Significance
Kinetochores are large protein networks located on centromeres that mediate chromosome segregation during mitosis and maintain genomic stability. Mis12 complex (Mis12C) functions as a scaffold that targets Ndc80 and Knl1 complexes to the centromere by associating with CENP-C. Here, we provide insights into the molecular mechanism underlying the CENP-C–dependent kinetochore recruitment of Mis12C, which is negatively regulated by Aurora B-dependent CENP-C phosphorylation. Replacement of Schizosaccharomyces pombe Cnp3 with a phosphorylation-mimicking mutant, Cnp3T28E, results in defective chromosome segregation caused by improper kinetochore assembly. These findings indicate that Aurora B-dependent phosphorylation of CENP-C plays a role in interrupting the connection between the inner and outer kinetochore and is thus involved in the error correction/spindle assembly checkpoint pathway to prevent chromosome missegregation during mitosis.
Keywords: CENP-C, Mis12, Aurora B, phosphorylation, error correction
Abstract
Kinetochores are superprotein complexes that orchestrate chromosome segregation via a dynamic interaction with spindle microtubules. A physical connection between CENP-C and the Mis12–Ndc80–Knl1 (KMN) protein network is an important pathway that is used to assemble kinetochores on CENP-A nucleosomes. Multiple outer kinetochore components are phosphorylated by Aurora B kinase to activate the spindle assembly checkpoint (SAC) and to ensure accurate chromosome segregation. However, it is unknown whether Aurora B can phosphorylate inner kinetochore components to facilitate proper mitotic chromosome segregation. Here, we reported the structure of the fission yeast Schizosaccharomyces pombe Mis12–Nnf1 complex and showed that N-terminal residues 26–50 in Cnp3 (the CENP-C homolog of S. pombe) are responsible for interacting with the Mis12 complex. Interestingly, Thr28 of Cnp3 is a substrate of Ark1 (the Aurora B homolog of S. pombe), and phosphorylation impairs the interaction between the Cnp3 and Mis12 complex. The expression of a phosphorylation-mimicking Cnp3 mutant results in defective chromosome segregation due to improper kinetochore assembly. These results establish a previously uncharacterized regulatory mechanism involved in CENP-C–Mis12-facilitated kinetochore attachment error correction to ensure accurate chromosome segregation during mitosis.
Kinetochores, large protein complexes that assemble on centromeres, connect chromosomes to spindle microtubules (1). The structure of kinetochores consists of an inner and an outer layer. The CCAN (constitutive centromere-associated network) complex resides in the kinetochore inner layer, while the KMN (Knl1–Mis12–Ndc80) protein network forms the outer layer. The CCAN complex is composed of 16 protein subunits and functions by forming different subcomplexes. Although the function of each CCAN subcomplex within the kinetochore needs to be further investigated, it is well established that the CCAN complex is the key linker responsible for the assembly of the KMN network in the kinetochore outer layer (2). The KMN network is composed of three subcomplexes: Knl1C (Knl1 and Zwint), Mis12C (Mis12, Nnf1, Dsn1, and Nsl1), and Ndc80C (Ndc80, Nuf2, Spc24, and Spc25) (3). Two molecular mechanisms have been demonstrated to be responsible for the assembly of the KMN network. One mechanism shows that CENP-T, within the CCAN complex, directly interacts with Ndc80C, and the other mechanism shows that CENP-C, also within the CCAN complex, recruits Ndc80C to kinetochores by physically interacting with Mis12C (4, 5). A short fragment located in the N terminus of CENP-C is responsible for its interaction with Mis12C (6, 7). In addition, the phosphorylation of Dsn1 by Aurora B promotes the recruitment of Mis12C by associating with CENP-C to assemble functional kinetochores (8–10).
Faithful chromosome segregation during mitosis requires the accurate interaction of kinetochores with microtubules in a bipolar manner (biorientation), which is established by the stable attachment of kinetochores in each sister chromatid to spindle microtubules from opposite poles (3). The KMN network of the outer kinetochore can capture dynamic microtubules to support the stable “end-on” attachment between kinetochores and microtubules. Both Ndc80C and Knl1C in the KMN network have basic surfaces that bind acidic tubulins via electrostatic interactions (11, 12). Mis12C functions as a platform to coordinate the assembly of the KMN network by bridging Ndc80C and Knl1C. Although Mis12C is unable to interact with microtubules directly, additional Mis12C, present in a complete KMN network, synergistically enhances its binding ability to microtubules (12, 13).
However, erroneous attachments between kinetochores and microtubules frequently occur during early mitosis, giving rise to chromosome segregation defects (14). To prevent such catastrophic consequences, eukaryotes have evolved a sophisticated system, including error correction (EC) and spindle assembly checkpoint (SAC) pathways, to monitor and correct aberrant kinetochore–microtubule attachments that occur during the establishment of kinetochore biorientation (15, 16). The EC pathway serves as a “local” mechanism to selectively stabilize correct attachments between kinetochores and microtubules and to weaken erroneous attachments between them (16). The SAC pathway can extend into “global” signals from unattached or incorrectly attached kinetochores, leading to anaphase delay so that all sister chromatids reach the “bi-orientation” state (15).
Aurora B kinase is one of the essential components of the aforementioned pathways. This kinase localizes to the centromere to facilitate the establishment of accurate kinetochore–microtubule attachments (16). It has been shown that Aurora B destabilizes kinetochore–microtubule attachments by phosphorylating multiple kinetochore proteins that reside in the outer layer of kinetochores (17). For example, phosphorylation of Ndc80 by Aurora B kinase weakens its microtubule binding affinity, and phosphorylation of the components within the Knl1C and Mis12C complexes by Aurora B also disrupts the interaction between kinetochores and microtubules (11, 12, 18, 19). Additional outer kinetochore-associated factors are phosphorylated by Aurora B to cooperate with the KMN network to modulate the kinetochore–microtubule attachment. For example, both the Dam1 complex in budding yeast and the Ska (spindle and kinetochore-associated) complex in higher eukaryotes are located on the outer kinetochore to interact with Ndc80C and to stabilize the kinetochore–microtubule attachment. Phosphorylation of these complexes by Aurora B negatively regulates their binding to Ndc80C and kinetochore–microtubule attachment (20, 21). Aurora B also regulates the localization and activity of MCAK (mitotic centromere-associated kinesin, also known as Kif2c), which is a microtubule depolymerase that plays important roles in the EC pathway (22). Although Aurora B phosphorylates various outer kinetochore proteins to destabilize kinetochore–microtubule attachment and to promote the repair of erroneous kinetochore–microtubule attachment, none of the inner kinetochore components has yet to be shown to modulate the interaction of kinetochores with microtubules involved in the correction of aberrant kinetochore–microtubule attachment in an Aurora B-dependent manner.
In this study, we determined the crystal structure of the Mis12–Nnf1 complex in the fission yeast S. pombe [Protein Data Bank (PDB) ID code 5WWL]. A minimal region containing residues 26–50 of Cnp3 (the CENP-C homolog of S. pombe) was mapped and found to bind directly to the Mis12–Nnf1 complex. Furthermore, we identified Thr28 as the Ark1 (the Aurora B homolog of S. pombe) phosphorylation site in Cnp3, which significantly disrupts the interaction between Cnp3 and Mis12C. In vivo experiments showed that a phosphorylation-mimicking mutation of Cnp3, T28E, decreases the kinetochore localization of Mis12C and results in delayed and defective chromosome segregation during mitosis. In addition to the substrates located on the outer kinetochore, our findings showed that Aurora B phosphorylates the inner kinetochore component CENP-C to destabilize kinetochore–microtubule attachment at the interface between the inner and outer kinetochore, implying that Aurora B-dependent phosphorylation of CENP-C may be involved in the correction of erroneous kinetochore–microtubule attachment.
Results
Overall Structure of Mis12–Nnf1.
To understand the molecular mechanism by which CENP-C–dependent kinetochores are located on Mis12C, we attempted to obtain crystals of Mis12C–Cnp3, Mis12C, and Mis12–Nnf1 complexes from S. pombe. Unfortunately, no crystals of the Mis12C–Cnp3 or the Mis12C complex were obtained that were suitable for X-ray diffraction. We finally obtained a crystal of the stable Mis12–Nnf1 core complex containing Mis12 (1–215)–Nnf1 (1–175) (named MNC). The MNC crystals diffracted to a resolution of 2.4 Å, and the phase was determined using the SAD (single-wavelength anomalous dispersion) method. Model building and structure refinement were carried out as described in Materials and Methods. X-ray diffraction data and structure refinement statistics are shown in Table 1.
Table 1.
Data collection and refinement statistics
| Statistics | MNC, PDB ID code 5WWL |
| Data collection | |
| Space group | P21212 |
| Unit cell parameters | |
| a, b, c, Å | 41.5, 85.61, 123.79 |
| α, β, γ, ° | 90, 90, 90 |
| Resolution, Å | 50–2.4 (2.55–2.4)* |
| Rmerge,† % | 8.9 (39.2) |
| I/σI | 10.4 (3.3) |
| Wilson plot B factors, Å2 | 36.9 |
| Completeness, % | 96.3 (91.9) |
| Redundancy | 5.0 (4.9) |
| Refinement | |
| Resolution, Å | 50.0–2.4 |
| No. reflections | 16,402 |
| Rwork‡/Rfree,§ % | 23.26/27.34 |
| rmsd | |
| Bond lengths, Å | 0.014 |
| Bond angles, ° | 1.56 |
| B factors, Å2 | |
| Protein | 51.97 |
| Water | 42.43 |
| Ramachandran plot¶ | |
| Most favored regions, % | 95.1 |
| Allowed regions, % | 4.9 |
| Outliers, % | 0 |
Each structure was determined from a single crystal through a helical data-collection method.
The values in parentheses refer to statistics in the highest shell.
Rsym = |Ii − <I>|/|Ii|, where Ii is the intensity of the ith measurement and <I> is the mean intensity for that reflection.
Rwork = |FP − FP(calc)|/FP.
Rfree was calculated with 5.1% of the reflections in the test set.
Statistics for the Ramachandran plot from an analysis using MolProbity.
