SUMMARY
We are far away from the days when tuberculosis (TB) accounted for 1 in 4 deaths during the 19th century. However, Mycobacterium tuberculosis complex (MTBC) strains are still the leading cause of morbidity and mortality by a single infectious disease, with 9.6 million cases and 1.5 million deaths reported. One-third of the world's population is estimated by the WHO to be infected with latent TB. During the last decade, several studies have aimed to define the characteristics of dormant bacteria in these latent infections. General features of the shift to a dormant state encompass several phenotypic changes that reduce metabolic activity. This low metabolic state is thought to increase the resistance of MTBC strains to host/environmental stresses, including antibiotic action. Once the stress ceases (e.g., interruption of treatment), dormant cells can reactivate and cause symptomatic disease again. Therefore, a proper understanding of dormancy could guide the rational development of new treatment regimens that target dormant cells, reducing later relapse. Here, we briefly summarize the latest data on the genetics involved in the regulation of dormancy and discuss new approaches to TB treatment.
KEYWORDS: Mycobacterium tuberculosis, pretomanid, teixobactin, toxin-antitoxin, bedaquiline, dormancy, lassomycin, latency, persistent
INTRODUCTION
Although treatment regimens for tuberculosis (TB) have evolved over the years, control of the disease is difficult, and in 2015, active TB still accounted for 1.5 million deaths (1). According to the World Health Organization (1), current guidelines for TB treatment consist of a 2-month period of four antibiotics, isoniazid (INH), rifampin (RIF), ethambutol (EMB), and pyrazinamide (PZA), followed by a 4-month period of INH and RIF (1). The duration of this treatment challenges patient adherence and consequently limits its overall effectiveness. Even more challenging is the therapy for multidrug-resistant (MDR) TB cases, which are defined as having resistance to RIF or INH, and extensively drug resistant (XDR) TB cases, defined as having MDR plus resistance to at least one fluoroquinolone and 1 second-line injectable drug (1, 2). Treatment of infection by MDR/XDR strains requires 9 to 20 months or even longer, and the rate of a successful outcome falls below 50% on a global level (1, 2). Overall, a reduction in the duration of conventional and MDR/XDR treatment is considered a prime objective in the development of future treatment regimens to better prevent emerging resistance and increase the rate of successful treatment (3).
In 1958, it was proposed that the underlying reason for long treatment durations was the presence of nonreplicating cells (4). These cells, during active infection, represent a small subpopulation that remains unaffected during standard antibiotic therapy, requiring the use of special drugs with sterilizing capacities, such as PZA and RIF (5). Even if the patient completes treatment, dormant cells can resuscitate after decades, leading to relapse, especially in immunocompromised patients or those with coinfection with human immunodeficiency virus (HIV) or merely due to ageing (6).
The development of more-effective, shorter TB treatment requires not only the rapid killing of actively dividing cells but also the effective elimination of dormant populations. Therefore, there is an urgent need to understand the cellular and molecular basis of mycobacterial dormant states. How do cells enter the dormant state? What are their vulnerable points in dormancy? How can these vulnerabilities be exploited as new drug targets? This knowledge will allow the rational development of more-effective and shorter TB treatment regimens (7). In this review, we present an overview of recent findings describing the dormant phenotype in Mycobacterium tuberculosis complex (MTBC) strains and potential therapeutic options to tackle this problem.
WHAT IS DORMANCY?
Dormancy is a reversible metabolic shutdown (8), defined here for simplicity as a reversible, nonreplicating cellular state that enables mycobacteria to circumvent and survive host defenses (9). The earliest attempts to reproduce in vitro the nonreplicative state of MTBC cells began in 1994, resulting in the so-called Wayne model, which relies on the observation that a two-phase reduction of O2 availability leads MTBC cells to a nonreplicating state (10). Since then, similar models have been studied further, and it is now commonly accepted that virtually any kind of nutrient restriction causes a reduction in metabolic activity and subsequent dormancy (6). However, the mechanisms involved in this reduced metabolism that protects bacteria against most antimycobacterial drugs remain unclear.
Most antibiotics interfere with processes that are vital for actively dividing microorganisms, such as DNA replication, translation, or cell wall formation. An interruption of any of these processes, an imbalance between cellular processes, or the creation of metabolic products for which the cells cannot compensate leads to either a halt in replication (bacteriostatic drugs) or cell death (bactericidal drugs). As most drug targets are essential to replicating cells only, a reduction in metabolic activity protects the cell. This is frequently the cause of a decrease in drug efficacy and hence a reduction or disappearance of the drug's killing effect. Critically, this protective effect from the metabolic state is general, applying to many antibiotics from different classes simultaneously (8).
If a given treatment fails to kill even a small proportion of bacteria, there is the potential for resuscitation and consequent relapse, even years after treatment completion (Fig. 1). This nongrowing or low-replicating state can be considered a critical component of the MTBC life cycle, allowing long-term survival in the host. Intriguingly, it has been proposed that this nonreplicating state is decisive for the success of MTBC strains, and it can be achieved by a number of different mechanisms.
FIG 1.
Progression of tuberculosis. Here we consider symptomatic disease the beginning of the disease. During infection, MTBC bacteria (red dots) actively colonize different areas of the lungs. During this phase, most cells are actively growing. Thus, they are potentially susceptible to treatment and the immune response. Macrophages (blue cells) phagocytose MTBC cells, but dormant cells are better able to survive inside macrophages. T cells (light green), B cells (dark green), and giant foam cells (orange) contain the infection in a structure called a granuloma. Thus, there are three possible outcomes after treatment starts: interruption or failure of treatment that leads to a relapse where MTBC bacteria break the barrier of the granuloma, reactivating infection (A); partially effective treatment that leads to a stable quiescent structure with contained viable cells, which persist after treatment stops (B); or eradication of the disease (MTBC cells are entirely removed from the healing granuloma) (C).
Ways to Dormancy
Entry into dormancy can be an active process in which specific signals trigger the activation of master regulatory genes to drive MTBC cells toward the low-replicating/nonreplicating state. The Wayne model (nonreplication induced by hypoxia) contributed to the identification of key genes related to in vivo dormancy (10). The principal transcription factor in this model is the regulator DosR (also DevR), which is activated in response to hypoxia as well as nitric oxide produced by macrophages (11). DosR directly modulates the expression of at least 48 genes, enabling MTBC cells to survive during hypoxia (12). This model was also used to identify the essential gene phoP, which mediates the early hypoxic response and participates in MTBC virulence (13).
Similar pathways based on amino acid starvation, carbon limitation, or virtually any nutrient stress can also lead MTBC cells into dormancy. This transition of phenotype is a result of the stringent response and is orchestrated by the metabolite (p)ppGpp, known as the alarmone signal, and polyphosphate (poly-P) (14, 15). The stringent response begins when stress signals trigger the rapid synthesis of poly-P catalyzed by polyphosphate kinase (PPK) (16) and simultaneously constrain poly-P degradation by blocking exopolyphosphatase (PPX) (17). The accumulation of this molecule increases the activity of the two-component transcription regulator MprA/MprB (16), which subsequently enhances the expression of M. tuberculosis Rel (RelMtb), the enzyme that synthesizes ppGpp, and sigma factor E (σE). The latter is a subunit of RNA polymerase that has a high affinity for promoters of stress-resistant genes, favoring the phenotypic shift to stress resistance.
