Abstract
Taxadiene-5α-Hydroxylase (CYP725A4) is a membrane-bound plant cytochrome P450 that catalyzes the oxidation of taxadiene to taxadiene-5α-ol. This oxidation is a key step in the production of the valuable cancer therapeutic and natural plant product, taxol. In this work, we report the bacterial expression and purification of six different constructs of CYP725A4. All six of these constructs are N-terminally modified and three of them are fused to cytochrome P450 reductase to form a chimera construct. The construct with the highest yield of CYP725A4 protein was then selected for substrate binding and kinetic analysis. Taxadiene binding followed type-1 substrate patterns with an observed KD of 2.1 μM ± 0.4 μM. CYP725A4 was further incorporated into nanoscale lipid bilayers (nanodiscs) and taxadiene metabolism was measured. Taxadiene metabolism followed Michaelis-Menten kinetics with an observed Vmax of 30 ± 8 pmol/min/nmolCYP725A4 and a KM of 123 ± 52 μM. Additionally, molecular operating environment (MOE) modeling was performed in order to gain insight into the interactions of taxadiene with CYP725A4 active site. Taken together, we demonstrate the successful expression and purification of the functional membrane-bound plant CYP, CYP725A4, in E. coli.
Keywords: Taxadiene, taxadiene-5α-ol, recombinant expression, E. coli, Nanodiscs
Introduction
Taxadiene-5α-hydroxylase (CYP725A4) is a membrane-bound cytochrome P450 (CYP) found in the Japanese yew tree, Taxus cuspidata, which catalyzes the oxidation of taxadiene to taxadiene-5α-ol. This is the first functionalization step in the biological synthesis of the valuable phytochemical cancer therapeutic, taxol [1-3]. Natural sources are incapable of fully satisfying the demand for taxol and current total chemical synthesis approaches are economically cost-prohibitive [4-6]. As a result, emphasis has been placed on developing alternative synthetic routes. One such method is to use easily cultivated organisms such as Escherichia coli or Saccharomyces cerevisiae to perform heterologous biological production, also known as metabolic engineering [7, 8]. To date, this strategy was used for producing approximately g/L titers of taxadiene using E. coli culture [9]. The next step in native taxol synthesis is the CYP725A4-mediated hydroxylation of taxadiene to taxadiene-5α-ol. Initial studies have shown that the incorporation of CYP725A4 into a bacterial expression system leads to the generation of many other byproducts [9, 10]. Therefore, it is apparent that further optimizations of the metabolic engineering of taxol will require a more complete understanding of the CYP725A4 enzyme mechanism. This necessitates the ability to express this protein in milligram quantities for biochemical studies.
CYP725A4 was initially identified in microsomes made from Taxus brevifolia saplings as well as suspended Taxus cuspidata cell cultures [1]. In 2004, Croteau and coworkers were able to isolate the CYP725A4 cDNA, successfully express CYP725A4 in the fall armyworm Spodoptera fugiperda, create microsomes, and characterize CYP725A4-mediated taxadiene hydroxylation to taxadiene-5α-ol using kinetic and binding assays [11]. In 2008, Tissier and coworkers expressed CYP725A4 in the tobacco plant N. sylvestris along with taxadiene synthase and observed the product 5(12)-Oxa-3(11)-cyclotaxane (OCT) in the leaf exudate instead of the expected taxadiene-5α-ol [12]. As part of the same study, CYP725A4 was expressed in yeast, and microsomes were prepared that also converted taxadiene to OCT rather than taxadiene-5α-ol [12]. In 2014, Yadav studied a CYP725A4 construct and found that CYP725A4 acts on 4 substrates to form 12 products, and that taxadiene-5α-ol was only a very minor product [13]. Additionally, other groups have suggested the presence of an epoxide intermediate in the conversion of taxadiene to taxadiene-5α-ol [14, 15]. In order to help resolve the ambiguity surrounding CYP725A4-mediated taxadiene hydroxylation, we conducted a study comparing the product distribution of different CYP725A4 constructs that are purified via E. coli fermentation [16]. The use of purified protein is important because it reduces the chances for non-specific protein-protein interactions and other contaminants in the microsomes that might influence the metabolism of taxadiene by CYP725A4.
In general, membrane proteins such as CYP725A4 are difficult to express recombinantly in bacterial systems and thereby lead to poor expression of functional protein [17]. One method of increasing expression of functional membrane proteins in E. coli is to modify the protein’s hydrophobic N-terminal domain, which increases the protein solubility during expression [17-20]. In this work, we prepare several different constructs of CYP725A4 with N-terminus modifications in order to increase the expression of these constructs in bacterial systems.
The redox partner of CYP725A4 is cytochrome P450 reductase (CPR). CPR is the obligate redox partner of most CYPs [21]. We have shown that different constructs, i.e. CYP725A4 fused to Taxus cuspidata CPR, CYP725A4 paired with mammalian CPR, and CYP725A4 paired with plant CPR, catalyzed the conversion of taxadiene to both taxadiene-5α-ol and OCT [16].
Further functional characterization was pursued with the CYP725A4 construct that yielded the highest amount of functional protein when expressed in E. coli. In order to provide a native-like lipid environment to the protein, purified CYP725A4 was reconstituted into nanodiscs to analyze the kinetics of CYP725A4 taxadiene metabolism. Nanodiscs are uniform-sized nanoscale lipid bilayers encompassed by membrane scaffold proteins (MSPs) and which mimic a native-like membrane environment for membrane protein stability (Figure 1) [22-28]. In addition, it has been shown that membrane proteins in nanodiscs maintain their native-like functionality [23, 25, 29-33]. In 2004, Schuler and coworkers successfully inserted purified plant CYP73A5 into nanodiscs demonstrating the potential of this technology in studying plant CYPs [33]. Therefore, the nanodisc is an ideal system to stabilize purified membrane-bound CYP725A4 in order to evaluate the reaction kinetics of taxadiene metabolism.
Figure 1.