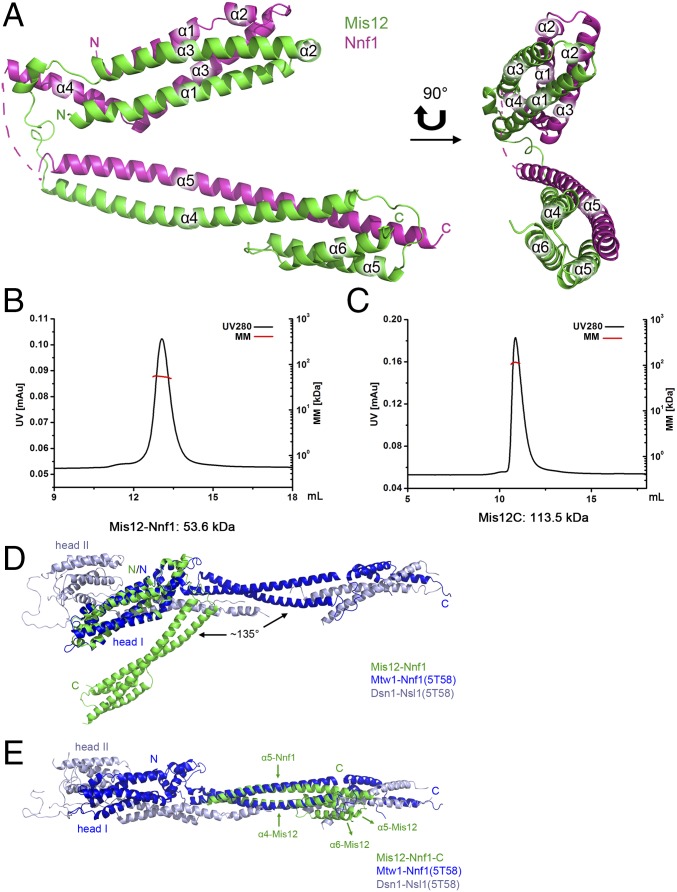
The crystallographic studies showed that the asymmetric unit contains one MNC complex. The fragments containing residues 90–105 and 173–175 of Nnf1 are not included in the final structure model because they are flexible and invisible in the electron density map. Mis12 and Nnf1 of MNC consist of six and five α-helices, respectively, which tightly pack against each other to form a “V”-shaped heterodimer. The V-shaped structure of MNC can be divided into two helix bundles (Fig. 1A): The N-terminal seven-helix bundle is composed of the helix α1–α3 of Mis12 and helix α1–α4 of Nnf1, while the helix α4–α6 of Mis12 and helix α5 of Nnf1 form a four-helix buddle at the C terminus. The buried surface area of the interface between Mis12 and Nnf1 is ∼4,549 Å2. Nineteen hydrogen bonds at the interface provide a strong interaction for the two subunits. Hydrophobic interactions and salt bridges also make significant contributions to stabilize the heterodimer.
Fig. 1.
Structure of MNC and stoichiometry of Mis12–Nnf1 and Mis12C components. (A) Ribbon model of MNC in two orientations. The Mis12 and Nnf1 are colored green and magenta, respectively. Residues 90–105 of Nnf1 missing in the final model are indicated by the dotted line. (B and C) Molecular masses of the Mis12–Nnf1 complex (B) and Mis12C (C) measured by SEC–MALS. Chromatograms show elution profiles measured in UV280 and calculated molecular mass (MM). The x axis shows elution volume in milliliters. (D) Comparison of our Mis12–Nnf1 subcomplex structure with K. lactis MIND-C1 structure (5T58). (E) Comparison of the C-terminal part of our Mis12–Nnf1 subcomplex structure with the K. lactis MIND–C1 structure (5T58). Mis12–Nnf1, Mtw1–Nnf1(5T58), and Dsn1–Nsl1(5T58) are colored green, blue, and gray, respectively.
Mis12 and Nnf1 were observed to form a heterodimer in the crystal structures, but a size exclusion chromatography (SEC) experiment indicated that there are two copies of each subunit in the Mis12–Nnf1 complex with an apparent molecular mass of 122.0 kDa versus a theoretical molecular mass of 53.9 kDa. The apparent molecular mass of Mis12C (244.0 kDa) compared with the theoretical molecular mass (114.8 kDa) also indicates that there is a dimer of Mis12C tetramers (Fig. S1). To clarify the oligomerization state and stoichiometry of the recombinant protein complexes, we performed SEC–MALS (multiangle light scattering) analysis of Mis12–Nnf1 and Mis12C. The molecular masses of the Mis12–Nnf1 subcomplex and Mis12C, determined by SEC–MALS, are 53.6 kDa and 113.5 kDa, respectively, indicating that each subunit is represented once in the purified protein complexes (Fig. 1 B and C and Table 2). The larger apparent molecular mass of Mis12C, estimated by the SEC method, suggests that Mis12C from S. pombe may adopt an elongated structure. This finding is consistent with previous reports of Mis12C from humans (23, 24), budding yeast (25), and Drosophila melanogaster (26, 27).
Table 2.
Molecular masses and stoichiometry of complexes measured by SEC–MALS
| Complex | Theoretical mass, kDa | Mass calculated by SEC–MALS, kDa | Stoichiometry |
| Mis12–Nnf1 | 53.9 | 53.6 | 1:1 |
| Mis12C | 114.8 | 113.5 | 1:1:1:1 |
During preparation of this manuscript, the structures of the human and budding yeast Kluyveromyces lactis Mis12/MIND complex were reported (28, 29). These two Mis12 complexes adopt a fold similar to each other by forming a long Y-shaped structure that is ∼200 Å in length along the long axis. The N-terminal parts of the Mis12–Nnf1 subcomplex and the Dsn1–Nsl1 subcomplex are termed “head I” and “head II,” respectively. Structural comparison of the S. pombe Mis12–Nnf1 complex with the two Mis12C/MIND complexes revealed that the helix buddle 1 of the S. pombe Mis12–Nnf1 complex can be overlaid on head I of both the human and K. lactis Mis12C/MIND complex (Fig. 1D). As for the helix buddle 2 of the S. pombe Mis12–Nnf1 complex, only helix α4 of Mis12 and helix α5 of Nnf1 can be superimposed well onto the corresponding helices in the Mis12C/MIND complex after a rotation of ∼135° relative to helix bundle 1 (Fig. 1 D and E). The helix α5 and helix α6 of Mis12 fold back to interact with helix α4 of Mis12 and helix α5 of Nnf1 in the S. pombe Mis12–Nnf1 complex, while in human and K. lactis Mis12C/MIND complexes, the corresponding helices extend out to associate with the C-terminal helices of Dsn1 and Nsl1 (Fig. 1E). The different conformation of the S. pombe Mis12–Nnf1 structure compared with those of K. lactis and human Mis12C might due to the flexibility of Mis12–Nnf1 in the absence of Dsn1–Nsl1.
The N-Terminal Region of Cnp3 Binds to a Negatively Charged Surface of the Mis12 Complex Through Electrostatic Interactions.
CENP-C is a conserved inner kinetochore protein that is required for kinetochore localization of Mis12C by directly interacting with Mis12C (30, 31). Human CENP-C tightly binds Mis12C in a 1:1 ratio at the N terminus (6). Further research defined an N-terminal fragment (∼20 residues) of CENP-C that mediates the interaction with Mis12C in humans (6) and Drosophila (26). However, the amino acid sequence of the N-terminal region of CENP-C is not conserved among other species. Meanwhile, characterization of the subunit of S. pombe Mis12C responsible for conducting interactions with CENP-C remains to be researched. To address these questions, we attempted to map the binding site of Cnp3 (CENP-C homolog of S. pombe) for Mis12–Nnf1.
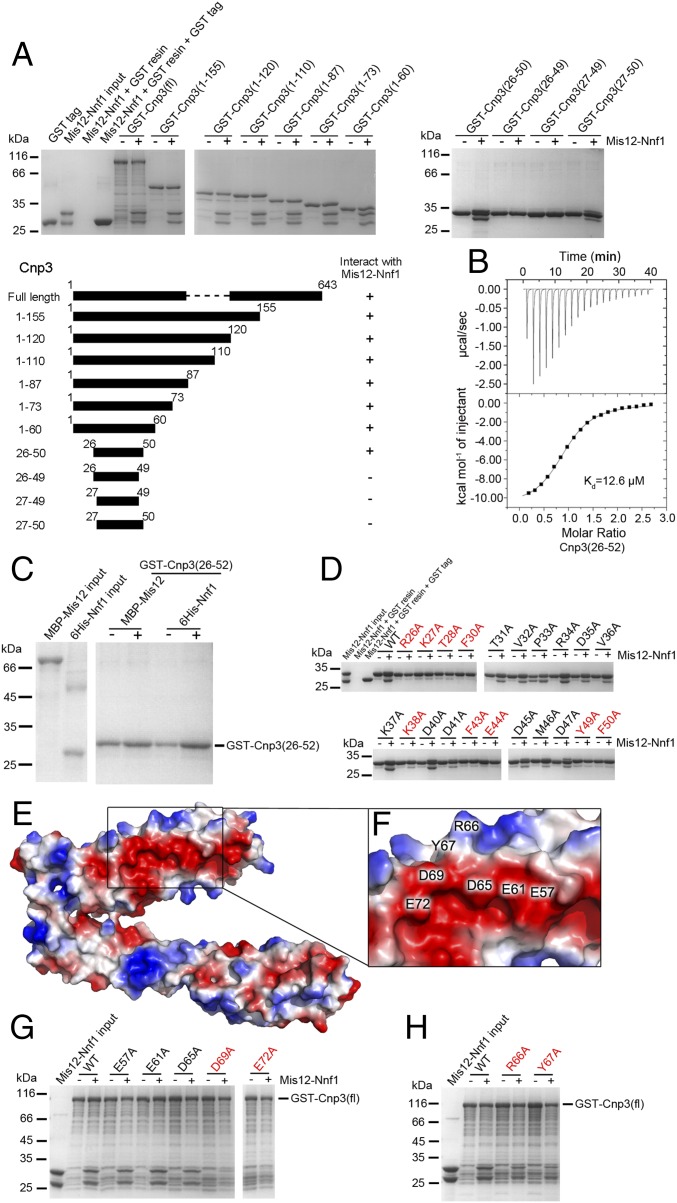
We used GST pull-down assays to confirm the interaction between Mis12–Nnf1 and full-length Cnp3 [Cnp3(fl)] (Fig. 2A). Next, we used several truncations of Cnp3 of different lengths to test its ability to bind Mis12–Nnf1. The results showed that the shortest fragment of Cnp3 tested, containing 25 amino acid residues (26–50), is sufficient to interact with Mis12-Nnf1 (Fig. 2A and Fig. S2). The binding of a synthesized Cnp3(26–52) peptide (Beijing Scilight Biotechnology Ltd. Co.) to Mis12–Nnf1 was verified by isothermal titration calorimetry (ITC) assays, and the measured dissociation constant (Kd) value was determined to be 12.6 μM (Fig. 2B). These results validate that the interaction between the N-terminal region of CENP-C and Mis12C is conserved in S. pombe and that the Mis12–Nnf1 complex alone is sufficient to bind directly to Cnp3(26–50) with high affinity.
Fig. 2.
