Abstract
Despite being time-consuming and costly, generating genome-edited pigs holds great promise for agricultural, biomedical, and pharmaceutical applications. To further facilitate genome editing in pigs, we report here establishment of a pig line with Cre-inducible Cas9 expression that allows a variety of ex vivo genome editing in fibroblast cells including single- and multigene modifications, chromosome rearrangements, and efficient in vivo genetic modifications. As a proof of principle, we were able to simultaneously inactivate five tumor suppressor genes (TP53, PTEN, APC, BRCA1, and BRCA2) and activate one oncogene (KRAS), achieved by delivering Cre recombinase and sgRNAs, which caused rapid lung tumor development. The efficient genome editing shown here demonstrates that these pigs can serve as a powerful tool for dissecting in vivo gene functions and biological processes in a temporal manner and for streamlining the production of genome-edited pigs for disease modeling.
The CRISPR-Cas9 system is a powerful genome editing technology that uses the endonuclease Cas9 and single-guide RNAs (sgRNAs) to create double-strand breaks in the genome, which stimulate homologous recombination (Cong et al. 2013; Jiang et al. 2013; Mali et al. 2013; Varshney et al. 2015). Successful genome editing has been achieved by using the CRISPR-Cas9 in numerous mammals, including mouse (Shen et al. 2013; Wang et al. 2013), rat (Li et al. 2013), rabbit (Yang et al. 2014), sheep (Crispo et al. 2015), pig (Yang et al. 2015, 2016; Zhou et al. 2015; Lai et al. 2016; Whitworth et al. 2016; Niu et al. 2017), dog (Zou et al. 2015), and monkey (Niu et al. 2014; Wan et al. 2015). The current generation of gene-edited mammals is mostly based on either pronuclear injection or somatic cell nuclear transfer (SCNT) approaches, which are both expensive and time-consuming. It has been reported that direct in vivo genome editing could be achieved by the delivery of the expression vectors of both the Cas9 gene and sgRNAs directly into selected tissues, such as the lung (Blasco et al. 2014; Maddalo et al. 2014; Sanchez-Rivera et al. 2014), liver (Cheng et al. 2014; Xue et al. 2014; Yin et al. 2014), pancreas (Chiou et al. 2015), brain (Swiech et al. 2015), heart (Xie et al. 2016), or muscle (Long et al. 2016; Nelson et al. 2016; Tabebordbar et al. 2016) of adult mice through hydrodynamic or orthotopic injection for single and multiplexed genetic modifications. However, the overall editing efficiency was low because this approach was mediated by lentivirus or adeno-associated virus (AAV), which is inefficient to produce due to the large size of the Streptococcus pyogenes Cas9 (SpCas9) endonuclease gene (∼4.2 kb) (Kumar et al. 2001; Wu et al. 2010). To overcome this problem, Zhang and Sharp laboratories generated a mouse model where the Cre-dependent Cas9-expressing cassette was specifically inserted into the Rosa26 locus (Platt et al. 2014). Subsequently, Cre and sgRNAs targeting genes of interest were introduced to specific somatic cell types and created oncogenic mutations causing rapid lung cancer development. This mouse model also allowed other in vivo genome editing to be conveniently and efficiently performed (Chiou et al. 2015; Chu et al. 2016; Chow et al. 2017).
Genetically modified pigs are important in agriculture and in biomedical and pharmaceutical research (Fan and Lai 2013). Efforts to create genetically modified pigs have been substantially accelerated using CRISPR-Cas9 (Yang et al. 2015, 2016; Zhou et al. 2015; Lai et al. 2016; Whitworth et al. 2016; Niu et al. 2017). On the other hand, a Cre-dependent Cas9-expressing pig would provide an easy and efficient way to produce inducible genetic modifications, which should substantially facilitate studying gene functions, modeling human diseases, and promoting agricultural productivity.
Results
Generation of the Cre-dependent Cas9-expressing pigs
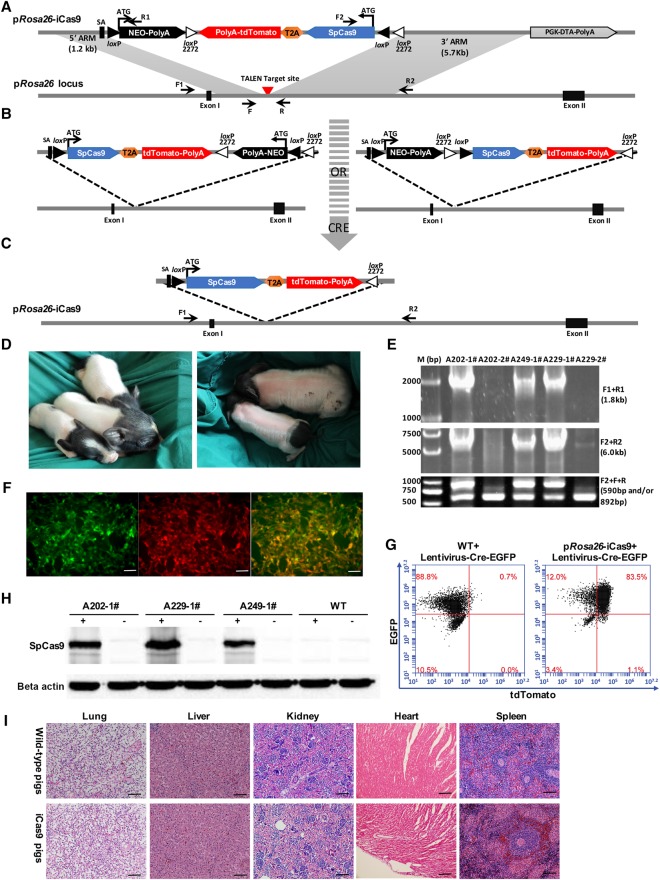
We aimed to express Cre-dependent Cas9 from the pig Rosa26 locus. We first constructed an expression cassette that included a pair of loxP sites, a pair of mutant loxP2272 sites, a viral splice acceptor (SA), a promoterless neomycin-resistance (Neo) gene, and an inverted SpCas9-T2A-tdTomato (iCas9) (Fig. 1A). To facilitate visualization of SpCas9-expressing cells, a tdTomato fluorescent protein was inserted downstream from SpCas9 via a self-cleaving T2A peptide. The loxP and mutant loxP2272 sites were arranged to flank the Neo and iCas9 genes, as shown in Figure 1A. Both loxP and loxP2272 sites are recognized by Cre recombinase but are incompatible with each other in recombination reactions. Given the specific position and orientation of pairs of loxP and loxP2272 sites, Cre recombinase-mediated recombination first induced inversion of the intervening DNA at either the loxP or loxP2272 site, thereby yielding a direct repeat of either two loxP or two loxP2272 sites (Fig. 1B). A further Cre-mediated excision will then irreversibly remove the Neo cassette along with its neighboring loxP or loxP2272 site and place the SpCas9 expression cassette under control of the endogenous pRosa26 promoter (pRosa26-iCas9) (Fig. 1C).
Figure 1.
