Abstract
Eukaryotic mRNA transcription initiation is directed by the formation of the megaDalton-sized pre-initiation complex (PIC). After PIC formation, double-stranded DNA is unwound to form a single-stranded DNA bubble and the template strand is loaded into the polymerase active site. DNA opening is catalyzed by Ssl2(XPB), the dsDNA translocase subunit of the basal transcription factor TFIIH. In yeast, transcription initiation proceeds through a scanning phase where downstream DNA is searched for optimal start-sites. Here, to test models for initial DNA opening and start-site scanning, we measure the DNA bubble sizes generated by Saccharomyces cerevisiae PICs in real time using single-molecule magnetic tweezers. We show that ATP hydrolysis by Ssl2 opens a 6 base-pair (bp) bubble that grows to 13 bp in the presence of NTPs. These observations support a two-step model wherein ATP-dependent Ssl2 translocation leads to a 6 bp open complex which RNA polymerase II expands via NTP-dependent RNA transcription.
Transcription initiation can be divided into phases of promoter recognition, PIC assembly, DNA unwinding, and promoter escape1,2. While the phases of initiation are conserved, specific DNA opening mechanisms vary across branches of life3–5 and between eukaryotic RNAPs I, II, and III6–8. In the RNAP II system, ATP hydrolysis by the Ssl2(XPB) subunit of TFIIH leads to the formation of an unstable open complex9–11. The position of Ssl2 in RNAP II PIC structures and protein-DNA cross-link maps suggest that it binds DNA downstream from the site where initial DNA unwinding occurs12–16. As such, it cannot function as a conventional helicase to promote open complex formation since it does not make direct contacts to the region of DNA to be unwound14. Consistent with this observation, in the context of TFIIH we have shown that Ssl2 functions as a double-stranded DNA translocase with an estimated processivity of ~10 bp17. These structural and functional data support the proposal of DNA unwinding mechanisms in which Ssl2 pumps downstream DNA towards the RNAP and generates torsional and mechanical strain that leads to DNA unwinding and formation of a DNA bubble12,14,17,18.
While much is known about the structure of the PIC and the activities of TFIIH and the other general transcription factors, relatively little is known regarding the mechanism of DNA opening and TSS scanning. Potassium permanganate DNA foot-printing in the human PIC has generated different estimates of the amount of open DNA during different phases of initiation ranging from 8 – 22 bp9–11,19,20. However, foot-printing measurements are ensemble averaged, only report on the relative accessibility of thymine bases, and do not directly report on the helical state of the DNA. A recent optical tweezers study of the yeast PIC observed the shortening of the distance between the polymerase and the downstream duplex DNA in the presence of dATP21. This data supports the compaction of the DNA template by the action of Ssl2, and was further interpreted in the context of the opening of an 85 bp DNA bubble (on average) in a TSS-position independent mechanism of DNA scanning21. However, the observed length change monitored by the optical tweezers in those experiments does not report on the helical state of the compacted DNA and can be interpreted as a fully unwound bubble, a fully dsDNA loop, or an intermediate structure. Here, we use single-molecule magnetic tweezers assays that directly detect DNA opening at high spatial and temporal resolution to measure the distribution of DNA bubble sizes produced during different phases of initiation.
Results
Single-molecule measurements of NTP-dependent promoter opening
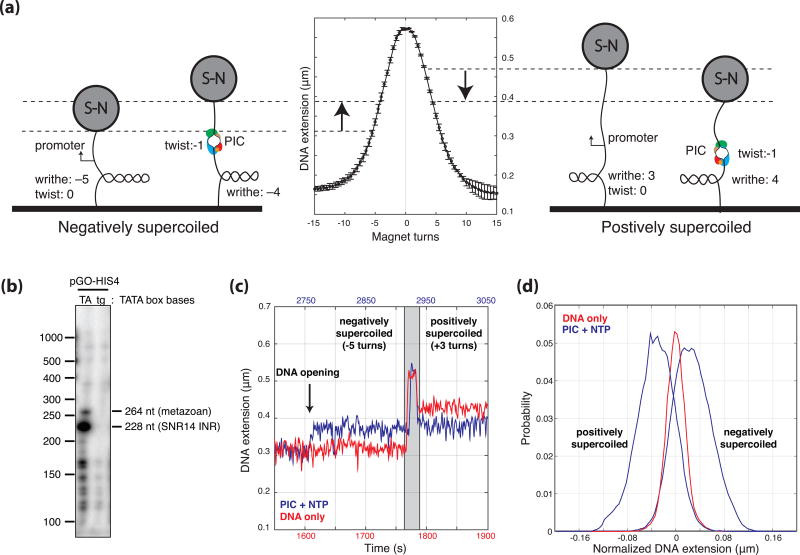
The magnetic tweezers allow for the detection of DNA opening due to linking number conservation (i.e. the interchange of twist and writhe) and the linear relationship between the number of writhes and the end-to-end distance of a DNA tether22–24 (Fig. 1a). A 2.1 kb DNA tether was designed to exhibit TATA-box-dependent transcription with a dominant start-site 70 bp downstream from TATA (Fig. 1b, Methods, and Supplemental Information). Transcription buffer with TBP, TFIIB, TFIIE, TFIIF, TFIIH, RNAP II, and 500 µM NTPs was introduced into the flow cell at a flow-rate of 10 µL/min for 2.5 minutes after which the template was supercoiled by turning the magnets and DNA extension was monitored over time. The flow rate and lack of template supercoiling during flow was designed to eliminate protein-independent DNA opening due to flow forces. On negatively supercoiled tethers, DNA opening was revealed by increases in the DNA extension (due to the loss of a negative writhe) and was only observed in the presence of all the general transcription factors, RNAP II, the TATA-box, and NTPs. As DNA compaction reduces the end-to-end length of the tether, any component of the signal due to DNA compaction acts to decrease the magnitude of the signal on negatively supercoiled templates and increase the magnitude of the signal on positively supercoiled templates23. NTP data collected on negatively and positively supercoiled DNA templates (Fig. 1c, d) exhibits equal and opposite extension changes within the resolution of the assay, suggesting that stable DNA compaction greater than 30 bp is not contributing to the observed DNA length change (30 bp of compaction would result in a difference in the signal between negative and positive supercoiled of approximately 10 nm which is the limit of detection at 0.3 pN). Thus, changes in DNA extension were interpreted as a result of DNA opening alone and converted to bubble size in base-pairs using the measured linear dependence between number of turns and DNA extension for individual templates (Fig. 1a and Supplemental Information).
Figure 1. Experimental Setup.
