Abstract
The centromere is the structural unit responsible for the faithful segregation of chromosomes. Although regulation of centromeric function by epigenetic factors has been well-studied, the contributions of the underlying DNA sequences have been much less well defined, and existing methodologies for studying centromere genomics in biology are laborious. We have identified specific markers in the centromere of 23 of the 24 human chromosomes that allow for rapid PCR assays capable of capturing the genomic landscape of human centromeres at a given time. Use of this genetic strategy can also delineate which specific centromere arrays in each chromosome drive the recruitment of epigenetic modulators. We further show that, surprisingly, loss and rearrangement of DNA in centromere 21 is associated with trisomy 21. This new approach can thus be used to rapidly take a snapshot of the genetics and epigenetics of each specific human centromere in nondisjunction disorders and other biological settings.
The centromere is a structural unit vital for faithful segregation of chromosomes during cell division, and destabilization of centromere function results in chromosomal missegregation, genomic instability, and aneuploidy, hallmarks of cancers as well as birth defects. Centromere sequences are mainly composed of 171-bp α-satellite repeat units, termed monomers, organized in a head-to-tail fashion to form high-order repeats (HORs). Centromere arrays consist of multiple HORs organized in tandem that can extend for several megabases (Verdaasdonk and Bloom 2011; Hayden 2012; Hayden and Willard 2012; Aldrup-Macdonald and Sullivan 2014; Supplemental Fig. S1). Interestingly, during human evolution, the HORs became homogeneous in each centromere core; today, HORs of a given centromere array are 98–100% similar/identical, but there is only ∼75% similarity among centromere arrays on different chromosomes (Choo et al. 1989; Vissel and Choo 1992; Jørgensen 1997; Roizès 2006). The α-satellite monomers are only 50–70% similar across the centromeres (Aldrup-MacDonald and Sullivan 2014). Although the sequences of HORs within a centromere array can be as much as 98% similar, the constituent monomers of each HOR may show 20–40% divergence among themselves (Waye and Willard 1986b). In general, the landscape of the centromere is delineated by the large α-repeat arrays. Repetitive sequences represent a major challenge when assembling centromere sequences; thus, human centromeres remain significant gaps in our knowledge of human genomics (Zeitlin 2010; Hood and Rowen 2013).
Toward the periphery of the centromere (the pericentromere), the sequence is more diverse, with smaller arrays or monomers of alphoid and other repeats, as well as transposon-like elements (Bersani et al. 2015; Miga 2015). Two of these elements are the human endogenous retroviruses K111 and K222, identified by our group (Supplemental Fig. S1; Contreras-Galindo et al. 2011, 2013; Zahn et al. 2015). It had previously been hypothesized that ∼30% of pericentric sequences originated from segmental duplications in centromeric regions within chromosomes, many of which then subsequently duplicated interchromosomally (Horvath et al. 2003; Kirsch et al. 2005), and we have recently provided evidence that this is indeed the case with K111 and K222 (Contreras-Galindo et al. 2013; Zahn et al. 2015). These elements have helped us understand the changes pericentromere sequences have undergone, shedding light on substantial recombination between different centromeres during evolution, areas of the genome often thought to be recalcitrant to the exchange of genetic material (Talbert and Henikoff 2010).
Studies aimed at understanding human centromeres to date have primarily focused on the epigenetic components that orchestrate centromere function (Amor and Choo 2002; Burrack and Berman 2012). CENPA, a histone-3-like protein that is only deposited at functional centromeres, is a key epigenetic factor that determines centromere identity and propagation. CENPA nucleosomes also serve as a platform for kinetochore formation (Supplemental Fig. S1). CENPA can establish functional neocentromeres at ectopic locations that lack canonical centromeric DNA sequence (e.g., human α-satellite repeats) (Amor and Choo 2002; Stimpson and Sullivan 2010; Burrack and Berman 2012; Scott and Sullivan 2014). However, neocentromere formation in humans has been documented only sporadically, in particular in a few developmental defects and cancers (Amor and Choo 2002; Mackinnon and Campbell 2011; Burrack and Berman 2012). CENPA nucleosomes at functional centromere sequences interact with CENPB, a protein that binds specifically to CENPB boxes, 17-nt sequences present in the majority of α-repeats, to stabilize CENPA nucleosomes (Supplemental Fig. S1; Ohzeki et al. 2002; Rosandić et al. 2006; Fachinetti et al. 2015, Fujita et al. 2015). The centromere protein CENPB also stabilizes and maintains cohesion between sister chromatids in pericentric heterochromatin, a separable function that is also required for proper chromosome segregation (Tanno et al. 2015).
Recent studies have suggested that centromere sequences, and potentially cognate RNA transcripts, contribute to centromere function and propagation, in addition to epigenetic mechanisms (Malik and Henikoff 2009; Maloney et al. 2012; Biscotti et al. 2015; Henikoff et al. 2015). Henikoff and colleagues have shown that in CENPA alphoid nucleosomes, CENPB/CENPC linkers bind to DNA boxes every other ∼340-bp of alphoid sequence, which is precisely wrapped around CENPA in two 100-bp segments, thus demonstrating a link between the genetic and epigenetic processes that directly affect centromere biology (Henikoff et al. 2015). The occupancy of CENPA and CENPB diminishes toward the pericentromere, where H3 trimethylated at lysine 9 (H3K9me3) becomes the major epigenetic mark and is found in the chromatin associated with pericentromeric elements such as K111 and K222 (Supplemental Fig. S1; Contreras-Galindo et al. 2013; Zahn et al. 2015). Centromeric protein marks have evolved to maintain their binding affinity for the constantly changing centromere α-repeats (Malik and Henikoff 2009). Thus, like epigenetic factors, canonical centromere DNA sequences indeed appear to be necessary for centromere function.
Methods for studying the variation of centromere arrays have relied primarily on Southern blot. Using this method, alphoid arrays have been shown to exist in different lengths in the human population (Alexandrov et al. 2001; Liehr 2013). However, Southern blots are laborious, time-consuming, and require large amounts of DNA, and thus are not well-suited to efficiently studying the evolution of multiple individual centromeres in health and disease. To overcome this obstacle, we developed rapid, PCR-based methods to specifically detect and quantitate the length and abundance of satellite arrays, and now can successfully identify the main alphoid arrays characteristic of 23 of 24 human centromeres simultaneously (centromere 19 being the exception). These 30-min assays can be used to successfully study the dynamics and evolution of human centromere sequences within a time frame that enables widespread analysis. We also developed PCR assays to study pericentromere-specific K111 and K222 sequences, which enable us to examine the evolution of human pericentromeres (Contreras-Galindo et al. 2013; Zahn et al. 2015).
