Abstract
Copper (Cu) was used in antiquity to prevent waterborne and food diseases because, as a broad-spectrum antimicrobial agent, it generates reactive oxygen species, ROS. New technologies incorporating Cu into low-cost biodegradable nanomaterials built on cellulose, known as cellulosic cupric nanoparticles or c-CuNPs, present novel approaches to deliver Cu in a controlled manner to control microbial growth. We challenged strains of Saccharomyces cerevisiae to soluble Cu and c-CuNPs to evaluate the potential of c-CuNPs as antifungal agents. Cells exposed to c-CuNPs demonstrated greater sensitivity to Cu than cells exposed to soluble Cu, although Cu-resistant strains were more tolerant than Cu-sensitive strains of c-CuNP exposure. At the same level of growth inhibition, 157 μM c-CuNP led to the same internal Cu levels as did 400 CuSO4, offering evidence for alternative mechanisms of toxicity, perhaps through β-arrestin dependent endocytosis, which was supported by flow cytometry and fluorescence microscopy of c-CuNPs distributed both on the cell surface and within the cytoplasm. Genes responsible for genetic variation to copper were mapped to the ZRT2 and the CUP1 loci. Through proteomic analyses, we found that the expression of other zinc (Zn) transporters increased in Cu-tolerant yeast compared to Cu-sensitive strains. Further, the addition of Zn at low levels increased the potency of c-CuNP to inhibit even the most Cu-tolerant yeast. Through unbiased systems biological approaches, we identified Zn as a critical component of yeast response to Cu and the addition of Zn increased potency of the c-CuNPs.
A. Introduction
Copper (Cu) has been used since ancient times to prevent microbial growth because of its broad-spectrum activity coupled with multiple modes of toxicity1. Excessive Cu induces damage by catalyzing the generation of reactive oxygen species (ROS)2. ROS generated by extracellular Cu oxidizes lipids and proteins of the cell membrane3 and, if cells survive these lesions, then DNA damage can occur from the high levels of internal Cu4. These mechanisms of toxicity and the response to Cu in microorganisms have been widely studied5. Importantly, the expression of genes in response to Cu is highly regulated to maintain a fine balance of this essential, yet toxic, trace metal within the cytoplasm6,7.
Nanotechnology is a transformative tool that can be used to develop and enhance high-value products from renewable and biocompatible raw materials such as wood8–12. Cellulose, as the predominant component of woody biomass, can be utilized not only commercially where bio-compatibility and biodegradability are relevant, but also as a support structure for nanoparticle formation in novel applications. The use of the cellulose nanostructure as a template and stabilizer for metallic nanoparticles expands the potential applications of nanomaterials13. Cellulosic cuprous nanoparticles (c-CuNPs) display effective antimicrobial activity against a non-pathogenic Escherichia coli DH5α14,15. c-CuNPs contain reduced copper (Cu(I)), which is thought to be slowly released as Cu(I) is oxidized making Cu(II) available to interact with biological molecules leading to toxicity.
Improving c-CuNPs depends on understanding its range of functions in order to manufacture products with increased antimicrobial effectiveness. We have developed a cost-effective hybrid nanomaterial built on a cellulosic scaffold with Cu(I) chemically linked to cellulose16. To understand how yeast respond to Cu delivered in c-CuNPs, we carried out toxicological and quantitative proteomic analyses on genetically diverse yeast strains. c-CuNPs delivered more Cu to yeast cells compared to soluble CuSO4, and this may result from the endocytic uptake of c-CuNPs. The levels of hundreds of proteins changed in response to Cu exposure, and yeast strains that do not tolerate Cu fail to increase expression of Zrt1, a high affinity zinc (Zn) transporter. Further, a genome-wide quantitative trait loci (QTL) analysis uncovered a role for Zn transporters in Cu response. The genetic variation in the low-affinity membrane transporter, Zrt2, directly contributed to variation of Cu tolerance, while yeast lacking Zrt1 or Zrt3 were sensitive to high levels of Cu. Supplementing Zn decreased growth of all yeast strains tested when also challenged with c-CuNPs. Understanding the interdependence of metal tolerances provides insights into improving antimicrobial properties of metal-based materials.
B. Materials and Methods
Yeast strains and plasmids
S288c strains used included strains S96 (MATa lys517) and GSY147 (MATa prototroph18). YJM789 (MATalpha lys217,19) was the parent of YJM789K5a (MATa prototroph). YJM789K5a was generated by transforming a PCR product containing LYS2 into YJM789 and then backcrossed to HS959 (MATa lys2) that is otherwise isogenic to YJM789. All S288c strains displayed the similar copper resistance. Both YJM789 strains were copper sensitive. Other strains used were AWRI1631, RM11, and YJM33920–22. The haploid recombinant segregant collection between YJM789 and S96 was previously generated17. Knockout strains were previously described23. Cloning of ZRT2 alleles was done by PCR amplified from genomic DNA using primers as follows 5′ZRT2 GTACCTGCAGGCGTCCTAATGAAAATGGAGG and 3′ZRT2 TTCCCGGGATTACTCTGCCCTTTTC. Endogenous promoter and terminator sequences within 580 nucleotides upstream of the start and 277 nucleotides downstream of the stop codons were included. Genes were cloned into the SbfI and XmaI sites of pGS36 plasmid with the hygromycin resistance marker18. Plasmids were kept under selection with hygromycin after LiAc chemical transformations.
Media and chemicals
All haploid yeast strains were grown in yeast minimal media (YM), which includes 2% dextrose, 6.7g/L yeast nitrogen base, and 20g/L agar in solid media. In liquid media, the agar was omitted. YM was, when needed, supplemented with lysine, methionine, uracil, leucine, and histidine. Plasmids were maintained by addition of 250 μg/ml hygromycin. All antioxidants were added to solid media when it had cooled to 55°C and used within 24 hours. Chronic exposure was defined as two days of exposure, while acute exposure was defined as two hours of exposure. Viability assays were done in liquid culture in biological triplicates. Yeast were grown to log-phase and split into subcultures. Subcultures served as controls, receiving no added Cu; soluble Cu-treated, receiving various concentrations of CuSO4.5H2O (Ricco Chemicals, CAS 7732-18-5); or receiving c-CuNPs. Carboxymethyl cellulose (CMC) copper nanoparticles (c-CuNPs) were created and physically characterized as previously reported16, where Cu is reduced and bound onto free hydroxyls of the cellulose scaffold. 10 μg/ml of c-CuNP corresponded to 157 μM of soluble Cu determined by atomic absorption spectrometry (AAS). Yeast media is acidic (pH 5), which would cause dissociation and oxidization of Cu from the c-CuNPs. Indeed, 27% more free copper was found in c-CuNPs suspended in YM than in water alone.
Atomic absorption spectrometry
Strains YJM789K5a, GSY147, RM11, and YJM339 were grown in YM media in triplicate into log phase. The control samples (YM) were collected. The remaining cultures were treated with 400 μM CuSO4 or 157 μM Cu as c-CuNP (10 μg/ml of CMC) for 2 hours. For cell collection, equal number of cells, as determined by optical density at 600 nm, was obtained (equivalent to 4 ODu). These samples were centrifuged and washed twice with distilled water. One ODu was separated for protein quantification. Samples were split, frozen in liquid nitrogen and stored at –80°C. Soluble protein concentration was determined via Bradford assay. For AAS, the cell pellets were treated with 600 μl of concentrated HNO3 and 200 μl of 30% H2O2 for digestion. These solutions were transferred to glass tubes and placed in a boiling water bath for 2 hours until clear. A 1:10 dilution with distilled water was analyzed for Cu at 324.8 nm. Copper concentrations were given in mg/L, adjusted for dilution, and normalized to soluble protein concentration for each sample. AAS was performed across four days, and a total average for the four repeated samples was determined for each day. From these values, the overall average was determined. To calculate the correction factor for each day’s analysis, the overall average was divided by the daily average. This correction factor was then applied to all the values for that particular day, prior to dilution adjustments.
Staining of yeast cells and cellulose copper nanoparticles for flow cytometry and microscopy
Cells of each strain were cultured from frozen stock in YM media to log phase and harvested by centrifugation. Cells were then transferred to 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) buffer (pH of 7.4) containing 5% dextrose. Rhodamine B was added to a final concentration of 100 nM, and cells were incubated at room temperature for 20 minutes in the dark. c-CuNPs were added to phosphate buffered saline (PBS) to a concentration of 400 μM with 5.136 mM FITC in dimethyl sulfoxide (DMSO) to a final concentration of 2.568 μM. The FITC-dyed PBS nanoparticle solution was incubated in the dark for 8 hours at 4°C. Because excess FITC could not be removed from the solution, 5.136 mM in DMSO was added to PBS to a final concentration of 2.568 μM and incubated in the dark for 8 hours at 4°C to serve as a control.
