SUMMARY
The recent development of commercial panel-based molecular diagnostics for the rapid detection of pathogens in positive blood culture bottles, respiratory specimens, stool, and cerebrospinal fluid has resulted in a paradigm shift in clinical microbiology and clinical practice. This review focuses on U.S. Food and Drug Administration (FDA)-approved/cleared multiplex molecular panels with more than five targets designed to assist in the diagnosis of bloodstream, respiratory tract, gastrointestinal, or central nervous system infections. While these panel-based assays have the clear advantages of a rapid turnaround time and the detection of a large number of microorganisms and promise to improve health care, they present certain challenges, including cost and the definition of ideal test utilization strategies (i.e., optimal ordering) and test interpretation.
KEYWORDS: syndromic testing, multiplex PCR, molecular methods
INTRODUCTION
The field of clinical microbiology has experienced significant changes over the past decade, due to new technologies that have improved the diagnosis of infectious diseases. These innovations include commercial molecular assays that simultaneously detect and identify multiple pathogens associated with clinical syndromes, such as bloodstream, respiratory, gastrointestinal (GI), or central nervous system (CNS) infections. These multiplex tests are revolutionary, enabling health care providers to rapidly diagnose certain infections and therefore allowing clinical management decisions (e.g., hospital admission, isolation, and antimicrobial treatment or lack thereof) to be made in a timely manner. These technologies have also, at times, introduced challenges. Multiplex tests are often expensive, requiring the development of utilization management strategies to guide their appropriate use. Current clinical practice guidelines may not yet address their utilization or provide guidance as to how results should be interpreted. Clinicians may not be familiar with all organisms and/or resistance genes detected, creating clinical confusion. This can lead to inappropriate treatment and unnecessary subsequent laboratory testing alongside provider and, potentially, patient anxiety. Panel compositions vary somewhat between manufacturers; their generally fixed panel composition may present challenges in certain circumstances. The design of these multiplex platforms, even those marketed to be closed systems, carries a risk of contamination, which may be challenging to recognize. Additional challenges include determining how multiplex panels should be integrated into laboratory workflows as well as how results should be monitored for accuracy following implementation. Although these tests clearly offer advantages, multiplex assays need to be thoughtfully integrated into clinical practice. Furthermore, their impact on public health laboratories should be considered.
It is anticipated that over time, syndromic testing will become increasingly common and will be performed outside clinical microbiology laboratories. The use of these assays in point-of-care settings will demand thoughtful implementation strategies, with guidance from both clinical and laboratory professionals.
Here, we review the current literature on multiplex molecular microbiology testing of positive blood culture bottles, respiratory specimens, stool, and cerebrospinal fluid (CSF) available in the United States, acknowledging that the field is in rapid evolution.
RAPID TESTING OF POSITIVE BLOOD CULTURE BOTTLES
Bacteremia and severe sepsis are major causes of mortality in hospitalized patients (1). There has been an increase in the number of hospitalizations for severe sepsis over the past decade, likely due to an aging population with chronic medical comorbidities and an increasing number of immunocompromised hosts (1, 2). Among patients with septic shock, delays in the administration of effective antimicrobial therapy are associated with increased mortality rates (3). Today, the efficacy of the early administration of antimicrobial treatment may be compromised by an increasing prevalence of bacterial drug resistance.
Although the introduction of automated, continuous-monitoring blood culture systems in the last century improved the diagnosis of bloodstream infections, there are still delays in the identification of pathogens, the detection of antimicrobial resistance, and the designation of contaminants. This can impact patient management decisions, directly contributing to morbidity and mortality and potentially leading to adverse outcomes (e.g., Clostridium difficile-associated diarrhea, selection of drug resistance, and increased patient costs) (4). The widespread implementation of matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) has expedited the identification of isolates compared to traditional biochemical methods, but this approach typically involves subculture, contributing to potential delays. Due to the significant impact of bacterial infections on national and global health, there are a number of initiatives to combat antimicrobial resistance that highlight the importance of innovative diagnostic tests to rapidly identify bacteria and detect resistance (5). Direct testing of positive blood culture bottles by MALDI-TOF MS yields accurate identification; however, this approach requires the processing of the blood culture bottle contents, is not U.S. Food and Drug Administration (FDA) approved/cleared, and has a lower success rate than testing of colony isolates (4, 6). As an alternative, the use of MALDI-TOF MS for the identification of bacterial isolates from positive blood cultures after short-term incubation (i.e., 2 to 6 h) of high-inoculum subcultures on solid media has been adopted by many laboratories, including our own (7–9). For example, we use this approach to test positive blood culture bottles showing Gram-negative bacilli upon Gram staining; in our hands, this method has excellent performance, enabling the identification of organisms in 92% (45/49) of blood culture bottles positive for Gram-negative bacilli following a 4-h incubation (our unpublished data). This offers a means of reducing the turnaround time compared to that of conventional testing, without adding substantial costs, given that MALDI-TOF MS is already available in many clinical microbiology laboratories and has a low per-test cost. Limitations of this approach are that it does not address antimicrobial susceptibility and that it is not useful for laboratories that do not have MALDI-TOF MS available.
FDA-Approved/Cleared Assays
Currently, there are three FDA-approved/cleared multiplex assays that simultaneously detect a number of microorganisms, as well as select resistance genes, directly from positive blood culture bottles: the FilmArray Blood Culture Identification (BCID) panel (BioFire Diagnostics, LLC) (which received FDA approval/clearance in 2013) and the Verigene Gram-positive blood culture (BC-GP) (which received FDA approval/clearance in 2012) and Gram-negative blood culture (BC-GN) (which received FDA approval/clearance in 2014) tests (Luminex Corporation) (Table 1) (10). The Accelerate Pheno system (Accelerate Diagnostics) is a newer, completely automated system that uses gel electrofiltration and fluorescence in situ hybridization (FISH) for the identification of a limited number of bacteria and yeast within 90 min directly from positive blood cultures. More importantly, this is the first FDA-approved/cleared assay to provide rapid (within 7 h) phenotypic antimicrobial susceptibility testing directly from positive blood cultures. This system extrapolates MIC values by analyzing bacterial growth in the presence of specific antibiotic concentrations using automated microscopy and time-lapse imaging. This assay it not further discussed here.
TABLE 1.
FDA-approved/cleared panel-based molecular assays for detection of select microorganisms and select resistance genes in positive blood culture bottles
| Parameter | FilmArray BCID | Verigene |
|
|---|---|---|---|
| Gram-positive blood culture | Gram-negative blood culture | ||
| Total no. of targets | 27 | 15 | 14 |
| Ability to detect pathogen | |||
| Gram-positive bacteria | |||
| Staphylococcus species | ✓ | ✓ | |
| Staphylococcus aureus | ✓ | ✓ | |
| Staphylococcus epidermidis | ✓ | ||
| Staphylococcus lugdunensis | ✓ | ||
| Streptococcus species | ✓ | ✓ | |
| Streptococcus agalactiae | ✓ | ✓ | |
| Streptococcus pyogenes | ✓ | ✓ | |
| Streptococcus pneumoniae | ✓ | ✓ | |
| Streptococcus anginosus group | ✓ | ||
| Enterococcus species | ✓ | ||
| Enterococcus faecalis | ✓ | ||
| Enterococcus faecium | ✓ | ||
| Listeria species | ✓ | ||
| Listeria monocytogenes | ✓ | ||
| Gram-negative bacteria | |||
| Klebsiella oxytoca | ✓ | ✓ | |
| Klebsiella pneumoniae | ✓ | ✓ | |
| Serratia marcescens | ✓ | ||
| Proteus species | ✓ | ✓ | |
| Acinetobacter species | ✓ | ||
| Acinetobacter baumannii | ✓ | ||
| Haemophilus influenzae | ✓ | ||
| Neisseria meningitis | ✓ | ||
| Pseudomonas aeruginosa | ✓ | ✓ | |
| Enterobacteriaceae | ✓ | ||
| Escherichia coli | ✓ | ✓ | |
| Enterobacter species | ✓ | ||
| Enterobacter cloacae complex | ✓ | ||
| Citrobacter species | ✓ | ||
| Yeasts | |||
| Candida albicans | ✓ | ||
| Candida glabrata | ✓ | ||
| Candida krusei | ✓ | ||
| Candida parapsilosis | ✓ | ||
| Candida tropicalis | ✓ | ||
| Ability to detect presence of resistance gene | |||
| mecA | ✓ | ✓ | |
| vanA | ✓ | ✓ | |
| vanB | ✓ | ✓ | |
| blaKPC | ✓ | ✓ | |
| blaNDM | ✓ | ||
| blaOXA | ✓ | ||
| blaVIM | ✓ | ||
| blaIMP | ✓ | ||
| blaCTX-M | ✓ | ||
| Time to result (h) | ∼1 | ∼2.5 | ∼2 |
The BCID test is a closed, multiplex PCR system that offers automated sample preparation, amplification, detection, and analysis and simultaneously tests for 27 targets (Table 1). It requires ∼2 min of hands-on time to add 200 μl of a sample from a positive blood culture bottle to sample buffer in a single pouch, which is then loaded into the FilmArray system. The test turnaround time is ∼1 h. The FilmArray Torch system received FDA clearance for use on all four existing FilmArray panels in 2016. This system provides higher throughput (configurable from 2 to 12 modules), as it allows the placement of multiple instrument modules in a tower configuration. The BC-GP and BC-GN panels use the Verigene system, consisting of two components, the Verigene Processor SP, which provides automated sample preparation and detection of bacterial DNA in a microarray format by using gold nanoparticle probe-based technology, and the Verigene reader, which generates results based on light scatter analysis from microarray spots. The test cartridge, sample (containing 350 μl of broth from a positive blood culture bottle), and test consumables are loaded into the Processor SP system for sample preparation and test processing. The slide from the test cartridge is then placed into the Verigene reader to yield results. A single Verigene reader can control up to 32 Processor SP units, allowing scalable workflow. The test has a hands-on time of ∼5 min and a run time of <2.5 h.
Assay Performance
Table S1 in the supplemental material shows the major studies evaluating the performance of FDA-approved/cleared multiplex molecular panels for testing positive blood culture bottles. Ward et al. (11) compared the accuracies and turnaround times of these multiplex assays to those of conventional, culture-based methods (primarily MALDI-TOF MS-based colony identification) using positive blood culture samples (n = 173). The Verigene and BCID tests reduced the turnaround time by 27.9 and 29.1 h, respectively, compared to conventional methods. The Verigene and BCID tests provided correct identification for 90.6 and 87.2% of samples, respectively, compared to conventional methods. The Verigene assay generated 6 false-positive results (among which were 2 viridans group streptococcal isolates that were falsely identified as Streptococcus pneumoniae), whereas the BCID test yielded 25 false-positive results. A subsequent investigation showed that the false-positive results by the BCID test were likely due to contamination of BacT/Alert standard anaerobic bottles (bioMérieux) with Pseudomonas aeruginosa DNA (11). This cautionary note is a reminder that quality control metrics need to be carefully developed when using multiplex molecular panels and especially so if the testing involves matrices (e.g., blood culture bottle contents) that are not themselves part of the tests. In a study by Bhatti et al. that assessed the performances of the BCID and Verigene assays in comparison to conventional, culture-based methods (colony identification by using the Vitek MS Ruo system [bioMérieux]), 95% and 99% of identifiable isolates in monomicrobial cultures (n = 118) were correctly identified by the BCID and Verigene assays, respectively. Both assays had shorter times to identification than those of conventional methods (1.15 to 2.5 h versus 25.6 h). The BCID panel detected mecA in 4 staphylococcal isolates (n = 3 for Staphylococcus aureus and n = 1 for coagulase-negative Staphylococcus species [CoNS]), which were susceptible to methicillin. The presence of an altered staphylococcal cassette chromosome mec element in these isolates was thought to account for the discrepant mecA results (12). Altun et al. evaluated the clinical performance of the BCID panel separately for monomicrobial and polymicrobial growth in blood culture bottles. Compared to conventional methods (panel of desktop spot tests and Vitek2 XL- and MALDI-TOF MS-based colony identification), the BCID test had sensitivities of 91.6% (153/167) and 71% (17/24) for monomicrobial and polymicrobial cultures, respectively. Of note, 7.8% (13/167) of the organisms in the monomicrobial group were not part of the BCID panel. Among polymicrobial specimens, while the panel failed to detect 2 isolates of Enterococcus faecalis and 1 isolate each of Escherichia coli and alpha-hemolytic Streptococcus species, the majority of organisms that were not identified were not part of the panel. The BCID panel could not accurately assess the methicillin susceptibility of S. aureus in a polymicrobial sample containing S. aureus alongside CoNS in which mecA was detected because the CoNS isolate was methicillin resistant and the S. aureus isolate was methicillin susceptible (13).
Clinical and Economic Impacts
Given the high costs of these assays, several studies have assessed their clinical and economic impacts (Table 2). Overall, these studies show a decrease in the time to organism identification and generally show a decrease in the time to optimization of antimicrobial therapy. However, there have been inconclusive results in terms of these panels' impacts on mortality rates and lengths of hospitalization, and interpretation of the data is complicated by suboptimal study design in many cases. In particular, data from pre/postintervention studies are hard to interpret due to nonstudy variations over time. Institution-specific variables, such as distinctive patient populations and local resistance rates, and the availability of antimicrobial stewardship programs likely affect the clinical impact of rapid molecular assays for testing positive blood culture bottles. These panels have the greatest impact when results are reported as quickly as possible and appropriately acted upon by providers caring for the patient. In the case of antibiotic deescalation, this is ideally accomplished in the context of delivery of results to an expert in antimicrobial stewardship (e.g., infectious diseases physician, infectious diseases pharmacist, or doctoral-level clinical microbiologist), who can then provide individualized and rapid guidance to providers caring for the patient (14).
TABLE 2.