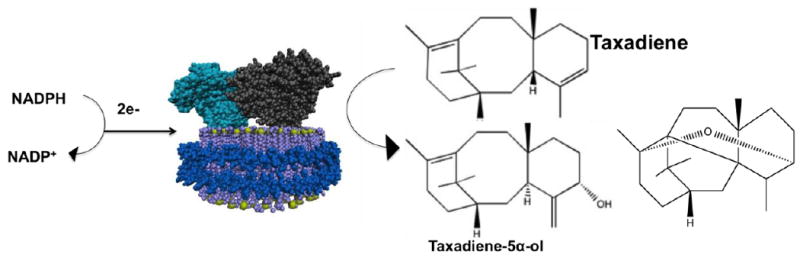
Schematic of CYP725A4 (grey) and CPR (cyan) inserted into the nanodisc (lipids are purple, phosphate atoms are yellow, and membrane scaffold protein is shown in blue). When NADPH is given to the system, the electrons are used in conjunction with molecular oxygen to convert taxadiene to taxadiene-5α-ol or OCT.
We studied the taxadiene binding to CYP725A4 through UV-Vis spectral shifts of the heme Soret band at 417 nm to lower wavelengths upon substrate binding. Concentration-dependent shifts in the heme absorption from 417 nm to 393 nm were observed, indicative of a type-1 substrate binding pattern.
Molecular Operating Environment (MOE) software was used to dock taxadiene into the CYP725A4 active site in order to understand the origin of the substrate binding. Several residues close to the active site were identified that are highly likely to influence substrate docking. Altogether, the best-docked conformation demonstrated favorable binding of taxadiene to CYP725A4.
Taken together, herein we present a method of expression and purification of four new constructs and two previously reported constructs of CYP725A4. Binding of taxadiene to CYP725A4 is analyzed using UV-vis spectroscopy. CYP725A4 is incorporated into nanodiscs in order to evaluate taxadiene metabolism reaction kinetics. Additionally, we perform MOE modeling studies to gain insight to the protein’s active site.
Materials and Methods
Materials
The reactants used were purchased from Fisher Scientific or Sigma-Aldrich unless stated otherwise. The yeast extract used in media was provided by Difco. Plasmid preparations were performed using Qiagen kits. Alfa Aesar provided the Ethylenediaminetetraacetic acid (EDTA) and Thiamine HCl. Gold Biotechnology was the source of the antibiotics (Spectinomycin and Ampicillin), arabinose, isopropyl β-D-1-thiogalactopyranoside (IPTG), PINKstain™ Protein Ladder, and Ni-NTA resin. Lipids were acquired from Avanti Polar Lipids and cholate was obtained from Affymetrix. Delta-ALA was bought from Frontier Scientific. EMD Millipore provided the Pall Nanosep MF (0.2 μM) and Amicon Ultra (50,000 and 10,000 MWCO) centrifugal filters. P212121.com was the source of NADPH and NADP. SimplyBlue™ SafeStain was purchased from Invitrogen. 7.5% Miniprotean® TGX™ PAGE was obtained from Bio-Rad. The taxadiene used in this project was synthesized and purified as described previously and dissolved in DMSO to 15 mM concentration [9].
Construction of his-tagged gene constructs for protein expression
Constructs were created as described before [9]. The histidine tags to construct 17-α-L-CPR and 17-α-CYP725A4 were added as published [16]. The remaining histidine tags were added using standard molecular cloning protocols. The plasmids p5Trc-(MA)CYP-l-CPR-6His (Construct CYP725A4-MA-L-CPR)and p5Trc-(2b1)CYP-l-CPR-6His (Construct CYP725A4-2b1-L_CPR) were created by amplification of the C-terminal part of CPR using primers Fw-CPRint-NcoI (5’-aagagcgcgagccatggttg) and Rv-CPR6His-SalI (5’-cggccgtcgacttagtgatggtgatgatgatgccaaatatcccgtaagtagc). A 6-His tag was added and subsequently cloned with the use of restriction enzyme sites NcoI and SalI. The plasmids p5Trc-(MA)CYP6His-o-(MA)CPR (Construct CYP725A4 MA) and p5Trc-(2b1)CYP6His-o-(2b1)CPR (Construct CYP725A4 2b1) were constructed by replacing the CYP fragment with an amplified CYP6His fragment with primers Fw-p5Trc (5’- atcatgccataccgcgaaag) and Rv-CYP6His-BamHI (5’- cgtcggatccttagtgatggtgatgatgatgcggacgagggaacag) using restriction enzyme sites NdeI and BamHI. An alignment of modifications made to the CYP is contained as Figure 2.
Figure 2. Sequence Alignment of CYP725A4 modifications in comparison to wild type CYP725A4.
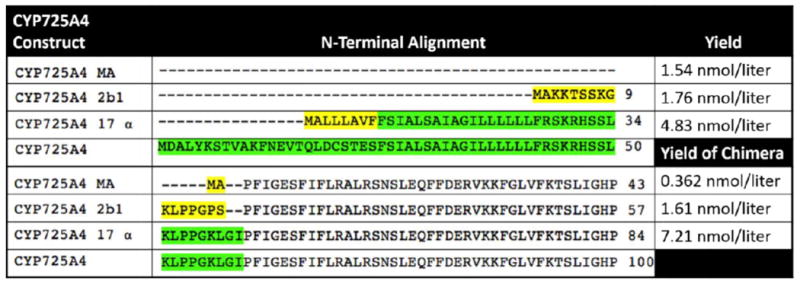
Yellow residues are amino acids that are added during the modification procedure. Green residues are amino acids that are part of wild type CYP725A4 but are deleted in one or more truncations. The yields of the purified protein from each CYP725A4 construct is shown to the right.
Fully Optimized CYP725A4 Expression
Colonies containing the CYP725A4 gene plasmid were cultured in 30 mL Luria Bertani broth supplemented with 60 μg/mL Spectinomycin or 0.1 mg/mL ampicillin antibiotics at 30° C, 250 rpm overnight (100 μg/mL ampicillin for unlinked constructs and 60 μg/mL spectinomycin for linked constructs). 5 mL of the overnight culture were used to inoculate 500 mL of Terrific Broth supplemented with 125 μL trace minerals and 60 μg/mL spectinomycin or 0.1 mg/mL ampicillin and grown at for 2 hours at 30° C and 250 rpm for 2.5 hours. Trace mineral composition is 166 mM FeCl3, 15 mM ZnCl2, 8.3 mM NaMoO4, 8.4 mM CoCl2, 9mM CaCl2, 7.3mM CuCl2, and 8.1 mM H3BO3 in 1.6 M HCl. After 2.5 hours, 250 μL of 0.1 mM delta-ALA was added to facilitate formation of the heme prosthetic group. When the OD600 approached 0.9, the addition of 500 μg IPTG and 2 g arabinose per 0.5 liter of media induced protein expression. The cells were grown for 44 hours at 250 rpm and 22° C. The bacteria cells were harvested by centrifugation for 30 minutes at 8000 rpm (10,000g) in a Sorvall GSA rotor (Thermo Scientific, Waltham, MA) at 4° C.
