Significance
All eukaryotes respond to DNA damage by polyubiquitylation and degradation of the largest subunit of RNA polymerase II (pol II), facilitated by the Elongin-Cullin ubiquitin ligase. In yeast, the recruitment of the ubiquitin ligase to pol II is dependent on Def1. We report a finding made in the course of isolating TFIIH-Def1 from yeast that suggests a potential novel function of Def1 as a transcription regulator in response to cellular stress. The function of Def1 in transcription regulation is demonstrably separable from its role in coordinating pol II stability in response to cellular stress.
Keywords: transcription, RNA polymerase II, TFIIH, stress response, Def1
Abstract
The DNA damage response is an essential process for the survival of living cells. In a subset of stress-responsive genes in humans, Elongin controls transcription in response to multiple stimuli, such as DNA damage, oxidative stress, and heat shock. Yeast Elongin (Ela1-Elc1), along with Def1, is known to facilitate ubiquitylation and degradation of RNA polymerase II (pol II) in response to multiple stimuli, yet transcription activity has not been examined. We have found that Def1 copurifies from yeast whole-cell extract with TFIIH, the largest general transcription factor required for transcription initiation and nucleotide excision repair. The addition of recombinant Def1 and Ela1-Elc1 enhanced transcription initiation in an in vitro reconstituted system including pol II, the general transcription factors, and TFIIS. Def1 also enhanced transcription restart from TFIIS-induced cleavage in a pol II transcribing complex. In the Δdef1 strain, heat shock genes were misregulated, indicating that Def1 is required for induction of some stress-responsive genes in yeast. Taken together, our results extend the understanding of the molecular mechanism of transcription regulation on cellular stress and reveal functional similarities to the mammalian system.
Rapid induction of stress response genes is an essential process for the survival of living cells. In mammalian cells, a key player is Elongin, which is composed of three subunits (A, B, and C) and was originally identified as a factor that stimulates RNA polymerase II (pol II) transcription (1). More recent in vivo studies have shown that Elongin is not a general regulator of gene expression, but rather is required for the expression of stress-responsive genes, including heat shock genes, in response to multiple stimuli, such as DNA damage, oxidative stress, and heat shock (2–4). Mammalian Elongin not only stimulates transcription activity, but also recruits Cullin-RING ligases via the suppressor of cytokine signaling (SOCS) box motif in Elongin A, leading to pol II ubiquitylation and degradation at sites of DNA damage (5–7). HeLa cells carrying a mutation in the SOCS box in Elongin A are defective in pol II ubiquitylation but are capable of ATF3 induction, indicating that recruitment of Cullin-RING ligases to Elongin A is dispensable for stress-responsive transcription activation (4).
In yeast, the proteins Ela1 and Elc1, homologs of Elongin A and C, form a heterodimer, but their transcriptional properties are unknown. Previous in vitro experiments have shown that Ela1 and Elc1 has no effect on transcription elongation, in contrast to mammalian Elongin (8). However, the yeast Elongin-Cullin-RING ligase is recruited to enhance pol II degradation; this process depends on Def1, whose C-terminal polyQ stretch is cleaved in half on exposure to multiple stimuli, allowing it to accumulate in the nucleus, where it recruits the Ela1-Elc1-Cullin complex for pol II polyubiquitylation (9).
In the course of purifying one of the general transcription factors, TFIIH, from yeast whole-cell extract, we resolved a stoichiometric amount of Def1-bound TFIIH, which prompted us to investigate the effects of Def1 on pol II transcription. Our findings identify the biochemical activity of Def1, along with Ela1-Elc1, in early transcription, suggesting potential dual functional roles in transcription and pol II degradation on DNA damage.
Results
Def1 Copurifies TFIIH from Yeast Whole Cell Extract.
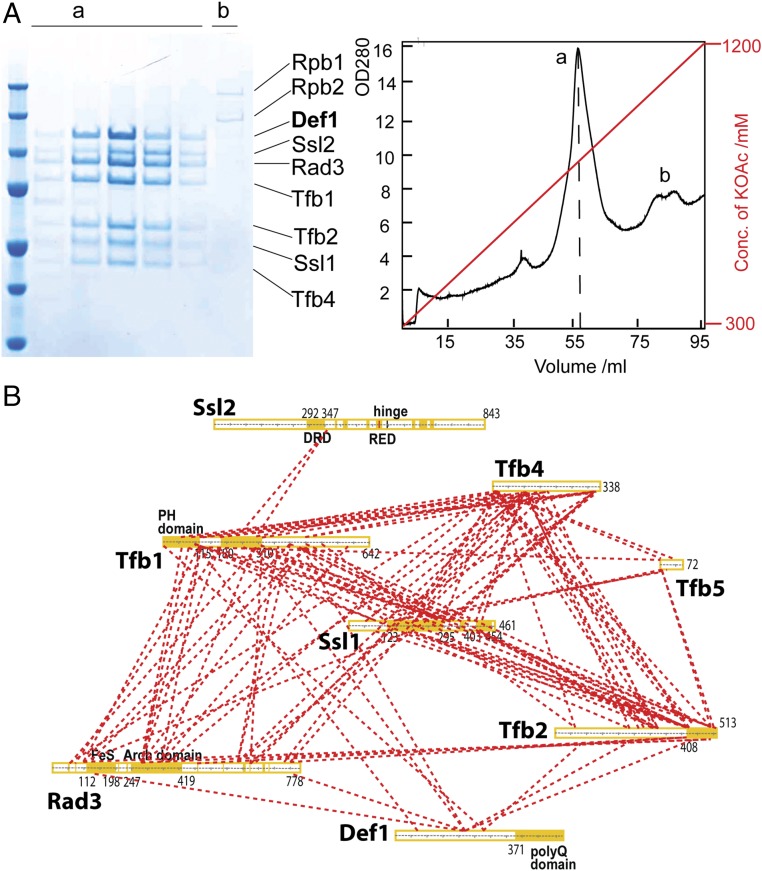
Eleven components of yeast holo-TFIIH have been well established (10–12). TFIIH was isolated using a TAP tag affinity purification with high salt (400 mM ammonium sulfate) from yeast cell extract, and a homogeneous TFIIH complex with an additional polypeptide with an apparent molecular mass of ∼100 kDa was obtained (Fig. S1A). This ∼100-kDa protein was excised from the gel and identified by mass spectrometry (MS) (Fig. S1A) as Def1, a protein previously known as a polyQ protein promoting ubiquitylation and degradation of pol II on DNA damage (13). Def1 directly binds the core TFIIH rather than the kinase module TFIIK; TFIIH was stripped of TFIIK when the Tap-tagged complex bound to the IgG column was washed with high salt (14).
