Significance
The findings in this manuscript report on the identification of a posttranslational modification in the huntingtin protein (phosphorylation on residue T3 in the N17 region of the protein), which can revert the conformational effects of the Huntington’s disease (HD) mutation itself on the huntingtin protein and inhibit its aggregation properties in vitro. Using the first ultrasensitive immunoassay for a posttranslational modification of huntingtin protein, we demonstrate that pT3 levels are decreased in mutant huntingtin in preclinical models as well as in clinically relevant samples from HD patients. These findings are of high significance to Huntington’s disease biology, provide insights into mechanisms of Huntington’s disease pathogenesis, and open new opportunities for the development of therapeutics and diagnostics for Huntington’s disease.
Keywords: huntingtin, posttranslational modification, immunoassay, phosphorylation, neurodegeneration
Abstract
Posttranslational modifications can have profound effects on the biological and biophysical properties of proteins associated with misfolding and aggregation. However, their detection and quantification in clinical samples and an understanding of the mechanisms underlying the pathological properties of misfolding- and aggregation-prone proteins remain a challenge for diagnostics and therapeutics development. We have applied an ultrasensitive immunoassay platform to develop and validate a quantitative assay for detecting a posttranslational modification (phosphorylation at residue T3) of a protein associated with polyglutamine repeat expansion, namely Huntingtin, and characterized its presence in a variety of preclinical and clinical samples. We find that T3 phosphorylation is greatly reduced in samples from Huntington’s disease models and in Huntington’s disease patients, and we provide evidence that bona-fide T3 phosphorylation alters Huntingtin exon 1 protein conformation and aggregation properties. These findings have significant implications for both mechanisms of disease pathogenesis and the development of therapeutics and diagnostics for Huntington’s disease.
Posttranslational modifications (PTMs) likely evolved as a strategy to flexibly modulate protein structure and function in response to developmental, homeostatic, or physiological requirements without the need to exert permanent changes at the genetic level (1). In recent years, it has become apparent that PTMs are extremely diverse and widespread and decorate proteins at multiple sites, thus increasing exponentially the capacity for adoption of multiple structural and functional states of a protein (2). Many neurodegenerative diseases are associated with accumulation and misfolding of specific proteins (3, 4), often associated with changes in PTM status. Some PTMs such as phosphorylation can have profound influences on the structure, function, and pathology-related properties of proteins associated with neurodegeneration [for example, TAU, Androgen Receptor, and Huntingtin (5–10)]. Their modulation therefore represents an attractive candidate therapeutic approach (7, 8, 11). However, the transient character of most PTMs presents a challenge for the development of assays to detect, monitor, and quantify PTMs, thus impeding the identification of mechanistically relevant PTMs, the establishment of their role in disease pathogenesis, and the discovery of their modulators.
Huntington’s disease (HD) is a progressive, autosomal-dominant neurodegenerative disease caused by a triplet repeat expansion mutation within exon 1 of the huntingtin gene (HTT) (12; reviewed in ref. 13). The mutation leads to the production of huntingtin protein (HTT) bearing a polyglutamine expansion (polyQ) within its amino terminus (N-term). Consistent with a determinant role in the pathology, the size of the polyQ expansion demonstrates a strong inverse correlation with the age-of-onset of the characteristic clinical (motor, psychiatric, and cognitive) symptoms (14, 15). The CAG repeat/polyQ expansion is believed to cause pathology primarily through a gain-of-function, which imparts novel characteristics on mutant HTT, including the production of N-terminal fragments, likely through aberrant mRNA splicing as well as proteolytic events (16, 17). Mutant HTT N-term fragments have been described to misfold and form detectable aggregates, and their overexpression has been reported to reproduce several aspects of HD in animal models (18–21; reviewed in ref. 22). The expansion of the polyQ domain associated with the HD mutation was recently shown to alter the conformation of the N-term region of HTT (23–25). Interestingly, regions flanking the polyQ domain, including the first 17 N-terminal amino acids of HTT (N17) and a polyproline-rich C-terminal to the polyQ region, have been shown to influence the propensity of short N-term HTT fragments to misfold and aggregate (26–30). Recent studies demonstrated that phosphomimetic mutations at specific residues (S13/S16) within the N17 region can modulate the subcellular localization, stability, aggregation, and toxicity properties of mutant HTT in different preclinical models (29, 31–33), raising interest in these PTMs as possible modifiers of mutant HTT biological and biophysical properties. Significantly, studies on HTT exon 1-like peptides bearing phosphorylated S13/16 residues or S/D mutations at the same residues demonstrated substantially reduced aggregation in vitro compared with the unphosphorylated or wild-type (WT) counterparts, suggesting that N17 phosphorylation modulates the capacity of mutant HTT N-term fragments to self-assemble and aggregate (34–36). Thus, increasing HTT N17 phosphorylation may represent a meaningful therapeutic approach aimed at ameliorating the toxic properties of mutant huntingtin. However, a critical requirement to address this hypothesis is the availability of quantitative, sensitive assays to measure N17 HTT phosphorylation and identify genetic and/or pharmacological modulators for proof-of-concept studies, as well as to monitor HTT N17 phosphorylation in different HD models and, ultimately, in HD patients. To date, HTT phosphorylation has been predominantly studied by either mass spectrometry approaches or Western blotting (WB) (32, 33, 37–42). Phosphorylation at T3 (pT3) appears to be the most readily observable N17 phosphorylation site, at least with current tools and in the preclinical models examined (39, 40). Additionally, pT3 has been reported to interfere with aggregation of purified mutant HTT exon 1 protein (36), suggesting that its modulation may impact a pathologically relevant aspect of HD, namely mutant HTT misfolding and aggregation. However, further analysis of this PTM’s relevance for HD and functional/mechanistic studies require the capacity to quantitatively and sensitively measure this PTM in preclinical and clinical samples, which is extremely challenging with current methods. Therefore, we developed a quantitative immunoassays to detect and quantify phosphorylation on HTT N17, starting with pT3 and leveraging the availability of semisynthetic HTT exon 1 proteins bearing a bona-fide pT3 modification (35) in both WT and mutant forms, as well as a corresponding anti-pT3 HTT antibody, which we previously characterized (35, 40). Importantly, these pg/mL assays uncovered a strong effect of polyQ expansion on pT3 HTT levels in mutant HTT in HD cells, HD mice, and HD patients and indicate that the presence of a mutant polyQ results in greatly decreased immunoreactive T3 phosphorylation on HTT. Mechanistically, using semisynthetic HTT exon 1 (HTT EX1) proteins, we provide evidence that phosphorylation on HTT T3 can influence the conformation and decrease the aggregation properties of mutant HTT in vitro, consistent with other findings reported elsewhere (36). Collectively, our data point to restoration of pT3 levels in mutant HTT as a therapeutically relevant candidate approach for HD and enable investigations aimed at identifying genetic and pharmacological modulators of phosphorylation on T3 in HTT protein.
Results
Development of a Singulex-Based Immunoassay for Huntingtin pT3 and Specific Detection of Huntingtin T3 Phosphorylation in Cells.