Results
Real-time PCR assays to study human centromeres
In order to develop specific markers for the major centromeric arrays and understand their structure and function, we performed a systematic analysis to identify specific DNA variations in every major α-repeat array and design primers for PCR assays. The PCR assays amplify sequences within the monomers of each array. By convention, the nomenclature of these α-repeat arrays starts with the letter D, followed by the chromosome number in which they reside (1–22, X or Y) as determined by fluorescent in situ hybridization (FISH), followed by a Z, and then a number indicating the order in which the sequence was discovered. We developed rapid qPCR assays for these unique markers, enabling us to detect and estimate the abundance of α-repeats in each array of the centromere of 23 of the 24 human chromosomes (Fig. 1; also see discussion below). These arrays have been previously assigned to the centromeres of specific chromosomes by FISH studies (Liehr 2013). We further developed an assay to detect the array p82H that is present in all human centromeres (Mitchell et al. 1985). To verify the specificity of the PCR assays, we used DNA from somatic rodent/human hybrids, each hybrid containing one single human chromosome. The primers and probes used in this study are given in Supplemental Tables 1 and 2. In some cases, the PCR assays detected arrays previously reported to exist in a unique chromosome on another chromosome, albeit with low levels of detection (Supplemental Table 2). This was particularly interesting in DNA from Y, where we detected α-repeat arrays from Chromosomes 1, 5, and 19 (D1Z7/D5Z2/D19Z3), Chr 4 (D4Z1), Chromosomes 5 and 19 (D5Z1/D19Z2), Chr 7 (D7Z1), Chr 9 (D9Z4), Chromosomes 13 and 21 (D13Z1/D21Z1), Chr 17 (D17Z1), and Chr 18 (D18Z1 and D18Z2) (Supplemental Fig. S2; Supplemental Table 2). The existence of these α-repeat arrays in Y was verified by sequencing the PCR products. These findings suggest that Chr Y may have exchanged material relatively frequently with the centromeres of somatic chromosomes during human evolution.
Figure 1.
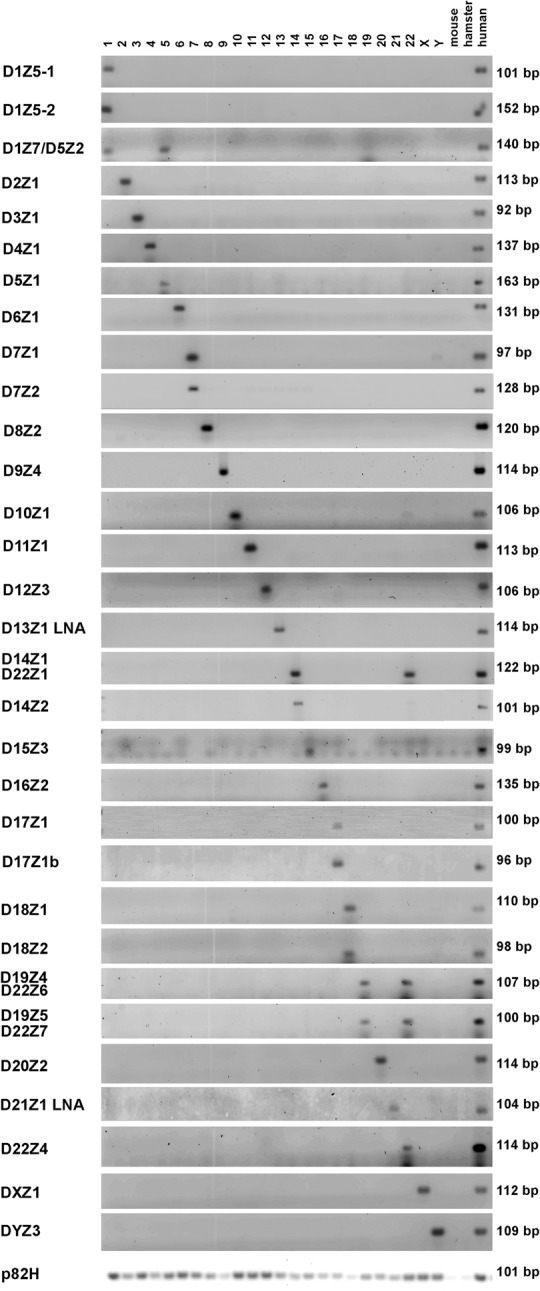
Detection of centromere α-repeat arrays in individual human chromosomes. Representative α-repeat arrays in each human chromosome (y-axis) were detected and the number of repeats quantitated by qPCR using specific primers. Gel electrophoresis of qPCR products amplified from DNA of human/rodent hybrid cells, with each hybrid cell containing only one human chromosome (displayed on the x-axis). DNA from rodent parental mouse or hamster cells is included to control for cross-species hybridization of repeats along with human DNA isolated from peripheral blood lymphocytes that served as a positive control. Water was used as an additional negative control. Using the primers and qPCR conditions described (Supplemental Tables 1, 2), specific centromeric α-repeat arrays were identified for each human chromosome (i.e., D2Z1, D3Z1, D4Z1…). Certain α-repeat arrays were found in two or more chromosomes (i.e., D1Z7/D5Z2 in Chromosomes 1 and 5, D14Z1/D22Z1 in Chromosomes 14 and 22, and D19Z4/D22Z6 and D19Z5/D22Z7 in Chromosomes 19 and 21). Primers specific for the ubiquitous α-repeat p82H amplified centromeres from all human chromosomes. Assays for the D13Z1 and D21Z1 arrays in this figure use LNA primers as shown in Supplemental Figures S4 and S5. The data shown in this figure are a composite from experiments run over time and demonstrate the results obtained once conditions for each chromosome had been optimized.
The identification of specific markers was challenging for certain human chromosomes with homologous centromeres. For example, Chromosomes 1, 5, and 19; 4 and 9; 13 and 21; and 14 and 22 contain centromere sequences with 99%–100% similarity (Hayden 2012). However, by targeting specific nucleotide sequences found in the monomers of a given array that are not found on other centromere arrays, optimizing PCR conditions, or using locked nucleic acid (LNA) technology (Ballantyne et al. 2008), we successfully designed assays for these centromeres. We further screened α-repeat arrays identified recently and annotated in the most recent human genome assembly, hg38, for centromeres 13, 14, 21, and 22 (Miga et al. 2014; Miga 2015). We identified some of these α-repeat arrays as uniquely existing on either centromere 14 or 22 (Supplemental Fig. S3), enabling us to design highly specific PCR reactions for the arrays of Chromosomes 14 and 22 also. We developed assays using LNA technology that successfully discriminate between the arrays D13Z1 and D21Z1 in centromeres 13 and 21 (Supplemental Figs. S4, S5).
Thus far, we have developed qPCRs to study α-repeat arrays for every human chromosome except Chromosome 19 (Liehr 2013; Miga et al. 2014; Miga 2015). Most of the PCR assays designed targeted the largest α-repeat array found in the centromere core of each chromosome, and therefore the one most likely to establish a kinetochore (arrays D1Z7/D5Z2, D2Z1, D3Z1, D4Z1, D5Z1, D6Z1, D7Z1, D7Z2, D8Z2, D9Z4, D10Z1, D11Z1, D12Z3, D13Z1/D21Z1, D14Z1/D22Z1, D15Z3, D16Z2, D17Z1, D17Z1b, D18Z1, D18Z2, D20Z2, DXZ1, and DYZ3) (Liehr 2013; Miga et al. 2014; Miga 2015). We also developed assays specific for pericentromeric arrays that surround the larger centromere arrays in the core (D1Z5, D13Z2, D13Z3, D13Z6, D13Z7, D13Z8, D13Z9, D14Z2, D14Z3, D19Z4, D19Z5, D22Z4, and D22Z5; see Supplemental Table 2) (Liehr 2013; Miga et al. 2014; Miga 2015).
K111 exists in the pericentromeres (likely at the centromere/pericentromere border as K111 associates with CENPA and CENPB, but also with H3K9 trimethylated chromatin) of 15 human chromosomes. K222 exists in the pericentromere of nine human chromosomes, which contain K111 proviruses as well. We developed PCR assays to quantitate these proviruses and use these markers to study pericentric regions. Therefore, we can now rapidly assess specific changes at the centromeric core and at the pericentromere in nearly all human centromeres.
