Significance
Cell communication is a precisely orchestrated mechanism in which cell receptors translate extracellular cues into intracellular signals. The Eph receptors act as a model guidance system steering cells to defined positions by their ligand ephrin. However, we still lack a mechanistic understanding of how membrane receptors can read a wide range of ligand concentrations and gradients and integrate them into coherent cellular responses. Here we reveal the evolution of Eph aggregation upon ephrin stimulation with unprecedented resolution by extending current imaging methods. The results fit biophysical models of protein aggregation. In these models, two protein oligomerization modes, polymerization and condensation, correlate with the “on/off” switching of the receptor activation, providing a precise, proportional, and dynamic response to variable ephrin inputs.
Keywords: Eph, ephrin, receptor tyrosine kinase, gradients, cell communication
Abstract
Eph receptor signaling plays key roles in vertebrate tissue boundary formation, axonal pathfinding, and stem cell regeneration by steering cells to positions defined by its ligand ephrin. Some of the key events in Eph-ephrin signaling are understood: ephrin binding triggers the clustering of the Eph receptor, fostering transphosphorylation and signal transduction into the cell. However, a quantitative and mechanistic understanding of how the signal is processed by the recipient cell into precise and proportional responses is largely lacking. Studying Eph activation kinetics requires spatiotemporal data on the number and distribution of receptor oligomers, which is beyond the quantitative power offered by prevalent imaging methods. Here we describe an enhanced fluorescence fluctuation imaging analysis, which employs statistical resampling to measure the Eph receptor aggregation distribution within each pixel of an image. By performing this analysis over time courses extending tens of minutes, the information-rich 4D space (x, y, oligomerization, time) results were coupled to straightforward biophysical models of protein aggregation. This analysis reveals that Eph clustering can be explained by the combined contribution of polymerization of receptors into clusters, followed by their condensation into far larger aggregates. The modeling reveals that these two competing oligomerization mechanisms play distinct roles: polymerization mediates the activation of the receptor by assembling monomers into 6- to 8-mer oligomers; condensation of the preassembled oligomers into large clusters containing hundreds of monomers dampens the signaling. We propose that the polymerization–condensation dynamics creates mechanistic explanation for how cells properly respond to variable ligand concentrations and gradients.
Cells constantly respond to other cells and their environments through receptor–ligand interactions, leading to fundamental cell decisions such as patterning or division (1). Ligand stimulation often induces receptor oligomerization, fostering transactivation (e.g., via phosphorylation) and transducing extracellular cues into intracellular signals (2, 3). Eph tyrosine kinase receptors represent a paradigmatic family of cell–cell communication molecules interacting with their ligand ephrin on the surface of neighboring cell membranes. Eph-ephrin signaling plays a central role in development, for example, during the establishment of vertebrate tissue boundaries (e.g., hindbrain cell segregation and somitogenesis) (4, 5). Ephrin cues are also presented in the form of concentration gradients, apparently guiding axonal patterning in retinotectal mapping or stem cell migration in the developing intestines (4, 6–8). Despite ample evidence for the precision of this signaling system in controlling cell patterning and positioning, the mechanism(s) by which different ephrin concentrations are interpreted by the Eph receptor into proportional responses is largely unknown.
The current model for Eph activation/clustering posits that the presentation of an ephrin dimer nucleates an Eph dimer, activating the receptor by the resulting transphosphorylation (9, 10). Activated receptors then propagate the signal horizontally by recruiting neighboring monomers into large-scale clusters, which leads to the endocytosis of the aggregate and termination of the signal (10–13). Receptor aggregation therefore has been interpreted as an “amplifier” that operates on the ligand signal and increases the receptor sensitivity for low ligand concentrations (3, 14). However, it is unclear how such simple signaling scheme, lacking an adaptation mechanism beyond endocytosis, offer the cell the ability to sense and transduce changes in ligand concentrations or gradients of ligands (15).
Here, we combine quantitative imaging and biophysical modeling to a model for the oligomerization and activation dynamics of the Eph receptor. Measuring the dynamic evolution of aggregates on living cells exceeds the capabilities of conventional imaging approaches; this requires molecular-level sensitivity over the area of an entire cell, and temporal scales ranging from millisecond-to-second times over which receptor dynamics take place, to the tens of minutes over which cell responses manifest. We meet these challenges by using a fluorescence fluctuation analysis of the short-term variations in the intensities of each pixel in an image, based on the powerful number and brightness (N&B) approach (16, 17) (Fig. 1). N&B analysis has been implemented to study the aggregation of transcription factors, focal adhesion proteins, or membrane-tethered proteins (18) during short acquisition times. Conventional N&B analysis yields the median concentration (number) and molecular aggregation state (brightness) of labeled proteins (16, 17) (Fig. 1C, gray bar). Our modeling showed that the median oligomerization (i-mer) state over a narrow time window of analysis was not adequate to test between different models of receptor activation. Here we define and deploy enhanced N&B (eN&B), which uses a more powerful statistical approach to reveal the distribution, rather than the mean, of Eph aggregation. Our eN&B analysis reveals a polymerization–condensation process mediating signal amplification and adaptation to the receptor signal.