To further characterize interactions of TFIIH with Def1 and other possible proteins, TFIIH was obtained without extensive washing during TAP tag affinity purification, and after being passed through a HiTrapQ column (Fig. 1), the crude eluate of Def1-TFIIH was subjected to MS-based semiquantitative proteomics analysis and cross-linking followed by MS (15). The MS analysis identified 170 yeast proteins with two or more unique peptides, including TFIIH, Def1, Ela1 (9), two subunits of pol II (Rpb4 and Rpb7), 21 subunits of Mediator (16–18), and all the known subunits of Ccr4-Not (19, 20) (Dataset S1). The 10-subunit pol II was separated from Def1-TFIIH by a HiTrapQ column (Fig. 1A) and was not detected in the MS analysis. Next, the HiTrapQ eluate was reacted with the cross-linker BS3 and cross-links were analyzed by MS. Possible cross-linked candidates were generated from sequences of the 50 most abundant proteins listed in Dataset S1, and were searched against cross-links under previously published matching criteria (15, 21). We were able to identify a total of 516 cross-links with moderate-to-high confidence (2.5% false-positive rate threshold) (Fig. 1B, Fig. S2, and Dataset S2). Among these were ∼170 cross-links between different proteins, with the majority between core TFIIH subunits. Observed cross-links between TFIIH subunits were in good agreement with those previously obtained from highly-purified TFIIH (21, 22). In addition, we also observed cross-links between TFIIH and Def1, Mediator, and Ccr4-Not. The nine identified cross-links between TFIIH and Def1 support our claim that Def1 physically interacts with the core TFIIH.
Fig. 1.
Coisolation of Def1 with TFIIH from yeast whole-cell extract. (A) TFIIH-Def1 was purified via affinity purification by means of a TAP tag, followed by chromatography on a HiTrapQ column. (Left) SDS/PAGE analysis of the HiTrapQ eluate. (Right) Elution profile of the HiTrapQ eluate. Peaks a and b correspond to Def1-TFIIH and pol II, respectively. (B) Cross-linking map of yeast Def1-TFIIH.
Amyloid Fibril Formation of Def1 Facilitates Dissociation of Def1 from TFIIH.
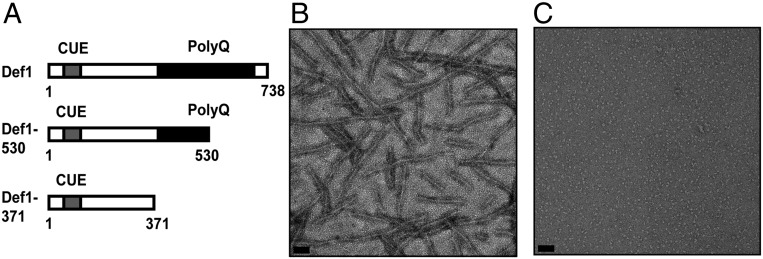
When the HiTrapQ fraction was loaded onto gel filtration through Superose 6, >80% of Def1 dissociated from TFIIH and eluted in the void volume (Fig. S1B). In agreement with previous evidence indicating that polyQ proteins tend to form amyloid fibrils or insoluble aggregates (23), our electron microscopy analysis of the void volume fraction revealed a uniform distribution of amyloid fibrils (Fig. 2B). No such amyloid fibril formations were obtained with either a shorter Def1 construct (residues 1–371; Def1-371) that lacks the entire C-terminal polyQ or a longer Def1 construct (residues 1–530; Def1-530) that lacks only one-half of the polyQ stretch (Fig. 2 A and C). The construct Def1-530 represents an endogenous functional form, as Def1 can be cleaved around amino acid 530 in response to multiple stimuli in vivo (9). These data suggest a relationship between the dissociation of Def1 from TFIIH and amyloid fibril formation.
Fig. 2.
Def1, but not Def1-530, forms amyloid fibrils. (A) Schematic diagram of the C-terminal deletion constructs of Def1, showing the CUE domain and polyQ region. Ela1 binds Def1 through the CUE domain (9). (B and C) Representative images of negatively stained endogenous Def1 forming amyloid fibrils (B) and recombinant soluble Def1-530 (C). Images were obtained at 40,000× magnification on a FEI Tecnai 12 microscope. (Scale bar: 50 nm.)
Amyloid fibril formation continued over the course of purification; thus, the ratio of Def1 that eluted at the void volume to that bound to TFIIH varied among independent experiments (n = 3). Def1 was not observed when TFIIH was isolated with 300 mM potassium acetate (12). Such a decreased salt concentration might have driven the amyloid fibril formation and prevented binding of TFIIH, and thus Def1 may have been previously undetectable.
Def1-Elongin Enhances Transcription Initiation in the Presence of TFIIS.
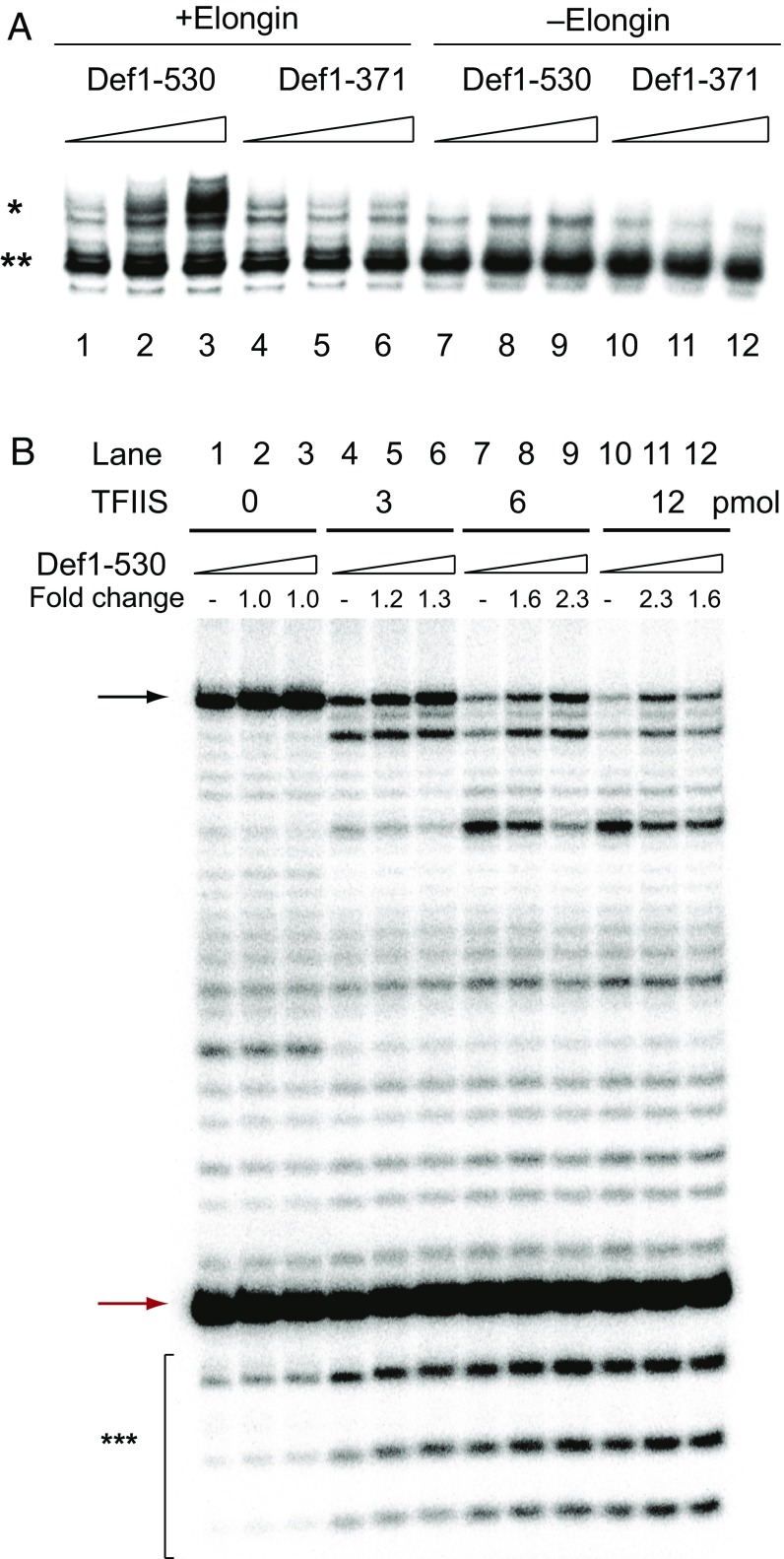
