Abstract
Increased physician prescribing of opioids to treat chronic nonprogressive pain has been accompanied by an increase in opioid addiction. Twin studies of opioid addiction are consistent with an inherited component of risk, approximately 50%. Several genome-wide association study (GWAS) reports indicate that genetic risk for opioid addiction is conveyed by many alleles of small effect (odds ratios <1.5). These reports have detected alleles in potassium-ion-channel genes (KCNC1 and KCNG2) and in a glutamate receptor auxiliary protein (CNIH3). Additionally, a variant at the µ-opioid receptor gene (OPRM1), which regulates OPRM1 expression appears promising. In pharmacogenetics of opioid addictions, methadone dose may be regulated by variants in cytochrome P450 2B6 (CYP2B6), a methadone-metabolizing enzyme, and by a locus 300 kb 5' to OPRM1. A δ-opioid-receptor gene single-nucleotide polymorphism may predict treatment response to methadone versus buprenorphine. To achieve better progress, larger sample sizes are needed for GWAS research, including controls with chronic opioid exposure, but no addiction. Large clinical trials comparing effective pharmacotherapies for opioid addiction (naltrexone, methadone, and buprenorphine) are needed for pharmacogenetic progress.
Keywords: addiction, dependence, genetics, opioid, pharmacogenetics
Abstract
El aumento de las prescripciones médicas de opioides, para tratar el dolor crónico no progresivo, se ha acompañado de un incremento en la adicción a opioides. Los estudios en gemelos de la adicción a opioides son consistentes con una herencia del componente de riesgo de aproximadamente el 50%. Algunos reportes de los estudios de asociación del genoma completo (GWAS, Genome Wide Association Study) señalan que el riesgo genético para la adicción a opioides se transmite por muchos alelos de pequeño efecto (con una probabilidad <1,5). Estos estudios han detectado alelos en genes para el canal de potasio (KCNC1 y KCNG2) y para una proteína auxiliar del receptor de glutamato (CNIH3). Además, parece promisoria una variante en el gen del receptor opioide mu (OPRM1), que regula la expresión de OPRM1. En la farmacogenética de las adicciones a opioides, la dosis de metadona puede ser regulada por variantes en el citocromo P450 2B6 (CYP2B6), por una enzima que metaboliza la metadona y por el locus 300kb 5' de OPMR1. Un polimorfismo del nucleótido único del gen del receptor opioide delta puede predecir la respuesta terapéutica a metadona versus buprenorfina. Para poder progresar más se necesitan muestras más grandes para investigar los GWAS, las cuales deben incluir controles con exposición crónica a opioides, pero sin adicción. Para un progreso en la farmacogenética se requiere de ensayos clínicos con muestras grandes que comparen farmacoterapias efectivas para la adicción a opioides (naltrexona, metadona y buprenorfina).
Abstract
L'augmentation des prescriptions médicales d'opioïdes pour traiter la douleur chronique stable s'accompagne d'une augmentation de l'addiction aux opioïdes. Les études de jumeaux sur l'addiction aux opioïdes sont cohérentes avec une composante héréditaire du risque, d'environ 50 %. D'après plusieurs rapports d'étude d'association pangénomique (GWAS : Genome Wide Association Study), le risque génétique pour l'addiction aux opioïdes est transmis par de nombreux allèles à l'effet minime (rapport de risque <1,5). Ces rapports ont détecté des allèles dans les gènes des canaux potassiques (KCNC1 et KCNG2) et dans une protéine auxiliaire du récepteur au glutamate (CNIH3). De plus, un variant du gène du récepteur aux opioïdes µ (OPRM1) régulant l'expression de OPRM1 semble prometteur. Dans la pharmacogénétique de l'addiction aux opioïdes, la dose de méthadone peut être régulée par des variants du cytochrome P450 2B6 (CYP2B6), une enzyme métabolisant la méthadone, et par le locus situé à 300 kb en en amont du gène OPRM1. Un polymorphisme nucléotidique du gène du récepteur δ aux opioïdes peut prédire la réponse au traitement à la méthadone versus buprénorphine. Afin de mieux progresser, les études d'association pangénomique nécessitent des échantillons plus grands, comportant des témoins ayant une exposition chronique aux opioïdes mais sans addiction. De grandes études cliniques comparant les traitements pharmacologiques efficaces pour l'addiction aux opioïdes (naltrexone, méthadone et buprénorphine) sont nécessaires pour le progrès pharmacogénétique.
Introduction
Opioid agonists are a group of heterogeneous compounds that have diverse actions, the medically most important of which is relief of acute pain. Opioid agonists are analgesic because they activate the µ-opioid receptor (MOR) and, to a minor extent, the δ-opioid receptor (DOR). Opioid agonists inhibit incoming pain signals in the pain-sensing spinothalamic tract and the spinoreticular tract by activating MORs at the spinal (a minor component) and supraspinal level. Most analgesic effects in these pathways are thought to occur in the thalamus, somatosensory cortex, and association cortex, which have an abundance of MORs. MORs in the periaqueductal gray matter and nucleus raphe magnus activate the descending pain-control pathway to enhance analgesia.
