Reads that did not pass the Illumina chastity filter were removed using Perl. Read mapping, feature counting, library normalization, quality checks, and differential gene-expression analysis were performed using R version 3.1.0, bowtie2 version 2.2.3, tophat2 version 2.0.12, SAMtools version 0.1.19, HTSeq version 0.6.1, and DESeq, as described in ref. 38. The C. elegans reference genome and gene transfer format files were downloaded from Ensembl release 75 and genome assembly WBcel235. Gene-dispersion estimates were obtained after pooling all sequenced samples. Each pairwise comparison for differential gene expression was performed at a Benjamini–Hochberg controlled false discovery rate <0.01.
RNA-seq data summaries.
PCA was performed using the DESeq package in R (37). Violin plotting was done in R using ggplot2 (95). KEGG biochemical pathway enrichment analysis was performed using clusterProfiler in R (96), at a cutoff of Benjamini–Hochberg corrected q-value < 0.01. GO enrichment analysis was performed using the PANTHER Overrepresentation Test for GO Biological Process, at a Bonferroni-adjusted P value cutoff of <0.05. GO and KEGG terms were ranked based on descending fold-enrichment for GO and ascending q-value for KEGG.
Soft clustering.
Soft clustering was performed using the mFuzz package in R (40, 41). Gene-expression data were standardized before clustering, and cluster numbers were chosen based on cluster stability, minimum cluster centroid distance, and visual clarity of the clusters. Overrepresented neuronal gene classes in clusters 1–6 were determined by hypergeometric test, using a cutoff of Bonferroni-corrected P value < 0.05.
Heatmaps.
Heatmaps were drawn using the gplots and RColorBrewer packages in R (97, 98). Mean count values were used for each time point, calculated by averaging the biological replicates. Heatmap dendrograms were drawn using correlation distances and average-linkage hierarchical clustering. Expression values were centered and scaled for each gene.
Gene-expression correlation analysis.
Spearman correlation scores were computed using Python 2.7.9 and the SciPy library (99) by ranking the transcripts per million (TPM) values in each RNA-seq experiment and then calculating the Pearson correlation on the ranked values for each pairwise combination of genes: ρ = covariance(gene1, gene2)/σgene1 σgene2, where σ represents the SD. P values were computed by comparing average correlation scores with the bootstrap distribution of average scores for random sets of 31 genes (mimicking the size of the flp family) and calculated as the fraction of times that bootstrapping produced a score greater than or equal to the score being tested. Because the ins and nlp families have more than 31 genes, the P value is an upper estimate. Bootstrapping was performed 10,000 times. The following RNA-seq datasets were used: our daf-9(dh6) RNA-seq data, requantified using kallisto (100) into TPM counts; and processed RNA-seq data from 246 publically available libraries (23) obtained using WormBase SPELL and converted to TPM counts (101). Genes detected in less than 80% of the experiments were discarded.
Dot plot analysis.
Dotplot analysis of the flp coding sequences was performed using Gepard (63). Coding sequences from the 31 total flp genes (taking only the a isoform for genes with multiple isoforms) were used in the analysis.
Dauer-Entry Assay on Pheromone Plates.
The preparation of crude pheromone and the dauer-entry assay were performed with modifications to previously described methods (102). Crude pheromone was extracted from exhausted liquid culture medium, resuspended with distilled water, and stored at −20 °C. Pheromone plates (0.3 g NaCl, 2.5 g agar, 97.5 mL H2O, 0.1 mL of 5 mg/mL cholesterol, 0.1 mL 1 M CaCl2, 0.1 mL 1 M MgSO4, 2.5 mL 1 M KPO4, and crude pheromone) were freshly prepared the day before each experiment and were dried overnight at room temperature. Heat-killed E. coli OP50 was used as a limited food source for the dauer-entry assays and was prepared by resuspending OP50 overnight cultures in S basal medium to 8 g/100 mL and heating at 100 °C for 5 min. On the day of the experiment, 7–10 young adults were selected, placed onto each plate, and allowed to lay ∼50–60 eggs before being removed. Heat-killed OP50 (20 μL) was added to the plates as a food source for the unhatched larvae. After 48 h of incubation at 25.5 °C, dauers and nondauers were counted on each plate based on their distinct morphologies.
