Abstract
Impaired skeletal muscle energy metabolism is linked to the pathogenesis of insulin resistance and glucose intolerance in type 2 diabetes. The contractile and metabolic properties of myofibers exhibit a high degree of heterogeneity and plasticity. The regulatory circuitry underpinning skeletal muscle energy metabolism is critically linked to exercise endurance and systemic homeostasis. Recent work has identified the Baf60 subunits of the SWI/SNF chromatin-remodeling complex as powerful regulators of the metabolic gene programs. However, their role in integrating myofiber energy metabolism with exercise endurance and metabolic physiology remains largely unknown. In this study, we conditionally inactivated Baf60a, Baf60c, or both in mature skeletal myocytes to delineate their contribution to muscle bioenergetics and metabolic physiology. Our work revealed functional redundancy between Baf60a and Baf60c in maintaining oxidative and glycolytic metabolism in skeletal myofibers and exercise endurance. Unexpectedly, mice lacking these two factors in skeletal muscle were protected from diet-induced and age-associated metabolic disorders. Transcriptional profiling analysis identified the muscle thermogenic gene program and myokine secretion as key pathways that integrate myofiber metabolism with systemic energy balance. As such, Baf60 deficiency in skeletal muscle illustrates a surprising disconnect between exercise endurance and systemic metabolic homeostasis.
Introduction
Impaired skeletal muscle energy metabolism contributes to the pathogenesis of insulin resistance and glucose intolerance in type 2 diabetes. Skeletal myofibers are highly heterogeneous with regard to their contractile function and metabolic properties (1–3). White muscle generates energy through glycolysis, whereas red muscle contains high mitochondrial content and relies primarily on oxidative phosphorylation. Myofiber energy metabolism exhibits a high degree of plasticity (4–6). Endurance exercise is known to stimulate mitochondrial biogenesis and oxidative metabolism and improve insulin sensitivity. Reprogramming of muscle energy metabolism also occurs in pathophysiological states, such as type 2 diabetes. The latter is associated with an oxidative to glycolytic metabolic shift in skeletal muscle (7–9). Although this metabolic shift has been proposed to contribute to muscle insulin resistance (10–12), the cause and effect relationship between skeletal myofiber metabolism and insulin sensitivity remains inconclusive (13,14). In fact, genetic activation of glycolytic metabolism in skeletal muscle improved insulin sensitivity and whole-body glucose metabolism in mice (15–18), suggesting that activation of glycolytic metabolism in diabetic muscles may serve an adaptive physiological role in the state of chronic hyperglycemia.
The transcriptional regulatory network that governs the specification and plasticity of myofiber energy metabolism is critically linked to exercise endurance and systemic metabolic physiology. Several transcriptional regulators have been identified to promote oxidative metabolism in skeletal muscle, including the nuclear receptors ERRα/γ, PPARα/δ, and Nur77 and their coactivators PGC-1α and PGC-1β (19–25). PGC-1α is highly inducible by exercise and is important for orchestrating the metabolic adaption of skeletal muscle following endurance exercise (26). These factors act in concert with key energy-sensing pathways, such as AMPK and SIRT1, to fine-tune the oxidative capacity of skeletal muscle to meet its energetic demand (19,27,28). RIP140 is a transcriptional corepressor that suppresses the gene program of oxidative metabolism (29,30). Much less is known about the factors that drive glycolytic muscle formation and metabolism. Recent studies identified Baf60c, a subunit of the SWI/SNF chromatin-remodeling complex, as a powerful regulator of glycolytic muscle metabolism (16,17). Baf60c expression is enriched in glycolytic muscles, such as plantaris and extensor digitorum longus (EDL) muscles. Muscle-specific transgenic expression of Baf60c promotes glycolytic metabolism in myofibers through induction of Deptor-mediated AKT activation; the latter has been previously shown to enhance muscle growth and glycolytic function (15). More recently, BAF60c was identified as a key component of myocyte glucose sensing that is indispensable for postprandial glucose metabolism (31).
Mammalian SWI/SNF complexes contain the catalytic ATPase subunit Brg1 or Brm and various combinations of subunits known as Brg/Brm-associated factors (BAFs) (32–34). The functional diversity of different SWI/SNF complexes is determined by the repertoire of BAFs within a given cell type. The Baf60 family members, including Baf60a, Baf60b, and Baf60c, are unique in that they exhibit restricted patterns of expression and physically interact with diverse transcription factors, thereby recruiting the SWI/SNF complexes to specific chromatin targets (35). Baf60a stimulates hepatic fatty acid oxidation through its interaction with PPARα and PGC-1α (36,37), whereas it forms a transcriptional complex with CAR to stimulate bile acid biosynthesis (38). Baf60c is abundantly expressed in skeletal muscle and heart in mice and has been implicated in the regulation of skeletal and cardiac myogenesis (39–41). Whereas Baf60b expression is nearly absent, Baf60a expression is readily detectable in skeletal muscle. Whether Baf60a and Baf60c are functionally redundant in the control of metabolic gene programs in mature skeletal myofibers remains unknown. In this study, we conditionally inactivated Baf60a, Baf60c, or both in skeletal muscle to address this question and define their role in integrating myofiber energy metabolism with exercise endurance and metabolic physiology. Our work revealed an unexpected function of muscle Baf60s in systemic energy balance through their regulation of the muscle thermogenic gene program and myokine secretion.
Research Design and Methods
Animal Experiments
For the generation of muscle-specific Baf60a knockout (AKO) mice, a targeting vector that carries two loxP sites flanking exons 3–5 of mouse Baf60a gene was constructed using BAC recombineering as previously described (38). For the generation of muscle-specific Baf60c knockout (CKO) mice, a targeting vector that carries two loxP sites flanking exon 2 of mouse Baf60c gene was constructed using BAC recombineering. A C57BL/6J embryonic stem cell line (Bruce 4) from the Transgenic Animal Model Core at the University of Michigan was chosen for homologous recombination to facilitate breeding into C57BL/6J genetic background for metabolic studies (31). MLC-Cre transgenic mice were used to cross with Baf60a or Baf60c flox/flox mice to achieve muscle-specific knockout of Baf60a or Baf60c. Muscle-specific Baf60a and Baf60c double knockout (ACKO) mice were obtained by crossing AKO with CKO mice. Mice were housed under a 12-h light/12-h dark cycle in a temperature- and humidity-controlled environment and were fed standard chow or high-fat diet (HFD) (D12492; Research Diets). Cold exposure studies were performed as previously described (42,43). All animal studies were performed according to procedures approved by the University Committee on Use and Care of Animals. All the mice used were on C57BL/6J genetic background. Littermates were used as controls for all animal studies. Unless otherwise indicated, male mice were used in this study.
mRNA and Western Blot Analysis
Total RNA from tissues was isolated using TRIzol reagent (Life Technologies), reverse transcribed, and analyzed by quantitative PCR (qPCR) using SYBR Green on a QuantStudio 6 Flex Real-Time PCR system (Applied Biosystems, Carlsbad, CA). Primers for ribosomal protein 36B4 were included for normalization. The list of qPCR primers is shown in Supplementary Table 1.