The same MOR is found in the ventral tegmental area (VTA). VTA neurons release dopamine at nerve terminals in ventral striatum and medial prefrontal cortex. Activation of this circuit is a common element of the experience of reward or euphoria, and this circuit is activated by abused drugs, including opioid agonists, ethanol, benzodiazepines, cocaine, nicotine, and cannabis (for review, see ref 1). Opioid agonists enhance dopaminergic transmission in ventral striatum and medial prefrontal cortex to produce reward. This enhanced dopaminergic transmission in the ventral striatum and medial prefrontal cortex is due to activation of MORs located on VTA inhibitory γ-aminobutyric acid (GABA)ergic interneurons. The activation of MORs results in inhibition of GABAergic inhibitory interneurons, so that the VTA dopaminergic neurons are free to release more dopamine in the ventral striatum and medical prefrontal cortex. Activation of this dopaminergic pathway is key to the rewarding properties of opioid agonists. MORs enhance VTA dopaminergic neuron activation by dampening the tonic inhibition of GABAergic interneurons. MORs are also found in the ventral striatum and medial prefrontal cortex, where they are thought to enhance the euphoria of opioids. Thus, MORs in two anatomically distinct circuits are responsible for analgesia and euphoria.1 This is a key reason why it has been so difficult to develop analgesic opioid agonists that are devoid of addictive potential.
Beginning 30 years ago, US physicians were encouraged to prescribe opioid agonists more widely for acute and chronic pain on the basis of weak evidence that suggested that risk for opioid addiction was negligible when opioids were prescribed for pain.2 In the decade ending in 2011, 25 million Americans used illicit prescription opioids (PO).3 The prevalence of a PO addiction among illicit users was 16.9% in the decade 2003-2013,4 and there were 16 651 PO-related fatalities; this is twofold greater than all the deaths from heroin and cocaine combined.5 Furthermore, increases in PO-related mortality paralleled increases in long-term PO treatment of chronic nonprogressive musculoskeletal pain,6-7 and it is well recognized that such treatment engenders substantial risk for PO disorder. However, the magnitude of PO addiction risk is controversial, in part because calculation of rates of opioid misuse (and its behavioral, vocational, medical, and legal consequences) in chronic pain patients have suffered from imprecise and poorly defined terminology.6 Estimates vary widely in the literature from 1% to more than 40% ,7-10 with a recent report suggesting the rate of PO addiction among those taking opioids for chronic pain was as high as 23 %.7
Epidemiologic studies in the United States indicate that chronic pain patients at highest risk for PO addiction are of European ancestry, on high doses of POs, younger, male, less educated, and have a personal/ family history of substance abuse (including smoking), childhood trauma, and/or psychiatric illness.10-12 Whereas the rate of nonmedical use of POs is highest among 18- to 25-year-olds,13 death rates from PO overdoses are highest in the 45-to-54-year age cohort, with non-Hispanic whites showing a threefold higher death rate than Hispanics and African Americans.14
From an international perspective, the 2010 Global Burden of Diseases, Injuries, and Risk Factors Study calculated the burden of illicit drug dependence separately for amphetamines, cocaine, opioids, and cannabis.15 Worldwide, suicide (as a risk of Diagnostic and Statistical Manual of Mental Disorders DSM)-IV—defined opioid dependence) explained 671 000 disability adjusted life years (DALYs).16 DALY is a measure of disease burden, expressed as the number of years of life lost due to disability, ill health, or early death. Opioid dependence was the largest contributor to the global direct burden of DALYs: 9.2 million.16 These data revealed that Western Europe had 1.3 million individuals with DSM-IV—defined opioid dependence.16
Twin studies have documented that a major fraction of the opioid-addiction risk is genetic in origin. Thus, Tsuang et al17 showed that 54% of the liability for opioid addiction was due to genetic variance and that 38% of the liability was explained by genetic variance specific to opioid addiction. Further, Kendler et al18 studied around 1200 male-male twin pairs for substance-abuse phenotypes and found that genetic liability for opioid addiction was 48%, consistent with an opioid-addiction model in which half the risk is genetic.
Genome-wide association studies in opioid addiction
Several genome-wide association studies (GWAS) of opioid addiction involving large sample sizes have been published. Gelernter et al19 studied three populations of US opioid-addiction individuals, recruited mostly from treatment facilities, including 5432 African Americans and 6788 European Americans. Both a DSM symptom count for opioid addiction (a quantitative trait) and a case-control analysis were done for each ethnicity. The investigators reported significant associations with single-nucleotide polymorphisms (SNPs) of small effect in the potassium- and calcium-channel subunit genes. Loci that contained some of the most significantly associated SNPs were the KCNC1 (potassium voltage-gated channel subfamily C member 1) and KCNG2 (potassium voltage-gated channel modifier subfamily G member 2) genes, which encode potassium voltage-gated-channel subunits. These findings are intriguing, but the MOR activates an inwardly rectifying potassium-ion channel. Pathway analysis of these genotypes implicated calcium signaling and synaptic long-term potentiation.