(a) The magnetic tweezers allow for the negative or positive supercoiling of single torsionally constrained dsDNA tethers and the measurement of the end-to-end distance of the tether via the optical tracking of the height of the 1 µm magnetic beads. Due to the conservation of linking number, any DNA opening that occurs (change in twist) is accompanied by an equal and opposite change in the number of writhes. As the height of the bead is linearly proportional to the number of writhes under the conditions of the experiment (i.e. 0.3 pN and between 2–8 turns), the change in twist, and thus the number of base-pairs unwound can be calculated. (b) Primer extension products generated after transcription reactions on the template used for the magnetic tweezers experiments. The template was designed with a dominant start-site 70 bp downstream from TATA driven by the strong initiator sequence from the yeast U4 small nuclear RNA gene SNR14 (228 bp band) and a minor start at the position of the predominant TSS distance in metazoans 34 bp downstream from TATA (264 bp band). (c) DNA extension plotted as a function of time showing a DNA only control (red) and a DNA opening trace (500 µM NTP, blue) with negative (−5 turns) and positive (+3 turns) super-helicity. After DNA opening occurred, the tethers were positively supercoiled by turning the magnets (gray area) to test for stability and to measure the corresponding DNA extension change. (d) A normalized distribution of DNA extensions with positively and negatively (blue) supercoiled extensions compared to DNA alone control traces (red).
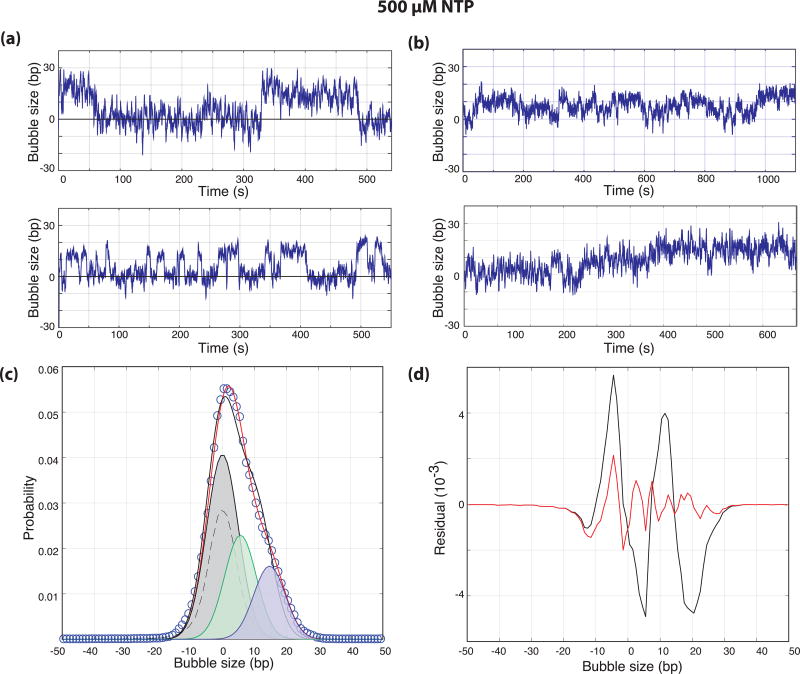
As DNA is negatively supercoiled in vivo and negative supercoiling promotes transcription, we focused our experiments on negatively super-helical templates. Visual inspection of traces collected in the presence of 500 µM NTPs revealed that while a third of the traces showed clear two-state DNA opening and closing transitions (Fig. 1c and 2a), the other two-thirds showed a preponderance of long-lived smaller bubbles (Fig. 2b). The probability distribution of the two-state-like traces reveal a DNA bubble size of 13.4 ± 0.2 bp consistent with structural measures of the elongation complex bubble (Supplemental Fig. 1)25.
Figure 2. NTP catalyzed DNA opening.
(a,b) Bubble size in base-pairs plotted as a function of time from the end of flow for negatively supercoiled templates in the presence of 500 µM NTP. (c) The normalized probability distribution of DNA bubble size (circles, N = 21 traces) with two-state (0.002 RMSD, black) and three-state (0.0014 RMSD, red) fits. The distribution of DNA only controls (dashed) were used to constrain the widths of the distributions during fitting and the individual distributions from the three-state fit are shown. The distributions have means of 0, 6.1, and 13.4 bp and represent 51%, 27%, and 22% of the overall probability distribution respectively. (d) Residuals from the two-state (black) and three-state (red) fits shown in (b).
While only a fraction of the DNA templates show activity (~5 %), those that do often show repeated openings and closings (Fig. 2). In addition, we have not observed traces consistent with the formation of multiple elongation complexes which would be evidenced by sequential steps of DNA opening that would be additive in nature. For example, the initiation of two polymerases would result in a 26 bp bubble signal consistent with the presence of two 13 bp bubbles. From these observations, we conclude that the repetitive opening and closing signals are indicative of reversible or abortive transitions between closed and open complexes in the context of the initiation of a single polymerase.
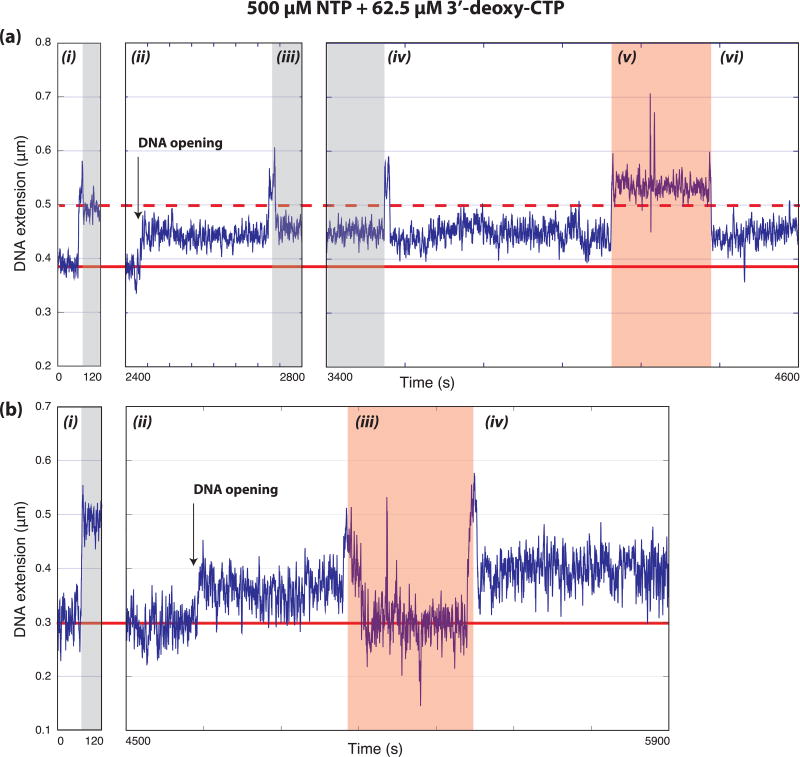
Due to the above, we asked whether the initiation reaction in the tweezers was proceeding to completion and producing stable elongation complexes by performing experiments in the presence of 62.5 µM 3’-deoxy-CTP to chain terminate the growing RNA transcript and stall elongating polymerases. These traces exhibit hyper-stable bubbles for the duration of the experiment as one would expect for a stalled elongation complex. Furthermore, these bubbles were stable even after removing NTPs and free protein factors by flowing buffer alone through the flow cell (Fig. 3). These observations are consistent with the formation of stable elongation complexes and the successful initiation of transcription during our single-molecule experiment.