For protein analysis, we homogenized tissues in a lysis buffer containing 50 mmol/L Tris-HCl (pH = 7.5), 137 mmol/L NaCl, 1 mmol/L EDTA, 1% Triton X-100, 10% glycerol, 10 mmol/L NaF, 10 mmol/L Na4P2O7, 1 mmol/L Na3VO4, and protease inhibitor cocktail (Roche) and centrifuged for 10 min at 14,000g and 4°C. The supernatant fraction was separated by SDS-PAGE and immunoblotted with the following antibodies. Rabbit polyclonal antibody against Baf60c was generated with the recombinant GST fusion mouse Baf60c protein, affinity-purified, and used at 1:1,000 dilution. Antibodies against phospho-AKT (Thr308) (1:1,000, #9275), phospho-AKT (Ser473) (1:3,000, #4060), AKT (1:3,000, #4691), phospho-AMPK (Thr172) (1:2,000, #2535), and AMPK (1:2,000, #2532) were from Cell Signaling. Antibody against α-tubulin (1:5,000, T6199) was from Sigma-Aldrich. Rabbit polyclonal antibodies against Sarcoplasmic/Endoplasmic Reticulum Calcium ATPase 2a (Serca2a) (1:3,000) and Serca1 (1:1,000) were kindly provided by Dr. Muthu Periasamy (Ohio State University). Antibody against Baf60a (1:1,000, #611728) was from BD Biosciences. Antibody against Hsp90 (1:500, #sc-7947) was from Santa Cruz Biotechnology. Antibody against Sarcolipin (Sln) (1:500, ABT13; Millipore) was kindly provided by Dr. Dave Bridges (University of Michigan).
Glucose and Insulin Tolerance Tests
Glucose and insulin tolerance tests (GTT and ITT) were performed as previously described (44,45). For GTT, mice were given an intraperitoneal injection of glucose (1 g/kg body weight for HFD-fed mice, 1.5 g/kg for 10-month-old [old] mice, 2 g/kg for 3-month-old [young] mice) after an overnight starvation. For ITT, mice were given an intraperitoneal injection of insulin (1.2 units/kg for HFD-fed mice, 0.4 units/kg for old mice, 0.6 units/kg for young mice) after a 3-h starvation. Blood glucose levels were measured at the indicated time points after injection by tail bleeding.
Plasma Insulin-Like Growth Factor 2 Measurement
For plasma preparation, 50 μL acid/ethanol reagent (12.5% 2 N HCl, 87.5% ethanol) was added to 12.5 μL plasma sample and incubated at room temperature for 30 min, followed by centrifugation at 10,000g for 10 min. Fifty microliters of the clear supernatant was harvested and neutralized with 20 μL neutralization buffer (1 mol/L Tris-HCl, pH 8.0 containing 300 ng IGF1). Ten minutes later, the sample was centrifuged in a SpeedVac concentrator for 1.5 h at ethanol mode to remove ethanol. Then, 200 μL reagent diluent was added to each sample to dissolve, from which 100 μL was used to measure insulin-like growth factor 2 (IGF2) levels using a Mouse IGF-II DuoSet ELISA Kit (R&D systems) according to the manufacturer's protocol.
Ex Vivo IGF2 Secretion Assay
For IGF2 secretion, intact plantaris and soleus muscles were dissected from 8-week-old control and ACKO mice and incubated in Krebs buffer (115 mmol/L NaCl, 2 mmol/L KCl, 25 mmol/L NaHCO3, 1 mmol/L MgCl2, 2 mmol/L CaCl2, pH 7.4) for 1 h. The Krebs buffer was then replaced with serum-free DMEM. Following incubation for 4 h at 37°C in a humidified atmosphere with 5% CO2, 100 μL culture medium was collected for IGF2 measurement using the Mouse IGF-II DuoSet ELISA Kit.
Histological Analysis
Skeletal muscle tissues were dissected and immediately embedded in optimal cutting temperature compound and then frozen with liquid nitrogen-cooled isopentane and sectioned on a cryostat microtome. Hematoxylin-eosin (H&E) staining was performed according to standard procedures. α-Glycerolphosphate dehydrogenase (α-GPDH) and succinate dehydrogenase (SDH) staining were performed as described (16).
Treadmill Running Study
We performed treadmill running studies using a motorized, speed-controlled treadmill system (Columbus Instruments). Mice were trained at a speed of 10 m/min for 10 min for three sessions to acquire running skills prior to the running tests. During the running test, mice ran for 5 min at 8 m/min followed by an increase of 2 m/min every 20 min to 16 m/min until exhaustion. A level inclination angle was used. Single or double knockout mice were allowed to run until exhaustion for the measurements of total running time. In a separate study, ACKO mice were subjected to run until exhaustion with running time-matched control mice used as controls. Blood glucose, lactate concentrations, and other metabolites were measured before the onset of the running study and immediately after exhaustion.
Chromatin Immunoprecipitation Assay
Chromatin immunoprecipitation (ChIP) assay using tissue nuclei was performed as previously described (38). Briefly, muscle nuclei were isolated and then cross-linked in 1% formaldehyde for 10 min followed by sonication. Chromatin lysates were prepared, precleared with Protein G agarose beads, and immunoprecipitated with antibodies against acetyl-H3 (06–599; Upstate), H3K9me2 (ab1220; Abcam), or control mouse IgG (A4416; Sigma-Aldrich) in the presence of BSA and salmon sperm DNA. Beads were extensively washed before reverse cross-linking. DNA was purified using a PCR purification kit (Invitrogen) and subsequently analyzed by qPCR. A list of ChIP qPCR primers is shown in Supplementary Table 1.
Metabolic Parameters
Blood samples were obtained by heart puncture, and the plasma samples were stored at −80°C until analysis. Blood glucose levels were determined using a Bayer Contour blood glucose meter. Blood lactate levels were determined using a Lactate Pro blood lactate test meter (ARKRAY). Plasma concentrations of triglycerides (TAGs) (Sigma-Aldrich), glycerol (Sigma-Aldrich), and nonesterified fatty acids (NEFA) (Wako Diagnostics) were measured using commercial assay kits according to the manufacturer’s protocols. Plasma insulin concentrations were measured using an ELISA kit from Crystal Chem. Liver TAGs were extracted and measured as previously described (37).
Comprehensive Lab Animal Monitoring System (CLAMS) Study
Metabolic rate was measured using Comprehensive Lab Animal Monitoring System (CLAMS) (Columbus Instruments) by University of Michigan Animal Phenotyping Core. Mice were housed individually and maintained at 22°C under a 12-h light/12-h dark cycle. Food and water were available ad libitum.
Statistics
Statistical analyses were carried out using GraphPad Prism 7. Statistical differences were evaluated using two-tailed unpaired Student t test for comparisons between two groups or ANOVA and appropriate post hoc analyses for comparisons of more than two groups. For ITT and GTT studies, two-way ANOVA with multiple comparisons was used for statistical analysis. Further, area under curve was calculated for each mouse and evaluated by two-tailed unpaired Student t test for statistical differences between two genotypes. A P value of less than 0.05 (*P < 0.05, **P < 0.01, and ***P < 0.001) was considered statistically significant.
Results
Conditional Inactivation of Baf60a and Baf60c Impairs Skeletal Muscle Energy Metabolism
Among three Baf60 members, Baf60a and Baf60c are expressed in skeletal muscle, with the latter exhibiting enriched expression in glycolytic muscles (Supplementary Fig. 1). Despite the emerging role of Baf60s in the control of metabolic gene programs, it remains unknown whether they contribute to metabolic adaptation and flexibility of skeletal muscle under stress conditions, such as exercise and obesity, and during aging. To address this, we generated mouse strains with conditional inactivation of Baf60a (AKO), Baf60c (CKO), or both (ACKO) in skeletal muscle using MLC-Cre, which is activated in mature skeletal myofibers and exhibits no leaky expression in cardiac muscle, thereby avoiding potential confounding effects of Baf60 inactivation on skeletal myogenesis and cardiac function.