Studies evaluating the effects of panel-based molecular assays for detection of select microorganisms and resistance genes in positive blood culture bottles on clinical and economic outcomes for patients with bacteremiaa
| Test(s) | Study design | Outcome(s) of rapid test compared to standard methods | Antimicrobial stewardship intervention(s) | Reference |
|---|---|---|---|---|
| BioFire BCID | Single-center, prospective, randomized, controlled trial of 617 subjects | Decreased TAT; decreased time to deescalation of antibiotics in BCID group with ASP intervention; no differences in LOS, mortality rates, adverse events, and cost | Audit and feedback by ASP 24 h a day/7 days a wk; treatment guidance comments included in microbiology result report | 14b |
| BioFire BCID | Single-center pre/postintervention study of 364 subjects | Decreased TAT; shorter time to effective therapy; no difference in mortality rate, ICU LOS, cost, or 30 day-readmission rate | Audit and feedback by ASP performed Monday to Friday during daytime; no templated comments in report | 22 |
| BioFire BCID | Single-center, pre/postintervention study of 336 subjects | Decreased TAT; shorter duration of empirical vancomycin use in patients with contaminated blood cultures and MSSA bacteremia, earlier effective therapy for VRE bacteremia, and shorter LOS and decreased costs for CoNS bacteremia | Audit and feedback by ASP performed once daily Monday to Friday during daytime | 23b |
| BioFire BCID | Single-center pre/postintervention study of 300 hospitalized children | Decrease in median time to optimal therapy; decrease in unnecessary antibiotic initiation for contaminated blood cultures | Audit and feedback by ASP performed in real time for postintervention group | 24 |
| Verigene BC-GP | Multicenter, pre/postintervention study with retrospective evaluation of preintervention group involving 167 subjects | Decreased TAT, LOS, cost, and duration of unnecessary antibiotic treatment; similar mortality rates between groups | Audit and feedback by ASP during daytime | 15b |
| Verigene BC-GP | Single-center, pre/postintervention study of 74 subjects with enterococcal bacteremia | Decreased TAT, LOS, and cost; no difference in mortality rates between groups | Audit and feedback by ASP Monday to Friday during daytime | 16b |
| Verigene BC-GP | Single-center, pre/postintervention study of 513 subjects | Reduction in time to deescalation of antimicrobial therapy; no difference in mortality rates or LOS | Audit and feedback by ASP Monday to Friday during daytime | 17 |
| Verigene BC-GP and BC-GN | Single-center, pre/postintervention study involving 235 hospitalized subjects | Decreased TAT, 30-day mortality rate, cost, and time to optimization of antimicrobial therapy | ID physicians reviewed and gave recommendations for all positive blood cultures on weekdays | 18b |
| Verigene BC-GP | Single-center, retrospective, pre/postintervention study involving 147 subjects with MSSA or VRE bacteremia | Decreased TAT and time to optimal antibiotics for MSSA and VRE bacteremia; no differences in LOS or mortality rates | Microbiology report included treatment guidance comments | 19 |
| Verigene BC-GN | Single-center, retrospective, pre/postintervention study involving 195 subjects | Decreased TAT, ICU LOS, 30-day mortality rate, mortality rate associated with MDR pathogens, and cost | Audit and feedback by ASP daily | 20b |
| Verigene BC-GN | Single-center, retrospective study with theoretical evaluation of antimicrobial therapy-related outcomes involving 132 subjects | Shorter times to effective and optimal antibiotic therapy | Theoretical ASP intervention by ID physician and pharmacist | 21 |
| Rapid blood culture diagnostics such as MALDI-TOF MS, PNA-FISH, PCR, and microarray technologies | Meta-analysis of 31 studies involving 5,920 subjects with bacteremia | Decreased mortality risk in the presence but not in the absence of ASP; lower mortality rates for infections by Gram-positive and Gram-negative organisms but not yeast; decreased time to effective therapy and LOS | Various | 25 |
Adapted from reference 4. TAT, turnaround time; ICU, intensive care unit; LOS, length of stay; ASP, antimicrobial stewardship program; MSSA, methicillin-susceptible Staphylococcus aureus; VRE, vancomycin-resistant Enterococcus species; MDR, multidrug resistant; ID, infectious diseases.
This study was included in the meta-analysis (25).
In a multicenter, pre/postintervention study involving 167 hospitalized patients with bacteremia, the use of the Verigene BC-GP assay was associated with decreased durations of unnecessary antibiotic treatment, median lengths of stay, and hospitalization costs compared to those of conventional organism identification using the BD Phoenix system (BD Diagnostics) (15). However, the overall mortality rates were similar between the groups. In a single-center, pre/postintervention study that evaluated the impact of the Verigene BC-GP assay on a cohort of 74 patients with enterococcal bacteremia, the authors observed reductions in the hospital length of stay by 21.7 days (P = 0.04) and mean hospital costs by $60,729 (P = 0.02) compared to those in the study period before the implementation of the multiplex assay (16). Another pre/postintervention study evaluated the impact of the Verigene BC-GP assay on 513 hospitalized patients with blood cultures positive for Gram-positive organisms (17). While there were no differences in overall mortality rates or hospital lengths of stay, a reduction in the time to deescalation of antimicrobial therapy compared to conventional methods (i.e., culture with MALDI-TOF MS identification) was observed for the postintervention group. Similarly, Suzuki et al. (18) reported reductions in 30-day mortality rates (3 versus 13%; P = 0.019), costs associated with antimicrobial treatment, and time to optimization of the antimicrobial regimen in hospitalized patients (control group, n = 147; intervention group, n = 88) with bacteremia after the implementation of the Verigene BC-GP and BC-GN assays compared to conventional identification (MicroScan WalkAway-96; Beckman Coulter, Inc.). Beal et al. reported increased deescalation of empirical antibiotics and a reduction in the time to optimal antibiotics for patients with methicillin-susceptible S. aureus and vancomycin-resistant enterococcal bacteremia after the implementation of the Verigene BC-GP assay paired with a defined result-reporting algorithm using electronic communications compared to a preintervention group for which traditional phenotypic methods and the Vitek 2 system (bioMérieux) were utilized for identification and antimicrobial susceptibility testing (19). In a retrospective analysis of Gram-negative bacteremia in 195 hospitalized patients over a 6-month period before and after the implementation of Verigene BC-GN test, the length of intensive care unit stay, 30-day mortality rates, and mortality rates associated with multidrug-resistant pathogens were lower in the postintervention group (P < 0.05). In this study, identification and susceptibility testing of isolated colonies in the preintervention group were performed by using the Vitek 2 system (bioMérieux) (20). Using a retrospective study design, Bork et al. reported that there could theoretically be shorter times to effective (3.3 versus 7.0 h; P < 0.01) and optimal (23.5 versus 41.8 h; P < 0.01) antibiotic therapy if the Verigene BC-GN assay and antimicrobial stewardship team review were used than with conventional identification using the Vitek 2 system (bioMérieux) (21). These findings show that the implementation of rapid multiplex assays may allow providers to optimize antimicrobial treatment and suggest that they may reduce durations of hospitalization.
In a pre/postintervention study by MacVane and Nolte involving 364 hospitalized adult patients with bacteremia, patients were categorized into three groups: conventional organism identification using phenotypic methods and the MicroScan WalkAway system (Beckman Coulter, Inc.) (control group), conventional organism identification with antimicrobial stewardship (antimicrobial stewardship group), and BCID with antimicrobial stewardship (BCID group). The BCID group had a shorter time to effective therapy (5 h; P < 0.001) than did the control group (15 h) or the antimicrobial stewardship group (13 h); however, there was no difference with respect to the mortality rate, 30-day readmission rate, intensive care unit length of stay, or cost of care (22). In a pre/postintervention study by Pardo et al., the implementation of the BCID panel led to a shorter duration of empirical vancomycin use for patients with contaminated blood cultures (P = 0.005) and methicillin-susceptible S. aureus bacteremia (P < 0.001), earlier effective therapy for patients with vancomycin-resistant enterococcal bacteremia (P = 0.047), and shorter postculture lengths of stay for those with CoNS bacteremia (P < 0.008) than with conventional identification and susceptibility testing using phenotypic methods and the Vitek 2 system (bioMérieux) (23). Another pre/postintervention study involving 300 hospitalized children with bacteremia noted decreases in the median times to optimal therapy (26.7 h versus 60.2 h; P = 0.001) and antibiotic initiation for cultures with contaminants (26% versus 76%; P < 0.001) in the BCID group compared to the preintervention group, for which MicroScan (Siemens Healthcare Diagnostics, Inc.), RapID NF, and RapID NH (Remel Inc.) were used for bacterial identification and the API 20C Aux system (bioMérieux) was used for the identification of yeasts, alongside rapid penicillin binding protein 2′ latex agglutination testing (Oxoid, Basingstoke, United Kingdom) (24).
Our group performed a randomized controlled clinical trial evaluating the clinical impact of BCID testing of positive blood cultures along with simultaneous antimicrobial stewardship guidance. A total of 617 patients was randomized to one of three arms, a standard processing or control arm, which included organism identification using MALDI-TOF MS performed on isolated colonies (n = 207); BCID results reported with templated comments (n = 198); or BCID results reported with templated comments and real-time audit with feedback by an antimicrobial stewardship team (n = 212). While there was no effect on the mortality rate, length of stay, or time to blood culture clearance among these groups, the time to deescalation of appropriate antibiotics was shortest for the BCID group with antimicrobial stewardship intervention. Antibiotic escalation occurred sooner in both BCID groups than in the control group (14). In a meta-analysis of 31 studies which involved 5,920 patients with bloodstream infections, the implementation of rapid blood culture bottle diagnostics such as MALDI-TOF MS, peptide nucleic acid FISH (PNA-FISH), PCR, or microarray technologies was associated with a lower mortality rate (odds ratio, 0.66; 95% confidence interval [CI], 0.54 to 0.8), a shorter time to implementation of effective therapy (mean difference, 5.03 h; 95% CI, −8.60 to −1.45 h), and a shorter length of stay (mean difference, 2.48 days; 95% CI, −3.90 to −1.06 days) than with conventional microbiological methods (25).
Advantages and Limitations
These multiplex assays offer minimal hands-on time and sample preparation and are highly automated. Another key advantage of these tests is their rapid turnaround time, enabling the identification of select pathogens within 1 to 3 h (depending on the platform), theoretically allowing the early optimization of antimicrobial therapy as well as the implementation of appropriate infection prevention and control measures. In order to enable the rapid escalation or deescalation of antimicrobial therapy, the results of these assays should, of course, be reported to providers as rapidly and directly as possible and should also ideally be communicated to an expert in antimicrobial stewardship who can work with the providers to optimize therapy (10).
A feature of these assays that may be helpful in the future is that it is theoretically possible to identify organisms and detect antibiotic resistance genes in blood culture bottles before the bottles signal positive in current blood culture systems (26). In a study that investigated the performance of BCID in identifying organisms from blood culture bottles prior to positivity using simulated BacT/Alert FA Plus BC bottles spiked with five isolates each of Escherichia coli and S. aureus, the BCID panel identified all 10 isolates before blood culture positivity, with 9/10 being detected 5 h and 1 being detected 7.5 h after incubation in the blood culture system (26). As blood culture systems that signal positive earlier than current systems are developed (27), panel-based molecular diagnostics will be able to be performed on these new positive blood culture bottles, enabling a faster diagnosis of bloodstream infection than possible today.
Although multiplex assays have potential benefits over routine testing, there are certain disadvantages to consider, primarily their relatively high cost. These tests are largely “add-on” tests, because conventional subcultures and antimicrobial susceptibility testing are still required, although reidentification of isolated colonies may be avoided if the colony morphology in question is consistent with the molecularly detected organism. To realize maximal benefits, these assays should be performed 24 h a day/7 days a week, adding logistical hurdles for both the laboratory and the stewardship system. In addition, the panels do not cover all causes of bloodstream infection and may not be capable of identifying all pathogens in mixed infections, even if the organisms are included in the panel (10). False-positive results may also occur. In our clinical practice, we observed that the BCID panel detected organisms that were not visualized upon Gram staining or recovered in culture in 1.7% of positive BD Bactec blood culture bottles (Becton Dickinson), with Candida parapsilosis and Proteus species being the most commonly involved organisms (28). The presence of nucleic acid from nonviable organisms in blood culture bottles could potentially explain this finding (29), as was mentioned for the detection of P. aeruginosa DNA in BacT/Alert standard anaerobic bottles in the study by Ward et al. (11). It is therefore important to correlate the results of the multiplex panel with Gram staining. Additionally, rare instances of species-level misidentification have been reported. For example, the Verigene BC-GP assay misidentified three of eight Streptococcus mitis isolates as S. pneumoniae in one study (30). Such discrepancies may be assay specific. While the Verigene assays offer customized ordering of different panels (BC-GP and BC-GN) based on Gram staining, this option is not available with the BCID panel. Finally, a narrow spectrum of genes associated with drug resistance in Gram-negative bacteria is included in these panels, especially in the case of the BCID panel, and therefore, their ability to predict the susceptibility of Gram-negative bacilli is imperfect. Hopefully, over time, cost will decrease and targets will be further refined to optimize performance of these assays.
Detection of Pathogens Directly from Blood
The sensitivity of blood culture-based diagnostics is decreased when antimicrobial therapy is initiated prior to culture. In addition, fastidious or noncultivable organisms (such as Coxiella burnetii, Tropheryma whipplei, and Rickettsia species) do not grow in routine blood cultures, often evading detection. Also, there are inherent delays in the time to identification of pathogens related to the time to growth in currently used systems (31). There is therefore an unmet need for the rapid identification of pathogens directly from blood without incurring the time loss associated with culture-based methods. Current limitations of multiplex molecular assays for this application include their modest sensitivity, ability to detect only a limited number of microbial targets, lack of standardization, potential for inhibition by human genomic DNA, and contamination of reagents. While there are no FDA-approved/cleared assays for the direct detection of bacteria from blood samples, the T2Candida panel (T2 Biosystems) is an in vitro diagnostic assay for the direct detection of Candida species from whole-blood specimens and has demonstrated high sensitivity compared to that of blood culture (32). This test runs on an automated platform (T2Dx) and uses PCR and T2 magnetic resonance for the multiplex detection of five Candida species (C. albicans, C. tropicalis, C. parapsilosis, C. krusei, and C. glabrata) directly from a whole-blood specimen (minimum volume of 3 ml), with an average turnaround time of 4.3 h. Mylonakis et al. performed a multicenter clinical trial to evaluate the sensitivity and specificity of the T2Candida panel to diagnose candidemia. Blood specimens from 1,801 hospitalized patients who had blood cultures ordered as standard care were evaluated. Among these specimens, 250 samples were manually supplemented (contrived specimens) with clinically relevant amounts of the five targeted Candida species, and 50 samples were studied as negative controls. The overall sensitivity and specificity of the T2Candida assay were 91.1 and 99.4%, respectively (compared to blood culture results, including those of the contrived specimens). The negative predictive value of this assay was 99.4% in a population with a 6% prevalence of Candida infection (33). In addition to panel-based diagnostics, other methods, such as 16S and 18S rRNA gene PCR/sequencing and metagenomic shotgun sequencing, are being developed and evaluated for the detection of pathogens in blood (31).
MULTIPLEX DETECTION OF RESPIRATORY PATHOGENS
A number of multiplex respiratory panels that simultaneously detect ≥5 pathogens have been FDA approved/cleared. These panels vary in the numbers of targets included (Table 3), performance characteristics, turnaround times, and levels of complexity. While the approved specimen type for these panels is a nasopharyngeal (NP) swab, some laboratories have validated testing on lower respiratory samples, such as bronchoalveolar lavage (BAL) fluid. The differential diagnosis of respiratory infections is often broad since the clinical presentation may be nonspecific. The use of multiplex panels to simultaneously detect and identify respiratory pathogens may simplify testing algorithms and improve the sensitivity and speed of diagnosis compared to those of conventional methods such as viral culture (34).
TABLE 3.
