Summary
Piezo2 is a mechanically activated ion-channel required for touch discrimination, vibration detection and proprioception. Here we discovered that Piezo2 is extensively spliced, producing different Piezo2 isoforms with distinct properties. Sensory neurons from both mice and humans express a large repertoire of Piezo2 variants, while non-neuronal tissues express predominantly a single isoform. Notably, even within sensory ganglia, we demonstrate the splicing of Piezo2 to be cell-type specific. Biophysical characterization revealed substantial differences in ion-permeability, sensitivity to calcium modulation, and inactivation kinetics among Piezo2 splice variants. Together our results describe, at the molecular level, a potential mechanism by which transduction is tuned permitting the detection of a variety of mechanosensory stimuli.
eTOC Blurb
Szczot et al. find that the mechanoreceptor Piezo2 is extensively alternatively spliced generating multiple distinct isoforms. Their findings indicate that these splice products have specific tissue and cell-type expression patterns and exhibit differences in receptor properties.
Introduction
Living organisms detect and respond to many types of mechanical force. For instance, our sensory systems allow us to identify objects by their tactile features (Gibson, 1962), coordinate controlled movement (Chesler et al., 2016), and enjoy the pleasure associated with interactive social touch (McGlone et al., 2014). Several gene families have been identified as sensors of mechanical stimuli; the Transient Receptor Potential (TRP), MSC channels, and DEG/ENaC channels in flies, bacteria and worms (Goodman et al., 2002; Sukharev et al., 1994; Walker et al., 2000), and the Piezo receptors in multiple phyla (Coste et al., 2010).
The Piezo genes, Piezo1 and Piezo2, encode exceptionally large mechanosensitive ion channels predicted to contain >14 transmembrane domains per monomer (Coste et al., 2015; Ge et al., 2015). Expression of Piezo proteins is sufficient to confer mechanically-evoked ionic currents to cells (Coste et al., 2010) and these molecules are believed to be intrinsically-gated by force (Syeda et al., 2016). Piezo2 is most highly expressed in sensory ganglia, although it has also been reported to be found in lung, bladder, skin, and bone (Coste et al., 2010; Ikeda et al., 2014; Woo et al., 2014). Mice and humans lacking functional Piezo2 exhibit severe deficits in detection of vibration, fine touch, hair movement, proprioception and breathing regulation (Chesler et al., 2016; Nonomura et al., 2017; Ranade et al., 2014).
These results point to Piezo2 as being crucial in the detection of a variety of mechanical stimuli known to be encoded by several classes of mechanosensory neurons. For example, gentle touch is mediated by Low Threshold Mechanoreceptors (LTMRs) with nerve endings in the skin while proprioceptors target muscles and tendons. LTMRs themselves are quite diverse, differing from one another based on the type of skin they innervate (e.g. glabrous and hairy skin), the size of receptive fields (wide and narrow field), their adaption properties (slow and rapid), and the morphology of their afferent end organs (Pacinian and Meissner’s corpuscles, Ruffini endings, and Merkel cell complexes, (Abraira and Ginty, 2013; Usoskin et al., 2015).
We were curious how a single molecule might function in such morphologically diverse settings to permit the detection of a wide range of mechanical stimuli. In particular, we focused on whether alternative splicing, a commonly used mechanism for genes to produce molecular and functional diversity (Lipscombe and Andrade, 2015; Pan et al., 2008; Wang et al., 2008), is used to regulate Piezo2 function. We find that Piezo2 undergoes a surprisingly extensive alternative splicing which is used to generate unique isoforms that are found in specific tissues and cell-types. We identify two previously unannotated exons and 16 isoforms of Piezo2 that are specifically enriched in mouse sensory neurons. Importantly, we demonstrate that Piezo2 variants exhibit major differences in three key biophysical properties: their rates of inactivation, ion permeability and modulation by intracellular calcium. Given that distinct classes of sensory neurons express select classes of Piezo2, we identify alternative splicing as an important determinant in mechanosensory specialization.
Results
Piezo2 is expressed by diverse types of mechanosensory cells
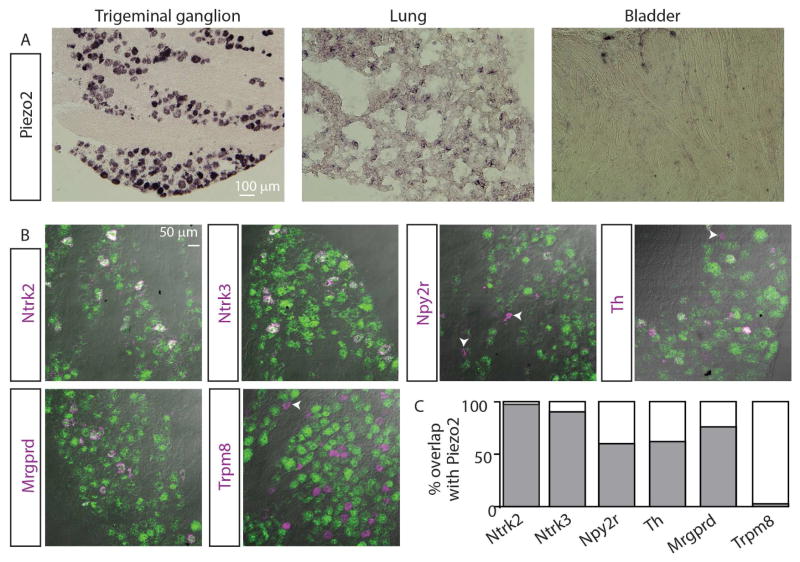
Piezo2 has been reported to be expressed in sensory ganglia and in non-neuronal tissues including lung and bladder (Coste et al., 2010). We therefore used in situ hybridization (ISH) to fully characterize the expression of Piezo2 in these tissues (Figure 1). As expected, Piezo2 was detectable in discrete cells in bladder and lung, whereas it is expressed at very high levels in a large proportion of neurons in the trigeminal ganglion (TG) (Figure 1A). Double-label ISH experiments showed that Piezo2 is co-expressed with genes found in LTMRs (Figure 1B, C). By contrast, Piezo2 is virtually absent from TRPM8-expressing neurons (2% TRPM8-neurons; Figure 1B–C) consistent with the proposed role of these neurons as dedicated cold-sensors (Knowlton et al., 2013; Pogorzala et al., 2013). Interestingly, despite the apparent lack of mechanical pain abnormalities in individuals with non-functional Piezo2 (Chesler et al., 2016) and similar absence of pain phenotypes in Piezo2 null mice (Ranade et al., 2014), the majority of non-peptidergic nociceptors marked by the expression of the Mas-related G-protein receptor D (Mrgprd) also express Piezo2 (76 % Mrgprd-neurons; Figure 1B–C). Piezo2 partially overlapped with TH, a proposed marker for C-LTMRs, supporting the idea that neurons expressing this gene are more heterogeneous than previously assumed (Nguyen et al., 2017). Furthermore, Piezo2 was also found in subsets of TRPV1 and TRPA1-positive neurons (32% and 60%, respectively, data not shown).
Figure 1. Piezo2 is highly expressed in heterogenous classes of peripheral sensory neurons.
A, Single label ISH shows that Piezo2 is expressed at high levels in TG neurons (left panel), it is also expressed at lower levels in scattered cells in the lung and bladder (middle and right panels). B, Double-label ISH reveals that Piezo2 (green) is found in neurons that are associated with sensation of low threshold mechanical stimuli, or are proprioceptive (Ntrk2, Ntrk3, Npy2r, and Th), and is present in HTMR-neurons (Mrgprd). However, Piezo2 is virtually absent from cold responsive neurons (Trpm8). Arrowheads indicate singly labeled neurons. C, Quantification of ISH data shows the extent of overlap in expression with Piezo2: 41/42 Ntrk2-neurons (280 Piezo2-cells), 114/126 Ntrk3-neurons (232), 27/45 Npy2r-neurons (242), 36/58 Th-neurons (464), 35/46 Mrgprd-neurons (464), and 1/38 Trpm8-neurons (232) 4–10 sections n=3 mice.