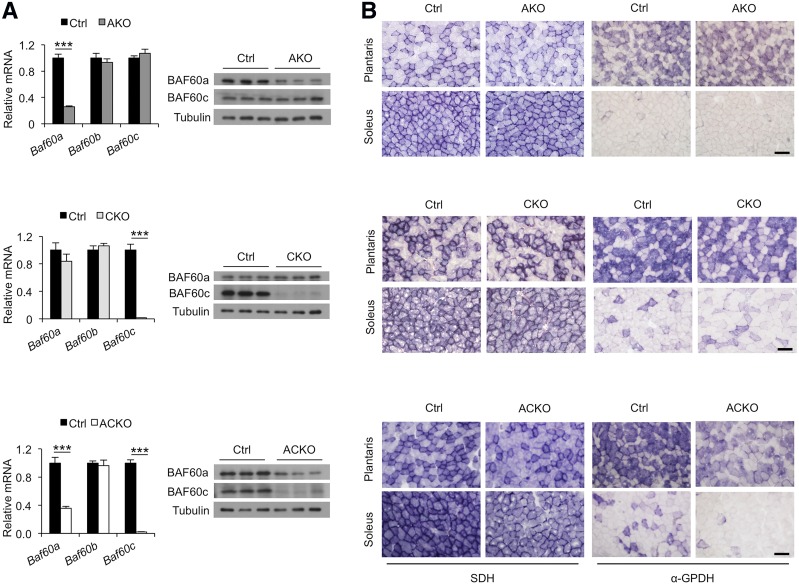
Gene expression analysis indicated that, as expected, Baf60c mRNA and protein expression was nearly abolished in quadriceps muscles from CKO and ACKO mice (Fig. 1A). Residual levels of Baf60a remained detectable in the whole-muscle lysates from AKO and ACKO mice, consistent with its less restricted expression profile among different cell types. We performed histochemical staining of SDH and α-GPDH on frozen muscle sections to assess the effects of Baf60 inactivation on oxidative and glycolytic functions, respectively. Whereas Baf60a or Baf60c inactivation alone had modest effects on the intensity of enzymatic staining, a combined deletion of these factors resulted in reduced SDH and α-GPDH activities in plantaris and soleus muscles from ACKO mice (Fig. 1B). Consistent with the enzymatic and gene expression data, gastrocnemius, quadriceps, and soleus muscles from ACKO mice appeared paler in color than control muscles (Fig. 2A). In contrast, muscle histology and myofiber size, as revealed by H&E and Laminin B immunofluorescent staining, respectively, were nearly indistinguishable between the two groups (Fig. 2B). Muscle mass and glycogen content remained similar (Fig. 2C and D), indicating that Baf60 inactivation in mature myocytes does not impair development and growth of skeletal myocytes. These results revealed some degree of redundancy for Baf60a and Baf60c in the control of myofiber energy metabolism.
Figure 1.
Role of Baf60a and Baf60c in skeletal muscle energy metabolism. A: Baf60a and Baf60c mRNA and protein expression in quadriceps muscles from AKO (n = 3), CKO (n = 6), and ACKO (n = 4–5) mice and their respective littermate controls (Ctrl). Data represent mean ± SEM. ***P < 0.001. B: Representative histochemical staining of SDH (left) and α-GPDH (right) enzymatic activity in plantaris and soleus muscles. Scale bars, 100 μm.
Figure 2.
Regulation of skeletal muscle transcriptome by Baf60a and Baf60c. A: Morphology of gastrocnemius (left), quadriceps (middle), and soleus (right) muscles from control (Ctrl) and ACKO mice. B: H&E (top) and Laminin B immunofluorescent (bottom) staining of tibialis anterior (TA) muscle sections. C and D: Weight (C) and glycogen content (D) of gastrocnemius (Gast), TA, and EDL muscles from chow-fed control and ACKO mice (n = 4–5). E: Heat map representation of Baf60-regulated genes in skeletal muscle. F: Enrichment score for the downregulated (cluster II, blue) and upregulated (cluster III, yellow) genes in panel E. G: Venn diagram representation of Baf60-regulated genes. H: qPCR analysis of myofiber type (left), lipid metabolism (middle), and glycolysis (right) gene expression in quadriceps muscles in control and ACKO mice (n = 4–5). Data in C, D, and H represent mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001.
Baf60s Coordinate Skeletal Muscle Transcriptome and Are Required for Exercise Endurance
We performed microarray studies to determine the relative contribution of Baf60a and Baf60c to global gene expression in skeletal muscle. Clustering analysis revealed that Baf60a deficiency impaired the expression of a small number of genes (cluster I, Fig. 2E). In contrast, Baf60c inactivation resulted in downregulation (cluster II) and upregulation (cluster III) of a large number of genes, many of which exhibited similar directions of regulation in ACKO muscle. Database for Annotation, Visualization and Integrated Discovery (DAVID) analysis indicated that genes in cluster II were enriched for lipid metabolism, mitochondrion, and decarboxylase, whereas those in cluster III were enriched for lipoprotein, actin binding, and cytoskeleton (Fig. 2F). We identified a cluster of 207 genes whose expression was significantly altered by inactivation of both Baf60s but not individual factors (Fig. 2G). These genes appeared to be regulated by Baf60a and Baf60c in a functionally redundant manner. qPCR analysis revealed that mRNA levels of a set of genes involved in lipid metabolism and glycolysis were significantly lower in quadriceps muscles from ACKO than control mice, whereas expression of myofiber contractile genes remained similar between the two groups (Fig. 2H). As such, Baf60s appear to be dispensable for the specification of muscle fiber types; instead, these two factors together are required for maintaining glycolytic and oxidative metabolic functions of skeletal muscle.
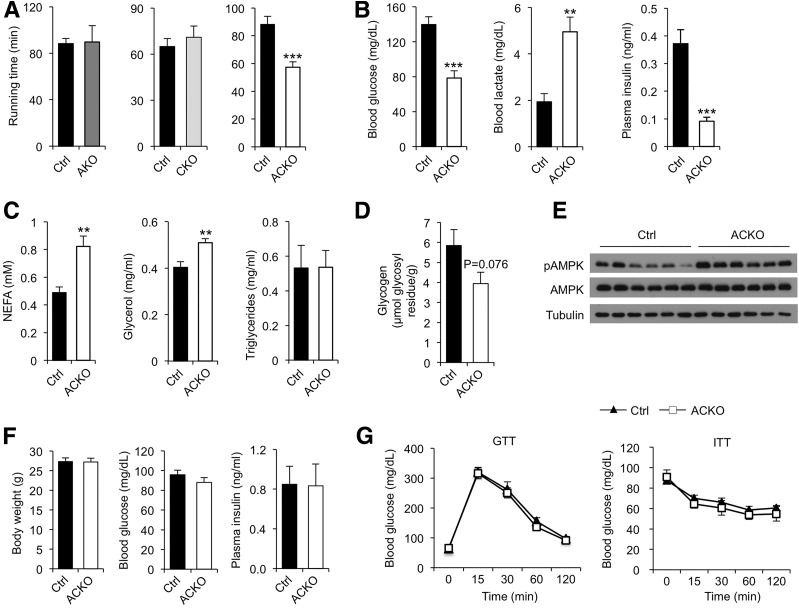
Having established that Baf60a and Baf60c are partially redundant in the control of muscle energy metabolism, we next focused our studies on ACKO mice to determine how Baf60s contribute to metabolic adaptation and flexibility of skeletal muscle. Treadmill running tests indicated that conditional inactivation of Baf60a and Baf60c individually did not significantly alter exercise endurance in AKO and CKO mice (Fig. 3A). In contrast, ACKO mice exhibited an approximately 35% decrease in their exercise capacity. Although blood glucose and plasma glycerol and TAG levels were similar between the two groups under resting conditions (Supplementary Fig. 2A), ACKO mice had significantly lower blood glucose and higher lactate levels than control mice following exercise (Fig. 3B). Postexercise plasma insulin concentration was also lower in ACKO mice. In addition, plasma concentrations of NEFA and glycerol, but not TAG, were higher in ACKO mice after exercise (Fig. 3C). Postexercise muscle glycogen content was moderately lower in ACKO muscle compared with controls (Fig. 3D). Defective exercise performance is likely due to impaired muscle energy metabolism in ACKO mice. In support of this, phosphorylation of AMPK, a cellular energy sensor, was elevated in quadriceps muscles from ACKO mice following treadmill running (Fig. 3E). These results demonstrate that Baf60s are required for maintaining myofiber energy metabolism to sustain the contractile activities of skeletal muscle during exercise.