N-terminal region of Cnp3 binds to a negatively charged surface of Mis12 through electrostatic interactions. (A) GST pull-down assays of Mis12–Nnf1 with different fragments of Cnp3 fused to GST tag (Top). Cnp3 fragments used for GST pull-down assays are indicated in the schematic diagram (Bottom). (B) The binding affinity of Mis12–Nnf1 with Cnp3(26–52) peptide analyzed by ITC. (C) GST pull-down assays of GST-Cnp3(26–52) with Mis12 (fused to MBP tag) and Nnf1 (fused to His tag) individually. (D) GST pull-down assays of Mis12–Nnf1 with Cnp3(26–52) bearing different point mutations. (E) Electrostatic surface depiction of MNC. (F) An enlarged view of the negatively charged groove on the MNC surface with key residues (E57, E61, D65, D69, E72, R66, and Y67) labeled in black. Surfaces with positive or negative electrostatic potential are colored blue or red, respectively. (G and H) GST pull-down assays of GST-Cnp3(fl) with mutants of Mis12–Nnf1 bearing point mutations of the residues shown in F to alanine. The mutants with decreased binding affinity to Cnp3 are shown in red. The grouping blots in A, D, and G were cropped from different gels and separated with white spaces. Two grouping blots in C were different parts cropped from the same gel.
Some studies showed that Drosophila CENP-C interacts with either Nnf1 (32) or Mis12 (26) individually, while others suggested that both Mis12 and Nnf1 are required for CENP-C binding (27). To elucidate the mechanism of CENP-C interaction with Mis12C in fission yeast, we overexpressed and purified S. pombe Mis12 and Nnf1 individually and performed in vitro pull-down assays with GST-Cnp3(26–52). We found that neither Mis12 nor Nnf1 could interact with Cnp3 alone, suggesting that both are required for Cnp3 recognition (Fig. 2C).
To further identify the interaction interface between Cnp3 and the Mis12–Nnf1 complex, we mutated all of the residues of Cnp3(26–52) to alanine individually, except alanine and glycine residues, to evaluate their binding ability to Mis12–Nnf1 using GST pull-down assays. As shown in Fig. 2D, nine residues (R26, K27, T28, F30, K38, F43, E44, Y49, and F50) of Cnp3(26–52) were identified to be crucial for the interaction. Three of these residues are positively charged, which raises the probability that Cnp3(26–52) binds a negatively charged pocket of Mis12–Nnf1. By analyzing the structure of MNC, we found a negatively charged surface area on the surface of the Mis12 subunit, which includes residues E57, E61, D65, D69, and E72 (Fig. 2 E and F). Analysis of the MNC structure suggested that the surface composed of acidic residues on Mis12 may be the binding site for Cnp3. As expected, mutagenesis combined with pull-down assays revealed that mutations D69A and E72A of Mis12 significantly reduce the binding affinity of Mis12–Nnf1 to Cnp3(26–52) (Fig. 2G). In addition, mutations R66A and Y67A of Mis12, which are located adjacent to the negatively charged pocket, decrease the binding of Mis12–Nnf1 to Cnp3(26–52). This result further confirms that negatively charged residues D69–E72 on the Mis12 surface are required for Cnp3(26–52) binding (Fig. 2H). Our findings suggest that the N terminus of Cnp3 binds to a negatively charged pocket of Mis12 mainly through electrostatic interactions.
Ark1 Phosphorylates Thr28 in Cnp3.
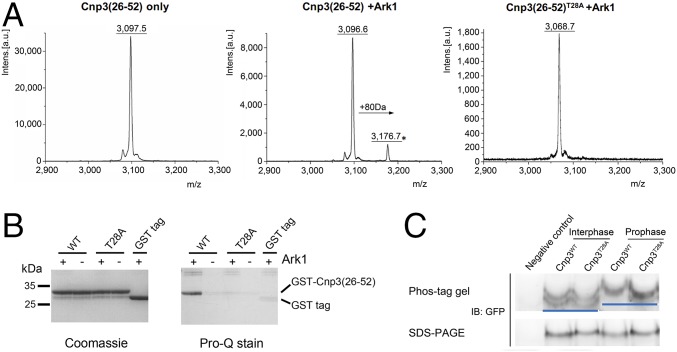
Previous work revealed that budding yeast CENP-C is phosphorylated by Aurora B (33); thus, we attempted to determine whether Cnp3(26–50) contains the potential Aurora B phosphorylation site by using the GPS webserver (34). According to the result, three residues of Cnp3(26–50)—namely, T28, T31, and Y49—could be possible phosphorylation sites. The sequence of residues 26–29 is RKTG, which is consistent with the consensus motif required by Aurora B: [RK]X[TS] (X is any amino acid residue) (35). In addition, Aurora B is a conserved kinase located at the centromere before anaphase and colocalized with CENP-C. Based on this analysis, we proposed that Thr28 of Cnp3 is a potential phosphorylation site for Aurora B. To verify this hypothesis, Ark1 (Aurora B homolog in S. pombe) was purified and incubated with the Cnp3(26–52) peptide. The reaction mixture was analyzed by MALDI-TOF MS. As expected, the Cnp3(26-52) peptide peaked at a molecular mass of 3,097 Da. Upon incubation with Ark1, a new peak appeared at a molecular mass of 3,177 Da, a +80 Da shift, indicating that the Cnp3(26–52) peptide was phosphorylated by Ark1 in vitro (Fig. 3A). When the Cnp3(26–52)T28A peptide (T28A mutation) was treated with Ark1, only one peak appeared at a molecular mass of 3,069 Da, which represents nonphosphorylated Cnp3(26–52)T28A (Fig. 3A). In addition, GST-Cnp3(26–52) and mutant GST-Cnp3(26–52)T28A were purified and used as the substrates of phosphorylation reactions in vitro. The phosphorylated proteins were then analyzed using Pro-Q Diamond Phosphoprotein Gel Stain. Consistent with the results of MALDI-TOF MS analysis, GST-Cnp3(26–52) is phosphorylated by Ark1, while the T28A mutation entirely eliminates Ark1-catalyzed phosphorylation (Fig. 3B). Thus, we identified an Ark1/Aurora B phosphorylation site at Thr28 of the inner kinetochore protein Cnp3 in vitro.
Fig. 3.
Thr28 of Cnp3 is phosphorylated by Ark1 in vitro and in vivo. (A) Phosphorylation of the Cnp3(26–52) peptide by Ark1 was measured by MALDI-TOF MS. The MS data of Cnp3(26–52) peptide, the products of Cnp3(26–52) peptide, and Cnp3(26–52)T28A peptide (with the mutation of Thr28 to alanine) catalyzed by Ark1 are shown in Left, Center, and Right, respectively. (B) Pro-Q Diamond phosphoprotein gel stain analysis of Cnp3(26–52) phosphorylation. After incubation with Ark1, the protein and phosphoprotein levels of GST-Cnp3(26–52) and GST-Cnp3(26–52)T28A were analyzed by Coomassie brilliant blue stain (Left) and Pro-Q Diamond phosphoprotein gel stain (Right). (C) Analysis of phosphorylation of Cnp3 in interphase and mitotic cells in SDS/PAGE and Phos-tag gel, which specifically retards the migration of phosphoproteins, followed by Western blotting. Negative control indicates the cell not expressing Cnp3-GFP. The Cnp3 band with and without phosphorylation at the Thr28 site is indicated with a blue line.
To examine whether Thr28 of Cnp3 is phosphorylated in vivo, we generated a site-specific anti-phosphoT28 (anti-pT28) Cnp3 antibody. An in vitro phosphorylation assay and Western blot data revealed that this antibody has a high specificity for phosphorylation at Thr28 of Cnp3 (Fig. S3A). This antibody was used to detect whether Cnp3 was phosphorylated at position Thr28 in vivo. We used the cold-sensitive mutant strain of S. pombe, nda3-KM311, which is arrested in prophase at the restrictive temperature because of a defect in β-tubulin (36). As shown in Fig. S3B, phosphorylation at Thr28 of Cnp3 occurs during prophase, while the synchronized cells with a mutation of Thr28 to alanine (T28A) eliminate the phosphorylation of this site. After being treated with λ-phosphatase (λ-PPase), the phosphorylation level at Thr28 of Cnp3 significantly decreased compared with the cnp3 without λ-PPase treatment (Fig. S3C). This result clearly showed that the phosphorylation takes places specifically at Thr28 of Cnp3 during early mitosis. To provide further evidence to support the claim that Cnp3T28 phosphorylation specifically occurs in mitosis, we first collected interphase Cnp3WT and Cnp3T28A cells (asynchronous cells) and mitotic Cnp3WT and Cnp3T28A cells (using the nda3-KM311 mutant arrested at 16 °C for 10 h). We then examined phosphorylation events by Phos-tag gel-analysis in which phosphorylated proteins generally display a noticeable band-shift in the gel. As shown in Fig. 3C, no band-shift was observed between Cnp3WT and Cnp3T28A in interphase, indicating that the Thr28 site is not phosphorylated in interphase. While in mitotic cells, Cnp3WT displayed significant band-shift compared with Cnp3T28A and the one in interphase cells, clearly proving that the phosphorylation specifically takes places at Cnp3T28 in mitosis. This result is consistent with the Western blot result shown in Fig. S3B, in which the phosphorylation of Cnp3WT in mitotic cells was detected by the antibody against phosphorylated Cnp3T28. Based on these two consistent results, we conclude that the phosphorylation of Cnp3 at Thr28 specifically occurs in mitosis but not in interphase.
Phosphorylation of Cnp3 by Ark1 Impairs the Kinetochore Localization of Mis12.