Nelson et al20 studied an Australian opioid-addiction sample, drawn from treatment facilities, and confirmed findings in two other populations, including populations analyzed by Gelernter et al.19 They reported genomewide significant association for multiple SNPs of small effect in the CNIH3 gene (cornichon family AMPA receptor auxiliary protein 3). The A allele of rs10799590, the most significant SNP, was protective against opioid addiction (P=4.30 x 10-9; odds ratio [OR]=0.64). The CNIH3 protein is an α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor auxiliary protein, and there is abundant evidence that AMPA receptors play a critical role in the evolution of opioid addiction.21,22 The results implicating the CNIH3 gene are interesting in part because the A allele of rs10799590 is associated with significantly greater habituation of the right amygdala to threatening facial images, a phenotype that may convey resilience to psychiatric disorders, generally.23 rs10799590 is within an H3 lysine 4 monomethylation (H3K4me1) peak in fetal brain, suggesting that epigenetic influences may play a role in the function of this SNP20 rs10799590 is also located within the binding site of TALI (T-cell acute lymphoblastic leukemia 1), a brain transcription factor regulating midbrain GABAergic neuronal differentiation.24 The A allele weakens the TALI binding potential, compared with the G allele.
In an alternative approach, Hancock et al25 employed SNP genotype and gene-expression data available on pathologically normal postmortem prefrontal cortex samples from more than 100 European Americans and 100 African Americans. They searched for expression quantitative trait loci (eQTLs) near the gene encoding MOR (OPRM1) on human chromosome 6. Those SNPs regulating OPRM1 messenger RNA (mRNA) levels in prefrontal cortex were then studied for association with opioid addiction in several cohorts, including European Americans from the United States and Australia, as well as African Americans. Of the 103 SNPs near the OPRM1 locus tested for eQTL status, nine SNPs met statistical significance as eQTLs, and 16 SNPs were tested for association with opioid addiction. One eQTL SNP, rs3778150, was statistically significant across the populations of opioid-addicted persons and controls, with the C alleles increasing risk (OR=1.3; P=4.3 x 10-8).
Of particular interest regarding a common OPRM1 exon 1 missense functional A/G SNP (rs1799971; A encodes the wild-type asparagine allele, whereas G encodes the aspartate allele), Hancock et al25 noted that a haplotype comprised of the C allele of rs3778150 and the A allele of rs1799971 was associated with opioid addiction. This observation may aid in understanding the multitude of association reports for rs1799971 in addictions.26,27
Copy number variations (CNVs) are duplications and deletions of chromosomal material, often spanning millions of base pairs, and are associated with many brain diseases, particularly schizophrenia and autism. For opioid addiction, Li et al28 studied CNVs in African Americans and European Americans, including 547 African Americans and 1054 European Americans with opioid addiction and 2944 African Americans and 607 European Americans that were screened controls with no addiction diagnoses. These samples were also studied by Gelernter et al.19 Li et al28 reported significance for an intergenic protective chromosome-18 deletion (P=2.2 x 10-8 OR=0.64) spanning 3 kb: 37118901-37122002, as well as a risk-increasing chromosome 1 duplication (P=9.4 x 10-7; OR=1.6) involving the genes LCE3B (late cornified envelope 3B) and LCE3C (late cornified envelope 3C). Deletion of these two genes is associated with various autoimmune diseases.29-31 Lastly, they noted an X chromosome risk-increasing intergenic deletion (P=2.6 x 10-6; OR=4.68) spanning around 1700 base pairs, 150019751-150021351. The CNV burden analysis suggests that CNVs in opioid addiction cases are more numerous, larger, and may involve a greater number of genes.
In summary there are GWAS reports in opioid addiction that are promising, genome-wide significant risk alleles with which to conduct additional research. Efforts to ascertain and evaluate much larger numbers of people with opioid addiction should be supported, particularly studies of PO addiction and opioid addiction in the context of chronic nonprogressive musculoskeletal pain.