The mycobacterial stringent response appears to be stabilized by two positive-feedback loops (Fig. 2): first, the transcription factor MprA regulates its operon positively, and second, as poly-P accumulates in the cell, it induces relMtb expression and ppGpp synthesis, with ppGpp inhibiting PPX, which degrades poly-P (18).
FIG 2.
Representation of the partial regulatory network of the MTBC stringent response. Stress signals trigger poly-P accumulation, activating the two-component Mpr complex. This increases SigE expression, which, in conjunction with the Mpr complex, activates the transcription of relMtb, thereby increasing the concentration of ppGpp. In a positive-feedback loop, ppGpp in turn blocks PPX activity, amplifying the signal and triggering the stringent response.
Many reports have recently related the stringent response to dormancy, both as a whole and as individual components. For example, it has been found that poly-P contributes to antibiotic tolerance. Infection of guinea pigs with an MTBC Δppk2 strain, with a deletion of the gene encoding the enzyme responsible for poly-P synthesis, led to a reduction in persistence and higher antibiotic susceptibility (19) (20). In addition, an MTBC strain lacking relMtb has been shown to be more vulnerable in vitro to nutrient deprivation and hypoxia (15), also resulting in a decreased ability of this strain to sustain chronic infection in mice (21) and in guinea pig lungs (22). Finally, σE is upregulated when the bacterium grows within human macrophages, and its deletion leads to attenuated viability (23).
The stringent-response hypothesis, therefore, may partly explain how cells enter into the dormant state once they sense stress in their environment, yet it does not clarify the nongrowing populations previous to any stress. A complementary hypothesis proposed by Balaban and colleagues suggests that pathogens can display a nongrowing phenotype, even in the absence of environmental stress as a phenotypic switch (24). This might involve the stochastic activation of a molecular mechanism that allows random switching between growing and nongrowing phenotypes. This phenomenon is known as persistence. In contrast to dormancy, persistence involves a preexisting subpopulation, defined here as a nongrowing fraction of the bacterial population, which displays a nonheritable ability to survive exposure to high concentrations of an antibiotic (24).
Although the nongrowth strategy could seem counterproductive for bacteria, Kussell and colleagues have shown that a stochastic switch between growing and nongrowing (persister) phenotypes can be an evolutionarily favorable strategy (25). Their model states that even during optimal growth (before sensing stress), there is a small fraction of the population in a nongrowing state (25, 26). These dormant cells act as an “insurance policy” against unpredictable risks, protecting the population as a whole (26). Incidentally, the fraction of persistent cells directly affects the growth rate, with a larger persistent fraction resulting in less growth. This fact could partially explain why MTBC cells grow slowly in vitro, even in rich growth medium (27). The immediate question that emerges here is what is the mechanism that MTBC cells use to “ignore” the external conditions to maintain or trigger persistence.
Toxin-Antitoxin in M. tuberculosis Complex Strains
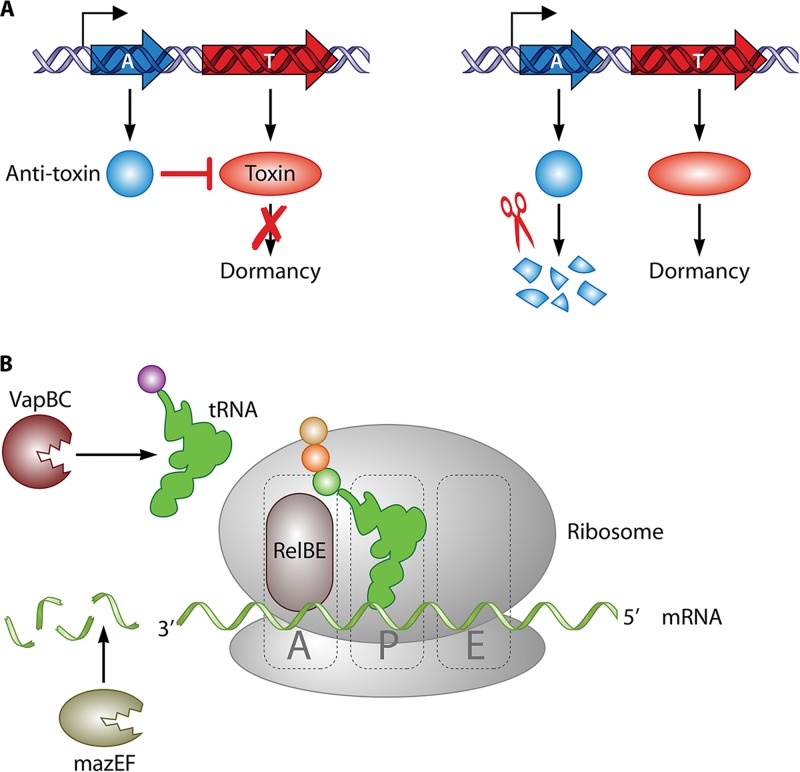
In the last decades, it has been proposed that toxin-antitoxin (TA) modules can control the stochastic switch between growing and nongrowing phenotypes (8). TA modules comprise two or more genes in the same operon (Fig. 3A): one has a “toxic” effect that leads to the dormant phenotype, and the other is an antitoxin that counteracts this effect to allow cell growth (8). The most common regulatory circuit of TA modules (TA type II) is based on two elements: a positive-feedback loop, where the toxin induces its expression, and the fragility of the antitoxin. In combination, this creates a stochastic equilibrium, which means that switching between the “growing” and “nongrowing” phenotypes is controlled partially by environmental signals and partially by random stochastic factors (26, 28). As a consequence, regardless of the external conditions, this mechanism generates a heterogeneous population in which most cells are actively dividing, but some are in a persistent state. This phenomenon is probably one of the main reasons why the eradication of MTBC bacteria in the host is so complicated. Remarkably, it has been determined that the MTBC has around 80 TA modules, while its nonpathogenic relatives Mycobacterium smegmatis and M. marinum have only 4 and 1 TA modules, respectively (28).
FIG 3.
Representation of TA functioning. (A) Typical structure of a TA operon. The toxin gene (T) produces a protein that leads to dormancy; however, an antitoxin (A) that counteracts the toxin's effects is also encoded in the same mRNA. The cell enters into dormancy only when antitoxin is degraded. (B) Mechanism of action of the major TA families in the MTBC. (i) VapBC attacks tRNA, (ii) RelBE binds to the ribosomal A site, and (iii) MazEF is an endonuclease that cleaves mRNA sequences.
To date, five TA families have been described, based on the mode of inhibition of the toxin. However, only TA type II has been reported to be present in MTBC strains (29). The type II TA modules are characterized by a specific mode of action where the antitoxin inhibits the toxin that would otherwise block protein translation (28), although this can be achieved by a different mechanism depending on the TA family (Fig. 3B). This halt in protein synthesis stops cell growth. Interestingly, the latest research indicated that the protein synthesis block is not random; it is instead part of a coherent response for establishing the dormant phenotype (29–31).
The two largest TA families in the MTBC, VapBC and MazEF, cleave RNA (28). VapBC, with more than 50 copies in MTBC strains, cleaves tRNA, causing a halt in protein translation (32). Several VapBC modules were found to be upregulated in a nutrient starvation model, and VapBC modules were found to be induced by stimuli present during infection, such as hypoxia or interferon (IFN) (33). These facts directly connect VapBC with the establishment of dormant infection. The MazEF toxin selectively cleaves mRNA, which ultimately promotes a specific change in the transcriptome (30, 31). The expression of toxins from the MazEF family has also been reported during infection of macrophages and antibiotic treatment in vitro and for MTBC cells in infected mouse lungs (34).
