SUMMARY
The emerging pathogen Candida auris has been associated with nosocomial outbreaks on five continents. Genetic analysis indicates the simultaneous emergence of separate clades of this organism in different geographical locations. Invasive infection and colonization have been detected predominantly in patients in high-dependency settings and have garnered attention due to variable antifungal resistance profiles and transmission within units instituting a range of infection prevention and control measures. Issues with the identification of C. auris using both phenotypic and molecular techniques have raised concerns about detecting the true scale of the problem. This review considers the literature available on C. auris and highlights the key unknowns, which will provide direction for further work in this field.
KEYWORDS: Candida auris, emerging infection, nosocomial transmission
INTRODUCTION
Candida auris, a novel Candida species first reported in Japan in 2009, is an emerging pathogen that has been isolated on five continents (1). There are separate clonal strains displaying distinct mechanisms of antifungal resistance. C. auris is associated with nosocomial outbreaks in intensive care settings, and transmission despite the implementation of enhanced infection prevention and control (IPC) measures is a particular concern. Variable antifungal susceptibility profiles and the development of resistance following antifungal exposure have been observed. In addition, difficulties in identification using conventional phenotypic and molecular techniques, the unknown population prevalence, the uncertain environmental niches, and the unclear mechanisms of spread have hindered control.
The increasing prevalence of colonization and infection with non-albicans Candida species in recent years is thought to be driven largely by the increasing use of prophylactic antifungal agents such as fluconazole (2). Previously, invasive candidiasis was caused predominantly by Candida albicans. As a result of the shift toward non-albicans Candida species with various susceptibility patterns, including multidrug-resistant species, fluconazole can no longer be the mainstay of empirical antifungal treatment. C. auris, with its propensity to spread rapidly in critically ill patients, has the potential to become a dominant opportunistic pathogen in these populations.
Given these uncertainties, we performed a literature review to identify the current state of knowledge on a variety of parameters such as epidemiology, genetics, identification, cell biology, and management, including prevention and control strategies. We also highlight the key unknowns and identify targeted areas for further work.
METHODS
We performed a search of the literature between January 2000 and September 2017 for data on C. auris using Medline, Embase, Scopus, NICE Evidence Search, Global Health, and CINAHL, limited to publications in the English language. The search terms Candida auris and C. auris were used. Abstracts were analyzed by two researchers (A.J.-S. and C.S.B.). Articles were deduplicated and excluded if there was no, or passing, reference to C. auris and if they did not contain information on epidemiology, diagnosis, treatment, or resistance patterns. Gray literature and international guidelines were included in a separate search based on discussions with international colleagues relating to public health responses.
RESULTS
After deduplication, 84 results were available until September 2017. Seventeen results were deemed to be not relevant. The findings were thematically grouped and are presented below.
Epidemiology and Genomic Analysis
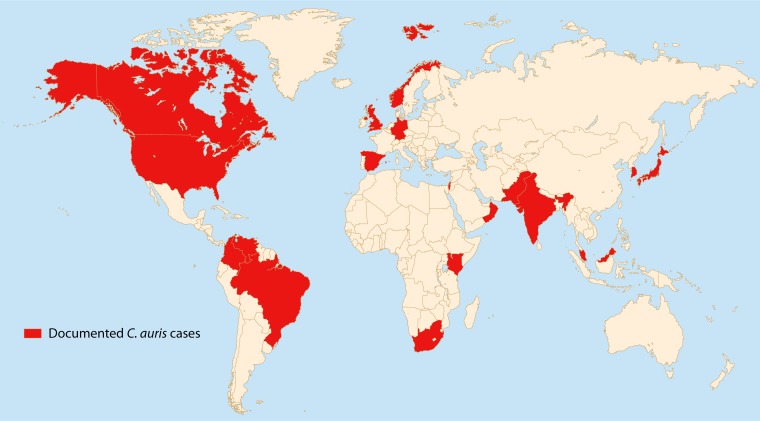
The Candida species Candida auris, so named as it was first described as an isolate from the ear canal of a patient in Japan in 2009, has subsequently been isolated from several body sites of patients in multiple countries on five continents (Fig. 1) (1). Infection and colonization have been detected mainly in critical care patients and affect both pediatric and adult populations (3, 4). Information regarding patients from whom C. auris has been isolated has now been reported globally from South Korea, India, Pakistan, Kuwait, Israel, Oman, South Africa, Colombia, Venezuela, the United States, Canada, and Europe, including the United Kingdom, Norway, Germany, and Spain (3–17). In addition, there have been a number of phenotypic and genotypic characterization studies comparing isolates from different regions, including samples from Brazil, Kenya, and Malaysia, which show distinct geographic clades (6, 18–20).
FIG 1.
Countries that have reported detection of C. auris (shown in red). C. auris has been detected in mainland Norway and Canada, a single Brazilian hospital, and the continental United States, excluding Alaska.
The haploid genome of C. auris is approximately 12.5 Mb, with a guanine-cytosine content of nearly 45% (21–23). Genome analysis suggests that there are between 6,500 and 8,500 protein-coding sequences, with a number of these genes coding for proteins characterized as virulence factors in other Candida species, such as biofilm formation (23). In addition, multiple transporter genes and protein kinases, which may facilitate the acquisition of drug resistance, have been identified (22).
C. auris may be responsible for a significant proportion of Candida infections in regions where it has been recognized for some time. A prospective multicenter study from India reviewing cases of candidemia acquired from an intensive care unit (ICU) found that C. auris was isolated in in 19 out of 27 ICUs, representing 5.2% of cases. There was a difference in prevalences in private (3.2%) versus public (8.2%) hospitals (24).
Genetic analyses have shown a striking divergence of C. auris from some Candida species, while it remains more closely related to C. lusitaniae and C. haemulonii (Table 1). There is also widespread variation between geographic clades, with thousands of single nucleotide polymorphism (SNP) differences. At present, C. auris is separated into four geographic clades: the South Asian, South African, South American, and East Asian clades (6, 23, 25). In India, clonal isolates have been detected over very widespread geographic regions (26). Within each geographic clade, however, there are minimal genetic differences (6).
TABLE 1.
Percent nucleotide identities of various yeast species compared to Candida auris (South Asian clade), calculated over the 285-bp D1-D2 portion of the C. auris 28S ribosomal DNA gene
| Organism | % identity |
|---|---|
| Candida auris (South Asian clade) | 100 |
| Candida auris (South African clade) | 99 |
| Candida auris (East Asian clade) | 99 |
| Candida lusitaniae | 82 |
| Candida haemulonii | 82 |
| Candida guilliermondii | 80 |
| Candida ciferrii | 80 |
| Candida pseudohaemulonii | 79 |
| Candida duobushaemulonii | 79 |
| Candida tropicalis | 79 |
| Candida kefyr | 79 |
| Candida pelliculosa | 78 |
| Saccharomyces cerevisiae | 77 |
| Candida utilis | 76 |
| Candida famata | 75 |
| Candida parapsilosis | 70 |
| Candida magnoliae | 46 |
| Candida albicans | 43 |
| Candida krusei | 43 |
| Candida glabrata | 42 |
| Candida inconspicua | 42 |
| Candida norvegensis | 42 |
| Candida rugosa | 39 |
Whole-genome sequencing (WGS) of U.S. isolates indicated links to two geographic clades: the South Asian clade, with fewer than 60 SNPs, and the South American clade, with fewer than 150 SNPs. The isolates linked to these different geographic clades in the United States showed minimal variation, with between 10 and 70 SNP differences (9). Further WGS analysis comparing isolates from the four geographic regions confirmed clade differences and the striking genetic similarity of isolates within regions (6). Fewer than 16 SNPs differentiated isolates from the South American clade, and fewer than 70 SNPs differentiated isolates from the South African clade. Interestingly, within the South Asian clade, a cluster within one hospital consisted of strains with fewer than 2 SNP differences, whereas isolates from the same patient have demonstrated up to 10 SNP differences (6).
C. auris was discovered to have been misidentified from a historical sample from a South Korean patient with fungemia, originally taken in 1996 (5). A previously unrecognized Pakistani isolate of C. auris from 2008 has also been identified (6). However, a review of the SENTRY isolate collection, with thousands of Candida isolates from four continents, did not reveal the presence of other misidentified C. auris samples prior to 2009 (6).
Identification and Typing
C. auris can often be misidentified in conventional diagnostic laboratories using biochemical typing (27–29). Several studies have examined the accuracy of phenotypic diagnostics in comparison with molecular techniques for the identification of Candida species. Chowdhary et al. recently tabulated the reported misidentifications of C. auris by different commercial methods (18).
With phenotypic and biochemical methods, including API 20C, Vitek 2 (bioMérieux), Phoenix (BD), and MicroScan (Beckman Coulter, Pasadena, CA), C. auris isolates have been misidentified as a range of other Candida species. Most commonly, these isolates have been misidentified as C. haemulonii, a rare cause of infection in humans, but also C. famata, C. sake, Rhodotorula glutinis, Rhodotorula mucilaginosa, and Saccharomyces species. Rarely, C. auris has been identified as C. catenulate, C. lusitaniae, C. guilliermondii, or C. parapsilosis or only to the Candida species level (Table 2) (3, 5, 7–9, 27, 29–32).
TABLE 2.