The addition of Def1-530 to our in vitro reconstituted transcription system, which includes the general transcription factors (TFIIA, TFIIB, TBP, TFIIE, TFIIF, and TFIIH), pol II, and a promoter DNA fragment, failed to enhance transcription initiation (Fig. 3A). However, with both Elongin (Ela1-Elc1) and TFIIS, Def1-530 supported transcription from an additional upstream transcription start site (indicated by an asterisk), resulting in approximately twofold more run-off transcripts (lane 3 in Fig. 3A). The shorter construct Def1-371 failed to enhance transcription initiation even in the presence of Elongin and TFIIS (lanes 4–6 in Fig. 3A). The N-terminal half (residues 372–530) of the polyQ region, where glutamine residues account for >50% of the amino acid residues, is indispensable for this transcription activation.
Fig. 3.
Def1 enhances transcription initiation and transcription restart from TFIIS-induced cleavage in a pol II transcribing complex. (A) Effects of Def1 and Elongin on transcription initiation. SNR20 promoter DNA (SNR20 91W) (14) was mixed with TFIIB, TFIIA, TBP, TFIIE, TFIIH, TFIIF, pol II, and TFIIS. Each reaction was supplemented with either 3 pmol (lanes 2 and 8) or 6 pmol (lanes 3 and 9) of Def1-530 and 3 pmol (lanes 5 and 11) or 6 pmol (lanes 6 and 12) of Def1-371. Transcription was initiated by adding an equal volume of 2× transcription mixture containing 1.6 mM ATP, 1.6 mM GTP, 1.6 mM CTP, 40 μM UTP, and 0.083 μM [α-32P]UTP. With the addition of 3 pmol of Elongin, an approximate twofold increase in activity was observed. Transcripts initiated from upstream and downstream TSSs are indicated by * and **, respectively. (B) Effect of Def1 on the transcribing complex. Pol II elongation complexes were assembled on a DNA-RNA hybrid containing a 9-nt radiolabeled nascent transcript and supplemented with increasing amounts of TFIIS (0, 3, 6, and 12 pmol) and Def1 (0, 3, and 6 pmol). The 9-nt RNA (red arrow) was elongated by the addition of NTPs (50 µM ATP, 50 µM GTP, and 50 µM CTP) and stalled at the end of T-less cassette (+32) by the omission of UTP (black arrow). The fold stimulation was calculated for each reaction based on the amount of 32-nt RNA product compared with a control reaction without Def1. TFIIS-induced small RNA cleavage products (6–8 nt) are indicated by ***. Red and black arrows indicate 9-nt and 32-nt RNAs, respectively.
Def1 Facilitates Transcription Restart from TFIIS-Induced Transcript Cleavage.
The requirement for TFIIS and the N-terminal half of the polyQ region of Def1 was further supported by assays using transcribing pol II assembled on a preannealed synthesized DNA-RNA hybrid (Fig. 3B and Fig. S3). The 9-nt RNA (red arrow in Fig. 3B) was elongated by the addition of ATP, GTP, and CTP, and then stalled by the T-stop at +32 (black arrow in Fig. 3B and Fig. S3A), causing backtracking (mainly ∼2–7 bp) of pol II, followed by TFIIS-induced cleavage of transcripts (compare lanes 1, 4, 7, and 10 in Fig. 3B). The addition of Def1-530, but not of Def1-371, increased the amount of 32-nt RNA product compared with a control transcription reaction without Def1 by as much as approximately threefold [1.70 ± 0.17-fold (n = 2) and 2.89 ± 0.48-fold (n = 2) with 3 pmol and 6 pmol of Def1, respectively, in the presence of 6 pmol of TFIIS]. Ela1-Elc1 negatively regulates and TFIIF positively regulates the activity of Def1 in the presence of TFIIS (Fig. S3 B and C). The effect of Def1 was observed regardless of whether the factor was preincubated with transcribing pol II or added after the reaction (Fig. S3A, Right), indicating that Def1 stimulated transcription restart from TFIIS-induced transcript cleavage and did not prevent pol II backtracking.
The requirement for TFIIS may be consistent with previous yeast genetic studies demonstrating that the def1 dst1 (encoding TFIIS) double mutant is extremely sensitive to the nucleotide-depleting drug 6-azauracil (6-AU), a common screening agent for transcription elongation factors (13). Notably, without TFIIS, virtually no significant effects were observed from the addition of any one of factors that we tested (Ela1-Elc1, TFIIF, or Def1) (Fig. S3B), and these factors could exert their activities only through TFIIS (Fig. S3C). This finding is in good agreement with a previous single-molecule optical-trapping study demonstrating that yeast TFIIF does not directly affect the catalytic rate of pol II, but can exert its activity through TFIIS (24). In contrast, mammalian Elongin (25) and TFIIF (26) are known to stimulate pol II elongation by TFIIS-independent mechanisms.
Def1 Is Required for Regulation of Heat Shock Genes in Response to DNA Damage and Heat Shock.