FDA-approved/cleared multiplex respiratory panelsa
| Parameter | FilmArray | Verigene | x-TAG RVP | x-TAG RVP Fast | NxTAG-RPP | eSensor RVP | ePlex |
|---|---|---|---|---|---|---|---|
| Analysis platform | FilmArray system or FilmArray Torch | Verigene system | Luminex 100/200 | Luminex 100/200 | Luminex Magpix | eSensor | ePlex system |
| No. of targets | 20 | 16 | 12 | 8 | 20 | 14 | 17 |
| Ability to detect pathogen | |||||||
| Viruses | |||||||
| Adenovirus | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ (differentiates subgroup B/E from C) | ✓ |
| Coronavirus | ✓ | ||||||
| Coronavirus HKU1 | ✓ | ✓ | |||||
| Coronavirus NL63 | ✓ | ✓ | |||||
| Coronavirus 229E | ✓ | ✓ | |||||
| Coronavirus OC43 | ✓ | ✓ | |||||
| Human bocavirus | ✓ | ||||||
| Human metapneumovirus | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| Influenza A virus | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| Subtype H1 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| Subtype H3 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| Subtype 2009 H1N1 | ✓ | ✓ | ✓ | ||||
| Influenza B virus | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| Parainfluenza virus 1 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |
| Parainfluenza virus 2 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |
| Parainfluenza virus 3 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |
| Parainfluenza virus 4 | ✓ | ✓ | ✓ | ✓ | |||
| Respiratory syncytial virus | ✓ | ✓ | |||||
| Respiratory syncytial virus A | ✓ | ✓ | ✓ | ✓ | ✓ | ||
| Respiratory syncytial virus B | ✓ | ✓ | ✓ | ✓ | ✓ | ||
| Rhinovirus/enterovirus | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| Bacteria | |||||||
| Chlamydophila pneumoniae | ✓ | ✓ | ✓ | ||||
| Mycoplasma pneumoniae | ✓ | ✓ | ✓ | ||||
| Bordetella pertussis | ✓ | ✓ | |||||
| Bordetella parapertussis-Bordetella bronchiseptica | ✓ | ||||||
| Bordetella holmesii | ✓ | ||||||
| Time to result (h) | ∼1 | ∼2–3 | ∼8 | ∼6 | ∼4 | ∼6 | ∼1.5 |
The acceptable specimen type for all panels is a nasopharyngeal swab. RVP, respiratory virus panel; RPP, respiratory pathogen panel.
FDA-Approved/Cleared Assays
Currently, there are seven multiplex panels that have received FDA approval/clearance for the detection and identification of >5 respiratory pathogens: (i) Luminex xTAG RVP v1 (Luminex Corporation) (which received FDA approval/clearance in 2008), (ii) Luminex xTAG RVP Fast (Luminex Corporation) (which received FDA approval/clearance in 2011), (iii) the FilmArray respiratory panel (FA-RP) (BioFire Diagnostics) (which received FDA approval/clearance in 2011), (iv) eSensor RVP (GenMark Diagnostics) (which received FDA approval/clearance in 2013), (v) Verigene Respiratory Pathogens Flex test (Luminex Corporation) (which received FDA approval/clearance in 2015), (vi) the Luminex xTAG respiratory pathogen panel (NxTAG-RPP) (Luminex Corporation) (which received FDA approval/clearance in 2015), and (vii) the ePlex respiratory pathogen panel (ePlex RPP) (GenMark Diagnostics) (which received FDA approval/clearance in 2017) (Table 3). The FA-RP includes the most targets (n = 20) and has the fastest turnaround time (∼1 h).
Luminex xTAG RVP v1 and xTAG RVP Fast detect 12 and 8 targets, respectively, and have turnaround times of 8 and 6 h, respectively. The Luminex system uses fluorescently labeled bead array technology, which includes a 45- to 60-min sample pretreatment step, nucleic acid extraction (∼45 min), multiplex reverse transcription, PCR amplification, fluorescent-bead hybridization to specific amplified targets (∼2.5 h), and analysis by using the Magpix or Luminex 100/200 system (∼10 min). It is an open-amplification platform and requires the transfer of extracted nucleic acids or amplified nucleic acids at two points in the workflow.
The eSensor RVP assay is performed on the eSensor XT-8 system and uses microarray hybridization and solid-phase electrochemical detection to detect 14 targets within 8 h. The FA-RP can simultaneously detect 20 targets and has a turnaround time of 1 h. It utilizes the BioFire individual FilmArray platform or FilmArray Torch, which are discussed in the section on blood culture, above. The Verigene Respiratory Pathogens Flex test uses the Verigene system (described in the section on blood culture, above), targets 16 potential pathogens, and takes about ∼2 to 3 h to perform.
The NxTAG RPP simultaneously detects 20 targets and has a turnaround time of 4 h for a batch of 96 samples. This test uses the Luminex Magpix platform, wherein an aliquot of extracted nucleic acid is directly added to preplated lyophilized reagents and multiplexed real-time PCR (RT-PCR)/bead hybridization is carried out as one single cycling program in a closed PCR vessel. No post-PCR sample handling is required.
The ePlex RPP is a cartridge-based assay that is performed on the ePlex instrument to simultaneously detect 17 targets with a turnaround time of 1.5 h. The assay has a hands-on time of <2 min. The ePlex system utilizes electrowetting technology to perform multiplexed nucleic acid amplification, followed by the detection of analyte targets using eSensor technology.
Assay Performance
Table S2 in the supplemental material shows the major studies evaluating the performance of FDA-approved/cleared multiplex respiratory panels. Popowitch et al. (35) compared the performances of four assays (FA-RP, eSensor RVP, xTAG RVPv1, and xTAG RVP Fast) using 300 specimens and reported overall sensitivities and specificities for each panel of 84.5 and 100% for FA-RP, 98.3 and 99.2% for eSensor RVP, 92.7 and 99.8% for Luminex xTAG RVPv1, and 84.4 and 99.9% for Luminex xTAG RVP Fast. Sensitivity and specificity were calculated by using the criterion “positive by at least two platforms” as the reference result. The greatest number of discrepancies between the multiplex panels and the reference standard was for adenovirus (for which the FA-RP had a low sensitivity of 57% compared to the other assays) and influenza B virus (for which xTAG RVP Fast and FA-RP had sensitivities of 45.5% and 77.3%, respectively, which were significantly lower than those of the other assays) (35). In another study, Chen et al. (36) compared the NxTAG-RPP with the FA-RP using 284 clinical respiratory samples. Complete concordance between the results of the two assays was noted for 98.8% of positive samples, but significant differences in agreement were found for human metapneumovirus (P = 0.001) and parainfluenza virus 3 (P = 0.031), with higher positivity rates for the NxTAG-RPP than for the FA-RP assay. Using the FA-RP and laboratory-developed real-time PCR as the reference standard, the NxTAG-RPP demonstrated >93% sensitivity and specificity for most targets, except human coronaviruses OC43 (66.7 and 99.6%, respectively) and HKU1 (66.7 and 100%, respectively). The NxTAG-RPP has a higher sample throughput than that of the FA-RP (96 samples versus 1 sample per run) and a turnaround time of ~5 h (for up to 96 samples, which may be misleading, as the assay is often batched) versus ~1 h (for 1 sample) for the FA-RP assay (36).
Nijhuis et al. compared the performance of the ePlex RPP to those of laboratory-developed RT-PCR assays and showed that the ePlex RPP had an overall agreement of 97.4% (452/464 respiratory pathogens from 323 positive clinical specimens). After discrepancy analysis, 10 out the 12 RT-PCR positive/ePlex RPP negative discordant targets were confirmed by a third assay to be human bocavirus (n = 3), rhinovirus (n = 2), human coronavirus (n = 3), parainfluenza virus 2 (n = 1), and human metapneumovirus (n = 1) (37).
Clinical and Economic Impacts
The clinical and economic impacts of multiplex respiratory testing have been evaluated in several studies. Rappo et al. (38) compared outcomes for adult patients with positive tests for respiratory viruses across two influenza seasons in a retrospective cohort study. During the first influenza season, conventional methods (i.e., viral cultures, rapid antigen testing, and direct fluorescent-antibody testing) were used, and during the second season, the FA-RP was used, as the primary tests. After the implementation of the FA-RP, there was a decrease in the time to diagnosis of influenza virus (1.7 versus 7.7 h) and noninfluenza viruses (1.5 versus 13.5 h). In addition, detection of influenza virus by the FA-RP was associated with lower odds ratios for admission (P = 0.046), numbers of chest radiographs (P = 0.005), lengths of stay (P = 0.040), and durations of antimicrobial use (P = 0.032) by using multivariate analysis (38). Subramony et al. (39) performed a retrospective, pre/postintervention study evaluating the impact of multiplex PCR testing for respiratory pathogens in pediatric inpatients. The preintervention group (n = 2,349 patients) was tested by enzyme immunoassays, direct fluorescent-antigen tests, PCR assays (other than the FA-RP), and/or viral cultures, whereas the postintervention group (n = 2,430) was tested by using the FA-RP. Forty-two percent of patients in the postintervention group had a positive result by the FA-RP, compared to 14% in the preintervention group. In addition, patients in the postintervention group were less likely to receive antibiotics for more than 2 days (odds ratio, 0.5) and to have a chest radiograph performed upon admission (odds ratio, 0.4) and were more likely to be in isolation for more than 2 days (odds ratio, 2.4) than those in the preintervention group (39). In a similar retrospective, pre/postintervention study by Rogers et al. (40), the impact of the FA-RP on clinical outcomes for pediatric inpatients with uncomplicated acute respiratory tract illness was assessed. In this study, the implementation of the FA-RP was associated with a shorter duration of antibiotic administration (P = 0.003) than for the preintervention group, in which nasopharyngeal specimens were tested for influenza virus, respiratory syncytial viruses (RSVs) (Focus Diagnostics, Cypress, CA), and parainfluenza viruses 1 through 3 (Prodesse; Hologic Gen-Probe, San Diego, CA) by PCR. Furthermore, among patients with a positive result(s) by the FA-RP, the inpatient length of stay (P = 0.03) and time in isolation (P = 0.03) were decreased (40). Brendish et al. conducted a single-center, randomized, controlled trial to assess the clinical impact of the use of rapid point-of-care molecular testing (POCT) for viruses among 720 adult patients who presented to their hospital with acute respiratory illness. Patients assigned to the POCT group (n = 362) were tested by using the FA-RP, and those in the control group (n = 358) were tested for respiratory viruses by laboratory PCR assays, at the discretion of the clinical team. While there was no difference in the proportions of patients treated with antibiotics and mean durations of antibiotic use between the two groups, more patients in the POCT group received single doses or brief courses (<48 h) of antibiotics than in the control group (17% versus 9%; P = 0.004). The use of POCT was associated with a shorter length of stay (5.7 versus 6.8 days; P = 0.04) and improved antiviral use for influenza virus-positive patients (91% versus 65%; P = 0.002) (41).
The cost of performing multiplex testing is an important consideration. Mahony et al. (42) performed a cost analysis study comparing the xTAG RVP assay to shell vial culture and direct fluorescent-antibody staining and reported that multiplex PCR was the least expensive approach if the prevalence of a respiratory viral illness was >11%. Overall, this study reported a cost savings of $291 per case with the use of multiplex PCR compared to conventional methods (42). Nelson et al. (43) developed a decision analytical model to evaluate the cost-effectiveness of multiplex PCR testing in the emergency department for children with influenza and reported that rapid multiplex PCR testing was more cost-effective than traditional PCR, direct fluorescent-antibody assays (DFAs) and rapid antigen tests (43).
Advantages and Limitations
Multiplex panels have a number of limitations, including the fact that most panels do not allow customized ordering. A few exceptions include the Verigene RP Flex panel (Nanosphere) and the Luminex xTAG respiratory pathogen panel (Luminex, Austin, TX), which offer flexible configurations for customized testing (34, 44). The Verigene RP Flex panel also allows the unmasking of suppressed results without running an additional test, at an extra cost. Multiplex panels that offer customized ordering may limit unnecessary testing, thereby minimizing costs to the patient. Although most data suggest that multiplex panels offer a performance comparable to those of conventional methods (e.g., viral culture and individual RT-PCR), some multiplex tests may have a lower sensitivity for the detection of certain pathogens. For example, in a study that compared the performances of four multiplex respiratory panels, the FilmArray assay was noted to have modest sensitivities for the detection of adenovirus (57%), influenza A virus H1/2009 (73%), and influenza B virus (77%) (35, 45). In addition, the clinical significance of the detection of multiple targets in these multiplex panels remains unclear. One study that evaluated the performances of four multiplex respiratory panels found a coinfection rate of 10%. Most coinfections involved enterovirus (EV) and rhinovirus; cross-reactivity between these two targets may have been a contributing factor (35). Positive results may not distinguish between colonization and active infection and may miss coinfection with bacteria or fungi. Nasopharyngeal specimen collection may cause discomfort to the patient and has the potential to miss lower respiratory tract infection in critically ill patients, thereby necessitating additional testing of BAL fluid samples. These panels do not offer exhaustive testing; for example, viruses such as cytomegalovirus (CMV), Middle East respiratory syndrome coronavirus (MERS-CoV), severe acute respiratory syndrome-associated coronavirus (SARS-CoV), and hantavirus are not detected. The use of multiplex respiratory assays may have clinical benefits, including the potentials to deescalate antibiotics if a viral pathogen(s) is detected, decrease the use of invasive sample collection procedures, and allow informed decisions to be made regarding infection control measures and timely outbreak investigations. For example, an EV D68 outbreak in 2014 was rapidly detected because of the use of multiplex respiratory panels (46). Although a clinical diagnosis of a viral respiratory infection should suffice (as many viral pathogens do not require specific therapy), the use of these assays may be associated with a sense of “fulfillment and closure” for the treating clinicians and their patients by providing a microbiological diagnosis and may allow the avoidance of further workup, including send-out testing.
Multiplex respiratory panels are likely to allow the epidemiology of certain pathogens to be better defined. In an epidemiological analysis of 44,230 patients with respiratory illness, multiplex molecular testing identified that infection with human coronavirus was more common during the influenza season than previously recognized (47). Multiplex testing may lead to the diagnosis of some infections that have been commonly missed due to a lack of clinical suspicion or available routine testing. For example, one study reported that 75% of Mycoplasma pneumoniae infections were detected unexpectedly by the use of multiplex PCR (48). This is important because it is an actionable (i.e., treatable) finding.
Immunocompromised hosts, in particular, may benefit from the use of these large respiratory panels, whereas otherwise healthy patients with mild, self-limited respiratory infections may benefit from more targeted diagnostic assays or no testing based on their clinical presentation and epidemiological exposures (44). However, there is a potential for prolonged shedding of microorganisms or nucleic acid in immunocompromised patients without necessarily causing clinical disease. It is therefore important that laboratory results be interpreted in the context of clinical findings. Furthermore, the array of potential pathogens in immunocompromised hosts may be broader than encompassed on these panels. Individualized utilization guidelines for specific patient populations (e.g., children, adults, immunocompromised patients, inpatients, and outpatients, not all of which are mutually exclusive) are needed for the proper use of these assays.