Fluorescein isothiocyanate (FITC), HEPES, Rhodamine B Hexyl Ester (RhodB), and dextrose were purchased from Fisher Scientific (Chicago, IL). DMSO was purchased from Biotium (Hayward, CA). A four laser LSR/Fortessa (BD Biosciences, San Jose CA) with BD FACS Diva software (BD Biosciences) located in the WVU Flow Cytometry & Single Cell Core Facility was used to collect flow cytometric data. Cells of each strain were transferred from HEPES buffer, treated, and transferred to PBS. To determine the extent of association of c-CuNPs with the yeast cell wall, 1mL of PBS containing 1 million dyed cells was added to 1mL of 400 μm dyed c-CuNPs PBS solution. To control for FITC labeling of cells, 1 mL of PBS containing 1 million dyed cells was added to 1 mL of 2.568 μM FITC in PBS. Solutions were allowed to incubate at 30°C for 2 hours. Samples in PBS were analyzed on a LSR Fortessa (BD Biosciences San Jose CA) using BD FACS Diva version 8.0 software (Fig S1). A total of 10,000 events were collected/sample and the median fluorescence intensity for each sample was calculated. Also, in order to ensure that the FITC signal only came from c-CuNPs that were inside the yeast, trypan blue was added to the samples prior to analysis on the flow cytometer.
After treatment, samples were harvested by centrifugation and re-suspended in 1ml PBS containing 4% paraformaldehyde and incubated for 30 minutes at 30°C. Cells were washed twice with PBS and re-suspended in 500 μl 80% glycerol before sonicating at low for 5 intervals of 1 second bursts on a Branson model SSE-1 Sonifier. Concanavalin A (Sigma – L7647) coated microscope slides were utilized to mount 10 μl of the samples and sealed with clear fingernail polish. Fluorescent yeasts were viewed with a laser scanning confocal microscope (Olympus FV1000) equipped with red/green HeNe and argon lasers. Images were acquired using a 60x oil immersion magnification optical objective. Distances between confocal planes were optimized for the objective using Fluoview software (FV10-ASW Version 04.00.02.09). Pixel resolution was adjusted for best resolution capable of performing a z-stack scan. Images were only modified for contrast enhancement.
Viability assay
After two hours with 400 μM of copper or 157 μM c-CuNPs, cultures were washed twice with water, and the OD600 was measured. 0.1 OD units (ODu) of each sample were diluted 100-fold and 100 μl plated on YPD. The number of untreated colonies that grew were considered 100% viability, and the viability of treated cells were taken as a percentage of untreated cells.
Proteomics
To measure the global proteomic changes of S288c and YJM789 in response to Cu, cells were treated for 90 minutes in YM with or without 800 μM CuSO4. Peptides were prepared using a modified FASP method24. Treated cells were harvested by briefly spinning down 5 ODu of cells, washing once with water and then freezing cell pellets in liquid nitrogen in 2 mL screw cap tubes. Cells were lysed in 4% SDS, 100 mM DTT, and 100 mM Tris pH 8.5 in a 1:10 v/v cell pellet to buffer. 0.5 mm zirconium beads were added and cells were lysed in a Fastprep at 3x 20 seconds at speed 5 with a 5-minute rest at room temperature. Using a 24-gauge needle, the bottom of the tube was punctured and placed into 2 ml low protein binding locking cap tubes. All liquid and cellular debris were then spun at 1000 rpm into a 2-ml tube. Tubes were incubated at 95°C for 4 minutes. Cellular debris was pelleted for 10 minutes at 14,000 rpm at room temperature. The supernatant was transferred to new tubes. Protein concentration was calculated using Bradford reagent on 1:100 diluted sample. 200 μg of protein was diluted with 8 M urea/100 mM Tris pH 8.5 to a final SDS concentration of 1% and transferred to a Microcon YM-30 column. Microcon columns were spun at 10,000 rpm. Proteins were washed five times with 8 M urea/100 mM Tris pH 8.5 and resuspended in 100 μl 0.05 M idoacetamide in 8 M urea/0.1 M Tris HCl 8.5. Samples were alkylated for 45 minutes in the dark. Liquid was spun through the Microcon. Buffer was exchanged into 200 mM TEAB by washing proteins five times. Protein concentrations were retested using a BCA kit (Thermo Fisher Scientific) on 1:100 diluted samples. Trypsin was freshly dissolved in 200 mM TEAB and incubated at 30°C for 20 minutes. Trypsin was incubated with proteins on the Microcon wrapped in parafilm overnight at 37°C at 1:20 dilution. Digested peptides were spun through the Microcon and collected into fresh tubes. Concentration of peptides were approximated, and 5 μg of peptides were labeled with Thermo Fisher Tandem Mass Tags 6plex in a 10:4.1 ratio according to manufacturer’s instructions for 1.5 hours at room temperature. The reaction was quenched with 5% hydroxylamine diluted in 200 mM TEAB. Equal amounts of labeled peptides were mixed and dried.
Peptides were resuspended in 100mM ammonia formate at pH 10 and separated by reverse phase chromatography at high pH in the first dimension, followed by an orthogonal separation at low pH in the second dimension. In the first dimension the mobile phases were buffer A:20mM ammonium formate at pH 10 and buffer B: Acetonitrile. Peptides were separated on a Xbridge 300μm x 5 cm C18 5.0μm column (Waters) using 15 discontinuous step gradient at 2 μl/min. Peptides eluted off from the first dimension were trapped by Symmetry 180μm x 2cm C18 5.0 μm trap column (Waters). In the second dimension, peptides were loaded to an in-house packed 75 μm ID/15 μm tip ID x 20cm C18-AQ 3.0 μm resin column with buffer A (0.1% formic acid in water). Peptides were separated with a linear gradient from 5% to 30% buffer B (0.1% formic acid in acetonitrile) at a flow rate of 300 nl/min over 180 minutes. Each sample separation was repeated three times.
Peptides were identified by a linear trap quadrupole (LTQ)-Orbitrap Velos instrument (Thermo Fisher Scientific). The mass spectrometer was run in a data dependent mode. One survey scan acquired in the Orbitrap mass analyzer with resolution 60,000 was followed by MS/MS of the 10 most intense peaks with charge state ≥ 2 and above an intensity threshold of 5,000. Peptides were fragmented in the high collisional Cell (HCD) with normalized collision energy of 40% and activation time of 0.1 s. The MS/MS scan was acquired in the Orbitrap at resolution of 7,500.
Peptides were identified and quantitated using Proteome Discoverer 1.4.1.14 (S1 Table). Spectra were searched against a protein database consisting of all the S288c (http://downloads.yeastgenome.org/sequence/S288C_reference/genome_releases/S288C_reference_genome_R64-1-1_20110203.tgz) and YJM789 (http://downloads.yeastgenome.org/sequence/strains/YJM789/YJM789_Stanford_2007_AAFW02000000) proteins available on Saccharomyces Genome Database (http://www.yeastgenome.org). Common contaminants such as human keratin and trypsin were also included in the database to prevent peptides of those contaminants from being falsely identified as yeast peptides (S2 Table). TMT6plex was used for quantification labeling according to manufacturer’s instructions and the six isobaric labels of 126–131 Da were assigned to samples as follows: 126 – S288c, YM, replicate 1; 127 – S288c, YM, replicate 2; 128 – S288c, Cu treated, replicate 1; 129 – S288c, Cu treated, replicate 1; 130 – YJM789, YM; 131 – YJM789, Cu treated.
The default method used by Proteome Discoverer to quantify protein fold change from peptide intensities is to calculate the median intensity ratio between two isobaric labels for all the peptide spectrum matches (PSMs) associated with the protein in question. This method is problematic when a peptide sequence is identified in multiple PSMs where ratios have large variation. For example, a peptide can have different phosphorylation states between two treatments, and consequently show very different ratios for the phosphorylated PSMs and the unphosphorylated PSMs. This type of variation can skew quantification results when there are only a few PSMs usable for quantification; therefore, we used different methods than the Proteome Discoverer default for protein quantification. For each peptide sequence identified by Proteome Discoverer, the reporter ion intensities of all PSMs associated with this peptide were summed into the total intensity for this peptide in each sample. For each protein, only its unique (not belonging to any other protein) peptides of high to middle confidence (false discovery rate 0.01–0.05) were considered usable for quantification. Four comparisons, each between two experimental scenarios, were performed by calculating intensity ratios of reporter ions: (1) S288c, Cu vs. YM – (128 + 129) / (126 + 127); (2) YJM789, Cu vs. YM – 131 / 130; (3) YJM789 vs. S288c, YM – 130 × 2 / (126 + 127); (4) YJM789 vs. S288c, Cu – 131 × 2 / (128 + 129). For the comparisons between strains, (3) and (4), only peptides showing no sequence polymorphisms between the two strains were considered usable for quantification. Four methods were used to calculate protein ratios for each comparison: (1) ‘means then ratio’ – the mean of all the usable peptides belonging to this protein was calculated for each experimental scenario, then the ratio between the two scenarios was calculated from the two means; (2) ‘medians then ratio’ – the median of usable peptides was calculated for each scenario, then the ratio was calculated from two medians; (3) ‘ratios then mean’ – the ratio between the two scenarios was calculated for each usable peptide, then the mean of all the ratios was calculated; (4) ‘ratios then median’ – ratio was calculated for each usable peptide, then the median of all the ratios was calculated. For each of the four methods and each comparison of two scenarios, the resulting ratios for all the proteins were divided by the median of the ratios for normalization. This normalization method assumes that the majority of reported proteins do not change much in each comparison, which was the case in our experiment (see Results). For each comparison, only the proteins showing ratios > 2 or < 0.5 by all the four methods were included in Table S1. The ratios calculated using method (1) ‘means then ratio’ were transformed into logarithms of 2 (two).