Figure 3.
Conditional inactivation of Baf60s impairs exercise endurance. A: Running time until exhaustion in control (Ctrl) and conditional knockout mice; AKO (n = 4–6), CKO (n = 4–6), ACKO (n = 7–9). B: Postrunning blood glucose, lactate, and plasma insulin levels in control and ACKO mice (n = 7–9). C: Postrunning plasma NEFA, glycerol, and TAG levels in control and ACKO mice (n = 6). D: Postrunning glycogen content in tibialis anterior muscles (n = 5–6). E: Immunoblots of quadriceps muscle total protein lysates from control and ACKO mice following exercise. F: Body weight (left, n = 5–7), fasting blood glucose (middle, n = 4–8), and ad libitum plasma insulin (right, n = 7–9) levels from chow-fed control and ACKO mice. G: GTT (left, n = 7) and ITT (right, n = 7–12) in mice fed with chow diet. Data in A–D, F, and G represent mean ± SEM. **P < 0.01, ***P < 0.001.
Muscle-Specific Baf60 Inactivation Protects Mice From Diet-Induced Metabolic Disorders
Skeletal muscle is responsible for the bulk of postprandial glucose disposal and serves as a major site of fuel oxidation in the body. Defective myofiber energy metabolism not only impairs endurance exercise capacity but also contributes to the disruption of whole-body energy balance and glucose homeostasis. To determine whether impaired muscle energy metabolism by Baf60 inactivation predisposes ACKO mice to diet-induced obesity and its associated disorders, we examined the metabolic parameters in control and ACKO mice fed with standard chow diet or HFD. Body weight, fasting blood glucose, and plasma insulin levels were similar between control and ACKO mice fed standard chow (Fig. 3F). GTT and ITT revealed that glucose tolerance and insulin sensitivity were similar between these two groups (Fig. 3G).
Conditional inactivation of Baf60a or Baf60c alone did not significantly alter HFD-induced weight gain in AKO and CKO mice (Fig. 4A). Unexpectedly, ACKO mice were remarkably resistant to diet-induced obesity. Whereas control mice gained an average of approximately 20 g of body weight, ACKO mice gained less than 5 g following 16 weeks of HFD feeding (Fig. 4B). Measurements of tissue weight upon dissection indicated that ACKO mice had significantly smaller liver, epididymal white adipose tissue (eWAT), inguinal white adipose tissue (iWAT), and brown adipose tissue (BAT) mass than control mice (Fig. 4C). Although plasma concentrations of TAG, glycerol, and NEFA remained similar between the two groups (Supplementary Fig. 2B), fasting blood glucose and plasma insulin concentrations were significantly lower in ACKO mice (Fig. 4D and E), suggesting that muscle-specific inactivation of Baf60a and Baf60c improves whole-body glucose metabolism. In support of this, GTT and ITT indicated that ACKO mice exhibited enhanced insulin sensitivity and glucose tolerance (Fig. 4F). Insulin action, as indicated by insulin-stimulated AKT phosphorylation, in skeletal muscle, liver, and eWAT was enhanced in ACKO mouse tissues (Fig. 4G). Histological staining revealed that BAT from ACKO mice had less lipid accumulation (Fig. 4H). The presence of crown-like structures in eWAT was also markedly reduced by Baf60 inactivation in skeletal muscle. ACKO mice were protected from HFD-induced hepatic steatosis, as indicated by improved liver histology and reduced hepatic fat content (Fig. 4H and I). Interestingly, ACKO and control mice gained comparable body weight following HFD feeding at thermoneutral temperature (Supplementary Fig. 3), suggesting that activation of thermogenesis may underlie the metabolic effects of Baf60 inactivation. Together, these studies illustrate an unexpected role of skeletal muscle Baf60 signaling in systemic energy balance and metabolic homeostasis.
Figure 4.
ACKO mice are protected from HFD-induced metabolic disorders. A: Body weight of AKO (left, n = 9–10) and CKO (right, n = 9–17) mice and their respective controls (Ctrl) before and after 16 weeks of HFD feeding. B: Body weight curve of control and ACKO mice (n = 10–14) fed with HFD for 16 weeks. C: Tissue weight of control and ACKO mice (n = 10–14) fed with HFD for 16 weeks. D and E: Fasting blood glucose (D) and plasma insulin (E) concentrations in mice fed with HFD for 12 weeks (n = 10–14). F: GTT (left) and ITT (right) in mice fed with HFD for 20 weeks (n = 6). G: Immunoblots of total protein lysates from HFD-fed mice injected with saline or insulin (Ins, 1 unit kg−1) for 10 min. H: H&E staining of tissue sections. Scale bars, 200 μm. I: Liver TAG content in HFD-fed mice (n = 7–9). Data represent mean ± SEM. AUC, area under the curve; Quad, quadriceps muscle. *P < 0.05, **P < 0.01, ***P < 0.001.
ACKO Mice Have an Improved Metabolic Profile During Aging
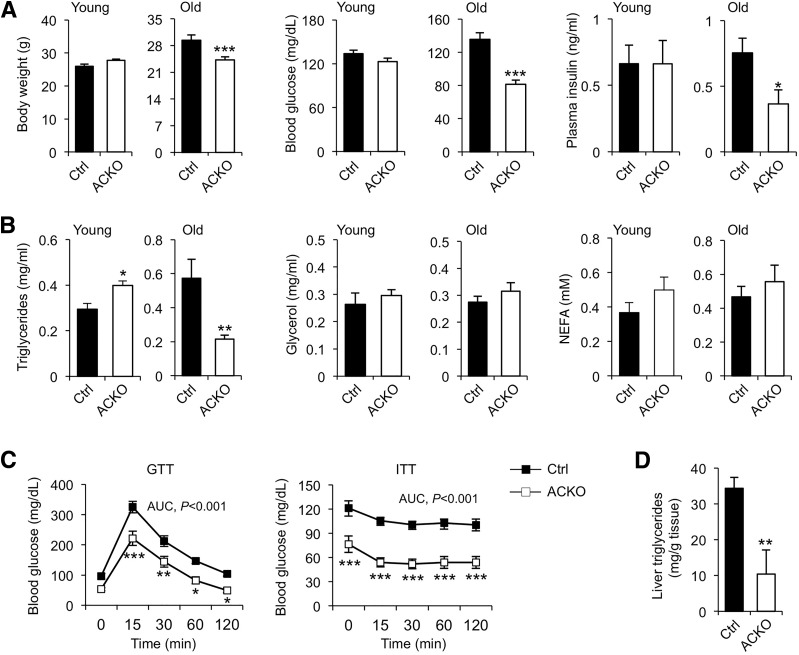
Aging is a risk factor for the development of insulin resistance and glucose intolerance. We next determined whether Baf60 inactivation in skeletal muscle alters whole-body glucose and energy metabolism during aging. Young ACKO mice were nearly indistinguishable from control littermates with regard to their body weight and blood glucose and insulin levels (Fig. 5A). ACKO mice weighed moderately but significantly less than old control mice. Blood glucose and plasma insulin levels were lower in old ACKO mice. Although plasma NEFA and glycerol concentrations remained similar between the two groups, plasma TAG levels were significantly lower in old ACKO mice (Fig. 5B). GTT and ITT indicate that ACKO mice were more insulin sensitive and glucose tolerant (Fig. 5C). Age-associated hepatic steatosis was also significantly improved in ACKO mice (Fig. 5D). As such, muscle-specific Baf60s inactivation improves metabolic parameters in mice in diet-induced obesity and during aging.
Figure 5.