Dauer entry can exhibit day-to-day variation caused by environmental conditions such as humidity or temperature (25). To control for this variation, wild-type controls were run in every trial. The wild-type results from the same batch of pheromone were pooled together for better statistical power, and each statistical analysis was done with samples treated with the same batch of pheromone.
Statistical Analysis for Dauer-Entry Assay.
The mean and 99% CI of the dauer-entry percentage were calculated nonparametrically for each genotype by pooling the data from all the plates and computing 10,000 bootstrap replicates (103). Pairwise comparisons were performed through a nonparametric permutation test with 10,000 replicates. The difference in dauer-entry probability between two genotypes was estimated using a Bayesian approach (104) to compute the posterior probability of dauer entry for each genotype. Bootstrapping, permutation, and Bayesian statistics were performed using Python 3.5 and the SciPy library (99). Detailed information about the statistical approach is given in SI Materials and Methods.
Dauer Behavior Assays.
Crude pheromone plates were used to induce synchronized dauers for behavior assays. For each pheromone plate, 20 μL of heat-killed OP50 (8 g/100 mL) were spotted, and 12–15 young adult animals were picked onto the plate to lay eggs at 20 °C for 12 h before being removed. After 2–4 d of incubation at 25.5 °C, dauers were identified by their morphology and isolated for the following assays.
Nictation assay.
The nictation assay was performed on microdirt chips with modifications to previously described methods (19). Dauers suspended in distilled water were transferred onto the microdirt chip (4% agarose in distilled water) and allowed to nictate. Each nictating dauer was observed for at least 3 min or until the end of nictation. The time of recording began when a dauer initiated nictation (by lifting the neck region of its body), and the total duration of each nictation event was also recorded. If the dauer stopped nictating and exhibited quiescence in a standing posture during the recording, the data were excluded from further analysis. The average nictation duration was calculated by dividing the total duration of nictation by the number of nictation events. At least 20 dauers were assayed for each genotype. The mean of average duration, 99% CI, and the pairwise P value were computed nonparametrically as described for the dauer-entry assay.
CO2 chemotaxis assay.
CO2 chemotaxis assays were performed on dauers with modifications to previously described methods (18). Dauers were washed three times with distilled water and were transferred to standard chemotaxis assay plates (105). Two gas mixtures were delivered to the plate at a rate of 0.5 mL/min through PVC tubing and holes drilled through the plate lid. The CO2 gas mixture was 10% CO2 and 21% O2 balanced with N2, and the control gas mixture was 21% O2 balanced with N2 (Airgas). The two holes were positioned on opposite sides of the plate along a diameter line, with each positioned 1 cm from the edge of the plate. The scoring regions were set as the areas of the plate 1 cm beyond a central line drawn orthogonally to the diameter on which the gas mixtures were presented. At the end of 1 h, the numbers of animals in each scoring region were counted, and the chemotaxis index was calculated as (N at the CO2 scoring region − N at the control gas scoring region)∕(N at the CO2 scoring region + N at the control gas scoring region). Statistical analysis (two-tailed t test) was performed using GraphPad Prism.
Acute CO2 avoidance assay.
CO2 avoidance assays were performed as previously described (62), with slight modifications. Dauers were washed three times with distilled water and transferred to unseeded NGM plates. A 50-mL gas-tight syringe was filled with either a CO2 gas mixture or a control gas mixture and was connected to a pipette tip using PVC tubing. Gas was pumped out through the pipette tip at a rate of 1.5 mL/min using a syringe pump, and the tip was presented to the head of forward-moving dauers. A response was scored if the animal reversed within 4 s. At least 20 animals on each plate were assayed per gas mixture, with each plate counted as a trial. The avoidance index was calculated as (N reversed to CO2/N presented with CO2) − (N reversed to control gas/N presented with control gas). Statistical analysis (one-way ANOVA with Bonferroni post correction test) was performed using GraphPad Prism.
flp Gene Expression in IJs.