Pharmacogenetics of opioid addiction
In this section, promising results regarding pharmacogenetics of therapeutics in opioid addiction are discussed. There are several medications in widespread use for opioid addiction, including naltrexone (tablets and long-acting injectable formulations), buprenorphine (compounded with naloxone to reduce abuse potential), and methadone. There are important pharmacologic differences among these pharmacotherapy options for opioid addiction. Naltrexone is a pure antagonist at MOR, and it will precipitate intense withdrawal symptoms if administered to an individual with active (current) opioid addiction. Buprenorphine is a partial MOR agonist, which acts as an antagonist in the presence of other opioid agonists, so that the buprenorphine-treated opioid addict experiences a markedly attenuated euphoria if a MOR agonist (such as heroin) is taken. Buprenorphine will precipitate intense withdrawal (similar to naltrexone) if administered to an opioid-addicted individual who has taken an opioid agonist in the past 12 to 24 hours. Lastly, methadone is a full MOR agonist, in widespread use for opioid addiction since the 1970s. It is given as a mixture of (R) and (S) enantiomers, the (R) isomer being responsible for the MOR effect. There are currently no biomarkers to identify optimal pharmacotherapy or efficacious doses for individuals with opioid addiction.
Genetic variability may be one factor in the complex phenotype of therapeutic response to pharmacotherapy for opioid addiction. It should be recalled that pharmacotherapy for opioid addiction is optimally provided in conjunction with regular (weekly) counseling, treatment of comorbid illnesses, and social assistance. Adequate treatment of opioid-addicted individuals should include efficacious therapy for comorbid illnesses (addiction to nicotine and alcohol, anxiety, and mood disorders).11
Methadone has been studied most intensively in opioid-addiction pharmacogenetics research (for review see Dennis et al32). Among the most often studied DNA variants for methadone dose requirements and therapeutic response are SNP rs1045642 in ABCB1 (adenosine triphosphate [ATP]-binding cassette subfamily B member 1) and the CYP2B6*6 haplotype SNPs. SNP rs1045642 did not influence trough (R), (S) plasma concentrations, methadone dose, or methadone response in a meta-analysis.32 CYP2B6*6 homozygotes had significantly higher trough (R)- and (S)-enantiomer methadone plasma concentrations than noncarriers.32 However, CYP2B6*6 homozygotes did not differ from noncarriers in therapeutic response or daily dose of methadone.32 There was no influence of rs1045642 genotype on trough (R) or (S) methadone plasma levels, methadone dose, or therapeutic methadone response.
A GWAS of methadone therapeutic dose was recently described in 1027 European American and 383 African American opioid-addicted individuals.33 Although there were no significant signals among the larger European American population, the minor C allele at rs73568641 in the African Americans was associated with higher oral methadone dose. It is uncertain why this effect is seen only among African Americans. This SNP is located 300 kb 5' to OPRM1 and was associated with higher postoperative opioid dose for pain among 241 African American children.33 It is unknown whether this SNP influences the therapeutic response (in terms of urine drug screen results) for opioid-addicted individuals.
A GWAS of (R)- and (S)-methadone plasma levels in around 280 Han Chinese34 revealed that a single intergenic SNP on chromosome 9, rs17180299, was statistically significant and explained nearly 10% of the variance in plasma (R)-methadone levels. This effect was also seen in several haplotypes tagged by rs17180299. It is essential that this report be confirmed independently.
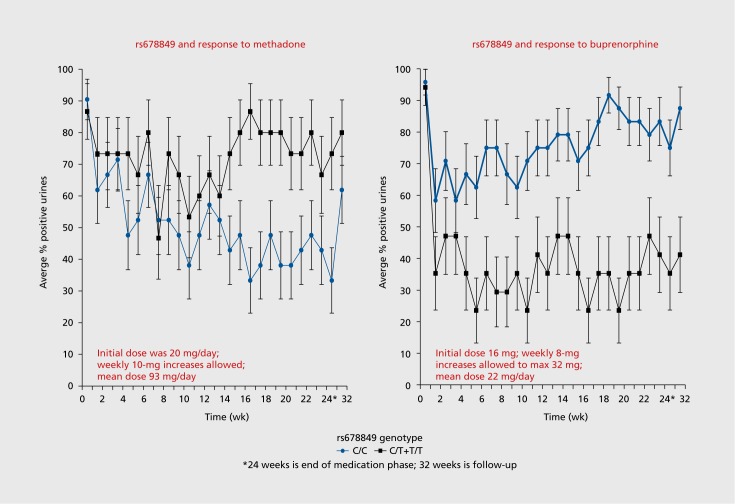
Fifteen years ago, case reports were published of worsening liver function test results in opioid-addicted individuals taking buprenorphine/naloxone.35 In response to this, an open-label, multicenter 6-month study of liver function in opioid-addicted persons, randomized to either buprenorphine/naloxone or methadone, was done across the United States at licensed methadone treatment programs, with the goal of detecting evidence for buprenorphine hepatotoxicity.36 There was no evidence of hepatotoxicity due to buprenorphine. Randomized participants provided weekly urine samples for detection of opioids other than the prescribed medication over the 24 weeks of the study. This therapeutic response (positive or negative urine drug screen for opioids other than the one prescribed) was the phenotype for a pharmacogenetic analysis of eight opioid-system genes.37 An intron 1 SNP, rs678849, in the δ-opioid receptor gene (OPRD1) had a substantial effect on therapeutic response to both methadone and buprenorphine/naloxone in African Americans (n=77), but not in European Americans (n=564). In African Americans, the C allele of rs678849 has a frequency of about 70%, so roughly half of all African Americans have a homozygous C genotype. As shown in Figure 1, African-American participants randomized to methadone had a poor therapeutic response (in terms of urine drug screen for opioids other than the one prescribed) if they were T-allele carriers at rs678849, whereas T-allele carriers had a good therapeutic response to buprenorphine. In parallel, African American participants who were C homozygotes had a good therapeutic response to methadone and a poor response to buprenorphine. In a gene-environment interaction analysis that used a generalized estimating equation approach, the effect was significant (P=5.9 x 10-5).37 It is uncertain why this effect is seen only among African Americans.