In addition, three other minor TA families have been identified in MTBC strains, although they are not as thoroughly characterized as those described above. The RelBE family is present in the MTBC in three copies: RelBE, RelFG, and RelJK (31) (35). The overexpression of toxin genes of this family inhibits cell growth (35); however, the deletion of individual cassettes does not affect virulence in a mouse model, suggesting that these toxins may not individually contribute to virulence in vivo (36). The HigBA family is among the 10 most upregulated TA systems in MTBC drug-tolerant persisters (37). Finally, TAC (toxin-antitoxin-chaperone) seems to be induced in vitro under several stress conditions, such as DNA damage or drug treatment (29).
Altogether, these data support the idea that TA modules have a key role in persistence (29). However, there are still open questions, such as the degrees of interaction between different TA systems or the difference in TA regulation between MTBC lineages.
From Genotype to Dormant Phenotype
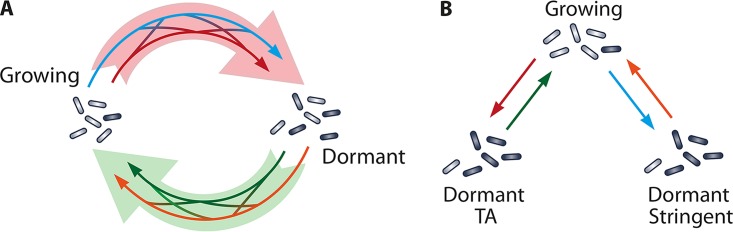
Novel approaches seek a systemic reconstruction of the MTBC regulatory network during dormancy to predict changes in gene expression and cellular metabolic conditions (38). While many genes, such as those involved in central carbon metabolism, rRNA synthesis, and cell division, are downregulated during dormancy, other genes are upregulated, actively contributing to the metabolic shift (Fig. 4).
FIG 4.
Schematic description of the cooperative-dormancy model (A) versus the unique-dormancy model (B). (A) It is possible that there is a single spectrum of dormant phenotypes with a network of different signals triggering entry and exit from this dormant phenotype (pale red and green arrows). This could happen through an interconnected system in which TA modules (red arrow) interact with the stringent response (blue arrow), which results in a distinctive dormant phenotype (black cells). Dormant cells may be resuscitated by resuscitation-promoting factor, nutrients (green arrow), the degradation of toxin modules, or other signals (orange) stimulating cells to restart growth. (B) It is also possible that there is only one growth phenotype and that, depending on the signal, cells switch by independent mechanisms to separate dormant phenotypes. TA modules (red arrow) could generate one distinctive kind of dormant cell, and these cells will resuscitate only when toxin modules are removed from the cell (green arrows). Growing cells could respond to environmental signals through the stringent response (blue arrow) to enter a distinct form of dormancy. These cells will resuscitate when the appropriate stimulus, for example, resuscitation-promoting factor, or nutrients (orange arrow) are present.
The transition to dormancy is facilitated by Clp proteases. These proteases are essential for MTBC viability, as they are responsible for the selective degradation of misfolded proteins that accumulate under stress (39). For example, ClpX has been shown to be upregulated during entry into dormancy induced by hypoxia (32). The resulting proteome and transcriptome are characterized by the downregulation of the glycolysis, DNA replication, and protein synthesis pathways (32, 33). However, energy metabolism and ATP synthesis remain essential processes in dormant cells (40). The maintenance of ATP production is achieved by an increase in Krebs cycle activity and the upregulation of the isocitrate lyase (icl-1) of the glycolate shunt (41). Other changes required to maintain ATP production during dormancy are the upregulation of NADH dehydrogenase 2 (NDH-2), nitrate reductase (NarG-I), and nitrate transporter (NarK2) complexes and the downregulation of the NDH-1 and cytochrome complexes (40).
Furthermore, dormant MTBC cells increase their cell wall thickness and, with it, fatty acid metabolism. Indeed, lipid metabolism has been demonstrated to be crucial for dormancy and subsequent reactivation. Genes such as mce4, related to cholesterol transport, or the triacyl glycerol synthase (tgs) gene that participate in fatty biosynthesis appear to be required for persistence (42). A tgs1 deletion mutant exhibited a lower degree of antibiotic tolerance, and complementation restored antibiotic tolerance (41). Interestingly, lipid body inclusions have been found to be a distinctive characteristic of persistent bacilli in sputum (43). Finally, the structural gene acr (α-crystallin) has been demonstrated to be a key factor for dormancy and is currently being studied for its use as a biomarker for latent MTBC infection (44).
Although it is not known if the genes and proteins identified through in vitro models, such as those for hypoxia, starvation, or antibiotic-TA, correspond directly to human disease, they represent very useful knowledge for further studies targeting this process. Research using these models has demonstrated their utility for finding new drug targets and understanding the basis of resistance.
STRATEGIES TO TARGET DORMANT CELLS
The design of new treatments targeting persistent populations in mycobacteria appears to be critical for the development of more-effective and shorter TB therapies. Currently, the four-drug standard treatment contains only one drug (PZA) that targets explicitly dormant cells, while the others act mainly on growing cells (5).
The replacement of streptomycin by PZA in combination with RIF shortened TB treatment from 12 to 6 months (short-course regimen) while keeping relapse rates low. Although this was thought to be due to the extraordinary capacity of PZA to kill dormant cells, the precise mechanism of action of pyrazinoic acid (the active form of PZA) is not fully understood. The conventional model proposed that a low pH was required for PZA activity, but this has recently been questioned (45). It was also proposed that PZA targets fatty acid synthase I (FAS-I), but a lack of correlation between FAS-I activity and the PZA dosage has stimulated the search for an alternative model (46). New insights proposed a double mechanism: PZA depletes coenzyme A (CoA) reservoirs and blocks the production of phthiocerol dimycocerosate (a virulence factor). This CoA pathway seems to be essential only in dormant cells (47), which might explain why PZA has greater sterilizing power on slower-growing cells. Another plausible mechanism of action for PZA may be its ability to bind the ribosomal protein S1 (RpsA), which halts protein translation (48).
Unfortunately, PZA is a prodrug. Thus, mutations in the gene responsible for PZA activation, the pyrazinamidase gene (pncA), represent a major cause of bacterial resistance to PZA. This is an emerging problem (49) that is rendering short-course treatment more difficult.
Taking into account all of the above-described data, novel approaches and new drugs targeting essential processes in dormant cells would be expected to be very efficient in shortening treatment. Here, we have categorized these approaches into two groups: (i) new drugs against dormant MTBC cells and (ii) alternative treatment concepts.
New Drugs Active against Dormant TB
Considering the global epidemic of drug-resistant strains, drug discovery and development are priorities, yet they are proving to be difficult and time-consuming endeavors. Thirty-nine drugs are currently in development (50), but only six of them aim to target cells in the dormant state (Table 1).
TABLE 1.