63,563 PSMs were reported with high to middle confidence (false discovery rate 0.01–0.05), representing 27,794 unique peptide sequences. Peptides were labeled with isobaric mass tags for quantification with Tandem Mass Tags (TMT). TMT labeling rates of those PSMs were 92.7–97.3%. 4,431 yeast proteins were reported containing at least one peptide with middle to high confidence (each case of two homologous proteins from S288c and YJM789 were counted as one protein), among which 2,991 were quantified with at least two unique peptides and considered for differential abundance quantification. 112 proteins showed greater than two-fold change (absolute value of log2 (fold change) or |log2fc| > 1) between strains and/or copper treatments (Table S1) by all the four calculation methods for fold change.
Quantitative trait loci studies on copper response
Strains from the recombinant haploid segregant collection17 from crossing S288c (S96) and YJM789 were grown to saturation in YM supplemented with lysine. Cells were then diluted to 0.2 OD600 in 200 μl in YM or YM with 50 μM CuSO4 in triplicate in 96 well plates. Plates were sealed and readings taken every hour in a TECAN M200, an automatic plate reader with no shaking to mimic growth on solid media. Culture density at 12-hours (late-log phase) and at 18-hours (endpoint or stationary phase) were used to map quantitative trait loci (QTL). All readings in Cu were subtracted from the same strain grown in YM for QTL values (Table S3). For each locus, we calculated a growth score and a significance score. To calculate the growth score, all strains that inherited the segregant YJM789 (1) allele were assigned to group 1, and all strains that inherited the segregant S96 (2) allele were assigned to group 2. Average growth was calculated for strains in group 1 and was subtracted from the average growth for strains in group 2. The calculation resulted in the growth score. A positive growth score indicates that the segregant 1 allele leads to high growth and Cu resistance while a negative growth score indicates the segregant 2 allele confers Cu resistance. The significance score was also calculated, which is the probability of these growth scores occurring if the alleles were assigned randomly. To calculate the significance score, each yeast strain was randomly assigned to group 1 or group 2 regardless of that allele was inherited. The growth score was calculated given these 2 randomly assigned groups and repeated 100,000 times using a Monte Carlo simulation. The resulting distribution over growth scores was approximately normal. This normal distribution was used to calculate the significance score. We then studied the loci that have large magnitude growth scores and high significance scores for 12-hour growth (Table S4) and 18-hour growth (Table S5).
C. Results
Clinical isolates are more sensitive to copper than agricultural isolates
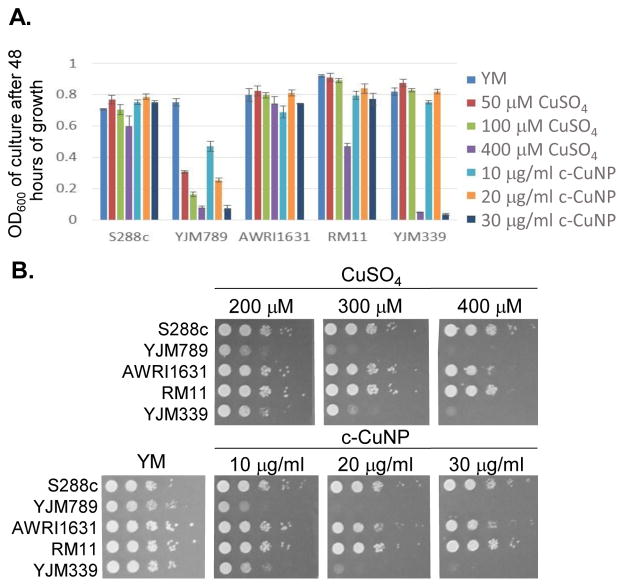
S. cerevisiae is among the oldest domesticated species and has evolved high tolerances to copper independently multiple times both in the wild and in lab-evolution experiments5,25–29. To assess antimicrobial potency of copper incorporated onto cellulosic nanoparticles, genetically diverse yeast were incubated in liquid culture with different concentrations of c-CuNP [16]. The agricultural isolates AWRI163120, RM1121,30,31, and S288c32,33 were resistant to Cu whereas YJM33919 and YJM78919,34, clinical isolates of yeast, had lower Cu tolerances (Fig 1A). The c-CuNPs were diluted to a level that would inhibit growth of YJM789 to a similar degree as 400 μM of soluble Cu when measured for growth in a plate reader with no shaking, mimicking their growth on solid media. Both 30 μg/ml (by CMC concentration) of c-CuNP and 400 μM of soluble copper decreased growth of YJM789 by 90% compared to YM media. Similarly, both the soluble Cu and c-CuNP embedded into solid agar media inhibited the growth of yeast, with the agricultural isolates being more resistant than the clinical isolates (Fig 1B). However, growth patterns on agar suggested that c-CuNPs may be more toxic than soluble Cu, with growth at 10 μg/ml c-CuNP inhibiting growth to the same extent as 200 μM Cu (Fig 1B).
Fig 1. Impact of genetic variation in S. cerevisiae to soluble copper and cellulosic hybrid copper nanoparticles.
A. Growth of yeast in liquid culture in synthetic media (YM) with an addition of dilutions of CuSO4 and c-CuNP. B. Serial dilutions of genetically diverse S. cerevisiae with dilutions of CuSO4 and c-CuNP on solid YM media.
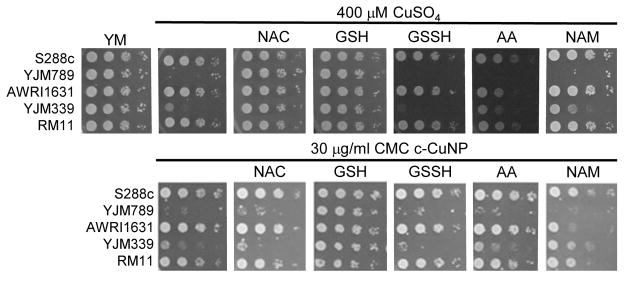
Because copper can catalyze the generation of reactive oxygen species2, we tested the ability of antioxidants to quench free radicals induced by soluble copper and c-CuNP. Both glutathione (GSH) and N-acetylcysteine (NAC), a glutathione precursor, rescued growth inhibition by soluble copper in clinical strains YJM789 and YJM339 (Fig 2). No growth rescue was detected with oxidized glutathione (GSSH) or ascorbic acid (another antioxidant) in the presence of Cu for YJM789, but there was a slight improvement for YJM339. When cells were treated with nicotinamide (NAM) and ascorbic acid (AA), the Cu tolerance of YJM339, but not YJM789, increased slightly, suggesting that, although YJM789 and YJM339 are Cu sensitive, there are other genetic differences that affect Cu response. However, NAC could not rescue growth inhibition caused by c-CuNPs. While NAC can function as an antioxidant when it is imported into cells, NAC is the precursor to glutathione, which is more potent at reducing ROS35. Therefore, while the pattern of c-CuNP sensitivity approximately mirrored soluble Cu sensitivity, the difference in NAC rescue suggests differences in Cu delivery, and likely, cellular internalization, between cells exposed to soluble Cu and c-CuNPs.
Fig 2. Rescue of copper nanoparticle sensitivity with antioxidants.
Serial dilutions of genetically diverse S. cerevisiae on synthetic media (YM) with an addition of 400 μM CuSO4 or c-CuNPs and supplemented with 100 μM of antioxidants as listed N-acetylcysteine (NAC), glutathione (GSH), oxidized glutathione (GSSH), ascorbic acid (AA) and nicotinamide (NAM). Haploid strains were serially diluted and spotted onto solid media. After 2 days of growth at 30°C plates were photographed.