Purification of CYP725A4
In order to create spheroplasts and to isolate the CPR from the bacterial proteases that are known to remove the N-terminus, the cell pellet was resuspended in 500 mL of Lysozyme buffer containing 75 mM Tris-HCl pH 8, 0.25 M Sucrose, 0.25 mM EDTA, and 0.02 mg/mL Lysozyme at 4° C for 30 minutes. Spheroplasts were pelleted using the same Sorvall rotor at 8000 rpm (10,000g) and 4° C for 30 minutes and resuspended in lysis buffer. Lysis buffer contains 0.1 M pH 7.4 potassium phosphate buffer, 20% glycerol, 6 mM MgCl2, 0.1 mM DTT, 0.2 mM PMSF, and 5 mg of DNase. Spheroplasts were lysed by sonication at 40 seconds on, 40 seconds off intervals for 6 cycles. The lysate was centrifuged at 4° C for 45 minutes at 35,000 rpm (140,000g) using a Beckman Ti-45 rotor (Beckman, Coulter, Brea, CA). The pelleted E. coli membrane fraction was then suspended in extraction buffer for 5-6 hours at 4° C. Extraction buffer is 0.1 M pH 7.4 potassium phosphate buffer, 20% glycerol, 200 mM NaCl, 0.1mM DTT, and 1% Cholate. After 5 hours the mixture was centrifuged at 140,000g for 45 minutes at 4° C. The supernatant containing CYP725A4 was loaded onto a Ni-NTA column pre-incubated with column buffer. CYP725A4 column buffer is 0.1 M pH 7.4 potassium phosphate buffer, 20% glycerol, 0.1% cholate, 1 mM EDTA, and 1 mM DTT. The loaded column was washed with 50 mL of column buffer and 50 mL of column buffer containing 20 mM imidazole to remove weakly bound nonspecific proteins. CYP725A4 was eluted with 50-100 mL of column buffer spiked with 200 mM imidazole and 200 mM NaCl. A schematic of this protocol is shown as Supplemental Figure S1. Unlinked CYP725A4 (N-terminally modified CYP725A4 construct not fused to CPR) was concentrated by centrifugation in Amicon Ultra 10,000 MWCO concentrators and chimera CYP725A4 was concentrated by centrifugation in Amicon Ultra 50,000 MWCO concentrators. Imidazole was removed by buffer exchanging the protein to 0.1 M pH 7.4 potassium phosphate buffer, 20% glycerol, 0.1% cholate, 1 mM DTT, and 200 mM NaCl. Concentration and quality were assessed by UV-VIS Spectroscopy using a 1-cm path length quartz cuvette. Concentration was calculated using ε417 = 116,000 M-1 cm-1.
SDS-PAGE and native PAGE Gel
10 μL (corresponding to 0.1-0.2 nmol) of CYP725A4 were mixed with 3 μL of 5X Laemmli Buffer (60 mM Tris-HCl pH 6.8, 10% glycerol, 5% β-mercaptoethanol, 0.01% bromophenol blue) with and without 2% SDS. Denaturing samples were boiled with 100° C water for 10 min prior to being loaded into the gel. The samples were loaded into a 7.5% Miniprotean® TGX™ PAGE gel and ran at 85 mV for approximately 1-2 hours. Running buffer used was standard 1x PAGE Buffer consisting of 25 mM Tris, 192 mM glycine, with and without 0.1% SDS. PINKstain™ Protein Ladder was used to determine the protein molecular mass estimates. After being washed thrice with water, the gel was stained with SimplyBlue™ SafeStain for one hour. The gel was then washed with water to remove background dye.
Substrate Titration
CYP725A4-17α-L-CPR was pre-incubated at 32° C in a quartz cuvette. Taxadiene was then titrated into the solution using concentrations ranging from 0.06 μM to 91.7 μM and the Soret band shift at 417 nm was recorded by UV-Vis spectrometry. After addition of each aliquot of taxadiene, the titration mixture was incubated for 5 min to equilibrate before the spectra was taken. The difference in the Soret band shift from 417 to 393 nm was measured and fitted to the one-site binding model.
Nanodisc Assembly
The gene for MSP1E3D1 expression was generously provided by Dr. Stephen G. Sligar. Membrane scaffold protein (MSP) was expressed, purified, and subsequently optimized for the nanodisc assembly as previously described [22-28]. A nanodisc mixture was used to stabilize CYP725A4-17α-L-CPR for kinetic metabolism studies. Briefly, POPC lipids were dried under nitrogen overnight and subsequently solubilized in 200 mM cholate. MSP was added to this mixture in a 1:130 MSP:lipid ratio and incubated for 60 minutes at 4° C on a rocker. CYP725A4-17α-L-CPR was then added in a 10:1 MSP:CYP ratio and the mixture was allowed to incubate as before for 1 hour. Detergent was removed with an equivolume amount of Amberlite Bio-Beads for four hours at 4° C. Finally the Bio-Beads were removed from the nanodisc mixture using a spin column.
High Performance Liquid Chromatography (HPLC) Analysis of Nanodiscs
Nanodisc HPLC was performed as described previously [34].
Taxadiene Metabolism Kinetics Assays
0.2 μM of CYP725A4-17α-L-CPR mixed nanodiscs was incubated with 50 μM POPC Lipids, 400 μM NADPH, and a range of taxadiene concentrations dissolved in DMSO (20μM, 40 μM, 60 μM, 90 μM, 120 μM, 160 μM or 200 μM) in 0.1 M potassium phosphate buffer pH 7.4. The total reaction volume was 2 mL and the volume of DMSO remained constant throughout. The reaction mixtures incubated at 32° C for 1 hour and were quenched with 1x reaction volume of 4 M NaCl. The product was then extracted twice with 2x reaction volume of 4:1 hexane:ether and the organic extracts were combined and dried under nitrogen gas. The dried organic extract was shipped on dry ice for GC-MS analysis that was performed as previously described [16]. Products were identified by matching retention time to the retention times of previously verified structures [16]. Data were fit to the Michaelis-Menten equation.