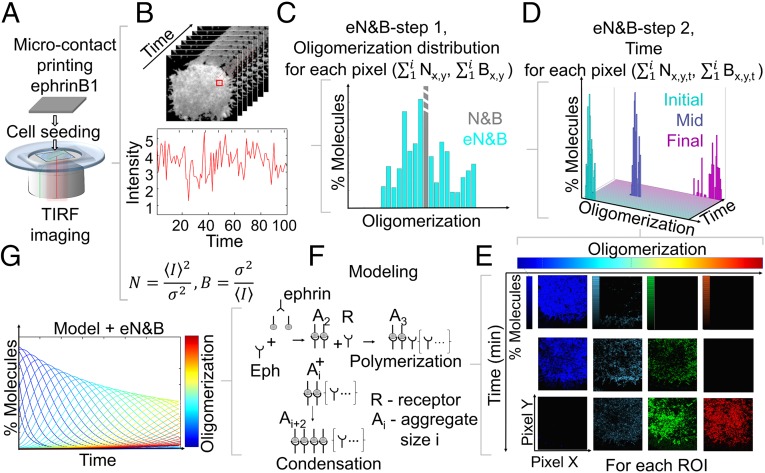
Fig. 1.
Experimental pipeline overview. (A) EphrinB1 is microcontact printed on imaging cell culture dishes. Cells are seeded and prepared for TIRF imaging. (B) Short-scale time imaging (seconds) is performed to capture fluorescence fluctuations, the basis of N&B analysis (B, brightness; N, number). (C) Statistical enhancement of N&B expands oligomerization information for each pixel from a median value (N&B, gray) to a distribution (eN&B, cyan). (D) Photobleaching compensation (19, 20) (SI Appendix, SI Materials and Methods) allows long-scale time imaging (minutes) providing a distribution of oligomers for each time point in each pixel. (E) Oligomerization map distribution for a ROI in a cell over long-scale imaging (minutes). Color-coding (jet color map) represents oligomerization level; each color is color-mapped to black to represent the relative percentage of molecules at a specific oligomer state. (F) Mathematical modeling is coupled to the eN&B analysis (G).
Results
Enhancement of N&B.
Eph receptor aggregation takes place in response to ligand interaction and, as a result, a variety of oligomeric species must coexist on the cell surface. Standard number and brightness (16) can interpret the fluctuations in an image to reflect the mean concentration and oligomerization in each pixel, but cannot offer insights into the full variety of oligomerization states (brightness) that can coexist in the same pixel. To answer this challenge, we created an eN&B analysis, which employs a statistical resampling method (SI Appendix, SI Materials and Methods and Fig. S1) and can obtain the histogram of the Eph receptor aggregation (i-mer distribution) within each pixel of an image (Fig. 1C, cyan bars). The distribution of aggregation states for all of the pixels in an image are determined from a rapid series of images acquired over a few seconds. Because Eph receptor aggregation requires minutes, we bridge this temporal gap by extending the eN&B analysis over time (19, 20) (Fig. 1D), measuring the oligomerization dynamics of proteins in each pixel for the full time-course of each cell’s response. The information-rich 4D space (x, y, oligomer distribution, time) offered by eN&B can be related to Eph receptor activation by closely coupling analysis with mathematical modeling (Fig. 1 F and G).
Aggregation Dynamics Using Microprinted Ephrin Presentation.
Receptor aggregation was studied in transgenic cells (HEK293T) stably expressing the fusion protein EphB2_mRuby; alternatively, we used the kinase-deficient (KD) construct KD_ EphB2_mRuby (Materials and Methods and SI Appendix, Fig. S2), which can be used to study the role of endocytosis, because it cannot activate the endocytic pathway and is not internalized by the cell upon stimulation by ephrinB1 (15, 21). The ephrin-Fc protein was microcontact printed on functionalized glass to present the ligand to the cells in a localized yet homogenous manner (SI Appendix, SI Materials and Methods); control experiments used functionalized glass printed with Fc alone or coated with poly-l-lysine (PLL).
Cells were imaged using total internal reflection fluorescence (TIRF) microscopy, which yields high signal-to-noise images of the cells’ membranes as they interacted with the ligand-functionalized surfaces enabling N&B analysis on membrane proteins (18, 22). This approach offered the needed pixel size and temporal resolution required for eN&B analysis. SI Appendix, Fig. S1 shows the fluorescence intensity fluctuations for two representative pixels, one from an ephrinB1-stimulated cell (red trace), and another from a control cell (black trace). Over the 60-min data collection period, the fluctuations in fluorescence intensity increased in size and decreased in frequency in the ligand-stimulated cell; such fluctuations yield the brightness (B) in N&B analysis and can be converted to mean oligomer sizes (i-mer) by multiplying the brightness value of the unstimulated monomer (Bmonomer = 1.17, relative Bbackground = 1.00; SI Appendix, SI Materials and Methods and Fig. S3).
Simple observation of the mean fluorescence intensity cannot distinguish stimulated and unstimulated cells over 60 min (Fig. 2A). The eN&B analysis, instead, reveals large differences by exploring oligomer distributions over time for each pixel. This multidimensional dataset, however, cannot be directly represented as an image. To intuitively visualize this information, we color-code the image based on the average oligomer size, scaling from monomer to 40-mer. This dimensionally reduced representation reveals a striking difference between the dramatic EphB2 clustering on the cells presented with ephrinB1 (Fig. 2A) and the near static oligomerization level of cells presented only with PLL or Fc (SI Appendix, Fig. S4).