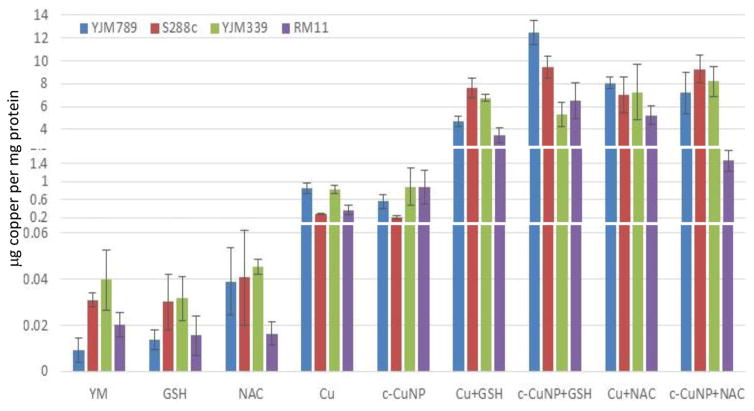
To determine how much Cu is imported into yeast, internal copper levels were measured by atomic absorption spectrometry (AAS). When grown in YM, YJM789 has three-fold less Cu than YJM339 or S288c. Furthermore, YJM789 accumulates as much Cu as RM11, a Cu-resistant strain (Fig 3). Thus, basal untreated internal Cu levels do not correspond to Cu sensitivity. Treatment with GSH or NAC did not significantly change internal copper levels except for YJM789, in which cellular Cu increased with NAC to levels comparable to S288c and YJM339 (Fig 3). Treatment with 400 μM Cu increased internal Cu concentrations in all strains, although to varying extent, by between 9- and 90-fold (Fig 3). YJM789 and YJM339 had ~0.8 μg of internal Cu per mg of protein, whereas Cu-resistant strains S288c and RM11 had levels 66% to 53% lower, respectively. To establish internal similar levels of copper to levels from c-CuNP as 400 μM soluble copper, the amount of c-CuNP was decreased to 157 μM of Cu bound to CMC (= 10 μg/ml CMC). The acute exposure of yeast to 157 μM Cu as c-CuNP led to higher Cu accumulation than levels measured when exposed to soluble Cu (Fig 3): internal Cu concentrations of yeast treated with 157 μM Cu as c-CuNP were similar to yeast treated with 2.5-times higher levels of CuSO4. Co-treatment of yeast with glutathione and soluble Cu or c-CuNP increased Cu uptake by 4- to 10-fold than copper treatments alone. The internal levels of Cu for GSH- and NAC-treated yeast, in general, were elevated and similar irrespective of copper source with two exceptions. YJM798 accumulated elevated Cu from c-CuNPs when supplemented with GSH and RM11 exposed to c-CuNP and NAC had less internal Cu levels.
Fig 3. Internal levels of copper in yeast treated with different sources of copper and antioxidants.
Internal levels of copper were measured by atomic absorption spectrometry and normalized to protein content. The following treatments are displayed in this graph: YM, 157 μM c-CuNP, 400 μM CuSO4, 100 μM GSH, 100 μM NAC, 157 μM c-CuNP + 100 μM GSH, 157 μM c-CuNP + 100 μM NAC, 400 μM CuSO4 + 100 μM GSH, and 400 μM CuSO4 + 100 μM NAC. The graph is rescaled to better show the concentrations of the controls. The average of three independent samples for each treatment was determined and plotted error bars.
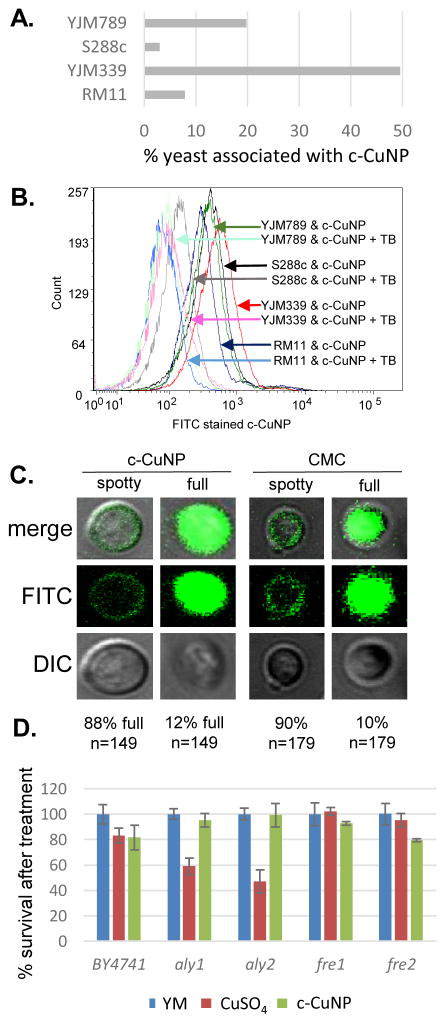
Because the c-CuNPs increased efficiency of Cu delivery and lacked the NAC rescue to c-CuNP toxicity, we turned our focus to additional paths of Cu delivery by c-CuNP. c-CuNP are 10–20 nm particles with a unique three-dimensional spherical structure built on a cellulose scaffold16. To assess the interaction of c-CuNP with the outer surface of yeast cells, yeast cells were measured by flow cytometry in presence of c-CuNP. Yeast were stained with Rhodamine B, which crosses cell membranes and is sequestered by mitochondria without inducing cell lysis, while c-CuNPs were stained with FITC. Because c-CuNPs labeled with FITC are too small to be directly detected in flow cytometry, FITC labeling can only be measured when c-CuNPs are associated with yeast cells (Fig S1). The fluorescence intensities of S288c, YJM789, YJM339, and RM11 cells incubated for two hours with 157 μM of FITC pre-stained c-CuNPs increased compared to yeast cells alone (Fig 4A), suggesting the binding or uptake of c-CuNPs by yeast cells. Additionally, a greater proportion of cells of YLM789 and YJM339 accumulated FITC-labeled c-CuNPs (Fig 4A) and accumulated these to a greater extent (Fig 4B) than S288 c or RM11 lines.
Fig 4. Physical interaction of copper nanoparticle and yeast cells.
A. Flow cytometry measuring interaction between FITC stained c-CuNP and Rhodamine B stained yeast strains, YJM789 (YJM789K5a), S288c (GSY147), RM11 and YJM339. 157 μM. Dual stained c-CuNP and yeast are graphed as percentage. Yeast were stained with Rhodamine B. c-CuNP and yeast were then incubated for two hours. B. Flow cytometry of FITC stained c-CuNP associated with yeast were quenched with trypan blue (TB). Arrows point to peaks as they shift in fluorescence. The number of cells measured is on the x-axis as count. Light lines represent yeast with c-CuNP and solid lines represent yeast and c-CuNP treated with trypan blue. C. Confocal microscopy of yeast (GSY147) and FITC stained c-CuNP. D. Cellular viability of yeast knockouts in the BY4741 (S288c) background normalized to yeast grown in YM supplemented with required amino acids treated with 400 μM CuSO4 or 157 μM c-CuNP.
To evaluate the physical location of c-CuNPs on yeast cells, cells exposed to FITC-labeled c-CuNPs were further treated with trypan blue, which quenches FITC fluorescence. Trypan blue does not enter living cells and can only quench FITC if the dye was physically located outside the yeast. This addition decreased fluorescence intensity FITC-stained c-CuNP associated with yeast (Fig 4B). To confirm that c-CuNPs were both outside and inside, the yeast were again treated with c-CuNP and CMC (cellulose backbone of the nanoparticles). Further examination revealed two populations of cells: most of the FITC stained c-CuNPs appeared as speckled spots on the outside of the yeast, while about 10% of the cells exhibited fluorescence inside the cells as seen through z-stack using confocal microscopy (Fig 4C). The extracellular and intracellular fluorescence was not dependent on the presence of Cu on CMC (Fig 4C).
To determine whether c-CuNPs were actively internalized by endocytosis, the viability of mutants in aly1 and aly2, arrestins that act as adapters for clatherin-mediated endocytosis36, were evaluated. Also tested were mutants of fre1 and fre237,38, which reduce oxidized Cu for transport by Ctr1. These knockout strains were constructed in the BY4741 strain background that is related to S288c23. Testing the viability of yeast under acute exposure can reveal smaller differences in sensitivity and determine if changes in viability to c-CuNP liquid culture is different than on plates. Cells were grown in similar conditions as when measuring internal copper levels (Fig 3). When endocytosis is affected by deletion of Aly1 and Aly2, cellular viability was restored in the presence of c-CuNP but not soluble copper (Fig 4D). Loss of Fre1 and Fre2 increased cellular viability in the presence of soluble copper but not c-CuNP (Fig 4D).
Previous assessment of growth inhibition with c-CuNP was done under chronic 2-day exposure measuring changes of growth on solid media (e.g., Figs 1 and 2). Physical interactions between yeast and c-CuNP were measured under acute exposure in liquid media (e.g., Figs 3 and 4). Viability assays were undertaken to evaluate the impact of soluble Cu and c-CuNPs on cell survival under acute exposures at comparable total Cu concentrations. Overall, the greater toxicity of c-CuNPs is evident across all isolates, with near complete loss of viability following exposure to 400 μM Cu as c-CuNP (Fig 5). Differential resistance of the isolates was also confirmed, with S288c and RM11 exhibiting greater viability than YJM789 or YJM339 regardless of Cu source (Fig 5).
Fig 5. Acute exposure of yeast to copper nanoparticles.