Generation and characterization of Cre-dependent Cas9-expressing pigs. (A) A diagram for TALEN-mediated knock-in of Cre-dependent Cas9-expressing cassette into the pRosa26 locus. Gray triangles, wild-type loxP site; white triangles, mutant loxP2272 site; SA, splice acceptor; TALEN target site and PCR primers (F1, R1, F2, R2, F, and R) are indicated. (B,C) Schematic of two alternative patterns of Cre-mediated activation of SpCas9 and tdTomato: (B, left) Cre recombinase induces inversion of both Neo and iCas9 expression cassettes flanked by two loxP sites, followed by excision of Neo expression cassette flanked by two loxP2272 sites (C); (B, right) Cre recombinase-induced inversion of iCas9 expression cassettes by two loxP2272 sites, followed by excision of Neo expression cassette between two loxP sites (C). After inversion of iCas9 expression cassette and removal of the Neo expression cassette, SpCas9 and tdTomato expression are controlled by the endogenous porcine Rosa26 promoter (C). (D) Morphologically normal piglets were born from SCNT with the pRosa26-iCas9 PFFs. (E) PCR analysis confirmed the correct homologous recombination at the pRosa26 locus in 3/5 cloned piglets. Three positive piglets were all monoallelic modifications, as detected by PCR (F2 + F + R), consistent with those of cells chosen as nuclear donors. Primer pairs are shown in A and in Supplemental Table 3. (F) SpCas9 and tdTomato activations using Cre recombinase in fibroblasts isolated from the ear tissues of cloned piglets shown in D. Cells were infected with Cre-EGFP lentivirus, and the expression of tdTomato and EGFP were observed after 48 h by using a fluorescence microscope. Scale bars, 50 µm. (G) FACS analysis of Cre recombinase-induced tdTomato activation in pRosa26-iCas9 fibroblasts. (H) Western blot analysis was used to directly verify SpCas9 expression in pRosa26-iCas9 fibroblasts infected with lentivirus containing Cre. Cells not infected with Cre lentiviruses and WT cells were used as negative control. (I) H&E staining of the lung, liver, kidney, heart, and spleen of sacrificed wild-type and Cre-dependent Cas9-expressing piglets.
For gene targeting, primary porcine fetal fibroblasts (PFFs) derived from a 35-d-old fetus were electroporated with the linear targeting donor and pRosa26-TALENs targeting the intron 1 (Supplemental Fig. 1A). The cell colonies were selected out in G418 (1.0 mg/mL from Day 10 to Day 14) and genotyped. A total of 75 out of 101 colonies (75/101, 74.3%) were correctly targeted on the basis of 5′- and 3′-junction fragment PCR analyses and tdTomato expression induced by Cre recombinase (Supplemental Fig. 1B–D). Further sequencing analysis revealed that, among these 75 correctly targeted colonies, all colonies carried the knock-in cassette at one allele and an NHEJ-mediated mutation or no change at the other allele. No homozygous knock-in colonies were identified. To validate the functionality of iCas9, the correctly targeted cells were infected with the Cre recombinase-lentivirus, which led to high levels of tdTomato expression (Supplemental Fig. 1C).
Nineteen correctly targeted PFF cell colonies were selected as donor cells for SCNT. We pooled 8–10 colonies for one nuclear transfer, and the reconstructed embryos from the pooled cells were transferred to a surrogate. A total of 2214 cloned embryos were generated and transferred into 10 surrogate mothers (Supplemental Table 1). Three surrogates were confirmed pregnant by ultrasound examination 1 mo after the embryo transfer. In total, five cloned male piglets were born at term from these three pregnant surrogates after 120–130 d of gestation (Fig. 1D). Genetic characterization confirmed that three piglets carried the Cre-dependent Cas9 allele, identical to the donor cells (pRosa26-iCas9) (Fig. 1A,E). We next tested whether the cells in the cloned piglets expressed Cre-induced SpCas9 expression. Fibroblasts from the three live piglets were cultured and infected with lentiviruses expressing Cre recombinase and EGFP. As soon as 48 h after infection, tdTomato+/GFP+ cells were visible under fluorescence microscopy and were subsequently detected by flow cytometry (Fig. 1F,G; Supplemental Fig. 2A). The tdTomato+/GFP+ cells were FACS-sorted and analyzed for SpCas9 protein expression, which revealed that Cre recombinase in pRosa26-iCas9 fibroblasts induced efficient recombination between the two loxPs and the two loxP2272 sites, respectively, and led to robust SpCas9 protein expression (Fig. 1H). In addition, SpCas9 could also be activated by Cre recombinase in primary cells isolated from the heart, kidney, liver, brain, lung, and spleen of pRosa26-iCas9 piglets (Supplemental Fig. 2B).
To exclude the off-target effects of pRosa26-TALENs, we computationally predicted potential off-target sites using TALENoffer (http://galaxy.informatik.uni-halle.de/) to scan the porcine genomic sequence (Grau et al. 2013). A total of eight loci (with TALENoffer score ≥−1.60) were identified as potential off-target sites (Supplemental Table 2). Genomic DNA extracted from all three cloned piglets was used as a PCR template to amplify the potential off-target regions. T7EN1 cleavage assay and DNA sequencing results suggested that no DNA sequence changes were caused by the TALENs in these potential off-target sites in any of the three cloned piglets (Supplemental Fig. 3A–F).
The Cre-dependent Cas9-expressing founder pigs were healthy and grew up to maturation age without overt abnormalities. To further perform the histological examination of the lung, liver, kidney, heart, and spleen sections, one 3-mo-old founder piglet and one age-matched wild-type piglet were sacrificed. Again, no obvious differences were detected between the cloned pig carrying the pRosa26-iCas9 allele and the wild-type control (Fig. 1I). The pRosa26-iCas9 pig colony was established by mating one founder male pig with two wild-type sows. In the first litters, one sow delivered 12 piglets, and the other delivered nine. Of the 21 offspring, 12 (eight male and four female) carried the pRosa26-iCas9 allele (Supplemental Fig. 4A–C).
Ex vivo genome editing in pRosa26-iCas9 fibroblasts
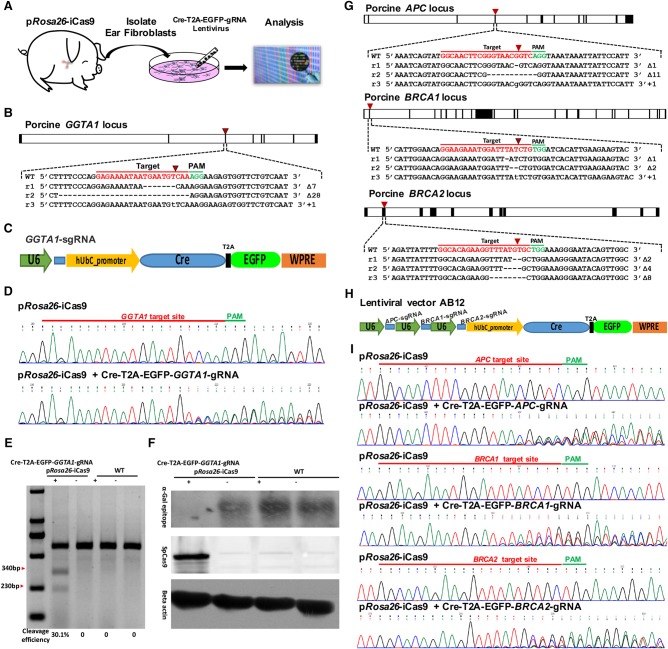
We next demonstrated the inactivation of endogenous genes in the primary porcine fibroblasts isolated from the ear tissue of the pRosa26-iCas9 allele piglets (Fig. 2A). The endogenous α-1,3-galactotransferase (GGTA1) gene, which is related to hyperacute rejection in xenotransplantation of pig organs into humans (Lai et al. 2002), was used as the first gene of interest. To target the GGTA1 locus, the Cre-U6-GGTA1-sgRNA lentivirus expressing Cre recombinase and the sgRNA specific for the exon 1 of porcine GGTA1 locus was used to infect the fibroblasts (Fig. 2B,C). At 7 d after transduction, genomic DNA was isolated and initially screened by PCR and T7EN1 cleavage assay for the presence of nucleotide changes surrounding the target sites at the GGTA1 locus (Fig. 2D,E). The genetic changes were further confirmed by sequencing the PCR products where 16 out of 20 sequenced subclones (80.0%) carried the nucleotide changes (Fig. 2B; Supplemental Fig. 6A). Western blot analysis showed that Gal-α-1,3-Gal (α-Gal) epitope expression in the collected fibroblasts significantly decreased (Fig. 2F).
Figure 2.