Immunoassay development requires the availability of antibodies capable of specifically recognizing the epitopes of interest and availability of the purified antigen protein to assess specificity and sensitivity in a controlled context. Recently, we reported the development of semisynthetic strategies that enabled the generation of HTT EX1 proteins with bona-fide phosphorylation on residue T3 (pT3) as well as a rabbit polyclonal antibody (pAb) specific for pT3 HTT (35, 40). We were therefore in a position to attempt the development of quantitative immunoassays to detect and measure levels of pT3 in HTT protein in different biological contexts using these reagents (Fig. 1 A and B). Because a specific PTM may affect only a fraction of the total cellular steady-state pool of the protein of interest, we turned to one of the most sensitive immunoassay platforms available (43), an application of which was recently employed to develop an ultrasensitive immunoassay for detection of mutant HTT in cerebrospinal fluid (44). The assay is essentially a quantitative fluorescent sandwich immunoassay coupled to single-molecule counting technology (Singulex Erenna immunoassay). Two antibodies are therefore required (one for capture and one for detection; Fig. 1D). As the purified antigen protein was the protein encoded by the first exon of HTT (35), antibody choice for assay development was limited to antibodies with epitopes within the protein encoded by HTT exon 1 (Fig. 1 C, i and ii). Importantly, an immunoassay capable of measuring pT3 levels needs to be coupled to a companion immunoassay for the measurement of overall HTT levels (“total HTT”) to allow for normalization of pT3 levels. The two immunoassays (pT3-specific and total HTT-specific) ideally need to be based on the same capture antibody, to enable meaningful comparisons between pT3 HTT and total HTT levels (Fig. 1D). A mouse monoclonal antibody (mAb) specific for a polyQ repeat, MW1 (45, 46), was selected as the capture antibody for both pT3 HTT and total HTT immunoassays. MW1 displays an apparently higher affinity for HTT bearing expanded (mutant) polyQ repeats, but can efficiently detect HTT bearing WT polyQ repeats under native conditions (24, 25, 46), a finding that we confirmed in the Singulex Erenna immunoassay (Fig. 2B and Fig. S1 A and B). Importantly, we observed an attenuated difference in apparent affinity for expanded (mutant) HTT protein relatively to unexpanded polyQ (wild type) HTT protein when the MW1 was used as a capture Ab (although a degree of selectivity for expanded polyQ is still retained) in the Singulex Erenna immunoassay, as opposed to the same Ab used as a detection Ab (as employed by ref. 44) (Fig. S1), thus validating the use of MW1 as an antibody allowing detection of both WT and mutant HTT in these experimental conditions. The anti-pT3 HTT pAb previously characterized was selected as the detection antibody for the pT3 HTT immunoassay, while 2B7 (a mouse mAb specific for an epitope within the N17 HTT domain) (47) was selected as the detection antibody for the total HTT immunoassay (Fig. 1C). We then asked if MW1/2B7 and MW1/pT3 combinations were able to specifically detect HTT EX1 proteins of known purity and concentration bearing no phosphorylation, a phosphorylated T3 residue (pT3), or the same pT3 protein subjected to phosphatase treatment. A serial dilution of each protein was produced and analyzed using the Singulex Erenna platform with the MW1/pT3 antibody pair (Fig. 2A) or with the MW1/2B7 pair (Fig. 2B). The MW1/pT3 pair was clearly able to detect HTT EX1 bearing a pT3 residue, but not the phosphatase-pretreated protein or its unphosphorylated equivalent, thus demonstrating specificity toward pT3 (Fig. 2A). Key parameters for the MW1/pT3 and MW1/2B7 Singulex immunoassays, namely Limit of Detection (LoD), Lower Limit of Quantification (LLoQ), and Upper Limit of Quantification (ULoQ) (48) are summarized in Fig. 2 A and B. Collectively, the data obtained using purified HTT EX1 proteins indicated that the MW1/pT3 and MW1/2B7 Singulex immunoassays were specific, sensitive, and robust. The specificity of the MW1/pT3 and MW1/2B7 Singulex immunoassays was next tested in HEK293T cells through RNAi (knockdown of HTT expression on pT3 HTT and total HTT levels under endogenous or transfected HTT expression) or through specific mutation (effects of T3A mutation on pT3 HTT and total HTT levels with HTT expressed from a plasmid construct). Endogenous HTT expression in HEK293T cells is detectable by WB (Fig. 3A), and these cells have been widely employed to study various huntingtin and mutant huntingtin characteristics, including its phosphorylation status (37, 40, 42). First, we investigated the effects of RNAi [using either a siRNA targeting a specific HTT sequence or a scrambled, control (CTRL) siRNA] on HTT and pT3 HTT protein levels in HEK293T expressed from the endogenous HTT locus or from a transiently transfected plasmid construct expressing a full-length HTT Q18 cDNA. The efficiency of knockdown of endogenous HTT mRNA levels was verified by quantitative real-time PCR (Fig. S2A). As shown in Fig. 3A, HTT protein expression was clearly detected in WB by MAB2166, a specific anti-HTT monoclonal antibody widely employed to detect HTT in different samples (49–53), and was specifically reduced in cells transfected with the HTT-specific siRNA, as confirmed by densitometric analysis (Fig. S2B). Endogenously expressed HTT protein is phosphorylated on T3 as detected in WB by the anti-pT3 pAb. A reduction was observed in cells transfected with the specific HTT siRNA, thus further confirming the specificity of the anti-pT3 pAb. We then interrogated these lysates using the MW1/pT3 and MW1/2B7 Singulex immunoassays to examine the effects of specific genetic knockdown of HTT expression. As shown in Fig. 3B, comparable reductions in pT3 HTT and total HTT protein levels were achieved using genetic knockdown of HTT expression, irrespective of the context (endogenous HTT or overexpressed HTT). These results indicated that the signal obtained from the MW1/pT3 and MW1/2B7 immunoassays in a biological matrix is specific for pT3 HTT and total HTT proteins, respectively.
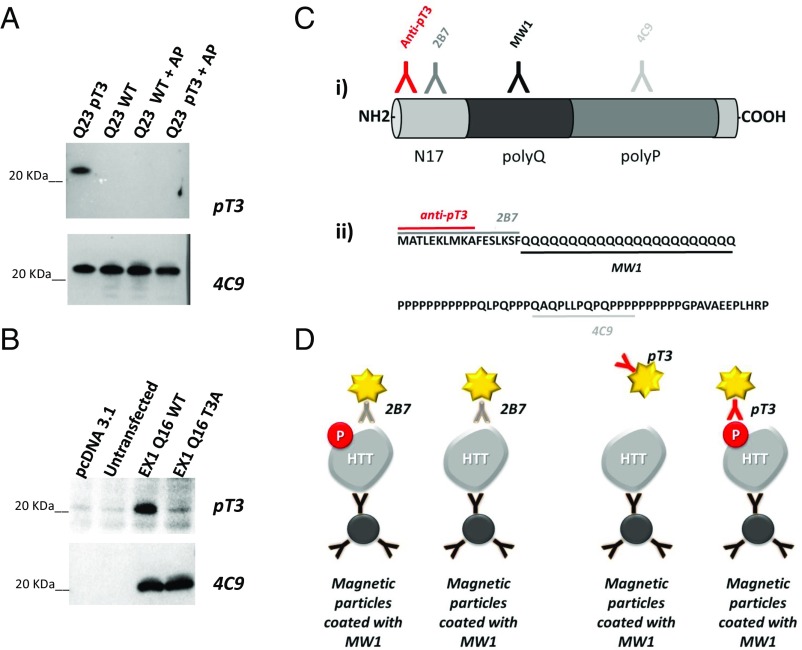
Fig. 1.
Profiling of anti-pT3 HTT antibody. (A) The pT3 signal is specific for the presence of a phosphorylated T3 residue. Western immunoblotting profiling of anti-pT3 antibody on semisynthetic HTT Q23 EX1 protein, HTT EX1 Q23 protein with phosphorylated T3 residue, and the same proteins treated with alkaline phosphatase. (B) The anti-pT3 antibody detects a specific signal on WT HTT EX1 Q16 overexpressed in HEK293T cells lysates, but not on the T3A mutant form by Western immunoblotting analysis. (C) Diagram illustrating the position (i) and epitopes (ii) of antibodies used in the present study. (D) Strategy adopted for the development of sandwich assays specific for total HTT and pT3 HTT, employing antibody MW1 as a capture antibody for both HTT forms.
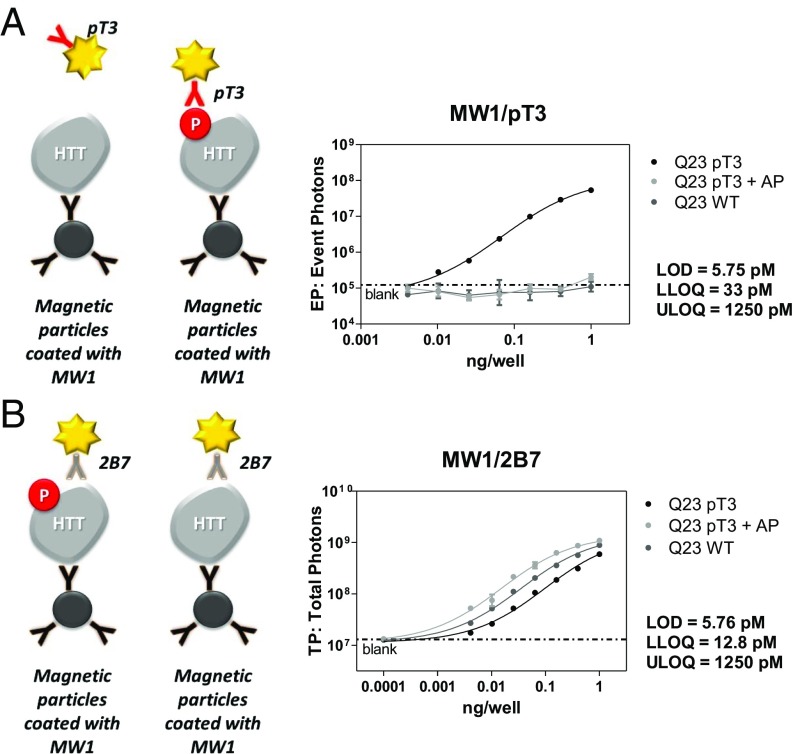
Fig. 2.
Performance of MW1/pT3 and MW1/2B7 Singulex immunoassays. (A) Performance of the MW1/pT3 Singulex immunoassay examined using serial dilutions of semisynthetic HTT EX1 Q23 protein, HTT EX1 Q23 bearing a phosphorylated T3 residue, and HTT EX1 Q23 bearing a phosphorylated T3 residue treated with alkaline phosphatase. The pT3 signal is proportional to the concentration of HTT EX1 Q23 pT3 protein and is sensitive to alkaline phosphatase treatment. Limits of detection and quantification are indicated. (B) Performance of the MW1/2B7 Singulex immunoassay examined using serial dilutions of the same proteins. Comparable levels of the three proteins are detected. Limits of detection and quantification are indicated.
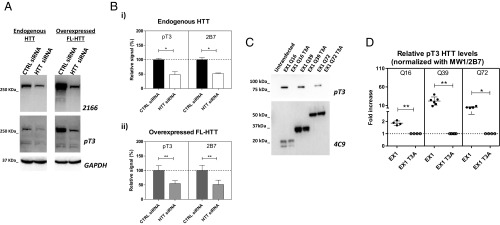
Fig. 3.
Specificity of the MW1/pT3 and MW1/2B7 Singulex immunoassays. (A) Western immunoblot of HEK293T protein lysates demonstrating the effects of HTT mRNA knockdown using a specific HTT siRNA on endogenous and overexpressed full-length (FL) HTT for both pT3 HTT and total HTT protein levels. (B) Singulex analysis using MW1/pT3 or MW1/2B7antibody combinations showing the effects of HTT RNA knockdown on endogenous HTT levels (i) and on HTT protein levels obtained in cells transfected with a plasmid encoding a full-length HTT Q18 cDNA (ii). Data demonstrate that signals from both assays are sensitive to specific HTT expression knockdown. Means and SD of three independent experiments, paired t-test (two-tailed; *P < 0.05; **P < 0.01). (C) Western immunoblot of Phos-Tag SDS/PAGE, representing HEK293T cells transfected with each HTT EX1 plasmid construct bearing different CAG repeats with or without a T3A mutation and probed either with anti-pT3 pAb or with mAb 4C9. The HTT pT3 signal is sensitive to the T3A mutation. (D) Singulex immunoassay detecting effects of the T3A mutation on relative HTT pT3 levels in lysates from HEK293T cells expressing HTT EX1 constructs bearing different polyQ repeats. Means and SD of at least four independent experiments, paired t-test (two-tailed; *P < 0.05; **P ≤ 0.01).