Figure 1. The average percent positive urine drug screen result (positive meaning that the urine tested positive for an opioid other than the one prescribed, ie, the participant relapsed) is given for African-American subjects randomized to methadone (N=left panel) or buprenorphine/naloxone (N=right panel) for each week from 0 through 24 weeks. Week 32 is a follow-up appointment, 8 weeks after the end of the study. The gray line indicates participants who are homozygous C (the major allele), and the black line indicates participants with at least one T allele. Generalized estimating equations were used to calculate the effect of this gene-environment interaction when the 24 weeks of urinalysis data were taken as repeated measures. A significant interaction between the genotype at rs678849 and treatment group was again observed in African Americans (relative risk=3.26; 95% confidence interval 2.66-4.19; P=5.9 x 10-5).
In the methadone arm of the study, the two genotype groups do not separate in response until week 12 to 14. A partial explanation for this may be the dose schedule used to treat the participants with methadone. Each methadone participant was started on 20 mg/day, with weekly increases in 10-mg increments, according to participant symptoms and physician recommendation.36 Thus, it required 10 to 12 weeks for participants in the methadone arm to arrive at an optimal dose: mean dose at the end of the trial was 93 mg daily.36 In contrast, the genotype groups in the buprenorphine/naloxone arm separated in response by week 4. Participants in this arm started at a daily dose of 16 mg, and they were eligible for weekly 8-mg increases, according to participant symptoms and physician recommendation, to a maximum of 32 mg daily.36 Thus, in the buprenorphine/ naloxone arm, participants arrived at an optimal dose in 3 weeks: mean dose at the end of the trial was 22 mg.36
In the parent clinical trial, polysubstance abuse was common among participants. For example, at baseline, one-third of participants' urine samples were positive for cocaine, 25% were positive for cannabis, and 20% were positive for benzodiazepines.36 There was no effect of genotype on abuse of these other substances.
If therapeutic response is dichotomized into responders (those with less than 50% of urine samples positive for opioids other than the one prescribed) and nonresponders (those with greater than 50% of urine samples positive), then a number-needed-to- treat (NNT) analysis can be done. This NNT analysis specifies the number of opioid-addicted persons who must be given buprenorphine—if they are T-allele carriers at rs678849—to gain one additional responder, as opposed to treating everyone with buprenorphine. The NNT in this instance is 2.8. This clinical effect is sufficiently large to be clinically meaningful if it can be confirmed in larger populations and in prospective randomized clinical trials.
The influence of rs678849 on OPRD1 gene function is unknown, and it exists in a linkage disequilibrium island, meaning that alleles at rs678849 do not predict alleles at adjacent SNPs. Located 6 kb 3' from the end of exon 1, it is unclear what influence this SNP might have on gene transcription or translation. This SNP has been associated with regional brain volumes of multiple structures,38 such that the C allele predicts smaller gyri in temporal, occipital, and frontal cortical regions (P=7 x 10-5). Further, the C allele of rs678849 increases risk for cocaine addiction in African Americans (P=8.5 x 10-7), but no such association was found in European Americans.39
Only one GWAS has been published regarding opioid requirements to treat pain. In this study, Nishizawa et al40 report that the C allele of rs2952768 was associated with use of greater amounts of opioid to control postoperative pain in a Japanese population. This was observed in both a facial surgery patient group (n=355) and in an abdominal surgery patient group (n=112), providing for confirmation of the effect.
Issues for the future and summary
Progress in the clinical genetics and pharmacogenetics of opioid addiction has been made over the past decade. Promising common alleles for opioid addiction have been identified, including potassium-ion-channel genes (KCNC1 and KCNG2)19, an eQTL near OPRM1 24 and an auxiliary glutamate-receptor protein (CNIH320). Perhaps the single greatest impediment to additional progress is the relative paucity of large, adequately phenotyped populations. For GWAS studies, case populations exceeding 50 000 individuals may be required to detect with genome-wide levels of significance alleles with odds ratios of 1.2 or less.