MULTIPLEX DETECTION OF GASTROINTESTINAL PATHOGENS
Infectious diarrhea occurs worldwide and can cause substantial morbidity and mortality. The World Gastroenterology Organization estimates that there are 2 billion new cases each year, leading to 1.9 million deaths among children under the age of 5 years (49). The majority of childhood fatalities occur in developing countries; however, approximately a thousand deaths in children under the age of 5 years are recorded annually in the United States (50–52). Timely detection and treatment of gastrointestinal (GI) pathogens may prevent adverse patient outcomes and mitigate disease transmission. Enteric pathogens can be transmitted from contaminated food and water sources or from close contact with an infectious person. Many infectious gastroenteritis cases in the United States are associated with improperly prepared food, with the increasing globalization of food distribution providing new opportunities for pathogens to spread. For example, Cyclospora cayetanensis outbreaks in the United States have been linked to cilantro and salad mixes imported from Mexico (53–55). Increases in international travel and immigration have also expanded the breadth of enteric pathogens that physicians and laboratorians need to consider in their patient population. Traditionally, diarrheal pathogens have been identified by using microscopy, culture, antigen detection, and individual PCR assays. Pathogen identification via culture can take several days, and some microscopy assays require multiple stool samples to be collected over a period of days to reach maximum sensitivity. Thus, there can be a substantial time lag between when a patient seeks clinical care and when results are reported. In recent years, commercial and laboratory-developed PCR assays have been increasingly used for the detection of specific pathogens. However, the use of these assays requires ordering clinicians to select the pathogens that are most likely to be associated with the disease, which may result in certain pathogens being missed. Furthermore, this approach can become expensive if a large number of individual assays are ordered. In many cases, only laboratory-developed tests are available for individual agents. Recently, syndromic testing through the use of multiplex GI panels has become available for the diagnosis of diarrheal illnesses.
FDA-Approved/Cleared Assays
There are currently three FDA-approved/cleared multiplex assays that detect >5 stool pathogens: the Luminex xTAG gastrointestinal pathogen panel (Luminex GPP; Luminex Corporation) (which received FDA approval/clearance in 2013), the BioFire FilmArray gastrointestinal panel (BioFire GIP; BioFire Diagnostics, LLC) (which received FDA approval/clearance in 2014), and the Verigene Enteric Pathogens (Verigene EP) test (Luminex Corporation) (which received FDA approval/clearance in 2014) (Table 4).
TABLE 4.
FDA-approved/cleared multiplex gastrointestinal panelsa
| Parameter | Verigene EP | Luminex GPP | BioFire GIP |
|---|---|---|---|
| Analysis platform | Verigene system | Magpix or Luminex 100/200 system | FilmArray system or FilmArray Torch |
| Acceptable specimen type | Stool in Cary-Blair medium | Fresh stool or stool in Cary-Blair medium | Stool in Cary-Blair medium |
| No. of targets | 9 | 14 | 22 |
| Ability to detect pathogen | |||
| Bacteria | |||
| Campylobacter species | ✓ | ✓ | ✓ |
| Salmonella species | ✓ | ✓ | ✓ |
| Shigella species/enteroinvasive E. colib | ✓ | ✓ | ✓ |
| Vibrio species | ✓ | ✓ | |
| Vibrio cholerae | ✓ | ✓ | |
| Yersinia enterocolitica | ✓ | ✓ | |
| Escherichia coli O157 | ✓ | ✓ | |
| Enterotoxigenic E. coli | ✓ | ✓ | |
| Enteropathogenic E. coli | ✓ | ||
| Enteroaggregative E. coli | ✓ | ||
| Plesiomonas shigelloides | ✓ | ||
| Shiga toxin-producing E. coli (stx1-stx2) | ✓c | ✓ | ✓ |
| Clostridium difficile (toxin A/B) | ✓ | ✓ | |
| Viruses | |||
| Norovirus GI/GII | ✓ | ✓ | ✓ |
| Rotavirus A | ✓ | ✓ | ✓ |
| Astrovirus | ✓ | ||
| Adenovirus 40/41 | ✓ | ✓ | |
| Sapovirus | ✓ | ||
| Parasites | |||
| Cryptosporidium species | ✓ | ✓ | |
| Entamoeba histolytica | ✓ | ✓ | |
| Giardia lamblia | ✓ | ✓ | |
| Cyclospora cayetanensis | ✓ | ||
| No. of samples (throughput) | 1–32 (scalable) | 24 | 1–12 (scalable) |
| Time to result (h) | <2 | ∼5 | ∼1 |
EP, enteric pathogens; GPP, gastrointestinal pathogen panel; GIP, gastrointestinal panel.
The Verigene EP and Luminex GPP do not specifically target enteroinvasive E. coli.
The Verigene EP has separate targets for stx1 and stx2.
The Luminex GPP detects 14 gastrointestinal pathogens (Table 4). The testing time for 96 samples is ∼5 h and consists of a 45- to 60-min sample pretreatment step, nucleic acid extraction (∼45 min), multiplex reverse transcription, PCR amplification, fluorescent-bead hybridization to specific amplified targets (∼2.5 h), and analysis by using the Magpix or Luminex 100/200 system (∼10 min). The Luminex GPP is an open-amplification platform and requires the transfer of extracted nucleic acids or amplified nucleic acids at two points in the workflow, resulting in a chance of contamination occurring. The sample input for the assay can be 100 mg of fresh stool, 100 μl of liquid stool, or 400 μl of stool in Cary-Blair medium.
The BioFire GIP is an automated, pouch-based assay that can identify 22 targets (Table 4). The sample input for the GIP is 200 μl of stool preserved in Cary-Blair medium, and results are available in ∼1 h. The BioFire GIP uses the BioFire FilmArray technology/instrumentation, which is discussed in detail in the section on blood culture, above.
The Verigene EP targets 9 potential pathogens (Table 4). The automated assay runs on the Nanosphere Verigene system, performing nucleic acid extraction, reverse transcription, PCR, microarray gold particle hybridization with silver enrichment to detect amplified DNA, and interpretation of results. One sample (200 μl of stool in Cary-Blair medium) can be run at a time, which takes ∼5 min of hands-on preparation time and a <2-h run time.
Assay Performance
Table S3 in the supplemental material shows the major studies evaluating the performance of FDA-approved/cleared multiplex gastrointestinal panels. A substantial body of research evaluating the BioFire GIP and Luminex GPP is available and demonstrates that both assays yield more positive results than conventional testing methods. A multicenter study involving 709 samples across 10 European countries reported that the BioFire GIP detected at least one organism in 54% of samples, whereas only 18% of samples were positive by conventional testing (56). These results were consistent with the 53% (832/1556) positivity rate reported in the BioFire GIP clinical trial data (57). A study conducted at our institution demonstrated that the BioFire GIP and Luminex GPP assays have positivity rates of 33 and 30%, respectively, compared to only 8% with routine testing (58, 59). Another study evaluating the Luminex GPP assay reported 22% positivity by the multiplex platform, compared to 12% by routine testing (60). Similarly, Rand et al. reported that an additional 22% of their specimens were positive for at least one target by using the BioFire GIP compared to routine clinical testing. Interestingly, 60% of the patients from whom a positive result was identified only by the multiplex assay were not under appropriate contact precautions during their hospital stay (61). The higher positivity rates are likely due to the increased number of targets that are included in the multiplex assays and an increased sensitivity for some targets compared to that with conventional testing. Mengelle et al. found that the Luminex GPP had a higher sensitivity than conventional methods for C. difficile, Campylobacter species, norovirus, and rotavirus (62). Suboptimal accuracy for Salmonella species and reduced sensitivity for Yersinia enterocolitica have been reported for the Luminex GPP (59, 63). Among studies assessing the performances of multiplex GI panels, the most commonly detected organisms have been C. difficile, enteropathogenic E. coli (EPEC), enteroaggregative E. coli (EAEC), Salmonella species, norovirus, rotavirus, sapovirus, and Cryptosporidium species (56, 59–62).
A consistent observation of studies evaluating multiplex GI panels is that detection of ≥2 targets occurs more frequently than with conventional testing. Spina et al. observed ≥2 pathogens in 16% of samples using the BioFire GIP, while conventional testing identified codetections in 1% of samples. Among the samples with multiple pathogens present, 84% were positive for EAEC or EPEC (56). High rates of detection of EAEC (5 to 9%) and EPEC (10 to 30%) by multiplex GI panels have been reported across multiple studies (56, 58, 59). Historically, most clinical laboratories have not specifically tested for EAEC and EPEC. This has created a clinical conundrum in that health care providers are now faced with results that were not previously reported and for which current guidelines provide no direction as to management (treatment, clinical significance, or the need for additional or repeat testing). Studies evaluating the clinical significance of the detection of EAEC and EPEC are needed. A study performed in the Netherlands assessed asymptomatic children (n = 5,197) in day care centers for enteric pathogens and found that 19.9% were positive for EPEC (64). Other studies involving symptomatic patients have found high rates of coinfections involving organisms other than EAEC or EPEC, such as norovirus or C. difficile (59). Of note, asymptomatic individuals can be colonized with C. difficile, making the interpretation of positive C. difficile results difficult. Overall, it is important for health care providers and laboratory professionals to consider all aspects of the patient's condition (e.g., symptom duration and severity and prior antimicrobial treatment) when interpreting the results of multiplex GI panels. Nevertheless, interpretation can be challenging; for example, C. difficile can be associated with both community-associated and nosocomial diarrhea, making it difficult to interpret the finding of a positive result for C. difficile in a patient in whom this organism may not have been historically looked for.
Fewer data are available on the Verigene EP, and to date, only one study has directly compared the three commercially available multiplex GI platforms to conventional testing (65). In this study, 152 stool samples (98 retrospective and 54 prospective) from pediatric patients with acute gastroenteritis were used to evaluate the BioFire GIP, Luminex GPP, and Verigene EP. The samples were positive for Campylobacter species (n = 12), Salmonella species (n = 24), Shigella species (n = 43), stx1-stx2 (n = 12), norovirus (n = 19), or rotavirus (n = 7), as determined by conventional testing or by being positive by at least two of the multiplex panels. Conventional testing consisted of stool cultures for Salmonella, Shigella, and Campylobacter species; rapid immunochromatographic tests for Shiga toxins 1 and 2 (ImmunoCard Stat! EHEC; Meridian Bioscience) and rotavirus (Sure-Vue Rota test; Fisher Scientific); and RT-PCR for norovirus (Xpert norovirus; Cepheid). Only these six targets were addressed in that study. The BioFire GIP demonstrated a sensitivity of 100% for Campylobacter species, Shigella species, stx1-stx2, and rotavirus, while lower sensitivities were observed for norovirus (94.7%) and Salmonella species (95.8%). The specificity of the BioFire GIP was 100% for all the tested targets except norovirus and rotavirus, for which the specificities were 99.3% and 98.6%, respectively. Sensitivities for the Luminex GPP were as follows: 91.7% for Campylobacter species, 79.2% for Salmonella species, 100% for Shigella species, 91.7% for stx1-stx2, 89.5% for norovirus, and 100% for rotavirus. The specificities for all six targets were 100%. The Verigene EP demonstrated good specificities for all six of the tested targets (99.1% for Shigella species, 99.3% for Campylobacter species, and 100% for Salmonella species, norovirus, and rotavirus) but had the lowest sensitivities of the three multiplex panels for certain targets, specifically, rotavirus, Campylobacter species, and Salmonella species, which showed sensitivities of 71.4, 83.3, and 83.3%, respectively. The Verigene assay demonstrated sensitivities of 95.4% for Shigella species, 91.7% for stx1-stx2, and 89.0% for norovirus. All three multiplex assays detected organisms and/or coinfections that were missed by conventional methods, including Campylobacter species (n = 1), Salmonella species (n = 1), and Shigella species (n = 2).
In a multicenter evaluation of the BioFire GIP involving 1,556 samples, Buss et al. reported sensitivities of 100% for 12 targets and ≥94.5% for 7 targets. The overall specificity of the BioFire GIP was ≥97% (58). Specificity and sensitivity were determined by comparing the results of the BioFire GIP to those of stool cultures and RT-PCR with amplicon sequencing in some cases for norovirus, Giardia lamblia, and sapovirus. (RT-PCR assays are described in references 66–68.) Although the overall specificity of the multiplex GI panels appears to be high, one recent report highlighted a potential issue with the specificity of the BioFire GIP norovirus assay. In this study, clinical stool samples (n = 100) were tested by a laboratory-developed norovirus RT-PCR assay, and the results were compared with those of the BioFire GIP. Among 18 samples that tested positive by the BioFire GIP and negative by the norovirus RT-PCR assay, 16/18 (88.9%) were negative for norovirus by alternate molecular methods at two outside laboratories (69). Together, these data suggest that commercially available, multiplex GI panels offer overall high sensitivity and specificity, with a few exceptions. More research is needed to assess the performance of these panels in routine clinical practice (56, 59–62).
Clinical and Economic Impacts
Cost-benefit and clinical outcome data are important to consider when developing test utilization strategies. Goldenberg et al. conducted a parallel diagnostic study on hospitalized patients (n = 800) to compare the costs of conventional enteric pathogen testing and the Luminex GPP assay. Laboratory costs and costs associated with patient isolation were measured or estimated for each patient. This study found that the implementation of the Luminex GPP increased costs to the clinical laboratory (additional £22,283) but reduced overall costs (£66,765 saved) by decreasing the number of days that patients were under isolation protocols (70). To assess the clinical impact of multiplex GI panels, Rand et al. evaluated how length-of-stay and patient isolation decisions were impacted through the use of the FilmArray GIP. This study included a cohort of 158 patients and found that 21 (13.3%) were positive for an organism(s) by the multiplex GI panel that was not identified by conventional methods. Collectively, these 21 patients spent 109 days in the hospital without appropriate isolation precautions due to incomplete coverage by routine testing. This study also identified 25 (16%) patients who were placed under contact precautions unnecessarily for a total of 181 days (61).
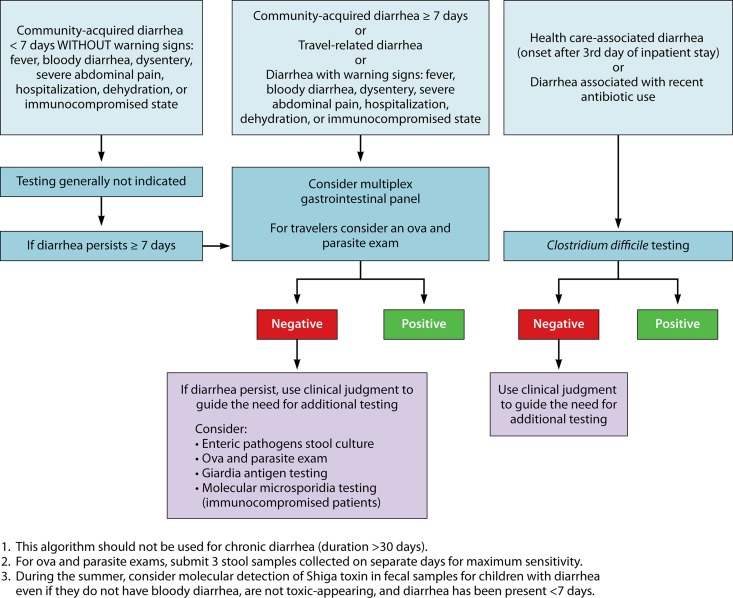
With the increasing use of multiplex panels, it is important for clinical laboratorians to be actively involved in the development of test utilization strategies focusing on the use and interpretation of the results of these tests. For example, community-acquired diarrhea often resolves in <7 days without treatment, so multiplex GI testing may not be necessary for this patient population. For travel-related or community-acquired diarrhea lasting ≥7 days, the American College of Gastroenterology suggests that multiplex testing may be useful (71). Patients experiencing diarrhea associated with antecedent antibiotic use or hospitalization are at risk for C. difficile infection; in such cases, specific testing for C. difficile is most cost-effective in this scenario (Fig. 1).