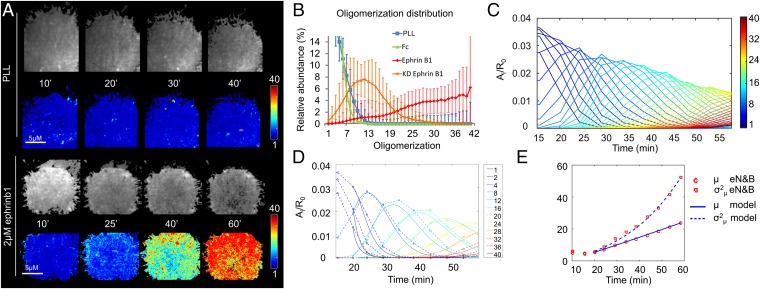
Fig. 2.
eN&B analysis of EphB2 clustering using microprinted ligand stimulation. (A) Time-lapse oligomerization map of HEK293T:EphB2_mRuby cells acquired with a TIRF microscope. The cells were seeded on plates coated with PLL with either no additional coating or 2 μM ephrinB1. For every coating condition, the top panels show gray-scale snapshots of the cells after photobleaching compensation (19, 20) at indicated time points. The bottom rows depict the oligomerization maps of the same images. Every pixel in the cell represents the weighted average i-mer species color-coded according to the color scale bar. PLL and 2-μM ephrinB1 experiments were replicated 47 and 61 times, respectively (Movies S1–S3). (B) Distribution of average and SD oligomerization values for multiple cells (NPLL = 10, NFC = 8, NEphrinB1 = 19, NKD = 16) presented with the relevant ligand for 60 min. (C) An i-mer evolution plot (SI Appendix). (C) Evolution of the concentration of each aggregate (Ai) over time from A, normalized by the initial concentration of free receptor (R0). The i-mer values are color-coded according to the color scale bar. (D) Model fitting to experimental data (SI Appendix, Dynamical Model and section 3). The experimental data from C (dashed lines) was used to fit 12 selected species into the mathematical model (solid lines). Additional fittings can be found in SI Appendix. (E) Mean (µ) and covariance (σ2) of the aggregate size for eN&B measurements and prediction of the polymerization–condensation model for microprinted ephrinB1.
The oligomerization state averaged across all pixels for ephrinB1-stimulated cells after 60 min (i-mer = 28.2 ± 0.9) was significantly higher (P < 0.01) than the oligomerization in control cells (i-mer = 3.7 ± 0.2 for PLL coating; i-mer = 2.9 ± 0.2 for microprinted Fc protein) (Fig. 2B). The KD-EphB2 mutant showed intermediate levels of aggregation when presented with ephrinB1 (i-mer = 12.0 ± 0.5). Such averaged results indicate that receptor clustering was strong and specific to cells presented with ephrinB1. Ephrin stimulation did not have any impact on GFP oligomerization in cells coexpressing membrane-tethered GFP and the Eph receptor (SI Appendix, Fig. S4H), indicating that brightness increase derives from specific EphB2 receptor oligomerization rather than spurious phenomena such as membrane ruffling or cell adhesion variability (18, 22). We have also performed an automated tracking of the top 10% brightest aggregates from several cells stimulated with microprinted ligand on one of the sequence of 200 frames. The results (SI Appendix, Fig. S4I) reveal high mobility of the clusters formed, suggesting that receptor mobility is not compromised by microprinting ligand delivery, probably due to the noncovalent adsorption of ephrin to the surface. Internalization of Eph clusters occurred normally as well after microprinted ligand delivery (SI Appendix, Fig. S4J).
The eN&B analysis over time revealed an orderly progression of Eph receptor aggregation after stimulation with ephrinB1 over the 60 min of observation (i-mer plot; Fig. 2C). Initially, low-order species dominate (monomer–pentamer), then decay rapidly (within the first 30 min). Each i-mer species increases in abundance in turn over the few minutes after its initial appearance; thereafter, each one decreases as higher i-mers form. Extended observation (75 min) did not reveal an upper limit to the i-mers being formed (SI Appendix, Fig. S4). The relatively fast depletion of the monomers revealed by eN&B analysis in the presence of progressive EphB2 clustering indicates that higher-order EphB2 oligomers cannot be assembled predominantly by the recruitment of monomers; instead, it seems that oligomer growth must involve the recruitment of smaller oligomers into larger complexes (23).
Polymerization–Condensation Model.
A mathematical model was used to interpret the rich information about oligomerization dynamics contained in the multiple eN&B distributions, and to validate the hypothesis that coalescence of oligomers contribute to aggregate growth beyond the point of monomer depletion. We built our model based on the Lumry–Eyring biophysical theory on protein aggregation (24, 25), assuming that two oligomerization mechanisms foster receptor aggregation—namely, polymerization by accretion of monomers and condensation by coalescence of oligomers into larger aggregates. The rich eN&B data allowed us to explore the parameter space of the polymerization–condensation model to study the relative impacts of polymerization and condensation in controlling oligomer formation and the strength of the ensuing signal (SI Appendix, Dynamical Model). The best fit model shows that EphB2 receptor oligomer growth is not a monotonic process, but instead results from the combined action of polymerization and condensation, which are mechanistically uncoupled but whose contribution overlaps in time (Fig. 2D). Two growth phases take place: a first phase in which free monomeric receptors form dimers by ephrin induction (nucleation) and incrementally higher oligomers independent of additional ligand binding (polymerization). The second growth phase involves both the accretion of any free monomers and the coalescence (condensation) of two aggregates to form a larger one.
The polymerization–condensation model predicts an initial phase in which the addition of monomers predominates (Fig. 2D) until observable monomer concentration falls to below 1%. Condensation then becomes more important with a contribution of monomers being mainly recruited from the unobservable part of the membrane. The excellent agreement between the model prediction and the eN&B data supports the hypothesis of a dual oligomerization mode (polymerization and condensation) contributing to receptor aggregation. In agreement, note that the variance in the EphB2 oligomer sizes increases around minute 30 (Fig. 2E), when monomer concentration is very low, as predicted if the oligomers grew by a condensation of previously formed oligomers (24). Processes in which oligomers grow only by adding monomers should reveal a variance that grows slowly with the mean aggregate size (μ) (SI Appendix). The stimulation of EphB2_KD cells with ephrinB1 (SI Appendix, Fig. S2) showed impaired receptor aggregation, in terms of a lower degree of oligomerization (measured by brightness maps) and slower aggregation dynamics, compared with the functional receptor. These results suggest a role of the tyrosine kinase domain of the Eph receptor in the formation of high-order clusters, possibly by harboring specific interfaces needed for condensation of oligomers (26).