Differential splicing of the Piezo2 gene
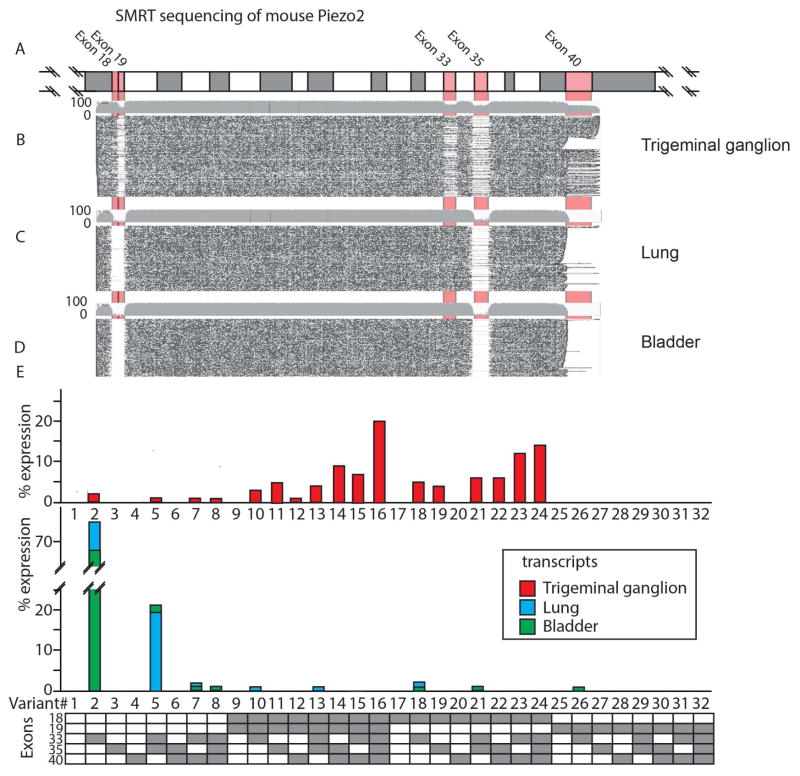
Piezo2 is found in both LTMRs that rapidly signal hair movement, proprioceptors that detect muscle contraction, and cells in lung and bladder that respond relatively slowly to radial stretch. Given the differences in these mechano-sensory tasks, we hypothesized that Piezo2 might, at the molecular level, be differential regulated by sensory and non-sensory tissues. To investigate the possibility of alternative splicing underlying this process, we sequenced Piezo2 mRNA from the lung and bladder and compared it with Piezo2 from the TG (Figure 2). The sequences of Piezo2 amplified from lung and bladder were the same as those reported previously (Coste et al., 2010). Remarkably, sequencing Piezo2 mRNA from TG revealed the existence of multiple novel variants that included two previously unannotated exons (Exons 18 and 35). In total, we found that five exons (that we named E18, E19, E33, E35, and E40) are alternatively spliced in mouse sensory ganglion neurons (Figure 3A). Importantly, usage of these exons does not generate translational frame shifts in the coding sequence of Piezo2 transcripts and, as expected for bone fide exons, the genomic sequences surrounding these exons contain consensus splice acceptor and donor sequences (Trapnell et al., 2009). In addition, ISH with probes designed to specifically detect these exons corroborated their expression in sensory neurons (Figure S1A).
Figure 2.
Multiple splice isoforms of mouse Piezo2 are alternatively expressed in neuronal and non-neuronal tissues. Alignment of sequencing reads of Piezo2 transcripts with the coding sequence of Piezo2 (A) reveals that 5 exons are alternatively spliced and there are profound differences in splicing between neuronal and non-neuronal tissues. Sequence reads from TG (B), lung (C), and bladder (D) are displayed as individual line plots, black indicates sequence identity and open areas regions of sequence lacking similarity (for clarity, junctions between exons are highlighted). Grey bars show the approximate percentage of sequence reads. Exons 17–41 of Piezo2 are colored in alternating blocks, and pink blocks for alternatively spliced exons. E, Analysis of reads from TG (Upper panel) showed that 16 variants are expressed, while lung and bladder (Middle panel) express one predominant form of Piezo2, V2. Lower panel is a schematic of the numbering system for splice variants.
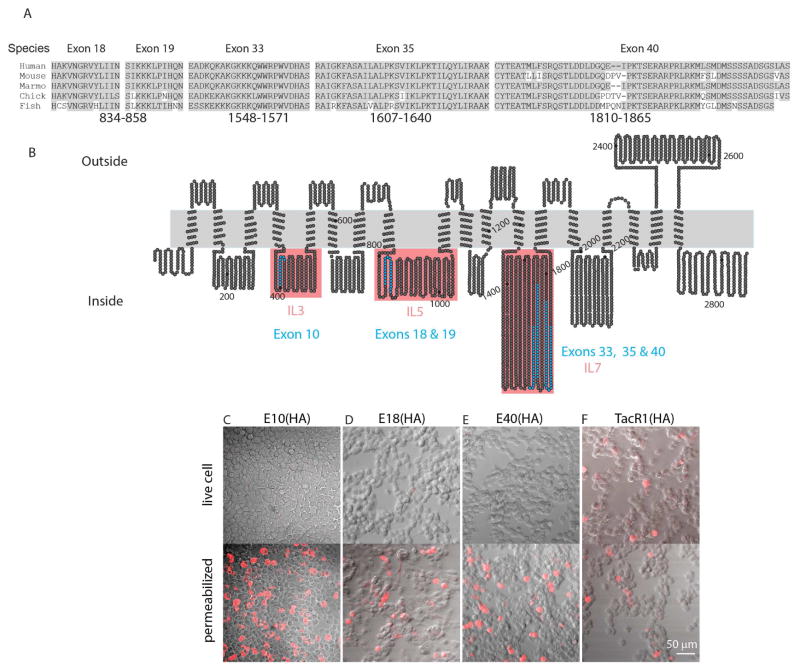
Figure 3. Sequences encoded by alternately spliced exons have intracellular locations.
A, Alignment of the coding sequence of alternatively spliced exons from multiple species show their high level of sequence similarity; amino-acids identical to those of human Piezo2 are shaded grey. Numbering below sequence refers to the position in V16 mouse Piezo2 sequence. B, Schematic representation of the proposed structure of Piezo2, based on sequence alignment of Piezo2 and Piezo1 and the predicted membrane topology of Piezo1 (Coste et al., 2015). The proposed positions of sequences encoded by alternatively spliced exons are indicated in blue as well as the approximate positions of amino-acid for V32 mouse Piezo2. B–E, The predicted intracellular location of alternatively spliced exons was confirmed by HA-epitope tagging experiments; C–E. Fields of HEK293-cells transfected with HA-epitope tagged Piezo2 constructs were immune-stained, live (upper panels) and following permeabilization (lower panels). HA epitope was engineered into E10 (C), E18–19 (D), and E40(E). HA epitope was detected (red) only after membrane disruption in cells expressing E10(HA), E18(HA), and E40(HA) confirming the sequences they encode have a intracellular location. F, As expected, cells expressing extracellular N-terminally tagged TacR1, produced detectable epitope staining in both permeabilized and non-permeabilized cells. Note, all epitope tagged Piezo2 constructs retained normal mechanically activated ion-channel activity.
To precisely quantify the extent of alternative splicing, we turned to a recently developed NextGen sequencing methodology (SMRT sequencing; (Eid et al., 2009)) which enabled long sequencing reads (3–3.5 KBp) of thousands of Piezo2 transcripts from multiple mouse tissues; TG (n=1721 full length reads of middle region), lung (n=4800) and bladder (n=2373). Figure 2 shows the sequence reads of Piezo2 from TG, lung, and bladder aligned against the coding sequence of Piezo2 (also see, Figure S1B and Table S1). Remarkably, this type of comparison revealed that lung and bladder principally express a single form of Piezo2, we name this V2 (approximately 80% of reads), which contains E33, but lacks the other 4 alternatively spliced exons (Figure 2C–E). By contrast, the same comparison showed that TG neurons express at least 17 different splice variants of Piezo2 (Figure 2B, E). Certain splice forms were absent from the TG, lung, and bladder demonstrating that splicing is selective (Figure 2E). Unbiased analysis of sequencing results for other regions of Piezo2 failed to reveal additional sites of alternative splicing except for a sixth alternatively spliced exon, E10, which is alternatively spliced in the lung and bladder (Figure S1B). E10 is expressed in all TG neurons and is apparently not alternatively spliced in this tissue.