Ex vivo single- and multigene knockout in pRosa26-iCas9 fibroblasts. (A) Schematic diagram of ex vivo genome editing experimental workflow. First, pRosa26-iCas9 fibroblasts were isolated from the ear tissues of Cre-dependent Cas9-expressing pigs; second, the isolated pRosa26-iCas9 fibroblasts were infected with lentivirus containing Cre, EGFP, and specific sgRNAs; finally, the genome modifications in infected cells were analyzed at 1 wk posttransduction. (B) Design of sgRNA targeting porcine GGTA1 locus and three representative Sanger sequencing reads of subclones into T-vector from pRosa26-iCas9 fibroblasts. (C) A diagram of lentiviral vectors for Cre recombinase, EGFP, and GGTA1-sgRNA expression. (D) Sanger sequencing of PCR products containing GGTA1-sgRNA targeting site. Upper: pRosa26-iCas9 fibroblasts uninfected with lentivirus; bottom: pRosa26-iCas9 fibroblasts infected with lentivirus containing Cre recombinase, EGFP, and GGTA1-sgRNA. (E) GGTA1-sgRNA-mediated cleavage in wild-type and pRosa26-iCas9 fibroblasts infected or uninfected with lentivirus was analyzed by using a T7EN1 cleavage assay. (F) Western blot analysis for verifying α-Gal epitope and SpCas9 expression in wild-type and pRosa26-iCas9 fibroblasts infected or uninfected with lentivirus. Beta actin was used as a control. (G) Design of sgRNAs targeting early exons of porcine APC, BRCA1, or BRCA2, and three representative Sanger sequencing reads of subclones into T-vector from pRosa26-iCas9 fibroblasts infected with lentivirus AB12. (H) A diagram of lentiviral vector AB12 containing Cre recombinase, EGFP, APC-sgRNA, BRCA1-sgRNA, and BRCA2-sgRNA. (I) Sanger sequencing results of PCR products containing APC-sgRNA, BRCA1-sgRNA, and BRCA2-sgRNA targeting sites.
We next examined whether multiple genetic alterations can be introduced simultaneously into the pRosa26-iCas9 fibroblasts. pRosa26-iCas9 fibroblasts were infected with a lentivirus (AB12) that expressed Cre, EGFP, and three sgRNAs targeting exons of the porcine APC, BRCA1, and BRCA2 loci (Fig. 2G,H). Efficient cleavage at the respective target loci was detected (Fig. 2I; Supplemental Fig. 5A–C). Sanger sequencing of the amplified products from the targeted genomic regions revealed the indel mutations rates: 18/20 (90.0%) at the APC, 17/20 (85.0%) at the BRCA1, and 17/20 (85.0%) at the BRCA2 (Fig. 2G; Supplemental Fig. 6B–D).
Ex vivo oncogenic chromosomal rearrangements in pRosa26-iCas9 fibroblasts
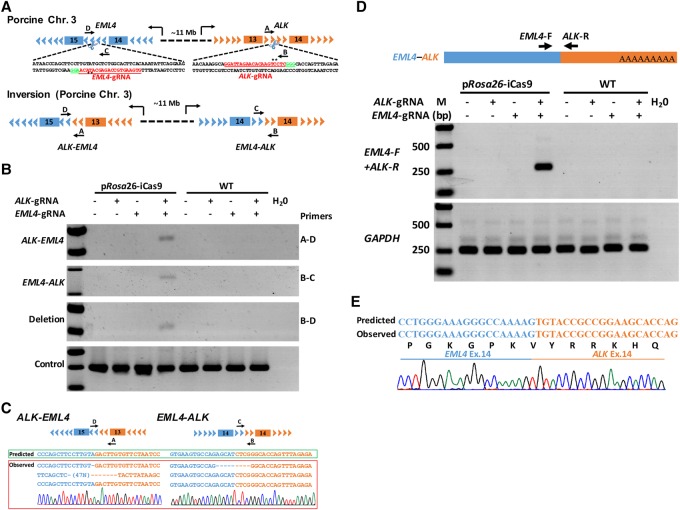
Chromosomal rearrangements between the genes encoding echinoderm microtubule associated protein like 4 (EML4) and anaplastic lymphoma kinase (ALK) were associated with the pathogenesis of human non-small cell lung cancer (NSCLC) and were among the most frequent rearrangements in solid human cancers (Soda et al. 2007). Modeling such genetic events in large animals has been proven challenging and requires complex manipulation in the germline. Although recent reports have described an efficient method to induce specific chromosomal rearrangements by using viral-mediated delivery of the CRISPR-Cas9 system to somatic cells in adult mice (Blasco et al. 2014; Maddalo et al. 2014), modeling of such genetic events in large animals has not been reported. We took advantage of the efficient pRosa26-iCas9 system to generate these chromosomal translocations in pigs. In the porcine genome, both EML4 and ALK are located on Chromosome 3, approximately 11 megabases (Mb) apart, in a region syntenic to human Chromosome 2 (Fig. 3A). We engineered two lentiviral vectors expressing Cre recombinase, EGFP, and the sgRNA for targeting intron 14 of the porcine EML4 gene (corresponding to intron 13 of the human EML4 gene and intron 14 of the mouse Eml4 gene), or the sgRNA for intron 13 of the porcine ALK gene (corresponding to intron 19 of the human ALK gene and the mouse Alk gene) (Fig. 3A; Supplemental Fig. 7A–C). pRosa26-iCas9 fibroblasts were transduced with lentiviruses expressing either individual sgRNA (EML4 or ALK sgRNA) or both (EML4 and ALK sgRNAs). One week postinfection, we identified and confirmed the EML4–ALK inversion (A-D and B-C primers), and the large deletion between the two cut sites (B-D primers) occurred in cells expressing both sgRNAs, but not in cells expressing only a single sgRNA (Fig. 3B,C). As predicted by chromosomal inversion, the EML4–ALK rearrangements should produce in-frame fusion of EML4–ALK mRNA transcripts with adjoined coding exons 1–14 of the EML4 gene and exons 14–23 of the ALK gene. The EML4–ALK mRNA fusion transcripts in the pig were expected to encode the same in-frame EML4–ALK chimeric protein as found in human NSCLC (Fig. 3D,E; Supplemental Fig. 7C). Therefore, large chromosomal rearrangements could be efficiently generated in the pRosa26-iCas9 fibroblasts and potentially achieved in vivo in the pRosa26-iCas9 pigs.
Figure 3.
Induction of EML4–ALK rearrangements in pRosa26-iCas9 fibroblasts. (A) Schematic representation of porcine EML4–ALK rearrangements induced by CRISPR-Cas9. EML4-sgRNA and ALK-sgRNA (red) were designed to target the mutation sites of the porcine EML4 gene intron 14 and porcine ALK gene intron 13. PCR primers are indicated (primers A, B, C, and D). (B) PCRs were performed to analyze ALK–EML4 (primers A and D were used) and EML4–ALK rearrangements (primers B and C were used) and large fragment deletion (primers B and D were used). The fragment amplified by primers A and B was used as positive control (bottom panel). (C) The ALK–EML4 and EML4–ALK PCR products were subcloned into T-vector, and the Sanger sequencing results of five independent clones and a representative chromatogram are shown in the left and right panels, respectively. (D) Diagram of EML4–ALK mRNA fusion transcripts (upper panel). Agarose gel electrophoresis analysis suggested that the RT-PCR products of EML4–ALK mRNA fusion transcripts only exist in pRosa26-iCas9 fibroblasts infected with both EML4-sgRNA and ALK-sgRNA; GAPDH was used as positive control (bottom panel). (E) The Sanger sequencing results of RT-PCR products showing that the sequences of EML4–ALK mRNA fusion transcripts are identical with predicted sequences (bottom panel).