Figure 3. Formation of stable elongation complexes.
Representative data (from 14 traces collected in 7 experiments) showing long-lived DNA bubbles formed in the presence of 500 µM NTP and 62.5 µM 3’-deoxy-CTP lasting over the remainder of the trace and stable after flowing out NTPs and protein factors. Regions of negatively supercoiled DNA (not shaded), positively supercoiled DNA (shaded grey), and where NTPs are being flowed out (shaded red) are indicated. The solid and dashed horizontal red lines indicate negatively and positively supercoiled DNA extensions in the absence of factors respectively. (a) A single tether monitored over 4600 s while super-helicity and buffer conditions are modified. The sections of the trace are as follows: (i) DNA only rotation-extension curve showing negatively supercoiled DNA and positively supercoiled DNA base-lines, (ii) a DNA opening signal observed after flowing in PIC components and NTPs (consistent with the large bubble described in the text) indicated by an increase in the negatively supercoiled tether length, (iii) the bubble is stable to positive supercoils (shaded grey), (iv) a return to negatively supercoiled conditions (bubble still open), (v) changes in extension caused by flowing buffer to remove factors and NTPs, (vi) the bubble is still open suggesting the formation of the stable elongation complex. (b) A single tether monitored over 5900 s while buffer conditions are modified. The sections of the trace are as follows: (i) DNA only rotation-extension curve showing negatively supercoiled DNA and positively supercoiled DNA base-lines, (ii) a DNA opening signal observed after flowing in PIC components and NTPs (consistent with the large bubble described in the text) indicated by an increase in the negatively supercoiled tether length, (iii) changes in extension caused by flowing buffer to remove factors and NTPs, (iv) the signal for an open bubble is still present suggesting the formation of the stable elongation complex.
A second open complex in the presence of NTPs
As mentioned above, two-thirds of the traces observed in the presence of 500 µM NTP showed evidence that the system does not always conform to a two-state model (Fig. 2b). In fact, the combined distribution of bubble sizes from all NTP traces (N = 21) is not well-fit by a closed state and a single open state (Fig. 2c, black fit). Fits of the complete experimental distribution with an additional open state improved the fit (Fig. 2c, red fit) and the fit residuals (Fig. 2d) and revealed the presence of a 6.1 ± 0.3 bp open state (Fig. 2c, green) in addition to the 13 bp bubble (Fig. 2c, magenta). Allowing the means of both open states to float during the fit resulted in estimates of bubble sizes of 5.6 ± 0.5 bp and 14.7 ± 0.4 bp. We postulated that while the 13–15 bp bubble represents fully formed open complexes (or escaped elongation complexes), the 5–6 bp bubble represents initial DNA opening generated by the ATPase activity of Ssl2 in TFIIH.
ATP hydrolysis by Ssl2 generates a partial open complex
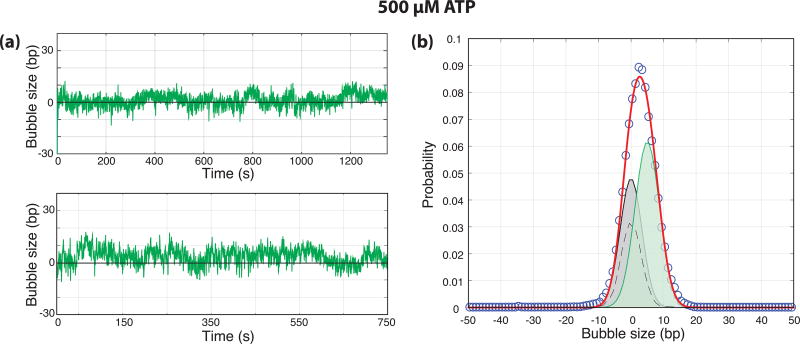
To test this hypothesis, we performed experiments in the presence of ATP alone where Ssl2 is active, presumably allowing for both DNA opening and start-site scanning, but the polymerase is unable to transcribe. Data acquired under both negative and positive supercoiling in the presence of 500 µM ATP (N = 23) show length changes consistent with the opening of approximately half a turn of DNA with no significant effect from DNA compaction or wrapping (Fig. 4a, Supplemental Fig. 2). Experiments in the presence of PIC factors, but in the absence of ATP did not show any DNA opening transitions (Supplemental Fig. 3). The distribution of bubble size in the presence of ATP is clearly distinct from that of DNA alone and can be fit with a closed state and a single open state of 5 bp (Fig. 4b). Importantly, larger bubbles were never observed under these conditions, illustrating the requirement of NTPs for the production of the 13–15 bp bubble. Furthermore, this bubble size is consistent with the smaller bubble size measured in the presence of 500 µM NTPs (Fig. 2) supporting the identification of that smaller bubble as ATP- and Ssl2-dependent.
Figure 4. ATP catalyzed DNA opening.
(a) Bubble size in base-pairs plotted as a function of time (s) from the end of flow for negatively supercoiled templates in the presence of 500 µM ATP. (b) Normalized probability distribution of bubble sizes (circles, N = 26 traces) along with a two-state fit (0.0012 RMSD, red). The distribution of DNA only controls (dashed) were used to constrain the widths of the distributions during fitting and the individual distributions from the fit are shown. The distributions have means of 0 and 5.0 and represent 44% and 56% of the overall probability distribution respectively.