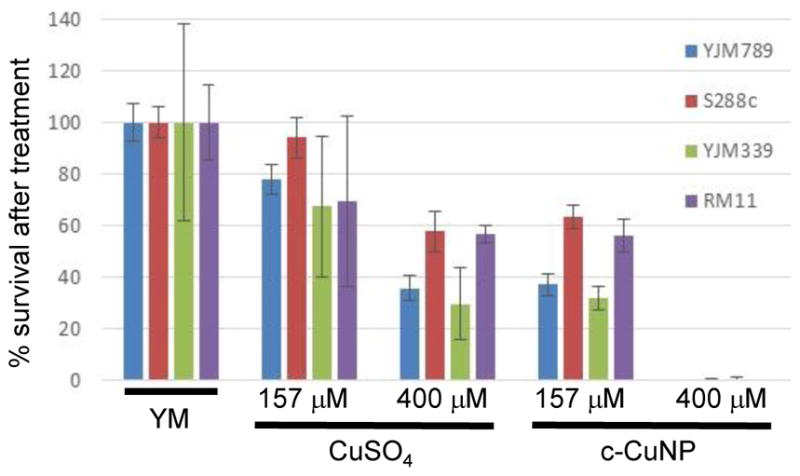
The number yeast forming colonies grown from cultures grown YM were normalized to 100%. The viability after yeast after two hours of exposure to or 400 μM and 157 μM of copper in the form of CuSO4 and c-CuNP is shown as a percentage compared to YM.
Genetic and proteomic responses to copper in genetically diverse yeast
The four diverse strains of yeast used in this study have underlying genetic differences that contribute to variation in Cu response. S288c and YJM789 exhibited the most extreme differences in their abilities to tolerate high doses of Cu and moderate doses of c-CuNP (Figs 1 and 2). To evaluate potential molecular underpinnings of these responses to Cu, proteome- and genome-wide measurements were carried out. While many mRNAs are known to change in response to copper7, we found only 22 proteins that showed at least 2-fold changes in S288c in response to 800 μM copper by quantitative mass spectrometry (Table 1 and Table S1). Many other proteins showed more than 2-fold changes between strains but not between copper treatments (Fig S2). 19 out of the 22 copper-responsive proteins showed very little change (less than 1.2-fold change) in Cu-sensitive YJM789 cells when treated with 800 μM CuSO4. Among the 19 proteins, Cup1, Fit2, Fet3, and Zrt1 were upregulated, all of which are involved in metal transport (Table 1). Cup1 is a protein that binds Cu intracellularly and mediates tolerance to high Cu concentrations39. The S288c genome has two CUP1 alleles annotated with two synonymous SNPs and identical protein sequences (Cup1-1 and Cup1-2), and they cannot be differentiated in proteomics27. Cup1YJM789 exhibited a higher abundance in YM relative to Cup1S288c. Following Cu exposure, Cup1YJM789 demonstrated lower abundance relative to Cup1S288 due to the upregulation of Cup1S288c combined with little change in Cup1YJM789 abundance (Table 1). Fit2 and Fet340,41 are involved in iron (Fe) transport, and Zrt1 is involved in Zn transport42. Fit2 and Zrt1 exhibited similar protein abundance between S288c and YJM789 in YM. After Cu exposure, Fit2 and Zrt1 showed higher abundance in S288c than in YJM789 because they were upregulated in S288c and changed little in YJM789 (Table 1). As for Fet3, S288c exhibited higher abundance than YJM789 in YM, and the upregulation by S288c in response to Cu exposure made strain-specific differences even greater (Table 1).
Table 1.
Examples of proteins differentially expressed (log2-fold ratio) between S288c and YJM789 with and without 800 μM CuSO4.
| Order in dendrogram | Systematic name | Protein name | S288cCu/ S288cYM | YJM789Cu/ YJM789YM | YJM789YM/ S288cYM | YJM789Cu/ S288cCu |
|---|---|---|---|---|---|---|
| 81 | YHR053C/ | Cup1 | 3.477 | 0.196 | 2.585 | −0.686 |
| YHR055C | ||||||
| 82 | YOR382W | Fit2 | 2.448 | −0.117 | 0.167 | −2.387 |
| 84 | YMR058W | Fet3 | 1.180 | −0.105 | −1.429 | −2.630 |
| 93 | YGL255W | Zrt1 | 1.141 | 0.123 | −0.268 | −0.848 |
| 63 | YDR424C | Dyn2 | −1.325 | −0.013 | 0.207 | 1.527 |
| 64 | YAL007C | Erp2 | −1.119 | 0.228 | 0.386 | 1.743 |
| 53 | YHR051W | Cox6 | −1.097 | −0.038 | 1.094 | 2.162 |
| 60 | YOL077W-A | Atp19 | −1.327 | −0.195 | 1.525 | 2.667 |
| 68 | YER057C | Hmf1 | −1.528 | 0.033 | 0.596 | 2.166 |
| 65 | YIL051C | Mmf1 | −1.328 | −0.043 | 0.445 | 1.739 |
| 58 | YBR230C | Om14 | −1.020 | −0.051 | 0.618 | 1.597 |
| 71 | YOR130C | Ort1 | −1.236 | −0.050 | 0.010 | 1.205 |
| 52 | YNL227C | Jjj1 | −1.050 | 0.015 | 0.796 | 1.869 |
| 72 | YDL081C | Rpp1a | −2.002 | −0.009 | −0.373 | 1.629 |
| 70 | YOR236W | Dfr1 | −1.134 | 0.574 | −0.346 | 1.429 |
| 80 | YNL190W | Ynl190w | 1.474 | −0.756 | 3.492 | 1.271 |
| 85 | YPR124W | Ctr1 | −1.966 | −0.494 | −1.380 | 0.101 |
| 35 | YAL046C | Aim1 | −0.168 | −1.334 | 1.136 | −0.021 |
| 96 | YMR173W | Ddr48 | 0.653 | 0.006 | −0.564 | −1.202 |
To further search for genetic loci contributing to Cu tolerance, we performed a genome-wide linkage study using a hybrid segregant collection17,43,44 to find association of genetic variation with differences in Cu tolerance between S288c and YJM789. 125 segregants from a S288c/YJM789 cross were grown at sub-lethal concentrations of copper (50 μM) in liquid culture (Fig S3 A–C). In contrast to previous experiments that used a acute exposure and high Cu concentration, we used a long exposure and lower dose of Cu to model the adaptive response to low level copper exposure. S288c grew better in low doses of Cu than in media with no additional Cu (Fig S3C) as opposed to YJM789, which was strongly inhibited by Cu (Fig S3B). In the presence of bathocuproinedisulfonic acid (BCS), a Cu chelator, S288c growth decreased (Fig S3D). The differences in growth of haploid segregants between YM and Cu treatment at 12-hours (mid-log phase) and 18-hours (endpoint) were used as quantitative trait loci (QTL). Using a threshold of LOD (logarithm of odds) > 5, three regions of the genome were identified at the mid-log phase (Table S4 and Fig S4A) and one region was identified at the 18-hour endpoint linked to Cu resistance (Table S5 and Fig S4B). The genetic variations within these regions are most likely associated with differences in copper tolerance between S288c and YJM789.
One region of the genome that was identified in the 12-hour, but not 18-hour, QTL mapping was on chromosome VIII between coordinates 190,664 and 223,431, which contains twenty-two genes (Fig. S4C). In the center of this region is the CUP1 locus, and its contribution to Cu tolerance has been well studied. The common laboratory strain S288c and many other Cu-tolerant strains contain independent amplifications of the CUP1 locus27,39. Other studies have also found that laboratory-evolved strains amplify this region of the genome45. The Saccharomyces Genome Database reference strain, S288c, has two alleles of CUP1 with YHR054c, a gene of unknown function, between them. This region is actually amplified seven times in S288c with fourteen copies of the CUP1 gene; YJM789 contains one copy of CUP1 in each repetitive unit that is amplified seven times27.
To pinpoint the specific genes that were contributing to Cu tolerance, we focused on the genes that had amino acid polymorphisms or copy number variation. The region on chromosome XI between 74,608 and 92,505 (Fig S4D) contained ten genes, among which MNN4, PEX1, MIA40, MST1, and DHP2 encoded amino acid polymorphisms between S288c and YJM789. Mia40 works with other proteins for the maturation of the mitochondrial fraction of Sod146. Sod1 is a major contributor to converting superoxides to hydrogen peroxide, which is further catalyzed into water and hydrogen47 and is important for tolerance to metals, including copper48. In our proteomic analysis, Sod1 levels were not significantly changed between strains or in response to Cu.
Another region that was identified in the endpoint linkage analysis was on chromosome II between coordinates 466,289 and 482,822 (Fig S4B). Within this interval is the LYS2 gene, which is mutated in YJM789 as an auxotrophic marker. As this peak was not seen at the 12-hour time point (Fig S4A), this may be interpreted as Lys2 becoming required as lysine present in the media was depleted as cells grew to saturation. Therefore, LYS2 may be selected by growth conditions instead of by Cu treatment.