ACKO mice exhibit an improved metabolic profile during aging. A: Body weight, ad libitum blood glucose, and plasma insulin levels in young mice (n = 4–5) or old mice (n = 6–7) fed with standard chow diet. B: Plasma lipid levels in young and old mice fed a standard chow diet. C: GTT and ITT assays in old control and ACKO mice (n = 6–9). D: Liver TAG content in old mice following overnight starvation (n = 6–9). Data represent mean ± SEM. AUC, area under the curve. *P < 0.05, **P < 0.01, ***P < 0.001.
Hypermetabolism in ACKO Mice Is Linked to Induction of Genes Involved in Muscle Calcium Cycling
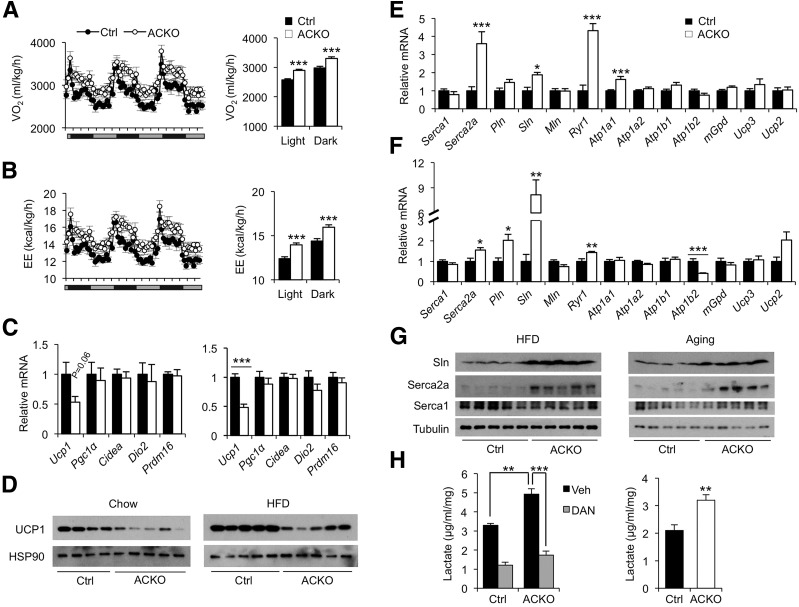
To explore how Baf60s in skeletal muscle regulate energy balance, we first performed metabolic cage studies on mice fed with chow diet and after 1 week of HFD feeding using CLAMS. Under chow feeding conditions, food intake and locomotor activity were similar between the two groups, but oxygen consumption rate and energy expenditure were moderately but significantly elevated in ACKO mice during the dark phase (Supplementary Fig. 4). Following 1 week of HFD feeding, food intake and respiratory exchange ratio appeared comparable in control and ACKO mice, but oxygen consumption rate and energy expenditure were significantly higher in ACKO mice (Fig. 6A and B and Supplementary Fig. 5A–C). Total locomotor activity was slightly reduced in ACKO mice during the dark phase. These results suggest that altered activity levels cannot fully account for the increased metabolic rate caused by muscle-specific inactivation of Baf60s.
Figure 6.
Conditional inactivation of Baf60a and Baf60c in skeletal muscle increases metabolic rate in ACKO mice. A and B: Oxygen consumption rate (VO2) (A) and energy expenditure (EE) (B) in control (Ctrl) and ACKO mice fed with HFD for 1 week (n = 8). C: qPCR analysis of BAT gene expression in mice fed with standard chow (left, n = 4–5) or HFD for 12 weeks (right, n = 5–11). D: Immunoblots of BAT total protein lysates from mice as described in C. E: qPCR analysis of quadriceps gene expression in HFD-fed mice (n = 6–7). F: qPCR analysis of quadriceps gene expression in chow-fed old mice (n = 6–7). G: Immunoblots of total muscle protein lysates from the HFD-fed (left) and aging (right) cohorts. H: Left, lactate production in ex vivo cultured EDL muscle treated with vehicle (Veh) or dantrolene (DAN) (100 μmol/L) for 3 h. Right, DAN-inhibitable lactate production rate calculated from data on the left. Data in A–C, E, F, and H represent mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001.
BAT is highly metabolically active and carries out thermogenesis via uncoupling protein 1 (UCP1) to defend against cold temperature. BAT thermogenesis also contributes to whole-body energy balance in rodents. Gene expression analysis indicated that Ucp1 mRNA levels were lower in BAT from ACKO mice than in control mice, whereas the expression of other genes involved in thermogenesis, including Pgc-1α, Prdm16, Cidea, and Dio2, remained similar between the two groups (Fig. 6C). Consistently, UCP1 protein levels were also lower in BAT from ACKO mice (Fig. 6D). Despite this, chow-fed ACKO mice did not exhibit increased sensitivity to cold temperature (Supplementary Fig. 5D). These data suggest that BAT thermogenesis is unlikely the cause of the hypermetabolic phenotype in ACKO mice.
Recent studies demonstrated that calcium cycling in skeletal myocytes contributes to thermogenesis and energy expenditure (46,47). As such, transgenic expression of Sarcolipin (Sln), a modulator of the SERCA pump, ameliorates HFD-induced weight gain in mice by enhancing muscle thermogenesis. We found that mRNA expression of several genes involved in calcium release and uptake was significantly elevated in HFD-fed ACKO mice, including Serca2a, Sln, and calcium release channel Ryanodine receptor 1 (Ryr1) (Fig. 6E). Similar induction of these genes was also observed in old ACKO mice fed standard chow (Fig. 6F). Interestingly, mRNA expression of phospholamban (Pln), but not myoregulin (Mln), both of which are homologous to Sln (48), was also elevated in muscle from old ACKO mice. Immunoblotting study confirmed that Serca2a and Sln protein levels were higher in quadriceps muscles from ACKO mice than in control mice (Fig. 6G). Because Sln has been implicated as a regulator of muscle thermogenesis, our results strongly suggest that Baf60 inactivation may increase energy expenditure through activation of myocyte calcium cycling and thermogenesis. In support of this, glucose utilization, as measured by lactate release, in ex vivo cultured EDL muscle from ACKO mice was significantly higher than in control mice (Fig. 6H). Treatment of muscle with dantrolene, an inhibitor of Ryr and calcium release from the sarcoplasmic reticulum, nearly abolished the effects of Baf60 deficiency on glucose utilization. Calcium cycling, as calculated by dantrolene-inhibitable lactate production, was significantly higher in ACKO muscle than controls (Fig. 6H).These results suggest that inactivation of BAF60 in skeletal muscle leads to elevated fuel use to support calcium release and reuptake by sarcoplasmic reticulum, perpetuating a futile cycle to alter energy balance.