Antibiotics described in this review
During the last decade, cyclopeptides emerged as a promising group of drug candidates. Using an original technique to grow uncultivable microorganisms, the Lewis group discovered lassomycin, which has potent specific activity against mycobacteria. The drug binds the protease ClpC1 without affecting related AAA ATPases. The Clp proteases are essential in mycobacteria, so they are an excellent target to eradicate nongrowing cells (39). When lassomycin interacts with ClpC proteases, it disrupts the proteolytic activity of the complex, and cells are believed to die because of the accumulation of misfolded proteins (51). In vitro assays showed that lassomycin has great killing activity against both exponentially growing cells and stationary-phase starved cells (51). Another cyclopeptide family, the acyldepsipeptides (ADEPs), shows a high efficacy of killing activity against persistent subpopulations in MTBC cultures, also by interfering with native Clp activity. In contrast to lassomycin, this compound increases the activity of ClpP1 in MTBC strains (52) and kills pathogens through the nonspecific degradation of over 400 proteins. When combined with RIF, this compound is capable of eradicating persistent subpopulations in either exponential- or stationary-phase cultures (53). Finally, a very promising compound in the preclinical stage is teixobactin (54). This molecule is a macrocyclic depsipeptide and targets lipid II and lipid III, which are precursors of the cell wall (55). Once the antibiotic binds to the lipids, it inhibits the maturation of the peptidoglycan layer, leading to cell lysis. As of today, it is not understood why this mechanism eradicates persistent populations, but the original study illustrates that after in vitro treatment of MTBC cells with teixobactin, no colonies were formed by aliquots plated onto fresh medium (54).
In phase I trials, only one compound exhibits activity against dormant MTBC bacteria. CPZEN-45 is a capreomycin derivate, and early assays showed that it had excellent activity against drug-sensitive strains as well as against nutrient-starved MTBC bacteria (56). CPZEN-45 seems to act by the inhibition of the undecaprenyl-P-GlcNAc-1-P transferase (WecA), which is responsible for the initiation of cell wall synthesis (57). Nevertheless, many studies are required to understand the role of WecA in dormant cells, and the exact mechanism that makes this compound effective against dormant MTBC cells needs to be clarified.
In a later stage of development, phase II, pretomanid (PA-824) represents a promising candidate with activity against both growing and nongrowing MTBC cells. When the drug was tested on nongrowing cells induced by hypoxia, MTBC cells showed susceptibility to micromolar concentrations of the drug. This might be because pretomanid attacks energy metabolism, which is essential for MTBC strains (40), by directly releasing NO and causing respiratory poisoning (58). However, pretomanid is a prodrug, requiring intracellular modifications for its biological activation, and it has already been found that a reduction in the level of glucose-6-phosphate dehydrogenase, or its deazaflavin cofactor F420, makes strains resistant (59).
Finally, in phase III, TMC207 or bedaquiline represents both a novel chemical class (diarylquinolines) as well as a novel MTBC-selective mechanism of action (60). Like pretomanid, bedaquiline targets energy metabolism. Bedaquiline binds ATP synthase with high affinity, interfering very efficiently with its activity (61). Because ATP synthesis is an essential process in dormant cells, treatment of hypoxic nonreplicating bacilli with bedaquiline reduces the cell count more dramatically than isoniazid (an inhibitor of mycolic acid synthesis) (62). Recently, clinical trials with bedaquiline achieved promising results, and in 2016, it acquired prior-authorization status under the trade name Sirturo.
Alternative Treatment Concepts
When cells enter dormancy, they become less vulnerable to most antibiotics, yet they must resuscitate to cause the symptomatic disease. Therefore, the use of intermittent drug dosing has been proposed as an alternative approach for killing dormant cell populations. The theory behind this assumes that during the time in which no drugs are given, cells resuscitate and become susceptible to drugs again. This was first proposed by the discoverer of dormant cells, J. Bigger (63), but despite working in vitro, this approach has several complications in vivo (Table 2). The withdrawal of antibiotics is not uniform in the body (64), and suboptimal dosing can stimulate the selection of resistant cells. Thus, there is considerable debate about the intermittency of the treatment. On the one hand, intermittent treatment with RIF, PZA, and EMB has an efficacy similar to that of conventional treatment (65), and this will reduce the number of visits to the doctor, which increases patient adherence to treatment. On the other hand, intermittent treatment has been proven to cause intolerance (66), and clinical studies showed adverse reactions to intermittent therapy with RIF (67). More importantly, there is a substantial risk of relapse with RIF resistance, mainly in HIV-infected patients (68). Because of the insufficient evidence to support the utility of the intermittent administration of antibiotics, this approach is currently discouraged by the WHO (69).
TABLE 2.
Approaches explored in order to optimize TB therapy
| Method | Pro(s) | Con(s) | References |
|---|---|---|---|
| Intermittent | Shortens treatment, reduces dosage | Emergence of resistance, requires supervision | 65–68 |
| High dose | Reduces visits to clinicians, eradicates persisters | Toxicity, tolerance | 71, 72 |
| Resuscitation | Tackles persistent cells directly | Uncontrolled resuscitation might cause relapse | 73, 74 |
| Immunotherapy adjuvants | Promising approach, reduces toxicity | Unexpected adverse reactions, variable success, more studies required | 75–78 |
Despite the inconclusive results for intermittent treatment regimens, this research line resulted in a new treatment approach based on the high-dose administration of antibiotics (70). During the initial stages of infection, the number of persistent cells is at a minimum; thus, a high dose of the antibiotic may eliminate a high number of cells faster, and the few survivors can be eliminated by the immune system. Tests in mice have indicated that a high-dose RIF regimen resulted in faster clearance (71). In addition, an increased dose of RIF shortened the duration of TB treatment (72). Based on these observations, another new phase II trial has been started, investigating the tolerability and toxicity problems of high-dose RIF treatment (72).
Other approaches involve substances that stimulate resuscitation from dormancy as coadjutants during conventional treatments. This hypothesis was first proposed by Seidi and Jahanban-Esfahlan (73), who suggested the delivery of resuscitation-promoting factor (RPF) to awaken dormant cells in combination with standard antibiotic treatment. After resuscitation, the cells should become susceptible, and thus, the infection should be eradicated. Because purified RPF lost most of its activity in vivo, Gan et al. proposed the use of mycobacterial phages to induce RPF expression, combined with standard therapy (74). However, none of these strategies have been tested in vivo yet.
Finally, host-directed therapy, which aims to stimulate the immune response to improve clinical outcomes, has received increased attention. The concurrent administration of adjuvants with antimicrobial chemotherapy has produced some positive preliminary results. For example, supplementation of standard treatment with tumor necrosis factor (TNF) blockers to reactivate latent MTBC infection has been to be found practical in patients with life-threatening tuberculosis infection (75). In addition, supplementation with high-dose corticosteroids to reduce TNF levels caused a striking reduction in sputum production. However, the adverse effects of corticosteroids (including hyperglycemia, hypertension, and fluid retention) make the continuation of studies with this therapy unlikely (76). Adjuvant therapy with IFN and interleukin-2 (IL-2) have been studied in only small numbers of clinical cases, so it is difficult to develop definitive conclusions (77, 78).
CONCLUSIONS
The success of the M. tuberculosis complex is largely attributable to its ability to escape the host immune response and persist in the host. Therefore, drugs that are rapidly bactericidal in vitro require prolonged administration to achieve comparable effects in vivo. This is most likely due to reservoirs of dormant MTBC cells that remain initially unaffected by the drugs and host immune system. A dormant state seems to be a critical component of the MTBC life cycle and a frequent reason for treatment failure or relapse, thus presenting an important challenge for improving TB therapies.