We next sought to determine the positions of alternately spliced exons within the Piezo2 protein. Little is known about the structure of Piezo2 so we took advantage of the fact that its sequence can be broadly aligned to Piezo1. Based on known and predicted membrane topologies of Piezo1 (Coste et al., 2015), we expected exons E10, E18–19, E33, E35, and E40 to be found within three intracellular loops (Figure 3B). To confirm our hypothesis, we performed live cell staining experiments using constructs where epitopes were inserted at exons E10, E18, or E40, locations found not to interfere with functional responses. Our results show that all the alternatively spliced exons are located in intracellular domains (Figure 3C–E). Intriguingly, exons 33, 35 and 40 are found in an exceptionally large loop toward the C-terminus of the receptor close to where the pore domain is located (Coste et al., 2015; Ge et al., 2015).
Alternative splicing alters the function of Piezo2
The preferential expression of multiple splice variants of Piezo2 in sensory neurons versus the highly-restricted splicing found in non-neuronal tissue suggested that there might be intrinsic functional differences between isoforms. To test this hypothesis, we compared the properties of the major Piezo2 isoform expressed in non-neuronal cells, V2 (which contains E33, but lacks E18, E19, E35, and E40; see Figures 2D and 4A), with the properties of the neuronal isoform that is most different from it, V14 (which contains E18, E19, E35, and E40, but lacks E33). We examined the electrophysiological properties of V2 and V14 by heterologously expressing them in HEK-293 cells, assessing mechanically activated inward currents generated by membrane indentation. Expression of both V2 and V14 was sufficient to generate large responses which increased proportionally to stimulus strength and which were not seen in control GFP transfected cells (Chesler et al., 2016). This control, an exclusion cut-off of <50 pA (unless stated otherwise), and other controls (see Methods and Figure S2A–C) were employed to ensure HEK293 endogenous Piezo1 currents did not contaminate recordings (Dubin et al., 2017a, b). Having confirmed that the splice variants were functional, we focused our experiments on three important channel properties which would be predicted to strongly influence neuronal excitability: ion-permeability, modulation by Ca2+, and inactivation kinetics.
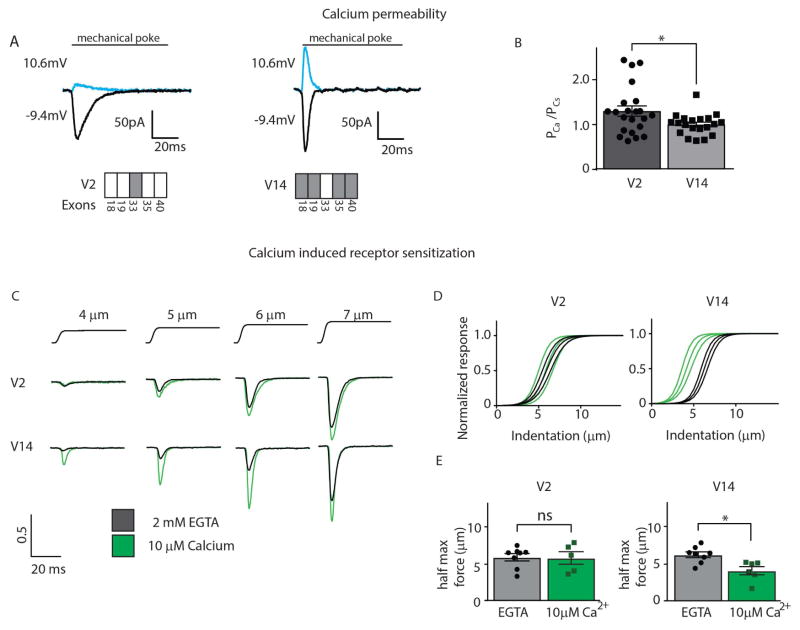
Figure 4.
Neuronal and non-neuronal splice forms of Piezo2 exhibit different calcium permeability and calcium-induced receptor sensitization. A, Mechanically-evoked responses of HEK293-cells expressing Piezo2 V2 and V14 show that the current of the non-neuronal splice form of Piezo2 differs from the neuronal variant at the indicated voltages, indicating a difference in relative calcium permeability. Cesium and calcium were the intracellular and extracellular cations, respectively. Lower panels schematize the exons usage of variant V2 and V14; these isoforms have opposite alternate splicing structures: grey boxes indicate presence of an exon and open boxes indicates the absence; all other regions in these constructs were identical. B, Quantification of the relative permeability of calcium versus cesium for V2 and V14. Data represent means ±SEM (n=22 for V2 n=20 for V14); significant difference in calcium permeability between variants * (Student’s t-test) p= 0.028. C, Sample traces demonstrating dose dependence of mechanically activated currents in cells expressing different variants in nominally calcium free intracellular solution (black) and high calcium intracellular solution (green). Traces are normalized to dose response fit maximums. D, The average fit of mechanically evoked currents for non-neuronal splice form, V2 (left panel) is not affected by intracellular calcium, while calcium decreases the indentation needed to gate V14 (right panel). Response profiles fits (middle trace represents the mean and outer traces SEM) were assessed by applying mechanical force (indentation of the plasma membrane) in the absence of intracellular calcium (black) and in the presence of 10 μM intracellular calcium (green). Note leftward shift in mechanical threshold of V14 in the presence of intracellular calcium (V2 calcium free n=8, V2 high calcium n=5, V14 calcium free n=8, V2 high calcium n=6) E, Quantification of the calcium induced shift in half maximal force showed that V2 is insensitive to intracellular calcium, while V14 is sensitized. Data are means ±SEM (V2 calcium free n=8, V2 high calcium n=5, V14 calcium free n=8, V2 high calcium n=6); significant difference in response sensitivity for V14 between no calcium and 10 μM calcium * (Student’s t-test) p= 0.0064, and for V2 it is not significant, p= 0.92.
Firstly, we examined ion permeability, looking for differences between V2 and V14. We measured permeability of Ca2+ by these Piezo2 isoforms relative to Cs+ (Coste et al., 2010). The relative permeability for Ca2+ of V2 was significantly higher than that of V14 (Figure 4A–B and Table S2). This difference in permeability was selective for Ca2+ since we did not find differences in Na+ or Mg2+ permeability (Figure S2D–E and Table S2). Splice V16 (Figure S2F–G) showed a calcium permeability similar to that of V2 consistent with the presence of exon 33 positively affecting calcium permeability despite being located in an intracellular loop. More importantly, these results provide evidence that alternate splicing of Piezo2 indeed generates receptors expressed by neurons and non-neuronal tissues with distinct pore properties.
A recent study reported that Piezo2 function is positively regulated by intracellular Ca2+ and that this may be important for alterations in touch sensitivity under pathological conditions (Eijkelkamp et al., 2013). Given the intracellular location of the alternatively spliced exons encoded parts of the loops (Figure 3), we hypothesized that, in addition to differences in ion permeability, sensory neuron specific variants might be more robustly modulated by intracellular calcium. Therefore, we compared the mechanically evoked currents from V2 and V14 either in the presence or in the absence of high intracellular Ca2+. When nominally divalent free intracellular solutions were used, V2 and V14 displayed similar responses to mechanical force. Strikingly, when intracellular solutions contained 10 μM Ca2+, we found a major decrease in the mechanical threshold of V14, while the threshold of V2 was unaffected (Figure 4C–E).