Misidentification of C. auris by different diagnostic methods
| Diagnostic method (manufacturer) | Misidentification example(s) (reference[s]) |
|---|---|
| Biochemical | |
| API 20CAUX | Rhodotorula glutinis (5, 31, 33) |
| C. sake (3, 15, 34) | |
| Unidentified (35) | |
| API Candida | C. famata (12) |
| Phoenix (BD Diagnostics) | C. haemulonii, C catenulate (31) |
| Vitek | C. haemulonii (3–5, 7, 12, 14, 15, 26, 27, 33–36) |
| C. lusitaniae (15) | |
| C. famata (3, 27) | |
| MicroScan (Beckman Coulter) | C. famata, C. lusitaniae, C. guilliermondii, C. parapsilosis, C. albicans, C. tropicalis (12, 31) |
| MALDI-TOF MS | |
| Vitek MS (bioMérieux) | C. albicans, C. haemulonii (29) |
| Not identified (28, 36) | |
| MALDI Biotyper (Bruker Daltonics) | Neisseria meningitides serogroup A, Pseudomonas rhizosphaerae (29)a |
Subsequently, samples were identified as containing C. auris by ITS sequencing of ear swab samples; the bacteria isolated by MALDI-TOF MS likely represent colonizing bacteria.
C. auris is phylogenetically closely related to the C. haemulonii species complex. These organisms were similarly rarely identified previously as causes of invasive infection but are being increasingly isolated. In particular, C. haemulonii complex species have been associated with deep-seated soft tissue and bone infections in diabetic patients and candidemia in immunosuppressed patients with prior antifungal exposure (33, 34). C. haemulonii complex species are less frequently detected than C. auris, although inaccuracies with the molecular identification of less common Candida species result in difficulties in characterizing the prevalences of these infections (24, 27). It is also possible that some of the reported isolates of C. haemulonii are misidentified as C. auris. The use of chromogenic agar to differentiate between C. auris and C. haemulonii isolates using growth characteristics has been suggested as a low-cost method to circumvent identification problems of commercial phenotypic assays (35). Although there are advantages to molecular techniques for microbiological identification, discrepancies can arise. Matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) compares spectra acquired for a sample to a database of spectra inputted for known isolates. Accurate identification is reliant on the spectra for the sample organism being present in the database. This has resulted in the misidentification of C. auris as C. haemulonii and C. albicans, among others, by MALDI-TOF MS (Table 2) (28, 29). Once spectra are obtained and added to the MALDI-TOF MS database, the identification of C. auris to the species level appears to be accurate, although differentiation between geographic strains is variable and relies on the number of spectra for different clades in the library (10, 20, 27, 28, 31, 36–39). Laboratories should check with the manufacturer regarding the presence of the C. auris reference strain spectra in their database. Confirmation of the laboratory detection capacity could then be tested by obtaining reference strains.
More recently, the development of PCR assays specific for C. auris and for C. auris-related species using cultured colonies has shown promise for the rapid and accurate identification of C. auris, which could prove particularly useful in outbreak situations (40). Confirmation of the sensitivity of these assays for the different clades of C. auris is warranted.
Sequencing of genetic loci, including D1/D2, RPB1, RPB2, and internal transcribed spacer (ITS) domains of the rRNA, has proven useful in the identification of C. auris, but it is not routinely used for the investigation of Candida species isolates and is unlikely to be available outside reference laboratories (3, 8, 21). However, the ability to easily differentiate between geographic clades has been demonstrated with this technology in the United Kingdom (19). Typing by amplified fragment length polymorphism (AFLP) analysis suggested that isolates from one United Kingdom hospital are somewhat distinct from those of previously identified geographical clades (10), although RNA sequencing places them within the South Asian clade, the East Asian clade, and the South African clade, indicating multiple introductions (19).
A range of molecular techniques, including AFLP analysis, pulsed-field gel electrophoresis (PFGE), M13 DNA fingerprinting, and sequencing of genetic loci, have been used for the typing of C. auris isolates. The utility of AFLP analysis in demarcating the geographical clusters of C. auris has been demonstrated (10, 20, 38, 41). One study discriminated both geographical clades and clusters of isolates in an outbreak investigation (37). AFLP analysis was used to demonstrate clonal outbreaks in critically ill patients in Venezuela and India. However, the clonality of temporally and spatially distinct isolates from India from hospitals hundreds of miles apart emphasizes the difficulty in using this technique to discriminate between separate introductions of the organism in possible outbreak situations (4, 26).
In South Korea, PFGE examination of 15 C. auris isolates from ear specimens of patients at three hospitals showed a variety of PFGE patterns and suggested clonal transmission in some of these cases (42). M13 DNA PCR analysis of C. auris candidemia samples from two hospitals in India showed that the Indian samples had a profile that was distinct from those of isolates from Japan and South Korea. Ten of the 12 samples had identical fingerprint patterns, indicating a single genotype (3).
While sequencing of genetic loci has proven useful in the differentiation of C. auris from other Candida species, its ability to discriminate between strains appears to be limited (21). Analysis of South African isolates showed 99% and 98% homologies to Kuwaiti and Indian isolates, respectively, when analyzing the ITS and D1/D2 alignments (8). In India, ITS sequencing of one C. auris isolate demonstrated 100% homology to an epidemiologically unrelated isolate and 98% homology to isolates from Japan and South Korea. Large ribosomal subunit sequences showed 100% homology to an unrelated isolate (3).
Cell Biology
C. auris forms pink to beige colonies on chromogenic agar Candida medium and grows well at 42°C but with variable growth at higher temperatures and no growth in the presence of 0.01% cycloheximide (1, 3, 10, 27, 43; A. Borman and E. M. Johnson, unpublished data). It forms oval or elongated yeast cells, which can occur singly, in pairs, or in groups. Importantly, no hyphal or pseudohyphal forms have been noted (1, 3, 27, 35, 43, 44). Carbon assimilation patterns on an analytical profile index (API) have varied, with isolates from South Africa and India, but not those from Japan or South Korea, showing assimilation of N-acetylglucosamine (1, 3, 8, 27).
An in vivo model comparing the pathogenic effects of C. auris isolates from the United Kingdom with other pathogenic Candida species in the invertebrate Galleria mellonella provided insights into the pathogenicity of this organism (44). That group found that C. auris isolates could behave differently, with some forming aggregates and others not. Non-aggregate-forming isolates demonstrated greater pathogenicity in larvae than did aggregate-forming isolates, to a level comparable to that of C. albicans. This was not linked to the formation of hyphae or pseudohyphae, which are not produced by C. auris except occasionally in a very rudimentary form.
Another group reviewed a range of virulence factors of C. auris isolates through comparison with C. albicans (45). Of the 16 C. auris isolates tested, 6 demonstrated phospholipase activity, and 9 showed secreted proteinase activity, in a strain-dependent manner. One C. auris isolate had phospholipase activity comparable to that of C. albicans.
The strong association of this organism with intensive care settings, especially patients with central venous catheters (CVCs) and long-term urinary catheters, suggests a potential role for biofilm formation (9, 10, 24). In one in vitro model, C. auris did not form biofilms, unlike the closely related species C. haemulonii and C. pseudohaemulonii (42). Recently, however, biofilm formation has been demonstrated with non-aggregate-forming strains and, to a lesser degree, aggregate-forming strains of C. auris (45, 46). C. auris biofilms demonstrated reduced biomass when compared with those of C. albicans but greater biomass than those of C. glabrata.
Resistance Profiles and Treatment
At present, there are no antifungal clinical breakpoints reported for C. auris. Studies examining the susceptibility of this organism to antifungals have used a variety of methods, including Clinical and Laboratory Standards Institute (CLSI) broth microdilution, Etest, and the Vitek 2 yeast susceptibility system. MICs obtained for C. auris isolates have been compared to the breakpoints determined for other Candida species (CLSI and EUCAST clinical breakpoint tables) (47–50). This approach appears to be supported by pharmacodynamic/pharmacokinetic (PK/PD) data from a C. auris candidemia mouse model, although a correlation with clinical outcomes is yet to be established (51). Increased fluconazole MICs, in a high proportion of cases (>64 mg/liter), have been demonstrated to be present in all geographic clusters (7, 8, 10, 20, 22, 27, 41, 43), but resistance is not ubiquitous (5, 6, 9). Treatment failure with fluconazole has been reported for fluconazole-sensitive isolates in the United States (9). Reduced susceptibility to other triazole antifungals, including voriconazole, itraconazole, and isavuconazole, has also been demonstrated (26, 41, 52, 53). In addition, there is variability in the susceptibilities of isolates to amphotericin B (4, 6, 8, 9, 17, 20, 22, 23, 30, 52, 54, 55).
The concern about resistance to triazole antifungal agents and amphotericin B has led to the recommendation for the use of echinocandins as empirical treatment prior to the availability of specific susceptibility testing results, as with invasive candidiasis in general in some regions (30, 56, 57–59). Micafungin demonstrated the highest efficacy in comparison to fluconazole and amphotericin B in a PK/PD study of C. auris candidemia in mice (51). However, as echinocandin use is becoming more widespread, C. auris isolates with reduced susceptibility to this class of drugs have been reported (6, 9, 22, 26).
In vitro investigations into the synergistic use of antifungal agents have resulted in initial promising data for the use of combination treatment of micafungin and voriconazole for multiresistant isolates. This was not reflected in other combinations of azole and echinocandins (60).
The site of infection plays a critical role in the choice of antifungal agent for invasive infections. Echinocandins have limited penetration into a number of sites, including cerebrospinal fluid, due to their high molecular weight, and very little active drug can be recovered from urine (61, 62). Therefore, other medications should be used for central nervous system (CNS) or renal tract infections with Candida species. The use of amphotericin B preparations with the possible addition of 5-flucytosine has been suggested for urinary tract infections (62). For CNS disease, as with other Candida species infections, empirical amphotericin B and 5-flucytosine have had some success, with optimization of therapy as informed by sensitivity testing (59).