The understanding of dormancy in MTBC cells remains incomplete, and further studies are required. Current data indicate that dormancy is caused by cells in a metabolic state (6, 9, 41). The reduced metabolic activity protects nongrowing cells against antibiotic action and the immune system, providing the possibility that any dormant cells can resuscitate if treatment is interrupted (8). The stringent response represents a reliable model to describe how cells switch from growth to a quiescent state (9), and TA modules might explain why, under optimal growth conditions, a small proportion of dormant cells is always present, even in an exponentially growing population. Interestingly, there is an association between some MTBC lineages and treatment failure as well as drug resistance; this may be related to larger proportions of dormant cells in a patient infected with bacteria from these lineages (79).
Overall, the pathway toward dormancy is complex. There is considerable evidence for an interaction between the stringent response and the TA modules: the ppGpp synthase interacts with the mazEF family toxins (80), VapBC also interacts with mazEF modules (81), and poly-P enhances the activity of Clp and Lon proteases, increasing the degradation of toxins (15), to give just a few examples. A more complete, holistic understanding of the genetic network explicitly controlling dormancy in MTBC strains is crucial for the determination of key targets for novel antimycobacterial agents. Therapy that is active against both growing and nongrowing cells would be expected to shorten the duration of treatment.
Currently, few options are available for shortened treatment regimens, especially in the case of MDR/XDR strains (3). The translation from a basic laboratory experiment to clinical application is long and arduous. However, strategic alliances, such as the TB Alliance, Stop TB (WHO), or the Aeras Global TB Vaccine Foundation, speed up this process to close the gap between basic and further drug development, including preclinical tests and phase I to III trials. New drugs such as cyclopeptides have been explicitly designed to target dormant cells, with a very precise knowledge of the molecular target. This is the result of a more profound comprehension of the genetic cell network in conjunction with the development of in vitro models with better concordance with in vivo conditions. Furthermore, gene-targeting approaches offer an exciting perspective, such as few interactions with other drugs, yet the associated genetic tools are still in a very early stage. Although their application might not be possible soon, if successful, it would open a new horizon in medicine.
Finally, the WHO Stop TB strategy aims to eliminate TB in 2050 (82). The latest WHO model predicts that current technology could achieve a 20% annual reduction in the incidence of TB. If just 8% of people infected with an MTBC strain were entirely and permanently protected each year, the incidence would fall to 90 per million population by 2050 (82). Therefore, an understanding of persistent MTBC infection represents a key research priority.
ACKNOWLEDGMENTS
All the authors contributed equally to the manuscript. We also express no conflict of interest.
Biographies

Santiago Caño-Muñiz received his degree in biology at the Autonomous University of Madrid, Spain. Following his graduate studies, he completed a master of science in Molecular Biology and Biotechnology at the University of Groningen, The Netherlands. Currently, he is pursuing a Ph.D. in Genetics at the University of Cambridge, United Kingdom. His major research interest is to elucidate the molecular mechanisms used by bacteria to enter a dormant state.

Richard Anthony obtained his Ph.D. from Kings College London in 1995 on the genotyping of the Malassezia yeasts. Shortly afterwards, he started working on methods to detect and track methicillin-resistant Staphylococcus aureus infections as well as improve the diagnosis of bacteremia. In 2001, he joined The Netherlands Royal Tropical Institute (KIT) in Amsterdam, where for 10 years he led a research group focused on simplifying and improving the diagnosis of tuberculosis and detection of drug resistance. This research group's output resulted in more than 50 papers and directly influenced national and international recommendations relating to tuberculosis control. In 2016, he joined the National Tuberculosis Reference Laboratory at the RIVM, where he now works on international collaborations and improving methods to control tuberculosis.

Stefan Niemann has studied Biology at the University of Bielefeld (Germany) and obtained his Dr. rer. nat. in 1996. From 1996 to 2006, he worked as a senior scientist at the National Reference Laboratory for Mycobacteria (NRL), Research Center Borstel (RCB) (Germany). In 2004, he became Assistant Professor (Privatdozent) at the University of Lübeck. Since 2006, he has been head of the Molecular and Experimental Mycobacteriology Group at RCB, and in 2012, he became Deputy Head of the Priority Area Infections and member of the extended board of directors of the RCB. In 2014, he was appointed W3 Professor for Molecular and Experimental Mycobacteriology at the University of Lübeck, and in 2017, he became one out of eight Schleswig-Holstein Excellence Chairs. In 2012, he received the Eva and Klaus Grohe Prize of the Berlin-Brandenburg Academy of Sciences and Humanities 2011, and in 2016, he received the Main Prize of the German Society for Hygiene and Microbiology. He coauthored more that 180 papers in peer-reviewed scientific papers, including high-impact journals such as The New England Journal of Medicine, Nature Genetics, The Lancet, and The Lancet Infectious Diseases.

Jan-Willem C. Alffenaar started his career in 2002 as pharmacist at the Department of Clinical Pharmacy and Pharmacology of the University Medical Center Groningen in the Netherlands. In 2008, he was board registered as a hospital pharmacist. In 2009, he finished his thesis entitled The Pharmacokinetics and TDM of Antimicrobial Agents, in Particular Antifungal Agents and Anti-TB Drugs. He has since been a Principal Investigator of many clinical studies evaluating the clinical pharmacology of antimicrobial drugs. In 2012, he finished his training in clinical pharmacology, and in 2013, he was appointed as associate professor at the Medical Faculty of the University of Groningen with the scope of optimizing antimicrobial therapy based on pharmacokinetics/pharmacodynamics (PK/PD) principles. He (co)authored over 150 papers in peer-reviewed scientific journals. He received the “young investigator award” from the International Association for Therapeutic Drug Monitoring and Clinical Toxicology in 2015.