The ATP-dependent bubble is an intermediate on the pathway to the fully open complex
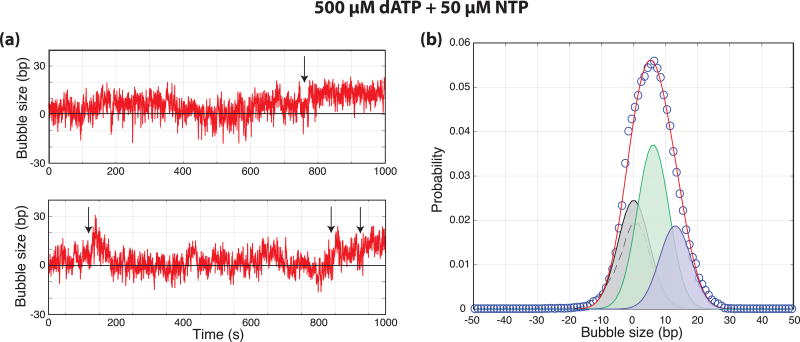
To further test the hypothesis that the ATP-dependent bubble is an on-pathway intermediate to the fully open complex, experiments were performed with high dATP (500 µM) and low NTP (50 µM) concentrations. As Ssl2(XPB) is known to utilize dATP in place of ATP for translocation and open complex formation10,17, the assay was designed to allow for robust Ssl2 activity while slowing the RNAP catalyzed extension of the RNA chain. If the intermediate bubble is directly expanded by RNAP-dependent NTP hydrolysis, the lower NTP concentration should result in longer lifetimes for the 6 bp bubble as one of the rates out of this state will be reduced. Thus, we expected that the relative population of the small bubble state should increase compared to 500 µM NTP. Data acquired under both negative and positive supercoiling again showed length changes consistent with the absence of significant amounts of DNA compaction or wrapping (Supplemental Fig. 4). Under these conditions, clear transitions between 6 and 13 bp bubbles were observed suggesting that the 6 bp bubble can be directly expanded to the 13 bp bubble and the 13 bp bubble can collapse directly to the 6 bp one (Fig. 5a). Furthermore, an unbiased fit of the distribution of bubble sizes observed under these conditions (N = 26) resulted in measures of the two bubble sizes (6.1 ± 0.8 bp and 13.0 ± 0.9 bp) consistent with the data from the other conditions (Fig. 5b). Lastly, the amplitude of the 6 bp bubble distribution increased by 60% compared to the 500 µM NTP data as predicted. Thus, the data fully support a model where the 6 bp bubble is an intermediate on the pathway to a fully open complex. As an additional note, these data contain examples of both forward and reverse transitions between the closed DNA and the 6 and 13 bp bubbles. As mentioned above, this reversibility strongly suggests that the initiation process passes through open complexes that do not always lead to the escape of an elongating polymerase and are suggestive of abortive initiation26.
Figure 5. A partially open complex is an intermediate in transcription initiation.
(a) Bubble size in base-pairs plotted as a function of time (s) from the end of flow for negatively supercoiled templates in the presence of 500 µM dATP and 50 µM NTP. Arrows indicate transitions between small and large bubble states. (b) Normalized probability distribution of bubble sizes (circles, N = 23 traces) along with a three-state fit (0.001 RMSD, red). The distribution of DNA only controls (dashed) were used to constrain the widths of the distributions during fitting and the individual distributions from the fit are shown. The distributions have means of 0, 6.1, and 13.0 bp and represent 30%, 47%, and 23% of the overall probability distribution respectively.
Sub1 and TFIIA do not change the size of the initiation bubbles
To directly compare our results to a previous single-molecule study, we repeated the above experiments in the presence of Sub127 and TFIIA28, two factors not strictly required for in vitro reactions21. In particular, as Sub1 likely has ssDNA binding activity, we asked if it would stabilize larger DNA bubble scanning intermediates on the order of 40–100 bp proposed in previous models21. However, the distribution of bubble sizes in the presence and absence of Sub1 and TFIIA were quite similar and no larger DNA opening signals were observed (Supplemental Fig. 5). We conclude that Sub1 and TFIIA do not modulate the amount of DNA that is unwound during initiation.
4–6 base-pair bubble templates abrogate the requirement for TFIIH
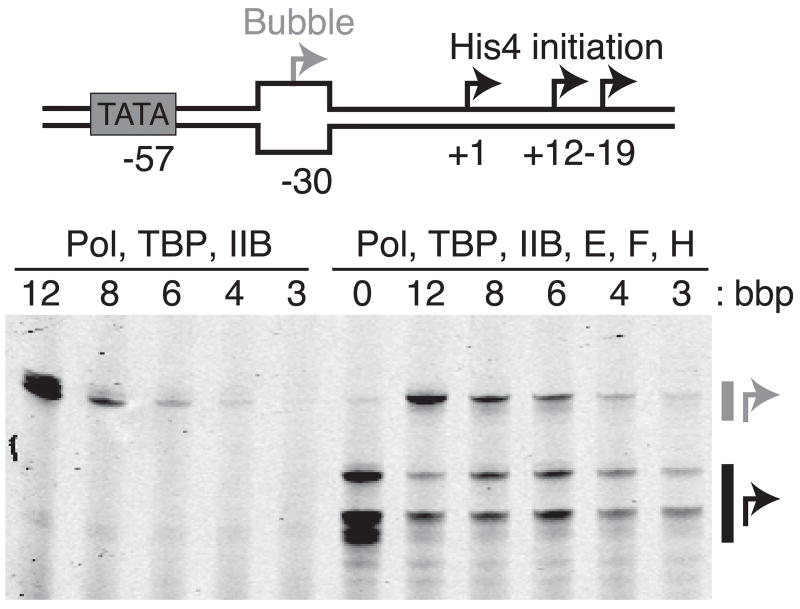
As an additional evaluation of our model, we performed transcription assays on DNA bubble templates. Initiation is TFIIH-independent on 12 bp bubble templates and can be performed in the presence of a subset of factors29. If the mechanistically important outcome of Ssl2 activity is a 6 bp bubble, then templates with unpaired bubbles of this size should result in initiation in the absence of TFIIH. Templates with bubbles of 3 – 12 bp were incubated for 30 minutes with TBP, TFIIB, RNAP II, and NTPs. Productive transcription increases with increasing bubble size (Fig. 6) and both the 4 and 6 bp bubbles are indeed sufficient for polymerase loading and transcription initiation in the absence of TFIIH. In addition, the low efficiency of initiation on these small bubbles is consistent with the hypothesis that ATP-hydrolysis by Ssl2 also plays roles in subsequent steps on the path to transcript production (i.e. initial transcription11 and promoter escape30). In fact, the sharp increase in transcription observed with the 12 bp bubble suggests that the stimulatory effect of TFIIH on the initial transcription needed to expand the bubble has now also been removed as the amount of open DNA is now comparable to the fully open 13 bp bubble observed in our single-molecule data.
Figure 6. A six base-pair bubble is sufficient to support initiation in the absence of TFIIH.
The His4-bubble construct is shown schematically along with a gel separating the products derived from bubble initiated start-sites (grey arrow) and WT downstream start-sites (black arrows) for different lengths of bubble base-pairs (bbp). In the absence of TFIIE, F, and H (left), downstream start-sites are not utilized and initiation efficiency increases as the bubble size is increased through 6 bp.