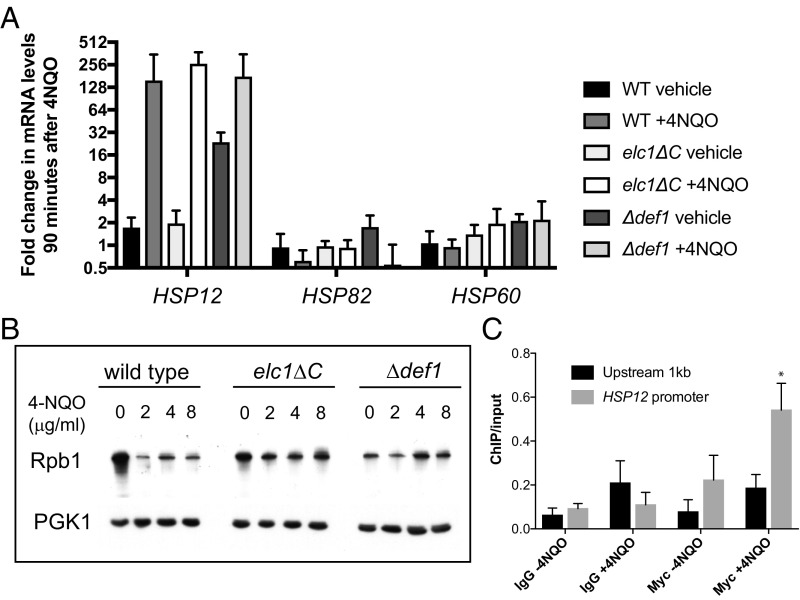
Elongin is required for maximal induction of stress-responsive genes (e.g., ATF3, p21, c-Myc) (4), including heat shock genes (e.g., HSP70) (3, 27), on exposure to multiple stimuli in vivo. Thus, we tested whether Def1 is involved in induction of gene expression in response to cellular stress. First, as Def1 had been demonstrated to be proteolytically cleaved and accumulated in the nucleus in response to DNA damage or other transcription stress (9), we tested Def1 dependency for the induction of stress-responsive genes (19) by DNA damage from UV-mimetic agent 4-nitroquinoline-1-oxide (4-NQO) as well as heat shock; critically, basal mRNA levels of heat shock genes were higher in the def1∆ strain compared with wild type (WT) (Fig. 4A and Fig. S5A). For example, basal levels of HSP12 (without stress) were ∼15-fold higher in the def1∆ strain (Discussion). Therefore, induction of HSP12 in response to 4-NQO and heat shock was reduced by ∼10-fold in the def1∆ strain compared with WT. Other genes that we tested (GLK1, MAG1, PHR1, and RNR1-3) were not significantly affected or only slightly affected (Fig. S5 B and C). We also performed chromatin immunoprecipitation (ChIP) assays and confirmed recruitment of Def1 to the promoter of HSP12, but not ∼1 kb upstream region of HSP12, in 4-NQO–treated cells (Fig. 4C).
Fig. 4.
mRNA levels of heat shock genes and pol II (Rpb1) degradation in response to the addition of DNA damage UV-mimetic agent 4-nitroquinoline-1-oxide (4-NQO). (A) mRNA levels of heat shock genes were determined at 90 min after the addition of 8 μg/mL 4-NQO. Mean ± SEM ∆∆Ct values (relative to mRNA levels in the untreated WT at time 0) are plotted (n = 3). Basal levels of HSP12 (P = 0.004) were increased in ∆def1 strains, but not in elc1∆C strains (P = 0.251, Welch’s t test). (B) Whole-cell extracts from WT, elc1∆C, and ∆def1 strains following 4-NQO treatment were resolved by SDS/PAGE. Rpb1, the largest subunit of pol II, and the loading control PGK1 were detected by Western blot analysis. An approximate 80% decline in the level of Rpb1 was observed after treatment with 4-NQO in WT, whereas Rpb1 levels remained unchanged in the elc1∆C and ∆def1 strains. (C) ChIP analysis of N-terminal Myc-Def1 on HSP12. Cells were untreated or treated with 8 μg/mL 4-NQO for 90 min. Negative control (IgG) was performed without loading c-Myc antibody. n = 3. Error bars represent mean ± SEM. *P < 0.05, two-tailed Student’s t test.
Previous studies have demonstrated that HeLa cells carrying a mutation in the SOCS box of Elongin A are defective in pol II ubiquitylation, but capable of supporting transcription activation in response to doxorubicin-induced DNA damage, suggesting that pol II degradation and stress-responsive transcription activation are separable (4). In yeast, the F-box (structurally analogous to the SOCS box) of Ela1 is not defined in detail, but we could uncouple these two functions by deleting the C-terminal 15 residues of Elc1, critical for recruiting Cullin-RING ligases to the F-box of Ela1 (28). Such a yeast mutant elc1∆C and def1∆ did not significantly degrade pol II, whereas the WT degraded ∼80% of pol II on DNA damage (Fig. 4B). Despite the defective activity in pol II degradation, the elc1∆C proved capable of maximal induction of HSP12 (Fig. 4A), suggesting that pol II degradation and stress-responsive transcription regulation function in separate pathways in response to DNA damage in a manner similar to that of the mammalian system.
Discussion
All eukaryotes respond to DNA damage by polyubiquitylation and degradation of the largest subunit of pol II, facilitated by the Elongin-Cullin ubiquitin ligase. In yeast, the recruitment of the ubiquitin ligase is dependent on Def1. Our finding in the course of TFIIH-Def1 isolation from yeast suggests a potential dual function of Def1. Two lines of evidence obtained in vitro and in vivo support a role for Def1 in pol II transcription regulation. First, Def1-Elongin was capable of enhancing transcription initiation and elongation in a defined transcription system reconstituted from highly purified proteins. Second, in the def1Δ strain, induction of some stress-responsive genes was misregulated.
The up-regulated basal mRNA levels of HSP genes in the def1Δ strain (Fig. 4A) were unexpected in light of studies demonstrating that full-length Def1 is cytoplasmic under normal conditions (9). This indicates an indirect role for cytoplasmic Def1 on transcription; for instance, it may sequester its interacting transcription factors, such as Ccr4-Not (Dataset S1), in the cytoplasm (29, 30). Strikingly, up-regulated basal mRNA levels of HSP12 were previously observed in Ccr4-Not mutants not5Δ (31) and not1-2 (32). In addition, we have observed effects of Def1 on MMS-induced transcription activation of RNR genes (Fig. S5B), which are regulated by Ccr4-Not (33). It will be of great interest to pursue possible interactions of Def1 with Ccr4-Not.
We have shown that Def1 has a strong stimulatory effect on pol II transcription, and that Elongin controls Def1 through positive activity during initiation, suggesting that Def1-Elongin functions predominantly in early transcription. This is in good agreement with previous in vivo fluorescence imaging studies showing that human Elongin A colocalized with CTD-Ser5 phosphorylated pol II better than with the CTD-Ser2 phosphorylated form (4). Additional factors may be required to facilitate the ubiquitylation and degradation of pol II at sites of DNA damage that arrests transcription during elongation,. For example, recent in vivo studies demonstrated that the CSB protein (a human homolog of Rad26) (34, 35) promotes recruitment of Elongin A and the ubiquitin ligase subunit CUL5 to sites of DNA damage (36). The CSB protein has been reported to be recruited to a subset of promoters after UV irradiation in vivo (37, 38).