One issue to consider for future population genetic GWAS investigations of opioid addiction is the question of exposure of the control group. Certainly, it would be ideal to use as a control group ethnicity- and gender-matched individuals who used opioids for euphoria on multiple occasions over years but never developed regular daily use leading to addiction, with its multiple adverse consequences in medical, legal, interpersonal, and vocational spheres of life. This ideal population would be very difficult to recruit.
As larger sample sizes are accrued for opioid addiction, it may be possible to develop a polygenic risk score for opioid addiction. This is typically done by assembling all the alleles in the GWAS that have a nominal P value for a higher allele frequency in the cases than in the controls (eg, ≤0.05 or 0.01). Certainly, the great majority of these roughly 10 000 to 50 000 alleles are false positives, but some are true positives. The polygenic risk score can be examined for specificity by testing for overlap in risk alleles with polygenic risk scores from unrelated conditions, eg, inflammatory bowel syndrome or Parkinson disease. In this case, the degree of overlap in risk alleles should be no higher than that expected randomly (ie, <5%). The polygenic risk score can also be validated by testing for significant overlap with GWAS data from related phenotypes, such as other addictions, recurrent major depression, or anxiety disorders, all of which are more common among opioid-addicted individuals. Eventually, after substantial fine tuning, with larger sample sizes, a polygenic risk score could be used to predict those individuals at increased risk for opioid addiction, given prolonged opioid-analgesic exposure for chronic pain. Such individuals could be monitored closely for early signs of opioid addiction, as well as being treated with alternative approaches for chronic pain, such as acupuncture, nerve blocks, and neuropathic pain medications.
Another issue for the future is the question of genetic studies of opioid addiction complicated by comorbidity. Common comorbidities include addiction to cocaine (~30%), benzodiazepines (20%), cannabis (25%), alcohol (30%), and nicotine (80%). The most common psychiatric comorbidities include mood disorders (40%) and anxiety disorders (25%). Analysis of substantial samples from individuals with opioid addiction plus these common comorbidities may provide more homogeneous phenotypes and thus improved power to detect risk alleles.
Progress in pharmacogenetics of opioid addiction has yielded some biological insights into methadone dose requirements, related to common variants in CYP2B6, a methadone metabolizing enzyme, and in a locus 5' to OPRM1. An OPRD1 common variant (rs678849) may be promising for prediction of therapeutic response to buprenorphine versus methadone.39
An issue for the future of pharmacogenetics of opioid addiction is the need for randomized, doubleblind, controlled, three-arm trials of naltrexone versus buprenorphine versus methadone, with a sufficiently large population that genome-wide significance may be achieved by genotyping participants for common SNPs. This experiment is necessary to optimize treatment of opioid-addicted individuals with the currently available pharmacotherapies.
Acknowledgments
The following National Institutes of Health grants supported the preparation of this manuscript: R21 DA036808-01 (Pharmacogenetics of Opioid Agonist Therapy) to Wade Berrettini, R01 DA032776 (Clinical and Genetic Characteristics of Opioid Addiction in Chronic Pain) to Martin Cheatle, PhD, and R01 DA044015 (Clinical and Genetic Study of Prescription Opioid Addiction) to Wade Berrettini, Vanessa Troiani, and Janet Robishaw. Dr Berrettini has no conflicts of interest to declare.
REFERENCES
- 1.Koob GF., Volkow ND. Neurobiology of addiction: a neurocircuitry analysis. Lancet Psychiatry. 2016;3(8):760–773. doi: 10.1016/S2215-0366(16)00104-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Madras BK. The surge of opioid use, addiction, and overdoses: responsibility and response of the US health care system. JAMA Psychiatry. 2017;74(5):441–442. doi: 10.1001/jamapsychiatry.2017.0163. [DOI] [PubMed] [Google Scholar]
- 3.Center for Behavioral Health Statistics and Quality. Results from the 2011 National Survey on Drug Use and Health: summary of national findings (HHS publication No. SMA 12-4713, NSDUH series H-44). Rockville, MD: Substance Abuse and Mental Health Services Administration; 2012. [Google Scholar]
- 4.Han B., Compton WM., Jones CM., Cai R. Nonmedical prescription opioid use and use disorders among adults aged 18 through 64 years in the United States, 2003-2013. JAMA. 2015;314(14):1468–1478. doi: 10.1001/jama.2015.11859. [DOI] [PubMed] [Google Scholar]