Discussion
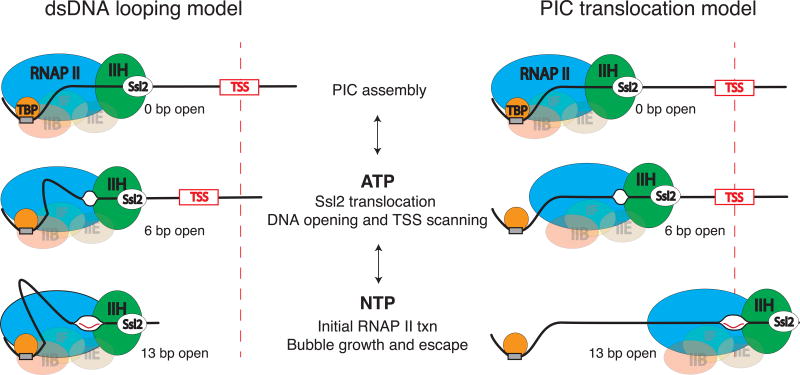
Given our results on the Ssl2-dependent dsDNA translocase activity of TFIIH, we hypothesize that formation of the 6 bp bubble is a direct consequence of Ssl2 translocation on downstream dsDNA17. Subsequently, initial transcription by RNAP II at a start-site sequence leads to the expansion of the bubble (Fig. 7). This model fits with structural work on the human pre-initiation complex suggesting that 10 bp of DNA is pumped into the complex by XPB (Ssl2) during initial DNA opening18 as well as with studies suggesting that TFIIH acts via a molecular wrench mechanism14. Within the context of our model, start-site scanning occurs via a translocating 6 bp bubble and is likely due to a competition between the rate of Ssl2 translocation and the rate of initial transcription by RNAP II. In the metazoan case where scanning does not occur on TATA-dependent promoters, we predict that this competition is tilted in favor of the polymerase so that initiation occurs on the first 6 bp bubble generated by XPB. Lastly, taking into account optical tweezers results showing the ATP-dependent compaction of downstream DNA21, we propose that scanning may occur within the context of a dsDNA loop.
Figure 7. Models of transcription initiation.
Two alternative models are proposed that are consistent with the magnetic tweezers data. After PIC assembly, ATP-hydrolysis by Ssl2 leads to translocation of downstream DNA into the PIC. This leads to initial unwinding of 5–6 bp and powers TSS scanning. In the dsDNA looping model, the PIC remains bound to TATA and the downstream DNA is looped out between TATA and the polymerase active site. In the PIC translocation model, the polymerase along with a subset of GTFs breaks contact with TATA and translocates downstream while scanning the DNA. In both models, NTP hydrolysis by RNAP II leads to the expansion of the bubble to 13 bp and subsequent promoter escape.
While the magnetic tweezers DNA unwinding assay is very sensitive to DNA melting, it is relatively poor at detecting compacted DNA. Although it has been used to detect large amounts of DNA wrapping (i.e. the ~100 bp that occur concomitantly with DNA melting in the bacterial system22), the degree of compaction that would be required to reach the downstream start site in a dsDNA looping model would result in a signal difficult to detect. The 40 bp from the initial site of unwinding to the downstream start site would result in a length change of only 6 nm under the conditions of our assay. That said, the data presented argue directly against a model of long range DNA compaction of the kind observed in the presence of ATP alone in the optical tweezers experiments21. The different activities observed in the two works may be related to differences in experimental methodology.
The optical tweezers data was acquired from pre-formed and purified pre-initiation complexes bound to polystyrene beads. In particular, multiple PICs on each individual bead bound to short pieces of DNA templates were used to increase the local concentration of PIC components (see Extended Data Fig. 2 in that work21). One concern is that this may result in the assay partially reporting on the activity of multiple PICs or multiple factors that are present and in close proximity to the tethered DNA (i.e. two TFIIHs). In addition, these assays were performed at 4 pN of DNA tension.
In our magnetic tweezers assay, PIC factors are flowed into the cell where they must assemble de novo on DNA tethers. This insures that we are observing the processes as it would occur in solution with a minimum of manipulation and away from any solid surfaces (i.e. a bead or a slide). Lastly, our experiments are performed at 0.3 pN which is more than ten times lower force and more similar to solution reactions where there is no force applied.
Although future work will aim at directly testing for the presence of a dsDNA loop, our data are currently consistent with either a dsDNA loop or the absence of significant compaction during scanning (Fig. 7). The PIC translocation model would necessarily require the detachment of the polymerase, TFIIH, and possibly other basal factors from the TATA-box to allow for translocation and scanning in the absence of compaction. If a looped structure does exist, we speculate that due to the structural constraints of the PIC, it would have to form via DNA kinks instead of a smoothly looped structure31–33. Based on the structures of the pre-initiation complex13,34,35, we propose that DNA may be extruded in between the TATA box where TBP is bound and the upstream edge of the polymerase footprint. Lastly, in both models, the potassium permanganate sensitivity of the entire scanning region observed in vivo36,37 is explained by a mobile bubble instead of a single extended bubble consisting of up to hundreds of base-pairs.
Eukaryotic transcription initiation is well conserved and the yeast pre-initiation complex is structurally homologous to the human complex18,38. Although there are important differences between the systems in terms of the locations of transcription start-sites and the prevalence of start-site scanning, our results are qualitatively similar to data acquired using potassium permanganate foot-printing20,39. As we observe here, these studies found that DNA opening occurs in two distinct steps. The first step is dependent on ATP and TFIIH(XPB) and results in the permanganate sensitivity of 6–10 bp of DNA while the second step is dependent on NTPs and results in the expansion of the sensitive region to 10–17 bp. While the size reported here is smaller, differences between the assays may explain the discrepancy. Specifically, footprints can be larger than the actual bubble if the bubble is mobile during the exposure to potassium permanganate. Conversely, while the magnetic tweezers assay cannot determine the position of the DNA bubble, it is not sensitive to bubble motion. Therefore, the existing data can be reconciled with a model where footprints are generated via a mobile 6 bp bubble. Furthermore, in bubble DNA template experiments similar to the ones reported here, the human polymerase can initiate on 5 bp bubbles in a TFIIH-independent manner20. Also, the formation of the first or second phosphodiester bond is thought to be the determining step for expansion of the smaller bubble20,39 which would both be possible in the context of the 6 bp bubble measured here. Lastly, once a 4 bp RNA-DNA hybrid is generated, ATP-hydrolysis by TFIIH is no longer required for productive initiation40. Thus, while the direct observation of the human system via single-molecule methods represents a potential future direction of this work, we propose that 6 bp is the actual bubble size of the TFIIH-dependent open complex and that this mechanism is conserved across all Eukaryotes.
Intriguingly, TFIIH also plays a role in nucleotide excision repair (NER) where current models describe an initial DNA opening of 5 bp catalyzed by XPB to provide a ssDNA binding region for the helicase XPD to further extend the bubble41,42. The similarity of this bubble size to the intermediate bubble we describe here makes it plausible that Ssl2(XPB) use a similar dsDNA translocation mechanism to unwind a similar amount of DNA in both processes.
Online Methods
Protein purification and DNA constructs
Recombinant S. cerevisiae general transcription factors: TBP, TFIIB, and TFIIF were over-expressed and purified from E. coli as previously described29. RNAP II29 and general transcription factors TFIIE and TFIIH were affinity purified from yeast17.
The S. cerevisiae HIS4 promoter was engineered to have a single strong initiation site 70 bp downstream of the TATA box using the transcription initiation site from the yeast U4 small nuclear RNA gene SNR14 which has a 90% initiation efficiency43 (Supplemental Information). Flanking sequences were designed to eliminate TATA-like sequences. The whole DNA template sequence (2.1 kb, Supplemental Information) was synthesized (Gene Oracle) and cloned into a plasmid vector (pGOHis4TATA). Additional engineering was required to remove TATA-independent initiation sites. A TATA-less DNA template was constructed by mutating the TATA sequence from TATATAATA to TGTGTGGTA.