Inducible genome editing in pRosa26-iCas9 fibroblasts
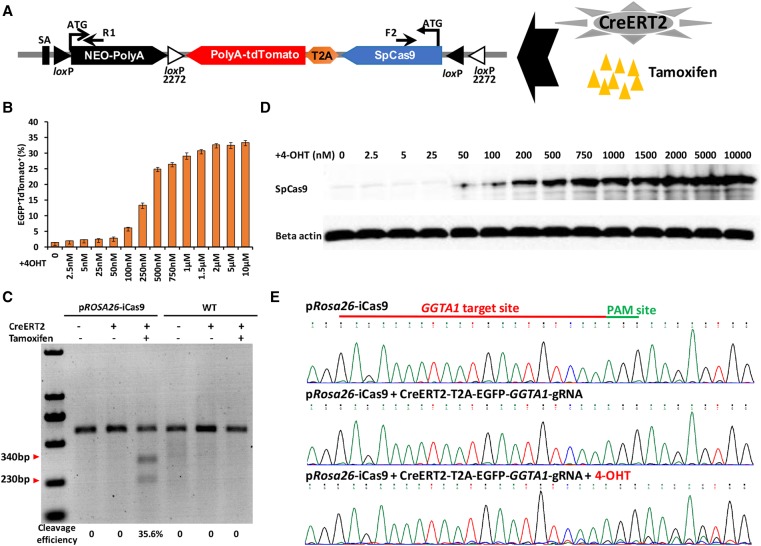
One major advantage of our pRosa26-iCas9 system is to allow temporal and conditional induction of expression of SpCas9, and thus, genome editing. We tested the system by using an external 4-hydroxytamoxifen (4-OHT) induction of Cre to tightly control SpCas9 expression (Fig. 4A). We infected pRosa26-iCas9 fibroblasts using a lentivirus expressing CreERT2 and EGFP. Seven days later, cells were treated with 4-OHT to induce Cre-loxP recombination, which led to SpCas9 and tdTomato expression. tdTomato+/GFP+ cells could be detected under a fluorescence microscope (Supplemental Fig. 8). In pRosa26-iCas9 fibroblasts, the activation efficiency of CreERT2 by 4-OHT was dependent on 4-OHT concentrations, with the optimal concentration at 2.0 µM (Fig. 4B,C; Supplemental Fig. 9). Importantly, the SpCas9 protein induced by 4-OHT catalyzed cleavages at the of GGTA1 locus (Fig. 4D,E). Therefore, the pRosa26-iCas9 system could permit temporal control of genome editing in the pig.
Figure 4.
Establishment and characterization of 4-OHT-inducible system in pRosa26-iCas9 fibroblasts. (A) Schematic of 4-OHT-induced SpCas9 and tdTomato expression in pRosa26-iCas9 fibroblasts infected with lentivirus containing CreERT2. (B) Percentage of EGFP- and tdTomato-positive cells under different concentrations of 4-OHT (0–10 µM). (C) Western blot analysis for verifying SpCas9 expression with different concentrations of 4-OHT inductions. (D) T7EN1 assays showing indel formation at the GGTA1 locus in pRosa26-iCas9 infected with lentivirus containing CreERT2, EGFP, and GGTA1-sgRNA and simultaneously supplied with 4-OHT, while not in uninfected or untreated fibroblasts. (E) Sanger sequencing analysis of the GGTA1-sgRNA targeting site. Top: pRosa26-iCas9 fibroblasts; middle, pRosa26-iCas9 fibroblasts infected with lentivirus containing CreERT2 and GGTA1-sgRNA, but not supplied with 4-OHT; bottom, pRosa26-iCas9 fibroblasts infected with lentivirus containing CreERT2 and GGTA1-sgRNA, simultaneously supplied with 4-OHT.
The pRosa26-iCas9 mediates efficient in vivo genome editing in a lung cancer model
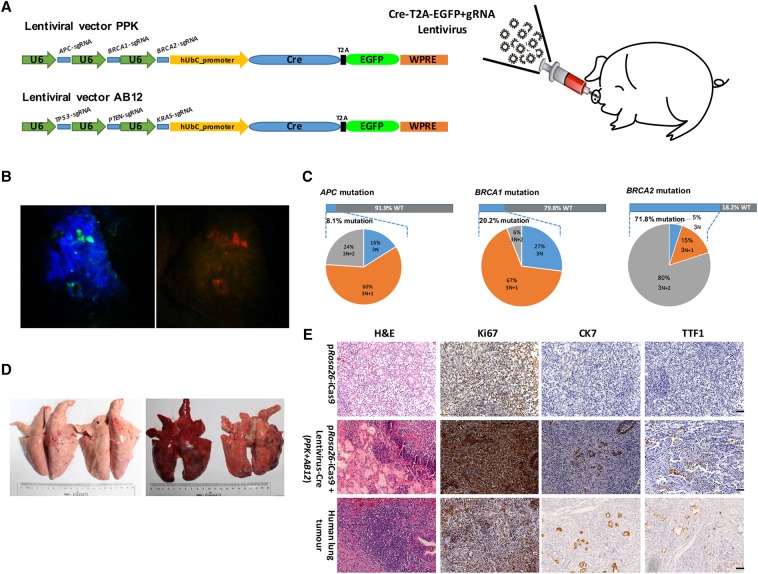
We next infected porcine ear tissues of a pRosa26-iCas9 pig with lentivirus (stereotactic injection subcutaneously) expressing Cre and sgRNAs targeting APC, BRCA1, and BRCA2 loci (Fig. 5A). After 3 wk of lentiviral inoculation, we used goggles to evaluate the Cre-loxP recombination in living piglets as previously reported (Deng et al. 2011) and observed high levels of tdTomato and EGFP fluorescence in the injected region (Fig. 5B). The ear fibroblasts were subsequently isolated from the tissue region positive for both tdTomato and EGFP fluorescence (Supplemental Fig. 10A). The frequency of cells expressing both tdTomato and EGFP determined by flow cytometry was ∼0.10% (Supplemental Fig. 10B). The FACS-sorted cells positive for both tdTomato and EGFP carried indel mutations near the predicted cleavage sites of all sgRNAs at the three loci at frequencies of 8.1% at the APC, 20.2% at the BRCA1, and 71.8% at the BRCA2 loci (Fig. 5C; Supplemental Fig. 11), but not in cells infected with empty lentiviruses.
Figure 5.
In vivo genome editing in the ear and lung tissues of Cre-dependent Cas9-expressing pigs. (A) Schematic of stereotactic delivery of lentiviruses AB12 and/or PPK into the ear and lung tissues of Cre-dependent Cas9-expressing piglets. (B) Florescence on ear tissues of Cre-dependent Cas9-expressing piglets infected with lentiviruses containing Cre, EGFP, and sgRNAs was directly observed using goggles. Left, EGFP fluorescence; right, tdTomato fluorescence. (C) Deep sequencing of sorted EGFP and tdTomato-positive cells show that all three sgRNAs could induce indel mutations near the predicted cleavage sites (8.10% of APC, 20.20% of BRCA1, and 71.80% of BRCA2), but not empty lentivirus infected cells. (D) Picture of the lungs from sacrificed Cre-dependent Cas9 expressing pigs infected (right) or uninfected (left) with lentivirus PPK and AB12. (E) Representative lung H&E staining and immunohistochemistry images of Cre-dependent Cas9-expressing pig injected with lentivirus PPK and AB12 at 3 mo posttransduction; tumor cells were stained positive for ki67 (an indicator of active cell cycle), CK7, and TTF1. Scale bar, 50 μm.