Molecular Operating Environment Modeling
Molecular Operating Environment (MOE) modeling was used to observe the docking of taxadiene into a PDB structure of wild type CYP725A4 developed using PHYRE2 homology modeling [35]. The initial model created by PHYRE2 does not contain a heme; therefore, a heme was added using MOE and an open conformation of the protein model (not energy minimized) was used as a starting point. Docking utilized the MOE docking module. Before docking, cavities around the heme oxygen were chosen for the substrates to dock in. Each cavity found in the homology model was given a propensity of ligand binding number (PLB) and MOE listed these cavities with their PLB score in order from greatest propensity for a ligand to bind to the least. The two cavities that were chosen were closest to the heme oxygen and were the first cavities listed. The taxadiene molecule was inserted into chosen cavities, and MOE generated a multitude of ligand conformations in these binding sites. Several residues were found within 4.5 Å of the substrate and were influential of positioning. The conformation selected for future modeling was the conformation with the shortest distance between the heme and the atom it hydroxylated, which is the fifth carbon on taxadiene. Once the conformation was chosen, MOE was used to conduct an energy minimization step causing the protein to clamp down on the substrate more tightly, illustrating induced fitting of a substrate to its enzyme.
After the substrate was docked and the energy minimization was performed the final distance between the heme oxygen and site of attack on the substrate was measured. The interaction energy number was also found using MOE, which shows if the binding state of the substrate in the protein is feasible and energetically favorable by indicating the change of free energy when the substrate is bound. The interaction energy number for the substrate bound was a negative number, indicating a negative free energy, which corresponds to favorable binding of the substrate with 5-α hydroxylase. All renderings were created using Virtual Molecular Dynamics (VMD) [36].
Results and Discussion
Construction of CYP725A4 N-terminal Mutants
The Kyte-Dolittle plot of wild-type CYP725A4 reveals that the N-terminus of the protein is highly hydrophobic (Supplemental Figure S2). Truncating and modifying the hydrophobic N-terminal regions of CYPs has been shown to increase expression without affecting protein activity [37-40]. Previously, several modified versions of CYP725A4 were created [9]. Herein we express only three of those CYP725A4 constructs. These three N-terminal modifications are denoted as MA, 2b1, and 17α. MA features a 59-residue truncation and an insertion of a Met-Ala sequence at the start of the N-terminus, which has previously shown to increase CYP expression levels. [41-43]. 2b1 is the N-terminal addition of a 16-residue peptide MAKKTSSKGKLPPGPS, adopted from the N-terminus of rabbit CYP 2b1, to a 59-residue N-terminally truncated CYP725A4. Addition of the MAKKTSSKGKLPPGPS peptide has been shown to increase solubility of an insect CYP [44].17α is the addition of the N-terminal 8-residue peptide MALLLAVF, adopted from the N- terminus of bovine steroid hydroxylase, to a 24-residue N-terminal truncation of CYP725A4. The 17α modification was used in our previous study and has been shown to increase recombinant protein expression in E. coli [41, 45]. An alignment of these N-terminal modifications was created using Clustal 2.1 Multiple Element Sequence Alignment on standard settings [46]. This alignment is shown in Figure 2. Complete sequences and definitions of the constructs are shown in the Supplemental Material (S3-S6). Additionally, a six-residue histidine tag was added to each construct to make them amenable to purification using a Ni-NTA column. The modified gene constructs were inserted into plasmid p5Trc for expression.
Construction of CYP725A4-taxus CPR-linked constructs
For each N-terminal modification to CYP725A4, a chimera construct linking the modified CYP to a similarly modified Taxus cuspidata CPR was also created. The CPR modifications are as follows. The CPR linked to the MA CYP has the MA peptide added to a 74-residue N-terminally truncated CPR. The CPR linked to the 2b1 CYP has the MAKKTSSKGKLPPGPS peptide added to an 89-residue N-terminally truncated CPR. The CPR linked to the 17α CYP has the MALLLAVF peptide added to a 74-residue N-terminally truncated CPR. In all chimera constructs the linker peptide is GSTGS. These constructs feature the same six-residue histidine tag to enable Ni-NTA column purification. Complete sequences of linked chimera constructs are shown in Supplemental Figures S7-S9.
Expression and Purification
Each gene was transformed into BL21 (DE3) E. coli cells. Initial growth was performed with the unlinked MA strain in order to optimize growth volume and temperature. Purification yields of these initial expressions are shown in Figure 2. Both 2b1 and 17α increased yields when compared to the MA construct. As the yield of the CYP725A4-17α-L-CPR was the highest, further studies were performed with this construct. In addition, separate membrane extractions were performed with 1% cholate for 5-6 hours, 1% cholate overnight, 1% Lubrol overnight, and 1% CHAPS overnight to determine the optimal detergent for further purifications. Purifications using all detergents but lubrol generated comparable amounts of CYP725A4 while the lubrol purification generated slightly less CYP725A4. Therefore, for further studies 1% cholate was used to extract the proteins from the membranes with an incubation time of 5-6 hours.
UV-Vis Spectroscopy Characterization
UV-Vis Spectroscopy was used to assess protein concentration and quality. The representative spectrum of purified CYP725A4-17α-L-CPR is shown in Figure 3A. The protein had an absorbance peak at 417 nm. This peak indicates successful incorporation of the heme to the protein and suggests a properly folded CYP. Furthermore, the absorbance spectrum of CYP linked to CPR features a prominent shoulder at 456 nm. This shoulder corresponds to the flavin co-factors of CPR and indicates that holo-CPR has been properly expressed and purified in this linked construct.
Figure 3. Characterization of CYP725A4.