Heteroduplex promoter templates were prepared as previously described29 and were modified with a biotin for immobilizing on beads for in vitro transcription as described below (Supplemental Information).
Magnetic tweezers microscope and flow cell construction
Single DNA tethers were observed on a magnetic tweezers built in house with temperature control as previously described44. Flow cells were constructed from drilled and un-drilled glass coverslips washed in piranha solution (3:1 H2SO4:H2O2) for 15 minutes and thoroughly rinsed with milliQ water. Cut parafilm spacers were sandwiched between the drilled and un-drilled coverslips and melted at 150 C for 10 s to seal the flow cell.
Flow cell inner surfaces are passivated and functionalized with a silane and H-hyroxysuccinimide (NHS) modified polyethylene glycol (PEG,) as described45. In brief, silane-PEG-NHS (14 mg/ml, Nanocs, PG2-NSSL-5K) in DMSO is pipetted into the flow cell and incubated at room temperature (~23 C) in the dark for 1 hr. The solution is removed and flow cell dried with N2 gas. Subsequently, the flow cell is washed with milliQ water (100 µl) followed by PBS buffer (100 µl) and then the cell is incubated with anti-digoxigenin sheep antibodies (Roche, 0.1 mg/mL in PBS) at room temperature for 30 min. The antibodies are covalently attached to the PEG via reaction of their primary amines with the NHS ester. To block un-reacted NHS esters the cell is incubated with 1 M Tris-HCl pH 8.0 (100 µl) for 30 min. After passivation and functionalization the flow cells are washed in DNA tether buffer (PBS, 0.1% Pluronic acid F127, 0.1 mg/ml BSA) and placed in humidified chamber at 4 C.
DNA tether and magnetic bead preparation
DNA tethers were constructed by ligating 1 kb biotin- and digoxigenin-labeled DNA handles generated by PCR in the presence of labeled dUTP nucleotides (biotin-16-dUTP or digoxigenin-11-dUTP, Roche) to a DNA template core (2.1 kb)46. The DNA template core was PCR amplified from an engineered promoter sequence consisting of a modified WT HIS4 promoter with a strong initiator sequence taken from the SNR14 promoter and a series of mutations to make transcription completely TATA-dependent (Fig. 1b and Supplemental Information).
Primers for generating the upstream and downstream handles had the restriction sites AscI, (GGCGCGCC) and NheI (GCTAGC), respectively. These restriction sites generate unique, but compatible overhangs (underlined) with the restriction sites engineered into the promoter DNA tether construct which were MluI (ACGCGT) and StyI (CCTAGG), allowing the restrictions sites to be destroyed once ligation of the handle DNA has occurred. Ligation performed in the presence of the restriction enzymes reduces handle:handle and promoter:promoter side products, resulting in a larger yield of the target 4.1 kb DNA tether product without the need of using large excess of handle DNA47. The 4.1kb DNA construct was gel purified, electro-eluted from the gel, concentrated, and stored at −20 C. Before storage, glycerol was added to a final concentration of 50% v/v.
Streptavidin-coated magnetic beads (5 µL, Dynabeads MyOne Streptavidin T1, Invitrogen, 1µm, 65601) were washed in 200 µl of blocking buffer (PBS, 1 mg/mL BSA), collected and re-suspended in 10 µL of blocking buffer. Polystyrene beads (2 µm, Spherotech, AP-20-10) were diluted 1:100 and 5 µL were added to the washed magnetic beads. DNA was diluted to 50 pM and 0.5 µL of the DNA was mixed with washed magnetic beads and diluted immediately with 90 µL of DNA tether buffer. The bead-DNA solution was pipetted into the flow cell and incubated for 20 min. Unbound beads were washed from the cell while the DNA tether buffer was exchanged for transcription buffer, 200 µL at 10 µL/min. Polystyrene beads nonspecifically stuck to the surface were tracked along with DNA tethered beads, serving as a stage tracking reference. DNA tethers in the magnetic tweezers were characterized to assure that they were single tethers with a length consistent with a 2 kb tether (0.5 – 0.7 µm) and were supercoiled both positively and negatively to generate a rotation extension curve at 0.3 pN (Fig. 1a) for converting changes in DNA extension to number of base-pairs opened23.
Single-molecule promoter opening assay
Prior to adding protein into the flow cell, DNA only traces were collected for both negative and positive supercoiled DNA at 0.3 pN. Pre-initiation complexes (PICs) were assembled on relaxed DNA tethers by flowing approximately 223 nM TBP, 31 nM TFIIB, 13 nM TFIIF, 4 nM TFIIE, 5 nM TFIIH, and 10 nM RNAP II in the presence or absence of 25–100 nM Sub1 and 100 nM TFIIA and in the presence of ATP, dATP, or NTP in transcription buffer (10 mM HEPES, pH 7.6, 100 mM potassium glutamate, 10 mM magnesium acetate, 3.5% (v/v) glycerol, 0.1% (v/v) Pluronic acid F127, and 0.1 mg/mL BSA) at 25 C and 10 µL/min. After flow, the DNA was negatively or positively supercoiled and DNA extension was monitored over 40 min. As extension could not be reliably monitored during flow and flowing on negatively supercoiled DNA could lead to artefactual DNA opening, experimental traces begin after flow had ceased and DNA opening may have already occurred.
Lab-written LABVIEW software was used to implement machine control48 and bead tracking was performed offline using the NanoBLOC software49. Experimental traces of DNA extension were generated by monitoring bead position at 50 Hz and were corrected for instrumental drift by subtracting the signal from a stuck reference bead over time23. The traces were converted to DNA basepairs opened using the measured rotation-extension profile for a given DNA tether mentioned above (Supplemental Information). To facilitate observing changes in DNA opening, the traces were filtered with a moving window average (1 s).
Calculation of base-pairs opened from DNA extension
In the case where no DNA compaction is observed (i.e. changes in DNA extension are the same for negatively and positively supercoiled DNA), the slope of the rotation-extension can be used to convert the change in DNA extension to the size of the DNA bubble in base-pairs. Rotation-extension curves were collected on individual tethers prior to flowing in protein and quantitated by fitting a line to the DNA extension versus turns data between turns 3 and 8 on either the negative or positive arm of the curve depending on the experiment. Each turn was assumed to be 10.5 bp to convert turns to base-pairs. The mean of the used conversion factors obtained under the experimental conditions (10 mM HEPES, pH 7.6, 100 mM potassium glutamate, 10 mM magnesium acetate, 3.5% (v/v) glycerol, 0.1% (v/v) Pluronic acid F127, 0.1 mg/mL BSA, 0.2 pN < force < 0.3 pN, 25 C) was 44 ± 6 nm/turn (standard deviation).