REFERENCES
- 1.World Health Organization. 2015. Global tuberculosis report 2015. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 2.Longo DL, Horsburgh CR, Barry CE, Lange C. 2015. Treatment of tuberculosis. N Engl J Med 373:2149–2160. doi: 10.1056/NEJMra1413919. [DOI] [PubMed] [Google Scholar]
- 3.Nimmo C, Lipman M, Phillips PPJ, McHugh T, Nunn A, Abubakar I. 2015. Shortening treatment of tuberculosis: lessons from fluoroquinolone trials. Lancet Infect Dis 15:141–143. doi: 10.1016/S1473-3099(14)70885-0. [DOI] [PubMed] [Google Scholar]
- 4.McDermott W, McCune RM, Tompsett R. 1974. Dynamics of antituberculous chemotherapy. Am Rev Tuberc 74:100–108. [DOI] [PubMed] [Google Scholar]
- 5.Hu Y, Coates AR, Mitchison DA. 2006. Sterilising action of pyrazinamide in models of dormant and rifampicin-tolerant Mycobacterium tuberculosis. Int J Tuberc Lung Dis 10:317–322. [PubMed] [Google Scholar]
- 6.Gengenbacher M. 2012. Mycobacterium tuberculosis: success through dormancy. FEMS Microbiol Rev 36:514–532. doi: 10.1111/j.1574-6976.2012.00331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koul A, Arnoult E, Lounis N, Guillemont J, Andries K. 2011. The challenge of new drug discovery for tuberculosis. Nature 469:483–490. doi: 10.1038/nature09657. [DOI] [PubMed] [Google Scholar]
- 8.Lewis K. 2010. Persister cells. Annu Rev Microbiol 64:357–372. doi: 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
- 9.Gomez JE, McKinney JD. 2003. M. tuberculosis persistence, latency, and drug tolerance. Tuberculosis (Edinb) 84:29–44. doi: 10.1016/j.tube.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 10.Wayne LG. 1994. Dormancy of Mycobacterium tuberculosis and latency of disease. Eur J Clin Microbiol Infect Dis 13:908–914. doi: 10.1007/BF02111491. [DOI] [PubMed] [Google Scholar]
- 11.Kumar A, Toledo JC, Patel RP, Lancaster JR, Steyn AJC. 2007. Mycobacterium tuberculosis DosS is a redox sensor and DosT is a hypoxia sensor. Proc Natl Acad Sci U S A 104:11568–11573. doi: 10.1073/pnas.0705054104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Voskuila MI, Viscontib KC, Schoolnik GK. 2004. Mycobacterium tuberculosis gene expression during adaptation to stationary phase and low-oxygen dormancy. Tuberculosis 84:218–227. doi: 10.1016/j.tube.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 13.Galagan JE, Minch K, Peterson M, Lyubetskaya A, Azizi E, Sweet L, Gomes A, Rustad T, Dolganov G, Glotova I, Abeel T. 2013. The Mycobacterium tuberculosis regulatory network and hypoxia. Nature 499:178–183. doi: 10.1038/nature12337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manganelli R. 2007. Polyphosphate and stress response in mycobacteria. Mol Microbiol 65:258–260. doi: 10.1111/j.1365-2958.2007.05819.x. [DOI] [PubMed] [Google Scholar]
- 15.Dalebroux ZD, Svensson SL, Gaynor EC, Swanson MS. 2010. ppGpp conjures bacterial virulence. Microbiol Mol Biol Rev 74:171–199. doi: 10.1128/MMBR.00046-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sureka K, Dey S, Datta P, Singh AK, Dasgupta A, Rodrigue S, Basu J, Kundu M. 2007. Polyphosphate kinase is involved in stress-induced mprAB-sigE-rel signalling in mycobacteria. Mol Microbiol 65:261–276. doi: 10.1111/j.1365-2958.2007.05814.x. [DOI] [PubMed] [Google Scholar]
- 17.Chuang YM, Bandyopadhyay N, Rifat D, Rubin H, Bader JS, Karakousis PC. 2015. Deficiency of the novel exopolyphosphatase Rv1026/PPX2 leads to metabolic downshift and altered cell wall permeability in Mycobacterium tuberculosis. mBio 6:e02428-14. doi: 10.1128/mBio.02428-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dutta NK, Karakousis PC. 2014. Latent tuberculosis infection: myths, models, and molecular mechanisms. Microbiol Mol Biol Rev 78:343–371. doi: 10.1128/MMBR.00010-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chuang YM, Dutta NK, Hung CF, Wu TC, Rubin H, Karakousis PC. 2016. Stringent response factors PPX1 and PPK2 play an important role in Mycobacterium tuberculosis metabolism, biofilm formation, and sensitivity to isoniazid in vivo. Antimicrob Agents Chemother 60:6460–6470. doi: 10.1128/AAC.01139-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singh R, Singh M, Arora G, Kumar S, Tiwari P, Kidwai S. 2013. Polyphosphate deficiency in Mycobacterium tuberculosis is associated with enhanced drug susceptibility and impaired growth in guinea pigs. J Bacteriol 195:2839–2851. doi: 10.1128/JB.00038-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dahl JL, Kraus CN, Boshoff HI, Doan B, Foley K, Avarbock D, Kaplan G, Mizrahi V, Rubin H, Barry CE. 2003. The role of RelMtb-mediated adaptation to stationary phase in long-term persistence of Mycobacterium tuberculosis in mice. Proc Natl Acad Sci U S A 100:10026–10031. doi: 10.1073/pnas.1631248100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klinkenberg L, Lee J, Bishai W, Karakousis PC. 2010. The stringent response is required for full virulence of Mycobacterium tuberculosis in guinea pigs. J Infect Dis 202:1397–1404. doi: 10.1086/656524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sachdeva P, Misra R, Tyagi AK, Singh Y. 2009. The sigma factors of Mycobacterium tuberculosis: regulation of the regulators. FEBS J 277:605–626. doi: 10.1111/j.1742-4658.2009.07479.x. [DOI] [PubMed] [Google Scholar]
- 24.Balaban N, Merrin J, Chait R, Kowalik L, Leibler S. 2004. Bacterial persistence as a phenotypic switch. Science 305:1622–1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- 25.Kussell E, Kishony R, Balaban NQ, Leibler S. 2005. Bacterial persistence a model of survival in changing environments. Genetics 169:1807–1814. doi: 10.1534/genetics.104.035352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kussell E, Leibler S. 2005. Phenotypic diversity, population growth, and information in fluctuating environments. Science 309:2075–2078. doi: 10.1126/science.1114383. [DOI] [PubMed] [Google Scholar]
- 27.Hett EC, Rubin EJ. 2008. Bacterial growth and cell division: a mycobacterial perspective. Microbiol Mol Biol Rev 72:126–156. doi: 10.1128/MMBR.00028-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loris R, Garcia-Pino A. 2014. Disorder- and dynamics-based regulatory mechanisms in toxin-antitoxin modules. Chem Rev 114:6933–6947. doi: 10.1021/cr400656f. [DOI] [PubMed] [Google Scholar]
- 29.Sala A, Bordes P, Genevaux P. 2014. Multiple toxin-antitoxin systems in Mycobacterium tuberculosis. Toxins 6:1002–1020. doi: 10.3390/toxins6031002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vesper O, Amitai S, Belitsky M, Byrgazov K, Kaberdina AC, Engelberg-Kulka H, Moll I. 2011. Selective translation of leaderless mRNAs by specialized ribosomes generated by MazF in Escherichia coli. Cell 147:147–157. doi: 10.1016/j.cell.2011.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramage HR, Connolly LE, Cox JS. 2009. Comprehensive functional analysis of Mycobacterium tuberculosis toxin-antitoxin systems: implications for pathogenesis, stress responses, and evolution. PLoS Genet 5:e1000767. doi: 10.1371/journal.pgen.1000767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gopinath V, Raghunandanan S, Gomez RL, Jose L, Surendran A, Ramachandran R, Pushparajan AR, Mundayoor S, Jaleel A, Kumar RA. 2015. Profiling the proteome of Mycobacterium tuberculosis during dormancy and reactivation. Mol Cell Proteomics 14:2160–2176. doi: 10.1074/mcp.M115.051151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keren I, Minami S, Rubin E, Lewis K. 2011. Characterization and transcriptome analysis of Mycobacterium tuberculosis persisters. mBio 2:e00100-11. doi: 10.1128/mBio.00100-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tiwari P, Arora G, Singh M, Kidwai S, Narayan OP, Singh R. 2015. MazF ribonucleases promote Mycobacterium tuberculosis drug tolerance and virulence in guinea pigs. Nat Commun 6:6059. doi: 10.1038/ncomms7059. [DOI] [PubMed] [Google Scholar]