PolyQ repeats are generally highly enriched in transcription factors, and polyQ repeat variation results in changes in expression of its target genes in associated diseases (39–41). The short polyQ stretches on Ccr4-Not (Ccr4, Not1, Caf1) and Mediator (Med15, Med3) are noteworthy. Our findings suggest that the polyQ region not only regulates nuclear/cytoplasmic localization as previously shown (9), but also directly serves as a positive transcription control.
Experimental Procedures
Protein Purification.
Saccharomyces cerevisiae harboring a TAP tag on Tfb4 was grown in 50 L of YPAD medium to OD 12.0. TFIIH was purified as described previously (12) with slight modifications. Whole cell lysate was prepared by bead beating in Buffer A (50 mM Hepes pH 7.6, 1 mM EDTA, 5% glycerol, 400 mM potassium acetate, 2-mercaptoethanol, and protease inhibitors). Following the addition of 100 mM ammonium sulfate and 0.1% PEI, lysed cells were stirred for 1 h and centrifuged, and then the cleared lysate was loaded onto an IgG column. After washing with 5–10 column volumes of Buffer A plus 400 mM ammonium sulfate, TFIIH was treated with TEV in buffer A, eluted from the IgG column, and loaded onto a HiTrap Q column (GE Healthcare). For extensive washing, the IgG column was washed with at least 50 column volumes of Buffer A plus 400 mM ammonium sulfate. TFIIH was eluted by salt gradient of concentration from 300 mM to 1.2 M potassium acetate, and further purified in a Superose 6 column (GE Healthcare) with 20 mM Hepes pH 7.5, 5% glycerol, 5 mM DTT, and 275 mM potassium acetate.
For the expression of recombinant Def1, the Escherichia coli Rosetta2 (DE3) strain (Stratagene) was transformed with pMAL-C4X (New England BioLabs) and pGEX-6P-1 (GE Healthcare) carrying the truncated Def1-530 gene. The transformed cells were grown at 37 °C and induced with 0.1 mM isopropyl-1-thio-β-d-galacto-pyranoside (IPTG) for 3 h at 30 °C. The cells were lysed by sonication, and the soluble fraction was purified using amylose resin (New England BioLabs) or glutathione Sepharose 4B resin (GE Healthcare), followed by anion exchange chromatography in a HiTrap Q column (GE Healthcare). MBP or GST was retained in assays to solubilize Def1-530. For the expression of Def1-371, the truncated Def1 gene was inserted into pCold II vector (Takara). E. coli Rosetta 2 (DE3) cells carrying the plasmid were grown at 37 °C and induced with 0.1 mM IPTG at 18 °C overnight. The protein was purified by Ni2+-affinity chromatography, followed by HiTrap Q and Superdex 200 columns (GE Healthcare). The gel filtration gave a single peak corresponding to the monomer.
Purification of recombinant Elongin from bacteria was done as described previously (8), with some modifications. In brief, Ela1 and SUMO-tagged Elc1 of S. cerevisiae were cloned into pCDF duet (Novagen) and pET28 (EMD Biosciences) vectors, and were coexpressed in E.coli Rosetta2 (DE3) cells (Novagen). Cells were grown at 37 °C up to OD 0.4 and then induced with 0.1 mM IPTG overnight at 18 °C. The cell pellet was resuspended in Buffer A (30 mM Hepes pH 7.6, 500 mM NaCl, 10 μM ZnSO4, 10% glycerol, 5 mM imidazole, and protease inhibitors). The cell suspension was sonicated and clarified by centrifugation at 26,074 × g for 60 min at 4 °C. The cleared lysate was loaded onto a Ni2+ affinity column pre-equilibrated with Buffer A and then extensively washed with Buffer A containing 25 mM imidazole. The bound protein was eluted by gradient against Buffer A containing 500 mM imidazole. The SUMO tag on Elc1 was cleaved during dialysis in Buffer B (30 mM Hepes pH 7.6, 300 mM NaCl, 10 μM ZnSO4, 10% glycerol, and 0.5 mM DTT) by incubation with Ulp1 overnight at 4 °C. The cleaved protein was passed through a Ni2+ affinity column and a cation exchange chromatograph (Capto-S HiScreen; GE Healthcare), and further purified by gel filtration (Superdex 200).
Three general transcription factors—TFIIF, TFIIE, TFIIH—and pol II were isolated from yeast, and the other factors—TFIIA, TFIIB, TBP, and TFIIS—were purified from bacteria (12, 14, 21, 42). Rad26 was a gift from the Dong Wang laboratory at the University of California, San Diego (43).
Cross-Linking and MS.
The Def1-TFIIH fraction of the HiTrapQ was reacted with BS3 and digested to peptides as described previously (44). Peptides and cross-linked peptides were desalted on a Pierce C18 Spin Column (Thermo Fisher Scientific) and eluted by 75% acetonitrile. One-fifth of the eluted peptides were dried in a SpeedVac, reconstituted in 0.1% formic acid, and measured immediately in the mass spectrometer. The rest of the sample was enriched for cross-linked peptides by either size exclusion chromatography (Superdex Peptide 3.2/300; GE Healthcare) or strong cation exchange chromatography (Mini S PC 3.2/3; GE Healthcare). Fractions containing enriched cross-linked peptides were retained for MS analysis. The peptides were analyzed by a 2-h gradient LC-MS on a Q-Exactive Plus mass spectrometer (Thermo Fisher Scientific). The resulting data files were analyzed by Proteome Discoverer (Thermo Fisher Scientific) for protein identification. The cross-links were identified using the FindXL software package (15). For each protein sample, the data from the nonenriched and enriched fractions were pulled together into one (nonredundant) list of cross-links. The false-positive rate for the cross-link in the final list was determined to be 2.5% by decoy analyses with erroneous cross-linker masses (15, 44).
Electron Microscopy.
Proteins were diluted to ∼250 μg/mL, and 2 μL were applied to continuous formvar/carbon-coated grids washed with 2% uranyl acetate solution, blotted, and dried. Images were collected at a magnification of 40,000× with a CCD camera (Gatan BM-Ultrascan) under low-dose conditions (each exposure ∼20 e−/Å2) on a FEI Tecnai 12 microscope operating at 120 kV.
In Vitro Transcription Initiation Assay.