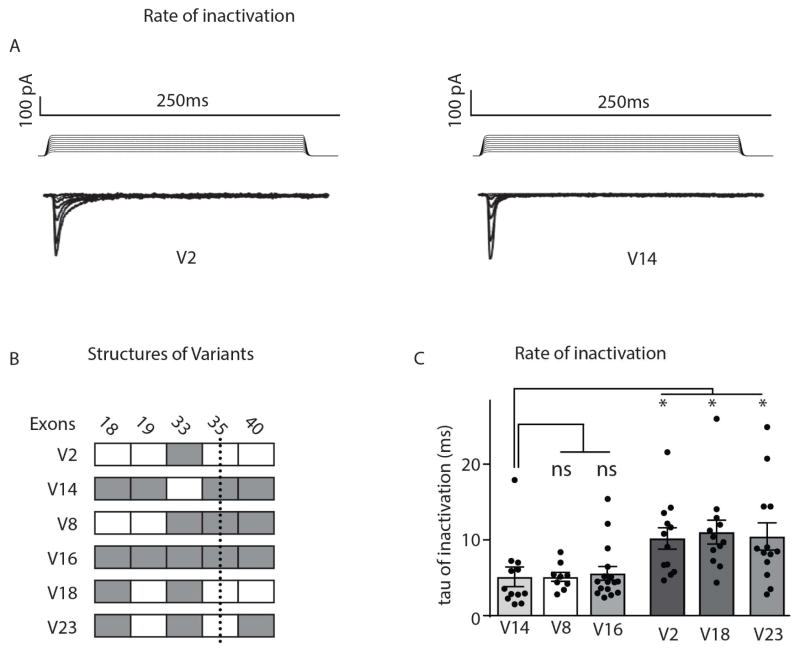
Piezo2 inactivates rapidly, within ~10 ms (Coste et al., 2010), to static indentation, presumably reflecting rapid entry of the channel into a non-conducting desensitized state. This allows Piezo2 to perform basic frequency filtering and is believed to be a key factor in the ability of sensory neurons to respond to rapid and/or vibrational stimuli (Lewis et al., 2017). Given that Piezo2 is required for the sensation of vibration and touch discrimination (Chesler et al., 2016; Ranade et al., 2014), we explored the possibility that splicing might fine tune inactivation. To quantify rates of inactivation of V2 and V14, we stimulated cells with increasing strengths of mechanical force, measured current decay, and calculated the tau of inactivation. Notably, we found that V2 inactivated significantly slower than V14 (Figure 5A–C). Comparing rates of inactivation of four additional Piezo2 isoforms (expressed by sensory neurons) revealed that the presence of E35 correlated with the increased rate of inactivation (Figure 5C). In summary, our results establish that alternate splicing selectively tunes the permeability, calcium mediated sensitivity, and rate of inactivation of Piezo2.
Figure 5.
Neuronal and non-neuronal splice forms of Piezo2 exhibit different rates of inactivation to mechanical stimulation. The neuronal variant V14 inactivates to mechanical stimulation significantly faster than the non-neuronal, V2 isoform of Piezo2. A, Mechanically evoked membrane currents for HEK293-cells transfected with the non-neuronal, V2 (left) and the neuronal, V14 (right) variants. Increasing mechanical stimulation (upper panel) elicits greater current (lower panel). B, Schematized exons usage of variants (see Figure 4 for details). C, Quantification of inactivation kinetics for different Piezo2 variants shows that V14 has significantly faster rates of inactivation than V2, V18, and V23. Data are means ±SEM V2 (n=12), V8 (n=16), V14 (n=12), V16 (n=9), V18 (n=12), and V23 (n=13) significant difference in rates of inactivation between variants * (one-way ANOVA with Dunnett’s multiple comparison test) V14 vs V2 p=0.009, V14 vs V8 p>0.99, V14 vs V16 p>0.99, V14 vs V18 p=0.0029, and V14 vs V23 p=0.0204.
Cell-type specific splicing of Piezo2
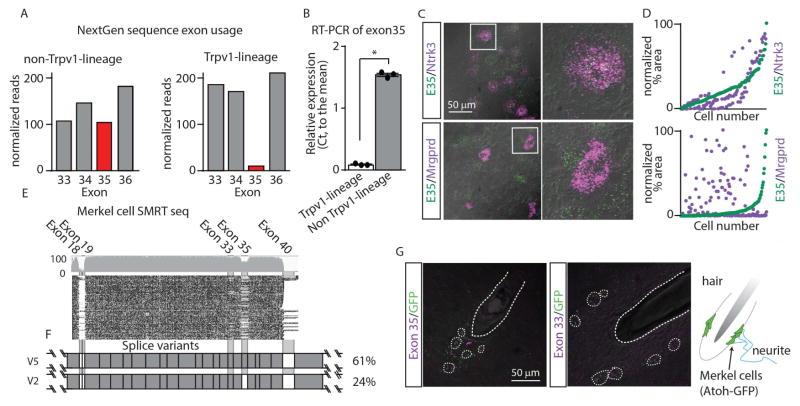
Neuronal sensory tissues express at least 17 isoforms of Piezo2 (Figure 2E and Table S1). We hypothesized that the functionally distinct Piezo2 isoforms might be differentially enriched in certain sub-types of sensory neuron. To test this hypothesis, we compared transcriptomic data from sensory neurons which developmentally express the Trpv1 ion-channel (Trpv1lineage neurons) with neurons that do not (non-Trpv1lineage neurons) (Hjerling-Leffler et al., 2007; Mishra et al., 2011). This strategy allowed us to compare the coding sequences of Piezo2 expressed in neurons that are broadly required for nociception, pruriception, and thermoreception (Le Pichon and Chesler, 2014) with those found in neurons involved in discriminative touch and proprioception. Interestingly, the relative abundance of alternative exons significantly differed between these two neuronal-lineages. More notably, we found that E35 was expressed at much higher levels in non-Trpv1lineage neurons than in Trpv1lineage neurons (Figure 6A). RT-PCR confirmed this difference (Figure 6B) and ISH validated that E35 is preferentially expressed in proprioceptive (Ntrk3+) and discriminative touch (Ntrk2+) neurons (Abraira and Ginty, 2013; Woo et al., 2015), while it was undetectable in mechanical pain Mrgprd-neurons (Figure 6C, (Cavanaugh et al., 2009)). These results indicate that proprioceptors and large diameter LTMRs selectively splice Piezo2 transcripts to include E35, the alternatively spliced exon that is sufficient to increases the rate of inactivation of the channel (Figure 5C).
Figure 6. Subsets of sensory neurons selectively splice exon 35 and Merkel cells express restricted classes of Piezo2 splice variants.
E35 is selectively spliced in Piezo2 transcripts expressed by LTMRs and proprioceptive neurons. A, Analysis of NextGen sequence reads reveals that E35 is expressed at high levels in transcripts from non-Trpv1lineage neurons and at a lower level in transcripts from Trpv1lineage neurons. Exon reads were normalized to individual length of exons and to the overall Piezo2 reads in non-Trpv1lineage neuron and Trpv1lineage neuron datasets. B, RT-PCR of cDNA from non-Trpv1lineage and Trpv1lineage neurons confirmed that there is significantly greater expression of E35 in non-Trpv1lineage neurons than Trpv1lineage neurons. Significant difference (Student’s t-test) in expression between cell-types *, were p<0.05 (means ±SEM, n=3 samples). C, Double-label ISH reveals that E35 (green) is expressed in many Ntrk3-positive neurons (purple; upper panel), but there is no co-expression of E35 with Mrgprd (purple; lower panel). D, Comparison of the density of staining of positive profiles of Ntrk3 cells (purple) relative to those for E35 (green) shows there is high correspondence in the expression profiles of these two molecules. In contrast, there is very poor correspondence in the profiles stained for Mrgprd (purple) and E35 (green). Sequencing of Piezo2 from purified Merkel cell preparations reveals that they predominantly express V5. E, plot of reads from Piezo2 transcripts aligned to the coding sequence of Piezo2 (exons 17–41). F, structures of V5 and V2 splice variants of Piezo2. Note that E10 is also variably spliced in Piezo2 mRNA from Merkel cells. G, Schematic of the location of Merkel cells illustrating their close association to hair-follicles; Merkel cells are indicated in green and surround the root of a hair-follicle. Double label ISH of tissue from Atoh1-EGFP mice show that E35 (left) and E33 (right), are expressed with GFP (green). Individual Merkel cells and the position of a hair follicle are outlined.
Given the differences in Piezo2 splicing we observed in peripheral sensory neurons, we wondered whether Merkel cells (a type of modified non-neuronal skin cell that is involved in fine touch), might express specific isoform(s) of Piezo2 (Ikeda et al., 2014; Woo et al., 2014). To investigate this, we purified Atoh1-GFP positive Merkel cells and used SMRT sequencing to determine the sequence(s) of Piezo2 expressed by these cells (n=2615). Interestingly, purified Merkel cells express a combination of Piezo2 transcripts distinct from those found in the lung, bladder or sensory ganglia (Figure 6E–F). Merkel cells predominantly express variant 5 of Piezo2 (61%) which is only found at low levels in sensory neurons. Consistent with our sequencing results, ISH demonstrates that Merkel cells express E33 and E35 (Figure 6G). Together these results support our proposal that discrete cell-types could attain distinct sensory characteristics by restricted expression of specific isoforms of Piezo2. Intriguingly, Merkel cells are enriched for the same exon (E35) as the sensory neurons involved in touch discrimination and proprioception.