We next investigated the induction of cancer development in pRosa26-iCas9 pigs by targeting multiple cancer gene loci. We chose targeting tumor suppressor genes TP53, PTEN, APC, BRCA1, and BRCA2 and oncogene KRAS to induce multilesion lung cancer as these genes are the most frequently mutated ones in human lung cancer. To generate mutations at these loci, we constructed another lentiviral vector, PPK, which was capable of simultaneously targeting TP53, PTEN, and KRAS loci, in addition to the lentiviral vector AB12 expressing sgRNAs targeting APC, BRCA1, and BRCA2 loci (Fig. 5A). Multiple sgRNAs were designed to target exon 4 of TP53, exon 5 of PTEN, and exon 2 of KRAS (Supplemental Fig. 13A–C). The lentiviral particles (PPK and AB12) were administered to the lungs of two 1-wk-old F1 pRosa26-iCas9 pigs by intra-nasal delivery (Fig. 5A). At 3 mo after the infection, the Cre-dependent Cas9-expressing pigs presented lung cancer symptoms including cough, breathing difficulty, and weight loss (pRosa26-iCas9 piglets: 14.14 ± 2.53 kg, pRosa26-iCas9 piglets infected with PPK and AB12 lentiviruses: 10.11 ± 0.86 kg). The two infected Cre-dependent Cas9-expressing pigs and the two controls were sacrificed to retrieve the lungs. Macroscopic examination upon necropsy detected large tumors on the surface of the lung (one with 25 tumors and the other with 13 tumors) (Fig. 5D). The tumors with large size were sectioned for hematoxylin and eosin (H&E) staining and immunohistochemistry (IHC) analysis. Histological and IHC analyses of putative alveolar adenomas indicated that the cells in the majority of tumors (86.8%, 33/38) were strongly positive for Ki67 and were proliferative. Furthermore, many cells in most tumors expressed cytokeratin 7 (CK7) (81.6%, 31/38) and thyroid transcription factor-1 (TTF1) (73.7%, 28/38), two pulmonary adenocarcinoma markers (Su et al. 2006), which was similar to the human adenocarcinoma patient samples (Fig. 5E).
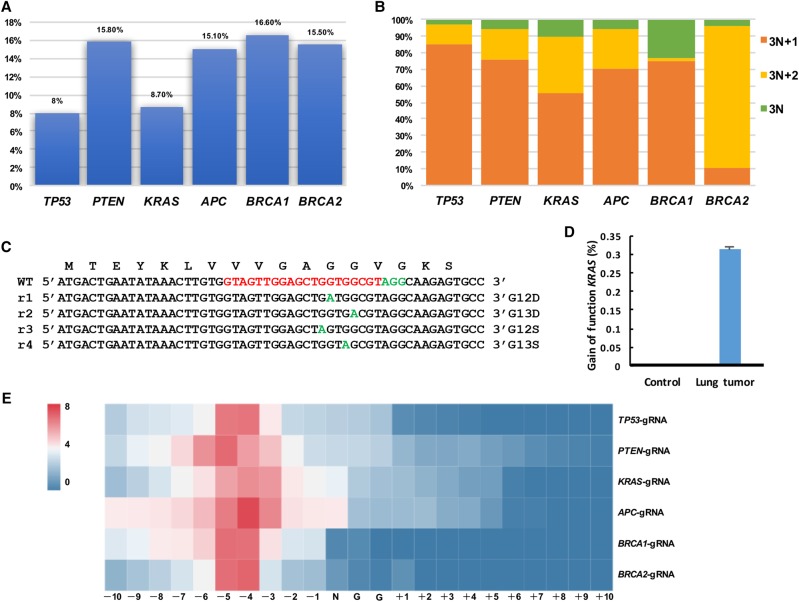
We next characterized genetic changes at the targeted loci in the lung tumors by performing captured Illumina deep sequencing of tumor genomic DNA and finding indels at the predicted cutting sites with the following frequencies: 8.0% at the TP53, 15.8% at the PTEN, 8.7% at the KRAS, 15.1% at the APC, 16.6% at the BRCA1, and 15.5% at the BRCA2 (Fig. 6A; Supplemental Figs. 12, 13A–F). Many of these indels caused potential frame shifts (i.e., 3n + 1 bp or 3n + 2 bp in length) and were located in the hotspot regions found in human cancers (Fig. 6B; Supplemental Fig. 13A–F; Supplemental Fig. 14A). Specific indels of tumor suppressor genes were also enriched in tumor cells, such as 76.7% indels of TP53 with +1-bp length, 72.2% indels of PTEN with +1- or −1-bp length, 68.9% indels of APC with +1- or −1-bp length, 59.8% indels of BRCA1 with +1-bp length, 81.7% indels of BRCA2 with −2-bp length, which suggested that these specific gene modifications may be the drivers for tumor formation (Supplemental Fig. 14B). Gain-of-function mutations of KRAS (GGT>GAT, GGT>AGT, GGC>GAC) were found with a frequency of ∼0.31% (0.31%/8.7%) (Fig. 6C,D). These missense mutations resulted in KRASG12D, KRASG12S, and KRASG13D mutations, which are frequently present in lung tumors and are very potent oncogenic mutations. To further confirm whether or not the selective expansion of the cancer cells contains gain-of-function KRAS mutations, ex vivo fibroblasts from pRosa26-iCas9 pigs were infected with lentiviruses PPK to test initial frequencies of these point mutations. At 1 wk postinfection, we collected the cells for deep sequencing. The initial frequency of these point mutations was 0.096% (0.096%/55.92%) (Supplemental Fig. 15A,B), a dozen times lower than that in lung cancer. These results suggested that the number of cancer cells with KRAS point mutations might undergo rapid expansion after initiating mutation. Furthermore, sequencing the control nontumor cells revealed that tumor samples were enriched with mutations within 7 bp upstream of the PAM sequences in the predicted cutting sites, thereby suggesting that the mutations detected in the tumor were unlikely to be secondary events or spontaneously arising mutations in tumor development (Fig. 6E). These data demonstrated that pRosa26-iCas9 pigs provide an efficient platform of generating mutations at multiple cancer gene loci in somatic cells for modeling human cancer.
Figure 6.
Mutation analysis in autochthonous lung tumors. (A) Efficiency of indel mutations in sectioned lung tumors was analyzed by deep sequencing. All six sgRNAs could induce indel mutations near the predicted cleavage sites (8.0% of TP53, 15.80% of PTEN, 8.70% of KRAS, 15.10% of APC, 16.60% of BRCA1, and 15.50% of BRCA2). (B) Calculation of mutation patterns (3N, 3N + 1, and 3N + 2) in sectioned lung tumors. (C,D) Gain-of-function mutations of KRAS in tumor cells. (E) Heat map analysis of the mutation efficiency at each position (−10 bp–+10 bp) around PAM sites with different sgRNAs.
Discussion
Genetically modified pigs have many potential applications in agriculture and biomedicine. However, the production of genetically modified pigs remains inefficient, expensive, and laborious, due to the unavailability of authentic germline-competent pluripotent stem cells (Fan and Lai 2013). In this study, Cre-dependent Cas9-expressing pigs were successfully generated via SCNT, providing a versatile large animal tool model to circumvent the current bottleneck and expand the CRISPR-Cas9 toolbox to facilitate powerful genome editing in pigs in vivo.
To guarantee consistently high Cas9 expression levels and overcome the silence of exogenous promoters (like CAG, CMV promoter) in transgenic pigs, the Cre-dependent Cas9-expression cassette was introduced into the porcine Rosa26 locus, which was previously identified as a safe harbor locus used for constitutive, ubiquitous gene expression (Li et al. 2014). To rigorously exclude potential problems associated with leaky transcription, instead of using the stop codons, we reversely oriented SpCas9-encoding cDNA relative to Rosa26 transcription (Irion et al. 2007; Li et al. 2007; Luche et al. 2007). Thus, the obtained Cre-dependent Cas9-expressing pigs did not show SpCas9 leaky expression, ensuring faithful SpCas9 and tdTomato activation for sophisticated gene modification and lineage-tracing experiments in vivo. The constitutive Cas9-expressing pigs appeared to be healthy and fertile. When the primary porcine fibroblasts isolated from the ear tissue of the Cre-dependent Cas9-expressing piglets were infected with lentivirus containing Cre recombinase and sgRNAs, single or multiple gene disruptions were efficiently generated, thereby indicating that the Cre-dependent Cas9-expressing construct can provide functional SpCas9 expression levels.