FIG 1.
Potential testing algorithm for evaluating patients with acute gastroenteritis. (Modified and used with permission of Mayo Foundation for Medical Education and Research. All rights reserved.)
Advantages and Limitations
In addition to broad coverage and the ability to identify a higher rate of coinfections, multiplex GI panels offer several benefits over conventional methods, including a reduced turnaround time and possible cost reduction. The multiplex panels allow results to be reported within hours of sample collection, instead of the 2 to 3 days needed for culture. Because the multiplex panels accept stool in Cary-Blair medium, transport of specimens is easy. If physicians order multiple individual stool pathogen assays, the total cost could be comparable to or more expensive than that of running a single multiplex assay, making multiplex testing more cost-effective. The reported literature on multiplex stool panels consistently demonstrates increased organism detection compared to that of conventional methods, and comprehensive testing allows diagnosis in more instances than à la carte testing alone. Using multiplex panels and sharing the results with public health laboratories could also provide public health benefits by defining circulating organisms (94).
Each of the commercially available systems has unique advantages and limitations. The Luminex GPP targets 14 pathogens and can run 24 samples in ∼5 h; however, the assay requires some hands-on preprocessing time (∼45 min), and the open-system design may increase the risk of contamination. The BioFire GIP is a closed, sample-to-answer system that tests for 22 pathogens in ∼1 h. The individual FilmArray instrument can run 1 sample at a time, and for laboratories requiring higher throughput, the FilmArray Torch can run up to 12 samples simultaneously. The Verigene EP is also designed as a closed, sample-to-answer system that can identify 9 pathogens in ∼2 h. Like the FilmArray system, the Verigene EP platform tests one sample per unit, but up to 32 Verigene Processor SP units can be connected to a single Verigene reader, allowing scalability. Minimal data are currently available for this platform, and future studies are needed to evaluate its performance. The Luminex GPP and the Verigene EP allow users to customize which targets on the panel are reported. All of the panel targets are still tested, but the user can mask any of the targets up front so that only the desired targets are reported. Under current configurations, masked results cannot be unmasked at a later time and reported to patients. To check additional targets, the sample needs to be rerun.
Conventional methods are still needed to detect pathogens that are not covered by the panels (e.g., Aeromonas species) and provide antimicrobial susceptibility information, when required. The most recent American College of Gastroenterology clinical guidelines for the diagnosis, treatment, and prevention of acute diarrheal infections in adults indicate that antibiotic susceptibility testing is typically not recommended because empirical treatment failure is uncommon. Antimicrobial susceptibility testing is currently most useful for outbreak investigations and community surveillance for resistance trends (71). To provide isolates for susceptibility testing, clinical laboratories utilizing multiplex GI panels may consider culturing for the organism(s) detected molecularly (i.e., reflexive culture) (72), a strategy that can also be adopted when public health laboratories need isolates (95). It is unknown whether repeat testing for Giardia species is needed if this organism is highly suspected and initial testing is negative. And, as mentioned, the “ideal” panel of organisms to be included remains to be defined. Hopefully, over time the cost of these assays will decrease.
MULTIPLEX DETECTION OF PATHOGENS ASSOCIATED WITH CENTRAL NERVOUS SYSTEM INFECTION
Meningitis and encephalitis are potentially devastating conditions and can be associated with significant morbidity and mortality. Although empirical treatment is often administered, establishing a specific diagnosis and initiating appropriate therapy, when possible, are needed to idealize patient outcomes. Meningitis is defined as inflammation of the meninges, encephalitis is defined as inflammation of the brain parenchyma, and meningoencephalitis is defined as inflammation at both locations. All these conditions can be caused by viruses, fungi, or bacteria, with encephalitis being more commonly associated with a viral etiology. Clinical presentations are usually nonspecific; patients often experience headache, altered mental status, and, in the case of meningitis, nuchal rigidity. White blood cell (WBC) counts and differentials, protein levels, and glucose concentrations in cerebrospinal fluid (CSF) provide insight into the type of infection (e.g., viral or bacterial). WBC counts are typically elevated, with a prevalence of neutrophils, in cases of bacterial meningitis (Table 5). In cases of fungal, viral, and tuberculous meningitis, WBC counts may also be elevated but generally to a lesser degree than with bacterial meningitis and often in the context of lymphocytosis (70–73). A detailed patient evaluation, including vaccination history, and consideration of seasonality, geography, and local epidemiology may aid in narrowing the differential diagnoses; however, microbiologic testing of CSF is generally required to establish a definitive diagnosis.
TABLE 5.
Cerebrospinal fluid parameters for patients with meningitisa
| Cerebrospinal fluid parameter | Normal | Viral infection | Bacterial infection | Tuberculous/fungal infection |
|---|---|---|---|---|
| Opening blood pressure (mm H2O) | 100–180 | Normal to elevated | 200–500 | 150–340 |
| Total white blood count (cells/μl) | 0–5 | 5–1,000 | 100–>1,000 | 5–1,000 |
| Protein level (mg/dl) | ≤30 | 30–300 | 60–500 | >60 |
| Glucose level (% of blood glucose) | ≥60 | ≥60 | ≤45 | ≤45 |
| CSF appearance | Clear | Clear | Turbid | Clear or fibrin web |
Traditionally, the laboratory diagnosis of bacterial meningitis has been made by using Gram staining and culture of CSF, with molecular testing for herpes simplex virus 1 (HSV-1), HSV-2, and enterovirus, in particular, playing an important role in the diagnosis of viral CNS infections (73, 74). For some causes of CNS infections, such as arboviruses, serology is considered the diagnostic method of choice. FDA-approved/cleared nucleic acid amplification tests for several viruses, including enteroviruses (Xpert EV; Cepheid) and HSV-1 and HSV-2 (Simplexa HSV 1&2 Direct [Focus Diagnostics] and MultiCode RTx HSV 1&2 kit [Luminex Corporation]), in CSF have been available for years, but until recently, there were no FDA-approved/cleared nucleic acid amplification tests for bacterial or fungal targets in CSF. Molecular detection of CNS pathogens can enable a more rapid diagnosis than with culture, and notably, cultures may be negative for patients receiving antimicrobial treatment.
FDA-Approved/Cleared Assays
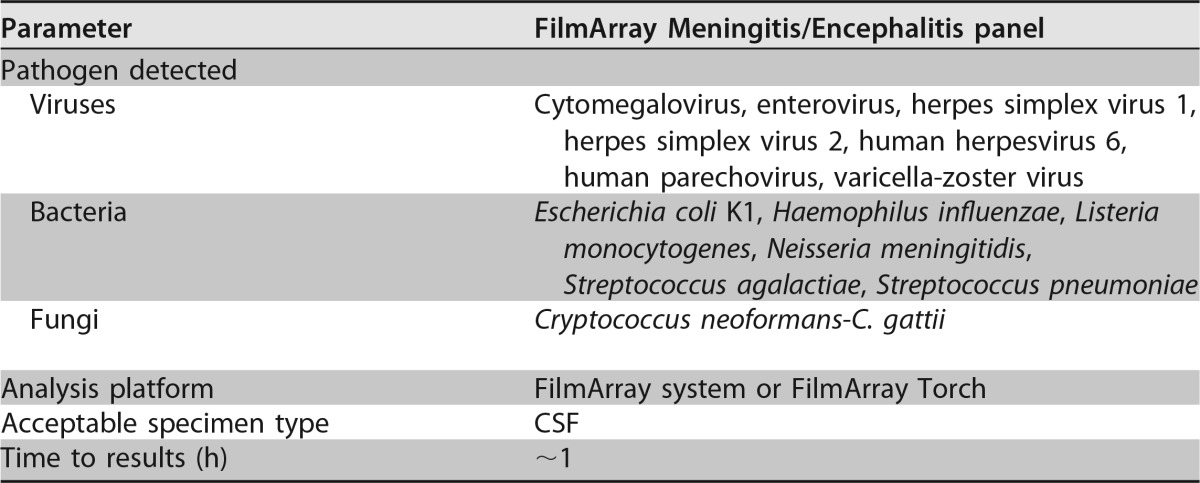
In 2015, the FilmArray Meningitis/Encephalitis panel (MEP) (BioFire Diagnostics, LLC) received FDA approval/clearance, offering the first rapid, multiplex assay for the diagnosis of CNS infection (Table 6). This PCR-based panel queries a total of 14 targets (Table 6). Similar to other multiplex assays performed on the FilmArray system, the MEP is a closed assay that performs nucleic acid extraction, purification, reverse transcription, PCR, and analysis in ∼1 h. Two hundred microliters of CSF is added directly to the MEP pouch and loaded into an individual FilmArray instrument or the FilmArray Torch system, which is capable of running up to 12 samples at a time, with these steps requiring <2 min of hands-on time.
TABLE 6.
Organisms targeted by the FilmArray Meningitis/Encephalitis panel
Assay Performance
Table S4 in the supplemental material summarizes the major studies evaluating the performance of the MEP. Leber et al. (75) conducted a large, prospective study of the MEP in which CSF samples (n = 1,560) collected at 11 U.S. sites were tested by the MEP and results were compared to those of conventional methods. The majority (93%) of samples were obtained from patients who were hospitalized or presented to an emergency department, and patients with a diverse range of ages were included (<2 months to ≥65 years). The MEP detected at least one pathogen in 136 (8.7%) samples, with the highest positivity rate of 12% (105/863) being observed among pediatric patients. In comparison, the MEP was positive in 4.4% (31/697) of patients >18 years old. Detection of ≥2 pathogens in the same sample was observed in five cases, with all samples considered to be falsely positive for one of the targets. Of the samples positive by conventional methods, the MEP demonstrated positive agreement with 9 of the 14 targets: CMV (n = 3), HSV-1 (n = 2), HSV-2 (n = 10), human parechovirus (HPeV) (n = 9), varicella-zoster virus (n = 4), E. coli K1 (n = 2), Haemophilus influenzae (n = 1), S. pneumoniae (n = 4), and Cryptococcus neoformans-C. gattii (n = 1). In total, 6 CSF samples were determined to be falsely negative by the MEP, yielding sensitivities of 95.7% (44/46) for EV, 85.7% (18/21) for human herpesvirus 6 (HHV-6), and 0% (0/1) for Streptococcus agalactiae. The MEP detected 43 pathogens that were not recovered by conventional testing, and supplemental testing by alternate methods (targeted PCR or clinical presentation) supported the MEP result in 21 of these 43 (43%) cases. There were 22 false-positive or unconfirmed MEP results after the resolution of discrepant test results. S. pneumoniae yielded the highest rate of false positivity; there were 9 true-positive and 7 false-positive results. Whether this was due to amplified product contamination, sample contamination, S. pneumoniae colonization of health care workers or laboratory staff handling the samples, cross-reactivity with other streptococci, or other factors is unknown. Results falsely positive for S. pneumoniae may lead to the overuse of inappropriate antibiotics and, of even more concern, failure to treat the actual cause of the involved patient's CNS infections. At least one false-positive result was identified among 10 of the panel targets (75). After discordance analysis, the authors reported an overall positive agreement rate of 84.4% and a negative agreement rate of >99.9% between the MEP and conventional testing.
In a separate study evaluating the research-use-only (RUO) version of the MEP, 342 CSF samples (197 adult and 145 pediatric) were analyzed, and results were compared to those of conventional testing (76). The performance of the MEP was generally good (>90% sensitivity and specificity), with the exception of the performances for CMV (57.1% sensitivity and 100% specificity), S. agalactiae (66.7% sensitivity and 98.6% specificity), and Epstein-Barr virus (EBV) (94.1% sensitivity and 84.2% specificity); EBV was ultimately not included in the FDA-approved/cleared version of the panel. Overall, the MEP missed 14 pathogens detected by conventional methods but identified 33 organisms that were not detected by routine testing. Of these 33 organisms, 19 were confirmed to be present by a second method. A single false-positive result for S. pneumoniae by the MEP was found.
In April 2016, an outbreak of encephalitis/encephalomyelitis occurred among children in Catalonia. Twenty children were found to have EV present in respiratory and/or fecal samples by using a laboratory-developed pan-EV real-time PCR assay. CSF samples from all 20 patients were negative by the same pan-EV PCR. Retrospectively, the CSF samples were tested by using the MEP, and 4 were positive for EV (77). In contrast, another study using 138 previously characterized CSF samples from patients at Children's Hospital Colorado with meningitis and/or encephalitis noted that the MEP detected EV in 68/72 (94.4%) samples that were positive by the Cepheid Xpert EV assay. The MEP identified one additional sample as being positive for EV that was negative by the Cepheid Xpert EV assay, and discrepancy analysis confirmed that the sample was positive for EV (78).
A recent study at the Children's Hospital of Pennsylvania evaluated the MEP for pediatric patients (n = 133) and found overall positive and negative agreement rates of 92% and 100%, respectively, compared to routine methods (e.g., PCR-based laboratory-developed tests and culture) (79). That study retrospectively tested CSF samples (n = 133), which included samples that were positive (n = 67) or negative (n = 66) by laboratory-developed PCR assays and/or culture. Six bacterial deletions (S. pneumoniae [n = 4], H. influenzae [n = 1], and S. agalactiae [n = 1]) were found by both the MEP and routine methods. Among the viral targets, the laboratory-developed PCR assays detected EV (n = 38), HPeV (n = 16), HSV-1 (n = 4), HSV-2 (n = 1), and HHV-6 (n = 2). Results of testing by the MEP correlated with the results of routine methods for 129/133 CSF specimens containing viral targets, with discordant results (laboratory-developed PCR assay positive/MEP negative) being observed for 4 samples. Among these four samples, the MEP was negative for samples that were positive by the laboratory-developed PCR assays for EV/HPeV (n = 1), EV (n = 1), and HSV-1 (n = 2).