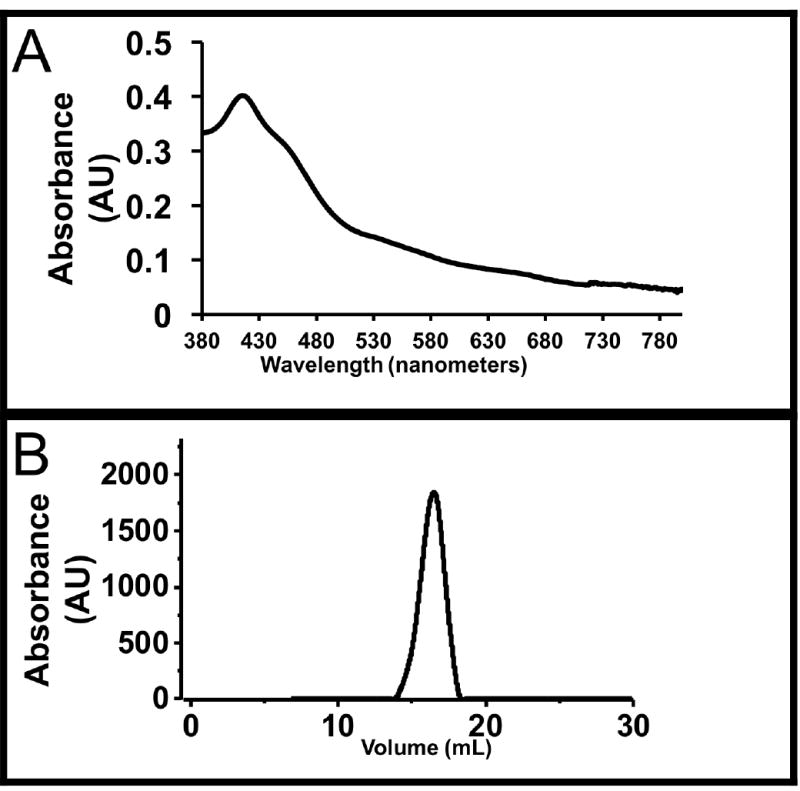
(A) UV-Vis spectra of CYP725A4-17-α-L-CPR. The peak at 417 nm is characteristic of Soret band of heme proteins. The shoulder at 456nm is due to the flavin cofactors of CPR that is linked to the CYP. (B) Size exclusion chromatogram of CYP725A4-17α-L-CPR Nanodisc mixture. The peak at 16.5 mL is indicative of CYP725A4-17α-L-CPR-nanodisc.
PAGE analysis of protein
SDS-PAGE and native-PAGE were performed to assess CYP725A4-17α-L-CPR purity. Under native conditions (Supplemental Figure S-10A), a major band at ~175 kDa was obtained. Since estimations of molecular mass are inexact using native gels, it is likely that this chimeric construct, with two large proteins connected by a linker peptide, was hindered by the gel matrix and migrated much more slowly than the ladder. This makes it appear to have an artificially high molecular mass. Using SDS-PAGE we obtain a better estimate of the molecular mass of the protein (Supplemental Figure S-10B). A clear band corresponding to a closer estimate of the molecular mass was obtained (~132 kDa observed and ~128 kDa calculated for the construct).
Assembly of Nanodisc Mixture
The nanodiscs mixture was assembled and the quality of the nanodiscs was assessed using size exclusion chromatography (Figure 3B). As seen by Figure 3B, there is an absorbance peak found at approximately 16.5 mL. Based on previously run standards of cytochrome P450-CPR-nanodiscs, a peak at 16.5 mL signifies reconstituted CYP-CPR nanodiscs. The lipid bilayers of nanodisc provides a homogenous, stable environment for measuring the activity of CYP725A4, free of other interacting proteins as compared to microsomes [22-28].
Taxadiene Titration
Typically for drug-metabolizing and steroid-synthesizing CYPs, binding of substrate near the active site heme produces a spin-state shift. This subsequently produces a Soret (heme absorbance) band shift from 417 to ~390 nm, which can be utilized to measure substrate binding. This is also termed as a type-1 shift [47]. CYP725A4-17α-L-CPR was titrated with taxadiene to determine the spin state shift of the heme iron upon substrate binding. Upon substrate binding, the CYP725A4 Soret band absorbance shifted from 417 nm to 393 nm. The binding fit to a one-site binding model with a calculated spin-state equilibrium constant, KS, of 2.1 ± 0.4 μM (Figure 4A). KS is taken to be approximately equal to KD during these experiments. Our KS (KD) of 2.1 ± 0.4 μM compares favorably to the previously reported one-binding site behavior observed by both Croteau and coworkers (KS varying between 5 and 8 μM) and Tissier and coworkers (KD of 6.9 ± 0.4 μM) [11, 12]. The work done by Croteau analyzed unmodified CYP725A4 in insect microsomes while Tissier analyzed CYP725A4 with a slight C-terminal modification in yeast microsomes. This suggests that the N-terminal modification and fusion to CPR has not affected the ability of CYP725A4 to bind taxadiene. However, these modifications have shown to slightly increase binding affinity.
Figure 4. CYP725A4-17α-L- CPR taxadiene titration.
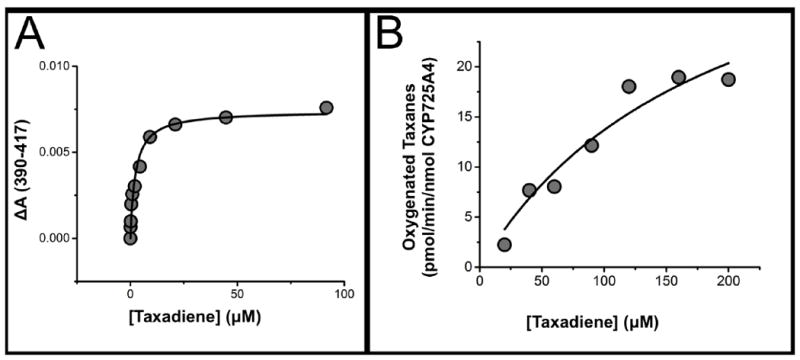
(A) Soret band at 417 nm wavelength shifts to lower wavelengths CYP725A4-17α-L-CPR upon taxadiene binding. The data was fit to the single binding isotherm and the average Ks was determined to be 2.1 ± 0.4. μM. Data shown is one trial. (B) Metabolism of taxadiene by CYP725A4-17-α-L-CPR follows Michaelis-Menten kinetics. The Vmax was 30 ± 8 pmol/min/nmol CYP725A4 and the Km was determined to be 123 ± 52 μM taxadiene. Data shown is one trial.