- 35.Korch SB, Contreras H, Clark-Curtiss JE. 2008. Three Mycobacterium tuberculosis Rel toxin-antitoxin modules inhibit mycobacterial growth and are expressed in infected human macrophages. J Bacteriol 191:1618–1630. doi: 10.1128/JB.01318-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singh R, Barry CE, Boshoff HIM. 2010. The three RelE homologs of Mycobacterium tuberculosis have individual, drug-specific effects on bacterial antibiotic tolerance. J Bacteriol 192:1279–1291. doi: 10.1128/JB.01285-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Albrethsen J, Agner J, Piersma SR, Højrup P, Pham TV, Weldingh K, Jimenez CR, Andersen P, Rosenkrands I. 2013. Proteomic profiling of Mycobacterium tuberculosis identifies nutrient-starvation-responsive toxin-antitoxin systems. Mol Cell Proteomics 12:1180–1191. doi: 10.1074/mcp.M112.018846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanz J, Navarro J, Arbués A, Martín C, Marijuán PC, Moreno Y. 2011. The transcriptional regulatory network of Mycobacterium tuberculosis. PLoS One 6:e22178. doi: 10.1371/journal.pone.0022178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raju RM, Unnikrishnan M, Rubin DH, Krishnamoorthy V, Kandror O, Akopian TN, Goldberg AL, Rubin EJ. 2012. Mycobacterium tuberculosis ClpP1 and ClpP2 function together in protein degradation and are required for viability in vitro and during infection. PLoS Pathog 8:e1002511. doi: 10.1371/journal.ppat.1002511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bald D, Koul A. 2010. Respiratory ATP synthesis: the new generation of mycobacterial drug targets? FEMS Microbiol Lett 308:1–7. doi: 10.1111/j.1574-6968.2010.01959.x. [DOI] [PubMed] [Google Scholar]
- 41.Zhang Y, Yew WW, Barer MR. 2012. Targeting persisters for tuberculosis control. Antimicrob Agents Chemother 56:2223–2230. doi: 10.1128/AAC.06288-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deb C, Lee CM, Dubey VS, Daniel J, Abomoelak B, Sirakova TD, Pawar S, Rogers L, Kolattukudy PE. 2009. A novel in vitro multiple-stress dormancy model for Mycobacterium tuberculosis generates a lipid-loaded, drug-tolerant, dormant pathogen. PLoS One 4:e6077. doi: 10.1371/journal.pone.0006077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.den Hertog AL, Visser DW, Ingham CJ, Fey FHAG, Klatser PR, Anthony RM. 2010. Simplified automated image analysis for detection and phenotyping of Mycobacterium tuberculosis on porous supports by monitoring growing microcolonies. PLoS One 5:e11008. doi: 10.1371/journal.pone.0011008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kaushik A, Singh UB, Porwal C, Venugopal SJ, Mohan A, Krishnan A, Goyal V, Banavaliker JN. 2012. Diagnostic potential of 16 kDa (HspX, α-crystalline) antigen for serodiagnosis of tuberculosis. Indian J Med Res 135:771–777. [PMC free article] [PubMed] [Google Scholar]
- 45.den Hertog AL, Menting S, Pfeltz R, Warns M, Siddiqi SH, Anthony RM. 2016. Pyrazinamide is active against Mycobacterium tuberculosis cultures at neutral pH and low temperature. Antimicrob Agents Chemother 60:4956–4960. doi: 10.1128/AAC.00654-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boshoff HI, Mizrahi V, Barry CE. 2002. Effects of pyrazinamide on fatty acid synthesis by whole mycobacterial cells and purified fatty acid synthase I. J Bacteriol 184:2167–2172. doi: 10.1128/JB.184.8.2167-2172.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peterson ND, Rosen BC, Dillon NA, Baughn AD. 2015. Uncoupling environmental pH and intrabacterial acidification from pyrazinamide susceptibility in Mycobacterium tuberculosis. Antimicrob Agents Chemother 59:7320–7326. doi: 10.1128/AAC.00967-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shi W, Zhang X, Jiang X, Yuan H, Lee JS, Barry CE, Wang H, Zhang W, Zhang Y. 2011. Pyrazinamide inhibits trans-translation in Mycobacterium tuberculosis. Science 333:1630–1632. doi: 10.1126/science.1208813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Winther KS, Gerdes K. 2011. Enteric virulence associated protein VapC inhibits translation by cleavage of initiator tRNA. Proc Natl Acad Sci U S A 108:7403–7407. doi: 10.1073/pnas.1019587108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wallis RS, Maeurer M, Mwaba P, Chakaya J, Rustomjee R, Migliori GB, Marais B, Schito M, Churchyard G, Swaminathan S, Hoelscher M. 2016. Tuberculosis—advances in development of new drugs, treatment regimens, host-directed therapies, and biomarkers. Lancet Infect Dis 16:e34–e46. doi: 10.1016/S1473-3099(16)00070-0. [DOI] [PubMed] [Google Scholar]
- 51.Gavrish E, Sit CS, Cao S, Kandror O, Spoering A, Peoples A, Ling L, Fetterman A, Hughes D, Bissell A, Torrey H. 2014. Lassomycin, a ribosomally synthesized cyclic peptide, kills Mycobacterium tuberculosis by targeting the ATP-dependent protease ClpC1P1P2. Chem Biol 21:509–518. doi: 10.1016/j.chembiol.2014.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Famulla K, Sass P, Malik I, Akopian T, Kandror O, Alber M, Hinzen B, Ruebsamen-Schaeff H, Kalscheuer R, Goldberg AL, Brötz-Oesterhelt H. 2016. Acyldepsipeptide antibiotics kill mycobacteria by preventing the physiological functions of the ClpP1P2 protease. Mol Microbiol 101:194–209. doi: 10.1111/mmi.13362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Conlon BP, Nakayasu ES, Fleck LE, LaFleur MD, Isabella VM, Coleman K, Leonard SN, Smith RD, Adkins JN, Lewis K. 2013. Activated ClpP kills persisters and eradicates a chronic biofilm infection. Nature 503:365–370. doi: 10.1038/nature12790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ling LL, Schneider T, Peoples AJ, Spoering AL, Engels I, Conlon BP, Mueller A, Schäberle TF, Hughes DE, Epstein S, Jones M. 2015. A new antibiotic kills pathogens without detectable resistance. Nature 517:455–459. doi: 10.1038/nature14098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Heijenoort J. 2007. Lipid intermediates in the biosynthesis of bacterial peptidoglycan. Microbiol Mol Biol Rev 71:620–635. doi: 10.1128/MMBR.00016-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Siricilla S, Mitachi K, Wan B, Franzblau SG, Kurosu M. 2014. Discovery of a capuramycin analog that kills nonreplicating Mycobacterium tuberculosis and its synergistic effects with translocase I inhibitors. J Antibiot (Tokyo) 68:271–278. doi: 10.1038/ja.2014.