Orthologs/analogs of C. elegans flp genes exist in other nematodes (69, 80, 106). The flp orthologs/analogs identified in refs. 69 and 106 were matched to transcript IDs in S. stercoralis, A. ceylanicum, G. pallida, and B. malayi using BLAST via the WormBase ParaSite. Published RNA-seq data were downloaded for S. stercoralis (73) and G. pallida (71) using the European Nucleotide Archive and for B. malayi (70) using WormBase SPELL; data for A. ceylanicum were used as published (72).
The read counts from the A. ceylanicum and B. malayi data were preprocessed into TPM counts. We obtained TPM counts for the S. stercoralis and G. pallida datasets using kallisto to align the read data and to quantify transcript abundances (100). kallisto was preferable to DESeq at this stage of our analysis, as it allowed us to quantify these large datasets quickly and accurately. To increase comparability among all the datasets, kallisto was used to requantify our own dauer RNA-seq data into TPM counts.
Supplementary Material
Acknowledgments
We thank Daehan Lee for providing the nictation microchip design; Elissa Hallem for advice on CO2 assays; Christopher Cronin for advice on behavioral assay setups; Mihoko Kato, Hillel Schwartz, Igor Antoshechkin, Ryoji Shinya, Han Wang, Animesh Ray, and Jonathan Liu for critically reading the manuscript; the Caenorhabditis Genetics Center, which is funded by the NIH Office of Research Infrastructure Programs (Grant P40 OD010440), and Adam Antebi, Mario de Bono, and H. Robert Horvitz for providing animal strains; WormBase and WormBase SPELL for tools, datasets, and invaluable information; and the Millard and Muriel Jacobs Genetics and Genomics Laboratory for support with library construction and sequencing.
Footnotes
The authors declare no conflict of interest.
Data deposition: The RNA-seq data in this paper have been deposited in the National Center for Biotechnology Information Sequence Read Archive (SRA) database (accession no. SRP116980). Codes used for data analysis have been deposited in GitHub and are available at https://github.com/WormLabCaltech/dauerRNAseq.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1710374114/-/DCSupplemental.
References
- 1.McLaughlin KA, et al. Causal effects of the early caregiving environment on development of stress response systems in children. Proc Natl Acad Sci USA. 2015;112:5637–5642. doi: 10.1073/pnas.1423363112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McQuaid RJ, McInnis OA, Stead JD, Matheson K, Anisman H. A paradoxical association of an oxytocin receptor gene polymorphism: Early-life adversity and vulnerability to depression. Front Neurosci. 2013;7:128. doi: 10.3389/fnins.2013.00128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reichmann F, Holzer P. Neuropeptide Y: A stressful review. Neuropeptides. 2016;55:99–109. doi: 10.1016/j.npep.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Teicher MH, Samson JA, Anderson CM, Ohashi K. The effects of childhood maltreatment on brain structure, function and connectivity. Nat Rev Neurosci. 2016;17:652–666. doi: 10.1038/nrn.2016.111. [DOI] [PubMed] [Google Scholar]
- 5.Gilbert SF. Developmental Biology. Sinauer Associates; Sunderland, MA: 2000. [Google Scholar]
- 6.Fusco G, Minelli A. Phenotypic plasticity in development and evolution: Facts and concepts. Introduction. Philos Trans R Soc Lond B Biol Sci. 2010;365:547–556. doi: 10.1098/rstb.2009.0267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Janzen FJ, Phillips PC. Exploring the evolution of environmental sex determination, especially in reptiles. J Evol Biol. 2006;19:1775–1784. doi: 10.1111/j.1420-9101.2006.01138.x. [DOI] [PubMed] [Google Scholar]