Data regarding the MICs of 5-flucytosine are minimal. Early reports from India and a recent study of United Kingdom isolates reported susceptibility of C. auris isolates to 5-flucytosine (10, 54). However, as with the other antifungal classes, there are also reports of isolates with raised MICs (26, 41). A number of isolates of C. auris have demonstrated raised MICs of multiple classes of antifungal agents, raising the possibility of pandrug resistance (6, 27).
The new 1,3-β-d-glucan synthesis inhibitor SCY-078 has in vitro and in vivo activity against a variety of Candida species and has oral bioavailability. Potent activity against C. auris isolates has been demonstrated in vitro, against all geographic clades, with exposed cells failing to divide (45, 63).
A study examining biofilm formation compared the effects of antifungal and disinfectant agents on planktonic cells and sessile cells of biofilms by measuring metabolic activity (46). Sessile cells were susceptible to only liposomal amphotericin B and amphotericin B, both at higher concentrations than those for planktonic cells, with the former being up to 16 mg/liter and the latter being 4 mg/liter. Echinocandins were ineffective against biofilms, although planktonic cells were susceptible. Both planktonic and sessile cells had raised MICs for fluconazole and voriconazole. Chlorhexidine was demonstrated to be active against both planktonic and sessile cells at concentrations below those used topically for disinfection (46). The significant reductions in the metabolic activity and thickness of C. auris biofilms in the presence of SCY-078 highlight the future potential of this new therapy (45). The current understanding of the C. auris genome gives insight as to how reduced susceptibility to multiple antifungal agents has arisen. Mutations in Erg11 associated with the development of fluconazole resistance in C. albicans have also been detected in C. auris isolates (6). Mutations conferring reduced susceptibility to fluconazole are strongly associated with geographic clades, adding support to the theory of separate genetic evolution (64). Although only a small proportion of the genome has been functionally annotated, a number of gene families encoding virulence factors and proteins associated with mechanisms of resistance orthologous to those of C. albicans have been suggested. Importantly, genes for enzymes such as protein kinases and transport proteins involved in efflux pumps, including the ATP-binding cassette (ABC) and major facilitator superfamilies (MFS), have been identified, and these may facilitate the acquisition of drug resistance (22, 23).
Colonization and Infection
British Society for Medical Mycology best-practice guidelines detail recommendations for the laboratory testing of samples (65). However, hospital practice policies for the investigation of isolates of Candida species vary globally. In the absence of a unified case definition for C. auris infection, and variable screening practices for Candida species, colonization rates and the significance of colonization in terms of the development of invasive infection are difficult to characterize.
Colonization with C. auris has been detected at multiple body sites, including nares, groin, axilla, and rectum, and has been isolated for 3 months or more after initial detection in spite of negative screens and echinocandin treatment in the intervening period (9, 10). These uncertainties suggest the need for multiple screens with ongoing patient isolation after treatment and upon readmission to health care facilities (57).
Risk factors for colonization include contact with patients known to harbor C. auris or their environment (66). The contact time for the acquisition of C. auris from a colonized patient or environment is suggested to be as little as 4 h (10), and invasive infections have been acquired within 48 h of admission to intensive care settings (54). The use of empirical antifungal therapy would need to be considered if a patient colonized with C. auris subsequently deteriorates.
C. auris has been associated with a variety of invasive fungal infections. The majority of the reported data regarding patient infections and outcomes have come from India, but there are also reports from small numbers of patients affected in South Korea, Venezuela, South Africa, the United Kingdom, the United States, Colombia, and Canada (Table 3) (4, 5, 8, 10, 12, 14–17, 26, 27, 67, 68). Invasive C. auris infection has been associated with candidemia to a high degree, including cases associated with CVC use, but also with pericarditis and respiratory tract and urinary tract infections (3–5, 9, 10, 26, 27, 64, 69). In the majority of cases, invasive infection with C. auris occurs in critically ill patients, i.e., those in intensive care facilities and undergoing invasive procedures (4, 5, 9, 24). These patients are generally those with serious underlying medical conditions, including hematological malignancies and other conditions resulting in immunosuppression (7, 10, 54). One report detailed a case of donor-derived C. auris infection following lung transplantation (70). Yeast was identified on bronchoalveolar lavage samples pre- and postimplantation, which was initially misidentified by both biochemical and molecular testing.
TABLE 3.
Candida auris infection cases by disease type reported in the literature
| Type of disease or location of isolationb | No. of cases (reference[s]) |
|---|---|
| Candidemia | 291 (3–5, 7, 8, 10, 12, 14–16, 26, 27, 57, 58, 70, 71) |
| Central venous catheter tip | 2 (70) |
| CNS | 1 (12) |
| ENT | 21a (1, 17, 58, 70, 72) |
| Respiratory tract | 18 (26, 27, 36, 70) |
| Urogenital system | 17 (12, 27, 56) |
| Abdominal | 13 (12, 27, 70) |
| Skin and soft tissue, including surgical wounds | 12 (3, 10, 27, 70) |
| Bone | 2 (12, 70) |
Two associated with otomastoiditis and 19 from ear swabs of patients with otitis externa.
CNS, central nervous system; ENT, ear, nose, and throat.
As might be expected, the majority of patients with invasive C. auris infections have received broad-spectrum antimicrobial agents and, in some cases, antifungal agents prior to the development of invasive candidiasis (6, 68). An association with medical devices such as CVCs and urethral catheters has also been reported, as anticipated for this patient group (3, 5, 9). A subgroup analysis of C. auris candidemia in Indian intensive care units indicated an association with lower acute physiology and chronic health evaluation II (APACHE II) scores, vascular surgery, and longer ICU stay prior to diagnosis than with other candidemias (68).
Mortality rates have varied significantly among geographic regions (64). Reports from Asia, the Far East, and the United States have detailed mortality rates of over 50% for those with invasive infections (5, 9, 54). This is in contrast to Venezuela, where the 30-day survival rate following candidemia was 72%. Similarly, in Colombia, the 30-day mortality rate associated with a delayed diagnosis of C. auris was 35.2% (12). However, the literature does not comment on the background case fatality rates in these cohorts of patients, many of whom have multiple comorbidities. As such, the overall attributable mortality rate is unclear. In the United Kingdom, all cases were reviewed, and no deaths were considered directly attributable to C. auris for 22 patients requiring antifungal treatment following the isolation of C. auris (4, 10). The number of deaths attributable to candidemia, as opposed to an underlying medical condition, may be difficult to quantify.
Infection Prevention and Control
Observations of rapid acquisition, an association with high mortality rates, and high levels of antifungal resistance highlight the importance of rapid implementation of IPC measures to curb transmission. Guidance has been released in the United Kingdom, the United States, Europe, and South Africa, with recommendations regarding the isolation of patients, contact precautions, and cleaning of equipment and environments in contact with affected patients (Table 4) (11, 57, 71–73). Due to the limited data on this emerging pathogen, much of this guidance is empirical, based on extrapolation from other resistant organisms, including methicillin-resistant Staphylococcus aureus (MRSA) and carbapenem-resistant Enterobacteriaceae (CRE).
TABLE 4.
Reported infection prevention and control recommendationsa
| Body | Recommendation(s) |
|||||
|---|---|---|---|---|---|---|
| Patient screening | Contact precaution(s) | Contact screening | Decolonization procedure(s) | Environmental management | Community management | |
| PHE (UK) | Recommended in units with ongoing cases or colonizations; those arriving from affected units (UK and abroad); screening sites such as groin, axilla, nose, throat, urine, perineal area, rectal area, and stool; consider screening, if indicated, LVS, sputum, endotracheal secretions, drain fluid, wounds, and cannula; rescreening of patients known to have been previously colonized; deisolation of screen-positive patients is not recommended apart from units with experience in managing C. auris | Side room with en suite facilities where possible; isolation of all patients from affected UK or international hospital until screening is available; strict adherence to hand hygiene using soap and water, followed by alcohol rub to dry hands; PPE with gloves and aprons or gowns if there is a high risk of body or body fluid contact; briefing of visitors regarding contact precautions; single-patient-use items such as blood pressure cuffs should be considered; for cleaning C. auris-exposed areas, glove and apron use with subsequent appropriate hand decontamination | If there is novel detection in a unit, close contacts should be screened and isolated or cohorted; if the index patient is isolated, identify all Candida species isolates from the same unit to the species level using a method able to detect C. auris; review Candida spp. detected in the same ward areas in the 4 wk prior to diagnosis of the index patient in case of unrecognized transmission; deisolation with 3 negative screens >24 h apart | Strict adherence to central and peripheral catheter care bundles, urinary catheter care bundle, care of the tracheostomy site; skin decontamination with chlorhexidine washes in critically ill patients; consider use of mouth gargles with chlorhexidine and use of topical nystatin and terbinafine for topical management of key sites | Use of chlorine-releasing agent at 1,000 ppm for cleaning contact environments; change privacy curtains; for equipment, consider single-use items or discarding less expensive items that are difficult to decontaminate; all equipment should be cleaned in accordance with the manufacturer's instructions; terminal cleaning when patient leaves the environment; schedule affected patients last for theater/procedures/imaging; for waste and linen disposal, follow local policy as for other multiresistance organisms; training and supervision of cleaning staff until competent | Nurse in a single room with en suite facilities when possible; if single room is not possible, the colonized individual should not share a room with an immunocompromised individual; thorough environmental cleaning with a chlorine-releasing agent at 1,000 ppm of available chlorine; follow standard infection control precautions; ensure that staff are trained in the use of PPE and hand hygiene; special care should be taken with wound, catheter, and device care |
| CDC (USA) | Axilla and groin screening; additional sites as directed clinically or by previously positive sites; periodic reassessment for presence of colonization at 1- to 3-mo intervals; for deisolation, 2 or more assessments 1 wk apart with negative results (off antifungals) | Single room with standard and contact precautions; gown and gloves; hand hygiene precautions | Wait 48 h after administration of topical chlorhexidine prescreening | Thorough daily and terminal cleaning/disinfection using Environmental Protection Agency-registered disinfectant effective against C. difficile spores | Do not restrict nursing home residents to rooms and perform hand hygiene; if receiving health input, gown and glove contact precautions; thorough cleaning of shared equipment | |
| ECDC (Europe-wide) | All patients from in-country or internationally affected units transferred in; conduct active surveillance in accordance with specified protocol; screening sites include urine, feces, wounds, drain fluid, respiratory samples | Contact precautions, single room isolation; patient cohorting; dedicated nursing staff for colonized or infected patients; hand hygiene | Cross-sectional patient screening in outbreak setting | Terminal cleaning of rooms using disinfectants and methods with certified antifungal activity; environmental sampling in outbreak setting | ||
| COTHI (South Africa) | Routine screening not advised | Single room with en suite or cohorting of patients; hand hygiene using soap and water or alcohol rub; gloves and aprons for patient contact; adherence to venous and urinary catheter and tracheostomy care bundles; advise visitors regarding contact precautions; notify receiving hospitals of positive status | Schedule affected patients last for theater/procedures/imaging; regular cleaning with chlorine-releasing agent at 1,000 ppm; terminal cleaning and disinfection of bed space; consider terminal cleaning with hydrogen peroxide vapor; clean multiuse equipment thoroughly; cleaning of all contact areas | |||
CDC, Centers for Disease Control and Prevention, USA; ECDC, European Centre for Disease Prevention and Control; COTHI, Centre for Opportunistic, Tropical, and Hospital Infections; LVS, low vaginal swab; PPE, personal protective equipment.