Validation of PCR-based techniques to identify specific centromeres
In order to validate the accuracy of our new approach, we first used our PCR assays that quantitate the abundance of α-repeats in each array to determine the size of each centromere array in DNA isolated from the peripheral blood lymphocytes of five individuals (Supplemental Fig. S6). The assays show that arrays at the centromere core are larger than arrays at pericentromere loci. The PCR assays also show that the lengths of these arrays vary in these individuals. An estimate of the average of the size of each array in these subjects is shown in Table 1. Interestingly, as previously noted by others, the sizes of the arrays do not correlate significantly with the size of the chromosomes (for review, see Liehr 2013).
Table 1.
Size of human centromere arrays
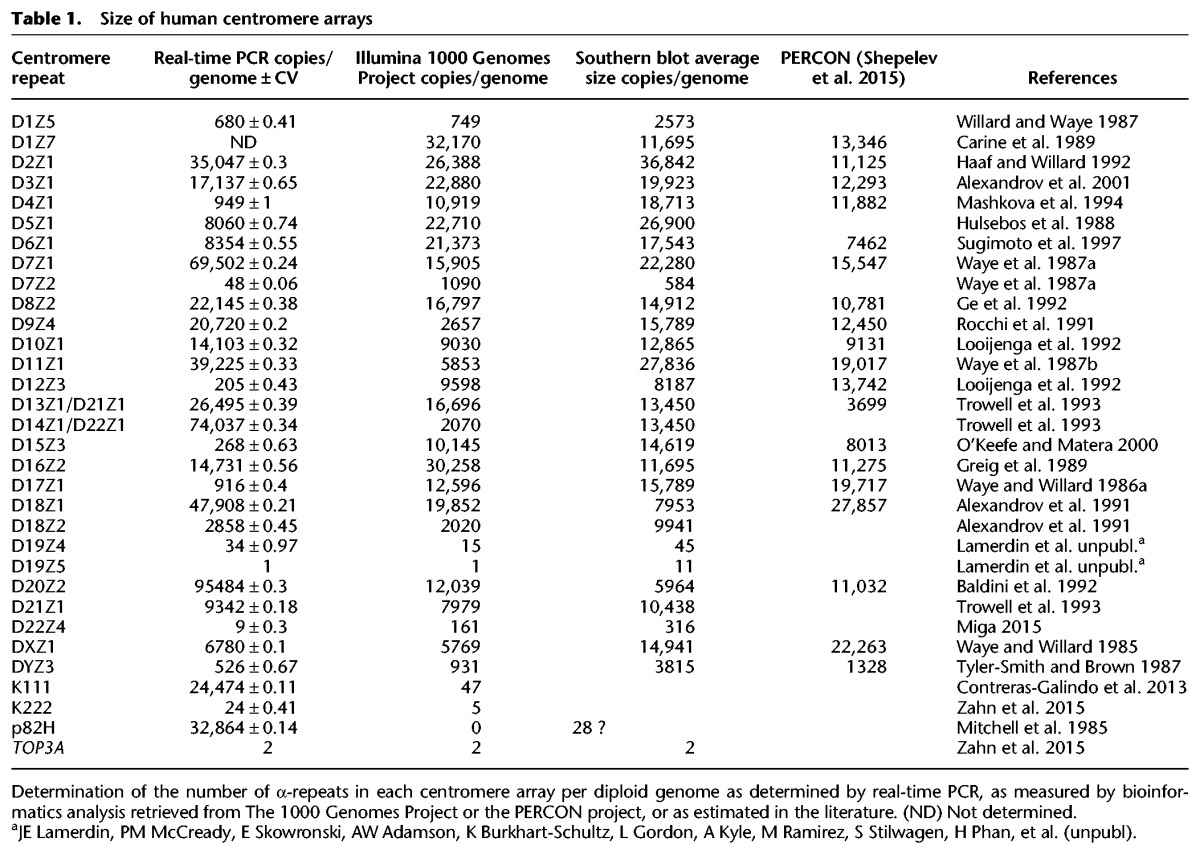
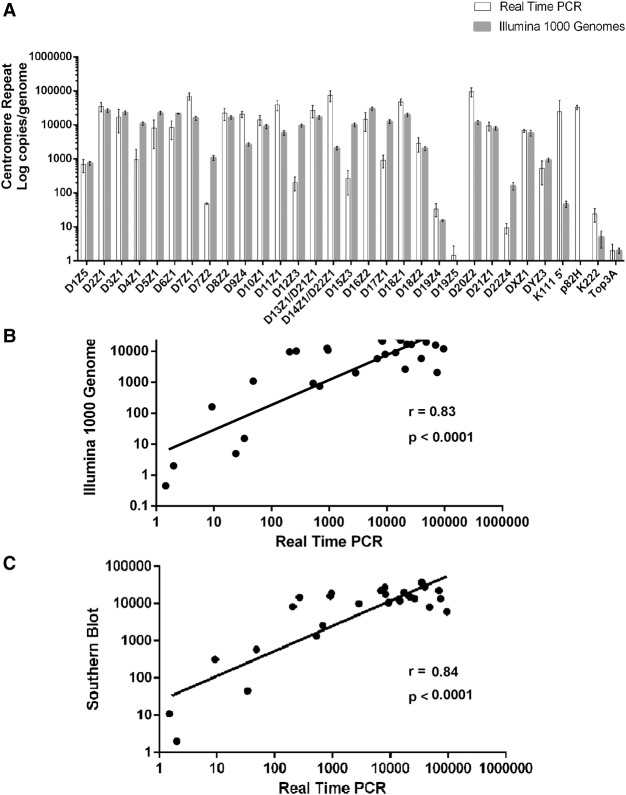
To gauge the size of these arrays using a parallel method, and to confirm the accuracy of the PCR methodology, we developed an in silico analysis approach using sequences generated by The 1000 Genomes Project (Supplemental Fig. S7; The 1000 Genomes Project Consortium 2015). These analyses validated our PCR data (Table 1). We observed a strong correlation between the average numbers of α-repeats in each array as determined by PCR and by in silico analysis (Fig. 2). We also correlated the numbers of α-repeats detected by PCR with the average number in each array as determined by Southern blotting analysis as reported in the literature (Table 1; Liehr 2013) and found a strong positive correlation as well (Fig. 2). We also found a positive correlation (r = 0.7345, P < 0.0001) (Supplemental Fig. S8) with the PCR data and estimates of centromere array size generated by the PERCON analysis, the most recent analysis of the human assembly (Table 1; Supplemental Text; Miga et al. 2014; Shepelev et al. 2015). Our PCR assays thus rapidly and accurately determine the size and variation of human centromeres and correlate well with next-generation sequencing (NGS) data and Southern blotting analysis.
Figure 2.
Positive correlation of copy number in each centromeric array as determined by qPCR assays, in silico analysis of The 1000 Genomes Project, and by Southern blotting hybridization. (A) A bar diagram representing the average log copy number of α-repeats in each centromeric array, of pericentromeric proviruses K111 and K222, and of single-copy genes per diploid genome as determined by either qPCR of the DNA from PBLs isolated from five individuals (Supplemental Fig. S6) or by in silico analysis of The 1000 Genomes Project (Supplemental Fig. S7). The error bars in the PCR analyses indicate the variation between the alpha repeat content of the DNA of five individuals, and in the bioinformatics analysis of The 1000 Genomes Project, the error bars show the variation between ethnicities. The average values are shown in Table 1. (B) Correlation of α-repeat copy number in each array, proviruses K111 and K222, and single-copy genes determined by qPCR and our bioinformatics analysis (see also Supplemental Figs. S6, S7). A discordant correlation was found in the number of K111 and p82H copies, meaning that these sequences were detected using PCR assays but not with bioinformatics analysis. We were unable to retrieve all p82H sequences using in silico analysis, either due to the stringency of our analytic parameters or technical limitations to sequencing these loci by Illumina. (C) Correlation of the copy number of α-repeats in each array determined by the qPCR assays to the estimated number reported in the literature. The average values are shown in Table 1. The Pearson's correlation coefficient and the P-value are shown.