Alternative splicing of Piezo2 in human
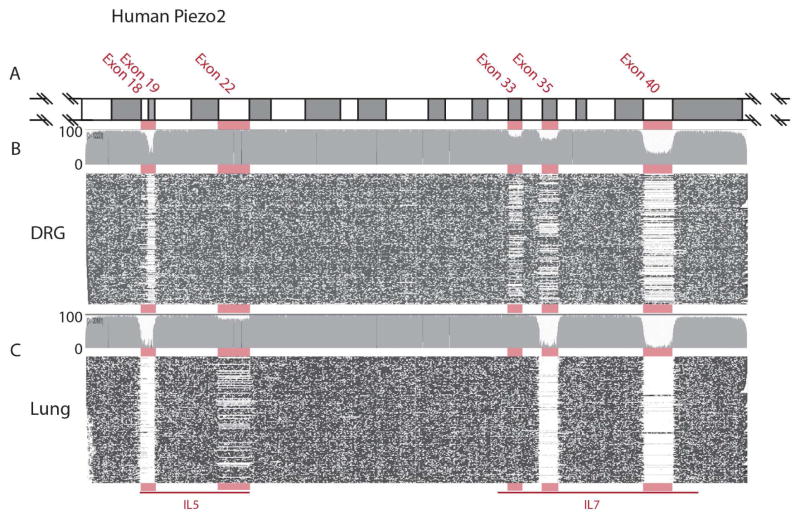
We hypothesized that splicing of Piezo2 is a general strategy employed to generate receptor diversity and therefore, we would predict that, if this process is biologically relevant, it should be evolutionarily conserved. Supporting this postulate, the alternatively spliced exons E18, E19, E33, E35, and E40, are found in Piezo2 genes from fish to human and the sequences encoded by these exons are highly conserved (Figure 3A). To investigate our hypothesis further, we SMRT sequenced and examined the sequences of Piezo2 transcripts expressed in human dorsal root ganglion (DRG) (n=2750) and lung (n=4729). As predicted, human sensory neurons express a mixture of spliced Piezo2 similar to those seen in mouse TG (Figure 7A–C, Table S3, and Figure S3B–C). Also, like in mice, the lung expresses a relatively small number of splice isoforms of Piezo2 and the predominant isoform is orthologous to mouse V2. Similar to mouse sensory neurons, there are multiple forms of Piezo2 expressed by human DRG (16 variants; Figure S3B–C). Comparison of data from sequencing lung and DRG revealed that human Piezo2 transcripts contain one additional alternatively spliced exon, E22, which is absent in many lung transcripts (Figure 7, Table S3, and Figure S3B). In the mouse tissues we examined, Piezo2 transcripts all contained exon 22. We also noted a difference in splicing of E10 in human and mouse; E10 is absent in approximately 20% of human DRG Piezo2 transcripts whereas it is present in all mouse TG transcripts (Figure S1B and S3B). Altogether, our results establish that the patterns of splicing of Piezo2 in human and mice are very similar confirming that alternative splicing is a conserved process.
Figure 7. Human Piezo2 is alternatively spliced.
Sequencing human Piezo2 transcripts reveals that six exons are alternatively spliced and that there are major differences in the splicing of Piezo2 in DRG and lung. A, E16–41 are colored in alternating grey and open blocks with the junctions of alternatively spliced exons highlighted in pink. Sequence reads from DRG (B), and from lung (C) are displayed as plots, black dots indicate sequence identity and open areas sequence lacking similarity. In DRG sensory neurons E19, E33, E35, and E40 are variably spliced while E18 is present in most transcripts. In contrast, in lung, transcripts E18, E19, E35, and E40 are present rarely. See Figure S3 and Table S3 for details.
Discussion
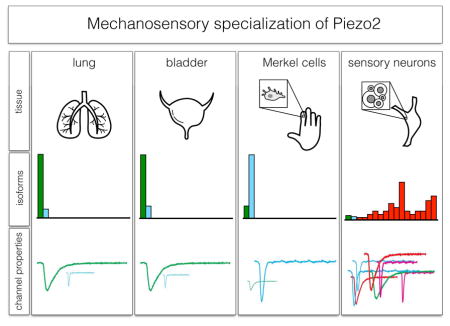
In this study, we found that splicing of the Piezo2 gene generates functionally diverse mechanosensitive ion channels that are expressed in a cell-type specific manner. We provide three lines of evidence supporting our proposal that splicing of Piezo2 enables cells with distinct functions to detect or respond to mechanical stimuli in different ways. First, we establish that alternative splicing is specifically regulated in both different tissues and neuron-types (Figures 2, 6, and 7). Second, we demonstrate that Piezo2 splice variants differ significantly in their functional response characteristics (Figures 4 and 5). Third, our results show that the differential splicing of Piezo2 is a conserved process (Figure 7).
The Piezo2 gene is extremely large and, based on our results, contains 56 exons. Six of these exons are alternatively spliced in mice and seven are alternatively spliced in humans. The existence of multiple isoforms of genes is a powerful way to generate functional complexity from a single gene. Intriguingly, we find that mice express 18 variants out of the possible 32 combinations (Figure 2) at a level of >1% of expressed sequences. The restricted number of isoforms expressed appears to be due to constraints on splicing specific sets of exons; when present, E19 is almost always found together with E18, and E40 is almost always paired with E33, E35, or both (Figure 2 and 8). Such cross-wise splicing specificity supports the concept that certain exons encode domains that influence distinct properties.
Our mapping of the locations of encoded spliced exons places them all within intracellular loops (Figure 3 and Figure S3D). We found that manipulating the loop containing E33, E35, and E40 confers functional differences (Figure 4 and 5). While the resolution of the Piezo1 cryo-electron microscopy structure (Ge et al., 2015) is insufficient to definitively position this loop within the overall channel architecture of the molecule, given its location near the C-terminus, it is possible that this loop interacts with the pore region of Piezo channels (Coste et al., 2015). If this is correct, it would fit with the role of splicing in modulating calcium permeability (Figures 4 and Figure S2). Calcium is a potent modulator of many intracellular pathways and its concentration is particularly dynamically regulated in neurons (Higley and Sabatini, 2012). The expression of certain splice variants in specific neurons would make specific afferent terminals more sensitive to gentle touch than others by changing the permeability and shifting the activation curve for Piezo2. In addition, conditions that promote mechanical allodynia and/or hyperalgesia (Andrew and Greenspan, 1999), might lead to changes in the expression of different splice variants of Piezo2 resulting in altered mechanical sensitivity.
We also found that Piezo2 variants containing E35 display two-fold faster rates of receptor inactivation than those variants without this exon (Figure 5). Interestingly, E35 is enriched in Ntrk2 and Ntrk3 large diameter neurons and Merkel cells (Figure 6 and 7), cells required for vibration detection and touch discrimination. Recently, it was shown that Piezo protein can act as a band pass filter, a process which is highly dependent on the rate of channel inactivation (Lewis et al., 2017). In addition, inherited mutations of Piezo2 linked to distal arthrogryposis type 5 (Coste et al., 2013; McMillin et al., 2014), accelerate, or reduce recovery from inactivation of Piezo2. Interestingly, our findings, that alternative splicing in and of itself produces similar magnitude changes in inactivation, suggest that alternative splicing should have significant influences on the properties of neurons. Our in vitro recordings demonstrate that distinct isoforms of Piezo2 display different functional properties. However, our analysis has not been exhaustive as we did not examine all splice variants of Piezo2 and we did not investigate the potential interaction of Piezo2 with other membrane proteins. Other intracellular signaling pathways (Borbiro et al., 2015; Dubin et al., 2012) and protein interactions (Narayanan et al., 2016; Poole et al., 2014) have been shown to modulate Piezo2 function. More importantly, to fully appreciate the contribution of receptor diversification, it will be important to determine the in vivo consequences of Piezo2 splicing.