Next, we interrogated the effect of a specific T3 mutation (T3A), abrogating the possibility of phosphorylation on HTT T3, on the signal obtained from the MW1/pT3 and MW1/2B7 immunoassays. As HEK293T cells bearing endogenous T3A HTT alleles are not available, we opted for a transient transfection context. Plasmid constructs encoding HTT EX1 with different polyQ repeats representing a WT expansion (Q16), an HD expansion (Q39), a juvenile-type HD expansion (Q72), or their T3A variants were transiently expressed in HEK293T cells. The specificity of the anti-pT3 pAb was first confirmed by Phos-Tag SDS/PAGE WB (Fig. 3C), as previously shown using HTT EX1-EGFP fusions in the same cell line (40). HTT EX1 protein levels were assessed using 4C9, a monoclonal Ab specific for the polyproline repeat domain (Fig. 3C) (47). Using these lysates, we determined the effect of the T3A mutation on pT3 HTT and total HTT signal obtained with the Singulex MW1/pT3 and MW1/2B7 immunoassays, respectively. The presence of the T3A mutation in the transiently transfected HTT EX1 constructs resulted in a strong reduction of pT3 HTT levels without any effect on total HTT levels (Fig. 3D and Fig. S2C), which was significant for all tested polyQ repeat lengths. Collectively, the data indicated that the Singulex MW1/pT3 and MW1/2B7 immunoassays were sufficiently specific and sensitive to allow investigations of pT3 HTT levels in biological samples of relevance to HD.
Huntingtin T3 Phosphorylation Is Strongly Decreased in HD Mice and in Human HD Samples.
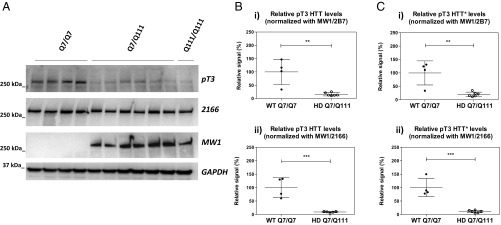
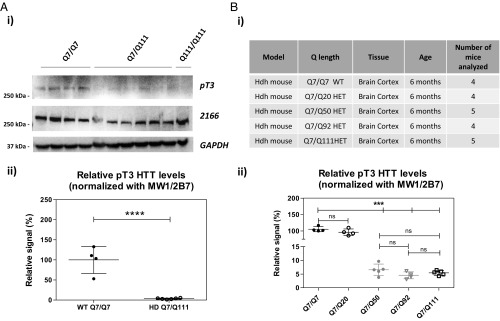
We first examined the effect of expanded polyQ on pT3 HTT levels in the brain, using the Hdh Q111 mouse model (54–56) where the mouse exon 1 sequence has been replaced with a human exon 1 sequence with either a Q7 or a Q111 repeat (Q7/Q111 and Q111/Q111; HD mouse strains). Mutant huntingtin is therefore expressed in a fully physiological manner. Two brain regions (cortex and cerebellum) were investigated from 6-mo-old mice (an age at which EM48-positive nuclear inclusions are not yet detectable) (54, 55). Animals included four homozygous WT (Q7/Q7) mice, six heterozygous mutant (Q7/Q111) mice, and one homozygous mutant (Q111/Q111) mouse. pT3 HTT and total HTT levels were interrogated by SDS/PAGE WB analysis and by Singulex immunoassays. Levels of pT3 HTT as detected by the anti-pT3 pAb in SDS/PAGE WB are clearly decreased in brain cortex from heterozygous Q7/Q111 and homozygous Q111/Q111 mice compared with WT (Q7/Q7) mice, despite comparable total soluble huntingtin protein levels as detected by MAB2166 (epitope amino acids 442–457; Fig. 4A) or the monoclonal antibody D7F7 (epitope around amino acid 1,220; Fig. S3A, i). The expanded polyQ allele was correctly detected by MW1 in brain cortex from heterozygous and homozygous mutant mice but not in cortex from WT mice (Fig. 4A) (46). Coherent results were obtained in pooled whole brains from WT (Q7/Q7) mice and mutant (Q7/Q111) mice (Fig. S3B). Next, pT3 and total HTT levels were assessed using the MW1/pT3 (Fig. S4 A, i) and the companion MW1/2B7 Singulex immunoassay (Fig. S4 A, ii) in a brain cortex sample. To exclude a possible confounding influence of known HTT PTMs on total HTT detection using 2B7, two additional total HTT Singulex immunoassays were employed, based on MAB2166 (epitope amino acids 442–457; Figs. S4 A, iii and S3C) or HDB4E10 (epitope amino acids 1,831–2,131; Figs. S3C and S5A). All three total HTT immunoassays (MW1/2B7, MW1/MAB2166, and MW1/HDB4E10), independent of the detection antibody employed, displayed higher total HTT signals in heterozygous mutant samples than in WT samples, consistent with a degree of selectivity for expanded polyQ displayed by MW1 when used as a capture antibody in these assays (Figs. S4 and S5). Importantly, upon normalization of pT3 signal for total HTT signal (irrespective of the anti-HTT detection antibody used to measure total HTT in the Singulex assay), the analysis confirms a robust decrease of pT3 immunoreactivity when polyQ is expanded (Fig. 4 and Fig. S5B), consistent with WB data. The specificity of the pT3 signal detected in Q7/Q7 and Q7/Q111 brain samples was confirmed by immunodepleting HTT protein using 2B7 (Fig. S6A) and then assessing pT3 HTT and total HTT levels in the Q7/Q7 and in Q7/Q111 contexts (Fig. S6 B and C, respectively) where immunodepletion resulted in a clear, comparable decrease of MW1/pT3 and MW1/MAB2166 signals relative to controls. We also assessed an independently produced and previously characterized rabbit pAb specific for pT3 HTT (39) in the same assay platform, using MW1 as a capture Ab and the anti-pT3 Ab as the detection Ab. This alternative pT3 HTT Singulex immunoassay behaved comparably to the MW1/pT3 Singulex immunoassay under all conditions tested (Fig. 4C and Fig. S4). To determine if the difference in pT3 HTT levels is observable in another brain area, we investigated the cerebellum from these mice by both WB (Fig. 5 A, i; see also Fig. S3A for total HTT detection using the alternative anti-HTT antibody, D7F7) and Singulex immunoassays (Fig. 5 A, ii and Fig. S7A) and found comparable results as those obtained in mouse cortex. The striatum was also investigated with similar results (Fig. S8). We also investigated pT3 HTT levels in cortex from knock-in Hdh mice bearing a humanized HTT exon 1 with different polyQ expansions (Q7, Q20, Q50, Q92, and Q111; Fig. 5 B, i and ii and Fig. S7B) to confirm data obtained in Q7/Q7 and Q7/Q111 mice. As shown in Fig. 5 B, ii, normalized pT3 levels are clearly decreased in the presence of a polyQ expansion of Q50 or greater. Therefore, pT3 levels in the brain of mice expressing a mutant huntingtin protein are decreased relatively to the same brain region of mice expressing WT huntingtin protein. As this effect is observed on the denatured protein (SDS/PAGE) as well as on the native protein (Singulex immunoassay), it is likely due to a bona-fide decrease in phosphorylation levels on the residue rather than a conformational change masking the pT3 epitope as a result of polyQ repeat expansion.
Fig. 4.
Mutant HTT is hypophosphorylated on residue T3 in Hdh Q111 knock-in HD mice cortex. (A) Western immunoblot of homogenates from Hdh knock-in mouse (6-mo cortex), analyzed using anti-pT3 (for HTT pT3), mAb 2166 (for total HTT), and MW1 (for expanded polyQ) antibodies, showing that mutant HTT is hypophosphorylated relative to its Q7 counterpart. (B) Relative HTT pT3 Singulex signal in brain cortex samples from WT (Q7/Q7) or heterozygous mutant (Q7/Q111) Hdh HD animals, normalized to total HTT levels measured either with the MW1/2B7 Singulex immunoassay (i) or with the MW1/2166 Singulex immunoassay (ii). Means and SD of four Q7/Q7 samples and six Q7/Q111 samples, unpaired t-test (two-tailed; **P < 0.01; ***P < 0.005). (C) Relative HTT pT3 (alternative anti-pT3 Ab from ref. 39) Singulex signal in brain cortex samples from WT (Q7/Q7) or heterozygous mutant (Q7/Q111) Hdh HD animals, normalized to total HTT levels measured either with the MW1/2B7 Singulex immunoassay (i) or with the MW1/2166 Singulex immunoassay (ii). Means and SD of four Q7/Q7 samples and six Q7/Q111 samples, unpaired t-test (two-tailed; **P < 0.01; ***P < 0.005).
Fig. 5.
Confirmation of mutant HTT hypophosphorylation on residue T3 in Hdh Q111 knock-in HD mice cerebellum and cortex from Hdh HD knock-in mice with different polyQ expansions. (A, i) Western immunoblot of homogenates from Hdh knock-in mouse (6 mo) cerebellum, analyzed using anti-pT3 (for HTT pT3) and mAb 2166 (for total HTT), and showing that mutant HTT is hypophosphorylated relative to its Q7 counterpart. (A, ii) Relative HTT pT3 Singulex signal in brain cerebellum samples from WT (Q7/Q7) and heterozygous mutant (Q7/Q111) Hdh HD animals, normalized to total HTT measured with the MW1/2B7 Singulex immunoassay. Means and SD of four Q7/Q7 samples and six Q7/Q111 samples, unpaired t-test (two-tailed; ****P < 0.001). (B, i) Analyzed brain cortex samples from Hdh Knock-in mice expressing mutant HTT bearing different expanded polyQ lengths. (B, ii) Relative HTT pT3 levels in these samples, confirming hypophosphorylation on residue T3 in mice expressing mutant HTT. Means and SD of four Q7/Q7, four Q7/Q20, five Q7/Q50, four Q7/Q92, and five Q7/Q111 samples; one-way ANOVA test (***P < 0.005).