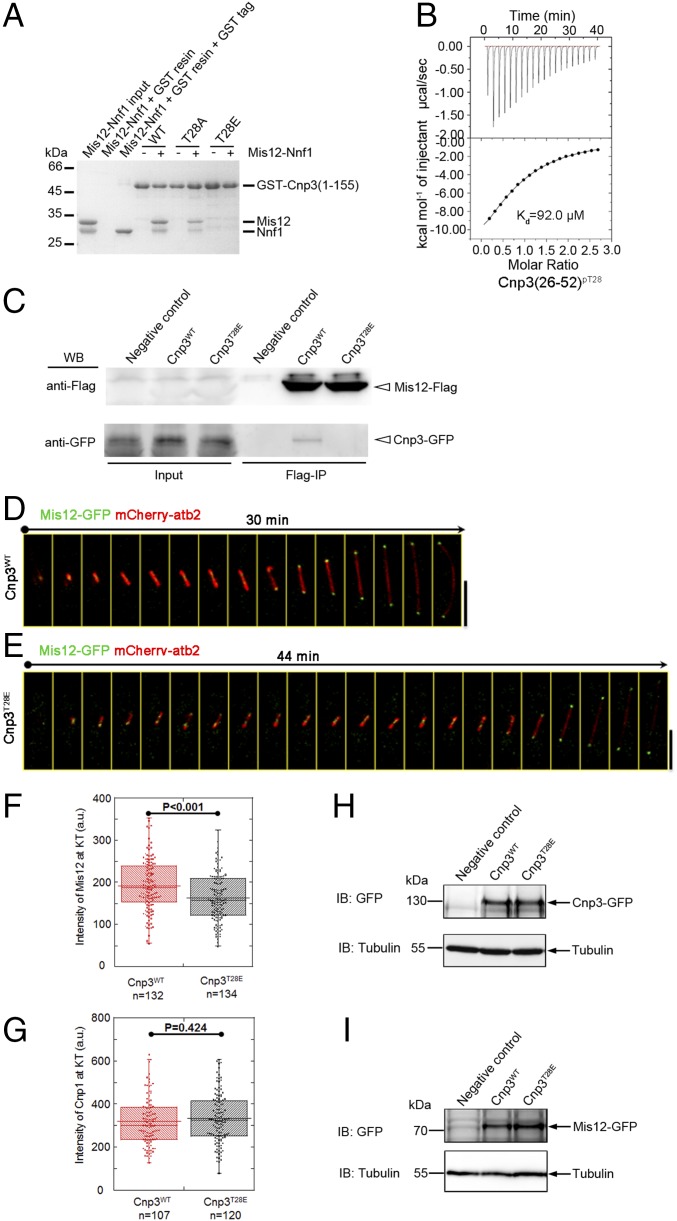
Our structural and biochemical analysis demonstrated that the N terminus of Cnp3 binds to a negatively charged region of Mis12 through electrostatic interactions. These observations provided clues that the interaction of Cnp3 with Mis12C might be regulated by the phosphorylation of Cnp3. In addition, Thr28 of Cnp3 was found to be phosphorylated both in vitro and in vitro. We speculated that phosphorylation at Thr28 of Cnp3 might be involved in regulation of the interaction between Cnp3 and Mis12–Nnf1. To verify this hypothesis, constructs consisting of the first 155 residues of Cnp3 [Cnp3(1–155)] with mutation of T28E or T28A were generated. The GST-tagged wild-type and mutant proteins were used for pull-down assays to evaluate their binding with Mis12–Nnf1. The results indicate that the T28E mutation, which mimics the phosphorylation of Thr28, significantly disrupts the interaction between Cnp3(1–155) and Mis12–Nnf1. By contrast, the phosphorylation-deficient mutant T28A has no effect on the interaction (Fig. 4A). We then synthesized a peptide that contains phosphorylated Thr28 [Cnp3(26–52)pT28] (Beijing Scilight Biotechnology Ltd. Co.) and performed ITC assays to verify its effects. The Kd value of the interaction between Cnp3(26–52)pT28 and Mis12–Nnf1 is 92.0 μM (Fig. 4B), which is ∼8 times higher than that of wild-type Cnp3(26–52) (Fig. 2B). To verify if the phosphorylation at Thr28 of Cnp3 attenuates its interaction with Mis12 in vivo, immunoprecipitation assays were performed by using Mis12-Flag and Cnp3-GFP. As shown in Fig. 4C, a dramatically decreased amount of Cnp3T28E was pulled down by Mis12 compared with Cnp3WT. These results indicate that the T28E mutant affects the association of Mis12 with Cnp3 both in vitro and in vivo. It has been reported that the kinetochore localization of Mis12C depends on CENP-C (31, 37). Based on the results of our in vitro and in vivo pull-down assays, we proposed that phosphorylation at Thr28 of Cnp3 might decrease the kinetochore localization of Mis12C. To test this hypothesis, we used living-cell imaging to investigate the kinetochore intensity of Mis12-GFP in mitosis in strains expressing Cnp3WT and Cnp3T28E, respectively. Consistent with our pull-down results, the kinetochore localization of Mis12 was decreased by about 10% in mutant Cnp3T28E compared with that of Cnp3WT, while Cnp1 (the CENP-A homolog of S. pombe) was not affected by the phosphorylation-mimicking mutant (Fig. 4 D–G). Our Western blotting results show that the levels of Mis12-GFP in both cells were comparable (Fig. 4I). In addition, we examined the in vivo expression levels of Cnp3WT and Cnp3T28E, and the Western blot results showed that the expression level of the phosphorylation-mimicking Cnp3 mutant is comparable to that of Cnp3WT (Fig. 4H). These results demonstrated that Ark1-dependent phosphorylation at Thr28 of Cnp3 disrupts the interaction between Cnp3 and Mis12C, which decreases the kinetochore localization of Mis12C.
Fig. 4.
Thr28 phosphorylation in Cnp3 decreases the kinetochore localization of Mis12. (A) GST pull-down assays of Mis12–Nnf1 with GST-Cnp3(1–155), with phosphorylation mimicking [GST-Cnp3(1–155)T28E] and eliminating [GST-Cnp3(1–155)T28A] mutants of GST-Cnp3(1–155). (B) The binding affinity of Mis12–Nnf1 with the Cnp3(26–52)pT28 peptide was analyzed by ITC. (C) Coimmunoprecipitation assay of Cnp3WT and Cnp3T28E with Mis12. Cells coexpressing Mis12-Flag and Cnp3-GFP were subjected to anti-FLAG resin immunoprecipitation and analyzed by Western blotting. Negative control indicates the cell not expressing Mis12-Flag. (D and E) Time-lapse live-cell images of Cnp3WT (D) and Cnp3T28E (E) cells expressing Mis12-GFP and mCherry-atb2 (α-tubulin marker) through mitosis. Kinetochore intensity of Mis12 in anaphase was analyzed in Cnp3WT and Cnp3T28E cells. Representative cells are shown. (Scale bars, 5 μm.) (F and G) Box plots comparing intensity of Mis12 (F) and Cnp1 (G) at the kinetochore in mitotic Cnp3WT and Cnp3T28E cells expressing Mis12-GFP or Cnp1-GFP and mCherry-atb2. Student’s t test was used to calculate P values. The analyzed samples number was indicated. a.u., arbitrary units. (H) Western blot analysis of the expression level of Cnp3-GFP in Cnp3WT and Cnp3T28E cells. Negative control indicates the cell not expressing Cnp3-GFP. (I) Western blot analysis of expression level of Mis12-GFP in Cnp3WT and Cnp3T28E cells. Negative control indicates the cell not expressing Mis12-GFP.
Phospho-Mimicking Cnp3 Mutation Results in Chromosome Segregation Defects.
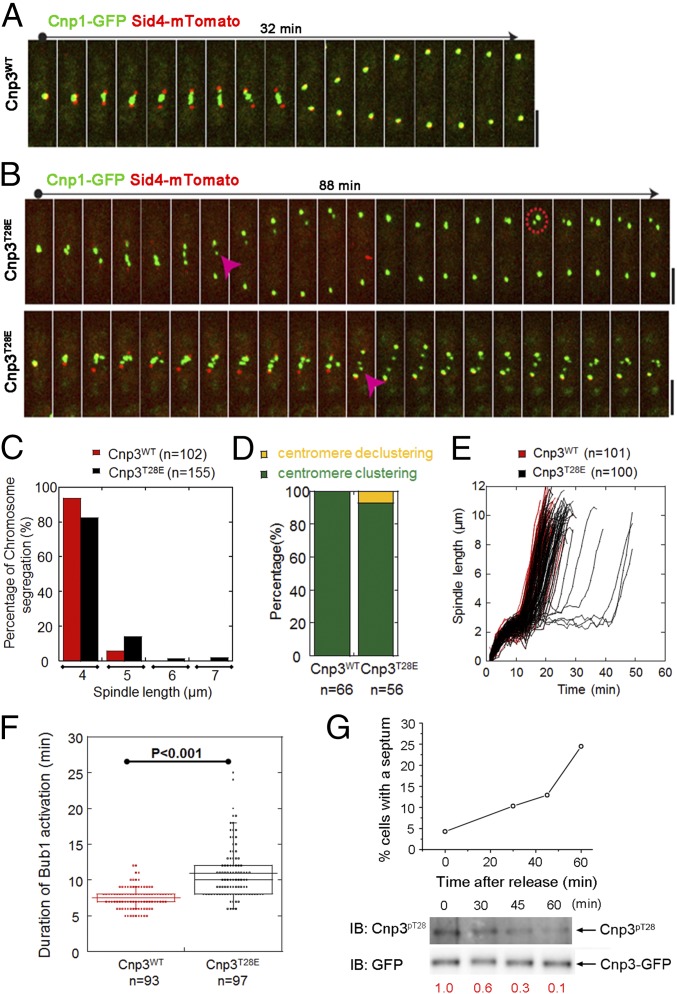
The interaction between CENP-C and Mis12C is one of the two reported pathways that attach the KMN network to kinetochores during mitosis. Although Mis12C is unable to bind microtubules directly, it functions as a “bridge” to attach the outer and inner kinetochore and plays a crucial role in kinetochore–microtubule attachment through its direct interactions with CENP-C and Ndc80C. Therefore, the decreased interaction between Mis12C and CENP-C might directly destabilize the kinetochore–microtubule attachment, which may facilitate EC, resulting in delayed or defective chromosome segregation. After we demonstrated that the phosphorylation at Thr28 of Cnp3 weakens the kinetochore localization of Mis12C, we sought to investigate whether it plays a role in chromosome segregation. As shown in Fig. 5, Cnp3T28E cells display lagging chromosomes during anaphase B. We quantified the phenotype by measuring the percentage of mitotic cells that accomplish the chromosome segregation at the spindle length at 4, 5, 6, and 7 μm (Fig. 5 B and C). The results show that more than 16% of Cnp3T28E cells display lagging chromosomes, whereas less than 6% of WT cells display lagging chromosomes with a spindle length longer than 4 µm (Fig. 5 A and C). In addition, Cnp3T28E also led to centromere declustering (38) when the cells entered a new cell cycle (Fig. 5 B and D). These results indicate that the phosphorylation at Thr28 of Cnp3 impairs the localization of Mis12C on the centromere, which in turn affects chromosome segregation. This result is consistent with previous studies that showed hMis12 knockdown in cells results in lagging chromosomes (39). Consistent with the results, we found that around 6% of Cnp3T28E cells showed prolonged metaphase (Fig. 5E). The delayed chromosome segregation in Cnp3T28E cells is likely caused by the activation of EC and SAC pathways, which is triggered by destabilized kinetochore–microtubule attachment. Consistently, the duration of the SAC protein Bub1 in Cnp3T28E cells is prolonged compared with wild-type cells, which represents the activation of the SAC (Fig. 5F). Together, these data confirm that mimicking phosphorylation at position Thr28 of Cnp3 destabilizes the interaction between Cnp3 and Mis12C, which may further lead to unstable kinetochore–microtubule attachment and result in delayed and defective chromosome segregation during mitosis.
Fig. 5.