Immunocompromised hosts, particularly HIV-infected and transplant patients with low CD4 cell counts, experience a broadened spectrum of potential meningitis and encephalitis etiologies, such as C. neoformans, thus increasing the appeal of multiplex panel testing (80). A diagnosis of cryptococcal meningitis may be made by culture (5 to 14 days), cryptococcal antigen (CrAg) testing, or India ink staining. CrAg testing is quick, easy, inexpensive, and sensitive, making it challenging for the MEP to replace this assay (81–83). Rhein et al. completed a study in Uganda evaluating the MEP in HIV-infected adults with meningitis/meningoencephalitis (84). That study included HIV-positive adults (n = 69) with suspected meningitis/meningoencephalitis. CSF samples were collected from 51 patients at the time of initial diagnosis, and another 68 CSF samples were collected during routine follow-up testing to monitor the response to therapy. At the time of initial presentation, patients were tested by using a CrAg lateral flow assay (IMMY, Norman, OK), and quantitative fungal cultures were performed; however, that study reported comparisons between only the MEP and quantitative culture. Among the 69 study patients, 44 (64%) were diagnosed with cryptococcal meningoencephalitis by conventional testing. Among CSF samples (n = 42) collected at the time of initial presentation, the MEP demonstrated 100% sensitivity (18/18) and specificity (24/24) compared with culture methods. Patients on cryptococcal treatment were monitored by culture for conversion to culture negativity to indicate successful treatment. The cultures collected to monitor when patients became culture negative were also tested by using the MEP, which demonstrated that the sensitivity of the MEP was directly proportional to the amount of the organism recovered in fungal culture. Using CSF samples collected during treatment monitoring, the MEP sensitivity was 96% (49/51) among samples with ≥100 CFU/ml in fungal culture. For the 28 patients on antifungal agents who had culture-negative CSF samples, the MEP panel identified 20 as being negative for Cryptococcus neoformans-C. gattii, providing a 71% negative predictive value for conversion to culture negativity. Several additional pathogens (CMV [n = 2], varicella-zoster virus [n = 2], HHV-6 [n = 6], and S. pneumoniae [n = 1]) were detected in this cohort by the MEP and confirmed by a second PCR method; the clinical significance of these organisms was not addressed.
Advantages and Limitations
Although the BioFire MEP includes a number of bacterial targets, bacterial meningitis caused by these organisms is now rare in the United States, due to the success of immunization programs. Therefore, the low prevalence of these infections in the United States may limit the clinical utility of the panel and introduce challenges with the interpretation of results. Additionally, those with CNS infections associated with a shunt or postneurosurgery may be infected with a spectrum of organisms, such as Staphylococcus species, Cutibacterium acnes, and Gram-negative bacilli, not included in the MEP. Immunocompromised hosts are at risk for infection by an innumerable list of opportunistic pathogens that would not be common enough to include in panel-based testing. Bacterial CSF culture may be negative for patients who have received antimicrobial therapy prior to lumbar puncture (although Gram stain is likely to remain positive). In these situations, molecular tests, including the MEP, may provide useful diagnostic information. Positive panel results for a viral CNS infection such as EV infection may help clinicians quickly discontinue unnecessary antibiotics, and rapid HSV results can help clinicians expedite the start or stop of acyclovir treatment; however, this information can also be accomplished with singleplex PCR assays.
Due to the potential severity of bacterial meningitis and the need to initiate antimicrobial therapy, the specificity of multiplex panels is especially important. Several studies have reported positive results by the MEP not confirmed by conventional methods. Leber et al. found potential false-positive results (7/1,560) for S. pneumoniae using the MEP (75). Those authors suggested that this may have been due to amplified DNA contamination or S. pneumoniae colonization of health care workers or laboratory staff handling the samples. The potential for health care providers and/or laboratory staff to introduce target nucleic acid (e.g., through colonization or transient shedding) into the testing process is an important issue to understand and address. Specific areas for specimen collection and sample loading may need to be defined, to minimize the possibility of contamination. Clinical and laboratory staff should also be educated as to the potential for contamination and how to avoid it and the significant adverse impact of false-positive results. In the future, rapid panel-based tests may be performed outside traditional clinical laboratories where processes and workflows are optimized to reduce the possibility of contamination. As molecular tests become more common in point-of-care settings, the issue of minimizing specimen and amplified-product contamination will need to be carefully considered. Environmental screening and routine monitoring of positivity rates will assist in identifying potential contamination events. An additional confounding factor with the MEP is the interpretation of HHV-6 positivity, as detection may be the result of germ line integration and not genuine CNS infection (85).
The FilmArray MEP provides rapid results and requires minimal hands-on time; however, it is not likely to replace conventional testing methods. CSF Gram staining, as well as routine cell counts with differentials, glucose level determinations, and protein level determinations, will continue to be part of the routine diagnostic algorithm. Furthermore, routine bacterial and fungal cultures of CSF will be essential to identify pathogens that are not included on the MEP and to provide isolates for antimicrobial susceptibility testing and for public health laboratories. The CrAg test is quick, inexpensive, and sensitive, making it challenging to replace. Because the availability of CNS syndromic testing is relatively recent, there is a dearth of reported information as to its clinical utility. Research evaluating its effect on patient outcomes, including length of hospital stay, morbidity, and mortality, as well as cost-benefit analyses and analyses of effects on antibiotic utilization and antibacterial resistance are needed. As the health care industry works to reduce overall costs and unnecessary testing, it will be important to establish appropriate utilization guidelines for multiplex CNS panels, which may vary from institution to institution and based on the patient populations tested and other tests available. A recent article reported a concerning case in which a diagnosis of tuberculous meningitis was delayed because the patient had a false-positive HSV-1 result with the MEP, providing a strong reminder about the impact that laboratory test results have on patient care (86).
MULTIPLEX DETECTION OF PATHOGENS FROM STERILE BODY FLUIDS
While there are no FDA-approved/cleared multiplex molecular assays for use on sterile body fluids (e.g., synovial, pleural, and peritoneal fluids), rapid detection of pathogens from these specimens may become useful in the near future. Michos et al. (87) reported two cases where the FilmArray BCID panel was used to identify Streptococcus pyogenes from synovial fluid in a child with septic arthritis and to identify S. pneumoniae from pleural fluid in another child with complicated pneumonia with empyema. Interestingly, routine bacterial cultures were negative for both of these children, likely due to prior antibiotic administration (87). Vasoo et al. (88) assessed the performance of the FilmArray BCID panel using sonication fluid (n = 216) (i.e., fluid into which biofilms on implant surfaces have been dislodged) for the diagnosis of prosthetic joint infection. The BCID panel and sonicate fluid culture had overall sensitivities of 53% and 69%, respectively (P = 0.04), for the diagnosis of prosthetic joint infection. Despite its modest sensitivity, the BCID panel detected an organism in six cases of culture-negative prosthetic joint infection (88). Mico et al. (89) evaluated the performance of the BCID panel using 88 clinical samples other than blood, including cerebrospinal, pleural, synovial, peritoneal, abscess, and bronchoalveolar lavage fluids. Compared to culture, the sensitivity and specificity of the BCID panel were 71 and 97%, respectively. Samples with low bacterial loads, such as pleural fluid, yielded a lower sensitivity (25%) by BCID than did specimens with higher bacterial loads, such as abscess fluid, which showed a sensitivity of 89% (89). It is worth bearing in mind that the analytical sensitivity (i.e., limit of detection) of BCID was optimized for positive blood culture bottles and may not be ideal for direct testing of patient specimens.
Altun et al. (90) evaluated the performance of the FilmArray BCID panel for the identification of organisms from positive blood culture bottles that had been inoculated with normally sterile body fluids (including pleural [n = 51], synovial [n = 38], abscess [n = 10], dialysis [n = 9], cerebrospinal [n = 7], and bile [n = 1] fluids). Those authors reported that the BCID panel accurately identified 100% (84/84) of organisms in monomicrobial infections and 75% (18/24) of targets if multiple organisms were recovered by routine culture (90). Of note, the FilmArray BCID panel is not FDA approved/cleared for direct testing of clinical specimens or of specimens other than blood in positive blood culture bottles.
CONCLUSIONS
Multiplex molecular assays targeting numerous pathogens directly from clinical specimens have led to a paradigm shift in the diagnosis of infectious diseases. Rather than ordering a series of individual, pathogen-specific assays, health care providers now have the option of ordering a single test designed to detect a number of organisms associated with an infectious syndrome. Syndromic multiplex panels are novel, powerful tools that may assist in the timely diagnosis of infectious diseases and influence decisions regarding patient management, including antimicrobial therapy, antimicrobial stewardship, and infection prevention and control. It is anticipated that syndromic testing will likely be utilized more in the future. It is important to have a clear understanding of the performance characteristics and limitations when implementing multiplex assays.
Concern has been raised about the medical appropriateness of multiplex assays, as they are “one-size-fits-all” tests instead of specific tests based on patients' exposures and clinical presentations. It should be noted, however, that traditional culture is a panel-based approach of sorts, with clinicians rarely ordering cultures for specific organisms. Furthermore, in many cases, it is not clinically possible to narrow down a differential diagnosis to one or two possibilities due to the significant overlap in clinical presentations of infection. The idea that clinicians should determine which specific pathogens might be associated with individual patient cases and select testing schemes that ensure that all the appropriate pathogens are included for each individual patient is a laudable one but is not in line with the reality of clinical practice. Failure to detect the causative agent due to reliance on insensitive methods can lead to delays in starting appropriate treatment and preventing the further spread of disease in the community (especially relevant in the case of respiratory and GI panels), thereby harming patients and their communities. In scenarios where physicians would normally order multiple individual tests, performing a multiplex panel can actually be more cost-effective, and the availability of easy-to-use multiplex panels helps standardize patient care, particularly in smaller hospitals and clinics. Offering individual molecular assays for a few key pathogens such as C. difficile, influenza virus, and RSV, in addition to multiplex panels, could help alleviate the overuse of syndromic testing. Importantly, the right targets that will make up each of these panels will need to be determined, as, given that they are currently differentially configured, there is not an established standard. The establishment of clear algorithms and guidelines for ordering and interpreting these panels, developed by laboratory and clinical professionals, will be necessary to inform their effective use. With time, clinicians can expect to see panel-based tests for additional syndromes. Hopefully, as technology improves, syndromic panel-based tests will become less expensive.
Supplementary Material
Biographies

Poornima Ramanan graduated from Coimbatore Medical College, India, with an M.B.B.S. in 2004 and then completed Internal Medicine residency and an Infectious Diseases fellowship in New York City. She completed fellowships in Transplant Infectious Diseases and Clinical Microbiology at the Mayo Clinic in Rochester, MN. Her research interests include antimicrobial drug resistance and antimicrobial susceptibility testing. Her clinical interests include managing infections in immunocompromised hosts (particularly among solid-organ transplant recipients) and tropical infectious diseases. She enjoys teaching microbiology and infectious diseases to medical students and residents.

Alexandra L. Bryson graduated from Texas A&M University with a B.S. in genetics and biochemistry. She then completed her Ph.D. at the University of Pennsylvania in microbiology and virology, where she studied the human gut microbiome and ways in which bacteria and phages interact though the clustered regularly interspaced short palindromic repeat (CRISPR) system. During graduate school, she received additional training and certification in medicine and translational research as a Howard Hughes Medical Institute Med-into-Grad Scholar. She is currently a Clinical Microbiology Fellow at Mayo Clinic in Rochester, MN. Her research focuses on metagenomics-based diagnostics and detection of human pathogens in global health settings.

Matthew J. Binnicker is an Associate Professor of Laboratory Medicine and Pathology and Director of the Clinical Virology Laboratory in the Division of Clinical Microbiology at Mayo Clinic in Rochester, MN. He completed his postdoctoral training in Clinical Microbiology at Mayo Clinic and now serves as the program director for the Ph.D. fellowship training program at Mayo Clinic. Dr. Binnicker's research interests focus on the molecular detection of viral infections, including the use of next-generation sequencing.

Bobbi S. Pritt is a Professor of Pathology and Director of the Clinical Parasitology Laboratory in the Division of Clinical Microbiology at Mayo Clinic in Rochester, MN. She is board certified in Anatomic Pathology, Clinical Pathology, and Medical Microbiology and holds a master's degree in Medical Parasitology from the London School of Hygiene and Tropical Medicine. Dr. Pritt's main areas of interest are laboratory detection of parasitic and vector-borne infections, medical education, and optimizing test utilization.

Robin Patel is a Professor of Medicine, Professor of Microbiology, the Elizabeth P. and Robert E. Allen Professor of Individualized Medicine, Co-director of the Clinical Bacteriology Laboratory, Director of the Infectious Diseases Research Laboratory, and Chair of the Division of Clinical Microbiology at Mayo Clinic in Rochester, MN. She is a Fellow of American Academy of Microbiology, is co-chair of the ASM Microbe Program Planning Committee, serves on the ASM Board of Directors, and is an associate editor for the Journal of Clinical Microbiology and Clinical Infectious Diseases. Her research focuses on clinical bacteriology diagnostic testing, antibacterial resistance, and microbial biofilms.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/CMR.00024-17.