We used pRosa26-iCas9 fibroblasts for producing oncogenic chromosomal rearrangements, specifically modeling human EML4–ALK fusion, which is one of the most frequent rearrangements in solid human cancers; both EML4–ALK inversion and a large deletion of the region were readily detected. Therefore, the pRosa26-iCas9 fibroblasts can be useful for engineering large deletions, inversions, and chromosomal translocations in the porcine genome. The pRosa26-iCas9 pig also permitted inducible expression of Cas9, thereby leading to genome editing in endogenous genes. In addition, by crossing the pRosa26-iCas9 pig line with transgenic pigs that express Cre-recombinase in specific cell types/tissues, the pig lines expressing Cas9 protein in specific cell types/tissues can be established. Then, by introducing exogenous sgRNAs targeting specific genes to those pigs, temporal, spatial, and locus-specific controls of gene expression can be achieved in pigs.
Generating multigene modifications in the pig are technically challenging (Yang et al. 2015; Niu et al. 2017). The use of Cre-dependent Cas9-expressing pigs in conjunction with multiplex sgRNA delivery allowed us to readily introduce multiple genetic lesions in the same animals, which recapitulated multimutational nature in tumor development. Previous reports found that the Cre-dependent Cas9-expressing mice were effective tools to model the dynamics of multiple cancer mutations; however, cancers in mice and human are biologically different. Consequently, studies on murine models often do not translate into clinical success. Only 5% of anticancer drugs developed in preclinical studies based on traditional mouse models demonstrated sufficient efficacy in phase-III testing (Flisikowska et al. 2016). In contrast, pigs share many similarities with humans; for example, for the immune system, the similarity between mouse and human is <10%, whereas more than 80% similarity exists between human and pig (Bode et al. 2010; Meurens et al. 2012). The Cre-dependent Cas9-expressing pigs with primary tumors will provide a new platform for developing novel models of human cancer, which eventually facilitates new diagnostic and therapeutic technologies.
In summary, we generated Cre-dependent Cas9-expressing pigs as a versatile large animal model. In some proof-of-principle experiments, we demonstrated for the first time in the pig, efficient development of lung cancer owing to multiple mutations in tumor suppressor and oncogene loci. We expect that the Cre-dependent Cas9-expressing pig models will attract broad interest and substantially facilitate in vivo functional investigation of genes relevant to human disease and productivity in agriculture.
Methods
Animals
A local strain of Chinese Bama mini-pigs from Southern China was used as experimental subjects for gene targeting. The pigs were maintained under conventional housing conditions in the Animal Center of Guangzhou Institutes of Biomedicine and Health. Protocols involving the use of animals complied with the guidelines of the Institutional Animal Care and Use Committee at Guangzhou Institute of Biomedicine and Health, Chinese Academy of Sciences (Animal Welfare Assurance #A5748-01). All surgical procedures were performed under anesthesia by using propofol (2 mg/kg) or under anesthesia machines for further anesthesia (O2 flux: 3 L/min, isoflurane concentration: 3%). All efforts were made to minimize animal suffering.
Construction of pRosa26-TALENs and targeting vector
TALENs targeting porcine Rosa26 locus were designed and constructed through Golden Gate TALEN Assembly, as previously described (Cermak et al. 2011). A pFlexibleDT-pRosa26-iCas9 targeting vector was constructed on the basis of the pFlexibleDT-pRosa26-iEGFP targeting vector (Addgene; #60952) that we reported previously (Li et al. 2014). Briefly, we removed the EGFP sequence, added multiple clone sites (MCSs), including SalI, NotI, and MluI, into the pFlexibleDT-pRosa26-iEGFP targeting vector and obtained a new intermediate vector named pFlexibleDT-pRosa26-LN. The SpCas9-T2A-tdTomato cassettes were digested with SalI and NotI from the plasmid pCAG-SpCas9-T2A-tdTomato and inserted into the SalI- and NotI-digested pFlexibleDT-pRosa26-LN vectors. We obtained the final pFlexibleDT-pRosa26-iCas9 targeting vector. In summary, the pFlexibleDT-pRosa26-iCas9 targeting vector contains a 1.2-kb 5′ arm and a 5.6-kb 3′ arm of pRosa26, a viral SA, a promoterless Neo gene with a SV40 PolyA signal sequence, and an inverted SpCas9-T2A-tdTomato with a SV40 polyA signal sequence. The different directions of loxP and mutant loxP2272 sites were arranged to flank the Neo and inverted SpCas9-T2A-tdTomato expression cassette, which can result in the removal of the Neo gene expression cassette and inversion of the inverted SpCas9-T2A-tdTomato expression cassette after Cre-mediated recombination. This Cre-mediated recombination would place the SpCas9-T2A-tdTomato expression cassette directly under the control of the endogenous porcine Rosa26 promoter.
Generation and identification of pRosa26-iCas9 targeted PFF colonies
Isolation and electroporation of porcine fetal fibroblasts (PFF) were performed as previously described (Li et al. 2014; Yang et al. 2016). The transfected cells were divided into 20 10-cm culture dishes and recovered for 24 h. After cell recovery, 1 mg/mL G418 (Merck) was added to the PFF culture medium. After 8–12 d of selection, G418-resistant colonies were picked and cultured in 24-well plates by using cloning cylinders. Upon 70%–80% confluency, the cell colonies were subcultured, and 10% of each colony was lysed individually in 10 µL of NP-40 lysis buffer (0.45% NP-40 plus 0.6% Proteinase K) for 60 min at 56°C and then for 10 min at 95°C. The lysate was used as a template for PCR screening, which was performed using Long PCR Enzyme Mix (Thermo Scientific), in accordance with the manufacturer's instructions. The positive cell colonies were expanded and cryopreserved in liquid nitrogen for further SCNT.
Somatic cell nuclear transfer and generation of Cre-dependent Cas9-expressing pigs
The protocol of SCNT was performed as previously described (Li et al. 2014; Yang et al. 2016). Before transferring embryos, the reconstructed embryos were maintained in an embryo-development medium covered with mineral oil at 38.5°C for 20 h. The reconstructed embryos were then surgically transferred into the oviducts of surrogates the day after the estrus was observed. An ultrasound scanner was used to monitor the pregnancy status of the surrogates weekly after 1 mo of implantation, and the cloned piglets were delivered through natural birth. The genomic DNA extracted from the ear tissue of newborn piglets was used as a PCR template. The primers used for PCR genotyping were similar to those for cell colony genotyping.
Lentivirus vector design, production, and purification
Design and construction of GGTA1-sgRNA-, APC-sgRNA-, BRCA1-sgRNA-, BRCA2-sgRNA-, EML4-sgRNA-, ALK-sgRNA-, TP53-sgRNA-, PTEN-sgRNA-, and KRAS-sgRNA-expressing vectors were performed in accordance with a previously reported protocol (Zhou et al. 2015; Yang et al. 2016). Cre recombinase and EGFP expression cassette and two BsmBI restriction enzyme sites were inserted into FUGW (Addgene; #14883), and a new intermediate vector named FUGW-Cre-T2A-EGFP was obtained. The U6-sgRNA expression cassettes were PCR-amplified from the constructed U6-sgRNA vectors and cloned into the lentiviral vector FUGW-Cre-T2A-EGFP. Lentiviral vectors PPK and AB12 were constructed through Golden Gate Assembly as previously described (Cermak et al. 2011). To produce the lentiviruses, HEK293T cells were seeded in 5 × 106 cells per 10-cm culture dish the day before transfection in HEK293T culture media (DMEM supplemented with 10% FBS). For each dish, 12.5 µg of lentiviral and auxiliary packaging vectors, i.e., 7.5 µg of psPAX2 (Addgene; #12260) and 5 µg of pMD2.G (Addgene; #12259), were cotransfected into HEK293T cells by using a calcium phosphate transfection method according to a previously reported protocol (Zou et al. 2014). Lentiviruses were harvested after 48 h of transfection and concentrated by ultracentrifugation at 50,000g for 2.5 h at 4°C. After centrifugation, the supernatant was aspirated, and the pellet was resuspended in 200 µL of sterile PBS (Gibco) or Opti-MEM reduced serum medium (Gibco). Aliquots were then stored at −80°C for future use.