- 5.Centers for Disease Controls National Vital Statistics System. Multiple cause of death file. Atlanta, GA: Centers for Disease Control and Prevention; 2012. Available at: www.cdc.gov/nchs/data/dvs/Record_Layout_2012. pdf. [Google Scholar]
- 6.Brady KT., McCauley JL., Back SE. Prescription opioid misuse, abuse, and treatment in the United States: an update. Am J Psychiatry. 2016;173(1):1–9. doi: 10.1176/appi.ajp.2015.15020262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vowles KE., McEntee ML., Julnes PE., Frohe T., Ney JP., van der Goes DN. Rates of opioid misuse, abuse, and addiction in chronic pain: a systematic review and data synthesis. Pain. 2015;156(4):569–580. doi: 10.1097/01.j.pain.0000460357.01998.f1. [DOI] [PubMed] [Google Scholar]
- 8.Smith SM., Dart RC., Katz NP., et al Classification and definition of misuse, abuse, and related events in clinical trials: ACTTION systematic review and recommendations. Pain. 2013;154(11):2287–2796. doi: 10.1016/j.pain.2013.05.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O'Connor AB., Turk DC., Dworkin RH., et al Abuse liability measures for use in analgesic clinical trials in patients with pain: IMMPACT recommendations. Pain. 2013;154(11):2324–2334. doi: 10.1016/j.pain.2013.06.035. [DOI] [PubMed] [Google Scholar]
- 10.Cheatle M. Facing the challenge of pain management and opioid misuse, abuse and opioid-related fatalities. Expert Rev Clin Pharmacol. 2016;9(6):751–754. doi: 10.1586/17512433.2016.1160776. [DOI] [PubMed] [Google Scholar]
- 11.Grant BF., Saha TD., Ruan WJ., et al Epidemiology of DSM-5 drug use disorder: results from the National Epidemiologic Survey on Alcohol and Related Conditions-III. JAMA Psychiatry. 2016;73(1):39–47. doi: 10.1001/jamapsychiatry.2015.2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilkerson RG., Kim HK., Windsor TA., Mareiniss DP. The opioid epidemic in the United States. Emerg Med Clin N Am. 2016;34(2):e1–e23. doi: 10.1016/j.emc.2015.11.002. [DOI] [PubMed] [Google Scholar]
- 13.Jones CM. Frequency of prescription pain reliever nonmedical use: 2002-2003 and 2009-2010. Arch Intern Med. 2012;172(16):1265–1267. doi: 10.1001/archinternmed.2012.2533. [DOI] [PubMed] [Google Scholar]
- 14.Paulozzi L., Jones C., Mack K., et al Centers for Disease Control and Prevention (CDC). Vital signs: overdoses of prescription opioid analgesics—United States, 1999-2008. MMWR Morb Mortal Wkly Rep. 2011;60(43):1487–1492. [PubMed] [Google Scholar]
- 15.Murray CJ., Vos T., Lozano R., et al Disability-adjusted life years (DALYs) for 291 diseases and injuries in 21 regions, 1990-2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380(9859):2197–2223. doi: 10.1016/S0140-6736(12)61689-4. [DOI] [PubMed] [Google Scholar]
- 16.Degenhardt L., Whiteford HA., Ferrari AJ., et al Global burden of disease attributable to illicit drug use and dependence: findings from the Global Burden of Disease Study 2010. Lancet. 2013;382(9904):1564–1574. doi: 10.1016/S0140-6736(13)61530-5. [DOI] [PubMed] [Google Scholar]
- 17.Tsuang MT., Lyons MJ., Meyer JM., et al Co-occurrence of abuse of different drugs in men. Arch Gen Psychiatry. 1998;55(11):967–972. doi: 10.1001/archpsyc.55.11.967. [DOI] [PubMed] [Google Scholar]
- 18.Kendler KS., Jacobson KC., Prescott CA., Neale MC. Specificity of genetic and environmental risk factors for use and abuse/dependence of cannabis, cocaine, hallucinogens, sedatives, stimulants, and opiates in male twins. Am J Psychiatry. 2003;160(4):687–695. doi: 10.1176/appi.ajp.160.4.687. [DOI] [PubMed] [Google Scholar]
- 19.Gelernter J., Kranzler HR., Sherva R., et al Genome-wide association study of opioid dependence: multiple associations mapped to calcium and potassium pathways. Biol Psychiatry. 2014;76(1):66–74. doi: 10.1016/j.biopsych.2013.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nelson EC., Agrawal A., Heath AC., et al Evidence of CNIH3 involvement in opioid dependence. Mol Psychiatry. 2016;21(5):608–614. doi: 10.1038/mp.2015.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van den Oever MC., Goriounova NA., Li KW., et al Prefrontal cortex AMPA receptor plasticity is crucial for cue-induced relapse to heroin-seeking. Nat Neurosci. 2008;11(9):1053–1058. doi: 10.1038/nn.2165. [DOI] [PubMed] [Google Scholar]
- 22.Cai YQ., Wang W., Hou YY., Zhang Z., Xie J., Pan ZZ. Central amygdala GluA1 facilitates associative learning of opioid reward. J Neurosci. 2013;33(4):1577–1588. doi: 10.1523/JNEUROSCI.1749-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Plichta MM., Grimm O., Morgen K., et al Amygdala habituation: a reliable fMRI phenotype. Neuroimage. 2014;103:383–390. doi: 10.1016/j.neuroimage.2014.09.059. [DOI] [PubMed] [Google Scholar]