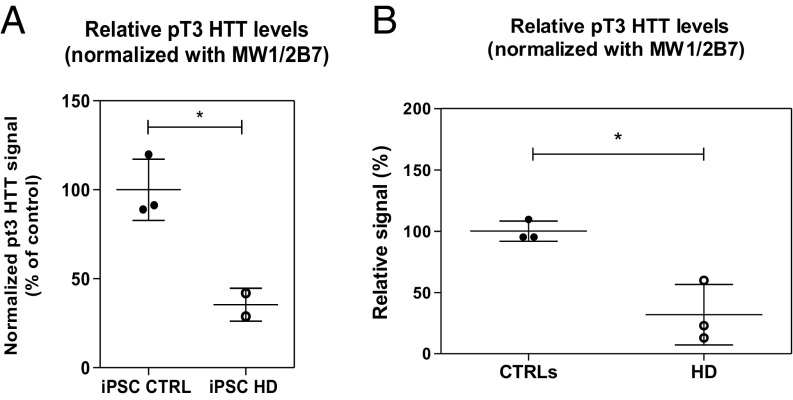
The findings in Hdh Q111 mice prompted us to interrogate HD samples of human origin to determine if the effect of mutant polyQ on HTT pT3 levels observed in animal models is relevant to human HD. Recently, the HD consortium reported the generation of induced pluripotent stem cell (iPSC) lines from HD patients and controls (57). A number of these iPSC lines can be differentiated into a neuronal phenotype, including striatal projection neurons (58). These cells may therefore represent a physiologically relevant human cellular model of HD. We decided to investigate if pT3 HTT levels differed significantly in neuronally differentiated cultures from two HD iPSC lines (Q109 and Q60) and three control iPSC lines (Q21, Q28, and Q33). Following Singulex immunoassay analysis of lysates from these samples (Fig. S9), relative pT3 HTT levels were significantly lower in differentiated cultures from HD iPSCs than in control iPSCs (Fig. 6A). This finding, obtained in one of the most translationally relevant preclinical HD models, led us to investigate HTT pT3 levels directly in clinical samples. To this extent, a small test cohort of peripheral blood mononuclear cell (PBMC) samples was collected from control individuals or from clinically characterized HD patients (Fig. 6B and Fig. S10), as described previously (41, 59). As shown in Fig. 6B, a clear relative decrease in HTT pT3 immunoreactivity was observed in HD PBMCs. Therefore, mutant HTT appears to be hypophosphorylated relative to its WT counterpart across different preclinical models and in human HD.
Fig. 6.
Mutant HTT is hypophosphorylated on residue T3 in HD iPSC-derived neuronal populations and in peripheral (blood) PBMCs isolated from HD individuals. (A) Relative HTT pT3 Singulex signal in iPSC-derived neuronal population protein lysate from WT and HD individuals, normalized to total HTT measured with the MW1/2B7 Singulex immunoassay. Means and SD of three CTRL iPSC lines (#CTRL21: CAG status 21/18; #CTRL28: CAG status 28/18; #CTRL33: CAG status 33/18) and two HD iPSC lines (#HD60: CAG status 60/18; #HD109: CAG status 109/19). Unpaired t-test (two-tailed; *P < 0.05). (B) HTT pT3 Singulex signal in PBMCs isolated from HD individuals, relative to PBMCs from normal individuals and normalized to total HTT measured with the MW1/2B7 Singulex immunoassay. Means and SD of three control and three HD samples, unpaired t-test (two-tailed; *P < 0.05).
T3 Phosphorylation Affects the Conformation and Decreases the Aggregation Properties of Mutant Huntingtin Exon 1.
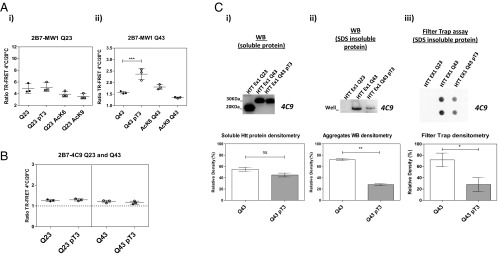
Having established an association between HTT pT3 levels and the HD mutation across multiple models, we asked if HTT T3 phosphorylation influences HD pathology-relevant properties, such as the capacity of mutant polyQ expansions to modulate HTT EX1 fragment conformational properties and aggregation (60–62). Recently, we and others reported on conformational effects imparted by the polyQ expansion on soluble HTT proteins (23–25), aspects of which can be meaningfully investigated with time-resolved fluorescence energy transfer (TR-FRET)–based immunoassays (24, 25). Briefly, the ratio of the signals produced by the TR-FRET antibody pair on HTT proteins at the two test temperatures is inversely proportional to the length of the polyQ repeat and is paralleled by a temperature-dependent change in the α-helicity of HTT’s N terminus, which increases with increasing polyQ length (24, 25). We therefore interrogated the effects of pT3 on the conformation of semisynthetic HTT EX1 with WT (Q23) or mutant (Q43) expansions using the 2B7/MW1 conformational immunoassay, taking advantage of the recent availability of semisynthetic HTT EX1 with Q43. In this assay, pT3 had no significant conformational effect on HTT EX1 Q23 as detected by the 2B7/MW1 TR-FRET immunoassay, with the temperature ratio of the 2B7/MW1 signals from the pT3-modified HTT EX1 Q23 and the unmodified HTT EX1 Q23 proteins being comparable (Fig. 7 A, i). However, pT3 modification of the mutant EX1 HTT Q43 resulted in a significant increase in the temperature ratio of the 2B7/MW1 signals obtained from the EX1 HTT Q43 protein, approximately halving the conformational constraint imposed by the mutant polyQ expansion (Fig. 7 A, ii). Interestingly, semisynthetic HTT Q23 and Q43 proteins bearing another PTM associated with HTT’s N17 region [acetylated K6, AcK6 or acetylated K9, AcK9 residues (32)] produced no effect irrespective of the length of the polyQ region, suggesting that the observed conformational effect is specific for pT3 (Fig. 7 A, i and ii). Additionally, a control TR-FRET immunoassay [2B7/4C9, which does not interrogate the polyQ region and is not sensitive to temperature and polyQ length (24, 25)] did not produce temperature- and polyQ-dependent differences in TR-FRET signals (Fig. 7B). These results suggest that pT3 can influence mutant HTT EX1 conformation, apparently ameliorating the conformational rigidity imparted by the mutant polyQ expansion. In this light, pT3 might therefore be a protective modification. To further investigate this hypothesis, we interrogated the effect of pT3 on semisynthetic mutant HTT EX1 aggregation using a standard filter trap assay (63). A notable difference in the SDS-insoluble fraction present in semisynthetic HTT EX1 Q43 and Q43 pT3 was detectable, suggesting that the presence of T3 phosphorylation may reduce aggregation in these semisynthetic proteins (Fig. 7C). A filter trap analysis confirmed these observations, with a reduction of HTT EX1 Q43 filter-retained material observed when the protein was phosphorylated (Fig. 7 C, iii), consistent with data obtained by other methodologies. As expected, HTT EX1 Q23 did not produce significant signal in the filter trap assay. Collectively, these data suggest that T3 phosphorylation can ameliorate the conformational constraint imposed by the HD mutation on HTT protein and reduce aggregation, at least in the in vitro context examined.
Fig. 7.
T3 phosphorylation impacts mutant HTT N-terminal conformational rigidity and decreases HTT EX1 aggregation. (A, i) T3 phosphorylation has no effect on the conformation of soluble semisynthetic HTT EX1 Q23 protein as measured by the 2B7/MW1 TR-FRET conformational immunoassay. (A, ii) T3 phosphorylation decreases the conformational rigidity of HTT EX1 Q43 protein assessed with the same TR-FRET. Other relevant N17 PTMs, specifically acetylation on residues K6 and K9, do not affect the conformational rigidity of HTT EX1 Q43 protein as measured in parallel using the same assay. Summary of three independent experiments (one-way ANOVA test; ***P < 0.005). (B) The control 2B7/4C9 TR-FRET immunoassay, the signal of which on HTT protein is not temperature- and polyQ-dependent, is not affected by T3 phosphorylation. (C) T3 phosphorylation reduces the aggregation propensity of mutant HTT protein. (i) WB and corresponding densitometric analysis of equivalent nominal amounts of semisynthetic HTT EX1 Q43 protein and its T3 phosphorylated counterpart. Means and SD of three independent experiments, paired t-test (two-tailed; ns, not significant). (ii) Detergent-insoluble material was present in lanes loaded with HTT EX1 Q43 protein, the presence of which was invariably reduced if this protein was phosphorylated on residue T3. Means and SD of three independent experiments, paired t-test (two-tailed; **P < 0.01). (iii) Filter trap analysis of semisynthetic HTT EX1 Q43 protein aggregation in the presence/absence of T3 phosphorylation, confirming reduction of insoluble HTT EX1 Q43 aggregates if residue T3 is phosphorylated. Note that a small difference in soluble Q43 and pT3 Q43 semisynthetic protein levels is observed as measured by densitometry; however, a much larger difference in insoluble protein levels is observed. Means and SD of three independent experiments, paired t-test (two-tailed; *P < 0.05).