- 8.Woodward DE, Murray JD. On the effect of temperature-dependent sex determination on sex ratio and survivorship in crocodilians. Proc R Soc Lond B Biol Sci. 1993;252:149–155. [Google Scholar]
- 9.Newman RA. Adaptive plasticity in amphibian metamorphosis. Bioscience. 1992;42:671–678. [Google Scholar]
- 10.Cassada RC, Russell RL. The dauerlarva, a post-embryonic developmental variant of the nematode Caenorhabditis elegans. Dev Biol. 1975;46:326–342. doi: 10.1016/0012-1606(75)90109-8. [DOI] [PubMed] [Google Scholar]
- 11.Fielenbach N, Antebi A. C. elegans dauer formation and the molecular basis of plasticity. Genes Dev. 2008;22:2149–2165. doi: 10.1101/gad.1701508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Golden JW, Riddle DL. The Caenorhabditis elegans dauer larva: Developmental effects of pheromone, food, and temperature. Dev Biol. 1984;102:368–378. doi: 10.1016/0012-1606(84)90201-x. [DOI] [PubMed] [Google Scholar]
- 13.Schaedel ON, Gerisch B, Antebi A, Sternberg PW. Hormonal signal amplification mediates environmental conditions during development and controls an irreversible commitment to adulthood. PLoS Biol. 2012;10:e1001306. doi: 10.1371/journal.pbio.1001306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wadsworth WG, Riddle DL. Developmental regulation of energy metabolism in Caenorhabditis elegans. Dev Biol. 1989;132:167–173. doi: 10.1016/0012-1606(89)90214-5. [DOI] [PubMed] [Google Scholar]
- 15.Oriordan VB, Burnell AM. Intermediary metabolism in the dauer larva of the nematode Caenorhabditis elegans. 2. The glyoxylate cycle and fatty-acid oxidation. Comp Biochem Physiol B. 1990;95:125–130. [Google Scholar]
- 16.Albert PS, Riddle DL. Developmental alterations in sensory neuroanatomy of the Caenorhabditis elegans dauer larva. J Comp Neurol. 1983;219:461–481. doi: 10.1002/cne.902190407. [DOI] [PubMed] [Google Scholar]
- 17.Schroeder NE, et al. Dauer-specific dendrite arborization in C. elegans is regulated by KPC-1/Furin. Curr Biol. 2013;23:1527–1535. doi: 10.1016/j.cub.2013.06.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hallem EA, et al. A sensory code for host seeking in parasitic nematodes. Curr Biol. 2011;21:377–383. doi: 10.1016/j.cub.2011.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee H, et al. Nictation, a dispersal behavior of the nematode Caenorhabditis elegans, is regulated by IL2 neurons. Nat Neurosci. 2011;15:107–112. doi: 10.1038/nn.2975. [DOI] [PubMed] [Google Scholar]
- 20.Vanfleteren JR, Braeckman BP. Mechanisms of life span determination in Caenorhabditis elegans. Neurobiol Aging. 1999;20:487–502. doi: 10.1016/s0197-4580(99)00087-1. [DOI] [PubMed] [Google Scholar]
- 21.Klass M, Hirsh D. Non-ageing developmental variant of Caenorhabditis elegans. Nature. 1976;260:523–525. doi: 10.1038/260523a0. [DOI] [PubMed] [Google Scholar]
- 22.Erkut C, et al. Molecular strategies of the Caenorhabditis elegans dauer larva to survive extreme desiccation. PLoS One. 2013;8:e82473. doi: 10.1371/journal.pone.0082473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gerstein MB, et al. modENCODE Consortium Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science. 2010;330:1775–1787. doi: 10.1126/science.1196914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang J, Kim SK. Global analysis of dauer gene expression in Caenorhabditis elegans. Development. 2003;130:1621–1634. doi: 10.1242/dev.00363. [DOI] [PubMed] [Google Scholar]