Transcription assays were performed as described previously (21) with minor modifications. In brief, 1.3 pmol of DNA template SNR20 91W (14) was mixed with 3.75 pmol TFIIA, 3 pmol TFIIB, 1.5 pmol TBP, 3 pmol TFIIE, 1.6 pmol TFIIF, 1.5 pmol TFIIH, 1.8 pmol TFIIK, 1.3 pmol Pol II, and 3 pmol TFIIS. The reaction was supplemented with other additional factors as required in 5 μL of Buffer 300 (50 mM Hepes pH 7.6, 300 mM potassium acetate, and 5% glycerol), diluted with 5 μL of Buffer 10 (20 mM Hepes pH 7.6, 10 mM potassium acetate, 5 mM MgSO4, and 5 mM DTT) and incubated on ice for at least 2 h to form the preinitiation complex (PIC). The transcription reaction was initiated with the addition of an equal volume of 2× NTP mix composed of 20 mM Hepes pH 7.6, 10 mM potassium acetate, 5 mM MgSO4, 5 mM DTT 1.6 mM ATP, 1.6 mM CTP, 1.6 mM GTP, 40 μM UTP, 1 U of RNaseOUT (Invitrogen), and 0.83 μM [α-32P] UTP (2.5 μCi) at 30 °C. The reaction was stopped after 45 min by adding 190 μL of stop buffer (300 mM sodium phosphate pH 5.5, 5 mM EDTA, 0.7% SDS, 0.1 mg/mL glycogen, and 0.013 mg/mL Proteinase K; Sigma-Aldrich). RNA was extracted by ethanol precipitation and then analyzed on a denaturing 6% polyacrylamide gel.
Preparation of Stalled Pol II Complexes.
Pol II elongation complexes were assembled as described previously (45) with slight modifications. In brief, elongation complexes were assembled by mixing 22 pmol template DNA fragment, 22 pmol 32P-labeled 9-nt RNA, 19 pmol pol II, and 22 pmol nontemplate DNA fragment, followed by purification in a MicroSpin G-50 column (GE Healthcare). Elongation complexes (3.75 pmol) were incubated with TFIIS, Def1, TFIIF, and Ela1-Elc1 as required in 6 μL of Buffer 10 for at least 1 h. The 9-nt RNA was elongated by the addition of 2 μL of 4× NTP mix containing 200 μM ATP, GTP, and CTP, 20 mM magnesium acetate, and 1 U of RNaseOUT (Invitrogen) for 1 min at 25 °C, and stopped by the addition of an equal volume of stop buffer [7 M urea, 1× Tris/borate/EDTA (TBE) buffer, 2 mg/mL bromophenol blue, and 10 mM EDTA], and incubated at 55 °C for 5 min. RNA transcripts were quantified on a 23% acrylamide gel.
Native Gel Assay.
The native gel assay was performed as described previously (20) with slight modifications. Pol II elongation complexes were assembled on templates containing 9-nt and 27-nt radiolabeled nascent transcripts, and then purified in a MicroSpin G-50 column. Elongation complexes (∼3.75 pmol) were mixed with increasing amounts of GST-Def1-530, as indicated above the lanes, incubated for 15 min, and then run on 1% agarose gel in 0.2× TBE. The 1.2 pmol PIC was formed on a radiolabeled SNR20 fragment as previously described (21), incubated with increasing amounts of GST-Def1-530, and run on 0.8% agarose gel in 0.2× TBE.
Strain Construction and Quantitative PCR Analysis.
The def1∆ strain was constructed in the FY602 background (MATa his3Δ200 leu2Δ1 lys2–128δ ura3–52 trp1Δ63) by supplementation of DEF1 with centromeric plasmid pRS413 under a GAL4 promoter with HIS3 marker (pRS413GALp•DEF1), followed by deletion of genomic DEF1 through homologous recombination using a URA3 marker. The elc1∆C strain was also constructed in the FY602 background by deleting the last 15 amino acid residues at the C-terminal end using the URA3 marker. A WT strain with empty vector (pRS413GALp) and the def1∆ strain were grown to OD 1.0 in YPAD medium and then treated with 8 µg/mL 4-NQO, heat shock at 37 °C, or 0.03% MMS. Cells were harvested after 0 and 90 min, RNA was extracted with TRizol (Life Technologies) in accordance with the manufacturer’s instructions, and cDNAs were synthesized using a random hexamer primer with SuperScript IV reverse transcriptase (Life Technologies).
Quantitative PCR (qPCR) was performed with primers for ACT1, HSP60, HSP12, and HSP82 with SYBR Green Master Mix with Rox (Roche) using an Applied Biosystems 7500 Real-Time PCR (Thermo Fisher Scientific) and analyzed with high-resolution melting software. qPCR was performed for two to three biological replicates and three technical replicates. Relative gene expression was calculated using the Livak method (∆∆Ct) with genes normalized to actin.
ChIP on Yeast.
The Myc-tagged Def1 yeast strain (9) was grown in 80 mL of YPAD medium to OD 1.0, and cells were then grown for another 90 min in the presence of 8 µg/mL 4NQO. The cells were cross-linked with a final concentration of 1% formaldehyde by shaking at 100 rpm for 15 min at 30 °C. The cross-linking reaction was quenched by the addition of 2.5 M glycine to a final concentration of 100 mM, followed by shaking at 100 rpm for an additional 5 min. Cells were then washed with 25 mL of 1× PBS, and centrifuged at 1,620 × g for 3 min to obtain the cell pellets. The pellets were then transferred to 2-mL screw top centrifuge tubes and stored at −80 °C. Frozen pellets were thawed on ice for 10–15 min and then resuspended in 250 µL of lysis buffer (50 mM Hepes pH 7.5, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% Na-deoxycholate, and 1× protease inhibitors), after which 200 µg of acid-washed, sterilized glass beads were added. The cells were disrupted using a Biospec Bead beater (five cycles of 1 min with 2 min on ice). The lysates were briefly centrifuged and then extracted using the stacked transfer technique, with a needle used to puncture the tube. The lysates were then transferred to another tube by centrifuging the stacked setup at 1,016 × g for 1 min at 4 °C. Chromatin was sheared to an average size of 250–300 bp with a Covaris S220 ultrasonicator, using 1-mL millitubes and a high cell number shearing protocol for 15 min. Lysates were cleared by centrifugation at 16,100 × g for 15 min. The concentration of lysates was determined by the Bradford assay, and ChiP analyses were conducted using equal concentrations (0.8–1.5 mg) of sheared chromatin.
ChIP was performed by incubating lysates with protein G magnetic Dynabeads preconjugated with 2 µg of antibody overnight at 4 °C with rotation. Immunocomplexes were then washed five times in ChIP wash buffer (50 mM Hepes-KOH pH 7.5, 500 mM LiCl, 1 mM EDTA, 1% Nonidet P-40, 0.7% Na-deoxycholate, and 0.1% N-lauroylsarcosine) and once in ChIP final wash buffer (10 mM Tris⋅HCl pH 8.0 at 4 °C, 1 mM EDTA, and 50 mM NaCl). Immunocomplexes were eluted by incubating at 65 °C for 30 min in ChIP elution buffer (50 mM Tris⋅HCl pH 8.0 at 25 °C, 10 mM EDTA, and 1% SDS), and cross-linking was reversed by overnight incubation at 37 °C.