Discussion
Mutant HTT self-association and toxicity are believed to be primary determinants of HD pathology (13). Accumulating evidence indicates that the first 17 amino acids of HTT (N17) play a key role in modulating mutant HTT toxicity (30, 64). This domain can adopt an α-helical structure (65–67) and appears to be a key cis-acting modifier of polyQ aggregation (28), subcellular localization, and toxicity (29, 31, 33, 68). The N17 domain can be posttranslationally modified at several residues including pT3 (39) and S13/S16 (32, 33). Introduction of mutations that abolish (S → A) or mimic (S → E/D) phosphorylation at S13 and S16 residues has been shown to affect the conformation of HTT’s N17 domain as well as subcellular localization, aggregation, and toxicity of mutant HTT (29, 33). Studies employing HTT exon 1-like peptides containing pS13/pS16 have confirmed the modulatory effects of phosphomimetic S13/S16 mutations on HTT aggregation and suggest that pS13/pS16 may impart a thermodynamic barrier to the initial phases of oligomerization by interfering with the packing and stabilization of oligomers or by affecting overall oligomer stability (34). The collective evidence therefore argues that increasing or mimicking phosphorylation within HTT N17’s domain reduces mutant HTT toxic properties (29, 33, 69), and significant potential exists for therapeutic strategies aimed at addressing HD through the modulation of these PTMs. Despite this, our capacity for testing the hypothesis that HD pathology can be modulated by leveraging HTT PTMs is presently hindered by the paucity of quantitative, sensitive, and robust assays capable of profiling these PTMs across disease models, of confirming their relevance to human HD, and of enabling the identification of genetic and pharmacological tools for proof-of-concept studies.
We therefore set out to develop HTT assays to interrogate HTT PTMs, focusing initially on phosphorylation of the N17 domain on residue T3. This HTT PTM was chosen for assay development for a number of reasons. First, by analogy with the effects of S13 and S16 modification on HTT’s N17 structure and function, it seemed plausible that phosphorylation of T3 may also play a role in regulating HTT N17’s structure and function, and indeed recent structural evidence argues that T3 is strategically positioned to regulate α-helical content in the N17 domain of HTT (70–72). Importantly, recent studies showed that pT3 inhibits the aggregation of mutant HTT Ex1 (36), and pT3 is the phosphorylation event most readily detected in the N terminus of HTT by mass spectrometry and WB, at least when HTT is overexpressed in cells (39, 40). Also, HTT T3 phosphorylation has been profiled previously in HD models by standard immunoblotting methods to a more detailed extent than other HTT N17 phosphorylations (39). Finally, relevant tools such as semisynthetic HTT proteins bearing a phosphorylated T3 residue and specific antibodies are available (35, 36, 39, 40).
The approach that we applied for pT3 immunoassay design involved the Erenna Singulex sandwich immunoassay, a version of which, based on 2B7 as a capture Ab and MW1 as a detection Ab, was recently employed to develop an ultrasensitive immunoassay for detection of mutant HTT in cerebrospinal fluid (44). Sandwich immunoassay design involved the concomitant development of immunoassays for the detection of HTT pT3 and “total” HTT based on the same capture antibody. Assay development followed a standard approach and involved preliminary assay qualification using reference standards of high purity and known concentration (semisynthetic HTT EX1 proteins with/without pT3), followed by a rigorous interrogation of assay specificity using genetic means (specific RNA knockdown and T3A mutations) in a simple cell-line context. Following development and validation, the MW1/pT3 and MW1/2B7 Singulex immunoassays were used to interrogate the relevance of T3 phosphorylation for HD pathology, starting with the effect of mutant polyQ expansion on pT3 levels. We have shown that MW1 can efficiently bind native HTT-bearing WT polyQ expansions when used as a capture Ab in the Singulex Erenna immunoassay and that under these conditions MW1’s reported higher affinity for expanded polyQ is significantly attenuated. When quantifying pT3 HTT levels in biological samples, we opted to normalize the signal obtained with the MW1/pT3 Singulex Erenna immunoassay with the signal obtained as a measure of total HTT protein present in the sample using the MW1/2B7 Ab combination, which employs the same Ab (MW1) as a capture IgG. We therefore obtained a pT3 HTT signal that is relative to total HTT measured in the same sample. Intersample comparisons are then performed between relative pT3 HTT levels, thus informing in a polyQ-normalized fashion on the proportion of HTT phosphorylated on residue T3. Naturally, several factors aside from absolute T3 phosphorylation levels may influence apparent PTM levels detected by immunoassays because of the intrinsic nature of the antibody–antigen interaction. These include conformational aspects as well as the presence of additional modifiers such as additional PTMs, which we have tried to address using orthogonal approaches (e.g., the use of WB and of multiple antibodies). Immunoreactive pT3 HTT levels were examined in one of the most widely employed mouse models of HD, the Hdh Q111 mouse model (54, 55). In brain cortex of Hdh mice, a thorough examination of pT3 HTT levels was performed, comprising different normalization approaches toward total HTT (the selected MW1/2B7 pair plus an alternative pair composed of the MW1/2166 combination), the use of an alternative, independently developed anti-pT3 pAb (39) for HTT pT3 detection, the demonstration of specificity using an immunodepletion approach, and orthogonal validation by WB. In brain cortex, immunoreactive pT3 HTT levels were clearly decreased in Hdh Q7/Q111 and Q111/Q111 mice relative to Q7/Q7 mice, a result that was mirrored in another brain area, the cerebellum, and observed also in Hdh mice bearing a humanized exon 1 with different mutant polyQ expansions (Q50, Q92, and Q111) relative to mice bearing the same transgene with WT polyQ expansions (Q7 or Q20). Next, we decided to investigate if pT3 HTT levels differed significantly in neuronally differentiated cultures from human iPSC lines derived from control and HD individuals, as patient-derived iPSC lines represent one of the most translationally relevant preclinical HD models (73). Coherent with results obtained in the mouse HD model, relative pT3 HTT levels were significantly lower in differentiated cultures from HD iPSC than in control iPSCs. Finally, we investigated HTT pT3 levels in a clinical sample, the most accessible of which is PBMCs isolated from peripheral blood. Although not representative of the CNS, these peripheral samples can be employed to probe aspects of mutant HTT pertinent to HD pathology (47). A clear decrease in immunoreactive HTT pT3 levels was observed in HD PBMCs relative to PBMCs from control individuals, thus confirming results obtained in preclinical HD models.
Given the lower levels of apparent T3 phosphorylation in mutant huntingtin relative to its WT counterpart in vivo, we investigated the role of pT3 on mutant HTT behavior. We and others have demonstrated that polyQ expansion results in conformational changes in HTT proteins, either purified or expressed in cells, which can be detected by TR-FRET immunoassays (24, 25). Using this conformational TR-FRET immunoassay, we interrogated purified HTT exon 1 proteins (Q23 or Q43) with or without bona-fide T3 phosphorylation and observed that the presence of a phosphorylated T3 residue appears to mitigate the effect of polyQ expansion on HTT exon 1 protein conformation. Although the effects of T3 phosphorylation on HTT N-terminal conformation require further studies, an N17 modification in HTT may alter the α-helicity of HTT’s N terminus (33, 36), and indeed a phosphorylated T3 residue was shown to modulate the α-helicity of HTT exon 1 protein (35, 36). As temperature-dependent α-helicity as well as overall α-helicity at any one temperature are greater in mutant HTT than in WT HTT (24, 25), an alteration in α-helicity would be more readily detected by the conformational TR-FRET immunoassay in mutant HTT than in WT HTT, as indeed was observed here (Fig. 7). Consistent with pT3 mitigating the conformational effects of polyQ expansion on an exon 1 HTT protein fragment (36), we also observed a decreased aggregation propensity for semisynthetic pT3 HTT exon 1 Q43 relative to its WT counterpart (Fig. 7). Although the phosphomimetic T3D mutation was previously reported to increase aggregation of mutant HTT exon 1 in ST14A cells and in a Drosophila HD model (39), recent biophysical studies showed that the T3D mutation does not reproduce the effect of T3 phosphorylation on the structure and aggregation of HTT EX1 (36). It is also plausible that the T3D and T3A mutations may result in structural changes that go beyond the simple phenocopy of the presence/absence of T3 phosphorylation. Consistent with this hypothesis is the observation of the identical protective effects of T3A and T3D mutations in the Drosophila HD model (39).
In conclusion, we have developed a quantitative, sensitive immunoassay for a posttranslational modification of HTT. A strong influence of mutant polyQ expansion on pT3-HTT levels was uncovered across a range of preclinical models, and, importantly, the presence of phosphorylation on T3 residue and polyQ sensitivity in clinical samples was proved, therefore supporting the significance of T3 phosphorylation in human HD. Additionally, we have provided initial evidence that pT3 may influence mutant HTT conformation and aggregation, at least in isolated proteins in vitro. Confirmation of these initial findings in more physiological settings requires the identification of genetic and/or pharmacological tools to modulate HTT T3 phosphorylation in preclinical HD models to interrogate HTT pT3 modulation effects on mutant HTT oligomerization/aggregation and toxicity. The availability of an immunoassay to detect HTT pT3, reported here, opens the way for screening approaches aimed at the identification of such tools and, potentially, for therapeutic approaches leveraging HTT PTM modulation.
Materials and Methods
Antibodies.