- 25.Karp X. Working with dauer larvae. WormBook. 2016 doi: 10.1895/wormbook.1.180.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Husson SJ, Schoofs L. Altered neuropeptide profile of Caenorhabditis elegans lacking the chaperone protein 7B2 as analyzed by mass spectrometry. FEBS Lett. 2007;581:4288–4292. doi: 10.1016/j.febslet.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 27.Li C, Kim K. Family of FLP peptides in Caenorhabditis elegans and related nematodes. Front Endocrinol (Lausanne) 2014;5:150. doi: 10.3389/fendo.2014.00150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pierce SB, et al. Regulation of DAF-2 receptor signaling by human insulin and ins-1, a member of the unusually large and diverse C. elegans insulin gene family. Genes Dev. 2001;15:672–686. doi: 10.1101/gad.867301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jia K, Albert PS, Riddle DL. DAF-9, a cytochrome P450 regulating C. elegans larval development and adult longevity. Development. 2002;129:221–231. doi: 10.1242/dev.129.1.221. [DOI] [PubMed] [Google Scholar]
- 30.Motola DL, et al. Identification of ligands for DAF-12 that govern dauer formation and reproduction in C. elegans. Cell. 2006;124:1209–1223. doi: 10.1016/j.cell.2006.01.037. [DOI] [PubMed] [Google Scholar]
- 31.Gerisch B, Weitzel C, Kober-Eisermann C, Rottiers V, Antebi A. A hormonal signaling pathway influencing C. elegans metabolism, reproductive development, and life span. Dev Cell. 2001;1:841–851. doi: 10.1016/s1534-5807(01)00085-5. [DOI] [PubMed] [Google Scholar]
- 32.Sulston JE, Horvitz HR. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol. 1977;56:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- 33.Marinov GK, et al. From single-cell to cell-pool transcriptomes: Stochasticity in gene expression and RNA splicing. Genome Res. 2014;24:496–510. doi: 10.1101/gr.161034.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miura F, et al. Absolute quantification of the budding yeast transcriptome by means of competitive PCR between genomic and complementary DNAs. BMC Genomics. 2008;9:574. doi: 10.1186/1471-2164-9-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aken BL, et al. The Ensembl gene annotation system. Database (Oxford) 2016;2016:baw093. doi: 10.1093/database/baw093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Howe KL, et al. WormBase 2016: Expanding to enable helminth genomic research. Nucleic Acids Res. 2016;44:D774–D780. doi: 10.1093/nar/gkv1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anders S, et al. Count-based differential expression analysis of RNA sequencing data using R and bioconductor. Nat Protoc. 2013;8:1765–1786. doi: 10.1038/nprot.2013.099. [DOI] [PubMed] [Google Scholar]
- 39.Jones SJ, et al. Changes in gene expression associated with developmental arrest and longevity in Caenorhabditis elegans. Genome Res. 2001;11:1346–1352. doi: 10.1101/gr.184401. [DOI] [PubMed] [Google Scholar]
- 40.Futschik ME, Carlisle B. Noise-robust soft clustering of gene expression time-course data. J Bioinform Comput Biol. 2005;3:965–988. doi: 10.1142/s0219720005001375. [DOI] [PubMed] [Google Scholar]
- 41.Kumar L, Futschik ME. Mfuzz: A software package for soft clustering of microarray data. Bioinformation. 2007;2:5–7. doi: 10.6026/97320630002005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45:D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44:D457–D462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mi H, et al. PANTHER version 11: Expanded annotation data from gene ontology and reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2017;45:D183–D189. doi: 10.1093/nar/gkw1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaplan F, et al. Ascaroside expression in Caenorhabditis elegans is strongly dependent on diet and developmental stage. PLoS One. 2011;6:e17804. doi: 10.1371/journal.pone.0017804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.von Reuss SH, et al. Comparative metabolomics reveals biogenesis of ascarosides, a modular library of small-molecule signals in C. elegans. J Am Chem Soc. 2012;134:1817–1824. doi: 10.1021/ja210202y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li C, Kim K. Neuropeptides. WormBook. 