Immunoprecipitated DNA was treated with RNase A (0.2 mg/mL final concentration) for 2 h at 37 °C, and then proteinase K (0.2 mg/mL final concentration) for 2 h at 55 °C. DNA was then purified by phenol:chloroform extraction and ethanol precipitation and resuspended in 1× TE buffer. qPCR was carried out with Power SYBR (Applied Biosystems) using primers for HSP12 promoter sequence or a 1-kb upstream nonbinding control.
Mouse IgG (I5381-1MG; Sigma Aldrich) and anti-MYC (9E10; University of Pennsylvania Cell Center) antibodies were used. The following primer pairs were used: Hsp12_promoter_Forward ACAACCCACAAACACAGACC, Hsp12_promoter _Reverse ACCGGAACCTCAAAGTTGAC, Upstream_1 kb_Forward ACGTTCTTGGACCCAAACAC, and Upstream_1 kb_Reverse GTTTGGTGCTGCATTTGGTG.
Rpb1 Degradation Assay.
Yeast cells were grown in YPAD medium at 30 °C to OD 1.0, treated with varying concentrations of 4-NQO (2, 4, 8 μg/mL), and then grown for another 90 min. Cultures were diluted to OD 0.6, and then 10 mL of cells were harvested and stored at −80 °C. The cells were treated with 0.1 M NaOH for 5 min at room temperature and then centrifuged at high speed. The resulting cell pellets were resuspended in 100 μL of preheated SDS sample buffer and then boiled for 5 min. Cell lysates were resolved by SDS/PAGE (4–12% Bis-Tris; NuPAGE Novex; Invitrogen) and then transferred to a PVDF membrane (Bio-Rad) for 1.5 h at 30 V at 4 °C. The membrane was blocked for 1 h in 5% milk in TBS plus 0.1% Tween 20, and then incubated at 4 °C overnight with 8WG16, mouse monoclonal antibody to polII CTD (ab817; Abcam) or 22C5D8, mouse monoclonal anti-phosphoglycerate kinase antibody (459250; Invitrogen) as a loading control. Blots were then further processed using anti-mouse secondary antibody (NA931; ECL mouse IgG, HRP-linked whole Ab from sheep; Amersham) for 1 h at room temperature, and then visualized using SuperSignal West Pico chemiluminescent substrate (Thermo Fisher Scientific).
Supplementary Material
Acknowledgments
We thank Craig Kaplan and Yunye Zhu (Texas A&M University) for their help with analysis of HSP12 expression; Hua-Ying Fan, Hillary Nelson, and Rina Fujiwara (University of Pennsylvania) and P.-J. Mattei and Ralph Davis (Stanford University) for critical reading of the manuscript and helpful discussions; Dong Wang (University of California, San Diego) for providing recombinant Rad26; and Jeremy Wilusz, Chie Arai, and Deirdre Tatomer (University of Pennsylvania) for technical support in qPCR and Western blot analyses. The N-terminal Myc-tagged Def1 yeast strain was a gift from the Svejstrup laboratory at the Francis Crick Institute. This research was supported by National Institutes of Health (NIH) Grant GM123233. J.Z. and N.K. are funded by Israel Science Foundation Grant 15/1768. T.V.E. is supported by Structural Biology and Molecular Biophysics Training Program Grant GM008275. E.L.-S. is funded by NIH Grant 1 F31 GM123744-01.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data Deposition: The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (www.proteomexchange.org) via the Proteomics Identifications (PRIDE) partner repository (PXD008223).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1707955114/-/DCSupplemental.
References
- 1.Aso T, Lane WS, Conaway JW, Conaway RC. Elongin (SIII): A multisubunit regulator of elongation by RNA polymerase II. Science. 1995;269:1439–1443. doi: 10.1126/science.7660129. [DOI] [PubMed] [Google Scholar]
- 2.Chopra VS, Hong JW, Levine M. Regulation of Hox gene activity by transcriptional elongation in Drosophila. Curr Biol. 2009;19:688–693. doi: 10.1016/j.cub.2009.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gerber M, et al. Regulation of heat shock gene expression by RNA polymerase II elongation factor, Elongin A. J Biol Chem. 2005;280:4017–4020. doi: 10.1074/jbc.C400487200. [DOI] [PubMed] [Google Scholar]
- 4.Kawauchi J, et al. Transcriptional properties of mammalian elongin A and its role in stress response. J Biol Chem. 2013;288:24302–24315. doi: 10.1074/jbc.M113.496703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ribar B, Prakash L, Prakash S. ELA1 and CUL3 are required along with ELC1 for RNA polymerase II polyubiquitylation and degradation in DNA-damaged yeast cells. Mol Cell Biol. 2007;27:3211–3216. doi: 10.1128/MCB.00091-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yasukawa T, et al. Mammalian Elongin A complex mediates DNA-damage-induced ubiquitylation and degradation of Rpb1. EMBO J. 2008;27:3256–3266. doi: 10.1038/emboj.2008.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harreman M, et al. Distinct ubiquitin ligases act sequentially for RNA polymerase II polyubiquitylation. Proc Natl Acad Sci USA. 2009;106:20705–20710. doi: 10.1073/pnas.0907052106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koth CM, et al. Elongin from Saccharomyces cerevisiae. J Biol Chem. 2000;275:11174–11180. doi: 10.1074/jbc.275.15.11174. [DOI] [PubMed] [Google Scholar]
- 9.Wilson MD, et al. Proteasome-mediated processing of Def1, a critical step in the cellular response to transcription stress. Cell. 2013;154:983–995. doi: 10.1016/j.cell.2013.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Svejstrup JQ, et al. Different forms of TFIIH for transcription and DNA repair: Holo-TFIIH and a nucleotide excision repairosome. Cell. 1995;80:21–28. doi: 10.1016/0092-8674(95)90447-6. [DOI] [PubMed] [Google Scholar]
- 11.Gibbons BJ, et al. Subunit architecture of general transcription factor TFIIH. Proc Natl Acad Sci USA. 2012;109:1949–1954. doi: 10.1073/pnas.1105266109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murakami K, et al. Tfb6, a previously unidentified subunit of the general transcription factor TFIIH, facilitates dissociation of Ssl2 helicase after transcription initiation. Proc Natl Acad Sci USA. 2012;109:4816–4821. doi: 10.1073/pnas.1201448109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Woudstra EC, et al. A Rad26-Def1 complex coordinates repair and RNA pol II proteolysis in response to DNA damage. Nature. 2002;415:929–933. doi: 10.1038/415929a. [DOI] [PubMed] [Google Scholar]
- 14.Murakami K, et al. Uncoupling promoter opening from start-site scanning. Mol Cell. 2015;59:133–138. doi: 10.1016/j.molcel.2015.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kalisman N, Adams CM, Levitt M. Subunit order of eukaryotic TRiC/CCT chaperonin by cross-linking, mass spectrometry, and combinatorial homology modeling. Proc Natl Acad Sci USA. 2012;109:2884–2889. doi: 10.1073/pnas.1119472109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Esnault C, et al. Mediator-dependent recruitment of TFIIH modules in preinitiation complex. Mol Cell. 2008;31:337–346. doi: 10.1016/j.molcel.2008.06.021. [DOI] [PubMed] [Google Scholar]