At present, PHE recommends the development of screening policies based on risk assessment within local units. It is recommended that patients transferred from affected units within the United Kingdom and abroad should be screened, as would be the case for MRSA and CRE. All patients known to be infected or colonized with C. auris should be isolated, preferably in en suite facilities. Screening to determine longitudinal carriage should be undertaken, including screening of all previously positive patients upon readmission to the hospital (57). With evidence of recurrent colonization subsequent to negative screens and antifungal use, there remains a significant issue around the question of deisolation. The CDC currently recommends that patients with at least two negative screens over a week apart, while not receiving antifungals, can be moved out of isolation (72). PHE has suggested that patients with a sample positive for C. auris should not be deisolated, apart from those in units with experience in managing C. auris (Table 4) (57).
One unit implemented a bundle of measures to reduce the spread of C. auris, including decolonization of patients with chlorhexidine gluconate body washes, chlorhexidine mouthwashes, and chlorhexidine-impregnated pads for CVC exit sites (10). Data on the inhibition of growth of C. auris with chlorhexidine body washes at contact times and concentrations representative of hand washing have shown that there is a several-log difference in inhibition compared to that of C. albicans. Povidone iodine, in contrast, appears effective at levels below those used for antiseptic preparations (46, 74, 75). The impact of skin disinfection measures on colonization and shedding is yet to be established.
Environmental screening is problematic because of probable transient, sporadic contamination and difficulties with the interpretation of results. One study did not detect any environmental contamination (54). Others found C. auris to be associated with samples from multiple patient contact areas, including mattresses, furniture, windowsills, and air settle plate sampling (9, 10, 67).
C. auris has been demonstrated to survive on a range of surface types, including dry, moist, and plastic surfaces, with organisms being viable for up to 14 days on plastic. The rate of recovery of C. auris over a period of 7 days was higher than that of C. albicans on both moist and dry surfaces, indicating the potential significance of environmental contamination (76, 77). A synthetic polymer with antimicrobial properties designed for potential use in medical devices showed promise against a number of organisms but did not demonstrate any efficacy against C. auris (78).
In a comparison of the efficacies of a range of disinfectants against Candida species and MRSA, sodium hypochlorite and hydrogen peroxide resulted in the greatest reduction in C. auris CFU. Acetic acid, ethyl alcohol, and quaternary ammonium compounds, in contrast, showed less of a reduction in CFU, far below that observed for MRSA (79).
Postdischarge environmental decontamination of patient areas with high-concentration chlorine solutions in combination with hydrogen peroxide vapor or UV light appears to effectively eliminate the organism (9, 10, 67). United Kingdom experience has also highlighted the importance of thorough terminal decontamination of patient contact items, such as pulse oximeter probes and axillary temperature probes (10, 66, 74, 80).
Where possible, it is recommended that the same isolation, contact, and cleaning precautions be utilized for patients being cared for in community settings. Where single rooms with en suite facilities are not available, it is advised that patients colonized with C. auris should not share facilities with those known to be immunosuppressed (Table 4) (73).
The possible role of health care workers (HCWs) in the transmission of organisms between patients is difficult to evaluate given the emotive, social, and financial implications. At one United Kingdom hospital, concerns over the continued detection of C. auris in spite of IPC measures led to the voluntary screening of 258 HCWs in contact with critical care settings. Multiple body sites, including hands, nose, throat, and groin, were screened, with only one individual being found to have a sample positive for C. auris, from a nose swab. Chlorhexidine washes, nasal ointment, and oral nystatin for 5 days resulted in successful decolonization, which was confirmed by repeat negative screens. The HCW involved was known to have cared for a patient who was heavily colonized with C. auris and was not implicated in any onward transmission (10).
Costs
It is important to understand the wide-ranging impact that outbreaks of emerging infections, such as C. auris infections, can have on those affected. As with any outbreak situation, costs can quickly increase, but these costs are not merely financial. With an emerging infection, there are the added costs associated with the development of diagnostics and research strategies to increase the understanding of the biology, pathogenicity, and transmission of the organism. These costs have not yet been quantified for C. auris outbreaks.
DISCUSSION
Our review highlights the considerable range of questions that remain to be answered regarding C. auris. This is often the case with emerging pathogens, where the initial priority is the local control of the organism. C. auris is being isolated from patients from an increasingly widespread geographical area, and it is probable that the number of patients affected is significantly higher than the literature suggests. Identification remains problematic: some countries may be unable to detect C. auris due to a lack of available laboratory technology. It is also likely that there are significant nonpublished data that could inform current practice and assist in the development of strategies for the management of C. auris. In the early stages of emerging infection situations, both informal and formal notification networks prove vital for the spread of information and to ensure awareness among the wider medical and public health communities.
The simultaneous detection of C. auris on multiple continents, the clonality of isolates from different regions, and the various geographic resistance mechanisms suggest independent clonal expansion and evolution. This could theoretically have occurred if C. auris has been circulating unrecognized, with historical separation from a common ancestral strain. However, this seems unlikely, as there are only two instances where the organism has been retrospectively identified from historical isolates, and a review of thousands of stored isolates from four continents did not identify any C. auris isolates prior to 2009 (5, 6). Further review of stored isolates may help elucidate this.
Another possibility is the development of a common environmental niche. The use of broad-spectrum antimicrobials and antifungal therapy for prophylaxis and treatment continues to increase in certain patient groups, including those who are immunosuppressed due to chemotherapy or HIV and those in intensive care settings. The natural flora of these patients is being dramatically altered. Fluconazole use in particular may alter the balance toward colonization and infection with non-albicans Candida species, contributing to the greater variety of Candida species now associated with invasive infections (2). The contribution of possible animal reservoirs to the recent emergence of C. auris should also be considered and investigated, given the range of growth characteristics observed.
Awareness of the difficulties in the identification of C. auris has resulted in the development and validation of MALDI-TOF MS in geographical areas currently known to be affected. In addition, the development of C. auris-specific PCR will aid in rapid, accurate diagnosis. However, the availability of these technologies may be limited. There are large parts of the globe without the infrastructure or facilities to perform testing and where health priorities are such that any funding available has to be diverted to other areas. This will impede the epidemiological understanding of C. auris, and it is likely that the number of other organisms that C. auris is misidentified as will continue to increase.
Differentiating geographic clades of C. auris strains with thousands of nucleotide differences between them can be achieved with molecular typing techniques. However, different methods give various results that are not comparable. WGS has demonstrated that within geographic clades, there is minimal genetic variation among strains. Therefore, discrimination between a novel introduction and the transmission of the same strain between patients in outbreak situations is unlikely to be achieved by using techniques that are reliant on distinguishing strains by molecular weight or differences within a small part of the genome. Clade-specific PCR for C. auris is in development and may be useful for the rapid identification of samples of C. auris in the future.
Invasive infection and colonization have been identified almost exclusively in patients in high-dependency areas with the highest degree of medical intervention. Prevalence studies will help clarify whether C. auris is associated mainly with this environment or whether there is widespread hospital and community carriage. Screening at one United Kingdom hospital over a period of 2 months suggested that C. auris is not widespread within the community or hospital setting in that area (K. Jeffery, unpublished data). Establishing prevalence is vital to the development of appropriate screening and control strategies; a point prevalence survey of hospitals serving multiethnic populations is currently being performed in the United Kingdom (81). It is important to establish sites of endogenous carriage through systematic screening for C. auris. Possibilities include colonization with C. auris in the gastrointestinal (GI) tract and subsequent overgrowth onto the skin under environmental pressure from antimicrobial and antifungal use. Alternatively, C. auris may predominantly be a skin dweller with transmission routes similar to those of MRSA, with axilla and groin carriage, as reported by many centers. Irrespective of the location of initial carriage, it appears that certain patients shed large amounts of this organism from their skin, contaminating the environment and resulting in onward transmission (10). As a consequence, effective strategies for environmental cleaning of patient areas following discharge are needed.