Taxadiene Metabolism Kinetics
To determine taxadiene metabolism kinetics, we measured the taxadiene hydroxylation activity of CYP725A4-17α-L-CPR in nanodiscs. In these experiments, CYP725A4-17α-L-CPR nanodiscs were incubated with 20-200 μM taxadiene concentrations and 400 μM NADPH for 1 hour. NADPH acts as an electron source by donating electrons to the linked CPR that then transfers them to CYP725A4. CYP725A4 subsequently uses these electrons to oxidize taxadiene into taxadiene-5α-ol or OCT [11, 12]. After extraction, the products of these incubations were identified and quantified by GC-MS [16]. Data from one of the trials is shown as Figure 4B. The metabolism of taxadiene by CYP725A4 followed Michaelis-Menten kinetics showing a Vmax of 30 ± 8 pmol/min/nmol and a KM of 123 ± 52 μM. Our KM is greater than the KM determined by Croteau’s microsomal assays (16 ± 3.2 μM) [11], but nevertheless our kinetics display reliable conversion of taxadiene to taxadiene-5α-ol indicating the expression of a functional protein. Additionally, taxadiene-5α-ol formation was not observed until CYP725A4-17α-L-CPR was incubated with 90 μM taxadiene. This suggests that metabolism was below the detecting limit or that OCT is made first. GC-MS chromatogram traces are displayed as Supplemental Figures S-11 to S-17.
CYP725A4 homology modeling with taxadiene
Molecular Operating Environment (MOE, Chemical Computing Group, Montreal, QC, Canada) software was used to identify substrate binding characteristics within the active site of the protein. A hybrid structure model was constructed from the crystal Protein Data Bank coordinates of similar CYP isozymes (class-dependent sequence alignment strategy), as described in Materials and Methods. This approach takes advantage of the conserved sequences that maintain the secondary and tertiary CYP folds to generate the core structures surrounding the buried catalytic sites. An MOE modeling system was used to dock taxadiene in the most preferred conformation (Figure 5A). This conformation placed the heme 2.36 Å away from Carbon 5 of the substrate taxadiene (Figure 5B). This distance is close enough for the hydroxylation to occur and closer than the 3.50 Å difference between the heme and carbon of attack of the best conformations of the furanocoumarins (plant toxins) xanthoxin and bergapten docked into the robustly investigated insect enzyme CYP6B1 [48]. Furthermore, the interaction potential energy number, a value calculated by MOE used to predict binding, was negative. Negative values for interaction potential energy correspond to favorable binding of the substrate to the protein active site. Several residues were influential in the docking of taxadiene to the active site. The hydrophobic residues Trp 125, Ala 129, Met 133, Ala 306, Pro 373, Val 374, Phe 375, Gly 376, Pro 482, and Leu 483 helped to stabilize and mediate the position of the highly hydrophobic taxadiene in the active site. These residues are displayed as Figure 5C. The polar residues Ser 302, His 305, Asp 309, Thr 310, and Thr 377 also influenced positioning and could function to stabilize reaction intermediates (Figure 5D). These residues would be part of future mutation studies or protein engineering of CYP725A4. The raw MOE data containing this information is shown in Supplemental Figure S18.
Figure 5. MOE docking of taxadiene to CYP725A4.
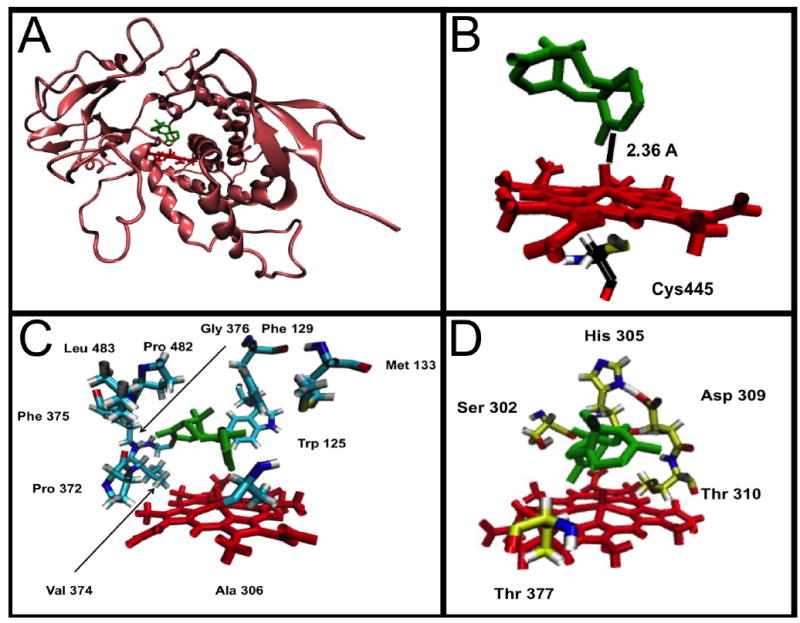
The images in this figure feature a molecule of taxadiene docked into the active site of CYP725A4 by the MOE Docking program. In all images the red structure is CYP725A4 Heme and the green structure is taxadiene. (A) CYP725A4 (champagne colored molecule) with taxadiene docked into the active site. (B) View of the CYP725A4 active site. From this view it is possible to see that the heme is 2.36 angstroms away from the carbon atom it hydroxylates. The catalytic cysteine is included as well with the following color scheme: Carbon-Black, Nitrogen-Blue, Oxygen-Red, Sulfur-Yellow, Hydrogen-Grey (C) The nonpolar residues used to position taxadiene in the active site. The residue color scheme is Carbon-Cyan, Nitrogen-Blue, Oxygen-Red, Sulfur-Yellow, Hydrogen-Grey. (D) Polar residues identified by MOE that could stabilize reaction intermediates. The residue color scheme is Carbon-Yellow, Nitrogen-Blue, Oxygen-Red, Hydrogen-Grey.
Conclusion
In this work we report the expression and purification of CYP725A4 from E. coli expression system. We then performed binding and kinetic studies of the highest yielding CYP725A4 construct (CYP725A4-17α-L-CPR) in nanodisc. To this end we titrated CYP725A4 with taxadiene and determined a binding affinity that agrees with previously published data. Furthermore, a kinetics analysis determined that the linked construct catalyzed the conversion of taxadiene to taxadiene-5α-ol and OCT following Michaelis-Menten kinetics. We therefore demonstrate that our expression and purification protocol produces functional CYP725A4. This expression and purification protocol will be useful for purifying other plant CYPs expressed in E. coli.
Supplementary Material
Highlights.
E. coli expression of six taxadiene-5α-hydroxylase (CYP725A4) constructs was achieved.
CYP725A4 constructs maintain enzymatic activity in Nanodiscs.