- 17.Robinson PJ, et al. Structure of a complete mediator-RNA polymerase II pre-initiation complex. Cell. 2016;166:1411–1422.e16. doi: 10.1016/j.cell.2016.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsai KL, et al. Mediator structure and rearrangements required for holoenzyme formation. Nature. 2017;544:196–201. doi: 10.1038/nature21393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaillard H, et al. Genome-wide analysis of factors affecting transcription elongation and DNA repair: A new role for PAF and Ccr4-Not in transcription-coupled repair. PLoS Genet. 2009;5:e1000364. doi: 10.1371/journal.pgen.1000364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kruk JA, Dutta A, Fu J, Gilmour DS, Reese JC. The multifunctional Ccr4-Not complex directly promotes transcription elongation. Genes Dev. 2011;25:581–593. doi: 10.1101/gad.2020911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murakami K, et al. Formation and fate of a complete 31-protein RNA polymerase II transcription preinitiation complex. J Biol Chem. 2013;288:6325–6332. doi: 10.1074/jbc.M112.433623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luo J, et al. Architecture of the human and yeast general transcription and DNA repair factor TFIIH. Mol Cell. 2015;59:794–806. doi: 10.1016/j.molcel.2015.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fiumara F, Fioriti L, Kandel ER, Hendrickson WA. Essential role of coiled coils for aggregation and activity of Q/N-rich prions and PolyQ proteins. Cell. 2010;143:1121–1135. doi: 10.1016/j.cell.2010.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schweikhard V, et al. Transcription factors TFIIF and TFIIS promote transcript elongation by RNA polymerase II by synergistic and independent mechanisms. Proc Natl Acad Sci USA. 2014;111:6642–6647. doi: 10.1073/pnas.1405181111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elmendorf BJ, Shilatifard A, Yan Q, Conaway JW, Conaway RC. Transcription factors TFIIF, ELL, and Elongin negatively regulate SII-induced nascent transcript cleavage by non-arrested RNA polymerase II elongation intermediates. J Biol Chem. 2001;276:23109–23114. doi: 10.1074/jbc.M101445200. [DOI] [PubMed] [Google Scholar]
- 26.Renner DB, Yamaguchi Y, Wada T, Handa H, Price DH. A highly purified RNA polymerase II elongation control system. J Biol Chem. 2001;276:42601–42609. doi: 10.1074/jbc.M104967200. [DOI] [PubMed] [Google Scholar]
- 27.Gerber M, et al. In vivo requirement of the RNA polymerase II elongation factor elongin A for proper gene expression and development. Mol Cell Biol. 2004;24:9911–9919. doi: 10.1128/MCB.24.22.9911-9919.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bullock AN, Debreczeni JE, Edwards AM, Sundström M, Knapp S. Crystal structure of the SOCS2-elongin C-elongin B complex defines a prototypical SOCS box ubiquitin ligase. Proc Natl Acad Sci USA. 2006;103:7637–7642. doi: 10.1073/pnas.0601638103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller JE, Reese JC. Ccr4-Not complex: The control freak of eukaryotic cells. Crit Rev Biochem Mol Biol. 2012;47:315–333. doi: 10.3109/10409238.2012.667214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Collart MA, Panasenko OO. The Ccr4–Not complex. Gene. 2012;492:42–53. doi: 10.1016/j.gene.2011.09.033. [DOI] [PubMed] [Google Scholar]
- 31.Deluen C, et al. The Ccr4-Not complex and yTAF1 (yTaf(II)130p/yTaf(II)145p) show physical and functional interactions. Mol Cell Biol. 2002;22:6735–6749. doi: 10.1128/MCB.22.19.6735-6749.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.James N, Landrieux E, Collart MA. A SAGA-independent function of SPT3 mediates transcriptional deregulation in a mutant of the Ccr4-Not complex in Saccharomyces cerevisiae. Genetics. 2007;177:123–135. doi: 10.1534/genetics.107.076299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mulder KW, Winkler GS, Timmers HT. DNA damage and replication stress induced transcription of RNR genes is dependent on the Ccr4-Not complex. Nucleic Acids Res. 2005;33:6384–6392. doi: 10.1093/nar/gki938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Gool AJ, et al. RAD26, the functional S. cerevisiae homolog of the Cockayne syndrome B gene ERCC6. EMBO J. 1994;13:5361–5369. doi: 10.1002/j.1460-2075.1994.tb06871.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hanawalt PC, Spivak G. Transcription-coupled DNA repair: Two decades of progress and surprises. Nat Rev Mol Cell Biol. 2008;9:958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 36.Weems JC, et al. Cockayne syndrome B protein regulates recruitment of the Elongin A ubiquitin ligase to sites of DNA damage. J Biol Chem. 2017;292:6431–6437. doi: 10.1074/jbc.C117.777946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Proietti-De-Santis L, Drané P, Egly JM. Cockayne syndrome B protein regulates the transcriptional program after UV irradiation. EMBO J. 2006;25:1915–1923. doi: 10.1038/sj.emboj.7601071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lake RJ, Boetefuer EL, Won KJ, Fan HY. The CSB chromatin remodeler and CTCF architectural protein cooperate in response to oxidative stress. Nucleic Acids Res. 2016;44:2125–2135. doi: 10.1093/nar/gkv1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fondon JW, 3rd, Garner HR. Molecular origins of rapid and continuous morphological evolution. Proc Natl Acad Sci USA. 2004;101:18058–18063. doi: 10.1073/pnas.0408118101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gemayel R, Vinces MD, Legendre M, Verstrepen KJ. Variable tandem repeats accelerate evolution of coding and regulatory sequences. Annu Rev Genet. 2010;44:445–477. doi: 10.1146/annurev-genet-072610-155046. [DOI] [PubMed] [Google Scholar]
- 41.Gemayel R, et al. Variable glutamine-rich repeats modulate transcription factor activity. Mol Cell. 2015;59:615–627. doi: 10.1016/j.molcel.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fazal FM, Meng CA, Murakami K, Kornberg RD, Block SM. Real-time observation of the initiation of RNA polymerase II transcription. Nature. 2015;525:274–277. doi: 10.1038/nature14882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang L, et al. Regulation of the Rhp26ERCC6/CSB chromatin remodeler by a novel conserved leucine latch motif. Proc Natl Acad Sci USA. 2014;111:18566–18571. doi: 10.1073/pnas.1420227112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murakami K, et al. Architecture of an RNA polymerase II transcription pre-initiation complex. Science. 2013;342:1238724. doi: 10.1126/science.1238724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Čabart P, Jin H, Li L, Kaplan CD. Activation and reactivation of the RNA polymerase II trigger loop for intrinsic RNA cleavage and catalysis. Transcription. 2014;5:e28869. doi: 10.4161/trns.28869. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.