For data to be comparable, the utilization of universal case definitions for invasive candidiasis is necessary (59). Unlike for other Candida species, which are not usually associated with outbreaks, detection of colonization and differentiation from invasive infection are vital for effective infection control. It is important to gain a greater understanding of the impact of different treatments and decolonization regimens on carriage and whether lifelong carriage is likely. The impact of skin cleansers, including soap and water, quaternary ammonium compounds, alcohol gel, and surgical skin preparation solutions, on C. auris viability requires evaluation.
Understanding the contribution of different transmission routes, including airborne spread via skin particles, HCW contact, and fomites in the patient microenvironment, is pivotal to preventing hospital outbreaks. Investigating the role of environmental contamination and the impact of decontamination measures will further inform IPC policies. However, the regional clonality of strains and the lack of discrimination between individual isolates by using a range of typing methods mean that it may be impossible to accurately determine where transmission has occurred.
The institution of broad-ranging IPC care bundles appears, from limited available data, to be effective at reducing the number of invasive infections (10). Effects on colonization, however, are unclear, as is the need to decolonize patients prior to surgical procedures and whether invasive infections can be prevented or at least significantly reduced with IPC measures. An increased understanding will also inform the development of guidance regarding the management of patients colonized with C. auris transferred into community environments.
Genomic analyses demonstrated the presence of a number of genes associated with virulence factors and reduced antifungal susceptibility in other Candida species. The possibility of the development of further antifungal resistance remains a significant concern and highlights the need for the development of novel antifungal agents (82). Further genome analysis to understand the development of resistance mechanisms and the impact upon the fitness of the organism is important to help in the development of appropriate antifungal recommendations for at-risk populations. Echinocandins are the recommended first-line therapy, as for other candidemias. New options on the horizon include SCY-078 and the use of combinations of antifungals in patients with multiresistant organisms.
The significance of C. auris as a human pathogen remains unclear. Mortality rates from initial studies were concerning, although C. auris-attributable mortality cannot be established from those studies. Underlying medical conditions and the availability of antifungal therapies will clearly have a heavy impact on outcomes, especially in developing countries, where infection control practices may not be able to prevent transmission, detection methods may be lacking, and echinocandin availability may be limited. Data from the United Kingdom are more reassuring and raise the possibility of differing pathogenicities among strains.
As for other emerging pathogens, laboratory costs associated with our increasing understanding of C. auris include those associated with increased sample throughput and the greater use of reference laboratory testing for confirmation and susceptibility testing. In affected hospitals, members of staff from multiple disciplines are required to deal with the evolving situation, with consequent effects on routine workflows. The need for the implementation of urgent infection prevention and control measures can have wide-ranging effects, from single-use equipment to increased cleaning and decontamination requirements. In addition, this can cause delays in patient investigations and procedures and extend hospital stays. Where there is a limited understanding of the mechanisms of transmissibility, as with C. auris, competing priorities of opportunity cost and alterations to service will need to be balanced against possible risks of spread.
CONCLUSION
With its predilection for the most vulnerable patients and concerns regarding antifungal resistance, C. auris has the potential to significantly impact morbidity, mortality, and health care infrastructure and finance. There are multiple unanswered questions regarding the natural environment of C. auris, the origin of its sudden emergence, population prevalence, environmental contamination, transmission dynamics, acquisition of antifungal resistance, effectiveness of IPC measures, and impact on patient mortality. It remains unclear as to whether this organism will continue to be a cause for global concern or if it will decline as quickly as it seems to have appeared. The increased number of cases detected in an ever larger number of countries suggests that the latter possibility is unlikely. The identification of increasingly resistant isolates is particularly concerning. Current research has the potential to have a significant impact on future outcomes for patients and institutions worldwide.
ACKNOWLEDGMENTS
We thank Lois Woods, Public Health England (PHE) Knowledge and Library Services, for conducting the literature search for this review.
The contributing members of the Candida auris Incident Management Team at the timing of writing of this review are as follows: Louise Bishop (PHE), Yimmy Chow (PHE), Fiona Cummings (PHE), Martina Cummins (Barts Health NHS Trust), Daniele Curtis (PHE), Dona Foster (Oxford University Hospitals NHS Foundation Trust [guest for review]), Rebecca Guy (PHE), Anne Hall (Royal Brompton and Harefield NHS Foundation Trust), Peter Hoffman (PHE), Katy Marden (Taunton and Somerset NHS Foundation Trust), Berit Muller-Pebody (PHE), Ginny Moore (PHE), Fiona Neely (PHE), Karthik Paranthaman (PHE), Bharat Patel (PHE), Richard Puleston (PHE), James Sedgwick (PHE), Nandini Shetty (PHE), Deborah Turbitt (PHE), and Jimmy Walker (PHE).
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Biographies

Anna Jeffery-Smith received her medical degree from the University of Oxford. She subsequently went on to clinical training in London, United Kingdom, and Auckland, New Zealand, before specializing in infectious diseases and virology in London. She currently works as an academic clinical fellow in infectious diseases and virology at Barts Health NHS Trust and Public Health England. In this role, she has become involved in the investigation and response to outbreak situations, leading to her involvement in the management of Candida auris in the United Kingdom. Continuing with her interests with public health, she is due to start a Ph.D. investigating the monitoring of patients with chronic hepatitis B virus infection.

Surabhi K. Taori received her undergraduate medical qualifications from India and postgraduate training, including a Ph.D., from Edinburgh University, United Kingdom. She has diverse experience in infection control, having worked in India and Edinburgh and with the Rare and Imported Pathogens department (PHE, Porton Down, United Kingdom) and has been studying new emerging infectious diseases and their transmission for over 15 years. She is currently the infection control doctor at King's College Hospital, London, where she was instrumental in successfully controlling one of the first outbreaks of C. auris in the United Kingdom. She takes a keen interest in education and training.

Silke Schelenz obtained her M.D. from the Free University of Berlin, Germany. She studied for her Ph.D. on the subject of the host response to aspergillosis and cryptococcosis at the London School of Hygiene and Tropical Medicine. She is now the consultant microbiologist and infection control doctor as well as Head of the Microbiology Department at the Royal Brompton Hospital and honorary senior lecturer at Imperial College. She is chair of the United Kingdom Clinical Mycology Networks/PHE, a member of the English Surveillance Programme for Antimicrobial Utilization and Resistance (ESPAUR) Antifungal Resistance and Consumption Subgroup (PHE/DoH), council member of the British Society for Medical Mycology, specialty advisory committee member at RCPath, and UK Standards in Microbiology Steering Committee Member for devising standard operating procedures (SOPs) for microbiology in the United Kingdom. She has published extensively in the field of infection and acts as a referee for peer-reviewed medical journals and grant-awarding bodies.

Katie Jeffery is a Consultant in Clinical Infection and the Deputy Director for Infection, Prevention, and Control for the Oxford University Hospitals NHS Foundation Trust. She trained in medicine at Cambridge, Oxford, and Imperial College, London. Her interests are infection prevention and control, molecular diagnosis, neurological infection, viral hepatitis, and infections in the immunocompromised host. She has published on a wide variety of infectious disease topics. She has managed one of the largest outbreaks to date of Candida auris in the United Kingdom, based on a neurological intensive care unit.

Elizabeth M. Johnson received a B.Sc. (Hons) in Medical Microbiology and a Ph.D. in Medical Mycology from the University of Bristol, United Kingdom, and has worked in the field of medical mycology for more than 30 years, first for the National Health Service and later for the Public Health Laboratory Service, Health Protection Agency, and Public Health England (PHE). For the last 15 years, she has been director of the PHE National Mycology Reference Laboratory and curator of the United Kingdom National Collection of Pathogenic Fungi. Dr. Johnson has a great interest in all pathogenic fungi and their treatment and is especially concerned by how Candida auris appears to have achieved global spread in a short time frame, is often resistant to the azole class of antifungal drugs and sometimes multiple classes, and has a propensity, unusual among yeast isolates, to rapidly spread from patient to patient.

Andrew Borman was educated at the Universities of Manchester and Cambridge. Dr. Borman was a senior research scientist and then deputy director of a research unit at the Pasteur Institute, Paris, France, from 1992 until 2003, when he joined the Public Health England United Kingdom National Mycology Reference Laboratory, Bristol, as principal clinical scientist and Deputy Director. His interests include emerging fungal pathogens, the diagnosis and management of fungal infections, and the molecular identification and taxonomy of pathogenic fungi.

Rohini Manuel is a Consultant Clinical Microbiologist at the Public Health Laboratory London, National Infection Service, Public Health England. She qualified in Medicine from the National University of Ireland, Galway, in 1994 and obtained her doctorate on the diagnosis of invasive aspergillosis at University College London (UCL) in 2007. She is a member of the Royal College of Pathologists (RCPath) London Regional Council and the public health champion for the North Thames NIHR Clinical Research Network in Infectious Diseases and Microbiology. Her specialist area of expertise is mycology, particularly infections affecting immunocompromised individuals. She is a member of the United Kingdom Clinical Mycology Network National Steering Group. She is a Senior Examiner in Medical Microbiology at the RCPath and sits on the UCL Medical Mycology Board of Examiners. She is an Editor for the Oxford Textbook in Medical Mycology and has over 50 publications on infection and public health-related topics.