Sensory afferents have highly specialized endings that are likely important for mechanosensation (Bai et al., 2015; Rutlin et al., 2014). In mammals, other forms of mechanosensory specialization have been observed. For instance, sensory ganglia with different representations of mechanosensory neurons (Schneider et al., 2014) and signaling components (Gerhold et al., 2013) have been reported. This suggests that several strategies have been used to allow sensory specialization. Our findings that functionally different cell-types express distinct Piezo2 isoforms suggest another way in which different mechanical stimuli can be distinguished. Indeed, the existence of functionally distinct isoforms of Piezo2 is highly reminiscent of the finding that alternate splicing of the Trpv1 ion-channel is associated with thermosensory specialization (Gracheva et al., 2011). Therefore, the splicing of Piezo2 may be another means by which the somatosensory system generates sensory diversification.
Experimental procedures
Animals
All experiments using mice followed NIH guidelines and were approved by the National Institute of Dental and Craniofacial ACUC. Atoh1-EGFP (Jax, cat # 013593) and C57BL/6N (Charles River) mice of both sexes were used. Merkel cells were purified from 5 day old pups as previously described (Piskorowski et al., 2008). Tissue for other experiments was harvested from adult C57BL/6N mice.
In Situ Hybridization
RNAscope, a multiplexed fluorescent in situ hybridization technique (Advanced Cell Diagnostics #320850), was performed according to the manufacturer’s instructions, on fresh frozen tissue sections. Images were acquired using an Olympus confocal microscope at 20X magnification and adjusted for overall brightness and contrast using FIJI (Image J) software. To identify and count positive cells, ROIs were manually selected from 4 sections. Binary thresholds were use used to determine signal over background for each probe. Values for each channel above threshold were compared and scored for overlap.
Single label ISH was performed at high stringency as described previously (Mishra et al., 2011).
Piezo2 sequencing and bioanalysis
RNeasy kit (Qiagen, USA) was used to isolate RNA from mouse tissues and SMARTScribe (Clontech, USA) was used to generate first stand cDNA. Human cDNAs were purchased (Promega Corp, USA). The full-length coding sequence of Piezo2 is 8.2–8.6 kb, we were unable to amplify the complete full-length sequence of Piezo2 and therefore we resorted to amplifying three long overlapping fragments (Figure S1) using LongAmp DNA polymerase (NEB, USA). PCR primers were designed to include unique barcodes to identify fragments amplified from different tissues (Table S4). After amplification (35 cycles), approximately equal amounts of individually bar-coded DNA products (1 μg each) were pooled and subjected to SMRT sequencing (PacBio sequencing; Eurofins Corp, USA). In addition, we sequenced cloned DNA fragments from mouse TG (~ 40 5′ fragments, ~100 middle fragments, and ~40 3′ fragments), mouse lung (~20 5′ fragments, ~20 middle fragments, and ~20 3′ fragments), and mouse bladder (~20 5′ fragments, ~20 middle fragments, and ~20 3′ fragments). DNA sequences were analyzed with Blast (NCBI) to determine splicing. SMRT sequence datasets were analyzed using PacBio SMRT. Analysis software to separate sequences by barcode, using the “RS_Resequencing_Barcode.1” protocol in the SMRT Analysis software package. Sequences with incomplete coverage of the amplified fragment, were discarded. Resulting sequences (Table S3–5 for numbers) were analyzed for patterns of exon usage; a strategy implemented to allow an unbiased analysis of exon usage for each transcript. In many cases aligned reads were longer than predicted exons, and we employed the BEDTools 2.22.0 (Quinlan and Hall, 2010) bamtobed splitD option to split alignments on the D CIGAR operator. Split reads shorter than twelve bases were filtered using awk (Aho et al., 1987); however, many of the split reads were still longer than the exons. Thus, the BEDTools intersect command was used to find the intersections between exons and split reads. A fraction of 0.1 overlap was the minimum required to define the intersection.
Analysis of NextGen Transcriptomic Trpv1-lineage and Non-Trpv1-lineage sequences
The generation of NextGen transcriptomic data for Trpv1lineage and non-Trpv1lineage neurons has previously been described; 36.7 million reads 35 base end-reads (Goswami et al., 2014). CLC genomics workbench (CLCbio) was used to align reads against Piezo2, and numbers of reads manually counted for each exon. Read numbers were normalized to the total Piezo2 reads in each sample, and secondly, against exon size.
RT PCR
RNeasy kit (Qiagen) was used to isolate RNA and SMARTScribe (Clontech, USA) was used to generate first stand cDNA. Real time Taqman PCR was performed on an ABI 7900HT with the following primers for mouse Piezo2 exon 35, CTCAGCCATTTTAGCCTTGC, forward, CAGATCCTTTGCGTCTCTCC, reverse, and 5′ FAM-TGATCCTAAAACGGCACTCC-3′ TAMRA. Results were normalized to GAPDH. Experiments were performed with triplicate experimental samples and controls, and fold increases were calculated using the comparative threshold cycle method.
Epitope tagging
HEK293 cells were transfected with construct encoding Piezo2 HA-tag (see HEK293 cell culture) and, plated on poly-L-lysine coated coverslips. For non-permeabilized staining, cells were incubated for 5–15 minutes with anti-HA preconjugated Dylight antibody (ThermoFisher, Halethorpe, USA), washed 4 times in PBS. For permeabilized staining, cells were fix with 4% paraformaldehyde for 10 minutes, washed 3 times in PBS + 0.1% triton X-100 and incubated for 1 hour with anti-HA antibody, washed in PBS.
HEK293 cell culture
HEK293-cells were cultured in DMEM+glutamine media (Lonza, Walkersville, MD, USA) supplemented with: 10% FBS (Gibco, Gaithersburg, MD, USA), 100 U/ml penicillin and 100 U/ml streptomycin (Gibco) in an incubator at 37°C in 5% CO2 atmosphere. cDNA encoding Piezo2 variants was sub-cloned in pCDNA3.1 vector (Epoch Life Sciences, USA). 24 hours prior to transfection HEK cells were plated on 24-well plates and transfected at 60–90% confluency with 400 ng of plasmid and 1 ng of EGFP-encoding plasmid to aid identification of transfected cells. A standard lipofectamine 2000 (ThermoFisher, Waltham, MA, USA) transfection protocol was used. The day following the transfection cells were re-plated on 12 mm glass coverslips. Experiments were performed 48–72 hours after transfection.
Whole cell recordings
Whole cell voltage clamp current responses were recorded with Multiclamp 700B amplifier (Molecular Devices, Sunnyvale, CA, USA) at a holding potential of −40 mV unless stated otherwise. Signals were digitized with Digidata 1550 (Molecular Devices) digitizer at 100kHz, low pass filtered at 10kHz and saved on a PC computer running Clampex 10.3 (Molecular Devices). Extracellular solutions consisted of (in mM): 133 NaCl, 3 KCl, 1 MgCl2, 10 HEPES, 2.5 CaCl2, 10 Glucose, 18.9 Sucrose, pH 7.3 with NaOH. Patch pipettes (resistance of 2–4 MΩ) were filled with intracellular recording solution consisting of (in mM) 133 CsCl, 10 HEPES, 5 EGTA, 1 CaCl2, 1 MgCl2, 4Mg-ATP, 0.4 Na-GTP, 10 Cs-gluconate, pH 7.3 with CsOH. For ion permeability experiments pipettes were filled with (in mM): 150 CsCl, 10 HEPES (pH 7.3 with CsOH) and extracellular solutions were composed of (in mM): 150 NaCl for sodium permeability experiments and 100 CaCl2 or MgCl2 for divalent cations permeability experiments, 10 HEPES (pH 7.3 with CsOH). For measurements of intracellular calcium impact on recombinant Piezo2 responses intrapipette solution consisted of (in mM) 133 CsCl, 10 HEPES, 1 MgCl2, 4 Mg-ATP, 0.4 Na-GTP, 10 Cs-gluconate, pH 7.3 with CsOH and was supplemented with (in mM): 2 EGTA (for Ca2+ free conditions), or, 0.01 CaCl2. All chemicals were from Sigma-Aldrich unless otherwise stated. Cells exhibiting a stationary leak current above 200 pA were excluded from analysis. For experiments with varying concentrations of intracellular ions, stimulation began 2–5 minutes after breaking into whole-cell configuration to allow for intra-pipette solution perfusion of the cell. Previously, ((Chesler et al., 2016) Figure 1C) we showed that the same HEK 293-cells used in this study, when transfected with EGFP, have endogenous currents well below 50pA.