CYP725A4 was shown to interact favorably with taxadiene using MOE modeling
Acknowledgments
We thank Prof. Mary Schuler, Mr. Brendon Colón, and Mr. Eryk Radziszewski for their training and assistance with the MOE modeling software. We thank Ms. Jahnabi Roy (Das Lab) and Holly Pondenis (Fan Lab at UIUC) for assistance with running the gel. We thank the Molecular and Cellular Biology Summer Fellowship for supporting John’s summer research.
Footnotes
AUTHOR CONTRIBUTIONS
JER, WRA, and BWB did the experiments. JER, WRA, and AD wrote the manuscript.
The authors declare no conflicts of interest and have approved the final submission.
References
- 1.Hefner J, et al. Cytochrome P450-catalyzed hydroxylation of taxa-4(5),11(12)-diene to taxa-4(20),11(12)-dien-5alpha-ol: the first oxygenation step in taxol biosynthesis. Chem Biol. 1996;3(6):479–89. doi: 10.1016/s1074-5521(96)90096-4. [DOI] [PubMed] [Google Scholar]
- 2.Li H, et al. Studies on Taxol Biosynthesis: Preparation of Taxadiene-diol- and triol-Derivatives by Deoxygenation of Taxusin. Tetrahedron. 2008;64(27):6561–6567. doi: 10.1016/j.tet.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lokeshwar BL, Ferrell SM, Block NL. Enhancement of radiation response of prostatic carcinoma by taxol: therapeutic potential for late-stage malignancy. Anticancer Res. 1995;15(1):93–8. [PubMed] [Google Scholar]
- 4.Xu P, Bhan N, Koffas MAG. Engineering plant metabolism into microbes: from systems biology to synthetic biology. Current Opinion in Biotechnology. 2013;24(2):291–299. doi: 10.1016/j.copbio.2012.08.010. [DOI] [PubMed] [Google Scholar]
- 5.Marienhagen J, Bott M. Metabolic engineering of microorganisms for the synthesis of plant natural products. Journal of Biotechnology. 2013;163(2):166–178. doi: 10.1016/j.jbiotec.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 6.Keasling JD, Mendoza A, Baran PS. Synthesis: A constructive debate. Nature. 2012;492(7428):188–189. doi: 10.1038/492188a. [DOI] [PubMed] [Google Scholar]
- 7.Keasling JD. Manufacturing Molecules Through Metabolic Engineering. Science. 2010;330(6009):1355–1358. doi: 10.1126/science.1193990. [DOI] [PubMed] [Google Scholar]
- 8.Pickens LB, Tang Y, Chooi YH. Annual review of chemical and biomolecular engineering. Metabolic Engineering for the Production of Natural Products. 2011;2:211–236. doi: 10.1146/annurev-chembioeng-061010-114209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ajikumar PK, et al. Isoprenoid pathway optimization for Taxol precursor overproduction in Escherichia coli. Science. 2010;330(6000):70–4. doi: 10.1126/science.1191652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Biggs BW, et al. Overcoming heterologous protein interdependency to optimize P450-mediated Taxol precursor synthesis in Escherichia coli. Proceedings of the National Academy of Sciences. 2016 doi: 10.1073/pnas.1515826113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jennewein S, et al. Cytochrome p450 taxadiene 5alpha-hydroxylase, a mechanistically unusual monooxygenase catalyzing the first oxygenation step of taxol biosynthesis. Chem Biol. 2004;11(3):379–87. doi: 10.1016/j.chembiol.2004.02.022. [DOI] [PubMed] [Google Scholar]
- 12.Rontein D, et al. CYP725A4 from yew catalyzes complex structural rearrangement of taxa-4(5),11(12)-diene into the cyclic ether 5(12)-oxa-3(11)-cyclotaxane. J Biol Chem. 2008;283(10):6067–75. doi: 10.1074/jbc.M708950200. [DOI] [PubMed] [Google Scholar]
- 13.Yadav VG. Unraveling the multispecificity and catalytic promiscuity of taxadiene monooxygenase. Journal of Molecular Catalysis B: Enzymatic. 2014;110(0):154–164. [Google Scholar]
- 14.Edgar S, et al. Mechanistic Insights into Taxadiene Epoxidation by Taxadiene-5α-Hydroxylase. ACS chemical biology. 2015 doi: 10.1021/acschembio.5b00767. [DOI] [PubMed] [Google Scholar]
- 15.Barton NA, et al. Accessing low-oxidation state taxanes: is taxadiene-4 (5)-epoxide on the taxol biosynthetic pathway? Chemical Science. 2016 doi: 10.1039/c5sc03463a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Biggs BW, et al. Orthogonal assays clarify the oxidative biochemistry of Taxol P450 CYP725A4. ACS Chemical Biology. 2016 doi: 10.1021/acschembio.5b00968. [DOI] [PubMed] [Google Scholar]
- 17.Zelasko S, Palaria A, Das A. Optimizations to achieve high-level expression of cytochrome P450 proteins using Escherichia coli expression systems. Protein Expr Purif. 2013;92(1):77–87. doi: 10.1016/j.pep.2013.07.017. [DOI] [PubMed] [Google Scholar]
- 18.McDougle DR, et al. Incorporation of charged residues in the CYP2J2 F-G loop disrupts CYP2J2-lipid bilayer interactions. Biochim Biophys Acta. 2015;1848(10 Pt A):2460–2470. doi: 10.1016/j.bbamem.2015.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McDougle DR, et al. Functional studies of N-terminally modified CYP2J2 epoxygenase in model lipid bilayers. Protein Science. 2013;22(7):964–79. doi: 10.1002/pro.2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hammarström M, et al. Effect of N-terminal solubility enhancing fusion proteins on yield of purified target protein. Journal of Structural and Functional Genomics. 2006;7(1):1–14. doi: 10.1007/s10969-005-9003-7. [DOI] [PubMed] [Google Scholar]
- 21.Wang M, et al. Three-dimensional structure of NADPH-cytochrome P450 reductase: prototype for FMN- and FAD-containing enzymes. Proc Natl Acad Sci U S A. 1997;94(16):8411–6. doi: 10.1073/pnas.94.16.8411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Denisov IG, Sligar SG. Cytochromes P450 in nanodiscs. Biochim Biophys Acta. 2011;1814(1):223–9. doi: 10.1016/j.bbapap.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bayburt TH, Sligar SG. Membrane protein assembly into Nanodiscs. FEBS Letters. 2010;584(9):1721–1727. doi: 10.1016/j.febslet.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leitz AJ, et al. Functional reconstitution of Beta2-adrenergic receptors utilizing self-assembling Nanodisc technology. Biotechniques. 2006;40(5):601–2, 604, 606. doi: 10.2144/000112169. passim. [DOI] [PubMed] [Google Scholar]