In vitro transcription
In vitro transcription using purified factors was carried out as previously described29 with the following changes: Twenty-microliter reaction mixtures contained 10 mM HEPES (pH 7.6), 100 mM potassium glutamate, 10 mM magnesium acetate, 3 mM dithiothreitol (DTT), 38 mM creatine phosphate, 0.03 units creatine phosphokinase, 2 µg bovine serum albumin (BSA), 4 units RNase Out (Invitrogen), 0.05 % NP40, and 10 nM immobilized DNA template. Pre-initiation complexes were allowed to form for 30 – 40 minutes at room temperature, and transcription initiated with 1 µl nucleoside triphosphates (10 mM each). Transcription was stopped after 30 – 40 minutes, and assayed by primer extension.
Data Availability
Source data for the distributions shown in Fig. 2, 4, and 5 are available with the paper online. Other data generated and analyzed in this study are available from the corresponding author on request. In addition, a Life Sciences Reporting Summary for this article is available.
Supplementary Material
Acknowledgments
The research was supported by National Science Foundation-Molecular and Cellular Biosciences Grant 1243918 (to EAG), National Institute of Health – General Medical Science Grant 5R01GM120559 (to EAG), and National Institute of Health – General Medical Science Grant 2R01GM053451 (to SH). EJT was partially supported by a Keck Postdoctoral Fellowship.
Footnotes
Author Contributions
Author contributions were as follows: EJT, JF, SH, and EAG designed the research. EJT performed the magnetic tweezers experiments. JF purified factors and performed the in vitro transcription reactions. EJT and EAG analyzed single-molecule data. JF and SH analyzed in vitro experiments. EJT, FT, SH and EAG wrote the paper.
References
- 1.Sainsbury S, Bernecky C, Cramer P. Structural basis of transcriptioninitiation by RNA polymerase II. Nat Rev Mol Cell Biol. 2015;16:129–143. doi: 10.1038/nrm3952. [DOI] [PubMed] [Google Scholar]
- 2.Grünberg S, Hahn S. Structural insights into transcriptioninitiation by RNA polymerase II. Trends in Biochemical Sciences. 2013:1–9. doi: 10.1016/j.tibs.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saecker RM, Record MT, Jr, deHaseth PL. Mechanism of Bacterial Transcription Initiation: Promoter Binding, Isomerization to Initiation-Competent Open Complexes, and Initiation of RNA Synthesis. J Mol Biol. 2011:1–18. doi: 10.1016/j.jmb.2011.01.018. doi:10.1016 j.jmb.2011.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qureshi SA, Bell SD, Jackson SP. Factor requirements for transcription in the Archaeon Sulfolobus shibatae. EMBO Journal. 1997;16:2927–2936. doi: 10.1093/emboj/16.10.2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bell SD, Jaxel C, Nadal M, Kosa PF, Jackson SP. Temperature, template topology, and factor requirements of archaeal transcription. Proc Natl Acad Sci USA. 1998;95:15218–15222. doi: 10.1073/pnas.95.26.15218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beckouet F, et al. Two RNA polymerase I subunits control the binding and release of Rrn3 during transcription. Mol Cell Biol. 2008;28:1596–1605. doi: 10.1128/MCB.01464-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keys DA, et al. Multiprotein transcription factor UAF interacts with the upstream element of the yeast RNA polymerase I promoter and forms a stable preinitiation complex. Genes Dev. 1996;10:887–903. doi: 10.1101/gad.10.7.887. [DOI] [PubMed] [Google Scholar]
- 8.Lassar AB, Martin PL, Roeder RG. Transcription of class III genes: formation of preinitiation complexes. Science. 1983 doi: 10.1186/1471-2148-4-26. [DOI] [PubMed] [Google Scholar]
- 9.Holstege FC, Timmers HT. Analysis of open complex formation during RNA polymerase II transcription initiation using heteroduplex templates and potassium permanganate probing. 1997;12:203–211. doi: 10.1006/meth.1997.0472. [DOI] [PubMed] [Google Scholar]
- 10.Wang W, Carey M, Gralla JD. Polymerase II promoter activation: closed complex formation and ATP-driven start site opening. Science. 1992;255:450–453. doi: 10.1126/science.1310361. [DOI] [PubMed] [Google Scholar]
- 11.Dvir A, et al. A role for ATP and TFIIH in activation of the RNA polymerase II preinitiation complex prior to transcription initiation. J Biol Chem. 1996;271:7245–7248. doi: 10.1074/jbc.271.13.7245. [DOI] [PubMed] [Google Scholar]
- 12.Grünberg S, Warfield L, Hahn S. Architecture of the RNA polymerase II preinitiation complex and mechanism of ATP-dependent promoter opening. Nat Struct Mol Biol. 2012;19:788–796. doi: 10.1038/nsmb.2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He Y, et al. Near-atomic resolution visualization of human transcription promoter opening. Nature. 2016;533:359–365. doi: 10.1038/nature17970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim TK, Ebright RH, Reinberg D. Mechanism of ATP-dependent promoter melting by transcription factor IIH. Science. 2000;288:1418–1422. doi: 10.1126/science.288.5470.1418. [DOI] [PubMed] [Google Scholar]
- 15.Miller G, Hahn S. A DNA-tethered cleavage probe reveals the path for promoter DNA in the yeast preinitiation complex. Nat Struct Mol Biol. 2006;13:603–610. doi: 10.1038/nsmb1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murakami K, et al. Architecture of an RNA Polymerase II Transcription Pre-Initiation Complex. Science. 2013 doi: 10.1126/science.1238724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fishburn J, Tomko E, Galburt E, Hahn S. Double-stranded DNA translocase activity of transcription factor TFIIH and the mechanism of RNA polymerase II open complex formation. Proceedings of the National Academy of Sciences. 2015 doi: 10.1073/pnas.1417709112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He Y, Fang J, Taatjes DJ, Nogales E. Structural visualization of key steps in human transcription initiation. Nature. 2013:1–8. doi: 10.1038/nature11991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pal M, Ponticelli AS, Luse DS. The role of the transcription bubble and TFIIB in promoter clearance by RNA polymerase II. Mol Cell. 2005;19:101–110. doi: 10.1016/j.molcel.2005.05.024. [DOI] [PubMed] [Google Scholar]
- 20.Holstege FC, van der Vliet PC, Timmers HT. Opening of an RNA polymerase II promoter occurs in two distinct steps and requires the basal transcription factors IIE and IIH. EMBO Journal. 1996;15:1666–1677. [PMC free article] [PubMed] [Google Scholar]