The MW1 antibody (developed by Paul Patterson, Caltech, Pasadena, CA and obtained from the Developmental Studies Hybridoma Bank) binds to the polyQ stretch of HTT; the purified form was obtained using Protein G HP Spin Trap (catalog #28–9031-34; GE Healthcare Life Science) following the manufacturer’s recommendations. The 2B7 and 4C9 antibodies recognize the HTT N-Term 17 amino acids and the polyP region in exon 1 of the HTT protein, respectively (74), and were obtained from the CHDI (Cure for Huntington's Disease Initiative) Foundation. The anti-pT3 antibody was previously described (35, 40). MAB2166 was supplied by a commercial source (catalog #MAB2166; Millipore) and binds to a 15-aa region spanning from amino acids 445 to 459 of the human HTT protein (50). HDB4E10 antibody was distributed by Thermo Fisher Scientific and binds the region from amino acid 1,831 to 2,131 of human HTT protein. D7F7 is a commercial antibody supplied by Cell Signaling Technology and recognizes a region surrounding Pro1220 of human HTT protein. The 8A4 antibody (catalog #sc-47759) was purchased from Santa Cruz Biotechnology and binds amino acids 2,703–2,911 of human HTT protein. Antibody against GAPDH was distributed by Sigma-Aldrich (catalog #G9545). Secondary antibodies used for WB were goat–anti-mouse IgG HRP-conjugated (catalog #12–349; Merck Millipore) and goat–anti-rabbit IgG HRP-conjugated (catalog #12–348; Merck Millipore). The D2 fluorophore and the terbium cryptate antibody labelings were custom made by CisBio. The Alexa-647 labeling was performed using the Alexa Fluor-647 Monoclonal Antibody Labeling Kit from Thermo Fisher Scientific (catalog #A20186) following the manufacturer’s instructions. MW1 antibody was conjugated to magnetic particles for Singulex assays, following the manufacturer’s recommendations (catalog #03–0077-02; Singulex).
Semisynthetic Proteins.
The production of semisynthetic HTT exon 1 proteins with relevant PTMs was described before (35, 36). Pure trifluoroacetic acid was added to the lyophilized protein powder for disaggregation, and proteins were then dissolved in TBS buffer (50 mM Tris 150 mM NaCl) to obtain a final concentration of 20 µM (pH adjusted to 7.2–7.4 using 1 M NaOH). Protein solutions were filtered through a 100-kDa membrane (Nanosep Centrifugal Devices 100K Omega, catalog #OD100C34; Pall). Each sample was supplied with 1% Tween-20. Dephosphorylation of protein was performed using 10 unit/µL concentrated alkaline phosphatase (calf intestinal phosphatase, catalog #M0290S; New England Biolabs), following the manufacturer’s instructions.
Plasmid and Constructs.
cDNAs encoding N-terminal HTT fragments (exon 1) bearing different polyQ lengths (Q16, Q39, or Q72) were synthesized by Genscript, quality-controlled by DNA sequencing, and subcloned into pCDNA3.1, and their expression in mammalian cells was validated as previously reported (24). N17 domain mutant HTT exon 1 constructs (threonine in position 3 mutated to alanine) were synthesized and validated as above. cDNAs encoding human full-length HTT (Q18) in pCDNA3.1 were reported previously (75).
HEK293T Cell Culture and Manipulation.
HEK293T cells were cultured and lysed as previously described (24, 40). Lysis was performed 24 h after transfection in lysis buffer (PBS, 0.4% Triton X100) supplemented with 1× protease inhibitor mixture (Roche). RNA interference (RNAi) was performed using specific Huntingtin short interfering RNAs (siRNAs) (MISSION Predesigned HTT siRNAs, Sigma-Aldrich). mRNA was extracted from HEK293T cells silenced with siRNAs using RNeasy Plus Mini (catalog #74134; Qiagen), and lysis of cells was carried out 48 h after transfection (as described above). mRNA was reverse transcribed using the SuperScript III First-Strand Synthesis System (catalog #18080–051; Thermo Fisher Scientific). Real-time PCR was performed using Power SYBR Green PCR Master Mix, Applied Biosystem (catalog# 4367659; Thermo Fisher Scientific) with specific HTT primers (FWD 5′-GTGGAGGTTTGCTGAGCTG-3′; REV GCAAAATTGCCAAAAGAAGC; Bio-Fab Research Srl), and an Applied Biosystems real-time PCR platform (AbiPrism7900 HT) was used for amplification and analysis.
iPSC Culture and Sample Processing.
Neuronally differentiated iPSC lines from control individual and HD patients were generated and characterized by the E. Cattaneo laboratory (University of Milan). The human iPSC lines used (#CTRL21: CAG status 21/18; #CTRL28: CAG status 28/18; #CTRL33: CAG status 33/18; #HD60: CAG status 60/18; #HD109: CAG status 109/19) are those reported in ref. 57 and have been differentiated applying the striatal differentiation protocol in ref. 58. The protocol was characterized for its reproducibility in human ES and iPSC lines as extensively described in ref. 76, and the cells were harvested and immediately stored as pellets at −80 °C. Frozen-cell pellets were lysed using lysis buffer (composition described above) and cleared after sonication.
Animal Tissues.
HD knock-in mice with increasing CAG repeat lengths were reported previously (56, 77). For tissue preparation, 6-mo-old mice were employed. Following dissection, mouse brain-tissue samples were homogenized using a Fast-Prep 96 (BioMedical) in 10 vol (wt/vol) of lysis buffer (composition described above) using PreCellys tubes, and two lysis cycles of 10 min at 6,000 × g were carried out. Homogenates were cleared and stored at −80 °C. Tissues from knock-in Hdh mice bearing a humanized exon 1 with different polyQ expansions (Q7, Q20, Q50, Q92, and Q111) were obtained from the CHDI Foundation.
Human Samples.
Peripheral blood lymphocytes from control and HD subjects were obtained as described (78), harvested by centrifugation, and snap-frozen as pellets. Pellets were resuspended in lysis buffer (composition previously described), sonicated, and finally clarified before the total protein quantification. A program to collect biological specimens, including blood samples, at Instituto CSS-Mendel, Istituto di Ricovero e Cura a Carattere Scientifico (IRCCS), Ospedale Casa Sollievo della Sofferenza for research purposes, was approved by the Ethical Committee from the Casa Sollievo della Sofferenza Foundation, section of Istituto Tumori Giovanni Paolo II in Bari. Informed consents were obtained from patients and healthy control subjects.
Western Blot Assay, Filter Trap Assay, and pTag Gel.
For WB, samples were denatured at 95 °C in 4× Loading Buffer (125 mM Tris⋅HCl, pH 6.8, 6% SDS, 4 M urea, 4 mM EDTA, 30% glycerol, 4% 2-mercaptoethanol and bromophenol blue) and loaded on NuPAGE 4–12% Bis–Tris Gel (catalog #WG1402BOX; Thermo Fisher Scientific). Proteins were transferred on PVDF membrane (catalog #162–0177; Bio-Rad Laboratories) using wet blotting. After fixing in 0.4% paraformaldehyde/0.4% sucrose solution and blocking with 5% nonfat milk in TBS/0.1% Tween-20, primary antibody incubation was carried out overnight at 4 °C, and secondary antibody incubations for 1 h at room temperature. Protein bands were detected using chemiluminescence substrate (Supersignal West Femto Maximum catalog #3406; Supersignal West Pico Maximum catalog #34087; Thermo Fisher Scientific) on Chemidoc XRS+ (Bio-Rad Laboratories).
For filter-trap assay, semisynthetic proteins were diluted in TBS/1% SDS. Vacuum filtration of samples was performed through a 96-well dot-blot apparatus (Bio-Dot Apparatus #1706545; Bio-Rad Laboratories) containing a 0.2-µM cellulose acetate membrane filter (catalog #10404180; Whatman) prewetted in TBS/1% SDS. After two washing steps with TBS/1% SDS, the membrane was fixed, blocked, and incubated with antibodies as previously described (63). For Phos-Tag gel analysis, cell lysates were prepared and run as previously described (40).
Immunoprecipitation.
Immunoprecipitation was performed using Dynabeads Protein G (catalog #10004D; Thermo Fisher Scientific) following the manufacturer’s instructions and using an HTT-specific antibody (2B7) or an unrelated antibody (GFAP, catalog #G9269; Sigma- Aldrich). The pulled-down material was loaded on a SDS/PAGE, and the supernatant (immunodepleted sample) was analyzed by Singulex assay.
Singulex Assays.
A total of 50 µL/well of dilution buffer (6% BSA, 0.8% Triton X-100, 750 mM NaCl, and complete protease inhibitor) was added to a 96-well plate (catalog #P-96–450V-C; Axygen). Samples to be tested were diluted in artificial cerebral spinal fluid (0.3 M NaCl; 6 mM KCl; 2.8 mM CaCl2-2H2O; 1.6 mM MgCl2-6H20; 1.6 mM Na2HPO4-7H2O; 0.4 mM NaH2PO4-H2O) supplemented with 1% Tween-20 and complete protease inhibitor in a final volume of 150 µL/well. Finally, 100 µL/well of the MW1 antibody coupled with magnetic particles (appropriately diluted in Erenna Assay buffer, catalog #02–0474-00; Singulex) was added to the assay plate and incubated for 1 h at room temperature under orbital shaking. The beads were then washed with Erenna System buffer (catalog #02–0111-00; Singulex) and resuspended using 20 µL/well of the specific detection antibody labeled with D2 fluorophore (or Alexa-647 fluorophore) appropriately diluted in Erenna Assay buffer. The plate was incubated for 1 h at room temperature under shaking. After washing, the beads were resuspended and transferred to a new 96-well plate. A total of 10 µL/well of Erenna buffer B (catalog #02–0297-00; Singulex) was added to the beads for elution and incubated for 5 min at room temperature under orbital shaking. The eluted complex was magnetically separated from the beads and transferred to a 384-well plate (Nunc catalog #264573; Sigma-Aldrich) where it was neutralized with 10 µL/well of Erenna buffer D (catalog #02–0368-00; Singulex). Finally, the 384-well plate was heat-sealed and analyzed with the Erenna Immunoassay System.
TR-FRET Assays.
A total of 5 µL/well of samples and 1 µL/well of antibody mixtures (2B7-Tb 1 ng/µL; MW1-D2 10 ng/µL; 4C9-Alexa647 10 ng/µL) were diluted in lysis buffer (composition described above), and the assay was performed as described (24).
Data Analysis.