We further validated the specificity of centromere PCR assays by assessing the content of arrays in Chromosomes X and Y, which of course vary between female and male populations (Supplemental Text; Supplemental Fig. S9), finding results consistent with the gender of each individual. We were also able to infer aneuploidy in trisomy 8, 18, and X (Supplemental Fig. S9). As centromere transcripts have been increasingly found to be of biological importance for centromere function (Biscotti et al. 2015), we additionally used our PCR technology to delineate the specific RNA transcripts that arise from individual centromeres (Supplemental Text; Supplemental Fig. S10). These findings further confirmed the utility of using the newly developed PCR assays to study centromeric genetics with potentially important biological implications.
Studying centromere epigenetics
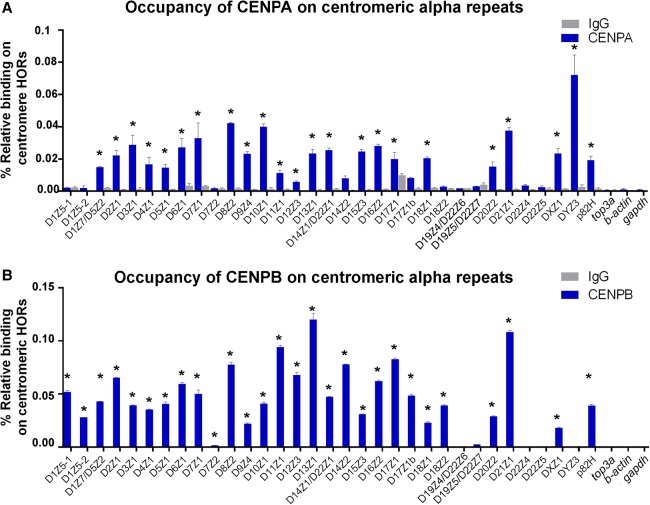
The affinity of centromere proteins for binding centromeric arrays has previously been studied in single stretched chromatin fibers or by ChIP-seq analysis (Malik and Henikoff 2009; Sullivan et al. 2011; Maloney et al. 2012; Henikoff et al. 2015; Ross et al. 2016). These studies measured the binding of CENPA to alphoid arrays from Chromosomes 1, 4, 5, 7, 11, 17, 19, X, and Y, and identified CENPB DNA boxes in all of them except for Chr Y. The question remains as to which specific α-repeat arrays throughout the entire genome are capable of binding CENPA and CENPB, a question that our simple PCR technology allowed us to address. We performed chromatin immunoprecipitation (ChIP) assays on the human LNCaP prostate cancer cell line using antibodies that recognize CENPA and CENPB and measured the quantity of centromere α-repeats. ChIP assays revealed that CENPA deposits to the centromere sequence of every single chromosome at least on one centromere array. CENPA deposits to the largest array in the centromere core of every chromosome (Fig. 3A, starred arrays); these α-repeats can therefore be classified as competent centromeric arrays as described by Henikoff et al. (2015). Similarly, our studies expand the list of centromere arrays that are not able to recruit CENPA. Interestingly, all competent centromere arrays that bind CENPA contain CENPB boxes and bind CENPB accordingly; DYZ3 in Chromosome Y does not have CENPB boxes (Fig. 3). When a given chromosome has more than one α-repeat array, for example in Chromosomes 1, 7, 17, 18, and 22, CENPA predominantly associates with the larger array (D1Z7 over D1Z5, D7Z1 over D7Z2, D14Z1 over D14Z2, D17Z1 over D17Z1b, D18Z1 over D18Z2, D22Z1 over D22Z4 or D22Z5). Although we cannot discriminate between some arrays that are present in Chromosomes 1/5, 13/21, 14/22, the ChIP assay was able to show that CENPA deposits to the largest core arrays in these chromosomes, which are D1Z7/D5Z2 in Chr 1 and Chr 5, D13Z1/D21Z1 in Chr 13 and 21, and D14Z1/D22Z1 in Chr 14 and 22. We also found that CENPA binds to a lesser extent, or not at all, to α-repeat arrays in pericentric areas (i.e., D14Z2, D19Z4/D22Z6, D19Z5/D22Z7, D22Z4, and D22Z5). Thus, using a different and rapid technique, our studies validated previous findings (Henikoff et al. 2015; Ross et al. 2016) and extend these observations to many more human chromosomes. Although previous studies have determined that CENPA occupies ∼40% of the centromere array DXZ1 in Chromosome X (Sullivan et al. 2011), our ChIP-PCR assays estimate the occupancy of CENPA on this and other centromere arrays to be substantially lower. This quantitative, but not qualitative, discrepancy might be due to limitations in the immunoprecipitation component of the ChIP approach.
Figure 3.
Functional capacity of α-repeat arrays in recruiting the centromere proteins CENPA and CENPB can be assessed by PCR-based assays of centromeric DNA. ChIP was performed on LNCaP prostate cancer cells using CENPA and CENPB antibodies or control mouse IgG antibody. The arrays to which centromere proteins bind were measured by qPCR in immunoprecipitated chromatin and compared to the input. (A) Occupancy of CENPA on specific centromere arrays. At least one array in the centromere of each human chromosome recruited CENPA. CENPA antibody precipitated between 0.1 and 0.4% of every centromere array other than the array DYZ3 in Chromosome Y (0.7%). Asterisks indicate dominant arrays in each chromosome that recruit CENPA; these arrays also represent the largest centromere core arrays in every chromosome. We do not yet have an assay to discriminate between the array D14Z1/D22Z1, which is present in both Chr 14 and 22. A PCR assay that measures both arrays shows that D14Z1/D22Z1 indeed dominates the recruitment of CENPA to centromeres 14 and 22. We do not have an assay for the larger array D19Z3 in Chr 19, which resembles D1Z7. Given that the latter recruits CENPA, it is likely that D19Z3 similarly recruits CENPA to centromere 19. (B) Occupancy of CENPB on centromeric arrays. At least one array in the centromere of each human chromosome (other than Y) recruited CENPB. CENPB did not bind the array DYZ3 in Y. There is a significant difference between the amounts of D7Z2 immunoprecipitated with CENPB antibody as compared to the IgG control (P < 0.001). The existence of only one CENPB box in a sequence of 16 alphoid repeats (Waye et al. 1987a) might explain the relatively low binding of CENPB to this array. Asterisks indicate arrays that contain CENPB box sequences.
ChIP assays of CENPB revealed that this protein binds to all centromere arrays, except for D19Z4/D22Z6, D19Z5/D22Z7, D22Z4, D22Z5, and DYZ3 (Fig. 3B). Previously, it was shown that CENPB does not bind the array DYZ3 in Chromosome Y (Ohzeki et al. 2002), which was confirmed by our results. In contrast to CENPA, we found that CENPB binds not only to the larger arrays but also to shorter ones. CENPB has been found to bind to a specific 17-nt CENPB DNA box. We verified the existence of four types of CENPB boxes along linear sequences of these arrays (Supplemental Sequences). We further determined from the sequence that in the α-repeat arrays to which CENPB binds, CENPB boxes exist every 340-nt sequence along the array, confirming and extending recent observations (Henikoff et al. 2015). Therefore, using our rapid PCR assays, we confirmed recent observations linking centromere genomics to epigenetics, and are able to extend these findings to almost any and all of the individual centromeres.