- 24.Colantuoni C., Lipska BK., Ye T., et al Temporal dynamics and genetic control of transcription in the human prefrontal cortex. Nature. 2011;478(7370):519–523. doi: 10.1038/nature10524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hancock DB., Levy JL., Gaddis NC., et al Cis-expression quantitative trait loci mapping reveals replicable associations with heroin addiction in OPRM1 . Biol Psychiatry. 2015;78(7):474–484. doi: 10.1016/j.biopsych.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwantes-An TH., Zhang J., Chen LS., et al Association of the OPRM1 variant rs1799971 (A118G) with non-specific liability to substance dependence in a collaborative de novo meta-analysis of European-ancestry cohorts. Behav Genet. 2016;46(2):151–169. doi: 10.1007/s10519-015-9737-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haerian BS., Haerian MS. OPRM1 rs1799971 polymorphism and opioid dependence: evidence from a meta-analysis. Pharmacogenomics. 2013;14(7):813–824. doi: 10.2217/pgs.13.57. [DOI] [PubMed] [Google Scholar]
- 28.Li D., Zhao H., Kranzler HR., et al Genome-wide association study of copy number variations (CNVs) with opioid dependence. Neuropsychopharmacology. 2015;40(4):1016–1026. doi: 10.1038/npp.2014.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Cid R., Riveira-Munoz E., Zeeuwen PL., et al Deletion of the late cornified envelope (LCE) 3B and 3C genes as a susceptibility factor for psoriasis. Nat Genet. 2009;41(2):211–215. doi: 10.1038/ng.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Docampo E., Rabionet R., Riveira-Munoz E., et al Deletion of the late cornified envelope genes, LCE3C Cand LCE3B, is associated with rheumatoid arthritis. Arthritis Rheum. 2010;62(5):1246–1251. doi: 10.1002/art.27381. [DOI] [PubMed] [Google Scholar]
- 31.Lu X., Guo J., Zhou X., et al Deletion of LCE3C_LCE3B is associated with rheumatoid arthritis and systemic lupus erythematosus in the Chinese Han population. Ann Rheum Dis. 2011;70(9):1648–1651. doi: 10.1136/ard.2010.148072. [DOI] [PubMed] [Google Scholar]
- 32.Dennis BB., Bawor M., Thabane L., Sohani Z., Samaan Z. Impact of ABCB1 and CYP2B6 genetic polymorphisms on methadone metabolism, dose and treatment response in patients with opioid addiction: a systematic review and meta-analysis. PLoS One. 2014;9(1):e86114. doi: 10.1371/journal.pone.0086114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith AH., Jensen KP., Li J., et al Genome-wide association study of therapeutic opioid dosing identifies a novel locus upstream of OPRM1. Mol Psychiatry. 2017;22(3):346–352. doi: 10.1038/mp.2016.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang HC., Chu SK., Huang CL., et al Genome-wide pharmacogenomic study on methadone maintenance treatment identifies SNP rs17180299 and multiple haplotypes on CYP2B6, SPON1, and GSG1L associated with plasma concentrations of methadone R- and S-enantiomers in heroindependent patients. PLoS Genet. 2016;12(3):e1005910.. doi: 10.1371/journal.pgen.1005910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berson A., Fau D., Fornacciari R., et al Mechanisms for experimental buprenorphine hepatotoxicity: major role of mitochondrial dysfunction versus metabolic activation. J Hepatol. 2001;34(2):261–269. doi: 10.1016/s0168-8278(00)00050-7. [DOI] [PubMed] [Google Scholar]
- 36.Saxon AJ., Ling W., Hillhouse M., et al Buprenorphine/naloxone and methadone effects on laboratory indices of liver health: a randomized trial. Drug Alcohol Depend. 2013;128(1-2):71–76. doi: 10.1016/j.drugalcdep.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Crist RC., Clarke TK., Ang A., et al An intronic variant in OPRD1 predicts treatment outcome for opioid dependence in African-Americans. Neuropsychopharmacology. 2013;38(10):2003–2010. doi: 10.1038/npp.2013.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roussotte FF., Jahanshad N., Hibar DP., et al A commonly carried genetic variant in the delta opioid receptor gene, OPRD1 is associated with smaller regional brain volumes: replication in elderly and young populations. Hum Brain Mapp. 2014;35(4):1226–1236. doi: 10.1002/hbm.22247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crist RC., Ambrose-Lanci LM., Vaswani M., et al Case-control association analysis of polymorphisms in the δ-opioid receptor, OPRD1, with cocaine and opioid addicted populations. Drug Alcohol Depend. 2013;127(1-3):122–128. doi: 10.1016/j.drugalcdep.2012.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nishizawa D., Fukuda K., Kasai S., et al Genome-wide association study identifies a potent locus associated with human opioid sensitivity. Mol Psychiatry. 2014;19(1):55–62. doi: 10.1038/mp.2012.164. [DOI] [PMC free article] [PubMed] [Google Scholar]