2008 doi: 10.1895/wormbook.1.142.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cohen M, et al. Coordinated regulation of foraging and metabolism in C. elegans by RFamide neuropeptide signaling. Cell Metab. 2009;9:375–385. doi: 10.1016/j.cmet.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 50.Lee D, Lee H, Kim N, Lim DS, Lee J. Regulation of a hitchhiking behavior by neuronal insulin and TGF-β signaling in the nematode Caenorhabditis elegans. Biochem Biophys Res Commun. 2017;484:323–330. doi: 10.1016/j.bbrc.2017.01.113. [DOI] [PubMed] [Google Scholar]
- 51.Braeckman BP, Houthoofd K, Vanfleteren JR. Intermediary metabolism. WormBook. 2009 doi: 10.1895/wormbook.1.146.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ghosh S, Sternberg PW. Spatial and molecular cues for cell outgrowth during C. elegans uterine development. Dev Biol. 2014;396:121–135. doi: 10.1016/j.ydbio.2014.09.028. [DOI] [PubMed] [Google Scholar]
- 53.Gupta BP, Hanna-Rose W, Sternberg PW. Morphogenesis of the vulva and the vulval-uterine connection. WormBook. 2012 doi: 10.1895/wormbook.1.152.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.L’Hernault SW. Spermatogenesis. WormBook. 2006 doi: 10.1895/wormbook.1.85.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hobert O. The neuronal genome of Caenorhabditis elegans. WormBook. 2013 doi: 10.1895/wormbook.1.161.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim Y, Bark S, Hook V, Bandeira N. NeuroPedia: Neuropeptide database and spectral library. Bioinformatics. 2011;27:2772–2773. doi: 10.1093/bioinformatics/btr445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lindberg I, Tu B, Muller L, Dickerson IM. Cloning and functional analysis of C. elegans 7B2. DNA Cell Biol. 1998;17:727–734. doi: 10.1089/dna.1998.17.727. [DOI] [PubMed] [Google Scholar]
- 58.Sieburth D, et al. Systematic analysis of genes required for synapse structure and function. Nature. 2005;436:510–517. doi: 10.1038/nature03809. [DOI] [PubMed] [Google Scholar]
- 59.Shen LL, Wang Y, Wang DY. Involvement of genes required for synaptic function in aging control in C. elegans. Neurosci Bull. 2007;23:21–29. doi: 10.1007/s12264-007-0003-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kiontke K, Sudhaus W. Ecology of Caenorhabditis species. WormBook. 2006 doi: 10.1895/wormbook.1.37.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Félix MA, Braendle C. The natural history of Caenorhabditis elegans. Curr Biol. 2010;20:R965–R969. doi: 10.1016/j.cub.2010.09.050. [DOI] [PubMed] [Google Scholar]
- 62.Hallem EA, Sternberg PW. Acute carbon dioxide avoidance in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2008;105:8038–8043. doi: 10.1073/pnas.0707469105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Krumsiek J, Arnold R, Rattei T. Gepard: A rapid and sensitive tool for creating dotplots on genome scale. Bioinformatics. 2007;23:1026–1028. doi: 10.1093/bioinformatics/btm039. [DOI] [PubMed] [Google Scholar]
- 64.Smith ES, Martinez-Velazquez L, Ringstad N. A chemoreceptor that detects molecular carbon dioxide. J Biol Chem. 2013;288:37071–37081. doi: 10.1074/jbc.M113.517367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim K, Li C. Expression and regulation of an FMRFamide-related neuropeptide gene family in Caenorhabditis elegans. J Comp Neurol. 2004;475:540–550. doi: 10.1002/cne.20189. [DOI] [PubMed] [Google Scholar]
- 66.Ringstad N, Horvitz HR. FMRFamide neuropeptides and acetylcholine synergistically inhibit egg-laying by C. elegans. Nat Neurosci. 2008;11:1168–1176. doi: 10.1038/nn.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Viney ME. How did parasitic worms evolve? BioEssays. 2009;31:496–499. doi: 10.1002/bies.200900010. [DOI] [PubMed] [Google Scholar]