Centromeric instability in trisomy 21
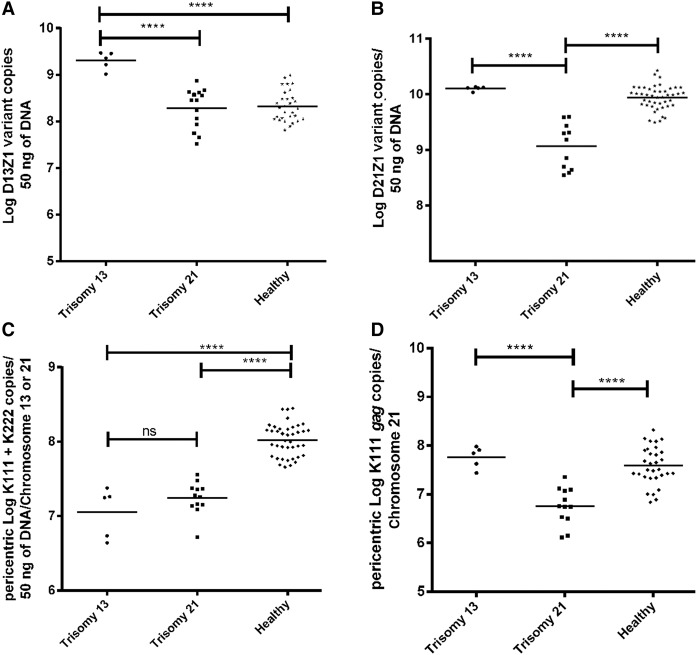
A key reason for developing rapid and comprehensive PCR assays to examine each distinct human centromere is the need to study whether these sequences might play a role in chromosomal nondisjunction disorders. We tested our PCR assays that detect nucleotide substitutions present in the array D13Z1 or D21Z1, optimized through the use of LNA technology and a molecular clamp (Supplemental Fig. S4), in genetically normal individuals and individuals with trisomy 13 (Patau syndrome) or 21 (Down syndrome). These arrays were shown to recruit CENPA and CENPB (Fig. 3) and therefore may have a role in chromosome segregation. As predicted, we detected the D13Z1 array at a level that is ∼1.5-fold higher in individuals with trisomy 13 than in those without the defect (Fig. 4A). Interestingly, a few individuals with trisomy 21, but not all, show lesser amounts of D13Z1 than does the normal population. The numbers of D21Z1 specific sequences is dramatically lower in individuals with trisomy 21, suggesting that a loss of D21Z1 α-repeat arrays or a partial loss of centromere 21 DNA exists in individuals with trisomy 21 (Fig. 4B). Although changes in the centromere of Chr 21, but not of Chr 13, appear to occur commonly in the general human population (Lo et al. 1999), our data suggest that defects in centromere 21 occur in trisomy 21.
Figure 4.
Centromere and pericentromere instability in individuals with trisomy 21. Detection of D13Z1 (A) and D21Z1 (B) variants in individuals with trisomy 13 or 21 by qPCR using LNA primers and clamps. The copy number of each repeat variant was determined in 50 ng of DNA. (C) Detection of K111 and K222 provirus sequences used as markers to study human pericentromeres. We have developed a PCR assay for K111 plus K222 env (C) and a PCR assay specific for K111 gag (D), in order to assess the structural variation (length) of pericentromeres 13 and 21 in DNA from healthy individuals and individuals with trisomy 13 or 21. The K111 + K222 assay (C) can predict the length of pericentromeres 13 and 21, whereas the K111-specific PCR (D) can predict the length mostly of pericentromere 21. In contrast to healthy individuals, loss of pericentromeric K111 sequences was seen in the DNA of individuals with trisomy 21. Statistical significance among the groups was calculated using the t-test. (****) P-values <0.0001 are shown.
IF-FISH to determine the extent of CENPB binding to Chr 21 in karyotypically normal and trisomy 21 cells
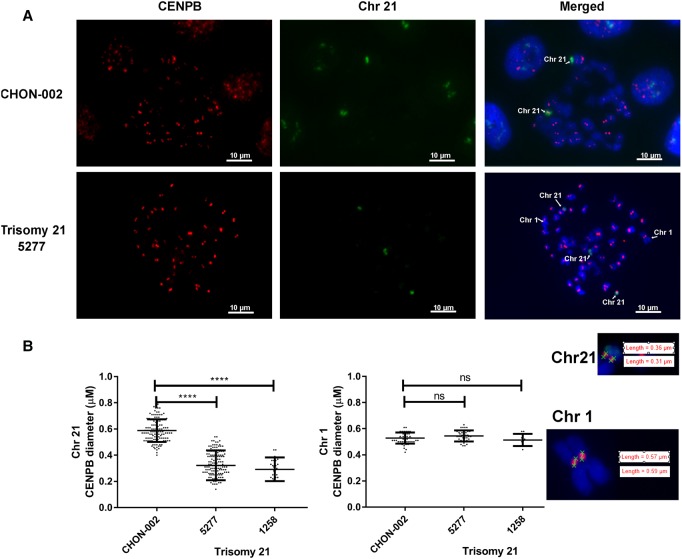
As we saw that the centromere alphoid repeat D21Z1 array in Chr 21 is shortened in trisomy 21 cells, we attempted to verify our PCR-based findings using a different methodology, IF-FISH. In chromosomal spreads, we detected Chr 21 by FISH analysis using a probe that paints Chr 21 and examined the binding of CENPB along the CENPB boxes of Chr 21 by IF using anti-CENPB antibodies (Fig. 5A). We then performed IF-FISH to evaluate the extent of CENPB binding over Chr 21 in cells that are karyotypically normal (CHON-002) and in two trisomy 21 cell lines (5277 and 1258). We also performed the same analysis on Chr 1 (identified on the basis of being clearly the largest chromosome). We measured the diameter of CENPB binding along the centromeres of Chr 1 and Chr 21 and observed that CENPB binding is reduced in Chr 21 of trisomy 21 cells, in contrast to the size of the CENPB stain in the karyotypically normal CHON-002 cells (Fig. 5B). Binding of CENPB was shown to be as much as 50% lower in trisomy 21 cells as compared to CHON-002 cells (P < 0.0001). No significant difference was found in the CENPB staining of Chr 1 in any of the cell lines evaluated. These data further support the observation made using our PCR assay that the centromeres of Chr 21 are shortened in the cells of individuals with trisomy 21 (Fig. 4B).
Figure 5.
Reduced CENPB binding to the centromere of Chr 21 in trisomy 21 cells. IF-FISH analysis to determine the binding of CENPB (IF) on Chr 21. (A) IF-FISH analysis of CENPB binding (red) on Chr 21 (identified by FISH, green) or Chr 1 (identified by large size) in chromosomal spreads stained with DAPI (blue). Karyotypically normal CHON-002 cells and trisomy 21 5277 cells are shown. (B) The diameter of CENPB binding sites along Chr 21 was measured using the NIS-Elements software in a NIKON microscope in CHON-002 cells and trisomy 21 cells isolated from Subjects A 1258 and B 5277. A statistically significant difference in CENPB binding along Chr 21 was found between the karyotypically normal cell line CHON-002 (n = 126) and the trisomy 21 cells lines 5277 (n = 141) or 1258 (n = 25) (P < 0.0001). No significant differences were found in CENPB binding in Chr 1. The insets show examples of the measurement of CENPB staining in Chromosomes 1 and 21 in trisomy 21 cells.