Aggregation Dynamics Using Ephrin in Solution.
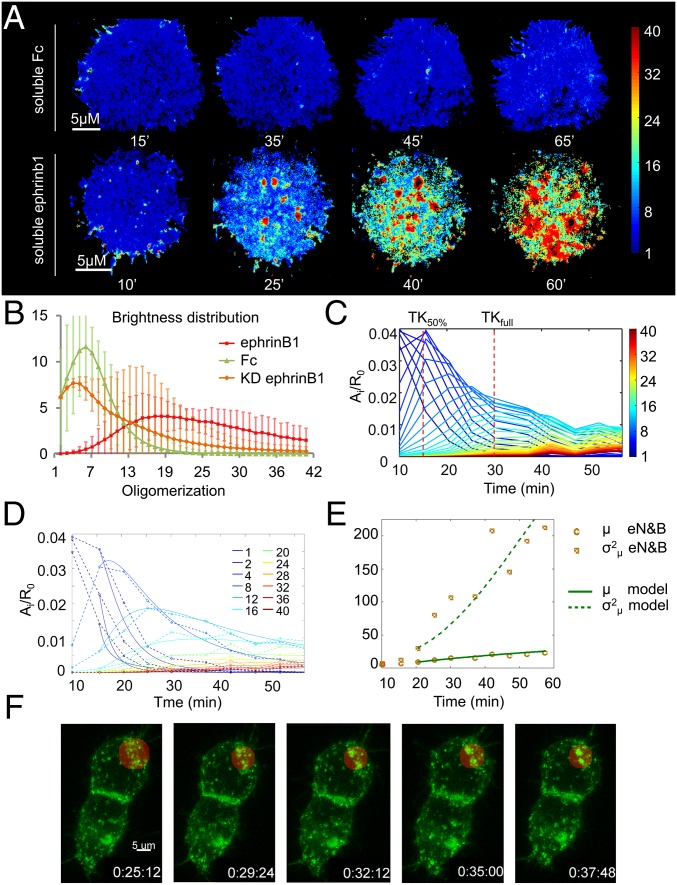
To show that the approach is valid for other means of ephrin presentation, we imaged cells that were stimulated by the addition of soluble preclustered ephrinB1. The soluble ligand, unlike the microprinted ephrin, has no restriction in mobility, allowing us to test the impact of ligand mobility, a feature shown to impact Eph receptor response (23, 27). Also, the soluble ligand bathes the entire surface of the cell, thus minimizing the effects of any unobservable receptor monomer populations, which can move from any unstimulated surfaces of the cells to the imaged microprinted surface and thereby obscure the impact of condensation (Fig. 3). eN&B revealed a qualitative difference in the temporal sequence of EphB2 oligomerization dynamics upon exposure to soluble ligand compared with microprinted ephrin. Oligomerization maps show aggregation as spatially heterogeneous, and a smaller number of larger clusters appeared (Fig. 3A). The average oligomerization of EphB2 cells stimulated with ephrinB1 for 60 min (Fig. 3B, i-mer = 21.7 ± 0.9) was significantly higher (P < 0.01) than cells stimulated with Fc (i-mer = 6.5 ± 0.4) or cells expressing KD EphB2 (i-mer = 10.6 ± 0.9). Despite the fact that the ligand concentration for the microprinted and soluble case are not directly comparable, the i-mer plot of Eph oligomerization using soluble ephrin (Fig. 3C) reveals a fast decay of the smaller oligomers and the sequential appearance of larger oligomers is less pronounced than in the microprinted case. Our model predicts a rapid depletion of the free monomer in the first minutes after ligand presentation to the entire cell surface (Fig. 3D). This reduction of available monomer and absence of a “hidden” reservoir shifts the clustering dynamics toward a strong dominance of condensation, which is reflected by the large separation between the curve of the variance of the cluster sizes and their average (μ) when using soluble ligand (Fig. 3E). Although most condensation events occur at the subpixel scale, some larger-scale events are observable with simple confocal microscopy (Fig. 3F and Movies S5–S7).
Fig. 3.
eN&B analysis of EphB2 clustering using soluble ligand stimulation. (A) Time-lapse oligomerization maps. HEK293T:EphB2_mRuby cells were stimulated with 0.2 μM Fc or 0.2 μM ephrinB1 in solution. The weighted average i-mer species is color-coded according to the color bar. Experiments for 0.2 μM Fc and 0.2 μM ephrinB1 were repeated 22 and 40 times, respectively (Movie S4). (B) Distribution of average and SD oligomerization values for multiple cells (NephrinB1 = 40, NFc = 22, NKD = 32) that have been presented with the relevant ligand for 60 min. (C) An i-mer evolution plot (Fig. 2 and SI Appendix). Ai, aggregate size i. R0, free receptor. The i-mer values are color-coded according to the color scale bar. TK50% and TKfull indicate time points 15 and 30 min, where 50% and the entire receptor population, respectively, is phosphorylated. (D) Mathematical model fitting of selected 12 species from C (SI Appendix). The dashed lines represent the experimental eN&B measurement, and the solid lines the model prediction (SI Appendix). (E) Mean (µ) and covariance (σ2) of the aggregate size for eN&B measurements and prediction of the polymerization–condensation model for ephrinB1 in solution. (F) Time-lapse 3D confocal reconstruction of HEK293T cells transfected with EphB2-GFP after stimulation (time = h:min:sec) with soluble ephrinB1 (Movies S5–S7). The red circle highlights Eph receptor clusters merging into larger aggregates.