REFERENCES
- 1.Mayr FB, Yende S, Angus DC. 2014. Epidemiology of severe sepsis. Virulence 5:4–11. doi: 10.4161/viru.27372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kumar G, Kumar N, Taneja A, Kaleekal T, Tarima S, McGinley E, Jimenez E, Mohan A, Khan RA, Whittle J, Jacobs E, Nanchal R, Milwaukee Initiative in Critical Care Outcomes Research Group of Investigators. 2011. Nationwide trends of severe sepsis in the 21st century (2000-2007). Chest 140:1223–1231. doi: 10.1378/chest.11-0352. [DOI] [PubMed] [Google Scholar]
- 3.Dellinger RP, Levy MM, Rhodes A, Annane D, Gerlach H, Opal SM, Sevransky JE, Sprung CL, Douglas IS, Jaeschke R, Osborn TM, Nunnally ME, Townsend SR, Reinhart K, Kleinpell RM, Angus DC, Deutschman CS, Machado FR, Rubenfeld GD, Webb SA, Beale RJ, Vincent JL, Moreno R, Surviving Sepsis Campaign Guidelines Committee Including the Pediatric Subgroup. 2013. Surviving sepsis campaign. International guidelines for management of severe sepsis and septic shock: 2012. Crit Care Med 41:580–637. doi: 10.1097/CCM.0b013e31827e83af. [DOI] [PubMed] [Google Scholar]
- 4.Banerjee R, Ozenci V, Patel R. 2016. Individualized approaches are needed for optimized blood cultures. Clin Infect Dis 63:1332–1339. doi: 10.1093/cid/ciw573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.White House. 2015. National action plan for combating antibiotic-resistant bacteria. White House, Washington, DC. [Google Scholar]
- 6.Saffert RT, Cunningham SA, Mandrekar J, Patel R. 2012. Comparison of three preparatory methods for detection of bacteremia by MALDI-TOF mass spectrometry. Diagn Microbiol Infect Dis 73:21–26. doi: 10.1016/j.diagmicrobio.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 7.Verroken A, Defourny L, Lechgar L, Magnette A, Delmee M, Glupczynski Y. 2015. Reducing time to identification of positive blood cultures with MALDI-TOF MS analysis after a 5-h subculture. Eur J Clin Microbiol Infect Dis 34:405–413. doi: 10.1007/s10096-014-2242-4. [DOI] [PubMed] [Google Scholar]
- 8.Altun O, Botero-Kleiven S, Carlsson S, Ullberg M, Ozenci V. 2015. Rapid identification of bacteria from positive blood culture bottles by MALDI-TOF MS following short-term incubation on solid media. J Med Microbiol 64:1346–1352. doi: 10.1099/jmm.0.000168. [DOI] [PubMed] [Google Scholar]
- 9.Kohlmann R, Hoffmann A, Geis G, Gatermann S. 2015. MALDI-TOF mass spectrometry following short incubation on a solid medium is a valuable tool for rapid pathogen identification from positive blood cultures. Int J Med Microbiol 305:469–479. doi: 10.1016/j.ijmm.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 10.Patel R. 2016. New developments in clinical bacteriology laboratories. Mayo Clin Proc 91:1448–1459. doi: 10.1016/j.mayocp.2016.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ward C, Stocker K, Begum J, Wade P, Ebrahimsa U, Goldenberg SD. 2015. Performance evaluation of the Verigene (Nanosphere) and FilmArray (BioFire) molecular assays for identification of causative organisms in bacterial bloodstream infections. Eur J Clin Microbiol Infect Dis 34:487–496. doi: 10.1007/s10096-014-2252-2. [DOI] [PubMed] [Google Scholar]
- 12.Bhatti MM, Boonlayangoor S, Beavis KG, Tesic V. 2014. Evaluation of FilmArray and Verigene systems for rapid identification of positive blood cultures. J Clin Microbiol 52:3433–3436. doi: 10.1128/JCM.01417-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Altun O, Almuhayawi M, Ullberg M, Ozenci V. 2013. Clinical evaluation of the FilmArray blood culture identification panel in identification of bacteria and yeasts from positive blood culture bottles. J Clin Microbiol 51:4130–4136. doi: 10.1128/JCM.01835-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Banerjee R, Teng CB, Cunningham SA, Ihde SM, Steckelberg JM, Moriarty JP, Shah ND, Mandrekar JN, Patel R. 2015. Randomized trial of rapid multiplex polymerase chain reaction-based blood culture identification and susceptibility testing. Clin Infect Dis 61:1071–1080. doi: 10.1093/cid/civ447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Box MJ, Sullivan EL, Ortwine KN, Parmenter MA, Quigley MM, Aguilar-Higgins LM, MacIntosh CL, Goerke KF, Lim RA. 2015. Outcomes of rapid identification for gram-positive bacteremia in combination with antibiotic stewardship at a community-based hospital system. Pharmacotherapy 35:269–276. doi: 10.1002/phar.1557. [DOI] [PubMed] [Google Scholar]
- 16.Sango A, McCarter YS, Johnson D, Ferreira J, Guzman N, Jankowski CA. 2013. Stewardship approach for optimizing antimicrobial therapy through use of a rapid microarray assay on blood cultures positive for Enterococcus species. J Clin Microbiol 51:4008–4011. doi: 10.1128/JCM.01951-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neuner EA, Pallotta AM, Lam SW, Stowe D, Gordon SM, Procop GW, Richter SS. 2016. Experience with rapid microarray-based diagnostic technology and antimicrobial stewardship for patients with gram-positive bacteremia. Infect Control Hosp Epidemiol 37:1361–1366. doi: 10.1017/ice.2016.175. [DOI] [PubMed] [Google Scholar]
- 18.Suzuki H, Hitomi S, Yaguchi Y, Tamai K, Ueda A, Kamata K, Tokuda Y, Koganemaru H, Kurihara Y, Ishikawa H, Yanagisawa H, Yanagihara K. 2015. Prospective intervention study with a microarray-based, multiplexed, automated molecular diagnosis instrument (Verigene system) for the rapid diagnosis of bloodstream infections, and its impact on the clinical outcomes. J Infect Chemother 21:849–856. doi: 10.1016/j.jiac.2015.08.019. [DOI] [PubMed] [Google Scholar]
- 19.Beal SG, Thomas C, Dhiman N, Nguyen D, Qin H, Hawkins JM, Dekmezian M, Benavides R. 2015. Antibiotic utilization improvement with the Nanosphere Verigene Gram-Positive Blood Culture assay. Proc (Bayl Univ Med Cent) 28:139–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walker T, Dumadag S, Lee CJ, Lee SH, Bender JM, Cupo Abbott J, She RC. 2016. Clinical impact of laboratory implementation of Verigene BC-GN microarray-based assay for detection of Gram-negative bacteria in positive blood cultures. J Clin Microbiol 54:1789–1796. doi: 10.1128/JCM.00376-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bork JT, Leekha S, Heil EL, Zhao L, Badamas R, Johnson JK. 2015. Rapid testing using the Verigene Gram-negative blood culture nucleic acid test in combination with antimicrobial stewardship intervention against Gram-negative bacteremia. Antimicrob Agents Chemother 59:1588–1595. doi: 10.1128/AAC.04259-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.MacVane SH, Nolte FS. 2016. Benefits of adding a rapid PCR-based blood culture identification panel to an established antimicrobial stewardship program. J Clin Microbiol 54:2455–2463. doi: 10.1128/JCM.00996-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pardo J, Klinker KP, Borgert SJ, Butler BM, Giglio PG, Rand KH. 2016. Clinical and economic impact of antimicrobial stewardship interventions with the FilmArray blood culture identification panel. Diagn Microbiol Infect Dis 84:159–164. doi: 10.1016/j.diagmicrobio.2015.10.023. [DOI] [PubMed] [Google Scholar]
- 24.Messacar K, Hurst AL, Child J, Campbell K, Palmer C, Hamilton S, Dowell E, Robinson CC, Parker SK, Dominguez SR. 2017. Clinical impact and provider acceptability of real-time antimicrobial stewardship decision support for rapid diagnostics in children with positive blood culture results. J Pediatr Infect Dis Soc 6:267–274. doi: 10.1093/jpids/piw047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Timbrook TT, Morton JB, McConeghy KW, Caffrey AR, Mylonakis E, LaPlante KL. 2017. The effect of molecular rapid diagnostic testing on clinical outcomes in bloodstream infections: a systematic review and meta-analysis. Clin Infect Dis 64:15–23. doi: 10.1093/cid/ciw649. [DOI] [PubMed] [Google Scholar]
- 26.Almuhayawi M, Altun O, Stralin K, Ozenci V. 2014. Identification of microorganisms by FilmArray and matrix-assisted laser desorption ionization–time of flight mass spectrometry prior to positivity in the blood culture system. J Clin Microbiol 52:3230–3236. doi: 10.1128/JCM.01084-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lim SH, Mix S, Xu Z, Taba B, Budvytiene I, Berliner AN, Queralto N, Churi YS, Huang RS, Eiden M, Martino RA, Rhodes P, Banaei N. 2014. Colorimetric sensor array allows fast detection and simultaneous identification of sepsis-causing bacteria in spiked blood culture. J Clin Microbiol 52:592–598. doi: 10.1128/JCM.02377-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramanan P, Gebrehiwot SA, Rucinski SL, Dylla BL, Wengenack NL, Hughes JG, Ihde SM, Patel R. 2016. Discrepancies between microbial detection and identification using the blood culture identification (BCID) FilmArray panel assay and standard subculture of positive blood culture bottles, abstr 57653, poster 188. Abstr IDWeek 2016, New Orleans, LA, 26 to 30 October 2016 http://www.idweek.org/. [Google Scholar]
- 29.Salimnia H, Fairfax MR, Lephart PR, Schreckenberger P, DesJarlais SM, Johnson JK, Robinson G, Carroll KC, Greer A, Morgan M, Chan R, Loeffelholz M, Valencia-Shelton F, Jenkins S, Schuetz AN, Daly JA, Barney T, Hemmert A, Kanack KJ. 2016. Evaluation of the FilmArray blood culture identification panel: results of a multicenter controlled trial. J Clin Microbiol 54:687–698. doi: 10.1128/JCM.01679-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dodemont M, De Mendonca R, Nonhoff C, Roisin S, Denis O. 2015. Evaluation of Verigene gram-positive blood culture assay performance for bacteremic patients. Eur J Clin Microbiol Infect Dis 34:473–477. doi: 10.1007/s10096-014-2250-4. [DOI] [PubMed] [Google Scholar]
- 31.Nieman AE, Savelkoul PH, Beishuizen A, Henrich B, Lamik B, MacKenzie CR, Kindgen-Milles D, Helmers A, Diaz C, Sakka SG, Schade RP. 2016. A prospective multicenter evaluation of direct molecular detection of blood stream infection from a clinical perspective. BMC Infect Dis 16:314. doi: 10.1186/s12879-016-1646-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pfaller MA, Wolk DM, Lowery TJ. 2016. T2MR and T2Candida: novel technology for the rapid diagnosis of candidemia and invasive candidiasis. Future Microbiol 11:103–117. doi: 10.2217/fmb.15.111. [DOI] [PubMed] [Google Scholar]
- 33.Mylonakis E, Clancy CJ, Ostrosky-Zeichner L, Garey KW, Alangaden GJ, Vazquez JA, Groeger JS, Judson MA, Vinagre YM, Heard SO, Zervou FN, Zacharioudakis IM, Kontoyiannis DP, Pappas PG. 2015. T2 magnetic resonance assay for the rapid diagnosis of candidemia in whole blood: a clinical trial. Clin Infect Dis 60:892–899. doi: 10.1093/cid/ciu959. [DOI] [PubMed] [Google Scholar]
- 34.Hanson KE, Couturier MR. 2016. Multiplexed molecular diagnostics for respiratory, gastrointestinal, and central nervous system infections. Clin Infect Dis 63:1361–1367. doi: 10.1093/cid/ciw494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Popowitch EB, O'Neill SS, Miller MB. 2013. Comparison of the Biofire FilmArray RP, Genmark eSensor RVP, Luminex xTAG RVPv1, and Luminex xTAG RVP fast multiplex assays for detection of respiratory viruses. J Clin Microbiol 51:1528–1533. doi: 10.1128/JCM.03368-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen JH, Lam HY, Yip CC, Wong SC, Chan JF, Ma ES, Cheng VC, Tang BS, Yuen KY. 2016. Clinical evaluation of the new high-throughput Luminex NxTAG respiratory pathogen panel assay for multiplex respiratory pathogen detection. J Clin Microbiol 54:1820–1825. doi: 10.1128/JCM.00517-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nijhuis RHT, Guerendiain D, Claas ECJ, Templeton KE. 2017. Comparison of ePlex respiratory pathogen panel with laboratory-developed real-time PCR assays for detection of respiratory pathogens. J Clin Microbiol 55:1938–1945. doi: 10.1128/JCM.00221-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rappo U, Schuetz AN, Jenkins SG, Calfee DP, Walsh TJ, Wells MT, Hollenberg JP, Glesby MJ. 2016. Impact of early detection of respiratory viruses by multiplex PCR assay on clinical outcomes in adult patients. J Clin Microbiol 54:2096–2103. doi: 10.1128/JCM.00549-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Subramony A, Zachariah P, Krones A, Whittier S, Saiman L. 2016. Impact of multiplex polymerase chain reaction testing for respiratory pathogens on healthcare resource utilization for pediatric inpatients. J Pediatr 173:196.e2–201.e2. doi: 10.1016/j.jpeds.2016.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rogers BB, Shankar P, Jerris RC, Kotzbauer D, Anderson EJ, Watson JR, O'Brien LA, Uwindatwa F, McNamara K, Bost JE. 2015. Impact of a rapid respiratory panel test on patient outcomes. Arch Pathol Lab Med 139:636–641. doi: 10.5858/arpa.2014-0257-OA. [DOI] [PubMed] [Google Scholar]
- 41.Brendish NJ, Malachira AK, Armstrong L, Houghton R, Aitken S, Nyimbili E, Ewings S, Lillie PJ, Clark TW. 2017. Routine molecular point-of-care testing for respiratory viruses in adults presenting to hospital with acute respiratory illness (ResPOC): a pragmatic, open-label, randomised controlled trial. Lancet Respir Med 5:401–411. doi: 10.1016/S2213-2600(17)30120-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mahony JB, Blackhouse G, Babwah J, Smieja M, Buracond S, Chong S, Ciccotelli W, O'Shea T, Alnakhli D, Griffiths-Turner M, Goeree R. 2009. Cost analysis of multiplex PCR testing for diagnosing respiratory virus infections. J Clin Microbiol 47:2812–2817. doi: 10.1128/JCM.00556-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nelson RE, Stockmann C, Hersh AL, Pavia AT, Korgenksi K, Daly JA, Couturier MR, Ampofo K, Thorell EA, Doby EH, Robison JA, Blaschke AJ. 2015. Economic analysis of rapid and sensitive polymerase chain reaction testing in the emergency department for influenza infections in children. Pediatr Infect Dis J 34:577–582. doi: 10.1097/INF.0000000000000703. [DOI] [PubMed] [Google Scholar]
- 44.Schreckenberger PC, McAdam AJ. 2015. Point-counterpoint: large multiplex PCR panels should be first-line tests for detection of respiratory and intestinal pathogens. J Clin Microbiol 53:3110–3115. doi: 10.1128/JCM.00382-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Doern CD, Lacey D, Huang R, Haag C. 2013. Evaluation and implementation of FilmArray version 1.7 for improved detection of adenovirus respiratory tract infection. J Clin Microbiol 51:4036–4039. doi: 10.1128/JCM.02546-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Midgley CM, Jackson MA, Selvarangan R, Turabelidze G, Obringer E, Johnson D, Giles BL, Patel A, Echols F, Oberste MS, Nix WA, Watson JT, Gerber SI. 2014. Severe respiratory illness associated with enterovirus D68—Missouri and Illinois, 2014. MMWR Morb Mortal Wkly Rep 63:798–799. [PMC free article] [PubMed] [Google Scholar]