Supplementary Material
Acknowledgments
We thank Daniel Lavery, Elizabeth Doherty, and Seung Kwak for advice and for reading the manuscript and providing comments. The CHDI Foundation is gratefully acknowledged for providing support and reagents. The research leading to these results has been funded in part by the Collezione Nazionale di Composti Chimici e Centro Screening (CNCCS Scarl).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1705372114/-/DCSupplemental.
References
- 1.Prabakaran S, Lippens G, Steen H, Gunawardena J. Post-translational modification: Nature’s escape from genetic imprisonment and the basis for dynamic information encoding. Wiley Interdiscip Rev Syst Biol Med. 2012;4:565–583. doi: 10.1002/wsbm.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khoury GA, Baliban RC, Floudas CA. Proteome-wide post-translational modification statistics: Frequency analysis and curation of the Swiss-Prot database. Sci Rep. 2011;1:srep00090. doi: 10.1038/srep00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Soto C. Unfolding the role of protein misfolding in neurodegenerative diseases. Nat Rev Neurosci. 2003;4:49–60. doi: 10.1038/nrn1007. [DOI] [PubMed] [Google Scholar]
- 4.Selkoe DJ. Cell biology of protein misfolding: The examples of Alzheimer’s and Parkinson’s diseases. Nat Cell Biol. 2004;6:1054–1061. doi: 10.1038/ncb1104-1054. [DOI] [PubMed] [Google Scholar]
- 5.Noble W, Hanger DP, Miller CC, Lovestone S. The importance of tau phosphorylation for neurodegenerative diseases. Front Neurol. 2013;4:83. doi: 10.3389/fneur.2013.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sabbagh JJD, Dickey CA. The metamorphic nature of the tau protein: Dynamic flexibility comes at a cost. Front Neurosci. 2016;10:3. doi: 10.3389/fnins.2016.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Basso M, Pennuto M. Serine phosphorylation and arginine methylation at the crossroads to neurodegeneration. Exp Neurol. 2015;271:77–83. doi: 10.1016/j.expneurol.2015.05.003. [DOI] [PubMed] [Google Scholar]
- 8.Ehrnhoefer DE, Sutton L, Hayden MR. Small changes, big impact: Posttranslational modifications and function of huntingtin in Huntington disease. Neuroscientist. 2011;17:475–492. doi: 10.1177/1073858410390378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Emamian ES, et al. Serine 776 of ataxin-1 is critical for polyglutamine-induced disease in SCA1 transgenic mice. Neuron. 2003;38:375–387. doi: 10.1016/s0896-6273(03)00258-7. [DOI] [PubMed] [Google Scholar]
- 10.Duvick L, et al. SCA1-like disease in mice expressing wild-type ataxin-1 with a serine to aspartic acid replacement at residue 776. Neuron. 2010;67:929–935. doi: 10.1016/j.neuron.2010.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marcus JN, Schachter J. Targeting post-translational modifications on tau as a therapeutic strategy for Alzheimer’s disease. J Neurogenet. 2011;25:127–133. doi: 10.3109/01677063.2011.626471. [DOI] [PubMed] [Google Scholar]
- 12.The Huntington’s Disease Collaborative Research Group A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 13.Ross CA, Tabrizi SJ. Huntington’s disease: From molecular pathogenesis to clinical treatment. Lancet Neurol. 2011;10:83–98. doi: 10.1016/S1474-4422(10)70245-3. [DOI] [PubMed] [Google Scholar]
- 14.Snell RG, et al. Relationship between trinucleotide repeat expansion and phenotypic variation in Huntington’s disease. Nat Genet. 1993;4:393–397. doi: 10.1038/ng0893-393. [DOI] [PubMed] [Google Scholar]
- 15.Brinkman RR, Mezei MM, Theilmann J, Almqvist E, Hayden MR. The likelihood of being affected with Huntington disease by a particular age, for a specific CAG size. Am J Hum Genet. 1997;60:1202–1210. [PMC free article] [PubMed] [Google Scholar]
- 16.Sathasivam K, et al. Aberrant splicing of HTT generates the pathogenic exon 1 protein in Huntington disease. Proc Natl Acad Sci USA. 2013;110:2366–2370. doi: 10.1073/pnas.1221891110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gipson TA, Neueder A, Wexler NS, Bates GP, Housman D. Aberrantly spliced HTT, a new player in Huntington’s disease pathogenesis. RNA Biol. 2013;10:1647–1652. doi: 10.4161/rna.26706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hazeki N, Nakamura K, Goto J, Kanazawa I. Rapid aggregate formation of the huntingtin N-terminal fragment carrying an expanded polyglutamine tract. Biochem Biophys Res Commun. 1999;256:361–366. doi: 10.1006/bbrc.1999.0337. [DOI] [PubMed] [Google Scholar]
- 19.Lunkes A, et al. Proteases acting on mutant huntingtin generate cleaved products that differentially build up cytoplasmic and nuclear inclusions. Mol Cell. 2002;10:259–269. doi: 10.1016/s1097-2765(02)00602-0. [DOI] [PubMed] [Google Scholar]
- 20.Wang CE, et al. Accumulation of N-terminal mutant huntingtin in mouse and monkey models implicated as a pathogenic mechanism in Huntington’s disease. Hum Mol Genet. 2008;17:2738–2751. doi: 10.1093/hmg/ddn175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Landles C, et al. Proteolysis of mutant huntingtin produces an exon 1 fragment that accumulates as an aggregated protein in neuronal nuclei in Huntington disease. J Biol Chem. 2010;285:8808–8823. doi: 10.1074/jbc.M109.075028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barbaro BA, et al. Comparative study of naturally occurring huntingtin fragments in Drosophila points to exon 1 as the most pathogenic species in Huntington’s disease. Hum Mol Genet. 2015;24:913–925. doi: 10.1093/hmg/ddu504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caron NS, Desmond CR, Xia J, Truant R. Polyglutamine domain flexibility mediates the proximity between flanking sequences in huntingtin. Proc Natl Acad Sci USA. 2013;110:14610–14615. doi: 10.1073/pnas.1301342110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fodale V, et al. Polyglutamine- and temperature-dependent conformational rigidity in mutant huntingtin revealed by immunoassays and circular dichroism spectroscopy. PLoS One. 2014;9:e112262. doi: 10.1371/journal.pone.0112262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cui X, et al. TR-FRET assays of Huntingtin protein fragments reveal temperature and polyQ length-dependent conformational changes. Sci Rep. 2014;4:5601. doi: 10.1038/srep05601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Darnell G, Orgel JP, Pahl R, Meredith SC. Flanking polyproline sequences inhibit beta-sheet structure in polyglutamine segments by inducing PPII-like helix structure. J Mol Biol. 2007;374:688–704. doi: 10.1016/j.jmb.2007.09.023. [DOI] [PubMed] [Google Scholar]
- 27.Bugg CW, Isas JM, Fischer T, Patterson PH, Langen R. Structural features and domain organization of huntingtin fibrils. J Biol Chem. 2012;287:31739–31746. doi: 10.1074/jbc.M112.353839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tam S, et al. The chaperonin TRiC blocks a huntingtin sequence element that promotes the conformational switch to aggregation. Nat Struct Mol Biol. 2009;16:1279–1285. doi: 10.1038/nsmb.1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gu X, et al. Serines 13 and 16 are critical determinants of full-length human mutant huntingtin induced disease pathogenesis in HD mice. Neuron. 2009;64:828–840. doi: 10.1016/j.neuron.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gu X, et al. N17 modifies mutant Huntingtin nuclear pathogenesis and severity of disease in HD BAC transgenic mice. Neuron. 2015;85:726–741. doi: 10.1016/j.neuron.2015.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng Z, Li A, Holmes BB, Marasa JC, Diamond MI. An N-terminal nuclear export signal regulates trafficking and aggregation of Huntingtin (Htt) protein exon 1. J Biol Chem. 2013;288:6063–6071. doi: 10.1074/jbc.M112.413575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thompson LM, et al. IKK phosphorylates Huntingtin and targets it for degradation by the proteasome and lysosome. J Cell Biol. 2009;187:1083–1099. doi: 10.1083/jcb.200909067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Atwal RS, et al. Kinase inhibitors modulate huntingtin cell localization and toxicity. Nat Chem Biol. 2011;7:453–460. doi: 10.1038/nchembio.582. [DOI] [PubMed] [Google Scholar]
- 34.Mishra R, et al. Serine phosphorylation suppresses huntingtin amyloid accumulation by altering protein aggregation properties. J Mol Biol. 2012;424:1–14. doi: 10.1016/j.jmb.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ansaloni A, et al. One-pot semisynthesis of exon 1 of the Huntingtin protein: New tools for elucidating the role of posttranslational modifications in the pathogenesis of Huntington’s disease. Angew Chem Int Ed Engl. 2014;53:1928–1933. doi: 10.1002/anie.201307510. [DOI] [PubMed] [Google Scholar]
- 36.Chiki A, et al. Mutant exon1 huntingtin aggregation is regulated by T3 phosphorylation-induced structural changes and crosstalk between T3 phosphorylation and acetylation at K6. Angew Chem Int Ed Engl. 2017;56:5202–5207. doi: 10.1002/anie.201611750. [DOI] [PubMed] [Google Scholar]
- 37.Schilling B, et al. Huntingtin phosphorylation sites mapped by mass spectrometry. Modulation of cleavage and toxicity. J Biol Chem. 2006;281:23686–23697. doi: 10.1074/jbc.M513507200. [DOI] [PubMed] [Google Scholar]
- 38.Warby SC, et al. Huntingtin phosphorylation on serine 421 is significantly reduced in the striatum and by polyglutamine expansion in vivo. Hum Mol Genet. 2005;14:1569–1577. doi: 10.1093/hmg/ddi165. [DOI] [PubMed] [Google Scholar]