Receptor Phosphorylation.
A dose–response curve was performed stimulating the cells with a 100-fold range of soluble ephrinB1 concentrations and measuring the Eph receptor response by Western blot densitometry of phosphorylated EphB2. The results revealed a uniform phosphorylation kinetics for all concentrations tested (Fig. 4A and SI Appendix, Fig. S5). The amount of phosphorylated receptor rapidly increased for 15 min, then slowed to an asymptote at ∼30 min poststimulation. The receptor response, however, was proportional to the ligand dose; the final amount of activation (phosphorylation) increased with larger concentrations of ephrinB1. Notably, these kinetics indicate that receptor activation and signaling primarily occur when low-order oligomers predominate (Fig. 3 C and D), implying that the condensation phase dominates after receptor activation. Moreover, the broad phosphorylation range of EphB2 (Fig. 4A) was confirmed for oligomerization measurements as well. Stimulating EphB2 with increasing ephrinB1 concentrations induced larger dynamic responses in aggregation as reported by eN&B analysis (Fig. 4B). These results suggest a direct link between oligomerization dynamics and receptor activation.
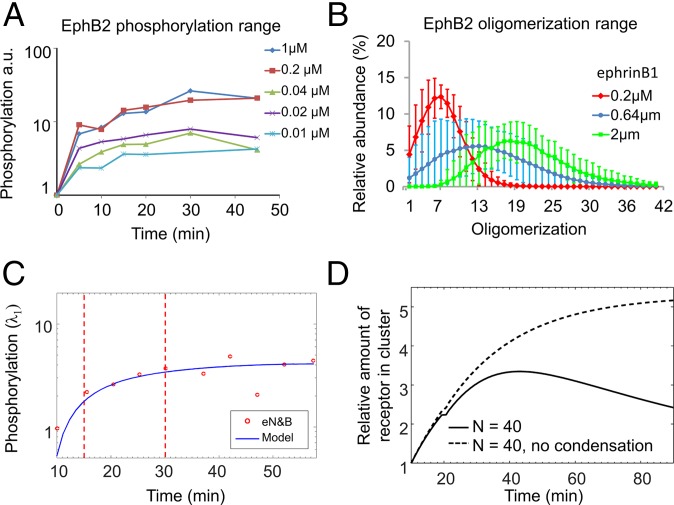
Fig. 4.
EphB2 activation kinetics. (A) EphB2 dose–response phosphorylation curve of cells stimulated with different ephrinB1 concentrations measured by Western blot densitometry. (B) EphB2 oligomerization range. Distribution of average and SD oligomerization values for multiple cells (N0.2 µM = 29, N0.64 µM = 37, N2 µM = 28) presented with the different ephrinB1 concentrations for 60 min. (C) Phosphorylation kinetics (blue line) from the cell in Fig. 3 A and C, as predicted from the polymerization–condensation model (SI Appendix). Red circles indicate the total monomer concentration obtained from eN&B. Vertical lines highlight 15- and 30-min time points. (D) The relative amount of receptors assembled in clusters (proportional to the receptor activation) was quantified for a truncation limit of n = 40-mer (SI Appendix, section 4.1) in the presence or absence of condensation.
We extended the polymerization–condensation model to predict EphB2 phosphorylation based on eN&B oligomerization data (SI Appendix, Dynamical Model). To do so we assume that tyrosine phosphorylation is mediated only by polymerization (the binding of free monomers to preexisting phosphorylated receptors) and not by condensation (11–13, 15). The model prediction shows good agreement with the monomer concentration calculated by eN&B (Fig. 4C) and also confirms the asymptotic kinetics reported by the Western blot measurements. Half-maximal or full (asymptotic) tyrosine kinase activation occur when 5-mers (maximum value after 15-min stimulation: TK50% = 5.2 ± 3.1) or 8-mers (maximum value after 30-min stimulation: TKfull = 8.5 ± 3.6), respectively, are the dominant species in the oligomer population. The later appearance of oligomers of 40-mers and beyond indicates that activation is decoupled from this high-order clustering.
Performing the simulations with and without condensation contributing to the dynamics offer ample evidence of the importance of condensation in the activation (Fig. 4D). For an example where 40-mer was the largest oligomer allowed to assemble before truncation (SI Appendix, Dynamical Model and Fig. S5), removing condensation from the system delayed the time to reach the maximum signal and increased the signal amplitude. These results suggest that with condensation, the time required to form large oligomers can be reduced (Fig. 5B), contributing to the signal adaptation and serving as a mechanism for dynamic range control.
Fig. 5.
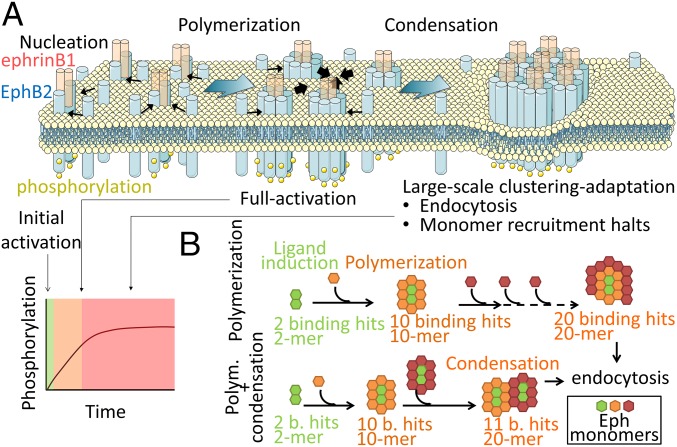
Schematic representation of the EphB2 polymerization–condensation model. (A) The model shows the nucleation of an EphB2 dimer upon interaction with ligand ephrinB1, which triggers the transactivation of the receptor. Lateral recruitment of receptors into low-order oligomers by polymerization (thin black arrows) leads to full activation. The coalescence of oligomers (condensation, thick black arrows) results in the formation of large-scale Eph aggregates, the recruitment of monomers slows down, and endocytosis leads to signal termination. (B) Condensation accelerates the formation of large aggregates. By introducing condensation, the same given size receptor cluster (i.e., 20-mer) can be assembled with fewer binding events compared with cluster growth by polymerization exclusively. Schematic modified with permission from https://servier.com/fr/accueil/ under Creative Commons license.