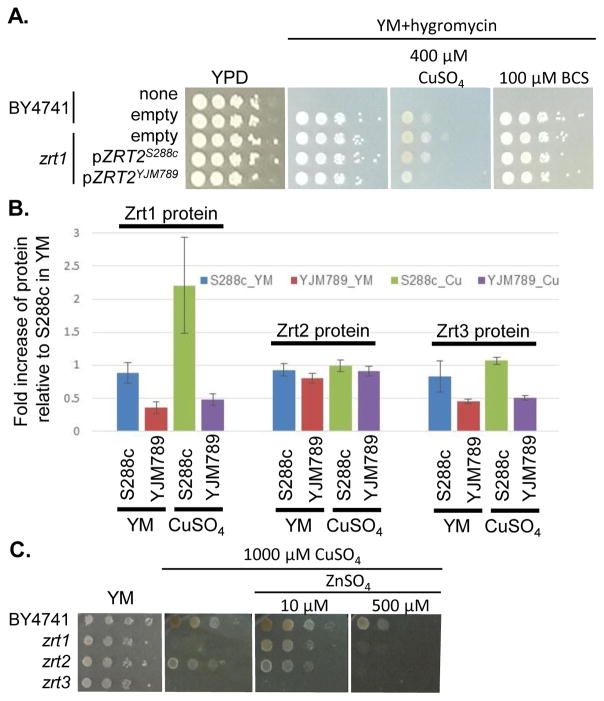
In both 12- and 18-hour association analyses, one strong peak was shared on chromosome XII between 393,415 and 414,994 (S6 Figs S4 A and B), which contained eleven genes (Fig S4 E). One of the genes known to be relevant to Cu tolerance was ACE2, which encodes the activating transcription factor of CUP1 expression49. However, ACE2 showed no significant change in mRNA abundance in YM50, no amino acid polymorphisms between S288c and YJM789, and minimal change in protein levels in response to Cu exposure. Near ACE2 was another gene of interest, ZRT2, which encodes a low affinity Zn transporter with five amino acid polymorphisms between S288c and YJM78933. Like Ace2, Zrt2 exhibited less than 2-fold changes in protein abundance between strains or treatments. However, we cannot rule out changes in post-translational modifications of Zrt2 in response to Cu or the impact of the polymorphisms on transporter function. Therefore, we cloned ZRT2 from each strain, including the endogenous promoters and terminators, into plasmids. When the plasmids were expressed in BY4741 zrt2 deleted yeast, only yeast expressing Zrt2YJM789 grew more slowly on media containing 400 μM Cu and showed no change in growth on BCS (Fig. 6A). The polymorphisms within Zrt2 protein apparently contribute to the Cu-sensitive phenotype of YJM789.
Fig 6. Impact of zinc transporters and zinc on copper response.
A. Zrt2 allele swaps in S288c (BY4741) in the presence of copper or BCS. ZRT2 plasmids were transformed and maintained in zrt2 mutants by the addition of hygromycin and strains were supplemented with necessary nutrients. B. Relative protein levels of three different zinc transporters in copper treated cells as measured from proteomic analysis. C. Serial dilution of S288c (BY4741) yeast with each of the zinc transporters deleted on increasing amounts of copper and zinc. After 2 days of growth at 30°C plates were photographed.
In the proteomic analysis, one other metal-responsive protein, Zrt1, was upregulated in S288c, but not in YJM789, when treated with Cu (Table 1). Zrt1 is a high affinity zinc transporter that is induced in Zn-depleted cells42. While Zrt1S288c was upregulated approximately 2-fold by Cu treatment, other Zn transporters, including Zrt2 and Zrt3, an intracellular Zn transporter, did not exhibit significant changes in response to Cu in either strain (Fig. 6B). YJM789 displayed slightly lower Zrt3 expression compared to S288c regardless of Cu. Due to this association between Zn transporters and resistance to Cu, we tested whether increased levels of exogenous Zn could the alter the Cu response of BY4741 and the Zn transporter mutant yeast lines. The zrt1 and zrt3 knockout lines were sensitive to 1000 μM Cu, whereas the zrt2 line was not (Fig 6C). The addition of 10 μM Zn improved the growth of zrt1, but not zrt3, mutants, and the zrt2 line was still resistant to Cu and unaffected by supplemental Zn (Fig 6C). These growth promotion effects were lost at 500 μM Zn in the presence of 1000 μM of Cu (Fig 6C).
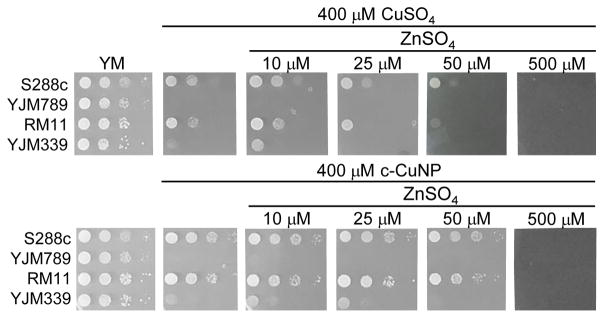
To determine if the relationship between Cu and Zn was a more general yeast response, additional yeast strains were tested. When S288c, YJM339, and RM11 were grown in 400 μM Cu in either the soluble form or from c-CuNP, adding 10 μM Zn resulted in modest growth increases, which were lost or became negative at and above 25 μM Zn (Fig. 7). In contrast, YJM789 did not respond to supplemental Zn (Fig 7). These results indicate that Zn, at low concentrations, plays a role in modulating Cu toxicity and this benefit may result from different Zn uptake capacities among yeast strains.
Fig 7. Addition of zinc to copper nanoparticles altered growth inhibition.
Serial dilution of genetically diverse yeast (S288c (GSY147), YJM789 (YJM789K5a), AWRI1631, YJM339 and RM11 prototrophic strains) in response to copper and c-CuNP on increasing amounts of zinc sulfate. After 2 days of growth at 30°C plates were photographed.
D. Discussion
We have characterized the interaction of the genetically diverse model eukaryotic organism, S. cerevisiae, with soluble Cu and c-CuNPs towards improving future application of hybrid nanomaterials for microbial control. Copper has been used extensively over history as an antimicrobial agent and microorganisms exposed to Cu gain the opportunity to develop resistance mechanisms to this metal. Metabolic mechanisms that reduce internal metal accumulation or free metal speciation, which will reduce the production of toxic ROS in the cytoplasm, are important in conferring heavy metal resistance. Overcoming existing microbial resistance pathways and altering the balance of metals in cells may enhance the efficacy of existing metal-based antimicrobial materials and aid in the development of novel antimicrobial agents.
Cellulosic copper nanoparticles exhibit enhanced toxicity
In the current study, the antimicrobial potency of Cu was increased by the delivery of Cu in the form of c-CuNPs. Exposure of 157 μM copper in NPs was as effective as 400 μM copper sulfate in yeast growth inhibition, and increase in toxicity resulted from enhanced delivery of Cu to the cytoplasm (Figs 3 and 5). Flow cytometry evidence and c-CuNP fluorescence suggested that CuNPs bound in large part to the cell walls of S. cerevisiae and such binding may presage the transport of Cu into the cell (Fig 4B and C). Variation in binding of yeast cells and c-CuNPs mirrored resistance, with low binding strains exhibiting greater resistance to Cu (Fig 4A). This differential binding may enhance exposure in Cu sensitive strains. Based on the size of c-CuNPs (10–20 nm)16, it is also possible that c-CuNPs were absorbed into S. cerevisiae by endocytosis and endocytotic activity varied between the strains. An endocytosis vesicle is 50–80 nm in S. cerevisiae51 and some cells in the current study did exhibit internal fluorescence (Fig 4C). Although most of the yeast with incubated c-CuNPs appeared speckled with c-CuNP on the cell surface, approximately 10% of the yeast took up c-CuNPs as representing intracellular FITC staining. We were unable to test if these two subpopulation had differences in survival, but 13% of the laboratory yeast had internal FITC stained c-CuNP which was not statistically different from 18% of the yeast that fail to form colonies after c-CuNP treatment (Fig. 4C). Arrestin mutants in the laboratory strain (S288c background) displayed rescued cellular viability in the presence c-CuNPs suggesting that endocytosis may be one mechanism of action of c-CuNPs, which would effectively deliver Cu to the cytoplasm.
Differential expression of metal transporters is linked to toxicity
Copper uptake and subsequent toxicity, regardless of the mechanism of enhanced delivery of copper via CuNPs, will most likely be governed by the reduction and transport of metals across the plasma membrane and their cellular compartmentation in different strains. In S288c, exposure to Cu down-regulated the Cu transporter, Ctr152, while expression of the Cu and Fe oxidizer, Fet353,54, the plasma membrane bound facilitator of iron transport, Fit3, and the high affinity Zn transporter, Zrt1, increased. These patterns suggest that Cu exposure perturbs trace metal homeostasis and the more resistant S288c strain responds to these perturbations40,55. YJM789 did not show significant changes of these proteins and we propose that the lack of regulation of Cu uptake underlies the sensitivity of YJM789 to Cu.
The expression levels of Crt1 tracked cell Cu concentrations in YJM789 and S288c. Expression is lower in YJM789 than S288c under control conditions, yet reverse under exposure to Cu (Table 1), leading to YJM789 exhibiting low basal Cu levels in YM media, but elevated Cu under exposure to Cu (Fig 3). S288c regulation of expression more tightly regulates cell Cu concentrations in these conditions. Thus, S288c apparently has more responsive metabolic regulation of Cu uptake. Changes in other metal transport genes may reflect mass balance displacement of Fe and Zn by Cu from transporters or secondary stresses requiring additional Fe or Zn to overcome toxicity. Fet3 is a ferroxidase required for the reduction of Fe(II) and subsequent transport across the plasma membrane, and its upregulation may reflect increased demand for Fe due to inhibition of Fe(II) transport by Cu or increased demand for redox metabolism within the cell. Similarly, Fit2 is an iron and iron-siderophore-binding protein expressed on the cell surface40 that facilitates Fe uptake and may be directly regulating Cu response by adjusting internal iron levels for Sod2, a mitochondrial superoxide dismutase56.