Pericentric instability in trisomy 13 and 21
We assessed whether instability at the pericentromere of Chromosome 21 exists in trisomy 21 individuals using pericentric markers K111 and K222. We quantitated pericentric K111 and K222 as a surrogate for pericentromere size and found that, in contrast to the DNA of healthy individuals, people with trisomy 13 and 21 show losses in the pericentromeres (Fig. 4C). These observations suggest that pericentromere instability or deletion of pericentric areas is present in individuals with trisomy 13 and 21. We additionally developed an assay using LNA-modified primers to specifically detect and quantitate K111, but not K222, to validate our findings. Indeed, K111 numbers are significantly reduced in individuals with trisomy 21 (Fig. 4D), suggesting once again that pericentric instability is seen in individuals with trisomy 21. We further validated these results by sequencing and found less diversity of K111 proviruses in trisomy 21 individuals (Supplemental Fig. S11). Interestingly, the DNA of trisomy 21 individuals contained novel K111 sequences that do not match to known K111 sequences (Supplemental Fig. S11). Phylogenetic analysis revealed that these sequences likely are the result of homologous recombination at pericentromere 21, further suggesting that the role of centromeric and pericentromeric instability in the pathogenesis of trisomy 21 requires investigation.
Discussion
The PCR-based methodology for rapidly studying centromere genomics offers substantial advantages over the previously existing technology. Southern blotting has been previously performed using probes that detect HORs, but not single monomers. Estimation of the size of centromere arrays varies with the Southern blot technique, in particular due to hybridization conditions. When using high stringency conditions, the probe detects fewer HORs with higher specificity, but at lower stringency the probe detects more HORs but with the possibility of nonspecifically detecting other arrays. Thus, Southern blot experiments performed on centromere arrays have estimated the size of the arrays within a window of variation roughly between 6000 and 24,000 α-repeats (Table 1; Liehr 2013). The centromere PCR assays described in this study detect monomers within the HORs. The PCR assays could successfully estimate the size of arrays that have monomers with high sequence similarity, but may underestimate the size of arrays that are more divergent. We optimized the PCR conditions in order to detect as many monomers as possible while retaining high specificity for a particular array. Our studies estimated the size of centromeres with a variation roughly between 0 and at most 1800 α-repeats. Current NGS technologies that also detect monomers still fall short in resolving the complexity of centromere sequences and are certainly inefficient. Data detailing centromere repeats are usually excluded from NGS analysis, as it is impossible to map small sequencing reads of these highly repetitive sequences unambiguously to the reference genome. NGS methods that rely on target enrichment also use blocking reagents that eliminate repetitive sequences such as centromeric α-repeats. Therefore, trying to understand the evolving pattern of individual centromere genetics in a biological setting would require a very large and independent bioinformatics analysis. In contrast, our PCR assays offer a rapid way to analyze individual centromere sequences simultaneously, and in real-time. One potential drawback to the PCR technology may be that it might not pick up α-repeats that have certain mutations. However, the data above from different ethnic populations thus far indicate that these PCR assays can be used specifically to study all centromere repeats in samples from diverse populations.
The PCR assays reported here can be used to study the centromeres of 23 of 24 human chromosomes comprehensively in rapid and simultaneous reactions. In contrast to the laborious techniques currently used to estimate the size of single centromere arrays, our assays offer the advantage of studying multiple, individual human centromere sequences in a given biological setting. Centromere-specific markers for Chromosome 19 remain unavailable to date, as the sequences identified so far in Chromosome 19 are identical to sequences in Chromosomes 1, 5, and 21. Identification of such markers awaits further sequencing of centromere 19.
We present a rapid and user-friendly way to study the genetics, and hence also epigenetics, of human centromeres. We have shown the utility of our PCR-based methodology in sensitively delineating centromere instability and chromosomal ploidy. We found evidence to propose the previously unsuspected hypothesis that centromeric and pericentromeric instability may underlie trisomy 21. We have also demonstrated that this approach can be used to understand the transcriptional profile of the alphoid repeats of each chromosome. Further, we show that this method can be applied to a specific and genome-wide approach to important questions in the vital field of centromere epigenetics. As these PCR-based techniques can address the fate of specific centromeres in a manner resembling real-time, they can be applied to understanding the role that specific centromeric genomic elements play in development and perhaps in the evolution of malignancy.
Methods
Real-time qPCR
Copy numbers for each centromeric array (number of α-repeats in each array), proviruses K111/K222, and single-copy genes were measured by qPCR using specific primers and PCR conditions as described in Supplemental Tables 1 and 2. PCR amplification products were confirmed by sequencing. The qPCR was carried out using the FastStart Universal SYBR Green Master mix (Rox) (Roche) with an initial enzyme activation step for 10 min at 95°C and 16–25 cycles consisting of 15 sec of denaturation at 95°C and 30 sec of annealing/extension at the temperature reported in Supplemental Table 2. The copy number was estimated using serial dilutions of plasmids containing the PCR amplicon. The DNA copy number of each plasmid was estimated by reading the DNA concentration of the plasmid at a wavelength of 260 nm using UV spectrophotometry. Serial dilution of these plasmids of known copy number served as calibrators for standard curves to estimate the number of copies of DNA per sample. The specificity of the qPCR assay detecting the centromere of unique chromosomes was assessed using DNA samples from human/rodent cell hybrids, each one containing a single human chromosome. Quantitation of the single-copy genes TOP3A, DEK, and CCR5 was assessed by qPCR in 40 cycles consisting of 15 sec of denaturation at 95°C and annealing/extension of 30 sec at 60°C. The copy number of single-copy genes was calculated using serial dilutions of purified plasmid containing the target PCR amplicon as described above. The relative copy number of α-repeats in each array per human diploid genome was estimated in reference to the quantitation of the gene TOP3A, which exists as a single copy in the human genome (Hanai et al. 1996). The relative copy number was calculated by dividing the number of copies obtained by qPCR by the number of copies of TOP3A detected in equal amounts of cellular DNA.
The Supplemental Methods includes additional information on the DNA and RNA samples, analysis of centromere transcription by qRT-PCR, description of ChIP and IF-FISH experiments, amplification and sequencing analysis of K111, qPCR analysis to differentiate centromere 13 from centromere 21 using LNA technology, and NGS analysis of centromere sequences.
Data access
The sequences generated in this study have been submitted to the NCBI Nucleotide database (http://www.ncbi.nlm.nih.gov/nuccore) under accession numbers MF624880–MF625017.
Supplementary Material
Acknowledgments
The authors thank Joseph Zahn for help in preparing this manuscript and Ting Wu for very helpful suggestions. This work was supported by grant K22 CA177824 from the National Cancer Institute to R.C.-G.; grant 05–5089 from the Concerned Parents for AIDS Research CPFA to M.H.K.; and by grant R01 CA144043 from the National Institutes of Health (NIH) to D.M.M. S.F. was supported by the fellowship MOV_CA_2013_1_10789 from the Programa de Desarrollo de Ciencias Básicas (PEDECIBA), Agencia Nacional de Investigación e Innovación (ANII) from the Uruguayan government. M.M. was supported by the University of Michigan Undergraduate Research Opportunity Program (UROP). A.K.S. was supported by a University of Michigan Cancer Biology Program Fellowship and the Medical Science Training Program (MSTP). P.W.C. was supported by NIH Postbaccalaureate Research Education Program (PREP) grant R25GM086262-07. G.S.O. acknowledges support from NIH grants RM-08-029 and P30U54ES017885.