- 25.Boldog T, et al. Nanodiscs separate chemoreceptor oligomeric states and reveal their signaling properties. Proceedings of the National Academy of Sciences. 2006;103(31):11509–11514. doi: 10.1073/pnas.0604988103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nath A, Atkins WM, Sligar SG. Applications of phospholipid bilayer nanodiscs in the study of membranes and membrane proteins. Biochemistry. 2007;46(8):2059–69. doi: 10.1021/bi602371n. [DOI] [PubMed] [Google Scholar]
- 27.Das A, Grinkova YV, Sligar SG. Redox potential control by drug binding to cytochrome P450 3A4. J Am Chem Soc. 2007;129(45):13778–9. doi: 10.1021/ja074864x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duan H, Schuler M. Heterologous expression and strategies for encapsulation of membrane-localized plant P450s. Phytochemistry Reviews. 2006;5(2-3):507–523. [Google Scholar]
- 29.Das A, Sligar SG. Modulation of the Cytochrome P450 Reductase Redox Potential by the Phospholipid Bilayer. Biochemistry. 2009;48(51):12104–12112. doi: 10.1021/bi9011435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bayburt TH, Sligar SG. Self-assembly of single integral membrane proteins into soluble nanoscale phospholipid bilayers. Protein Science : A Publication of the Protein Society. 2003;12(11):2476–2481. doi: 10.1110/ps.03267503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bayburt TH, Sligar SG. Single-molecule height measurements on microsomal cytochrome P450 in nanometer-scale phospholipid bilayer disks. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(10):6725–6730. doi: 10.1073/pnas.062565599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Civjan NR, et al. Direct solubilization of heterologously expressed membrane proteins by incorporation into nanoscale lipid bilayers. BioTechniques. 2003;35(3):556–60. 562–3. doi: 10.2144/03353rr02. [DOI] [PubMed] [Google Scholar]
- 33.Duan H, et al. Co-incorporation of heterologously expressed Arabidopsis cytochrome P450 and P450 reductase into soluble nanoscale lipid bilayers. Archives of Biochemistry and Biophysics. 2004;424(2):141–153. doi: 10.1016/j.abb.2004.02.010. [DOI] [PubMed] [Google Scholar]
- 34.McDougle DR, et al. Functional studies of N-terminally modified CYP2J2 epoxygenase in model lipid bilayers. Protein Science. 2013;22(7):964–979. doi: 10.1002/pro.2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kelley LA, et al. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protocols. 2015;10(6):845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. Journal of molecular graphics. 1996;14(1):33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 37.Gonzalez FJ, Korzekwa KR. Cytochromes P450 expression systems. Annu Rev Pharmacol Toxicol. 1995;35:369–90. doi: 10.1146/annurev.pa.35.040195.002101. [DOI] [PubMed] [Google Scholar]
- 38.Larson JR, Coon MJ, Porter TD. Purification and properties of a shortened form of cytochrome P-450 2E1: deletion of the NH2-terminal membrane-insertion signal peptide does not alter the catalytic activities. Proceedings of the National Academy of Sciences of the United States of America. 1991;88(20):9141–9145. doi: 10.1073/pnas.88.20.9141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.von Wachenfeldt C, et al. Microsomal P450 2C3 Is Expressed as a Soluble Dimer inEscherichia coliFollowing Modifications of Its N-terminus. Archives of Biochemistry and Biophysics. 1997;339(1):107–114. doi: 10.1006/abbi.1996.9859. [DOI] [PubMed] [Google Scholar]
- 40.Richardson TH, et al. A Universal Approach to the Expression of Human and Rabbit Cytochrome P450s of the 2C Subfamily inEscherichia coli. Archives of Biochemistry and Biophysics. 1995;323(1):87–96. doi: 10.1006/abbi.1995.0013. [DOI] [PubMed] [Google Scholar]
- 41.Barnes HJ, Arlotto MP, Waterman MR. Expression and enzymatic activity of recombinant cytochrome P450 17 alpha-hydroxylase in Escherichia coli. Proc Natl Acad Sci U S A. 1991;88(13):5597–601. doi: 10.1073/pnas.88.13.5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miki Y, Asano Y. Biosynthetic Pathway for the Cyanide-Free Production of Phenylacetonitrile in Escherichia coli by Utilizing Plant Cytochrome P450 79A2 and Bacterial Aldoxime Dehydratase. Applied and Environmental Microbiology. 2014;80(21):6828–6836. doi: 10.1128/AEM.01623-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guo ZY, et al. Expression of Modified Human Cytochrome P450 1A1 in Escherichia coli: Effects of 5′ Substitution, Stabilization, Purification, Spectral Characterization, and Catalytic Properties. Archives of Biochemistry and Biophysics. 1994;312(2):436–446. doi: 10.1006/abbi.1994.1330. [DOI] [PubMed] [Google Scholar]
- 44.Budriang C, R P, Y J. An Expression of an Insect Membrane-bound Cytochrome P450 CYP6AA3 in the Escherichia coli in Relation to Insecticide Resistance in a Malarial Vector. Pakistan Journal of Biological Sciences. 2011;14(8):466–475. doi: 10.3923/pjbs.2011.466.475. [DOI] [PubMed] [Google Scholar]
- 45.Müller P, et al. Field-Caught Permethrin-Resistant Anopheles gambiae Overexpress CYP6P3, a P450 That Metabolises Pyrethroids. PLoS Genetics. 2008;4(11):e1000286. doi: 10.1371/journal.pgen.1000286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 47.Al-Gailany KAS, Houston JB, Bridges JW. The role of substrate lipophilicity in determining type 1 microsomal P450 binding characteristics. Biochemical Pharmacology. 1978;27(5):783–788. doi: 10.1016/0006-2952(78)90521-x. [DOI] [PubMed] [Google Scholar]
- 48.Baudry J, et al. Molecular docking of substrates and inhibitors in the catalytic site of CYP6B1, an insect cytochrome P450 monooxygenase. Protein Engineering. 2003;16(8):577–587. doi: 10.1093/protein/gzg075. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