In vivo lentivirus transduction
Intra-nasal delivery and stereotactic injection of lentivirus were performed following a previous protocol (DuPage et al. 2009). In brief, 1-wk-old pRosa26-iCas9 piglets were anesthetized using isoflurane. For intra-nasal delivery, purified lentivirus solution was pipetted directly over the opening of one piglet nostril to dispense the virus dropwise until the entire virus volume has been inhaled. A titer of 5 × 106 infectious particles was administered to each pig. For stereotactic delivery into the ear, the 1 × 104 packaged lentiviruses were delivered into the subcutaneous tissue of the Cre-dependent Cas9-expressing porcine ears by using a 30G needle and syringe. The postoperative piglets were housed in a temperature-controlled environment (37°C) until ambulatory recovery.
Captured Illumina sequencing and indel analysis
Genomic DNA was extracted from cells and cancerlike tissues after being infected with lentivirus carrying Cre recombinase, EGFP, and sgRNAs-targeting specific locus using TIANGEN genomic DNA extraction kit in accordance with the recommended protocol. The extracted genomic DNA was used as PCR template for captured Illumina sequencing. Genomic PCR products were subjected to library preparation by using the Nextera XT DNA Sample Prep kit (Illumina) or customized barcoding methods. Briefly, low-cycle, first-round PCR was performed to amplify the target site. Second-round PCR was performed to add generic adapters, which were then used for third-round PCR for sample barcoding. Samples were pooled in equal amounts and purified using QiaQuick PCR Cleanup (QIAGEN) and then quantified using Qubit (Life Technologies). The mixed barcoded library was sequenced on an Illumina MiSeq System.
H&E and immunohistochemistry staining
The lung, liver, kidney, heart, and spleen tissues from the sacrificed wild-type and Cre-dependent Cas9-expressing piglets were fixed in 4% paraformaldehyde for 2 d. The fixed tissues were subsequently dissected, embedded in paraffin wax, and cross-sectioned at 3 µM. The sections were de-paraffinized with xylene and rehydrated using a graded series of alcohol (100%, 90%, 80%, 70%, and 50%), followed by H2O. For H&E staining, the rehydrated sections were stained with hematoxylin and eosin, differentiated, and then cover-slipped. For immunohistochemistry staining, sections were stained using standard IHC staining protocols as previously described. The following antibodies were used for IHC: anti-Ki67 (Novus; #NB500-170; 1:100), anti-CK7 (ZSGB-BIO; #ZA-0573), and anti-TTF (ZSGB-BIO; #ZM-0250).
Off-target analysis
For pRosa26-TALENs, the TALENoffer (http://galaxy.informatik.uni-halle.de/) was used to identify potential off-target sites in the porcine genome. The criteria for identifying off-target sites were TALENoffer score ≥ −1.60. A total of eight loci were identified as potential off-target sites. The sites with off-target sites were amplified and sequenced.
Data access
Sanger sequencing results are provided in Supplemental Material 2. The deep sequencing data from this study have been submitted to the NCBI Sequence Read Archive (SRA; https://www.ncbi.nlm.nih.gov/sra) under accession numbers SRX3029494, SRX3029499, SRX3029500, and SRX3029506.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (81672317, 81671121, 31601187, 31401271), the Bureau of International Cooperation, The Chinese Academy of Sciences (154144KYSB20150033), the Science and Technology Planning Project of Guangdong Province, China (2013B060300017, 2014B020225003, 2014B030301058, 2015A030310119, 2016A020216023, 2016B030229006), and the Bureau of Science and Technology of Guangzhou Municipality (201505011111498).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.222521.117.
References
- Blasco RB, Karaca E, Ambrogio C, Cheong TC, Karayol E, Minero VG, Voena C, Chiarle R. 2014. Simple and rapid in vivo generation of chromosomal rearrangements using CRISPR/Cas9 technology. Cell Rep 9: 1219–1227. [DOI] [PubMed] [Google Scholar]
- Bode G, Clausing P, Gervais F, Loegsted J, Luft J, Nogues V, Sims J, Project R. 2010. The utility of the minipig as an animal model in regulatory toxicology. J Pharmacol Toxicol Methods 62: 196–220. [DOI] [PubMed] [Google Scholar]
- Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia NV, Bogdanove AJ, Voytas DF. 2011. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res 39: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng R, Peng J, Yan Y, Cao P, Wang J, Qiu C, Tang L, Liu D, Tang L, Jin J, et al. 2014. Efficient gene editing in adult mouse livers via adenoviral delivery of CRISPR/Cas9. FEBS Lett 588: 3954–3958. [DOI] [PubMed] [Google Scholar]
- Chiou SH, Winters IP, Wang J, Naranjo S, Dudgeon C, Tamburini FB, Brady JJ, Yang D, Gruner BM, Chuang CH, et al. 2015. Pancreatic cancer modeling using retrograde viral vector delivery and in vivo CRISPR/Cas9-mediated somatic genome editing. Genes Dev 29: 1576–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow RD, Guzman CD, Wang G, Schmidt F, Youngblood MW, Ye L, Errami Y, Dong MB, Martinez MA, Zhang S, et al. 2017. AAV-mediated direct in vivo CRISPR screen identifies functional suppressors in glioblastoma. Nat Neurosci 20: 1329–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu VT, Graf R, Wirtz T, Weber T, Favret J, Li X, Petsch K, Tran NT, Sieweke MH, Berek C, et al. 2016. Efficient CRISPR-mediated mutagenesis in primary immune cells using CrispRGold and a C57BL/6 Cas9 transgenic mouse line. Proc Natl Acad Sci 113: 12514–12519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin SL, Barretto R, Habib N, Hsu PD, Wu XB, Jiang WY, Marraffini LA, et al. 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crispo M, Mulet AP, Tesson L, Barrera N, Cuadro F, dos Santos-Neto PC, Nguyen TH, Creneguy A, Brusselle L, Anegon I, et al. 2015. Efficient generation of myostatin knock-out sheep using CRISPR/Cas9 technology and microinjection into zygotes. PLoS One 10: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Yang DS, Zhao BT, Ouyang Z, Song J, Fan NN, Liu ZM, Zhao Y, Wu QH, Nashun B, et al. 2011. Use of the 2A peptide for generation of multi-transgenic pigs through a single round of nuclear transfer. PLoS One 6: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuPage M, Dooley AL, Jacks T. 2009. Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nat Protoc 4: 1064–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan NN, Lai LX. 2013. Genetically modified pig models for human diseases. J Genet Genomics 40: 67–73. [DOI] [PubMed] [Google Scholar]
- Flisikowska T, Kind A, Schnieke A. 2016. Pigs as models of human cancers. Theriogenology 86: 433–437. [DOI] [PubMed] [Google Scholar]
- Grau J, Boch J, Posch S. 2013. TALENoffer: genome-wide TALEN off-target prediction. Bioinformatics 29: 2931–2932. [DOI] [PubMed] [Google Scholar]
- Irion S, Luche H, Gadue P, Fehling HJ, Kennedy M, Keller G. 2007. Identification and targeting of the ROSA26 locus in human embryonic stem cells. Nat Biotechnol 25: 1477–1482. [DOI] [PubMed] [Google Scholar]
- Jiang WY, Bikard D, Cox D, Zhang F, Marraffini LA. 2013. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol 31: 233–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M, Keller B, Makalou N, Sutton RE. 2001. Systematic determination of the packaging limit of lentiviral vectors. Hum Gene Ther 12: 1893–1905. [DOI] [PubMed] [Google Scholar]
- Lai LX, Kolber-Simonds D, Park KW, Cheong HT, Greenstein JL, Im GS, Samuel M, Bonk A, Rieke A, Day BN, et al. 2002. Production of alpha-1,3-galactosyltransferase knockout pigs by nuclear transfer coning. Science 295: 1089–1092. [DOI] [PubMed] [Google Scholar]