Author contributions: R.C.-G., M.H.K., and D.M.M. conceptualized the research. R.C.-G., S.F., A.K.S., J.D.L., P.W.C., M.M., C.W., B.Q., and M.H.K. designed and performed the experiments. R.C.-G., S.F., A.K.S., P.W.C., M.D., F.M., and D.M.M. analyzed the data. R.C.-G., A.C., G.S.O., M.H.K., and D.M.M. provided supervision. R.C.-G., A.K.S., M.H.K., G.S.O., and D.M.M. wrote the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.219709.116.
References
- The 1000 Genomes Project Consortium. 2015. A global reference for human genetic variation. Nature 526: 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldrup-Macdonald ME, Sullivan BA. 2014. The past, present, and future of human centromere genomics. Genes (Basel) 5: 33–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov IA, Mashkova TD, Akopian TA, Medvedev LI, Kisselev LL, Mitkevich SP, Yurov YB. 1991. Chromosome-specific α satellites: two distinct families on human chromosome 18. Genomics 11: 15–23. [DOI] [PubMed] [Google Scholar]
- Alexandrov I, Kazakov A, Tumeneva I, Shepelev V, Yurov Y. 2001. α-Satellite DNA of primates: old and new families. Chromosoma 110: 253–266. [DOI] [PubMed] [Google Scholar]
- Amor DJ, Choo KH. 2002. Neocentromeres: role in human disease, evolution, and centromere study. Am J Hum Genet 71: 695–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldini A, Archidiacono N, Carbone R, Bolino A, Shridhar V, Miller OJ, Miller DA, Ward DC, Rocchi M. 1992. Isolation and comparative mapping of a human chromosome 20-specific α-satellite DNA clone. Cytogenet Cell Genet 59: 12–16. [DOI] [PubMed] [Google Scholar]
- Ballantyne KN, van Oorschot RA, Mitchell RJ. 2008. Locked nucleic acids in PCR primers increase sensitivity and performance. Genomics 91: 301–305. [DOI] [PubMed] [Google Scholar]
- Bersani F, Lee E, Kharchenko PV, Xu AW, Liu M, Xega K, MacKenzie OC, Brannigan BW, Wittner BS, Jung H, et al. 2015. Pericentric satellite repeat expansions through RNA-derived DNA intermediates in cancer. Proc Natl Acad Sci 112: 15148–15153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biscotti MA, Canapa A, Forconi M, Olmo E, Barucca M. 2015. Transcription of tandemly repetitive DNA: functional roles. Chromosome Res 23: 463–477. [DOI] [PubMed] [Google Scholar]
- Burrack LS, Berman J. 2012. Neocentromeres and epigenetically inherited features of centromeres. Chromosome Res 20: 607–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carine K, Jacquemin-Sablon A, Waltzer E, Mascarello J, Scheffler IE. 1989. Molecular characterization of human minichromosomes with centromere from chromosome 1 in human-hamster hybrid cells. Somat Cell Mol Genet 15: 445–460. [DOI] [PubMed] [Google Scholar]
- Choo KH, Vissel B, Earle E. 1989. Evolution of α-satellite DNA on human acrocentric chromosomes. Genomics 5: 332–344. [DOI] [PubMed] [Google Scholar]
- Contreras-Galindo R, Kaplan MH, Contreras-Galindo AC, Gonzalez-Hernandez MJ, Ferlenghi I, Giusti F, Lorenzo E, Gitlin SD, Dosik MH, Yamamura Y, et al. 2011. Characterization of human endogenous retroviral elements in the blood of HIV-1-infected individuals. J Virol 86: 262–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras-Galindo R, Kaplan MH, Contreras-Galindo AC, Gonzalez-Hernandez MJ, Ferlenghi I, Giusti F, Lorenzo E, Gitlin SD, Dosik MH, Yamamura Y, et al. 2013. HIV infection reveals widespread expansion of novel centromeric human endogenous retroviruses. Genome Res 23: 1505–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fachinetti D, Han JS, McMahon MA, Ly P, Abdullah A, Wong AJ, Cleveland DW. 2015. DNA sequence-specific binding of CENP-B enhances the fidelity of human centromere function. Dev Cell 33: 314–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita R, Otake K, Arimura Y, Horikoshi N, Miya Y, Shiga T, Osakabe A, Tachiwana H, Ohzeki J, Larionov V, et al. 2015. Stable complex formation of CENP-B with the CENP-A nucleosome. Nucleic Acids Res 43: 4909–4922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Y, Wagner MJ, Siciliano M, Wells DE. 1992. Sequence, higher order repeat structure, and long-range organization of α satellite DNA specific to human chromosome 8. Genomics 13: 585–593. [DOI] [PubMed] [Google Scholar]
- Greig GM, England SB, Bedford HM, Willard HF. 1989. Chromosome-specific α satellite DNA from the centromere of human chromosome 16. Am J Hum Genet 45: 862–872. [PMC free article] [PubMed] [Google Scholar]
- Haaf T, Willard HF. 1992. Organization, polymorphism, and molecular cytogenetics of chromosome-specific α-satellite DNA from the centromere of chromosome 2. Genomics 13: 122–128. [DOI] [PubMed] [Google Scholar]
- Hanai R, Caron PR, Wang JC. 1996. Human TOP3: a single-copy gene encoding DNA topoisomerase III. Proc Natl Acad Sci 93: 3653–3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden KE. 2012. Human centromere genomics: Now it's personal. Chromosome Res 20: 621–633. [DOI] [PubMed] [Google Scholar]
- Hayden KE, Willard HF. 2012. Composition and organization of active centromere sequences in complex genomes. BMC Genomics 13: 324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff JG, Thakur J, Kasinathan S, Henikoff S. 2015. A unique chromatin complex occupies young α-satellite arrays of human centromeres. Sci Adv 1: e1400234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood L, Rowen L. 2013. The Human Genome Project: Big science transforms biology and medicine. Genome Med 5: 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath JE, Gulden CL, Bailey JA, Yohn C, McPherson JD, Prescott A, Roe BA, de Jong PJ, Ventura M, Misceo D, et al. 2003. Using a pericentric interspersed repeat to recapitulate the phylogeny and expansion of human centromeric segmental duplications. Mol Biol Evol 20: 1463–1479. [DOI] [PubMed] [Google Scholar]
- Hulsebos T, Schonk D, van Dalen I, Coerwinkel-Driessen M, Schepens J, Ropers HH, Wieringa B. 1988. Isolation and characterization of alphoid DNA sequences specific for the pericentric regions of chromosomes 4, 5, 9, and 19. Cytogenet Cell Genet 47: 144–148. [DOI] [PubMed] [Google Scholar]
- Jørgensen AL. 1997. Alphoid repetitive DNA in human chromosomes. Dan Med Bull 44: 522–534. [PubMed] [Google Scholar]
- Kirsch S, Weiß B, Miner TL, Waterston RH, Clark RA, Eichler EE, Münch C, Schempp W, Rappold G. 2005. Interchromosomal segmental duplications of the pericentric region on the human Y chromosome. Genome Res 15: 195–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liehr T. 2013. Benign & pathological chromosomal imbalances. Academic Press, Elsevier, San Diego, CA. [Google Scholar]
- Lo AW, Liao GC, Rocchi M, Choo KH. 1999. Extreme reduction of chromosome-specific α-satellite array is unusually common in human chromosome 21. Genome Res 9: 895–908. [DOI] [PubMed] [Google Scholar]
- Looijenga LH, Oosterhuis JW, Smit VT, Wessels JW, Mollevanger P, Devilee P. 1992. α Satellite DNAs on chromosome 10 and 12 are both members of the dimeric suprachromosomal subfamily, but display little identity at the nucleotide sequence level. Genomics 13: 1125–1132. [DOI] [PubMed] [Google Scholar]