Phosphorylation at Thr28 of Cnp3 leads to chromosome segregation defects. (A and B) Time-lapse live-cell images of Cnp3WT (A) and Cnp3T28E (B) cells expressing Cnp1-GFP and sid4-mTomato (spindle pole body marker). Magenta triangles mark the lagging chromosomes. Dotted red circle marks the declustered centromere. (Scale bars, 5 μm.) (C) Histogram of the percentage of chromosome segregation of Cnp3WT and Cnp3T28E cells that accomplish chromosome segregation within different spindle length (4, 5, 6, and 7 μm, respectively). Cell numbers analyzed are indicated. (D) Diagrams showed the percentage of centromere clustering/declustering of Cnp3WT and Cnp3T28E cells. The analyzed cell numbers are indicated. (E) Representative plots of spindle length against time for Cnp3WT and Cnp3T28E cells. (F) Dot plots of Bub1-GFP duration of Cnp3WT and Cnp3T28E cells that knocked out the endogenous Cnp3 gene. Cell numbers analyzed are indicated. (G) Phosphorylation level of Cnp3 (pT28) along with cell cycle progression. Cells were arrested at the mitosis by shifting to the restrictive temperature (16 °C) and then released at different time intervals. The percentages of septate cells were counted at each time point. The cell extracts of each time point were analyzed by Western blotting using the GFP antibody and anti-phosphoThr28 antibody. The relative intensity of Cnp3pT28 is indicated at the bottom.
For further investigation, we examined the levels of phosphorylated Cnp3 (pT28) with cell-cycle progression. Cells were synchronized to prophase and then released at different time intervals. As shown in Fig. 5G, the phosphorylation level of Cnp3T28 peaks at prophase before releasing (0 min) when the nda3-KM311 cells establish and maintain a robust spindle checkpoint arrest due to the absence of microtubules. A high phosphorylation level weakens the interaction between Mis12 and Cnp3, which impairs the assembly of kinetochores and facilitates the activation of the spindle checkpoint. When the arrested cells were shifted to the permissive temperature (30 °C), cell-cycle progression was reinitiated and the phosphorylation level of Cnp3T28 reduced gradually after release. Along with the occurrence of bio-orientation, the phosphorylation level of Cnp3T28 is reduced to enhance the Mis12C–Cnp3 interaction and when SAC was deactivated (Fig. 5G). This result suggested that the phosphorylation at Thr28 of Cnp3 is cell cycle-dependent and functions in SAC and EC pathways.
Ark1-Dependent Phosphorylation of Dsn1 and Cnp3 Has Distinct Roles in Mitosis.
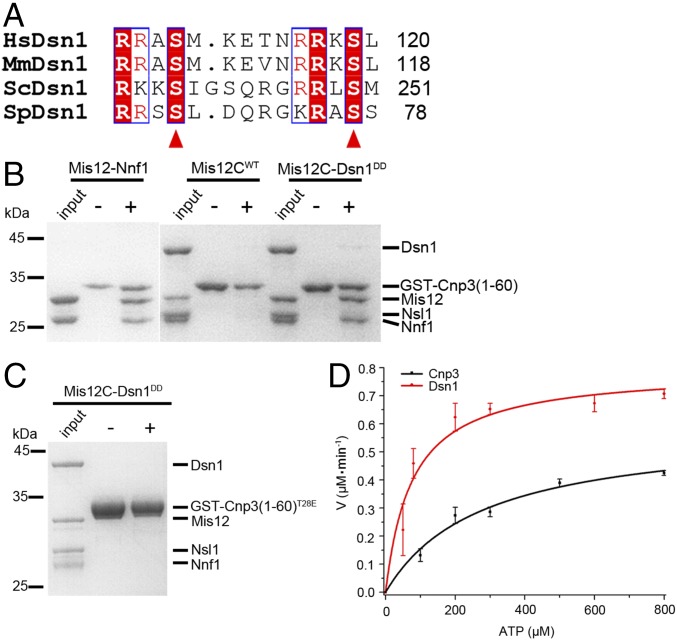
Aurora B phosphorylates S100 and S109 of Dsn1 to promote the interaction between human Mis12C and CENP-C, which is conserved in budding yeast (9, 10). Two human Dsn1 mutants—Dsn1(△91–113), which lacks these sites, and Dsn1-EE, which mimics the phosphorylation of these sites—can bind to CENP-C more efficiently (4). The region of Dsn1 containing these phosphorylation sites was proposed to block the interaction of Mis12C with CENP-C by a self-inhibition mechanism (28, 29). Because sequence alignment showed that the two Aurora B phosphorylation sites of human Dsn1 are conserved in S. pombe Dsn1 (corresponding to S68 and S77) (Fig. 6A), we wondered whether the same self-inhibition mechanism is conserved in S. pombe. A phosphorylation-mimicking mutant of S. pombe Dsn1 with mutations of S68D and S77D (named Dsn1DD) was generated. The binding of GST-Cnp3(1–60) to Mis12C containing either wild-type Dsn1 (Mis12CWT) or Dsn1DD mutant (Mis12C–Dsn1DD) was assessed by pull-down assays. As shown in Fig. 6B, the Mis12–Nnf1 complex binds to Cnp3(1–60), while Mis12CWT has no binding. After Dsn1 was replaced by Dsn1DD, the Mis12–Nnf1 subcomplex of Mis12C–Dsn1DD could associate with Cnp3(1–60). These results reveal that the self-inhibition mechanism is conserved in S. pombe and that the phosphorylation of Dsn1 relieves the autoinhibition of Mis12C for interacting with Cnp3. Surprisingly, for some unknown reasons, the Dsn1DD–Nsl1 subcomplex is missing, although the SEC–MALS shows that the Mis12C–Dsn1DD retains a full complex with each subunit represented once in solution (Fig. S4). One possibility is that Ndc80C or Knl1C may be involved in the maintenance of the integrity of Mis12C when Dsn1 is phosphorylated, since they form protein networks with Mis12C in vivo. Alternatively, full-length Cnp3 may be required to capture the integral Mis12C. The short N-terminal fragment of Cnp3 [i.e., Cnp3(1–60)] we used in this case might only bind the Mis12–Nnf1 subcomplex. The Dsn1–Nsl1 subcomplex would interact with other regions of Cnp3 located outside the N-terminal 60-amino acid residues.
Fig. 6.
Thr28 phosphorylation in Cnp3 destroys the Dsn1-mediated interaction between Mis12C and Cnp3. (A) Sequence alignment of the two Aurora B phosphorylation sites in Dsn1 homologs (Hs, Homo sapiens; Mm, Mus musculus; Sc, Saccharomyces cerevisiae; Sp, Schizosaccharomyces pombe). Red triangles indicate two conserved serine residues that were phosphorylated by Aurora B. (B) GST pull-down assay of Mis12–Nnf1, Mis12CWT, and Mis12C–Dsn1DD with GST-Cnp3(1–60)WT. (C) GST pull-down assay of Mis12C–Dsn1DD with GST-Cnp3(1–60)T28E. (D) The kinetics curves of Ark1 on the Dsn1 peptide (red) and the Cnp3 peptide (black) were generated by fluorometry-based kinase assay. Data were fit to the Michae-lis-Menten equation to calculate the Km and Kcat values (Table S1).
In contrast to Dsn1, phosphorylation at Thr28 of Cnp3 disrupts its interaction with Mis12C. While considering the opposing effects of the phosphorylation of these two proteins on Mis12C localization, we attempted to determine whether Cnp3 is capable of binding Mis12C when both Cnp3 and Dsn1 are phosphorylated. We determined whether Cnp3(1–60)T28E binds to Mis12C–Dsn1DD using a pull-down assay and found that T28E, the mutation that mimics phosphorylation, in Cnp3(1–60) disrupts its interaction with Mis12C–Dsn1DD (Fig. 6C). Moreover, the minimal optimal consensus phosphorylation motif for Aurora B is reported to be RX(S/T)Φ, where Φ stands for hydrophobic residues and X stands for any residue (40). We predicted that Ark1 phosphorylates Dsn1 more efficiently than Cnp3 according to the sequences of the phosphorylation sites in Dsn1 and Cnp3 (Fig. 6 and Fig. S5). Consistently, in vitro phosphorylation assays showed that Ark1 has ∼5 times higher catalytic efficiency toward the Dsn1(66–92) peptide (with a Kcat/Km value of 7.0 × 10−5 s−1·μM−1) than the Cnp3(26–52) peptide (with a Kcat/Km value of 1.4 × 10−5 s−1·μM−1) (Fig. 6D and Table S1). Furthermore, the protein level of Aurora B is dynamically regulated during cell-cycle progression, which begins to increase in prophase, reaches maximum protein levels during M phase, and decreases dramatically upon M phase release (41–43).
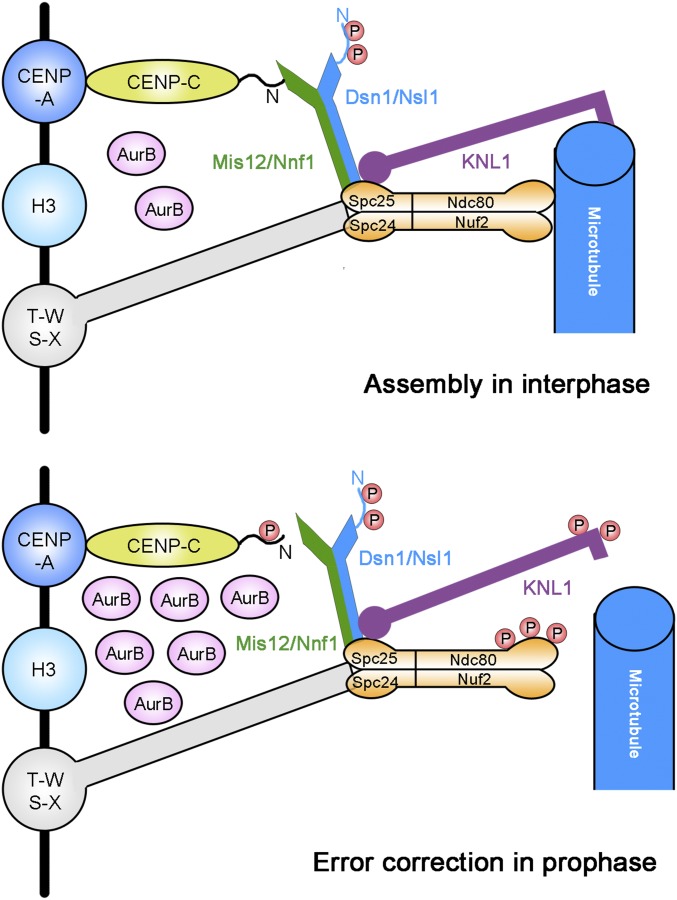
Our observations, in combination with previous reports, suggest that Aurora B-dependent phosphorylation of Dsn1 and Cnp3 is regulated temporally with distinct functions. In interphase, low levels of Aurora B are sufficient to phosphorylate the optimal substrate Dsn1 to promote the interaction between Mis12C and CENP-C, leading to successful kinetochore assembly. Because the phosphorylation site in CENP-C is not the optimal consensus motif for Aurora B and because the catalytic activity toward CENP-C is much lower, it can only be phosphorylated when the amount of Aurora B reaches the peak in M phase when EC and SAC pathways are activated, implying that Cnp3 Thr28 phosphorylation, in addition to Knl1 and Ndc80 phosphorylation, plays a role in correcting erroneous kinetochore–microtubule attachments (Fig. 7).