- Lai SS, Wei S, Zhao BT, Ouyang Z, Zhang QJ, Fan NN, Liu ZM, Zhao Y, Yan QM, Zhou XQ, et al. 2016. Generation of knock-in pigs carrying Oct4-tdTomato reporter through CRISPR/Cas9-mediated genome engineering. PLoS One 11: e0146562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Tognon CE, Godinho FJ, Yasaitis L, Hock H, Herschkowitz JI, Lannon CL, Cho E, Kim SJ, Bronson RT, et al. 2007. ETV6-NTRK3 fusion oncogene initiates breast cancer from committed mammary progenitors via activation of AP1 complex. Cancer Cell 12: 542–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li DL, Qiu ZW, Shao YJ, Chen YT, Guan YT, Liu MZ, Li YM, Gao N, Wang LR, Lu XL, et al. 2013. Heritable gene targeting in the mouse and rat using a CRISPR-Cas system. Nat Biotechnol 31: 681–683. [DOI] [PubMed] [Google Scholar]
- Li X, Yang Y, Bu L, Guo X, Tang C, Song J, Fan N, Zhao B, Ouyang Z, Liu Z, et al. 2014. Rosa26-targeted swine models for stable gene over-expression and Cre-mediated lineage tracing. Cell Res 24: 501–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long CZ, Amoasii L, Mireault AA, McAnally JR, Li H, Sanchez-Ortiz E, Bhattacharyya S, Shelton JM, Bassel-Duby R, Olson EN. 2016. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science 351: 400–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luche H, Weber O, Nageswara Rao T, Blum C, Fehling HJ. 2007. Faithful activation of an extra-bright red fluorescent protein in “knock-in” Cre-reporter mice ideally suited for lineage tracing studies. Eur J Immunol 37: 43–53. [DOI] [PubMed] [Google Scholar]
- Maddalo D, Manchado E, Concepcion CP, Bonetti C, Vidigal JA, Han YC, Ogrodowski P, Crippa A, Rekhtman N, de Stanchina E, et al. 2014. In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system. Nature 516: 423–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang LH, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. 2013. RNA-guided human genome engineering via Cas9. Science 339: 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meurens F, Summerfield A, Nauwynck H, Saif L, Gerdts V. 2012. The pig: a model for human infectious diseases. Trends Microbiol 20: 50–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson CE, Hakim CH, Ousterout DG, Thakore PI, Moreb EA, Rivera RMC, Madhavan S, Pan XF, Ran FA, Yan WX, et al. 2016. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science 351: 403–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu YY, Shen B, Cui YQ, Chen YC, Wang JY, Wang L, Kang Y, Zhao XY, Si W, Li W, et al. 2014. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell 156: 836–843. [DOI] [PubMed] [Google Scholar]
- Niu D, Wei HJ, Lin L, George H, Wang T, Lee IH, Zhao HY, Wang Y, Kan Y, Shrock E, et al. 2017. Inactivation of porcine endogenous retrovirus in pigs using CRISPR-Cas9. Science 357: 1303–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt RJ, Chen S, Zhou Y, Yim MJ, Swiech L, Kempton HR, Dahlman JE, Parnas O, Eisenhaure TM, Jovanovic M, et al. 2014. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 159: 440–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Rivera FJ, Papagiannakopoulos T, Romero R, Tammela T, Bauer MR, Bhutkar A, Joshi NS, Subbaraj L, Bronson RT, Xue W, et al. 2014. Rapid modelling of cooperating genetic events in cancer through somatic genome editing. Nature 516: 428–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B, Zhang J, Wu HY, Wang JY, Ma K, Li Z, Zhang XG, Zhang PM, Huang XX. 2013. Generation of gene-modified mice via Cas9/RNA-mediated gene targeting. Cell Res 23: 720–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, Fujiwara S, Watanabe H, Kurashina K, Hatanaka H, et al. 2007. Identification of the transforming EML4–ALK fusion gene in non-small-cell lung cancer. Nature 448: 561–566. [DOI] [PubMed] [Google Scholar]
- Su YC, Hsu YC, Chai CY. 2006. Role of TTF-1, CK20, and CK7 immunohistochemistry for diagnosis of primary and secondary lung adenocarcinoma. Kaohsiung J Med Sci 22: 14–19. [DOI] [PubMed] [Google Scholar]
- Swiech L, Heidenreich M, Banerjee A, Habib N, Li Y, Trombetta J, Sur M, Zhang F. 2015. In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9. Nat Biotechnol 33: 102–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabebordbar M, Zhu KX, Cheng JKW, Chew WL, Widrick JJ, Yan WX, Maesner C, Wu EY, Xiao R, Ran FA, et al. 2016. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science 351: 407–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney GK, Pei WH, LaFave MC, Idol J, Xu LS, Gallardo V, Carrington B, Bishop K, Jones M, Li MY, et al. 2015. High-throughput gene targeting and phenotyping in zebrafish using CRISPR/Cas9. Genome Res 25: 1030–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan HF, Feng CJ, Teng F, Yang SH, Hu BY, Niu YY, Xiang AP, Fang WZ, Ji WZ, Li W, et al. 2015. One-step generation of p53 gene biallelic mutant Cynomolgus monkey via the CRISPR/Cas system. Cell Res 25: 258–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HY, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. 2013. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153: 910–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitworth KM, Rowland RR, Ewen CL, Trible BR, Kerrigan MA, Cino-Ozuna AG, Samuel MS, Lightner JE, McLaren DG, Mileham AJ, et al. 2016. Gene-edited pigs are protected from porcine reproductive and respiratory syndrome virus. Nat Biotechnol 34: 20–22. [DOI] [PubMed] [Google Scholar]
- Wu ZJ, Yang HY, Colosi P. 2010. Effect of genome size on AAV vector packaging. Mol Ther 18: 80–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C, Zhang YP, Song L, Luo J, Qi W, Hu J, Lu D, Yang Z, Zhang J, Xiao J, et al. 2016. Genome editing with CRISPR/Cas9 in postnatal mice corrects PRKAG2 cardiac syndrome. Cell Res 26: 1099–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W, Chen S, Yin H, Tammela T, Papagiannakopoulos T, Joshi NS, Cai W, Yang G, Bronson R, Crowley DG, et al. 2014. CRISPR-mediated direct mutation of cancer genes in the mouse liver. Nature 514: 380–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang DS, Xu J, Zhu TQ, Fan JL, Lai LX, Zhang JF, Chen YE. 2014. Effective gene targeting in rabbits using RNA-guided Cas9 nucleases. J Mol Cell Biol 6: 97–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang LH, Guell M, Niu D, George H, Lesha E, Grishin D, Aach J, Shrock E, Xu WH, Poci J, et al. 2015. Genome-wide inactivation of porcine endogenous retroviruses (PERVs). Science 350: 1101–1104. [DOI] [PubMed] [Google Scholar]
- Yang Y, Wang K, Wu H, Jin Q, Ruan D, Ouyang Z, Zhao B, Liu Z, Zhao Y, Zhang Q, et al. 2016. Genetically humanized pigs exclusively expressing human insulin are generated through custom endonuclease-mediated seamless engineering. J Mol Cell Biol 8: 174–177. [DOI] [PubMed] [Google Scholar]
- Yin H, Xue W, Chen S, Bogorad RL, Benedetti E, Grompe M, Koteliansky V, Sharp PA, Jacks T, Anderson DG. 2014. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat Biotechnol 32: 551–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou XQ, Xin JG, Fan NN, Zou QJ, Huang J, Ouyang Z, Zhao Y, Zhao BT, Liu ZM, Lai SS, et al. 2015. Generation of CRISPR/Cas9-mediated gene-targeted pigs via somatic cell nuclear transfer. Cell Mol Life Sci 72: 1175–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou QJ, Yan QM, Zhong J, Wang KP, Sun HT, Yi XL, Lai LX. 2014. Direct conversion of human fibroblasts into neuronal restricted progenitors. J Biol Chem 289: 5250–5260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou QJ, Wang XM, Liu YZ, Ouyang Z, Long HB, Wei S, Xin JG, Zhao BT, Lai SS, Shen J, et al. 2015. Generation of gene-target dogs using CRISPR/Cas9 system. J Mol Cell Biol 7: 580–583. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.