Colin S. Brown is an Infectious Disease and Medical Microbiology consultant at Public Health England (PHE) and is the national incident director for the United Kingdom's Candida auris response. He works on a portfolio of respiratory, vaccine-preventable, and emerging infections and global health strengthening. He has a Medical Research Council-funded Epidemiology Masters from the London School of Hygiene and Tropical Medicine and held an Academic Clinical Fellowship in Infectious Diseases at King's College London. His main professional interests are tuberculosis; HIV; emerging and reemerging infections; and global health development, education, and volunteering. He is the Infectious Diseases Advisor for King's Sierra Leone Partnership (KSLP) and was heavily involved in the clinical and public health response to the Ebola virus disease outbreak in West Africa in 2014 to 2016. He is also an honorary consultant in Infectious Diseases and Medical Microbiology at the Royal Free Hospital.
REFERENCES
- 1.Satoh K, Makimura K, Hasumi Y, Nishiyama Y, Uchida K, Yamaguchi H. 2009. Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol Immunol 53:41–44. doi: 10.1111/j.1348-0421.2008.00083.x. [DOI] [PubMed] [Google Scholar]
- 2.Deorukhkar SC, Saini S, Mathew S. 2014. Non-albicans Candida infection: an emerging threat. Interdiscip Perspect Infect Dis 2014:615958. doi: 10.1155/2014/615958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chowdhary A, Sharma C, Duggal S, Agarwal K, Prakash A, Singh PK, Jain S, Kathuria S, Randhawa HS, Hagen F, Meis JF. 2013. New clonal strain of Candida auris, Delhi, India. Emerg Infect Dis 19:1670–1673. doi: 10.3201/eid1910.130393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calvo B, Melo AS, Perozo-Mena A, Hernandez M, Francisco EC, Hagen F, Meis JF, Colombo AL. 2016. First report of Candida auris in America: clinical and microbiological aspects of 18 episodes of candidemia. J Infect 73:369–374. doi: 10.1016/j.jinf.2016.07.008. [DOI] [PubMed] [Google Scholar]
- 5.Lee WG, Shin JH, Uh Y, Kang MG, Kim SH, Park KH, Jang HC. 2011. First three reported cases of nosocomial fungemia caused by Candida auris. J Clin Microbiol 49:3139–3142. doi: 10.1128/JCM.00319-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lockhart SR, Etienne KA, Vallabhaneni S, Farooqi J, Chowdhary A, Govender NP, Lopes Colombo A, Calvo B, Cuomo CA, Desjardins CA, Berkow EL, Castanheira M, Magobo REE, Jabeen K, Asghar RJ, Meis JF, Jackson B, Chiller T, Litvintseva AP. 2017. Simultaneous emergence of multidrug resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin Infect Dis 64:134–140. doi: 10.1093/cid/ciw691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Emara M, Ahmad S, Khan Z, Joseph L, Al-Obaid I, Purohit P, Bafna R. 2015. Candida auris candidemia in Kuwait, 2014. Emerg Infect Dis 21:1091–1092. doi: 10.3201/eid2106.150270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Magobo RE, Corcoran C, Seetharam S, Govender NP. 2014. Candida auris-associated candidemia, South Africa. Emerg Infect Dis 20:1250–1251. doi: 10.3201/eid2007.131765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vallabhaneni S, Kallen A, Tsay S, Chow N, Welsh R, Kerins J, Kemble SK, Pacilli M, Black SR, Landon E, Ridgway J, Palmore TN, Zelzany A, Adams EH, Quinn M, Chaturvedi S, Greenko J, Fernandez R, Southwick K, Furuya EY, Calfee DP, Hamula C, Patel G, Barrett P, Lafaro P, Berkow EL, Moulton-Meissner H, Noble-Wang J, Fagan RP, Jackson BR, Lockhart SR, Litvintseva AP, Chiller TM. 2016. Investigation of the first seven reported cases of Candida auris, a globally emerging invasive, multidrug-resistant fungus—United States, May 2013-August 2016. MMWR Morb Mortal Wkly Rep 65:1234–1237. doi: 10.15585/mmwr.mm6544e1. [DOI] [PubMed] [Google Scholar]
- 10.Schelenz S, Hagen F, Rhodes JL, Abdolrasouli A, Chowdhary A, Hall A, Ryan L, Shackleton J, Trimlett R, Meis JF, Armstrong-James D, Fisher MC. 2016. First hospital outbreak of the globally emerging Candida auris in a European hospital. Antimicrob Resist Infect Control 5:35. doi: 10.1186/s13756-016-0132-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.European Centre for Disease Prevention and Control. 2016. Candida auris in healthcare settings—Europe. European Centre for Disease Prevention and Control, Stockholm, Sweden. [Google Scholar]
- 12.Morales-Lopez SE, Parra-Giraldo CM, Ceballos-Garzon A, Martinez HP, Rodriguez GJ, Alvarez-Moreno CA, Rodriguez JY. 2017. Invasive infections with multidrug-resistant yeast Candida auris, Colombia. Emerg Infect Dis 23:162–164. doi: 10.3201/eid2301.161497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finn T, Novikov A, Zakin S, Bash E, Berman J, Ben-Ami R. 2016. Candida haemulonii and Candida auris: emerging multidrug-resistant species with distinct virulence and epidemiological characteristics. Open Forum Infect Dis 3(Suppl 1:124. doi: 10.1093/ofid/ofw194.37. [DOI] [Google Scholar]
- 14.Ben-Ami R, Berman J, Novikov A, Bash E, Shachor-Meyouhas Y, Zakin S, Maor Y, Tarabia J, Schechner V, Adler A, Finn T. 2017. Multidrug-resistant Candida haemulonii and C. auris, Tel Aviv, Israel. Emerg Infect Dis 23:195–203. doi: 10.3201/eid2302.161486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ruiz Gaitan AC, Moret A, Lopez Hontangas JL, Molina JM, Aleixandre Lopez AI, Cabezas AH, Mollar Maseres J, Arcas RC, Gomez Ruiz MD, Chiveli MA, Canton E, Peman J. 2017. Nosocomial fungemia by Candida auris: first four reported cases in continental Europe. Rev Iberoam Micol 34:23–27. doi: 10.1016/j.riam.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 16.Al-Siyabi T, Al Busaidi I, Balkhair A, Al-Muharrmi Z, Al-Salti M, Al'Adawi B. 2017. First report of Candida auris in Oman: clinical and microbiological description of five candidemia cases. J Infect 75:373–376. doi: 10.1016/j.jinf.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 17.Schwartz I, Hammond G. 2017. First reported case of multidrug-resistant Candida auris in Canada. Can Commun Dis Rep 43:150–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chowdhary A, Sharma C, Meis JF. 2017. Candida auris: a rapidly emerging cause of hospital-acquired multidrug-resistant fungal infections globally. PLoS Pathog 13:e1006290. doi: 10.1371/journal.ppat.1006290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Borman AM, Szekely A, Johnson EM. 2017. Isolates of the emerging pathogen Candida auris present in the UK have several geographic origins. Med Mycol 55:563–567. doi: 10.1093/mmy/myw147. [DOI] [PubMed] [Google Scholar]
- 20.Prakash A, Sharma C, Singh A, Kumar Singh P, Kumar A, Hagen F, Govender NP, Colombo AL, Meis JF, Chowdhary A. 2016. Evidence of genotypic diversity among Candida auris isolates by multilocus sequence typing, matrix-assisted laser desorption ionization time-of-flight mass spectrometry and amplified fragment length polymorphism. Clin Microbiol Infect 22:277.e1–277.e9. doi: 10.1016/j.cmi.2015.10.022. [DOI] [PubMed] [Google Scholar]
- 21.Sharma C, Kumar N, Meis JF, Pandey R, Chowdhary A. 2015. Draft genome sequence of a fluconazole-resistant Candida auris strain from a candidemia patient in India. Genome Announc 3:e00722-. doi: 10.1128/genomeA.00722-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sharma C, Kumar N, Pandey R, Meis JF, Chowdhary A. 2016. Whole genome sequencing of emerging multidrug resistant Candida auris isolates in India demonstrates low genetic variation. New Microbes New Infect 13:77–82. doi: 10.1016/j.nmni.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chatterjee S, Alampalli SV, Nageshan RK, Chettiar ST, Joshi S, Tatu US. 2015. Draft genome of a commonly misdiagnosed multidrug resistant pathogen Candida auris. BMC Genomics 16:686. doi: 10.1186/s12864-015-1863-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chakrabarti A, Sood P, Rudramurthy SM, Chen S, Kaur H, Capoor M, Chhina D, Rao R, Eshwara VK, Xess I, Kindo AJ, Umabala P, Savio J, Patel A, Ray U, Mohan S, Iyer R, Chander J, Arora A, Sardana R, Roy I, Appalaraju B, Sharma A, Shetty A, Khanna N, Marak R, Biswas S, Das S, Harish BN, Joshi S, Mendiratta D. 2015. Incidence, characteristics and outcome of ICU-acquired candidemia in India. Intensive Care Med 41:285–295. doi: 10.1007/s00134-014-3603-2. [DOI] [PubMed] [Google Scholar]