Discussion
The regulation of receptor dynamics is critical for the fidelity information flow in cell–cell communication (1). Previous studies have suggested that, in the absence of modulation, unlimited receptor clustering would amplify any given ligand input to the same maximum level of activation (3, 14, 28). Although highly sensitive, such transduction dynamics would seem to be unnecessarily slow because complete activation must await the assembly of large-scale clusters. Uncontrolled receptor clustering would also blunt the dynamic response of the receptor to integrate the information encoded in ligand gradients, because active signaling involves a winner-takes-all formation of high-order aggregates (29). To obtain a combination of sensitivity and range of response, the cell must control the degree to which the Eph receptor activity induced by the ligand can propagate toward free neighbor monomers.
We tackle the question on how receptor clustering dynamics can be regulated by using the eN&B analysis. This powerful tool allowed us to time-resolve the evolution of a wide spectrum of EphB2 species during ephrin-induced oligomerization, overcoming the previous limitation of measuring only the weighted averages of species (15, 24, 25). The fine-grained results enable fitting rich oligomerization data into standard biophysics models. The eN&B method offers unique space and time resolution and could be implemented to study different receptor and cellular responses, such as neuronal differentiation or immune response, induced by space-structured ligands (30).
The eN&B data can be largely explained by a Lumry–Eyring biophysics process of protein aggregation, in which polymerization and condensation run in parallel and combine with each other (24, 25). Activation takes place during nucleation and polymerization of monomers in the immediate 15–30 min following ephrin stimulation, reaching the maximum activation when pentamers to octamers predominate (Figs. 3C and 4). Similar timing has been reported for other receptor tyrosyne kinases (RTKs) (31). Moreover, previous studies using artificial dimerization of the Eph receptor suggested that complete activation can be reached without the assembly of large-scale clusters, in agreement with our measurements (15). After reaching maximal activation, our results show that aggregation of receptors is mainly driven by condensation of oligomers.
Our model also provides an explanation on how condensation can contribute to the receptor dynamic response to a broad range of ephrin concentrations (Fig. 4 B–D). The polymerization–condensation model suggests that coalescence of oligomers into larger aggregates reduces the overall recruitment of free monomers by accelerating the formation of large-scale (slow-diffusing) clusters and subsequent induction of endocytosis and signal termination (21) (Figs. 4D and 5B). Hence condensation can adapt signal propagation by dampening the lateral recruitment of free receptors, thus creating a fast and transitory response to the ligand (32, 33). This amplification adaptation strategy provides a simple mechanistic explanation on how receptor clustering combines the sensitivity and the dynamic range needed for the cell to respond the range of ephrin concentrations and gradients found in animal tissues (34, 35). Salaita et al. (23) demonstrated that high-order oligomerization plays a central role in cytoskeleton remodeling and cell invasiveness. We think our complementary models suggest a dual role of large Eph clusters as space concentrators of the signal (local cytoskeleton remodeling) and signal terminators (endocytosis induction).
Materials and Methods
Lentiviral Constructs.
mRuby was first excised from a pCDNA3.1 construct (36) using BamHI and BsrGI sites and cloned into pCDNA3_EphB2_GFP construct, a generous gift from the Rüdiguer Klein laboratory, Max Plank Institute of Neurobiology, Munich (21). Lentiviral constructs driving the expression of EhpB2_mRuby receptor were generated by cloning a PCR amplified cassette containing wild-type or mutated EhpB2_mRuby between BamHII and SalI sites of pLenti CMV Puro (Thermo Fisher). The KD receptor was first generated in the pCDNA3_EphB2_mRuby expression vector by amplification of the whole vector containing the wild-type EhpB2_mRuby construct with specific 5′ phosphorylated primers designed to generate an A-to-G point mutation (KD-EhpB2_mRuby) in the EhpB2_mRuby receptor: KD-FW_℗-ATGACCCCAGGCATGAGGATCTATATAGATCCT; KD_RV_℗-AGGATCTATATAGATCCTCATGCCTGGGGTCAT. Then wild-type or mutated EhpB2mRuby were amplified from pCDNA3_EhpB2_mRuby vector with the following primers: FW_CGCGGGCCCGGGATCCGCCACCATGAACTTTATCCCAGTCGA; RV_ GAGGTTGATTGTCGACTCAAACCTCTACAGACTGG. PCR products were cloned into pLenti CMV Puro using In-Fusion HD Cloning Kit (Clontech). EphB2 constructs and membrane GFP were kind gifts from the Klein laboratory and Le Trihn, University of Southern California, Los Angeles, respectively. Lentiviruses were a kind gift from Rusty Lansford, University of Southern California, Los Angeles.
Production of Lentiviruses.
HEK293T cells (standard cell line in the field; Thermo Fisher) (13) were grown on gelatin-coated plates and transfected with pLenti.CMV:EphB2_mRuby using Lipofectamine 2000 along with the ViraPower Lentiviral Packaging Mix (Thermo Fisher) according to the manufacturer’s protocol. Supernatants were collected 48 and 72 h after transfection, pulled together, filtered at 0.45 µm, and ultracentrifuged at 50,000 × g for 2 h at 4 °C to obtain virus concentration.