The up-regulation of Zrt1, the high affinity Zn transporter, also points to Cu-induced Zn limitation in S. cerevisiae. High affinity transporters are typically activated by mineral deficiency, and the low km for Zrt1 in yeast (~10 nM) suggests it would be effective in overcoming deficiency 57. That supplemental Zn rescued zrt1 knockouts (Fig 6) and, to a lesser degree, native strains under high Cu exposure (Fig 7) suggests that excess Cu ions at the plasma membrane surface may be impeding the binding of Zn to Zn transporters. It is interesting that the more Cu resistant strain S288c upregulates Zrt1 whereas the sensitive YJM789 does not, suggesting that metal homeostasis responses may be limited in cases where excessive Cu accumulation does not lead to loss of cellular signaling systems or excessive ROS damage as would occur in strains that cannot limit Cu accumulation (Fig 3).
Intracellular copper binding reduces copper toxicity
Transporters, intracellular sequestration, and chaperonins/chelation maintain metal concentrations within strict bounds and tightly regulate the balance and potential toxicity of metals within the cytoplasm. In the current study, S288c up-regulated expression of Cup1, the main metallothionein in S. cerevisiae, in response to Cu. YJM789 did not show significant changes in levels of this metal-binding protein in the presence of Cu and we propose that the lack of response to increased cytoplasmic Cu increased YJM789 sensitivity to this metal. Unregulated free Cu concentrations in the cytoplasm will increase the production of ROS and lead to broad disruptions in cell homeostasis.
In support of this, providing certain antioxidants increased the copper levels of all treated strains (Fig 2). Glutathione can bind copper transported into cells by Ctr1 before delivery to copper chaperones58. More importantly, glutathione and NAC may serve to prime phytochelatin and metallothionein production within S. cerevisiae59,60. These peptides/proteins function to regulate internal free metal concentrations and maintain cytoplasmic metal homeostasis. The large increases in Cu accumulation from both soluble Cu and CuNPs may reflect enhanced production of these metal-binding systems, the chelation of Cu intercellularly, and internal storage of nontoxic metal complexes.
Addition of zinc increases copper toxicity
Although the toxicity of Cu was alleviated in some strains by the addition of low concentrations of Zn as discussed above, the antimicrobial activity of Cu was enhanced in the current study by the application of higher Zn concentrations (Fig 7). As with Cu, Zn plays critical roles as a catalytic and structural cofactor in a wide array of proteins, and maintenance of free Zn2+ concentrations within the cytoplasm is important in maintaining metal homeostasis61. Zinc is transported in the cell by the membrane-bound transporters Zrt1 and Zrt2 and is sequestered in the vacuole by Zrt362. Given that exposure of yeast to Cu induces the expression of Zrt1 in Cu-tolerant strains (Table 1), the addition of Zn may into the environment may lead to the over-accumulation of Zn and subsequent toxicity to S. cerevisiae.
When in excess, Zn will induce many of the same lesions in S. cerevisiae as does Cu, including oxidative damage and negative interactions with Fe transport. Thus, the enhanced microbial activity of Cu and Zn combined may be due to the additional sympatric lesions induced by Zn. However, there is evidence that the stresses induced by Cu and Zn are distinctly different. In a unique multi-metal study of S. cerevisiae response to stress,63,64 found significant differences between the response of yeast to Cu and Zn. While there was some overlap in the common stress responses, such as metal transport and oxidative stress, significant groups of differentially expressed genes did not overlap and only two of the 10 most metal-responsive protein-protein and protein-DNA interacting networks were shared64. Thus, there may be limited overlap in the cell’s metabolic rescue capacity when, under Cu exposure, a second divergent metal is introduced. The addition of Zn to the production of Cu-based nanomaterials would improve the antimicrobial properties of these novel antimicrobial systems and may represent a new approach for public health and food safety.
Supplementary Material
Significance to metallomics.
Saccharomyces cerevisiae has developed resistance to copper independently multiple times and provides an ideal model to assess response of fungi to Cu. Previously, we demonstrated that c-CuNPs are an effective antibacterial material. In an effort to understand the mode of action of c-CuNPs, improve potency, ensure broad spectrum, and limit development of antimicrobial resistance, the response of S. cerevisiae to c-CuNPs was characterized using proteomic and genetic approaches. c-CuNPs expose yeast to higher Cu levels and, by comparing the response of a copper resistant to copper sensitive yeast, the role of Zn was uncovered. Through these interactions, we suggest that the antimicrobial effectiveness of metallic hybrid nanomaterials will enhance microbial control, which may offer a new and wide range of antimicrobial materials for public health and food safety.
Acknowledgments
We thank Mike Snyder for use of the Orbitrap used in these studies. Randall Mann generously shared his common protein contaminant list. Angela Lee generously gave us the yeast knockout collection. Jenay Grant assisted in confirming the genomic amplification of the CUP1 locus. Flow cytometry was carried out on the WVU Flow Cytometry & Single Cell Core Fortessa. Microscopy was done with the aid of Phil Chapman.
Footnotes
Author contributions
XRM carried out the proteomics analysis and LJ developed proteomics workflow. MJW carried out the microscopy, characterized Zrt2, and flow cytometry with JBL. JBL and VAP carried out viability assays. LMW and JRC measured internal copper concentrations. TWC analyzed QTL data. ZRL characterized Cup1 levels. ZRL, JRC, and JEGG wrote the manuscript. JEGG conceived and directed the experiments.
Notes and references
- 1.Dollwet HHA, Sorenson JRJ. Trace elements in Medicine. 1985;2:80–87. [Google Scholar]
- 2.Lloyd RV, Hanna PM, Mason RP. Free Radic Biol Med. 1997;22:885–888. doi: 10.1016/s0891-5849(96)00432-7. [DOI] [PubMed] [Google Scholar]
- 3.Avery SV, Howlett NG, Radice S. Appl Environ Microbiol. 1996;62:3960–3966. doi: 10.1128/aem.62.11.3960-3966.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Quaranta D, Krans T, Santo CE, Elowsky CG, Domaille DW, Chang CJ, Grass G. Appl Environ Microbiol. 2011;77:416–426. doi: 10.1128/AEM.01704-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Samanovic MI, Ding C, Thiele DJ, Darwin KH. Cell Host Microbe. 2012;11:106–115. doi: 10.1016/j.chom.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lyons TJ, Gasch AP, Gaither LA, Botstein D, Brown PO, Eide DJ. Proc Natl Acad Sci U S A. 2000;97:7957–7962. doi: 10.1073/pnas.97.14.7957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gross C, Kelleher M, Iyer VR, Brown PO, Winge DR. J Biol Chem. 2000;275:32310–32316. doi: 10.1074/jbc.M005946200. [DOI] [PubMed] [Google Scholar]
- 8.Wegner T, Jones P. Wood Fiber Sci. 2005;37:549–551. [Google Scholar]
- 9.Wegner TH, Jones PH. Adv Cellul nanotechnology Cellulose. 2006;13:115–118. [Google Scholar]
- 10.Wegner TH, Jones EP. Nanosci Technol Renew Biomater. 2009;1:1–41. [Google Scholar]
- 11.McCrank J. Nanotechnology applications in the forest sector. Natural Resources Canada; 2009. [Google Scholar]
- 12.Moon RJ, Martini A, Nairn J, Simonsen J, Youngblood J. Chem Soc Rev. 2011;40:3941–3994. doi: 10.1039/c0cs00108b. [DOI] [PubMed] [Google Scholar]
- 13.Cai J, Kimura S, Wada M, Kuga S. Biomacromolecules. 2008;10:87–94. doi: 10.1021/bm800919e. [DOI] [PubMed] [Google Scholar]