- 68.Okumura E, Yoshiga T. Host orientation using volatiles in the phoretic nematode Caenorhabditis japonica. J Exp Biol. 2014;217:3197–3199. doi: 10.1242/jeb.105353. [DOI] [PubMed] [Google Scholar]
- 69.McCoy CJ, et al. New insights into the FLPergic complements of parasitic nematodes: Informing deorphanisation approaches. EuPA Open Proteom. 2014;3:262–272. doi: 10.1016/j.euprot.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Choi YJ, et al. A deep sequencing approach to comparatively analyze the transcriptome of lifecycle stages of the filarial worm, Brugia malayi. PLoS Negl Trop Dis. 2011;5:e1409. doi: 10.1371/journal.pntd.0001409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cotton JA, et al. The genome and life-stage specific transcriptomes of Globodera pallida elucidate key aspects of plant parasitism by a cyst nematode. Genome Biol. 2014;15:R43. doi: 10.1186/gb-2014-15-3-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schwarz EM, et al. The genome and transcriptome of the zoonotic hookworm Ancylostoma ceylanicum identify infection-specific gene families. Nat Genet. 2015;47:416–422. doi: 10.1038/ng.3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stoltzfus JD, Minot S, Berriman M, Nolan TJ, Lok JB. RNAseq analysis of the parasitic nematode Strongyloides stercoralis reveals divergent regulation of canonical dauer pathways. PLoS Negl Trop Dis. 2012;6:e1854. doi: 10.1371/journal.pntd.0001854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bain O, Babayan S. Behaviour of filariae: Morphological and anatomical signatures of their life style within the arthropod and vertebrate hosts. Filaria J. 2003;2:16. doi: 10.1186/1475-2883-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Félix MA, Duveau F. Population dynamics and habitat sharing of natural populations of Caenorhabditis elegans and C. briggsae. BMC Biol. 2012;10:59. doi: 10.1186/1741-7007-10-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nusbaum MP, Blitz DM, Swensen AM, Wood D, Marder E. The roles of co-transmission in neural network modulation. Trends Neurosci. 2001;24:146–154. doi: 10.1016/s0166-2236(00)01723-9. [DOI] [PubMed] [Google Scholar]
- 77.Salio C, Lossi L, Ferrini F, Merighi A. Neuropeptides as synaptic transmitters. Cell Tissue Res. 2006;326:583–598. doi: 10.1007/s00441-006-0268-3. [DOI] [PubMed] [Google Scholar]
- 78.Landgraf R, Neumann ID. Vasopressin and oxytocin release within the brain: A dynamic concept of multiple and variable modes of neuropeptide communication. Front Neuroendocrinol. 2004;25:150–176. doi: 10.1016/j.yfrne.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 79.Bargmann CI. Beyond the connectome: How neuromodulators shape neural circuits. Bioessays. 2012;34:458–465. doi: 10.1002/bies.201100185. [DOI] [PubMed] [Google Scholar]
- 80.Jékely G. Global view of the evolution and diversity of metazoan neuropeptide signaling. Proc Natl Acad Sci USA. 2013;110:8702–8707. doi: 10.1073/pnas.1221833110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dockray GJ. The expanding family of -RFamide peptides and their effects on feeding behaviour. Exp Physiol. 2004;89:229–235. doi: 10.1113/expphysiol.2004.027169. [DOI] [PubMed] [Google Scholar]
- 82.Elphick MR, Mirabeau O. The evolution and variety of RFamide-type neuropeptides: Insights from deuterostomian invertebrates. Front Endocrinol (Lausanne) 2014;5:93. doi: 10.3389/fendo.2014.00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cardoso JC, Félix RC, Fonseca VG, Power DM. Feeding and the rhodopsin family g-protein coupled receptors in nematodes and arthropods. Front Endocrinol (Lausanne) 2012;3:157. doi: 10.3389/fendo.2012.00157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Guillermin ML, Castelletto ML, Hallem EA. Differentiation of carbon dioxide-sensing neurons in Caenorhabditis elegans requires the ETS-5 transcription factor. Genetics. 