- 47.Nickbakhsh S, Thorburn F, Von Wissmann B, McMenamin J, Gunson RN, Murcia PR. 2016. Extensive multiplex PCR diagnostics reveal new insights into the epidemiology of viral respiratory infections. Epidemiol Infect 144:2064–2076. doi: 10.1017/S0950268816000339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dalpke A, Zimmermann S, Schnitzler P. 2016. Underdiagnosing of Mycoplasma pneumoniae infections as revealed by use of a respiratory multiplex PCR panel. Diagn Microbiol Infect Dis 86:50–52. doi: 10.1016/j.diagmicrobio.2016.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Farthing M, Salam MA, Lindberg G, Dite P, Khalif I, Salazar-Lindo E, Ramakrishna BS, Goh KL, Thomson A, Khan AG, Krabshuis J, LeMair A. 2013. Acute diarrhea in adults and children: a global perspective. J Clin Gastroenterol 47:12–20. doi: 10.1097/MCG.0b013e31826df662. [DOI] [PubMed] [Google Scholar]
- 50.Esposito DH, Holman RC, Haberling DL, Tate JE, Podewils LJ, Glass RI, Parashar U. 2011. Baseline estimates of diarrhea-associated mortality among United States children before rotavirus vaccine introduction. Pediatr Infect Dis J 30:942–947. doi: 10.1097/INF.0b013e3182254d19. [DOI] [PubMed] [Google Scholar]
- 51.Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, Jones JL, Griffin PM. 2011. Foodborne illness acquired in the United States—major pathogens. Emerg Infect Dis 17:7–15. doi: 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scallan E, Griffin PM, Angulo FJ, Tauxe RV, Hoekstra RM. 2011. Foodborne illness acquired in the United States—unspecified agents. Emerg Infect Dis 17:16–22. doi: 10.3201/eid1701.P21101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Centers for Disease Control and Prevention. 2015. Cyclosporiasis outbreak investigations—United States, 2015. Centers for Disease Control and Prevention, Atlanta, GA: https://www.cdc.gov/parasites/cyclosporiasis/outbreaks/2015/index.html Accessed 27 January 2017. [Google Scholar]
- 54.Centers for Disease Control and Prevention. 2016. Cyclosporiasis update. Centers for Disease Control and Prevention, Atlanta, GA: https://www.cdc.gov/parasites/cyclosporiasis/outbreaks/2016/index.html Accessed 27 January 2017. [Google Scholar]
- 55.Centers for Disease Control and Prevention. 2013. Cyclosporiasis outbreak investigations—United States, 2013 (final update). Centers for Disease Control and Prevention, Atlanta, GA: https://www.cdc.gov/parasites/cyclosporiasis/outbreaks/investigation-2013.html Accessed 27 January 2017. [Google Scholar]
- 56.Spina A, Kerr KG, Cormican M, Barbut F, Eigentler A, Zerva L, Tassios P, Popescu GA, Rafila A, Eerola E, Batista J, Maass M, Aschbacher R, Olsen KE, Allerberger F. 2015. Spectrum of enteropathogens detected by the FilmArray GI Panel in a multicentre study of community-acquired gastroenteritis. Clin Microbiol Infect 21:719–728. doi: 10.1016/j.cmi.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 57.Food and Drug Administration. 30 May 2014. 510(k) substantial equivalence determination decision summary. Number K140407. Food and Drug Administration, Washington, DC: http://www.accessdata.fda.gov/cdrh_docs/reviews/k140407.pdf Accessed 28 February 2017. [Google Scholar]
- 58.Buss SN, Leber A, Chapin K, Fey PD, Bankowski MJ, Jones MK, Rogatcheva M, Kanack KJ, Bourzac KM. 2015. Multicenter evaluation of the BioFire FilmArray gastrointestinal panel for etiologic diagnosis of infectious gastroenteritis. J Clin Microbiol 53:915–925. doi: 10.1128/JCM.02674-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Khare R, Espy MJ, Cebelinski E, Boxrud D, Sloan LM, Cunningham SA, Pritt BS, Patel R, Binnicker MJ. 2014. Comparative evaluation of two commercial multiplex panels for detection of gastrointestinal pathogens by use of clinical stool specimens. J Clin Microbiol 52:3667–3673. doi: 10.1128/JCM.01637-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Halligan E, Edgeworth J, Bisnauthsing K, Bible J, Cliff P, Aarons E, Klein J, Patel A, Goldenberg S. 2014. Multiplex molecular testing for management of infectious gastroenteritis in a hospital setting: a comparative diagnostic and clinical utility study. Clin Microbiol Infect 20:O460–O467. doi: 10.1111/1469-0691.12476. [DOI] [PubMed] [Google Scholar]
- 61.Rand KH, Tremblay EE, Hoidal M, Fisher LB, Grau KR, Karst SM. 2015. Multiplex gastrointestinal pathogen panels: implications for infection control. Diagn Microbiol Infect Dis 82:154–157. doi: 10.1016/j.diagmicrobio.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 62.Mengelle C, Mansuy JM, Prere MF, Grouteau E, Claudet I, Kamar N, Huynh A, Plat G, Benard M, Marty N, Valentin A, Berry A, Izopet J. 2013. Simultaneous detection of gastrointestinal pathogens with a multiplex Luminex-based molecular assay in stool samples from diarrhoeic patients. Clin Microbiol Infect 19:E458–E465. doi: 10.1111/1469-0691.12255. [DOI] [PubMed] [Google Scholar]
- 63.Wessels E, Rusman LG, van Bussel MJ, Claas EC. 2014. Added value of multiplex Luminex Gastrointestinal Pathogen Panel (xTAG GPP) testing in the diagnosis of infectious gastroenteritis. Clin Microbiol Infect 20:O182–O187. doi: 10.1111/1469-0691.12364. [DOI] [PubMed] [Google Scholar]
- 64.Enserink R, Scholts R, Bruijning-Verhagen P, Duizer E, Vennema H, de Boer R, Kortbeek T, Roelfsema J, Smit H, Kooistra-Smid M, van Pelt W. 2014. High detection rates of enteropathogens in asymptomatic children attending day care. PLoS One 9:e89496. doi: 10.1371/journal.pone.0089496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huang RS, Johnson CL, Pritchard L, Hepler R, Ton TT, Dunn JJ. 2016. Performance of the Verigene enteric pathogens test, Biofire FilmArray gastrointestinal panel and Luminex xTAG gastrointestinal pathogen panel for detection of common enteric pathogens. Diagn Microbiol Infect Dis 86:336–339. doi: 10.1016/j.diagmicrobio.2016.09.013. [DOI] [PubMed] [Google Scholar]
- 66.Trujillo AA, McCaustland KA, Zheng DP, Hadley LA, Vaughn G, Adams SM, Ando T, Glass RI, Monroe SS. 2006. Use of TaqMan real-time reverse transcription-PCR for rapid detection, quantification, and typing of norovirus. J Clin Microbiol 44:1405–1412. doi: 10.1128/JCM.44.4.1405-1412.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Verweij JJ, Blange RA, Templeton K, Schinkel J, Brienen EA, van Rooyen MA, van Lieshout L, Polderman AM. 2004. Simultaneous detection of Entamoeba histolytica, Giardia lamblia, and Cryptosporidium parvum in fecal samples by using multiplex real-time PCR. J Clin Microbiol 42:1220–1223. doi: 10.1128/JCM.42.3.1220-1223.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Oka T, Katayama K, Hansman GS, Kageyama T, Ogawa S, Wu FT, White PA, Takeda N. 2006. Detection of human sapovirus by real-time reverse transcription-polymerase chain reaction. J Med Virol 78:1347–1353. doi: 10.1002/jmv.20699. [DOI] [PubMed] [Google Scholar]
- 69.Ramanan P, Espy MJ, Khare R, Binnicker MJ. 20 January 2017. Detection and differentiation of norovirus genogroups I and II from clinical stool specimens using real-time PCR. Diagn Microbiol Infect Dis doi: 10.1016/j.diagmicrobio.2017.01.009. [DOI] [PubMed] [Google Scholar]
- 70.Goldenberg SD, Bacelar M, Brazier P, Bisnauthsing K, Edgeworth JD. 2015. A cost benefit analysis of the Luminex xTAG gastrointestinal pathogen panel for detection of infectious gastroenteritis in hospitalised patients. J Infect 70:504–511. doi: 10.1016/j.jinf.2014.11.009. [DOI] [PubMed] [Google Scholar]
- 71.Riddle MS, DuPont HL, Connor BA. 2016. ACG clinical guideline: diagnosis, treatment, and prevention of acute diarrheal infections in adults. Am J Gastroenterol 111:602–622. doi: 10.1038/ajg.2016.126. [DOI] [PubMed] [Google Scholar]
- 72.Gebrehiwot SA, Rucinski SL, Schwab JJ, Patel R, Snippes P. 2016. “Reflexive culture”—a strategy for laboratories adopting molecular testing for enteric pathogens, poster 188. Abstr ASM Microbe 2016, Boston, MA. [Google Scholar]
- 73.Walker B, Powers-Fletcher MV, Schmidt RL, Hanson KE. 2016. Cost-effectiveness analysis of multiplex PCR with magnetic resonance detection versus empiric or blood culture-directed therapy for management of suspected candidemia. J Clin Microbiol 54:718–726. doi: 10.1128/JCM.02971-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tunkel AR, Hartman BJ, Kaplan SL, Kaufman BA, Roos KL, Scheld WM, Whitley RJ. 2004. Practice guidelines for the management of bacterial meningitis. Clin Infect Dis 39:1267–1284. doi: 10.1086/425368. [DOI] [PubMed] [Google Scholar]
- 75.Leber AL, Everhart K, Balada-Llasat JM, Cullison J, Daly J, Holt S, Lephart P, Salimnia H, Schreckenberger PC, DesJarlais S, Reed SL, Chapin KC, LeBlanc L, Johnson JK, Soliven NL, Carroll KC, Miller JA, Dien Bard J, Mestas J, Bankowski M, Enomoto T, Hemmert AC, Bourzac KM. 2016. Multicenter evaluation of BioFire FilmArray meningitis/encephalitis panel for detection of bacteria, viruses, and yeast in cerebrospinal fluid specimens. J Clin Microbiol 54:2251–2261. doi: 10.1128/JCM.00730-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hanson KE, Slechta ES, Killpack JA, Heyrend C, Lunt T, Daly JA, Hemmert AC, Blaschke AJ. 2016. Preclinical assessment of a fully automated multiplex PCR panel for detection of central nervous system pathogens. J Clin Microbiol 54:785–787. doi: 10.1128/JCM.02850-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Launes C, Casas-Alba D, Fortuny C, Valero-Rello A, Cabrerizo M, Munoz-Almagro C. 2017. Utility of FilmArray meningitis/encephalitis panel during outbreak of brainstem encephalitis caused by enterovirus in Catalonia in 2016. J Clin Microbiol 55:336–338. doi: 10.1128/JCM.01931-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Messacar K, Breazeale G, Robinson CC, Dominguez SR. 2016. Potential clinical impact of the FilmArray meningitis encephalitis panel in children with suspected central nervous system infections. Diagn Microbiol Infect Dis 86:118–120. doi: 10.1016/j.diagmicrobio.2016.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Graf EH, Farquharson MV, Cardenas AM. 2017. Comparative evaluation of the FilmArray meningitis/encephalitis molecular panel in a pediatric population. Diagn Microbiol Infect Dis 87:92–94. doi: 10.1016/j.diagmicrobio.2016.09.022. [DOI] [PubMed] [Google Scholar]
- 80.Bamba S, Lortholary O, Sawadogo A, Millogo A, Guiguemde RT, Bretagne S. 2012. Decreasing incidence of cryptococcal meningitis in West Africa in the era of highly active antiretroviral therapy. AIDS 26:1039–1041. doi: 10.1097/QAD.0b013e328352d1d8. [DOI] [PubMed] [Google Scholar]
- 81.Jarvis JN, Meintjes G, Williams A, Brown Y, Crede T, Harrison TS. 2010. Adult meningitis in a setting of high HIV and TB prevalence: findings from 4961 suspected cases. BMC Infect Dis 10:67. doi: 10.1186/1471-2334-10-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Siddiqi OK, Ghebremichael M, Dang X, Atadzhanov M, Kaonga P, Khoury MN, Koralnik IJ. 2014. Molecular diagnosis of central nervous system opportunistic infections in HIV-infected Zambian adults. Clin Infect Dis 58:1771–1777. doi: 10.1093/cid/ciu191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Durski KN, Kuntz KM, Yasukawa K, Virnig BA, Meya DB, Boulware DR. 2013. Cost-effective diagnostic checklists for meningitis in resource-limited settings. J Acquir Immune Defic Syndr 63:e101–e108. doi: 10.1097/QAI.0b013e31828e1e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rhein J, Bahr NC, Hemmert AC, Cloud JL, Bellamkonda S, Oswald C, Lo E, Nabeta H, Kiggundu R, Akampurira A, Musubire A, Williams DA, Meya DB, Boulware DR, ASTRO-CM Team. 2016. Diagnostic performance of a multiplex PCR assay for meningitis in an HIV-infected population in Uganda. Diagn Microbiol Infect Dis 84:268–273. doi: 10.1016/j.diagmicrobio.2015.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hall CB, Caserta MT, Schnabel K, Shelley LM, Marino AS, Carnahan JA, Yoo C, Lofthus GK, McDermott MP. 2008. Chromosomal integration of human herpesvirus 6 is the major mode of congenital human herpesvirus 6 infection. Pediatrics 122:513–520. doi: 10.1542/peds.2007-2838. [DOI] [PubMed] [Google Scholar]
- 86.Gomez CA, Pinsky BA, Liu A, Banaei N. 2016. Delayed diagnosis of tuberculous meningitis misdiagnosed as herpes simplex virus-1 (HSV-1) encephalitis with the FilmArray syndromic PCR panel. Open Forum Infect Dis 4:ofw245. doi: 10.1093/ofid/ofw245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Michos A, Palili A, Koutouzis EI, Sandu A, Lykopoulou L, Syriopoulou VP. 2016. Detection of bacterial pathogens in synovial and pleural fluid with the FilmArray blood culture identification system. IDCases 5:27–28. doi: 10.1016/j.idcr.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Vasoo S, Cunningham SA, Greenwood-Quaintance KE, Mandrekar JN, Hanssen AD, Abdel MP, Osmon DR, Berbari EF, Patel R. 2015. Evaluation of the FilmArray blood culture ID panel on biofilms dislodged from explanted arthroplasties for prosthetic joint infection diagnosis. J Clin Microbiol 53:2790–2792. doi: 10.1128/JCM.01333-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mico M, Navarro F, de Miniac D, Gonzalez Y, Brell A, Lopez C, Sanchez-Reus F, Mirelis B, Coll P. 2015. Efficacy of the FilmArray blood culture identification panel for direct molecular diagnosis of infectious diseases from samples other than blood. J Med Microbiol 64:1481–1488. doi: 10.1099/jmm.0.000180. [DOI] [PubMed] [Google Scholar]
- 90.Altun O, Almuhayawi M, Ullberg M, Ozenci V. 2015. Rapid identification of microorganisms from sterile body fluids by use of FilmArray. J Clin Microbiol 53:710–712. doi: 10.1128/JCM.03434-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Whiteley W, Al-Shahi R, Warlow CP, Zeidler M, Lueck CJ. 2006. CSF opening pressure: reference interval and the effect of body mass index. Neurology 67:1690–1691. doi: 10.1212/01.wnl.0000242704.60275.e9. [DOI] [PubMed] [Google Scholar]
- 92.Bronnestam R, Dencker SJ, Swahn B. 1961. Fibrinogen in cerebrospinal fluid: fibrinogen demonstrated by microimmunoelectrophoresis. Arch Neurol 4:288–290. doi: 10.1001/archneur.1961.00450090054008. [DOI] [Google Scholar]
- 93.Ryan KJ, Ray CG, Sherris JC. 2004. Sherris medical microbiology: an introduction to infectious diseases, 4th ed McGraw-Hill, New York, NY. [Google Scholar]
- 94.Fang FC, Patel R. 19 October 2017. 2017 Infectious Diseases Society of America infectious diarrhea guidelines: a view from the clinical laboratory. Clin Infect Dis doi: 10.1093/cid/cix730. [DOI] [PubMed] [Google Scholar]
- 95.Shane AL, Mody RK, Crump JA, Tarr PI, Steiner TS, Kotloff K, Langley JM, Wanke C, Warren CA, Cheng AC, Cantey J, Pickering LK. 19 October 2017. 2017 Infectious Diseases Society of America clinical practice guidelines for the diagnosis and management of infectious diarrhea. Clin Infect Dis doi: 10.1093/cid/cix669. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.