- Mackinnon RN, Campbell LJ. 2011. The role of dicentric chromosome formation and secondary centromere deletion in the evolution of myeloid malignancy. Genet Res Int 2011: 643628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik HS, Henikoff S. 2009. Major evolutionary transitions in centromere complexity. Cell 138: 1067–1082. [DOI] [PubMed] [Google Scholar]
- Maloney KA, Sullivan LL, Matheny JE, Strome ED, Merrett SL, Ferris A, Sullivan BA. 2012. Functional epialleles at an endogenous human centromere. Proc Natl Acad Sci 109: 13704–13709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashkova TD, Akopian TA, Romanova LY, Mitkevich SP, Yurov YB, Kisselev LL, Alexandrov IA. 1994. Genomic organization, sequence and polymorphism of the human chromosome 4-specific α-satellite DNA. Gene 140: 211–217. [DOI] [PubMed] [Google Scholar]
- Miga KH. 2015. Completing the human genome: the progress and challenge of satellite DNA assembly. Chromosome Res 23: 421–426. [DOI] [PubMed] [Google Scholar]
- Miga KH, Newton Y, Jain M, Altemose N, Willard HF, Kent WJ. 2014. Centromere reference models for human chromosomes X and Y satellite arrays. Genome Res 24: 697–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell AR, Gosden JR, Miller DA. 1985. A cloned sequence, p82H, of the alphoid repeated DNA family found at the centromeres of all human chromosomes. Chromosoma 92: 369–377. [DOI] [PubMed] [Google Scholar]
- Ohzeki J, Nakano M, Okada T, Masumoto H. 2002. CENP-B box is required for de novo centromere chromatin assembly on human alphoid DNA. J Cell Biol 159: 765–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Keefe CL, Matera AG. 2000. α Satellite DNA variant-specific oligoprobes differing by a single base can distinguish chromosome 15 homologs. Genome Res 10: 1342–1350. [DOI] [PubMed] [Google Scholar]
- Rocchi M, Archidiacono N, Ward DC, Baldini A. 1991. A human chromosome 9-specific alphoid DNA repeat spatially resolvable from satellite 3 DNA by fluorescent in situ hybridization. Genomics 9: 517–523. [DOI] [PubMed] [Google Scholar]
- Roizès G. 2006. Human centromeric alphoid domains are periodically homogenized so that they vary substantially between homologues. Mechanism and implications for centromere functioning. Nucleic Acids Res 34: 1912–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosandić M, Paar V, Basar I, Glunčić M, Pavin N, Pilaš I. 2006. CENP-B box and pJα sequence distribution in human α satellite higher-order repeats (HOR). Chromosome Res 14: 735–753. [DOI] [PubMed] [Google Scholar]
- Ross JE, Woodlief KS, Sullivan BA. 2016. Inheritance of the CENP-A chromatin domain is spatially and temporally constrained at human centromeres. Epigenetics Chromatin 9: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott KC, Sullivan BA. 2014. Neocentromeres: a place for everything and everything in its place. Trends Genet 30: 66–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepelev VA, Uralsky LI, Alexandrov AA, Yurov YB, Rogaev EI, Alexandrov IA. 2015. Annotation of suprachromosomal families reveals uncommon types of α satellite organization in pericentromeric regions of hg38 human genome assembly. Genome Data 5: 139–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stimpson KM, Sullivan BA. 2010. Epigenomics of centromere assembly and function. Curr Opin Cell Biol 22: 772–780. [DOI] [PubMed] [Google Scholar]
- Sugimoto K, Furukawa K, Kusumi K, Himeno M. 1997. The distribution of binding sites for centromere protein B (CENP-B) is partly conserved among diverged higher order repeating units of human chromosome 6-specific alphoid DNA. Chromosome Res 5: 395–405. [DOI] [PubMed] [Google Scholar]
- Sullivan LL, Boivin CD, Mravinac B, Song IY, Sullivan BA. 2011. Genomic size of CENP-A domain is proportional to total α satellite array size at human centromeres and expands in cancer cells. Chromosome Res 19: 457–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert PB, Henikoff S. 2010. Centromeres convert but don't cross. PLoS Biol 8: e1000326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanno Y, Susumu H, Kawamura M, Sugimura H, Honda T, Watanabe Y. 2015. The inner centromere-shugoshin network prevents chromosomal instability. Science 349: 1237–1240. [DOI] [PubMed] [Google Scholar]
- Trowell HE, Nagy A, Vissel B, Choo KH. 1993. Long-range analyses of the centromeric regions of human chromosomes 13, 14 and 21: identification of a narrow domain containing two key centromeric DNA elements. Hum Mol Genet 2: 1639–1649. [DOI] [PubMed] [Google Scholar]
- Tyler-Smith C, Brown WR. 1987. Structure of the major block of alphoid satellite DNA on the human Y chromosome. J Mol Biol 195: 457–470. [DOI] [PubMed] [Google Scholar]
- Verdaasdonk JS, Bloom K. 2011. Centromeres: unique chromatin structures that drive chromosome segregation. Nat Rev Mol Cell Biol 12: 320–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissel B, Choo KH. 1992. Evolutionary relationships of multiple α satellite subfamilies in the centromeres of human chromosomes 13, 14, and 21. J Mol Evol 35: 137–146. [DOI] [PubMed] [Google Scholar]
- Waye JS, Willard HF. 1985. Chromosome-specific α satellite DNA: nucleotide sequence analysis of the 2.0 kilobasepair repeat from the human X chromosome. Nucleic Acids Res 13: 2731–2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waye JS, Willard HF. 1986a. Molecular analysis of a deletion polymorphism in α satellite of human chromosome 17: evidence for homologous unequal crossing-over and subsequent fixation. Nucleic Acids Res 14: 6915–6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waye JS, Willard HF. 1986b. Structure, organization, and sequence of α satellite DNA from human chromosome 17: evidence for evolution by unequal crossing-over and an ancestral pentamer repeat shared with the human X chromosome. Mol Cell Biol 6: 3156–3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waye JS, England SB, Willard HF. 1987a. Genomic organization of α satellite DNA on human chromosome 7: evidence for two distinct alphoid domains on a single chromosome. Mol Cell Biol 7: 349–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waye JS, Greig GM, Willard HF. 1987b. Detection of novel centromeric polymorphisms associated with α satellite DNA from human chromosome 11. Hum Genet 77: 151–156. [DOI] [PubMed] [Google Scholar]
- Willard F, Waye JS. 1987. Hierarchical order in chromosome-specific human α satellite DNA. Trends Genet 3: 192–198. [Google Scholar]
- Zahn J, Kaplan MH, Fischer S, Dai M, Meng F, Saha AK, Cervantes P, Chan SM, Dube D, Omenn GS, et al. 2015. Expansion of a novel endogenous retrovirus throughout the pericentromeres of modern humans. Genome Biol 16: 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlin SG. 2010. Centromeres: the wild west of the post-genomic age. Epigenetics 5: 34–40. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.