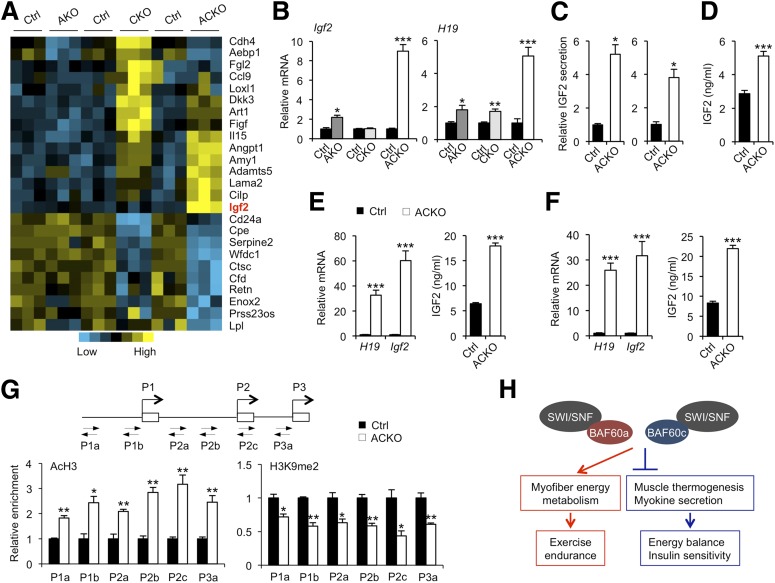
Baf60 Deficiency Activates the Expression and Secretion of IGF2 by Skeletal Muscle
Skeletal muscle is an important source of secreted factors that regulate systemic metabolism via endocrine, paracrine, and autocrine mechanisms (49–54). To determine whether Baf60a and Baf60c may regulate skeletal muscle secretome, we analyzed the microarray data set focusing on the expression of genes predicted to encode secreted proteins. Our analysis identified a cluster of genes encoding secreted proteins that exhibited differential regulation by Bad60s (Fig. 7A). Similar to the global transcriptional profile (Fig. 2), Baf60c inactivation strongly perturbed the expression of the secreted factor gene cluster in skeletal muscle, whereas Baf60a inactivation had a modest effect. Because inactivation of Baf60a or Baf60c alone does not alter systemic energy balance, we sought to identify myokines whose expression is altered in skeletal muscle from ACKO mice, but not AKO and CKO mice. This analysis identified Igf2 as a gene that was strongly induced by simultaneous inactivation of Baf60a and Baf60c in skeletal muscle. qPCR analysis indicated that Igf2 mRNA expression was markedly induced in quadriceps muscles from ACKO mice (Fig. 7B). Interestingly, the expression of H19, a neighboring noncoding transcript from the Igf2 locus, also exhibited a similar pattern of regulation in skeletal muscle (Fig. 7B). IGF2 is an imprinted gene that exerts pleiotropic effects on tissue growth and metabolism by signaling through the IGF1 and insulin receptors (55). Gene expression analysis indicated that mRNA levels of Igf1, insulin receptor (Ir), IGF1 receptor (Igf1r), and Igf2r remained similar between ACKO and control mice (Supplementary Fig. 6A).
Figure 7.
Baf60 deficiency enhances the expression and secretion of IGF2. A: Heat map representation of Baf60-regulated genes encoding secreted factors. B: qPCR analysis of gene expression in quadriceps muscle from AKO (n = 3), CKO (n = 6), ACKO (n = 4–5) mice and their littermate controls (Ctrl) on standard chow diet. C: IGF2 secretion in ex vivo cultured soleus (left) or plantaris (right) muscles. D: Plasma IGF2 concentration in chow-fed control and ACKO mice (n = 4–5). E: qPCR analysis of quadriceps gene expression in HFD-fed mice (left, n = 5–11) and plasma IGF2 concentrations in HFD-fed mice (right, n = 7–9). F: qPCR analysis of quadriceps gene expression (left) and plasma IGF2 levels (right) in old control and ACKO mice (n = 6–7). G: ChIP assays on control and ACKO muscle nuclei using antibodies against histone 3 acetylation (AcH3) and H3K9 demethylation (H3K9me2). Location of primers on the proximal regions of three IGF2 promoters is shown. H: Schematic model demonstrating the regulation of exercise preformation and systemic energy homeostasis by Baf60a and Baf60c. Data in B–G represent mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001.
Previous studies demonstrated that plasma IGF2 levels were inversely associated with future weight gain in humans (56,57). Additionally, transgenic expression of IGF2 protected mice from HFD-induced weight gain and metabolic disorders (58–60), suggesting that increased IGF2 levels in circulation may serve an adaptive role in maintaining energy homeostasis during chronic energy excess. ELISA measurements indicated that, compared with control mice, IGF2 protein expression was markedly induced in muscle lysates from ACKO mice (Supplementary Fig. 6B). Importantly, the secretion of IGF2 by soleus and plantaris muscles cultured ex vivo was approximately four- to fivefold higher in ACKO than control muscles (Fig. 7C). Accordingly, circulating IGF2 levels were significantly higher in ACKO mice than control mice (Fig. 7D). Expression of Igf2 in the liver, an important source of IGF2 in circulation, was similar between control and ACKO mice under both chow- and HFD-fed conditions (Supplementary Fig. 6C), ruling out the possibility that elevation of plasma IGF2 in ACKO mice is due to increased IGF2 production by the liver. We next examined IGF2 regulation in control and ACKO mice following HFD feeding. Compared with control mice, Igf2 mRNA and protein expression was increased by approximately 60- to 80-fold in muscle from ACKO mice following HFD feeding. Plasma IGF2 levels were also significantly elevated in HFD-fed ACKO mice (Fig. 7E). Similar increase of skeletal muscle Igf2 mRNA expression and circulating IGF2 levels was also observed in old ACKO mice (Fig. 7F). These results strongly suggest that activation of the IGF2 myokine pathway may contribute to improved systemic metabolic profile in ACKO mice.
The Baf60 subunits play an important role in mediating the recruitment of the SWI/SNF chromatin-remodeling complexes to the target genomic loci. To test whether inactivation of Baf60a and Baf60c alters the epigenetic status of chromatin, we performed ChIP assay on nuclei isolated from control and ACKO muscles using antibodies that recognize specific histone modifications. As shown in Fig. 7G, Baf60 inactivation caused a significant increase in histone H3 acetylation, a chromatin maker associated with transcriptional activation, and a decrease in demethylated H3K9, a repressive chromatin marker. This shift of histone marks from a repressive to an active state was observed for chromatin proximal to three Igf2 promoters known to be regulated by imprinting (61). It remains currently unknown whether Baf60 deficiency increased expression of Igf2 from both paternal and maternal alleles or selectively from one allele.
Discussion
Skeletal muscle fuel metabolism provides energy that supports its contractile function and serves an important role in systemic homeostasis. The regulatory network that governs the specification of oxidative and glycolytic metabolic functions of muscle fibers remains incompletely understood. The Baf60 subunits of the SWI/SNF chromatin-remodeling complex are emerging as important regulators of glucose and lipid metabolism. Baf60a promotes hepatic fatty acid β-oxidation through its interaction with PPARα and PGC-1α (37), whereas Baf60c stimulates glycolytic metabolism in skeletal muscle in part through Deptor-mediated AKT activation (16). Despite this, the relative contribution of Baf60a and Baf60c in myofiber energy metabolism has not been critically assessed. In this study, we demonstrated that Baf60a and Baf60c are indispensable for skeletal muscle fuel metabolism and exercise endurance. We uncovered an unexpected disconnect between myocyte energy metabolism and systemic energy balance. Despite impaired capacity for endurance exercise, mice lacking Baf60a and Baf60c in skeletal muscle were protected from diet-induced and age-associated metabolic disorders. At the mechanistic level, we identified muscle thermogenesis and myokine secretion as two novel target pathways of Baf60s that link myofiber fuel metabolism to systemic physiology (Fig. 7H).
The Baf60 family members are important for recruiting the SWI/SNF chromatin-remodeling complexes to transcription factors to regulate target gene expression. Both Baf60a and Baf60c are expressed in skeletal muscle with Baf60c exhibiting a more restricted pattern to glycolytic muscles. Transcription profiling studies revealed a prominent role of Baf60c, and to a lesser extent Baf60a, in the regulation of global gene expression in skeletal muscle. Despite this, neither factors appeared to be essential for maintaining glycolytic and oxidative functions and endurance exercise capacity. These observations illustrate remarkable functional redundancy between Baf60a and Baf60c in the regulation of myofiber energy metabolism. Unlike AKO and CKO mice, ACKO mice had significantly reduced exercise capacity that was associated with more pronounced AMPK activation in skeletal muscle following treadmill running. Impaired fuel metabolism in skeletal muscle has been linked to the pathogenesis of metabolic disorders. Surprisingly, ACKO mice were remarkably resistant to HFD-induced weight gain, insulin resistance, and hepatic steatosis and exhibited an improved metabolic profile during aging. Insulin sensitivity was also enhanced in peripheral tissues from ACKO mice following HFD feeding, including skeletal muscle, WAT, and liver. These observations support an emerging concept that exercise endurance can be uncoupled from systemic metabolic homeostasis. In fact, muscle-specific transgenic expression of Baf60c rendered mice exercise-intolerant while improving whole-body glucose metabolism (16). Similarly, inactivation of HDAC3 in skeletal muscle dissociated muscle insulin sensitivity from exercise endurance (62).