- 25.Sarma S, Upadhyay S. 2017. Current perspective on emergence, diagnosis and drug resistance in Candida auris. Infect Drug Resist 10:155–165. doi: 10.2147/IDR.S116229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chowdhary A, Anil Kumar V, Sharma C, Prakash A, Agarwal K, Babu R, Dinesh KR, Karim S, Singh SK, Hagen F, Meis JF. 2014. Multidrug-resistant endemic clonal strain of Candida auris in India. Eur J Clin Microbiol Infect Dis 33:919–926. doi: 10.1007/s10096-013-2027-1. [DOI] [PubMed] [Google Scholar]
- 27.Kathuria S, Singh PK, Sharma C, Prakash A, Masih A, Kumar A, Meis JF, Chowdhary A. 2015. Multidrug-resistant Candida auris misidentified as Candida haemulonii: characterization by matrix-assisted laser desorption ionization–time of flight mass spectrometry and DNA sequencing and its antifungal susceptibility profile variability by Vitek 2, CLSI broth microdilution, and Etest method. J Clin Microbiol 53:1823–1830. doi: 10.1128/JCM.00367-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wattal C, Oberoi JK, Goel N, Raveendran R, Khanna S. 2017. Matrix-assisted laser desorption ionization time of flight mass spectrometry (MALDI-TOF MS) for rapid identification of micro-organisms in the routine clinical microbiology laboratory. Eur J Clin Microbiol Infect Dis 36:807–812. doi: 10.1007/s10096-016-2864-9. [DOI] [PubMed] [Google Scholar]
- 29.Kim TH, Kweon OJ, Kim HR, Lee MK. 2016. Identification of uncommon Candida species using commercial identification system. J Microbiol Biotechnol 26:2206–2213. doi: 10.4014/jmb.1609.09012. [DOI] [PubMed] [Google Scholar]
- 30.Kindo AJ, Sivaranjini A, Rajyoganandh V, Vijayakumar R. 2015. Antifungal susceptibility testing by micro-broth dilution of rare Candida species isolated from blood—a study from a tertiary care center in South India. Abstr P037 Mycoses 58:68. doi: 10.1111/myc.12380. [DOI] [Google Scholar]
- 31.Mizusawa M, Miller H, Green R, Lee R, Durante M, Perkins R, Hewitt C, Simner PJ, Carroll KC, Hayden RT, Zhang SX. 2016. Can multidrug-resistant Candida auris be reliably identified in clinical microbiology laboratories? J Clin Microbiol 55:638–640. doi: 10.1128/JCM.02202-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharma C, Masih A, Singh PK, Meis JF, Chowdhary A. 2015. Candida haemulonii complex: the true scenario by sequencing and MALDI-TOF among clinical isolates in India. Abstr P057 Mycoses 58:75–76. doi: 10.1111/myc.12380. [DOI] [Google Scholar]
- 33.Kumar A, Prakash A, Singh A, Kumar H, Hagen F, Meis JF, Chowdhary A. 2016. Candida haemulonii species complex: an emerging species in India and its genetic diversity assessed with multilocus sequence and amplified fragment-length polymorphism analyses. Emerg Microbes Infect 5:e49. doi: 10.1038/emi.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cendejas-Bueno E, Kolecka A, Alastruey-Izquierdo A, Theelen B, Groenewald M, Kostrzewa M, Cuenca-Estrella M, Gomez-Lopez A, Boekhout T. 2012. Reclassification of the Candida haemulonii complex as Candida haemulonii (C. haemulonii group I), C. duobushaemulonii sp. nov. (C. haemulonii group II), and C. haemulonii var. vulnera var. nov.: three multiresistant human-pathogenic yeasts. J Clin Microbiol 50:3641–3651. doi: 10.1128/JCM.02248-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumar A, Sachu A, Mohan K, Vinod V, Dinesh K, Karim S. 2017. Simple low cost differentiation of Candida auris from Candida haemulonii complex using CHROMagar Candida medium supplemented with Pal's medium. Rev Iberoam Micol 34:109–111. doi: 10.1016/j.riam.2016.11.004. [DOI] [PubMed] [Google Scholar]
- 36.Ghosh AK, Paul S, Sood P, Rudramurthy SM, Rajbanshi A, Jillwin TJ, Chakrabarti A. 2015. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry for the rapid identification of yeasts causing bloodstream infections. Clin Microbiol Infect 21:372–378. doi: 10.1016/j.cmi.2014.11.009. [DOI] [PubMed] [Google Scholar]
- 37.Sandrine M, Marion C, Geraldine D, Alex VB, Ferry H, Jacques M, Anurhada C, Victoria C. 2015. Identification and typing of an emerging pathogen, Candida auris, by MALDI TOF MS using the vitek MS platform. Clin Chem Lab Med 53:S1321. doi: 10.1515/cclm-2015-5033. [DOI] [Google Scholar]
- 38.Girard V, Mailler S, Chetry M, Vidal C, Durand G, van Belkum A, Colombo AL, Hagen F, Meis JF, Chowdhary A. 2016. Identification and typing of the emerging pathogen Candida auris by matrix-assisted laser desorption ionisation time of flight mass spectrometry. Mycoses 59:535–538. doi: 10.1111/myc.12519. [DOI] [PubMed] [Google Scholar]
- 39.Grenfell RC, da Silva AR Jr, Del Negro GM, Munhoz RB, Gimenes VM, Assis DM, Rockstroh AC, Motta AL, Rossi F, Juliano L, Benard G, de Almeida JN Jr. 2016. Identification of Candida haemulonii complex species: use of ClinProTools to overcome limitations of the Bruker Biotyper, VITEK MS IVD, and VITEK MS RUO databases. Front Microbiol 7:940. doi: 10.3389/fmicb.2016.00940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kordalewska M, Zhao Y, Lockhart SR, Chowdhary A, Berrio I, Perlin DS. 2017. Rapid and accurate molecular identification of the emerging multidrug resistant pathogen Candida auris. J Clin Microbiol 55:2445–2452. doi: 10.1128/JCM.00630-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sharma C, Singh A, Singh PK, Prakash A, Meis JF, Chowdhary A. 2015. Genotyping of multidrug resistant Indian Candida auris isolates by multi locus sequence typing, amplified fragment length polymorphism and MALDI-TOF-MS and their antifungal susceptibility profile. Mycoses 58:119–120. doi: 10.1111/myc.12284. [DOI] [PubMed] [Google Scholar]
- 42.Oh BJ, Shin JH, Kim MN, Sung H, Lee K, Joo MY, Shin MG, Suh SP, Ryang DW. 2011. Biofilm formation and genotyping of Candida haemulonii, Candida pseudohaemulonii, and a proposed new species (Candida auris) isolates from Korea. Med Mycol 49:98–102. doi: 10.3109/13693786.2010.493563. [DOI] [PubMed] [Google Scholar]
- 43.Rudramurthy SM, Chakrabarti A, Ahmad R, Capoor M, Kindoo A, Marak R, Patel A, Sardana R, Arora A, Biswas S. 2013. Candida auris, emerging yeast causing candidemia in intensive care units; a multicentre study. Mycoses 56:102–103. doi: 10.1111/j.1439-0507.2012.02197.x. [DOI] [Google Scholar]
- 44.Borman AM, Szekely A, Johnson EM. 2016. Comparative pathogenicity of United Kingdom isolates of the emerging pathogen Candida auris and other key pathogenic Candida species. mSphere 1:e00189-. doi: 10.1128/mSphere.00189-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Larkin E, Hager C, Chandra J, Mukherjee PK, Retuerto M, Salem I, Long L, Isham N, Kovanda L, Borroto-Esoda K, Wring S, Angulo D, Ghannoum M. 2017. The emerging pathogen Candida auris: growth phenotype, virulence factors, activity of antifungals, and effect of SCY-078, a novel glucan synthesis inhibitor, on growth morphology and biofilm formation. Antimicrob Agents Chemother 61:e02396-. doi: 10.1128/AAC.02396-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sherry L, Ramage G, Kean R, Borman A, Johnson EM, Richardson MD, Rautemaa-Richardson R. 2017. Biofilm-forming capability of highly virulent multidrug-resistant Candida auris. Emerg Infect Dis 23:328–331. doi: 10.3201/eid2302.161320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.European Committee on Antimicrobial Susceptibility Testing. 2015. Clinical breakpoints—fungi (v 8.0). [Google Scholar]
- 48.Clinical and Laboratory Standards Institute. 2008. M27-A3 reference method for broth dilution antifungal susceptibility testing of yeasts, 3rd ed. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 49.Arendrup MC, Prakash A, Meletiadis J, Sharma C, Chowdhary A. 2017. Comparison of EUCAST and CLSI reference microdilution MICs of eight antifungal compounds for Candida auris and associated tentative epidemiological cutoff values. Antimicrob Agents Chemother 61:e00485-. doi: 10.1128/AAC.00485-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lockhart SR, Berkow EL, Chow N, Welsh RM. 2017. Candida auris for the clinical microbiology laboratory: not your grandfather's Candida species. Clin Microbiol Newsl 39:99–103. doi: 10.1016/j.clinmicnews.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lepak AJ, Zhao M, Berkow EL, Lockhart SR, Andes DR. 2017. Pharmacodynamic optimization for treatment of invasive Candida auris infection. Antimicrob Agents Chemother 61:e00791-. doi: 10.1128/AAC.00791-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Magobo RE, Govender NP, Corcoran C. 2016. Molecular typing of multidrug-resistant Candida auris strains in South Africa, poster 89 ASLM2016 conference programme book ASLM, Addis Ababa, Ethiopia. [Google Scholar]