Fig. 7.
A model for Ark1/AuroraB-dependent phosphorylation to regulate the connection between Mis12C and Cnp3/CENP-C to control kinetochore assembly. From interphase (Top), Ark1/Aurora B phosphorylates Dsn1 and enhances the interaction between Mis12C and CENP-C to promote Mis12C assembly. During early mitosis when erroneous kinetochore–microtubule attachment occurs frequently (Bottom), Cnp3/CENP-C is phosphorylated by Ark1/Aurora B to attenuate its binding to Mis12C and destabilize the connection between kinetochore and microtubule. N indicates the amino terminus of CENP-C or Dsn1. P indicates phosphorylation by Aurora B.
Discussion
In this study, we determined the structure of the Mis12–Nnf1 complex in fission yeast and found that helix buddle 1 of the S. pombe Mis12–Nnf1 complex can be well overlaid on head I of the recently reported human and budding yeast Mis12C/MIND complexes (Fig. 1 D and E). Head I of both Mis12C/MIND complexes contains a negatively charged surface, which was shown to be the binding site for the N-terminal fragment of CENP-C (28, 29). In fission yeast, we also identified a minimal fragment at the N terminus of Cnp3 [Cnp3(26–50)] that has several positively charged residues that are essential for the interaction with the Mis12–Nnf1 complex. Moreover, a negatively charged pocket on the surface of helix bundle 1 of S. pombe Mis12–Nnf1 that is responsible for Cnp3(26–50) binding was identified. In line with previous observations in humans and Drosophila (5, 6, 32), we found that disruption of the interaction between CENP-C and Mis12C results in reduced kinetochore localization of Mis12C. These findings suggest that the mechanism of electrostatic interaction between Mis12C and CENP-C for Mis12C recruitment and kinetochore assembly is conserved from yeast to humans.
Thr28 of S. pombe Cnp3 is phosphorylated by Ark1 both in vitro and in vivo (Fig. 3). Cnp3 binds Mis12C via electrostatic interactions with the negatively charged surface of Mis12–Nnf1. D69 and E72 of Mis12 located in this pocket are critical for the interaction. Ark1-dependent phosphorylation at Thr28 of Cnp3 might disrupt the electrostatic interaction between Cnp3 and Mis12C, which significantly attenuates the interaction between Cnp3 and Mis12C and leads to reduced kinetochore localization of Mis12 (Fig. 4). Sequence alignment of the N-terminal regions of CENP-C homologs showed that the Aurora B consensus motif “RKTG” is conserved in different yeast species (Fig. S5A), which suggests that Aurora B-dependent phosphorylation at the Mis12C binding site of CENP-C is conserved. Furthermore, structural comparison of helix bundle 1 of S. pombe Mis12–Nnf1 with head I of the K. lactis MIND complex revealed a similar negatively charged surface in both complexes, which is responsible for CENP-C binding of the K. lactis MIND complex (Fig. S6A). Two acidic residues, D69 and E72 of S. pombe Mis12, which are critical for Cnp3 binding, are conserved in different yeast species (Fig. S5B) and correspond to D81 and E84 of K. lactis Mis12 located in the CENP-C binding pocket (Fig. S6B). The side chain of D81 forms a hydrogen bond with T13 of K. lactis CENP-C, which is the counterpart of Thr28 in S. pombe Cnp3 (Fig. 6B and Fig. S5A). These data suggest that T13 of K. lactis CENP-C might also be a potential phosphorylation site of Aurora B and that this phosphorylation may disrupt the interaction between CENP-C and Mis12C. Based on these observations, we propose that the Aurora B-dependent phosphorylation of CENP-C is a conserved regulatory mechanism for Mis12C–CENP-C interaction in yeasts.
Previous work revealed that Aurora B kinase plays multiple roles in mitosis and is essential for ensuring correct kinetochore–microtubule attachment. In early mitosis, Aurora B is located at the kinetochore and plays an important role in SAC activation and correcting kinetochore–microtubule attachment errors (16, 17). When aberrant attachment of kinetochore–microtubules occurs, Aurora B phosphorylates multiple outer kinetochore proteins, including Ndc80C, Knl1, the Dam1 complex, the SKA complex, MCAK, and CENP-E, to destabilize the erroneous attachments and to activate the EC or SAC pathways (12, 18, 20–22, 44). Although more substrates of Aurora B continue to be identified, none of the inner kinetochore components has been identified as an Aurora B substrate that plays a role in the regulation of kinetochore–microtubule attachment. In this study, we showed that the Aurora B-dependent phosphorylation at Thr28 of Cnp3 in fission yeast impairs the interaction between Mis12C and Cnp3. In vivo experiments revealed that the phosphorylation-mimicking mutant T28E decreases the kinetochore localization of Mis12, resulting in delayed and defective chromosome segregation (Figs. 4 and 5). Moreover, phosphorylated Cnp3 was detected by the site-specific anti-pT28 antibody during early mitosis in cold-sensitive nda3-KM311 cells in which the SAC is fully activated (45, 46) (Fig. S3B), and the duration of the Bub1 protein in the Cnp3T28E cell was prolonged compared with WT cells (Fig. 5F). In addition, after releasing the nda3-KM311 cells from prophase, the phosphorylation level of Cnp3T28 reduced gradually to enhance the Mis12C–Cnp3 interaction for complete kinetochore assembly (Fig. 5G). These observations suggest that phosphorylation at Thr28 of Cnp3 is involved in EC and the SAC pathways by destabilizing the interaction between CENP-C and Mis12C, which may further cause unstable kinetochore–microtubule attachment at the interface between the inner and outer kinetochore.
In summary, we demonstrated that Aurora B phosphorylates Cnp3 to attenuate its interaction with Mis12C and to decrease kinetochore localization of Mis12C, which regulates kinetochore assembly and chromosome segregation. Based on the results from our in vitro and in vivo assays, we propose that the phosphorylation of CENP-C is involved in the destabilization of kinetochore assembly and therefore plays a role in the EC and SAC pathways. This example shows that an inner kinetochore component, in addition to the outer kinetochore proteins reported previously, is involved in fine-tuning the mechanism for correcting erroneous kinetochore–microtubule attachments elicited by Aurora B kinase.
Materials and Methods
Cloning, Expression, and Purification.
The gene encoding full-length Mis12 and Dsn1 from S. pombe was cloned into pET-28a(+) vector (Novagen) with a SUMO (small ubiquitin-related modifier) and a Trx tag fused to its N terminus, respectively. The full-length Nnf1 and Nsl1 were cloned into the pET-22b(+) vector (Novagen) with no tag. The maltose binding protein (MBP)-Mis12 plasmid was constructed by cloning Mis12 into the pET-28a(+) vector with a N-terminal (His)8-MBP tag. Nnf1 was cloned into the pET-22b(+) vector with a (His)6 tag fused to its N terminus to yield the His-Nnf1 plasmid. All of the plasmids were transformed into Escherichia coli Rosetta 2 (DE3) cells (Novagen) for protein expression.
The Mis12 and Nnf1 plasmids were cotransformed into E. coli Rosetta 2 (DE3) cells to express the Mis12–Nnf1 subcomplex. The expression of the Dsn1–Nsl1 subcomplex was performed in the same way as that of Mis12–Nnf1. The Mis12–Nnf1 complex was first purified by Ni-NTA affinity chromatography, and the His-SUMO tag was removed by Ulp1 (SUMO protease 1). The Mis12–Nnf1 samples were then purified by a Mono S 5/50 GL column (GE Healthcare) and further by Superdex 200 SEC column (GE Healthcare) with 20 mM Tris⋅HCl, pH 8.0, and 200 mM NaCl. Mis12–Nnf1 mutants were expressed and purified in the same way as wild type. Mis12–Nnf1 and Dsn1–Nsl1 complexes were expressed individually and copurified to yield the Mis12C. The His-SUMO tag and the His-Trx tag were removed using SUMO protease and Prescission protease at 4 °C overnight, respectively. Further purification was conducted by a Mono S 5/50 GL column and a Superdex 200 column sequentially.
MBP-Mis12 and His-Nnf1 were purified by amylose resin (New England Biolabs Inc.) and Ni-NTA resin, respectively. A selenomethionine derivative of Mis12–Nnf1 and Mis12(1–215) –Nnf1(1–175) [Se–Mis12–Nnf1, Se–Mis12(1–215) –Nnf1(1–175)] were expressed in E. coli Rosetta 2 (DE3) cells from M9 medium supplemented with selenomethionine (Sigma-Aldrich) at a final concentration of 60 mg/L.
Crystallization, Data Collection, Structure Determination, and Refinement.
Se–Mis12(1–215) –Nnf1(1–175) was concentrated to 2 and 4 mg/mL for initial crystallization trials using the sitting-drop vapor diffusion method at 16 °C. Crystals obtained from the condition containing 30% (vol/vol) PEG400, 0.1 M cacodylate, pH 6.5, and 0.2 M Li2SO4 were cryoprotected for data collection in a solution composed of the mother liquor supplemented with 20% (vol/vol) glycerol. X-ray diffraction data were collected at 100 K at SSRF beamline BL17U1 (47) with a crystal-to-detector distance of 300 mm. Individual frames were collected using 1 s for each 1.0° oscillation over a range of 210°. Diffraction data were indexed, integrated, and scaled using the HKL-2000 program (48).
The structure of Mis12(1–215) –Nnf1(1–175) complex was determined by the SAD method. The calculation of initial phases was performed using AutoSol, and automatic model building was carried out using AutoBuild from the PHENIX software suite (49). Molecular replacement was carried out with Phaser (50) using the initial phases of the Mis12(1–215)–Nnf1(1–175) complex as a search model. After several rounds of refinement with COOT (51) and REFMAC5 (52), the structure of Mis12(1–215)–Nnf1(1–175) was refined to 2.4 Å resolution with an Rfactor of 23.26% and an Rfree of 27.34%, ultimately. The quality of the final model was validated with programs MolProbity (53). Structural statistics are shown in Table 1.
SEC–MALS.