Lentiviral Transduction and Cell Lines.
The 1 × 106 HEK293T cells were infected in suspension and then plated in 10-cm plate. After two passages, the cells were infected a second time following the same protocol. After two additional passages the cells were trypsinized, and mRuby-positive cells were selected by cell sorting. Two lines were generated: HEK293T:EphB2_mRuby and HEK293T:KD-EphB2_mRuby. All cells lines were routinely tested for mycoplasma by real-time PCR. The plasmid pCS2-eGFP-CtermHras encodes for a GFP targeted to the membrane by the fusion to the C-terminal domain of HRas.
Western Blot.
Protein extracts, separated by SDS/PAGE and transferred onto PVDF membranes, were probed with antibodies against anti-phosphoY594-Eph receptor B1/B2 (ab61791, 1:500; Abcam) or actin (A1978, 1:5,000; Sigma) or anti-EphB2 (AF467, 1:2,000; R&D) (SI Appendix, Fig. S5). Proteins of interest were detected with anti-rabbit IgG antibody (NA934, 1:10,000; GE Healthcare) or anti-mouse IgG antibody (NA931, 1:5,000; GE Healthcare) or anti-goat IgG antibody (P0160, 1:2,000; Dako) and visualized with the Amersham ECL Western blotting detection reagents (RPN2209; GE Healthcare), according to the provided protocol.
Ethics Statement.
The experiments presented in this study were conducted following protocols approved by the Institutional Review Board of the Center of Regenerative Medicine in Barcelona.
Supplementary Material
Acknowledgments
The authors thank Giulia Ossato, William Dempsey, and Nicolas Plachta for useful discussions, and acknowledge the Nikon Center of Excellence at ICFO-The Institute of Photonic Sciences. S.O. was supported by Marie Curie International Outgoing Fellowship 276282 within the EU Seventh Framework Programme FP7/2007-2013 and Postdoctoral Fellowships LT000109/2011 from the Human Frontier Science Program Organization and EX2009-1136 from the Ministerio de Educación, Programa Nacional de Movilidad de Recursos Humanos del Plan Nacional de I-D+i 2008-2011. F.C. was supported by grants from the Moore Foundation and NIH (R01 HD075605 and R01 OD019037). J.J.O. acknowledges financial support from ICFONEST+, funded by the Marie Curie COFUND (FP7-PEOPLE-2010-COFUND) action of the European Commission and by the MINECO Severo Ochoa action at ICFO-The Institute of Photonic Sciences. Additional funding was provided by the Generalitat de Catalunya (2014-SGR-1442 and 2014-SGR-1460); the Spanish Ministry of Economy and Competitiveness (SAF2015-69706-R, MINAHE5, TEC2014-51940-C2-2-R, SEV-2015-0522); Instituto de Salud Carlos III (ISCIII)/FEDER (RD16/0011/0024); the European Union (GLAM project GA-634928; System’s Microscopy Network of Excellence consortium FP-7-HEALTH.2010.2.1.2.2); the European Research Council (337191-MOTORS and 647863-COMIET); the Fundació Privada Cellex; and CERCA Programme/Generalitat de Catalunya.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1713564114/-/DCSupplemental.
References
- 1.Lauffenburger DA, Linderman J. Receptors: Models for Binding, Trafficking, and Signaling. Oxford Univ Press; New York: 1993. [Google Scholar]
- 2.Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Endres RG. Physical Principles in Sensing and Signaling: With an Introduction to Modeling in Biology. Oxford Univ Press; Oxford: 2013. [Google Scholar]
- 4.Rohani N, Parmeggiani A, Winklbauer R, Fagotto F. Variable combinations of specific ephrin ligand/Eph receptor pairs control embryonic tissue separation. PLoS Biol. 2014;12:e1001955. doi: 10.1371/journal.pbio.1001955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kulesa PM, Fraser SE. Cell dynamics during somite boundary formation revealed by time-lapse analysis. Science. 2002;298:991–995. doi: 10.1126/science.1075544. [DOI] [PubMed] [Google Scholar]
- 6.Klein R. Eph/ephrin signalling during development. Development. 2012;139:4105–4109. doi: 10.1242/dev.074997. [DOI] [PubMed] [Google Scholar]
- 7.Genander M, Frisén J. Ephrins and Eph receptors in stem cells and cancer. Curr Opin Cell Biol. 2010;22:611–616. doi: 10.1016/j.ceb.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 8.Sheffler-Collins SI, Dalva MB. EphBs: An integral link between synaptic function and synaptopathies. Trends Neurosci. 2012;35:293–304. doi: 10.1016/j.tins.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Day B, et al. Three distinct molecular surfaces in ephrin-A5 are essential for a functional interaction with EphA3. J Biol Chem. 2005;280:26526–26532. doi: 10.1074/jbc.M504972200. [DOI] [PubMed] [Google Scholar]
- 10.Nikolov DB, Xu K, Himanen JP. Eph/ephrin recognition and the role of Eph/ephrin clusters in signaling initiation. Biochim Biophys Acta. 2013;1834:2160–2165. doi: 10.1016/j.bbapap.2013.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Himanen JP, et al. Architecture of Eph receptor clusters. Proc Natl Acad Sci USA. 2010;107:10860–10865. doi: 10.1073/pnas.1004148107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seiradake E, Harlos K, Sutton G, Aricescu AR, Jones EY. An extracellular steric seeding mechanism for Eph-ephrin signaling platform assembly. Nat Struct Mol Biol. 2010;17:398–402. doi: 10.1038/nsmb.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wimmer-Kleikamp SH, Janes PW, Squire A, Bastiaens PI, Lackmann M. Recruitment of Eph receptors into signaling clusters does not require ephrin contact. J Cell Biol. 2004;164:661–666. doi: 10.1083/jcb.200312001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bray D, Levin MD, Morton-Firth CJ. Receptor clustering as a cellular mechanism to control sensitivity. Nature. 1998;393:85–88. doi: 10.1038/30018. [DOI] [PubMed] [Google Scholar]