2011;189:1327–1339. doi: 10.1534/genetics.111.133835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- 86.Dalzell JJ, McMaster S, Fleming CC, Maule AG. Short interfering RNA-mediated gene silencing in Globodera pallida and Meloidogyne incognita infective stage juveniles. Int J Parasitol. 2010;40:91–100. doi: 10.1016/j.ijpara.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 87.Morris R, et al. A neuropeptide modulates sensory perception in the entomopathogenic nematode Steinernema carpocapsae. PLoS Pathog. 2017;13:e1006185. doi: 10.1371/journal.ppat.1006185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sudhaus W. Preadaptive plateau in Rhabditida (Nematoda) allowed the repeated evolution of zooparasites, with an outlook on evolution of life cycles within Spiroascarida. Palaeodiversity. 2010;3:117–130. [Google Scholar]
- 89.De Ley P. A quick tour of nematode diversity and the backbone of nematode phylogeny. WormBook. 2006 doi: 10.1895/wormbook.1.41.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Han Z, Boas S, Schroeder NE. Unexpected variation in neuroanatomy among diverse nematode species. Front Neuroanat. 2016;9:162. doi: 10.3389/fnana.2015.00162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mello C, Fire A. DNA transformation. Methods Cell Biol. 1995;48:451–482. [PubMed] [Google Scholar]
- 93.Maduro M, Pilgrim D. Identification and cloning of unc-119, a gene expressed in the Caenorhabditis elegans nervous system. Genetics. 1995;141:977–988. doi: 10.1093/genetics/141.3.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Arribere JA, et al. Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics. 2014;198:837–846. doi: 10.1534/genetics.114.169730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wickham H. ggplot2: Elegant Graphics for Data Analysis. Springer; New York: 2009. [Google Scholar]
- 96.Yu G, Wang LG, Han Y, He QY. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Warnes GR, et al. 2016 gplots: Various R Programming Tools for Plotting Data, Version 2.14.1. Available at https://CRAN.R-project.org/package=gplots. Accessed November 10, 2017.
- 98.Neuwirth E. 2014 RColorBrewer: ColorBrewer Palettes. R Package, Version 1.1-2. Available at https://CRAN.R-project.org/package=RColorBrewer. Accessed November 10, 2017.
- 99.Jones E, Oliphant T, Peterson P. 2001 SciPy: Open Source Scientific Tools for Python. Available at http://www.scipy.org/. Accessed November 10, 2017.
- 100.Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 101.Pachter L. 2011. Models for transcript quantification from RNA-Seq. arXiv:1104.3889.
- 102.Golden JW, Riddle DL. A pheromone-induced developmental switch in Caenorhabditis elegans: Temperature-sensitive mutants reveal a wild-type temperature-dependent process. Proc Natl Acad Sci USA. 1984;81:819–823. doi: 10.1073/pnas.81.3.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Efron B, Tibshirani RJ. An Introduction to the Bootstrap. CRC; Boca Raton, FL: 1994. [Google Scholar]
- 104.Sivia DS, Skilling J. Data Analysis: A Bayesian Tutorial. Oxford Univ Press; Oxford: 2006. [Google Scholar]
- 105.Bargmann CI, Hartwieg E, Horvitz HR. Odorant-selective genes and neurons mediate olfaction in C. elegans. Cell. 1993;74:515–527. doi: 10.1016/0092-8674(93)80053-h. [DOI] [PubMed] [Google Scholar]
- 106.McVeigh P, et al. Analysis of FMRFamide-like peptide (FLP) diversity in phylum Nematoda. Int J Parasitol. 2005;35:1043–1060. doi: 10.1016/j.ijpara.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 107.Blaxter M, Koutsovoulos G. The evolution of parasitism in Nematoda. Parasitology. 2015;142(Suppl 1):S26–S39. doi: 10.1017/S0031182014000791. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.