A surprising finding here is that ACKO mice had increased energy expenditure and were resistant to diet-induced obesity. We examined whether inactivation of Baf60s in myocytes may promote BAT thermogenesis indirectly through altering myokine secretion. The expression of Ucp1 was slightly downregulated in BAT from ACKO mice, whereas several other genes involved in thermogenesis appeared largely unchanged. In contrast, the expression of several genes involved in skeletal muscle calcium cycling and thermogenesis was significantly elevated in ACKO mice, including calcium release (Ryr1) and uptake (Serca2a) channels and Sln, a regulator of sarcolemma calcium uptake. Sln has been shown to be an important regulator of calcium cycling in skeletal muscle that influences whole-body energy balance (46,47). Our studies demonstrate that glucose utilization attributed to calcium cycling was higher in ex vivo cultured muscles from ACKO mice than control mice. However, whether the induction of muscle thermogenesis drives the metabolic phenotype in ACKO mice remains an important question for future studies. In addition to its role in fuel metabolism, skeletal muscle also releases diverse myokines that act locally and/or systematically to modulate metabolic physiology. We identified Igf2 as a myokine that was drastically induced in skeletal muscle from ACKO mice, but not AKO or CKO mice. Previous studies demonstrated that transgenic elevation of IGF2 levels in circulation improved metabolic parameters, including adiposity and blood glucose (58–60). These observations strongly suggest that elevated IGF2 levels in ACKO mice are likely a major contributor to the overall metabolic phenotype. Together, our work illustrates a surprisingly broad role of Baf60-mediated chromatin remodeling in the control of muscle bioenergetics, exercise endurance, and systemic metabolic homeostasis.
Supplementary Material
Article Information
Acknowledgments. The authors thank the staff at the University of Michigan Transgenic Animal Model Core for the generation of Baf60a and Baf60c floxed mice.
Funding. This work was supported by the National Institute of Diabetes and Digestive and Kidney Diseases (DK112800 and DK102456 to J.D.L.). This work used core services supported by the University of Michigan Diabetes Research Center and the Nutrition Obesity Research Center with grants from the National Institute of Diabetes and Digestive and Kidney Diseases (P30-DK020572 and P30-DK089503). Z.-X.M. was supported by the Scientist Development Grant from American Heart Association, the National Natural Science Foundation of China (No. 81670740), the Thousand Young Talents Plan of China, and the National Key Research and Development Programme of China (No. 2016YFC1305303).
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. Z.-X.M. and J.D.L. conceived the project and designed research. Z.-X.M., W.T., J.S., Q.W., and L.M. performed the studies. Z.-X.M. and J.D.L. analyzed the data and wrote the manuscript. Z.-X.M. and J.D.L. are the guarantors of this work and, as such, had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db17-0367/-/DC1.
References
- 1.Schiaffino S. Fibre types in skeletal muscle: a personal account. Acta Physiol (Oxf) 2010;199:451–463 [DOI] [PubMed] [Google Scholar]
- 2.Bassel-Duby R, Olson EN. Signaling pathways in skeletal muscle remodeling. Annu Rev Biochem 2006;75:19–37 [DOI] [PubMed] [Google Scholar]
- 3.Berchtold MW, Brinkmeier H, Müntener M. Calcium ion in skeletal muscle: its crucial role for muscle function, plasticity, and disease. Physiol Rev 2000;80:1215–1265 [DOI] [PubMed] [Google Scholar]
- 4.Kupr B, Schnyder S, Handschin C. Role of nuclear receptors in exercise-induced muscle adaptations. Cold Spring Harb Perspect Med 2017;7:a029835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Egan B, Zierath JR. Exercise metabolism and the molecular regulation of skeletal muscle adaptation. Cell Metab 2013;17:162–184 [DOI] [PubMed] [Google Scholar]
- 6.Rowe GC, Safdar A, Arany Z. Running forward: new frontiers in endurance exercise biology. Circulation 2014;129:798–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lillioja S, Young AA, Culter CL, et al. . Skeletal muscle capillary density and fiber type are possible determinants of in vivo insulin resistance in man. J Clin Invest 1987;80:415–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simoneau JA, Colberg SR, Thaete FL, Kelley DE. Skeletal muscle glycolytic and oxidative enzyme capacities are determinants of insulin sensitivity and muscle composition in obese women. FASEB J 1995;9:273–278 [PubMed] [Google Scholar]
- 9.Simoneau JA, Kelley DE. Altered glycolytic and oxidative capacities of skeletal muscle contribute to insulin resistance in NIDDM. J Appl Physiol (1985) 1997;83:166–171 [DOI] [PubMed] [Google Scholar]
- 10.Mootha VK, Lindgren CM, Eriksson KF, et al. . PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 2003;34:267–273 [DOI] [PubMed] [Google Scholar]
- 11.Patti ME, Butte AJ, Crunkhorn S, et al. . Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: Potential role of PGC1 and NRF1. Proc Natl Acad Sci U S A 2003;100:8466–8471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petersen KF, Befroy D, Dufour S, et al. . Mitochondrial dysfunction in the elderly: possible role in insulin resistance. Science 2003;300:1140–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muoio DM. Intramuscular triacylglycerol and insulin resistance: guilty as charged or wrongly accused? Biochim Biophys Acta 2010;1801:281–288 [DOI] [PMC free article] [PubMed]
- 14.Turner N, Heilbronn LK. Is mitochondrial dysfunction a cause of insulin resistance? Trends Endocrinol Metab 2008;19:324–330 [DOI] [PubMed] [Google Scholar]
- 15.Izumiya Y, Hopkins T, Morris C, et al. . Fast/glycolytic muscle fiber growth reduces fat mass and improves metabolic parameters in obese mice. Cell Metab 2008;7:159–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meng ZX, Li S, Wang L, et al. . Baf60c drives glycolytic metabolism in the muscle and improves systemic glucose homeostasis through Deptor-mediated Akt activation. Nat Med 2013;19:640–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meng ZX, Wang L, Xiao Y, Lin JD. The Baf60c/Deptor pathway links skeletal muscle inflammation to glucose homeostasis in obesity. Diabetes 2014;63:1533–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilkes JJ, Lloyd DJ, Gekakis N. Loss-of-function mutation in myostatin reduces tumor necrosis factor alpha production and protects liver against obesity-induced insulin resistance. Diabetes 2009;58:1133–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Narkar VA, Downes M, Yu RT, et al. . AMPK and PPARdelta agonists are exercise mimetics. Cell 2008;134:405–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arany Z, Lebrasseur N, Morris C, et al. . The transcriptional coactivator PGC-1beta drives the formation of oxidative type IIX fibers in skeletal muscle. Cell Metab 2007;5:35–46 [DOI] [PubMed] [Google Scholar]
- 21.Lin J, Wu H, Tarr PT, et al. . Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature 2002;418:797–801 [DOI] [PubMed] [Google Scholar]
- 22.Gan Z, Burkart-Hartman EM, Han DH, et al. . The nuclear receptor PPARβ/δ programs muscle glucose metabolism in cooperation with AMPK and MEF2. Genes Dev 2011;25:2619–2630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huss JM, Imahashi K, Dufour CR, et al. . The nuclear receptor ERRalpha is required for the bioenergetic and functional adaptation to cardiac pressure overload. Cell Metab 2007;6:25–37 [DOI] [PubMed] [Google Scholar]