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ishizaki Y, Hayashi C, Inoue K, Igarashi M, Takahashi Y, Pujari V, Crick DC, Brennan PJ, Nomoto A. 2013. Inhibition of the first step in synthesis of the mycobacterial cell wall core, catalyzed by the GlcNAc-1-phosphate transferase WecA, by the novel caprazamycin derivative CPZEN-45. J Biol Chem 288:30309–30319. doi: 10.1074/jbc.M113.492173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Singh R, Manjunatha U, Boshoff HIM, Ha YH. 2008. PA-824 kills nonreplicating Mycobacterium tuberculosis by intracellular NO release. Science 322:1392–1395. doi: 10.1126/science.1164571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Haver HL, Chua A, Ghode P, Lakshminarayana SB, Singhal A, Mathema B, Wintjens R, Bifani P. 2015. Mutations in genes for the F420 biosynthetic pathway and a nitroreductase enzyme are the primary resistance determinants in spontaneous in vitro-selected PA-824-resistant mutants of Mycobacterium tuberculosis. Antimicrob Agents Chemother 59:5316–5323. doi: 10.1128/AAC.00308-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Diacon AH, Pym A, Grobusch M, Patientia R, Rustomjee R, Page-Shipp L, Pistorius C, Krause R, Bogoshi M, Churchyard G, Venter A. 2009. The diarylquinoline TMC207 for multidrug-resistant tuberculosis. N Engl J Med 360:2397–2405. doi: 10.1056/NEJMoa0808427. [DOI] [PubMed] [Google Scholar]
- 61.Haagsma AC, Abdillahi-Ibrahim R, Wagner MJ, Krab K, Vergauwen K, Guillemont J, Andries K, Lill H, Koul A, Bald D. 2009. Selectivity of TMC207 towards mycobacterial ATP synthase compared with that towards the eukaryotic homologue. Antimicrob Agents Chemother 53:1290–1292. doi: 10.1128/AAC.01393-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Koul A, Vranckx L, Dendouga N, Balemans W, Van den Wyngaert I, Vergauwen K, Göhlmann HW, Willebrords R, Poncelet A, Guillemont J, Bald D. 2008. Diarylquinolines are bactericidal for dormant mycobacteria as a result of disturbed ATP homeostasis. J Biol Chem 283:25273–25280. doi: 10.1074/jbc.M803899200. [DOI] [PubMed] [Google Scholar]
- 63.Bigger J. 1944. Treatment of staphylococcal infections with penicillin by intermittent sterilisation. Lancet 244:497–500. doi: 10.1016/S0140-6736(00)74210-3. [DOI] [Google Scholar]
- 64.Cogan NG. 2006. Effects of persister formation on bacterial response to dosing. J Theor Biol 238:694–703. doi: 10.1016/j.jtbi.2005.06.017. [DOI] [PubMed] [Google Scholar]
- 65.Menzies D, Benedetti A, Paydar A, Martin I, Royce S, Pai M, Vernon A, Lienhardt C, Burman W. 2009. Effect of duration and intermittency of rifampin on tuberculosis treatment outcomes: a systematic review and meta-analysis. PLoS Med 6:e1000146. doi: 10.1371/journal.pmed.1000146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reves R, Heilig CM, Tapy JM, Bozeman L, Kyle RP, Hamilton CD, Bock N, Narita M, Wing D, Hershfield E, Goldberg SV. 2014. Intermittent tuberculosis treatment for patients with isoniazid intolerance or drug resistance. Int J Tuberc Lung Dis 18:571–580. doi: 10.5588/ijtld.13.0304. [DOI] [PubMed] [Google Scholar]
- 67.Zierski M. 1979. Intermittent treatment regimens in pulmonary tuberculosis. Lung 156:17–32. doi: 10.1007/BF02713988. [DOI] [PubMed] [Google Scholar]
- 68.Burman W, Benator D, Vernon A, Khan A, Jones B, Silva C, Lahart C, Weis S, King B, Mangura B, Weiner M. 2006. Acquired rifamycin resistance with twice-weekly treatment of HIV-related tuberculosis. Am J Respir Crit Care Med 173:350–356. doi: 10.1164/rccm.200503-417OC. [DOI] [PubMed] [Google Scholar]
- 69.World Health Organization, UNAIDS. 2009. The treatment of tuberculosis: guidelines, 4th ed World Health Organization, Geneva, Switzerland. [Google Scholar]
- 70.Heemskerk D, Day J, Chau TT, Dung NH, Yen NT, Bang ND, Merson L, Olliaro P, Pouplin T, Caws M, Wolbers M. 2011. Intensified treatment with high dose rifampicin and levofloxacin compared to standard treatment for adult patients with tuberculous meningitis (TBM-IT): protocol for a randomized controlled trial. Trials 12:25. doi: 10.1186/1745-6215-12-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hu Y, Liu A, Ortega-Muro F, Alameda-Martin L, Mitchison D, Coates A. 2015. High-dose rifampicin kills persisters, shortens treatment duration, and reduces relapse rate in vitro and in vivo. Front Microbiol 6:641. doi: 10.3389/fmicb.2015.00641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Milstein M, Lecca L, Peloquin C, Mitchison D, Seung K, Pagano M, Coleman D, Osso E, Coit J, Vasquez DE, Garavito ES. 2016. Evaluation of high-dose rifampin in patients with new, smear-positive tuberculosis (HIRIF): study protocol for a randomized controlled trial. BMC Infect Dis 16:453. doi: 10.1186/s12879-016-1790-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Seidi K, Jahanban-Esfahlan R. 2013. A novel approach to eradicate latent TB: based on resuscitation promoting factors. J Med Hypotheses Ideas 7:69–74. doi: 10.1016/j.jmhi.2013.04.002. [DOI] [Google Scholar]
- 74.Gan Y, Yao Y, Guo S. 2015. The dormant cells of Mycobacterium tuberculosis may be resuscitated by targeting-expression system of recombinant mycobacteriophage-rpf: implication of shorter course of TB chemotherapy in the future. Med Hypotheses 84:477–480. doi: 10.1016/j.mehy.2015.01.046. [DOI] [PubMed] [Google Scholar]
- 75.Wallis R, van Vuuren C, Potgieter S. 2009. Adalimumab treatment of life-threatening tuberculosis. Clin Infect Dis 48:1429–1432. doi: 10.1086/598504. [DOI] [PubMed] [Google Scholar]
- 76.Mayanja-Kizza H, Jones-Lopez E, Okwera A, Wallis RS, Ellner JJ, Mugerwa RD. 2005. Immunoadjuvant prednisolone therapy for HIV-associated tuberculosis: a phase 2 clinical trial in Uganda. J Infect Dis 191:856–865. doi: 10.1086/427995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Giosue S, Casarini M, Ameglio F, Zangrilli P, Palla M, Altieri AM, Bisetti A. 2000. Aerosolized interferon-alpha treatment in patients with multi-drug-resistant pulmonary tuberculosis. Eur Cytokine Netw 11:99–104. [PubMed] [Google Scholar]
- 78.Johnson JL, Ssekasanvu E, Okwera A, Mayanja H, Hirsch CS, Nakibali JG, Jankus DD, Eisenach KD, Boom WH, Ellner JJ, Mugerwa RD. 2003. Randomized trial of adjunctive interleukin-2 in adults with pulmonary tuberculosis. Am J Respir Crit Care Med 168:185–191. doi: 10.1164/rccm.200211-1359OC. [DOI] [PubMed] [Google Scholar]
- 79.den Hertog AL, Menting S, van Soolingen D, Anthony RM. 2014. Mycobacterium tuberculosis Beijing genotype resistance to transient rifampin exposure. Emerg Infect Dis 20:1932–1933. doi: 10.3201/eid2011.130560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Betts JC, Lukey PT, Robb LC, McAdam RA, Duncan K. 2002. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol Microbiol 43:717–731. doi: 10.1046/j.1365-2958.2002.02779.x. [DOI] [PubMed] [Google Scholar]
- 81.Zhu L, Sharp JD, Kobayashi H, Woychik NA, Inouye M. 2010. Noncognate Mycobacterium tuberculosis toxin-antitoxins can physically and functionally interact. J Biol Chem 285:39732–39738. doi: 10.1074/jbc.M110.163105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dye C, Glaziou P, Floyd K, Raviglione M. 2013. Prospects for tuberculosis elimination. Annu Rev Public Health 34:271–286. doi: 10.1146/annurev-publhealth-031912-114431. [DOI] [PubMed] [Google Scholar]