- 14.Zhong T, Oporto GS, Jaczynski J, Jiang C. Biomed Res Int. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang C, Oporto GS, Zhong T, Jaczynski J. Cellulose. 2015:1–10. doi: 10.1155/2015/456834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhong T, Oporto GS, Jaczynski J, Tesfai AT, Armstrong J. Wood Fiber Sci. 2013;45:1–8. 1. [Google Scholar]
- 17.Steinmetz LM, Sinha H, Richards DR, Spiegelman JI, Oefner PJ, McCusker JH, Davis RW. Nature. 2002;416:326–330. doi: 10.1038/416326a. [DOI] [PubMed] [Google Scholar]
- 18.Wenger JW, Schwartz K, Sherlock G. PLoS Genet. 2010;6:e1000942. doi: 10.1371/journal.pgen.1000942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCusker JH, Clemons KV, Stevens DA, Davis RW. Genetics. 1994;136:1261–1269. doi: 10.1093/genetics/136.4.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Borneman AR, Forgan AH, Pretorius IS, Chambers PJ. FEMS Yeast Res. 2008;8:1185–1195. doi: 10.1111/j.1567-1364.2008.00434.x. [DOI] [PubMed] [Google Scholar]
- 21.Mortimer RK, Romano P, Suzzi G, Polsinelli M. Yeast. 1994;10:1543–1552. doi: 10.1002/yea.320101203. [DOI] [PubMed] [Google Scholar]
- 22.McCullough MJ, Clemons KV, Farina C, McCusker JH, Stevens DA. J Clin Microbiol. 1998;36:557–562. doi: 10.1128/jcm.36.2.557-562.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tong AH, Evangelista M, Parsons AB, Xu H, Bader GD, Page N, Robinson M, Raghibizadeh S, Hogue CW, Bussey H, Andrews B, Tyers M, Boone C. Science (80- ) 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- 24.Wisniewski JR, Zougman A, Mann M. J Proteome Res. 2009;8:5674–5678. doi: 10.1021/pr900748n. [DOI] [PubMed] [Google Scholar]
- 25.Gerstein AC, Ono J, Lo DS, Campbell ML, Kuzmin A, Otto SP. Genetics. 2015;199:555–571. doi: 10.1534/genetics.114.171124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hodgins-Davis A, Adomas AB, Warringer J, Townsend JP. Genome Biol Evol. 2012;4:1061–1079. doi: 10.1093/gbe/evs084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao Y, Strope PK, Kozmin SG, McCusker JH, Dietrich FS, Kokoska RJ, Petes TD. G3 (Bethesda) 2014;4:2259–2269. doi: 10.1534/g3.114.012922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fay JC, McCullough HL, Sniegowski PD, Eisen MB. Genome Biol. 2004;5:R26. doi: 10.1186/gb-2004-5-4-r26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kvitek DJ, Will JL, Gasch AP. PLoS Genet. 2008;4:e1000223. doi: 10.1371/journal.pgen.1000223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Torok T, Mortimer RK, Romano P, Suzzi G, Polsinelli M. J Ind Microbiol. 1996;17:303–313. [Google Scholar]
- 31.Brem RB, Yvert G, Clinton R, Kruglyak L. Science (80- ) 2002;296:752–755. doi: 10.1126/science.1069516. [DOI] [PubMed] [Google Scholar]
- 32.Mortimer RK, Johnston JR. Genetics. 1986;113:35–43. doi: 10.1093/genetics/113.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cherry JM, Hong EL, Amundsen C, Balakrishnan R, Binkley G, Chan ET, Christie KR, Costanzo MC, Dwight SS, Engel SR, Fisk DG, Hirschman JE, Hitz BC, Karra K, Krieger CJ, Miyasato SR, Nash RS, Park J, Skrzypek MS, Simison M, Weng S, Wong ED. Nucleic Acids Res. 2012;40:D700–5. doi: 10.1093/nar/gkr1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tawfik OW, Papasian CJ, Dixon AY, Potter LM. J Clin Microbiol. 1989;27:1689–1691. doi: 10.1128/jcm.27.7.1689-1691.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sadowska AM. Ther Adv Respir Dis. 2012;6:127–135. doi: 10.1177/1753465812437563. [DOI] [PubMed] [Google Scholar]
- 36.Lin CH, MacGurn JA, Chu T, Stefan CJ, Emr SD. Cell. 2008;135:714–725. doi: 10.1016/j.cell.2008.09.025. [DOI] [PubMed] [Google Scholar]
- 37.Georgatsou E, Mavrogiannis LA, Fragiadakis GS, Alexandraki D. J Biol Chem. 1997;272:13786–13792. doi: 10.1074/jbc.272.21.13786. [DOI] [PubMed] [Google Scholar]
- 38.Yamaguchi-Iwai Y, Serpe M, Haile D, Yang W, Kosman DJ, Klausner RD, Dancis A. J Biol Chem. 1997;272:17711–17718. doi: 10.1074/jbc.272.28.17711. [DOI] [PubMed] [Google Scholar]
- 39.Karin M, Najarian R, Haslinger A, Valenzuela P, Welch J, Fogel S. Proc Natl Acad Sci U S A. 1984;81:337–341. doi: 10.1073/pnas.81.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Protchenko O, Ferea T, Rashford J, Tiedeman J, Brown PO, Botstein D, Philpott CC. J Biol Chem. 2001;276:49244–49250. doi: 10.1074/jbc.M109220200. [DOI] [PubMed] [Google Scholar]
- 41.De Silva DM, Askwith CC, Eide D, Kaplan J, De Silva DM, Askwith CC, Eide D, Kaplan J. J Biol Chem. 1995;270:1098–1101. doi: 10.1074/jbc.270.3.1098. [DOI] [PubMed] [Google Scholar]
- 42.Zhao H, Eide D. Proc Natl Acad Sci. 1996;93:2454–2458. doi: 10.1073/pnas.93.6.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gallagher JEG, Zheng W, Rong X, Miranda N, Lin Z, Dunn B, Zhao H, Snyder MP. Genes Dev. 2014;28:409–421. doi: 10.1101/gad.228940.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mancera E, Bourgon R, Brozzi A, Huber W, Steinmetz LM. Nature. 2008;454:479–485. doi: 10.1038/nature07135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Adamo GM, Lotti M, Tamas MJ, Brocca S. Microbiology. 2012;158:2325–2335. doi: 10.1099/mic.0.058024-0. [DOI] [PubMed] [Google Scholar]
- 46.Varabyova A, Topf U, Kwiatkowska P, Wrobel L, Kaus-Drobek M, Chacinska A. FEBS J. 2013;280:4943–4959. doi: 10.1111/febs.12409. [DOI] [PubMed] [Google Scholar]
- 47.Tamai KT, Gralla EB, Ellerby LM, Valentine JS, Thiele DJ. Proc Natl Acad Sci. 1993;90:8013–8017. doi: 10.1073/pnas.90.17.8013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pena MM, Koch KA, Thiele DJ. Mol Cell Biol. 1998;18:2514–2523. doi: 10.1128/mcb.18.5.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Butler G, Thiele DJ. Mol Cell Biol. 1991;11:476–485. doi: 10.1128/mcb.11.1.476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zheng W, Zhao H, Mancera E, Steinmetz LM, Snyder M. Nature. 2010;464:1187–1191. doi: 10.1038/nature08934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Buser C, Drubin DG. Microsc Microanal. 2013;19:381–392. doi: 10.1017/S1431927612014304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Knight SA, Labbe S, Kwon LF, Kosman DJ, Thiele DJ. Genes Dev. 1996;10:1917–1929. doi: 10.1101/gad.10.15.1917. [DOI] [PubMed] [Google Scholar]
- 53.de Silva D, Davis-Kaplan S, Fergestad J, Kaplan J. J Biol Chem. 1997;272:14208–14213. doi: 10.1074/jbc.272.22.14208. [DOI] [PubMed] [Google Scholar]
- 54.Askwith C, Eide D, Van Ho A, Bernard PS, Li L, Davis-Kaplan S, Sipe DM, Kaplan J. Cell. 1994;76:403–410. doi: 10.1016/0092-8674(94)90346-8. [DOI] [PubMed] [Google Scholar]
- 55.Stadler JA, Schweyen RJ. J Biol Chem. 2002;277:39649–54. doi: 10.1074/jbc.M203924200. [DOI] [PubMed] [Google Scholar]
- 56.Naranuntarat A, Jensen LT, Pazicni S, Penner-Hahn JE, Culotta VC. J Biol Chem. 2009;284:22633–22640. doi: 10.1074/jbc.M109.026773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Eide DJ. Biochim Biophys Acta - Mol Cell Res. 2006;1763:711–722. doi: 10.1016/j.bbamcr.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 58.Maryon EB, Molloy SA, Kaplan JH. Am J Physiol Physiol. 2013;304:C768–79. doi: 10.1152/ajpcell.00417.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kneer R, Kutchan TM, Hochberger A, Zenk MH. Arch Microbiol. 1992;157:305–310. doi: 10.1007/BF00248673. [DOI] [PubMed] [Google Scholar]
- 60.Wünschmann J, Beck A, Meyer L, Letzel T, Grill E, Lendzian KJ. FEBS Lett. 2007;581:1681–1687. doi: 10.1016/j.febslet.2007.03.039. [DOI] [PubMed] [Google Scholar]
- 61.MacDiarmid CW, Gaither LA, Eide D. EMBO J. 2000;19:2845–2855. doi: 10.1093/emboj/19.12.2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Winston F, Dollard C, Ricupero-Hovasse SL. Yeast. 1995;11:53–55. doi: 10.1002/yea.320110107. [DOI] [PubMed] [Google Scholar]
- 63.Liu X, Jia J, Popat R, Ortori CA, Li J, Diggle SP, Gao K, Cámara M. BMC Microbiol. 2011;11:26. doi: 10.1186/1471-2180-11-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jin YH, Dunlap PE, McBride SJ, Al-Refai H, Bushel PR, Freedman JH. PLoS Genet. 2008;4:e1000053. doi: 10.1371/journal.pgen.1000053. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.