Mechanical stimulation
Mechanical stimulation of the cells was performed by a heat-polished blunt pipette (tip size ~3–5μm), driven by a micromanipulator-mounted P841.20 piezoelectric translator (Physik Instrumente, Karlsruhe, Germany). The stimulating pipette was mounted at an angle of ~60°. For stimulation, the pipette was placed as close as possible to the cell without causing visible indentation and a series of increasing indentation stimuli was applied (typically 1–10 μm, with 1 μm increments) for 200 ms with a 2 ms ramp. Stimuli were separated by 10 s intervals, to avoid accumulation of channels in a desensitized state. Only cells with a good surface attachment (e.g. visible pseudopodia, spindle-shape morphology) to the coverslip surface throughout the stimulation period were included in the analysis.
Electrophysiology data analysis
Data were analyzed using Clampfit 10.3 (Molecular Devices). Data were digitally filtered at 3 kHz. Responses were fitted with a mono-exponential function: I(t) = Amp · exp (−t/τ) and inactivation was measured as an average of at the most first three mechanically evoked responses greater than 50 pA excluding responses above 2000 pA. Traces with severe mechanical distortions (i.e. large oscillations) or visible loss of cell attachment to the surface were excluded from analysis.
For permeability measurements, the response to a test pulse of a set amplitude was recorded at a range of nominal holding voltages (typically from −60 to +60 mV with 20 mV steps) and the last and first positive responses were used to calculate reversal potential after correcting for liquid junction potentials. Cation permeability relative to Cs+ was calculated from the GHK equation, for monovalent Na+ cations: , and divalent cations . To calculate the dose-response relationship and its dependence on the intracellular milieu, responses to subsequent indentations were fitted with the Boltzmann equation , yielding half-maximal activation indentation (p50) and sensitivity (s). To ensure that the fits of responses correctly reflect the underlying dose dependence of Piezo2 channels, stringent inclusion criteria were applied. Only cells with at least 4 responses at different indentations were analyzed. Cells in which the Boltzmann fit indicated that the maximal elicited current was <75% of the theoretical maximum were excluded as well as cells producing curves with a theoretical maximal current below 200 pA or above 2000 pA. All datasets were tested for normality. Statistical significance was tested with parametric Student’s t-test for datasets with normal distribution and nonparametric Mann-Whitney test when data differed from normal distribution, p=0.05.
Statistical analysis
Data are expressed as mean ± SEM. Statistical analysis used Prism 7.0 (GraphPad Software). Differences between 2 groups were examined using a Student’s t test, with p<0.05 was considered significant and p>0.05 non-significant. When comparisons were between more than two groups, one-way ANOVA was used. All relevant data are available.
Data availability
Sequences for mouse and human V16 Piezo2 have been deposited in Genbank, accession # MG254546 and MG254547.
Supplementary Material
Highlights.
Piezo2 undergoes alternative splicing.
Sensory neurons express multiple Piezo2 variants, non-neuronal tissues express one.
Different classes of touch neurons express different Piezo2 splice forms.
Alternatively splicing confers functional differences on Piezo2 properties.
Acknowledgments
This work was supported by the intramural research program of the National Institute of Dental and Craniofacial Research (NIDCR) (MAH), the National Center for Complementary and Integrative Health (NCCIH) (ATC), and Office of the Director National Institutes of Health (BLY). Authors declare no financial or non-financial competing interests. We thank members of the Hoon, Chesler and Le Pichon labs, Nick Ryba, Kenton Swartz, Julio Cordero, and Valeria Vasquez for the valuable comments and advice.
Footnotes
Author contributions
Investigation and Methodology, M.S., L.P., H.S., P.Y., C.L.P., A.C, M.H., Data Curation, M.S., L.P., L.Y., Writing-Original Draft, A.C. and M.H., Supervision, A.C., C.L.P., and M.H.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abraira VE, Ginty DD. The sensory neurons of touch. Neuron. 2013;79:618–639. doi: 10.1016/j.neuron.2013.07.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aho AV, Kernighan BW, Weinberger BPJ. The AWK Programming Language. Boston, MA, USA: Addison-Wesley Longman Publishing Co; 1987. [Google Scholar]
- Andrew D, Greenspan JD. Mechanical and heat sensitization of cutaneous nociceptors after peripheral inflammation in the rat. J Neurophysiol. 1999;82:2649–2656. doi: 10.1152/jn.1999.82.5.2649. [DOI] [PubMed] [Google Scholar]
- Bai L, Lehnert BP, Liu J, Neubarth NL, Dickendesher TL, Nwe PH, Cassidy C, Woodbury CJ, Ginty DD. Genetic Identification of an Expansive Mechanoreceptor Sensitive to Skin Stroking. Cell. 2015;163:1783–1795. doi: 10.1016/j.cell.2015.11.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borbiro I, Badheka D, Rohacs T. Activation of TRPV1 channels inhibits mechanosensitive Piezo channel activity by depleting membrane phosphoinositides. Sci Signal. 2015;8:ra15. doi: 10.1126/scisignal.2005667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanaugh DJ, Lee H, Lo L, Shields SD, Zylka MJ, Basbaum AI, Anderson DJ. Distinct subsets of unmyelinated primary sensory fibers mediate behavioral responses to noxious thermal and mechanical stimuli. Proc Natl Acad Sci U S A. 2009;106:9075–9080. doi: 10.1073/pnas.0901507106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesler AT, Szczot M, Bharucha-Goebel D, Ceko M, Donkervoort S, Laubacher C, Hayes LH, Alter K, Zampieri C, Stanley C, et al. The Role of PIEZO2 in Human Mechanosensation. N Engl J Med. 2016;375:1355–1364. doi: 10.1056/NEJMoa1602812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coste B, Houge G, Murray MF, Stitziel N, Bandell M, Giovanni MA, Philippakis A, Hoischen A, Riemer G, Steen U, et al. Gain-of-function mutations in the mechanically activated ion channel PIEZO2 cause a subtype of Distal Arthrogryposis. Proc Natl Acad Sci U S A. 2013;110:4667–4672. doi: 10.1073/pnas.1221400110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coste B, Mathur J, Schmidt M, Earley TJ, Ranade S, Petrus MJ, Dubin AE, Patapoutian A. Piezo1 and Piezo2 are essential components of distinct mechanically activated cation channels. Science. 2010;330:55–60. doi: 10.1126/science.1193270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coste B, Murthy SE, Mathur J, Schmidt M, Mechioukhi Y, Delmas P, Patapoutian A. Piezo1 ion channel pore properties are dictated by C-terminal region. Nat Commun. 2015;6:7223. doi: 10.1038/ncomms8223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubin AE, Murthy S, Lewis AH, Brosse L, Cahalan SM, Grandl J, Coste B, Patapoutian A. Editorial Note to: Endogenous Piezo1 Can Confound Mechanically Activated Channel Identification and Characterization. Neuron. 2017a;94:265. doi: 10.1016/j.neuron.2017.03.041. [DOI] [PubMed] [Google Scholar]
- Dubin AE, Murthy S, Lewis AH, Brosse L, Cahalan SM, Grandl J, Coste B, Patapoutian A. Endogenous Piezo1 Can Confound Mechanically Activated Channel Identification and Characterization. Neuron. 2017b;94:266–270. e263. doi: 10.1016/j.neuron.2017.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubin AE, Schmidt M, Mathur J, Petrus MJ, Xiao B, Coste B, Patapoutian A. Inflammatory signals enhance piezo2-mediated mechanosensitive currents. Cell Rep. 2012;2:511–517. doi: 10.1016/j.celrep.2012.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eid J, Fehr A, Gray J, Luong K, Lyle J, Otto G, Peluso P, Rank D, Baybayan P, Bettman B, et al. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323:133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- Eijkelkamp N, Linley JE, Torres JM, Bee L, Dickenson AH, Gringhuis M, Minett MS, Hong GS, Lee E, Oh U, et al. A role for Piezo2 in EPAC1-dependent mechanical allodynia. Nat Commun. 2013;4:1682. doi: 10.1038/ncomms2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge J, Li W, Zhao Q, Li N, Chen M, Zhi P, Li R, Gao N, Xiao B, Yang M. Architecture of the mammalian mechanosensitive Piezo1 channel. Nature. 2015;527:64–69. doi: 10.1038/nature15247. [DOI] [PubMed] [Google Scholar]