SEC–MALS measurements were performed by an in-line MALS detector (DAWN HELEOS–II; Wyatt Technology) and differential refractometer (Optilab T-rEX; Wyatt Technology) at room temperature. The samples were run on a Superdex 200 increase 10/300 GL column (GE Healthcare) in SEC–MALS buffer (20 mM Tris⋅HCl, pH 8.0, 200 mM NaCl) at a flow rate of 0.8 mL/min. The data of SEC–MALS are analyzed by ASTRA 6.1 software.
GST Pull-Down Assay.
Full-length, N-terminal constructs and mutants of Cnp3 were cloned into pGEX-6p-1 (GE Healthcare) vector and overexpressed in E. coli BL21 (DE3) cells (Novagen). Cell pellets from 5-mL culture were resuspended and lysed by sonication in 1 mL binding buffer (PBS buffer with 0.1% Triton X-100, PBS buffer, 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 2 mM KH2PO4). The lysate was then cleared by centrifugation at 35,000 ×g for 10 min at 4 °C. Supernatants were incubated with 20 μL of GST beads and pre-equilibrated with binding buffer. The beads were washed with 1 mL of binding buffer twice to remove impurities. GST-Cnp3 and mutants immobilized with GST beads were incubated with 20 μg Mis12–Nnf1, Mis12C, or mutants for 1 h at 4 °C. The beads were washed with 1 mL binding buffer three times to remove nonspecific binding proteins and then boiled with SDS sample buffer. The proteins retained on the GST beads were analyzed by SDS/PAGE.
ITC Assay.
ITC assay was performed by MicroCal ITC 200 at 20 °C. Experiments were carried out by injecting 40 μL Cnp3(26–52)/(26–52)pT28 (1,500 μM) into a sample cell containing 200 μL Mis12–Nnf1 (120 μM) in binding buffer (20 mM Tris⋅HCl, pH 8.0, and 200 mM NaCl). Cnp3(26–52)/(26–52)pT28 (1,500 μM) was titrated into binding buffer as a control. The ITC data were analyzed using Origin Software (MicroCal Inc.).
In Vitro Kinase Assays.
Full-length Ark1 with a (His)6-tag fused to its C terminus (Ark1–6His) was overexpressed in E. coli BL21 (DE3) cells and purified on Ni-NTA resin in the same manner described for the Mis12–Nnf1 subcomplex. The in vitro kinase activity assays were carried out via two methods:
MALDI-TOF MS.
Ark1–6His (10 μM) was incubated with 100 μM Cnp3(26–52) wild-type or mutant peptide in 100 μL buffer with 50 mM Tris⋅HCl (pH 7.5), 100 mM NaCl, 20 mM MgCl2, 1 mM DTT, and 1 mM ATP at 30 °C for 30 min. The reaction was terminated by adding 10 μL 1% formaldehyde, and the insoluble material was removed by centrifugation. The supernatant was desalted using C18Zip-Tip (Millipore) as described previously (54) and analyzed directly by MALDI-TOF MS.
Pro-Q.
Ark1–6His (200 ng) was incubated with 4 μg GST-Cnp3(26–52) wild-type or mutant protein in 20 μL buffer with 20 mM Hepes (pH 7.8), 1 mM EGTA, 5 mM MgCl2, 1 mM DTT, and 0.1 mM ATP at 30 °C for 30 min. The reaction was terminated by adding SDS loading buffer and separated by SDS/PAGE. The phosphorylated substrates were detected using Pro-Q Diamond Phosphoprotein Gel Stain (Invitrogen).
Kinetic Studies of Ark1.
The kinetic parameters of Ark1 were determined by detecting the ADP production using Fluorometric Kinase Assay Kits (31001; ATT Bioquest) following the manufacturer’s instructions. The purified Ark1 (1 μg) kinase, 80 μM Dsn1 or Cnp3 peptides, and ATP with various concentrations (0 μM to 800 μM) were added into reaction buffer (50 mM Tris⋅HCl, pH 7.5, 100 mM NaCl, 10 mM MgCl2, 0.5 mM TCEP) to a final reaction volume of 20 μL and incubated at 37 °C for 30 min. The ADP sensor buffer and sensor were added and incubated for another 15 min at room temperature. Finally, the fluorescence intensity was monitored with a fluorescence plate reader at an Ex/Em of 540/590 nm to determine ADP formation. Data were fit to the Michae-lis-Menten equation to calculate Km and Kcat values.
Phosphorylation in Vivo.
To assess the in vivo phosphorylation of Thr28 in Cnp3, an antibody that specifically recognizes phosphorylated Thr28 in Cnp3 was raised. The Cnp3WT/T28A-GFP nda3–KM311 strain was cultured in YE5S medium at 30 °C overnight and then incubated at 16 °C for 12 h to arrest the cells in mitosis. In brief, the cells were harvested and lysed in PBS buffer with 0.1% Triton X-100 using a Retsch RM200 motorized mortar grinder (Retsch). The cell lysates were centrifuging at 35,000 ×g for 30 min, and the supernatants were collected for further immunoprecipitation. For immunoprecipitation, the supernatant was incubated with the anti-GFP antibody and protein G-coupled Dynabeads (Novex; Thermo-Fisher Scientific) for 10 min at room temperature to immunoprecipitate the Cnp3-GFP. After incubation, the pellets were washed with PBS (with the addition of 0.1% Triton X-100) four times. The pellets were boiled in SDS sample buffer and resolved on an SDS/PAGE gel. Western blot with anti-GFP (1:2,000; Rockland) and anti-phosphoThr28 antibody (1:1,000; GL Biochem Ltd) was used to detect the total amount of Cnp3-GFP and Thr28-phosphorylated Cnp3-GFP. The method was applied to the Cnp3 phosphorylation analysis during cell cycle. The cells were released at 30 °C for 30 min, 45 min, and 60 min, respectively, after 12 h arresting at 16 °C. Quantification of the relative intensity of Cnp3pT28 was measured with MetaMorph. For the dephosphorylation assay with λ-PPase, 40 pmol λ-PPase (New England Biolabs) was incubated with the immunoprecipitated Cnp3-GFP in 1× NEBuffer for Protein MetalloPhosphatases, supplemented with 1 mM MnCl2 at 30 °C for 15 min. The phosphatase reaction was analyzed by Western blotting using anti-GFP and anti-phosphoThr28 antibody. Protein phosphorylation was also analyzed by phospho-affinity gel electrophoresis using SDS–6% polyacrylamide gels containing 50 μM Mn2+-Phos-tag acrylamide (AAL-107; NARD Institute) (55).
Immunoprecipitation.
For immunoprecipitation, the supernatant was incubated with EZview Red Anti-FLAG M2 Affinity Gel (Sigma-Aldrich) and incubated for 4 h at 4 °C. After incubation, the gel was washed with PBS containing 0.1% Triton X-100 three times. The pellets were boiled in SDS sample buffer and resolved on an SDS/PAGE gel. Western blot with anti-Flag antibody (1:2,000; Sigma-Aldrich) and anti-GFP (1:2,000; Rockland) was used to detect Cnp3-GFP and Mis12-Flag.
Yeast Strains and Plasmids Construction.
Standard yeast genetic techniques were used (56). Fluorescence tagging and deletion of cnp3+ were carried out using a PCR-based method for S. pombe (57). To generate cnp3 mutant strains (cnp3T28E), endogenous cnp3 was cloned into a PJK148 vector with an ase1 promoter, and the Thr28 in cnp3 was mutated to glutamate using a QuikChange II XL Site-Directed Mutagenesis Kit (Agilent Technologies). The resulting plasmids were linearized and integrated at the leu1-32 locus. The endogenous cnp3 was then deleted. Yeast strains and plasmids used in this study are listed in Tables S2 and S3, respectively.
Microscopy and Data Analysis.
Yeast cells were imaged with a spinning-disk confocal microscope (Ultraview LCI; Perkin-Elmer) equipped with 100× N.A. 1.40 Plan-Apochromat oil immersion objective, and an electron-multiplying charge-coupled device camera (ORCA-ER; Hamamatsu Photonics) was used to capture the real-time images. All imaging was carried out in a 26 °C incubator. Detailed imaging conditions are provided in Table S4.
The imaging data were processed by MetaMorph 7.7 (Molecular Devices). The intensity of Mis12 and Cnp1 at kinetochores was measured using the Region Measurement function in Metamorph 7.7. In brief, a maximum intensity projection of the images was performed, and the intensities of Cnp1 and Mis12 at the moment of chromosome segregation were measured. The spindle lengths were measured using Metamorph 7.7 and the MTrackJ plug-in in ImageJ (National Institutes of Health). Detailed information regarding data analysis was previously described (58).
Box plots and graphs were generated with KaleidaGraph 4.5 (Synergy Software). Student’s t test was performed using Microsoft Excel for comparison between means for data analysis.
Supplementary Material
Acknowledgments
We thank the staff at beamline BL17U1 and BL18U1 of the Shanghai Synchrotron Radiation Facility (SSRF) for assistance with the data collection and Y. Pan (National Synchrotron Radiation Laboratory, University of Science and Technology of China) for instrumentation supports. We are grateful to National Centre for Protein Science Shanghai (Protein Expression and Purification system) for MALS. This work was supported by National Key Research and Development Program of China Grants 2017YFA0503600 and 2016YFA0400903, Strategic Priority Research Program of the Chinese Academy of Sciences Grant XDB 08010101, and Foundation for Innovative Research Groups of the National Natural Science Foundation of China Grant 31621002. This work was also supported by National Natural Science Foundation of China Grants U1532109, 31370756, and 31361163002 and Scientific Research Grant of Hefei Science Center of CAS Grants 2015SRG-HSC043 and 2015HSC-UP019 (to J.Z.). This work was also supported by Anhui Provincial Natural Science Foundation Grant 1608085QC52 and National Natural Science Foundation of China Grant 31700671 (to Xuan Zhang), National Natural Science Foundation of China Grant 31601095 (to F.Z.), and National Natural Science Foundation of China Grant 31400627 (to M.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The coordinate for the Schizosaccharomyces pombe Mis12–Nnf1 complex has been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID code 5WWL).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1710506114/-/DCSupplemental.