- 21.Fazal FM, Meng CA, Murakami K, Kornberg RD, Block SM. Real-time observation of the initiation of RNA polymerase II transcription. Nature. 2015;525:274–277. doi: 10.1038/nature14882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Revyakin A, Ebright RH, Strick TR. Promoter unwinding and promoter clearance by RNA polymerase: detection by single-molecule DNA nanomanipulation. Proc Natl Acad Sci USA. 2004;101:4776–4780. doi: 10.1073/pnas.0307241101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Revyakin A, Liu C, Ebright RH, Strick TR. Abortive initiation and productive initiation by RNA polymerase involve DNA scrunching. Science. 2006;314:1139–1143. doi: 10.1126/science.1131398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Howan K, et al. Initiation of transcription-coupled repair characterized at single-molecule resolution. Nature. 2012:1–4. doi: 10.1038/nature11430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barnes CO, et al. Crystal Structure of a Transcribing RNA Polymerase II Complex Reveals a Complete Transcription Bubble. Mol Cell. 2015;59:258–269. doi: 10.1016/j.molcel.2015.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Majovski RC, Khaperskyy DA, Ghazy MA, Ponticelli AS. A functional role for the switch 2 region of yeast RNA polymerase II in transcription start site utilization and abortive initiation. J Biol Chem. 2005;280:34917–34923. doi: 10.1074/jbc.M502932200. [DOI] [PubMed] [Google Scholar]
- 27.Conesa C, Acker J. Sub1/PC4 a chromatin associated protein with multiple functions in transcription. RNA Biol. 2010;7:287–290. doi: 10.4161/rna.7.3.11491. [DOI] [PubMed] [Google Scholar]
- 28.Buratowski S, Hahn S, Guarente L, Sharp PA. Five intermediate complexes in transcription initiation by RNA polymerase II. Cell. 1989;56:549–561. doi: 10.1016/0092-8674(89)90578-3. [DOI] [PubMed] [Google Scholar]
- 29.Fishburn J, Hahn S. Architecture of the yeast RNA polymerase II open complex and regulation of activity by TFIIF. Mol Cell Biol. 2012;32:12–25. doi: 10.1128/MCB.06242-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spangler L, Wang X, Conaway JW, Conaway RC, Dvir A. TFIIH action in transcription initiation and promoter escape requires distinct regions of downstream promoter DNA. Proc Natl Acad Sci USA. 2001;98:5544–5549. doi: 10.1073/pnas.101004498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crick FH, Klug A. Kinky helix. Nature. 1975;255:530–533. doi: 10.1038/255530a0. [DOI] [PubMed] [Google Scholar]
- 32.Wiggins PA, Phillips R, Nelson PC. Exact theory of kinkable elastic polymers. Physical review E, Statistical, nonlinear, and soft matter physics. 2005;71:021909. doi: 10.1103/PhysRevE.71.021909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vafabakhsh R, Ha T. Extreme Bendability of DNA Less than 100 Base Pairs Long Revealed by Single-Molecule Cyclization. Science. 2012;337:1097–1101. doi: 10.1126/science.1224139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Plaschka C, et al. Transcription initiation complex structures elucidate DNA opening. Nature. 2016;533:353–358. doi: 10.1038/nature17990. [DOI] [PubMed] [Google Scholar]
- 35.Robinson PJ, et al. Structure of a Complete Mediator-RNA Polymerase II Pre-Initiation Complex. Cell. 2016;166:1411–1422.e16. doi: 10.1016/j.cell.2016.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Giardina C, Lis JT. DNA melting on yeast RNA polymerase II promoters. Science. 1993;261:759–762. doi: 10.1126/science.8342041. [DOI] [PubMed] [Google Scholar]
- 37.Zhang D-Y, Carson DJ, Ma J. The role of TFIIB-RNA polymerase II interaction in start site selection in yeast cells. Nucleic Acids Res. 2002;30:3078–3085. doi: 10.1093/nar/gkf422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mühlbacher W, et al. Conserved architecture of the core RNA polymerase II initiation complex. Nature Communications. 2014;5:4310. doi: 10.1038/ncomms5310. [DOI] [PubMed] [Google Scholar]
- 39.Yan M, Gralla JD. The use of ATP and initiating nucleotides during postrecruitment steps at the activated adenovirus E4 promoter. J Biol Chem. 1999;274:34819–34824. doi: 10.1074/jbc.274.49.34819. [DOI] [PubMed] [Google Scholar]
- 40.Holstege FC, Fiedler U, Timmers HT. Three transitions in the RNA polymerase II transcription complex during initiation. EMBO Journal. 1997;16:7468–7480. doi: 10.1093/emboj/16.24.7468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fuss JO, Tainer JA. XPB and XPD helicases in TFIIH orchestrate DNA duplex opening and damage verification to coordinate repair with transcription and cell cycle via CAK kinase. DNA Repair (Amst) 2011;10:697–713. doi: 10.1016/j.dnarep.2011.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Compe E, Egly JM. TFIIH: when transcription met DNA repair. Nat Rev Mol Cell Biol. 2012;13:343–354. doi: 10.1038/nrm3350. [DOI] [PubMed] [Google Scholar]
- 43.Kuehner JN, Brow DA. Quantitative analysis of in vivo initiator selection by yeast RNA polymerase II supports a scanning model. J Biol Chem. 2006;281:14119–14128. doi: 10.1074/jbc.M601937200. [DOI] [PubMed] [Google Scholar]
- 44.Galburt EA, Tomko EJ, Stump WT, Ruiz Manzano A. Force-dependent melting of supercoiled DNA at thermophilic temperatures. Biophys Chem. 2014;187–188C:23–28. doi: 10.1016/j.bpc.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 45.Schlingman D, Mack A, Mochrie S. A new method for the covalent attachment of DNA to a surface for single-molecule studies. Colloids and Surfaces B: …. 2010 doi: 10.1016/j.colsurfb.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Revyakin A, Allemand JF, Croquette V, Ebright RH, Strick TR. Single-molecule DNA nanomanipulation: detection of promoter-unwinding events by RNA polymerase. Meth Enzymol. 2003;370:577–598. doi: 10.1016/S0076-6879(03)70049-4. [DOI] [PubMed] [Google Scholar]
- 47.Cost GJ. Enzymatic ligation assisted by nucleases: simultaneous ligation and digestion promote the ordered assembly of DNA. Nat Protoc. 2007;2:2198–2202. doi: 10.1038/nprot.2007.325. [DOI] [PubMed] [Google Scholar]
- 48.Seol Y, Neuman KC. Magnetic tweezers for single-molecule manipulation. Methods Mol Biol. 2011;783:265–293. doi: 10.1007/978-1-61779-282-3_15. [DOI] [PubMed] [Google Scholar]
- 49.Cnossen JP, Dulin D, Dekker NH. An optimized software framework for real-time, high-throughput tracking of spherical beads. Rev. Sci. Instrum. 2014;85:103712. doi: 10.1063/1.4898178. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source data for the distributions shown in Fig. 2, 4, and 5 are available with the paper online. Other data generated and analyzed in this study are available from the corresponding author on request. In addition, a Life Sciences Reporting Summary for this article is available.