- 39.Aiken CT, et al. Phosphorylation of threonine 3: Implications for huntingtin aggregation and neurotoxicity. J Biol Chem. 2009;284:29427–29436. doi: 10.1074/jbc.M109.013193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bustamante MB, et al. Detection of huntingtin exon 1 phosphorylation by Phos-Tag SDS-PAGE: Predominant phosphorylation on threonine 3 and regulation by IKKβ. Biochem Biophys Res Commun. 2015;463:1317–1322. doi: 10.1016/j.bbrc.2015.06.116. [DOI] [PubMed] [Google Scholar]
- 41.Di Pardo A, et al. FTY720 (fingolimod) is a neuroprotective and disease-modifying agent in cellular and mouse models of Huntington disease. Hum Mol Genet. 2014;23:2251–2265. doi: 10.1093/hmg/ddt615. [DOI] [PubMed] [Google Scholar]
- 42.Huang B, et al. Scalable production in human cells and biochemical characterization of full-length normal and mutant huntingtin. PLoS One. 2015;10:e0121055. doi: 10.1371/journal.pone.0121055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Todd J, et al. Ultrasensitive flow-based immunoassays using single-molecule counting. Clin Chem. 2007;53:1990–1995. doi: 10.1373/clinchem.2007.091181. [DOI] [PubMed] [Google Scholar]
- 44.Wild EJ, et al. Quantification of mutant huntingtin protein in cerebrospinal fluid from Huntington’s disease patients. J Clin Invest. 2015;125:1979–1986. doi: 10.1172/JCI80743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ko J, Ou S, Patterson PH. New anti-huntingtin monoclonal antibodies: Implications for huntingtin conformation and its binding proteins. Brain Res Bull. 2001;56:319–329. doi: 10.1016/s0361-9230(01)00599-8. [DOI] [PubMed] [Google Scholar]
- 46.Klein FA, et al. Linear and extended: A common polyglutamine conformation recognized by the three antibodies MW1, 1C2 and 3B5H10. Hum Mol Genet. 2013;22:4215–4223. doi: 10.1093/hmg/ddt273. [DOI] [PubMed] [Google Scholar]
- 47.Weiss A, et al. Mutant huntingtin fragmentation in immune cells tracks Huntington’s disease progression. J Clin Invest. 2012;122:3731–3736. doi: 10.1172/JCI64565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Armbruster DA, Pry T. Limit of blank, limit of detection and limit of quantitation. Clin Biochem Rev. 2008;29:S49–S52. [PMC free article] [PubMed] [Google Scholar]
- 49.Chen M, et al. Minocycline inhibits caspase-1 and caspase-3 expression and delays mortality in a transgenic mouse model of Huntington disease. Nat Med. 2000;6:797–801. doi: 10.1038/77528. [DOI] [PubMed] [Google Scholar]
- 50.Cong SY, Pepers BA, Roos RA, Van Ommen GJ, Dorsman JC. Epitope mapping of monoclonal antibody 4C8 recognizing the protein huntingtin. Hybridoma (Larchmt) 2005;24:231–235. doi: 10.1089/hyb.2005.24.231. [DOI] [PubMed] [Google Scholar]
- 51.Kaltenbach LS, et al. Huntingtin interacting proteins are genetic modifiers of neurodegeneration. PLoS Genet. 2007;3:e82. doi: 10.1371/journal.pgen.0030082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Macdonald D, et al. Quantification assays for total and polyglutamine-expanded huntingtin proteins. PLoS One. 2014;9:e96854. doi: 10.1371/journal.pone.0096854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yao Y, et al. A striatal-enriched intronic GPCR modulates huntingtin levels and toxicity. Elife. 2015;4:e05449. doi: 10.7554/eLife.05449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wheeler VC, et al. Early phenotypes that presage late-onset neurodegenerative disease allow testing of modifiers in Hdh CAG knock-in mice. Hum Mol Genet. 2002;11:633–640. doi: 10.1093/hmg/11.6.633. [DOI] [PubMed] [Google Scholar]
- 55.Wheeler VC, et al. Long glutamine tracts cause nuclear localization of a novel form of huntingtin in medium spiny striatal neurons in HdhQ92 and HdhQ111 knock-in mice. Hum Mol Genet. 2000;9:503–513. doi: 10.1093/hmg/9.4.503. [DOI] [PubMed] [Google Scholar]
- 56.Brito V, et al. Neurotrophin receptor p75(NTR) mediates Huntington’s disease-associated synaptic and memory dysfunction. J Clin Invest. 2014;124:4411–4428. doi: 10.1172/JCI74809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mattis VB, et al. HD iPSC-derived neural progenitors accumulate in culture and are susceptible to BDNF withdrawal due to glutamate toxicity. Hum Mol Genet. 2015;24:3257–3271. doi: 10.1093/hmg/ddv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Delli Carri A, et al. Human pluripotent stem cell differentiation into authentic striatal projection neurons. Stem Cell Rev. 2013;9:461–474. doi: 10.1007/s12015-013-9441-8. [DOI] [PubMed] [Google Scholar]
- 59.Di Pardo A, et al. Changes of peripheral TGF-β1 depend on monocytes-derived macrophages in Huntington disease. Mol Brain. 2013;6:55. doi: 10.1186/1756-6606-6-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.DiFiglia M, et al. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997;277:1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- 61.Li SH, Li XJ. Aggregation of N-terminal huntingtin is dependent on the length of its glutamine repeats. Hum Mol Genet. 1998;7:777–782. doi: 10.1093/hmg/7.5.777. [DOI] [PubMed] [Google Scholar]
- 62.Martindale D, et al. Length of huntingtin and its polyglutamine tract influences localization and frequency of intracellular aggregates. Nat Genet. 1998;18:150–154. doi: 10.1038/ng0298-150. [DOI] [PubMed] [Google Scholar]
- 63.Wanker EE, et al. Membrane filter assay for detection of amyloid-like polyglutamine-containing protein aggregates. Methods Enzymol. 1999;309:375–386. doi: 10.1016/s0076-6879(99)09026-6. [DOI] [PubMed] [Google Scholar]
- 64.Arndt JR, Chaibva M, Legleiter J. The emerging role of the first 17 amino acids of huntingtin in Huntington’s disease. Biomol Concepts. 2015;6:33–46. doi: 10.1515/bmc-2015-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kelley NW, et al. The predicted structure of the headpiece of the huntingtin protein and its implications on huntingtin aggregation. J Mol Biol. 2009;388:919–927. doi: 10.1016/j.jmb.2009.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Atwal RS, et al. Huntingtin has a membrane association signal that can modulate huntingtin aggregation, nuclear entry and toxicity. Hum Mol Genet. 2007;16:2600–2615. doi: 10.1093/hmg/ddm217. [DOI] [PubMed] [Google Scholar]
- 67.Kim MW, Chelliah Y, Kim SW, Otwinowski Z, Bezprozvanny I. Secondary structure of Huntingtin amino-terminal region. Structure. 2009;17:1205–1212. doi: 10.1016/j.str.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Maiuri T, Woloshansky T, Xia J, Truant R. The huntingtin N17 domain is a multifunctional CRM1 and Ran-dependent nuclear and cilial export signal. Hum Mol Genet. 2013;22:1383–1394. doi: 10.1093/hmg/dds554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Di Pardo A, et al. Ganglioside GM1 induces phosphorylation of mutant huntingtin and restores normal motor behavior in Huntington disease mice. Proc Natl Acad Sci USA. 2012;109:3528–3533. doi: 10.1073/pnas.1114502109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Arndt JR, Brown RJ, Burke KA, Legleiter J, Valentine SJ. Lysine residues in the N-terminal huntingtin amphipathic α-helix play a key role in peptide aggregation. J Mass Spectrom. 2015;50:117–126. doi: 10.1002/jms.3504. [DOI] [PubMed] [Google Scholar]
- 71.De Genst E, et al. Structure of a single-chain Fv bound to the 17 N-terminal residues of huntingtin provides insights into pathogenic amyloid formation and suppression. J Mol Biol. 2015;427:2166–2178. doi: 10.1016/j.jmb.2015.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Elbaum MB, Zondlo NJ. OGlcNAcylation and phosphorylation have similar structural effects in α-helices: Post-translational modifications as inducible start and stop signals in α-helices, with greater structural effects on threonine modification. Biochemistry. 2014;53:2242–2260. doi: 10.1021/bi500117c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ross CA, Akimov SS. Human-induced pluripotent stem cells: Potential for neurodegenerative diseases. Hum Mol Genet. 2014;23:R17–R26. doi: 10.1093/hmg/ddu204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Paganetti P, et al. Development of a method for the high-throughput quantification of cellular proteins. Chembiochem. 2009;10:1678–1688. doi: 10.1002/cbic.200900131. [DOI] [PubMed] [Google Scholar]
- 75.Baldo B, et al. TR-FRET-based duplex immunoassay reveals an inverse correlation of soluble and aggregated mutant huntingtin in Huntington’s disease. Chem Biol. 2012;19:264–275. doi: 10.1016/j.chembiol.2011.12.020. [DOI] [PubMed] [Google Scholar]
- 76.Delli Carri A, et al. Developmentally coordinated extrinsic signals drive human pluripotent stem cell differentiation toward authentic DARPP-32+ medium-sized spiny neurons. Development. 2013;140:301–312. doi: 10.1242/dev.084608. [DOI] [PubMed] [Google Scholar]
- 77.Langfelder P, et al. Integrated genomics and proteomics define huntingtin CAG length-dependent networks in mice. Nat Neurosci. 2016;19:623–633. doi: 10.1038/nn.4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gantner F, Kupferschmidt R, Schudt C, Wendel A, Hatzelmann A. In vitro differentiation of human monocytes to macrophages: Change of PDE profile and its relationship to suppression of tumour necrosis factor-alpha release by PDE inhibitors. Br J Pharmacol. 1997;121:221–231. doi: 10.1038/sj.bjp.0701124. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.