References
- 1.Cleveland DW, Mao Y, Sullivan KF. Centromeres and kinetochores: From epigenetics to mitotic checkpoint signaling. Cell. 2003;112:407–421. doi: 10.1016/s0092-8674(03)00115-6. [DOI] [PubMed] [Google Scholar]
- 2.Perpelescu M, Fukagawa T. The ABCs of CENPs. Chromosoma. 2011;120:425–446. doi: 10.1007/s00412-011-0330-0. [DOI] [PubMed] [Google Scholar]
- 3.Cheeseman IM, Desai A. Molecular architecture of the kinetochore-microtubule interface. Nat Rev Mol Cell Biol. 2008;9:33–46. doi: 10.1038/nrm2310. [DOI] [PubMed] [Google Scholar]
- 4.Kim S, Yu H. Multiple assembly mechanisms anchor the KMN spindle checkpoint platform at human mitotic kinetochores. J Cell Biol. 2015;208:181–196. doi: 10.1083/jcb.201407074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rago F, Gascoigne KE, Cheeseman IM. Distinct organization and regulation of the outer kinetochore KMN network downstream of CENP-C and CENP-T. Curr Biol. 2015;25:671–677. doi: 10.1016/j.cub.2015.01.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Screpanti E, et al. Direct binding of Cenp-C to the Mis12 complex joins the inner and outer kinetochore. Curr Biol. 2011;21:391–398. doi: 10.1016/j.cub.2010.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klare K, et al. CENP-C is a blueprint for constitutive centromere-associated network assembly within human kinetochores. J Cell Biol. 2015;210:11–22. doi: 10.1083/jcb.201412028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Emanuele MJ, et al. Aurora B kinase and protein phosphatase 1 have opposing roles in modulating kinetochore assembly. J Cell Biol. 2008;181:241–254. doi: 10.1083/jcb.200710019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang Y, et al. Phosphorylation of HsMis13 by Aurora B kinase is essential for assembly of functional kinetochore. J Biol Chem. 2008;283:26726–26736. doi: 10.1074/jbc.M804207200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Akiyoshi B, Nelson CR, Biggins S. The Aurora B kinase promotes inner and outer kinetochore interactions in budding yeast. Genetics. 2013;194:785–789. doi: 10.1534/genetics.113.150839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ciferri C, et al. Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell. 2008;133:427–439. doi: 10.1016/j.cell.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–997. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- 13.Kudalkar EM, et al. Regulation of outer kinetochore Ndc80 complex-based microtubule attachments by the central kinetochore Mis12/MIND complex. Proc Natl Acad Sci USA. 2015;112:E5583–E5589. doi: 10.1073/pnas.1513882112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kops GJ, Weaver BA, Cleveland DW. On the road to cancer: Aneuploidy and the mitotic checkpoint. Nat Rev Cancer. 2005;5:773–785. doi: 10.1038/nrc1714. [DOI] [PubMed] [Google Scholar]
- 15.Foley EA, Kapoor TM. Microtubule attachment and spindle assembly checkpoint signalling at the kinetochore. Nat Rev Mol Cell Biol. 2013;14:25–37. doi: 10.1038/nrm3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krenn V, Musacchio A. The Aurora B kinase in chromosome bi-orientation and spindle checkpoint signaling. Front Oncol. 2015;5:225. doi: 10.3389/fonc.2015.00225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carmena M, Wheelock M, Funabiki H, Earnshaw WC. The chromosomal passenger complex (CPC): From easy rider to the godfather of mitosis. Nat Rev Mol Cell Biol. 2012;13:789–803. doi: 10.1038/nrm3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DeLuca KF, Lens SM, DeLuca JG. Temporal changes in Hec1 phosphorylation control kinetochore-microtubule attachment stability during mitosis. J Cell Sci. 2011;124:622–634. doi: 10.1242/jcs.072629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Welburn JP, et al. Aurora B phosphorylates spatially distinct targets to differentially regulate the kinetochore-microtubule interface. Mol Cell. 2010;38:383–392. doi: 10.1016/j.molcel.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tien JF, et al. Cooperation of the Dam1 and Ndc80 kinetochore complexes enhances microtubule coupling and is regulated by Aurora B. J Cell Biol. 2010;189:713–723. doi: 10.1083/jcb.200910142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chan YW, Jeyaprakash AA, Nigg EA, Santamaria A. Aurora B controls kinetochore-microtubule attachments by inhibiting Ska complex-KMN network interaction. J Cell Biol. 2012;196:563–571. doi: 10.1083/jcb.201109001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knowlton AL, Lan W, Stukenberg PT. Aurora B is enriched at merotelic attachment sites, where it regulates MCAK. Curr Biol. 2006;16:1705–1710. doi: 10.1016/j.cub.2006.07.057. [DOI] [PubMed] [Google Scholar]
- 23.Petrovic A, et al. The MIS12 complex is a protein interaction hub for outer kinetochore assembly. J Cell Biol. 2010;190:835–852. doi: 10.1083/jcb.201002070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petrovic A, et al. Modular assembly of RWD domains on the Mis12 complex underlies outer kinetochore organization. Mol Cell. 2014;53:591–605. doi: 10.1016/j.molcel.2014.01.019. [DOI] [PubMed] [Google Scholar]
- 25.Hornung P, et al. Molecular architecture and connectivity of the budding yeast Mtw1 kinetochore complex. J Mol Biol. 2011;405:548–559. doi: 10.1016/j.jmb.2010.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Y, et al. Insights from the reconstitution of the divergent outer kinetochore of Drosophila melanogaster. Open Biol. 2016;6:150236. doi: 10.1098/rsob.150236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Richter MM, et al. Network of protein interactions within the Drosophila inner kinetochore. Open Biol. 2016;6:150238. doi: 10.1098/rsob.150238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petrovic A, et al. Structure of the MIS12 complex and molecular basis of its interaction with CENP-C at human kinetochores. Cell. 2016;167:1028–1040.e15. doi: 10.1016/j.cell.2016.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dimitrova YN, Jenni S, Valverde R, Khin Y, Harrison SC. Structure of the MIND complex defines a regulatory focus for yeast kinetochore assembly. Cell. 2016;167:1014–1027.e12. doi: 10.1016/j.cell.2016.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kwon MS, Hori T, Okada M, Fukagawa T. CENP-C is involved in chromosome segregation, mitotic checkpoint function, and kinetochore assembly. Mol Biol Cell. 2007;18:2155–2168. doi: 10.1091/mbc.E07-01-0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Milks KJ, Moree B, Straight AF. Dissection of CENP-C-directed centromere and kinetochore assembly. Mol Biol Cell. 2009;20:4246–4255. doi: 10.1091/mbc.E09-05-0378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Przewloka MR, et al. CENP-C is a structural platform for kinetochore assembly. Curr Biol. 2011;21:399–405. doi: 10.1016/j.cub.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 33.Westermann S, et al. Architecture of the budding yeast kinetochore reveals a conserved molecular core. J Cell Biol. 2003;163:215–222. doi: 10.1083/jcb.200305100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xue Y, et al. GPS 2.0, a tool to predict kinase-specific phosphorylation sites in hierarchy. Mol Cell Proteomics. 2008;7:1598–1608. doi: 10.1074/mcp.M700574-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koch A, Krug K, Pengelley S, Macek B, Hauf S. Mitotic substrates of the kinase Aurora with roles in chromatin regulation identified through quantitative phosphoproteomics of fission yeast. Sci Signal. 2011;4:rs6. doi: 10.1126/scisignal.2001588. [DOI] [PubMed] [Google Scholar]
- 36.Hiraoka Y, Toda T, Yanagida M. The NDA3 gene of fission yeast encodes beta-tubulin: A cold-sensitive nda3 mutation reversibly blocks spindle formation and chromosome movement in mitosis. Cell. 1984;39:349–358. doi: 10.1016/0092-8674(84)90013-8. [DOI] [PubMed] [Google Scholar]
- 37.Przewloka MR, et al. Molecular analysis of core kinetochore composition and assembly in Drosophila melanogaster. PLoS One. 2007;2:e478. doi: 10.1371/journal.pone.0000478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hiraoka Y, et al. Inner nuclear membrane protein Ima1 is dispensable for intranuclear positioning of centromeres. Genes Cells. 2011;16:1000–1011. doi: 10.1111/j.1365-2443.2011.01544.x. [DOI] [PubMed] [Google Scholar]
- 39.Goshima G, Kiyomitsu T, Yoda K, Yanagida M. Human centromere chromatin protein hMis12, essential for equal segregation, is independent of CENP-A loading pathway. J Cell Biol. 2003;160:25–39. doi: 10.1083/jcb.200210005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alexander J, et al. Spatial exclusivity combined with positive and negative selection of phosphorylation motifs is the basis for context-dependent mitotic signaling. Sci Signal. 2011;4:ra42. doi: 10.1126/scisignal.2001796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bischoff JR, et al. A homologue of Drosophila Aurora kinase is oncogenic and amplified in human colorectal cancers. EMBO J. 1998;17:3052–3065. doi: 10.1093/emboj/17.11.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kimura M, et al. Cell cycle-dependent expression and spindle pole localization of a novel human protein kinase, Aik, related to Aurora of Drosophila and yeast Ipl1. J Biol Chem. 1997;272:13766–13771. doi: 10.1074/jbc.272.21.13766. [DOI] [PubMed] [Google Scholar]
- 43.Terada Y, et al. AIM-1: A mammalian midbody-associated protein required for cytokinesis. EMBO J. 1998;17:667–676. doi: 10.1093/emboj/17.3.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ditchfield C, et al. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J Cell Biol. 2003;161:267–280. doi: 10.1083/jcb.200208091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garcia MA, Vardy L, Koonrugsa N, Toda T. Fission yeast ch-TOG/XMAP215 homologue Alp14 connects mitotic spindles with the kinetochore and is a component of the Mad2-dependent spindle checkpoint. EMBO J. 2001;20:3389–3401. doi: 10.1093/emboj/20.13.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vanoosthuyse V, Valsdottir R, Javerzat JP, Hardwick KG. Kinetochore targeting of fission yeast Mad and Bub proteins is essential for spindle checkpoint function but not for all chromosome segregation roles of Bub1p. Mol Cell Biol. 2004;24:9786–9801. doi: 10.1128/MCB.24.22.9786-9801.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang QS, et al. The macromolecular crystallography beamline of SSRF. Nucl Sci Tech. 2015;26:12–17. [Google Scholar]
- 48.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Macromol Crystallogr. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 49.Adams PD, et al. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 50.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 52.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Davis IW, et al. MolProbity: All-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen Z, et al. Structural insights into histone demethylation by JMJD2 family members. Cell. 2006;125:691–702. doi: 10.1016/j.cell.2006.04.024. [DOI] [PubMed] [Google Scholar]
- 55.Kinoshita-Kikuta E, Aoki Y, Kinoshita E, Koike T. Label-free kinase profiling using phosphate affinity polyacrylamide gel electrophoresis. Mol Cell Proteomics. 2007;6:356–366. doi: 10.1074/mcp.T600044-MCP200. [DOI] [PubMed] [Google Scholar]
- 56.Forsburg SL, Rhind N. Basic methods for fission yeast. Yeast. 2006;23:173–183. doi: 10.1002/yea.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bähler J, et al. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 58.Zheng F, et al. Csi1p recruits alp7p/TACC to the spindle pole bodies for bipolar spindle formation. Mol Biol Cell. 2014;25:2750–2760. doi: 10.1091/mbc.E14-03-0786. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.