- Gibson JJ. Observations on active touch. Psychol Rev. 1962;69:477–491. doi: 10.1037/h0046962. [DOI] [PubMed] [Google Scholar]
- Goodman MB, Ernstrom GG, Chelur DS, O’Hagan R, Yao CA, Chalfie M. MEC-2 regulates C. elegans DEG/ENaC channels needed for mechanosensation. Nature. 2002;415:1039–1042. doi: 10.1038/4151039a. [DOI] [PubMed] [Google Scholar]
- Goswami SC, Mishra SK, Maric D, Kaszas K, Gonnella GL, Clokie SJ, Kominsky HD, Gross JR, Keller JM, Mannes AJ, et al. Molecular signatures of mouse TRPV1-lineage neurons revealed by RNA-Seq transcriptome analysis. J Pain. 2014;15:1338–1359. doi: 10.1016/j.jpain.2014.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gracheva EO, Cordero-Morales JF, Gonzalez-Carcacia JA, Ingolia NT, Manno C, Aranguren CI, Weissman JS, Julius D. Ganglion-specific splicing of TRPV1 underlies infrared sensation in vampire bats. Nature. 2011;476:88–91. doi: 10.1038/nature10245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higley MJ, Sabatini BL. Calcium signaling in dendritic spines. Cold Spring Harb Perspect Biol. 2012;4:a005686. doi: 10.1101/cshperspect.a005686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjerling-Leffler J, Alqatari M, Ernfors P, Koltzenburg M. Emergence of functional sensory subtypes as defined by transient receptor potential channel expression. J Neurosci. 2007;27:2435–2443. doi: 10.1523/JNEUROSCI.5614-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda R, Cha M, Ling J, Jia Z, Coyle D, Gu JG. Merkel cells transduce and encode tactile stimuli to drive Abeta-afferent impulses. Cell. 2014;157:664–675. doi: 10.1016/j.cell.2014.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowlton WM, Palkar R, Lippoldt EK, McCoy DD, Baluch F, Chen J, McKemy DD. A sensory-labeled line for cold: TRPM8-expressing sensory neurons define the cellular basis for cold, cold pain, and cooling-mediated analgesia. J Neurosci. 2013;33:2837–2848. doi: 10.1523/JNEUROSCI.1943-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Pichon CE, Chesler AT. The functional and anatomical dissection of somatosensory subpopulations using mouse genetics. Front Neuroanat. 2014;8:21. doi: 10.3389/fnana.2014.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis AH, Cui AF, McDonald MF, Grandl J. Transduction of Repetitive Mechanical Stimuli by Piezo1 and Piezo2 Ion Channels. Cell Rep. 2017;19:2572–2585. doi: 10.1016/j.celrep.2017.05.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipscombe D, Andrade A. Calcium Channel CaValpha(1) Splice Isoforms - Tissue Specificity and Drug Action. Curr Mol Pharmacol. 2015;8:22–31. doi: 10.2174/1874467208666150507103215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlone F, Wessberg J, Olausson H. Discriminative and affective touch: sensing and feeling. Neuron. 2014;82:737–755. doi: 10.1016/j.neuron.2014.05.001. [DOI] [PubMed] [Google Scholar]
- McMillin MJ, Beck AE, Chong JX, Shively KM, Buckingham KJ, Gildersleeve HI, Aracena MI, Aylsworth AS, Bitoun P, Carey JC, et al. Mutations in PIEZO2 cause Gordon syndrome, Marden-Walker syndrome, and distal arthrogryposis type 5. Am J Hum Genet. 2014;94:734–744. doi: 10.1016/j.ajhg.2014.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra SK, Tisel SM, Orestes P, Bhangoo SK, Hoon MA. TRPV1-lineage neurons are required for thermal sensation. EMBO J. 2011;30:582–593. doi: 10.1038/emboj.2010.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan P, Sondermann J, Rouwette T, Karaca S, Urlaub H, Mitkovski M, Gomez-Varela D, Schmidt M. Native Piezo2 Interactomics Identifies Pericentrin as a Novel Regulator of Piezo2 in Somatosensory Neurons. J Proteome Res. 2016;15:2676–2687. doi: 10.1021/acs.jproteome.6b00235. [DOI] [PubMed] [Google Scholar]
- Nguyen MQ, Wu Y, Bonilla LS, von Buchholtz LJ, Ryba NJP. Diversity amongst trigeminal neurons revealed by high throughput single cell sequencing. PLoS One. 2017;12:e0185543. doi: 10.1371/journal.pone.0185543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonomura K, Woo SH, Chang RB, Gillich A, Qiu Z, Francisco AG, Ranade SS, Liberles SD, Patapoutian A. Piezo2 senses airway stretch and mediates lung inflation-induced apnoea. Nature. 2017;541:176–181. doi: 10.1038/nature20793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- Piskorowski R, Haeberle H, Panditrao MV, Lumpkin EA. Voltage-activated ion channels and Ca(2+)-induced Ca (2+) release shape Ca (2+) signaling in Merkel cells. Pflugers Arch. 2008;457:197–209. doi: 10.1007/s00424-008-0496-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogorzala LA, Mishra SK, Hoon MA. The Cellular Code for Mammalian Thermosensation. J Neurosci. 2013;33:5533–5541. doi: 10.1523/JNEUROSCI.5788-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole K, Herget R, Lapatsina L, Ngo HD, Lewin GR. Tuning Piezo ion channels to detect molecular-scale movements relevant for fine touch. Nat Commun. 2014;5:3520. doi: 10.1038/ncomms4520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranade SS, Woo SH, Dubin AE, Moshourab RA, Wetzel C, Petrus M, Mathur J, Begay V, Coste B, Mainquist J, et al. Piezo2 is the major transducer of mechanical forces for touch sensation in mice. Nature. 2014;516:121–125. doi: 10.1038/nature13980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutlin M, Ho CY, Abraira VE, Cassidy C, Bai L, Woodbury CJ, Ginty DD. The cellular and molecular basis of direction selectivity of Adelta-LTMRs. Cell. 2014;159:1640–1651. doi: 10.1016/j.cell.2014.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider ER, Mastrotto M, Laursen WJ, Schulz VP, Goodman JB, Funk OH, Gallagher PG, Gracheva EO, Bagriantsev SN. Neuronal mechanism for acute mechanosensitivity in tactile-foraging waterfowl. Proc Natl Acad Sci U S A. 2014;111:14941–14946. doi: 10.1073/pnas.1413656111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukharev SI, Blount P, Martinac B, Blattner FR, Kung C. A large-conductance mechanosensitive channel in E. coli encoded by mscL alone. Nature. 1994;368:265–268. doi: 10.1038/368265a0. [DOI] [PubMed] [Google Scholar]
- Syeda R, Florendo MN, Cox CD, Kefauver JM, Santos JS, Martinac B, Patapoutian A. Piezo1 Channels Are Inherently Mechanosensitive. Cell Rep. 2016;17:1739–1746. doi: 10.1016/j.celrep.2016.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usoskin D, Furlan A, Islam S, Abdo H, Lonnerberg P, Lou D, Hjerling-Leffler J, Haeggstrom J, Kharchenko O, Kharchenko PV, et al. Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat Neurosci. 2015;18:145–153. doi: 10.1038/nn.3881. [DOI] [PubMed] [Google Scholar]
- Walker RG, Willingham AT, Zuker CS. A Drosophila mechanosensory transduction channel. Science. 2000;287:2229–2234. doi: 10.1126/science.287.5461.2229. [DOI] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo SH, Lukacs V, de Nooij JC, Zaytseva D, Criddle CR, Francisco A, Jessell TM, Wilkinson KA, Patapoutian A. Piezo2 is the principal mechanotransduction channel for proprioception. Nat Neurosci. 2015;18:1756–1762. doi: 10.1038/nn.4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo SH, Ranade S, Weyer AD, Dubin AE, Baba Y, Qiu Z, Petrus M, Miyamoto T, Reddy K, Lumpkin EA, et al. Piezo2 is required for Merkel-cell mechanotransduction. Nature. 2014;509:622–626. doi: 10.1038/nature13251. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequences for mouse and human V16 Piezo2 have been deposited in Genbank, accession # MG254546 and MG254547.