- 53.Kumar D, Banerjee T, Pratap CB, Tilak R. 2015. Itraconazole-resistant Candida auris with phospholipase, proteinase and hemolysin activity from a case of vulvovaginitis. J Infect Dev Ctries 9:435–437. doi: 10.3855/jidc.4582. [DOI] [PubMed] [Google Scholar]
- 54.Sarma S, Kumar N, Sharma S, Govil D, Ali T, Mehta Y, Rattan A. 2013. Candidemia caused by amphotericin B and fluconazole resistant Candida auris. Indian J Med Microbiol 31:90–91. doi: 10.4103/0255-0857.108746. [DOI] [PubMed] [Google Scholar]
- 55.Shin JH, Kim MN, Jang SJ, Ju MY, Kim SH, Shin MG, Suh SP, Ryang DW. 2012. Detection of amphotericin B resistance in Candida haemulonii and closely related species by use of the Etest, Vitek-2 yeast susceptibility system, and CLSI and EUCAST broth microdilution methods. J Clin Microbiol 50:1852–1855. doi: 10.1128/JCM.06440-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chowdhary A, Voss A, Meis JF. 2016. Multidrug-resistant Candida auris: ‘new kid on the block’ in hospital-associated infections? J Hosp Infect 94:209–212. doi: 10.1016/j.jhin.2016.08.004. [DOI] [PubMed] [Google Scholar]
- 57.Public Health England. 2017. Guidance for the laboratory investigation, management and infection prevention and control for cases of Candida auris. Public Health England, United Kingdom. [Google Scholar]
- 58.Cornely OA, Bassetti M, Calandra T, Garbino J, Kullberg BJ, Lortholary O, Meersseman W, Akova M, Arendrup MC, Arikan-Akdagli S, Bille J, Castagnola E, Cuenca-Estrella M, Donnelly JP, Groll AH, Herbrecht R, Hope WW, Jensen HE, Lass-Florl C, Petrikkos G, Richardson MG, Roilides E, Verweij PE, Viscoli C, Ullmann AJ. 2012. ESCMID guideline for the diagnosis and management of Candida diseases 2012: non-neutropenic adult patients. Clin Microbiol Infect 18(Suppl 7:19–37. doi: 10.1111/1469-0691.12039. [DOI] [PubMed] [Google Scholar]
- 59.Pappas PG, Kauffman CA, Andes DR, Clancy CJ, Marr KA, Ostrosky-Zeichner L, Reboli AC, Schuster MG, Vazquez JA, Walsh TJ, Zaoutis TE, Sobel JD. 2016. Clinical practice guideline for the management of candidiasis: 2016 update by the Infectious Diseases Society of America. Clin Infect Dis 62:e1–e50. doi: 10.1093/cid/civ1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fakhim H, Chowdhary A, Prakash A, Vaezi A, Dannaoui E, Meis JF, Badali H. 28 August 2017. In vitro interactions of echinocandins with triazoles against multidrug-resistant Candida auris. Antimicrob Agents Chemother doi: 10.1128/AAC.01056-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kofla G, Ruhnke M. 2011. Pharmacology and metabolism of anidulafungin, caspofungin and micafungin in the treatment of invasive candidosis: review of the literature. Eur J Med Res 16:159–166. doi: 10.1186/2047-783X-16-4-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fisher JF, Sobel JD, Kauffman CA, Newman CA. 2011. Candida urinary tract infections—treatment. Clin Infect Dis 52(Suppl 6:457–466. doi: 10.1093/cid/ciq144. [DOI] [PubMed] [Google Scholar]
- 63.Berkow EL, Angulo D, Lockhart SR. 2017. In vitro activity of a novel glucan synthase inhibitor, SCY-078, against clinical isolates of Candida auris. Antimicrob Agents Chemother 61:e00435-. doi: 10.1128/AAC.00435-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Britz E, Govender NP. 2016. Global emergence of a multi-drug resistant fungal pathogen, Candida auris. South Afr J Epidemiol Infect 31:3–4. [Google Scholar]
- 65.Schelenz S, Barnes RA, Barton RC, Cleverley JR, Lucas SB, Kibbler CC, Denning DW. 2015. British Society for Medical Mycology best practice recommendations for the diagnosis of serious fungal diseases. Lancet Infect Dis 15:461–474. doi: 10.1016/S1473-3099(15)70006-X. [DOI] [PubMed] [Google Scholar]
- 66.Shackleton J, Schelenz S, Rochon M, Hall A, Ryan L, Cervera-Jackson R. 2016. The impact of environmental decontamination in a Candida auris outbreak. J Hosp Infect 94(Suppl 1:S24–S134. doi: 10.1016/S0195-6701(16)30516-3. [DOI] [Google Scholar]
- 67.Tsay S, Welsh RM, Adams EH, Chow NA, Gade L, Berkow EL, Poirot E, Lutterloh E, Quinn M, Chaturvedi S, Kerins J, Black SR, Kemble SK, Barrett PM, Barton K, Shannon DJ, Bradley K, Lockhart SR, Litvintseva AP, Moulton-Meissner H, Shugart A, Kallen A, Vallabhaneni S, Chiller TM, Jackson BR. 2017. Notes from the field: ongoing transmission of Candida auris in health care facilities—United States, June 2016-May 2017. MMWR Morb Mortal Wkly Rep 66:514–515. doi: 10.15585/mmwr.mm6619a7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rudramurthy SM, Chakrabarti A, Paul RA, Sood P, Kaur H, Capoor MR, Kindo AJ, Marak RSK, Arora A, Sardana R, Das S, Chhina D, Patel A, Xess I, Tarai B, Singh P, Ghosh A. 2017. Candida auris candidaemia in Indian ICUs: analysis of risk factors. J Antimicrob Chemother 72:1794–1801. doi: 10.1093/jac/dkx034. [DOI] [PubMed] [Google Scholar]
- 69.Khillan V, Rathore N, Kathuria S, Chowdhary A. 2014. A rare case of breakthrough fungal pericarditis due to fluconazole-resistant Candida auris in a patient with chronic liver disease. JMM Case Rep doi: 10.1099/jmmcr.0.T00018. [DOI] [Google Scholar]
- 70.Azar MM, Turbett SE, Fishman JA, Pierce VM. 2017. Donor-derived transmission of Candida auris during lung transplantation. Clin Infect Dis 65:1040–1042. doi: 10.1093/cid/cix460. [DOI] [PubMed] [Google Scholar]
- 71.Centre for Opportunistic, Tropical, and Hospital Infections. 2016. Interim guidance for management of Candida auris infections in South African hospitals. Centre for Opportunistic, Tropical, and Hospital Infections, Johannesburg, South Africa. [Google Scholar]
- 72.Centers for Disease Control and Prevention. 2016. Candida auris interim recommendations for healthcare facilities and laboratories. Centers for Disease Control and Prevention, Atlanta, GA. [Google Scholar]
- 73.Public Health England. 2017. Candida auris: infection control in community settings. Public Health England, United Kingdom. [Google Scholar]
- 74.Abdolrasouli A, Armstrong-James D, Ryan L, Schelenz S. 2017. In vitro efficacy of disinfectants utilised for skin decolonisation and environmental decontamination during a hospital outbreak with Candida auris. Mycoses doi: 10.1111/myc.12699. [DOI] [PubMed] [Google Scholar]
- 75.Moore G, Schelenz S, Borman AM, Johnson EM, Brown CS. 2017. The yeasticidal activity of chemical disinfectants and antiseptics against Candida auris. J Hosp Infect doi: 10.1016/j.jhin.2017.08.019. [DOI] [PubMed] [Google Scholar]
- 76.Piedrahita CT, Cadnum JL, Jencson AL, Shaikh AA, Ghannoum MA, Donskey CJ. 2017. Environmental surfaces in healthcare facilities are a potential source for transmission of Candida auris and other candida species. Infect Control Hosp Epidemiol 38:1107–1109. doi: 10.1017/ice.2017.127. [DOI] [PubMed] [Google Scholar]
- 77.Welsh RM, Bentz ML, Shams A, Houston H, Lyons A, Rose LJ, Litvintseva AP. 2017. Survival, persistence, and isolation of the emerging multidrug-resistant pathogenic yeast Candida auris on a plastic healthcare surface. J Clin Microbiol 55:2996–3005. doi: 10.1128/JCM.00921-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chauhan R, Loonker S. 2017. Synthesis, characterization and biological evaluation of chitosan epoxy n-methyl piperazine as antimicrobial agent. Int J Pharm Sci Rev Res 45:266–270. [Google Scholar]
- 79.Cadnum JL, Shaikh AA, Piedrahita CT, Sankar T, Jencson AL, Larkin EL, Ghannoum MA, Donskey CJ. 2017. Effectiveness of disinfectants against Candida auris and other Candida species. Infect Control Hosp Epidemiol 38:1240–1243. doi: 10.1017/ice.2017.162. [DOI] [PubMed] [Google Scholar]
- 80.Madder H, Moir I, Moroney R, Butcher L, Newnham R, Sunderland M, Clarke T, Foster D, Hoffman P, Moore G, Brown CS, Jeffery KJM. 2017. Multiuse patient monitoring equipment as a risk factor for acquisition of Candida auris. bioRxiv 149054. doi: 10.1101/149054. [DOI] [Google Scholar]
- 81.Sharp A, Brown C, Charlett A, Cummins M, Guy R, Hall A, Jeffery K, Muller-Pebody B, Patel B, Schelenz S, Manuel R. 2017. Prevalence and risk factors for Candida auris colonisation among intensive care patients in English hospital: protocol for a field study, poster 0963. 27th Eur Congr Clin Microbiol Infect Dis, Vienna, Austria, 22 to 25 April 2017. [Google Scholar]
- 82.McCarthy MW, Walsh TJ. 2017. Drug development challenges and strategies to address emerging and resistant fungal pathogens. Expert Rev Anti Infect Ther 15:577–584. doi: 10.1080/14787210.2017.1328279. [DOI] [PubMed] [Google Scholar]