- 24.Mootha VK, Handschin C, Arlow D, et al. . Erralpha and Gabpa/b specify PGC-1alpha-dependent oxidative phosphorylation gene expression that is altered in diabetic muscle. Proc Natl Acad Sci U S A 2004;101:6570–6575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zechner C, Lai L, Zechner JF, et al. . Total skeletal muscle PGC-1 deficiency uncouples mitochondrial derangements from fiber type determination and insulin sensitivity. Cell Metab 2010;12:633–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin J, Handschin C, Spiegelman BM. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab 2005;1:361–370 [DOI] [PubMed] [Google Scholar]
- 27.Cantó C, Gerhart-Hines Z, Feige JN, et al. . AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature 2009;458:1056–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lagouge M, Argmann C, Gerhart-Hines Z, et al. . Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell 2006;127:1109–1122 [DOI] [PubMed] [Google Scholar]
- 29.Nautiyal J, Christian M, Parker MG. Distinct functions for RIP140 in development, inflammation, and metabolism. Trends Endocrinol Metab 2013;24:451–459 [DOI] [PubMed] [Google Scholar]
- 30.Seth A, Steel JH, Nichol D, et al. . The transcriptional corepressor RIP140 regulates oxidative metabolism in skeletal muscle. Cell Metab 2007;6:236–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meng ZX, Gong J, Chen Z, et al. Glucose sensing by skeletal myocytes couples nutrient signaling to systemic homeostasis. Mol Cell 2017;66:332–344.e4 [DOI] [PMC free article] [PubMed]
- 32.Wang W, Xue Y, Zhou S, Kuo A, Cairns BR, Crabtree GR. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev 1996;10:2117–2130 [DOI] [PubMed] [Google Scholar]
- 33.Sudarsanam P, Winston F. The SWI/SNF family nucleosome-remodeling complexes and transcriptional control. Trends Genet 2000;16:345–351 [DOI] [PubMed] [Google Scholar]
- 34.Wu JI, Lessard J, Crabtree GR. Understanding the words of chromatin regulation. Cell 2009;136:200–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang RR, Pan R, Zhang W, Fu J, Lin JD, Meng ZX. The SWI/SNF chromatin-remodeling factors BAF60a, b, and c in nutrient signaling and metabolic control. Protein Cell. 7 July 2017. [Epub ahead of print]. https://doi.org/10.1007/s13238-017-0442-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tao W, Chen S, Shi G, Guo J, Xu Y, Liu C. SWItch/sucrose nonfermentable (SWI/SNF) complex subunit BAF60a integrates hepatic circadian clock and energy metabolism. Hepatology 2011;54:1410–1420 [DOI] [PubMed] [Google Scholar]
- 37.Li S, Liu C, Li N, et al. . Genome-wide coactivation analysis of PGC-1alpha identifies BAF60a as a regulator of hepatic lipid metabolism. Cell Metab 2008;8:105–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meng ZX, Wang L, Chang L, et al. . A diet-sensitive BAF60a-mediated pathway links hepatic bile acid metabolism to cholesterol absorption and atherosclerosis. Cell Reports 2015;13:1658–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Puri PL, Mercola M. BAF60 A, B, and Cs of muscle determination and renewal. Genes Dev 2012;26:2673–2683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Forcales SV, Albini S, Giordani L, et al. . Signal-dependent incorporation of MyoD-BAF60c into Brg1-based SWI/SNF chromatin-remodelling complex. EMBO J 2012;31:301–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lickert H, Takeuchi JK, Von Both I, et al. . Baf60c is essential for function of BAF chromatin remodelling complexes in heart development. Nature 2004;432:107–112 [DOI] [PubMed] [Google Scholar]
- 42.Li S, Mi L, Yu L, et al. . Zbtb7b engages the long noncoding RNA Blnc1 to drive brown and beige fat development and thermogenesis. Proc Natl Acad Sci U S A 2017;114:E7111–E7120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li S, Yu Q, Wang GX, Lin JD. The biological clock is regulated by adrenergic signaling in brown fat but is dispensable for cold-induced thermogenesis. PLoS One 2013;8:e70109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang GX, Cho KW, Uhm M, et al. . Otopetrin 1 protects mice from obesity-associated metabolic dysfunction through attenuating adipose tissue inflammation. Diabetes 2014;63:1340–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang GX, Zhao XY, Meng ZX, et al. . The brown fat-enriched secreted factor Nrg4 preserves metabolic homeostasis through attenuation of hepatic lipogenesis. Nat Med 2014;20:1436–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bal NC, Maurya SK, Sopariwala DH, et al. . Sarcolipin is a newly identified regulator of muscle-based thermogenesis in mammals [published correction appears in Nat Med 2012;18:1857]. Nat Med 2012;18:1575–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pant M, Bal NC, Periasamy M. Sarcolipin: a key thermogenic and metabolic regulator in skeletal muscle. Trends Endocrinol Metab 2016;27:881–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Anderson DM, Anderson KM, Chang CL, et al. . A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell 2015;160:595–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boström P, Wu J, Jedrychowski MP, et al. . A PGC1-α-dependent myokine that drives brown-fat-like development of white fat and thermogenesis. Nature 2012;481:463–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Izumiya Y, Bina HA, Ouchi N, Akasaki Y, Kharitonenkov A, Walsh K. FGF21 is an Akt-regulated myokine. FEBS Lett 2008;582:3805–3810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim KH, Jeong YT, Oh H, et al. . Autophagy deficiency leads to protection from obesity and insulin resistance by inducing Fgf21 as a mitokine. Nat Med 2013;19:83–92 [DOI] [PubMed] [Google Scholar]
- 52.McPherron AC, Lawler AM, Lee SJ. Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature 1997;387:83–90 [DOI] [PubMed] [Google Scholar]
- 53.Ostrowski K, Rohde T, Zacho M, Asp S, Pedersen BK. Evidence that interleukin-6 is produced in human skeletal muscle during prolonged running. J Physiol 1998;508:949–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pal M, Febbraio MA, Whitham M. From cytokine to myokine: the emerging role of interleukin-6 in metabolic regulation. Immunol Cell Biol 2014;92:331–339 [DOI] [PubMed] [Google Scholar]
- 55.Livingstone C, Borai A. Insulin-like growth factor-II: its role in metabolic and endocrine disease. Clin Endocrinol (Oxf) 2014;80:773–781 [DOI] [PubMed] [Google Scholar]
- 56.Heald AH, Karvestedt L, Anderson SG, et al. Low insulin-like growth factor-II levels predict weight gain in normal weight subjects with type 2 diabetes. Am J Med 2006;119:167.e9–15 [DOI] [PubMed]
- 57.Sandhu MS, Gibson JM, Heald AH, Dunger DB, Wareham NJ. Low circulating IGF-II concentrations predict weight gain and obesity in humans. Diabetes 2003;52:1403–1408 [DOI] [PubMed] [Google Scholar]
- 58.Da Costa TH, Williamson DH, Ward A, et al. . High plasma insulin-like growth factor-II and low lipid content in transgenic mice: measurements of lipid metabolism. J Endocrinol 1994;143:433–439 [DOI] [PubMed] [Google Scholar]
- 59.Rogler CE, Yang D, Rossetti L, et al. . Altered body composition and increased frequency of diverse malignancies in insulin-like growth factor-II transgenic mice. J Biol Chem 1994;269:13779–13784 [PubMed] [Google Scholar]
- 60.Rossetti L, Barzilai N, Chen W, Harris T, Yang D, Rogler CE. Hepatic overexpression of insulin-like growth factor-II in adulthood increases basal and insulin-stimulated glucose disposal in conscious mice. J Biol Chem 1996;271:203–208 [DOI] [PubMed] [Google Scholar]
- 61.Monk D, Sanches R, Arnaud P, et al. . Imprinting of IGF2 P0 transcript and novel alternatively spliced INS-IGF2 isoforms show differences between mouse and human. Hum Mol Genet 2006;15:1259–1269 [DOI] [PubMed] [Google Scholar]
- 62.Hong S, Zhou W, Fang B, et al. . Dissociation of muscle insulin sensitivity from exercise endurance in mice by HDAC3 depletion. Nat Med 2017;23:223–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.