- 15.Schaupp A, et al. The composition of EphB2 clusters determines the strength in the cellular repulsion response. J Cell Biol. 2014;204:409–422. doi: 10.1083/jcb.201305037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Digman MA, Dalal R, Horwitz AF, Gratton E. Mapping the number of molecules and brightness in the laser scanning microscope. Biophys J. 2008;94:2320–2332. doi: 10.1529/biophysj.107.114645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Unruh JR, Gratton E. Analysis of molecular concentration and brightness from fluorescence fluctuation data with an electron multiplied CCD camera. Biophys J. 2008;95:5385–5398. doi: 10.1529/biophysj.108.130310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith EM, Macdonald PJ, Chen Y, Mueller JD. Quantifying protein-protein interactions of peripheral membrane proteins by fluorescence brightness analysis. Biophys J. 2014;107:66–75. doi: 10.1016/j.bpj.2014.04.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hellriegel C, Caiolfa VR, Corti V, Sidenius N, Zamai M. Number and brightness image analysis reveals ATF-induced dimerization kinetics of uPAR in the cell membrane. FASEB J. 2011;25:2883–2897. doi: 10.1096/fj.11-181537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trullo A, Corti V, Arza E, Caiolfa VR, Zamai M. Application limits and data correction in number of molecules and brightness analysis. Microsc Res Tech. 2013;76:1135–1146. doi: 10.1002/jemt.22277. [DOI] [PubMed] [Google Scholar]
- 21.Zimmer M, Palmer A, Köhler J, Klein R. EphB-ephrinB bi-directional endocytosis terminates adhesion allowing contact mediated repulsion. Nat Cell Biol. 2003;5:869–878. doi: 10.1038/ncb1045. [DOI] [PubMed] [Google Scholar]
- 22.Hur KH, Mueller JD. Quantitative brightness analysis of fluorescence intensity fluctuations in E. Coli. PLoS One. 2015;10:e0130063. doi: 10.1371/journal.pone.0130063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salaita K, et al. Restriction of receptor movement alters cellular response: Physical force sensing by EphA2. Science. 2010;327:1380–1385. doi: 10.1126/science.1181729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y, Roberts CJ. Lumry-eyring nucleated-polymerization model of protein aggregation kinetics. 2. Competing growth via condensation and chain polymerization. J Phys Chem B. 2009;113:7020–7032. doi: 10.1021/jp8083088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pallitto MM, Murphy RM. A mathematical model of the kinetics of beta-amyloid fibril growth from the denatured state. Biophys J. 2001;81:1805–1822. doi: 10.1016/S0006-3495(01)75831-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Egea J, et al. Regulation of EphA 4 kinase activity is required for a subset of axon guidance decisions suggesting a key role for receptor clustering in Eph function. Neuron. 2005;47:515–528. doi: 10.1016/j.neuron.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 27.Lohmüller T, Xu Q, Groves JT. Nanoscale obstacle arrays frustrate transport of EphA2-Ephrin-A1 clusters in cancer cell lines. Nano Lett. 2013;13:3059–3064. doi: 10.1021/nl400874v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Zon JS, Kienle S, Huelsz-Prince G, Barkoulas M, van Oudenaarden A. Cells change their sensitivity to an EGF morphogen gradient to control EGF-induced gene expression. Nat Commun. 2015;6:7053. doi: 10.1038/ncomms8053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Verveer PJ, Wouters FS, Reynolds AR, Bastiaens PI. Quantitative imaging of lateral ErbB1 receptor signal propagation in the plasma membrane. Science. 2000;290:1567–1570. doi: 10.1126/science.290.5496.1567. [DOI] [PubMed] [Google Scholar]
- 30.Conway A, et al. Multivalent ligands control stem cell behaviour in vitro and in vivo. Nat Nanotechnol. 2013;8:831–838. doi: 10.1038/nnano.2013.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Regot S, Hughey JJ, Bajar BT, Carrasco S, Covert MW. High-sensitivity measurements of multiple kinase activities in live single cells. Cell. 2014;157:1724–1734. doi: 10.1016/j.cell.2014.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ferguson SS, Caron MG. G protein-coupled receptor adaptation mechanisms. Semin Cell Dev Biol. 1998;9:119–127. doi: 10.1006/scdb.1997.0216. [DOI] [PubMed] [Google Scholar]
- 33.Endres RG, Wingreen NS. Precise adaptation in bacterial chemotaxis through “assistance neighborhoods”. Proc Natl Acad Sci USA. 2006;103:13040–13044. doi: 10.1073/pnas.0603101103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lang S, von Philipsborn AC, Bernard A, Bonhoeffer F, Bastmeyer M. Growth cone response to ephrin gradients produced by microfluidic networks. Anal Bioanal Chem. 2008;390:809–816. doi: 10.1007/s00216-007-1363-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gebhardt C, Bastmeyer M, Weth F. Balancing of ephrin/Eph forward and reverse signaling as the driving force of adaptive topographic mapping. Development. 2012;139:335–345. doi: 10.1242/dev.070474. [DOI] [PubMed] [Google Scholar]
- 36.Kredel S, et al. mRuby, a bright monomeric red fluorescent protein for labeling of subcellular structures. PLoS One. 2009;4:e4391. doi: 10.1371/journal.pone.0004391. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.