Abstract
The epigenome is sensitive to the availability of metabolites that serve as substrates of chromatin-modifying enzymes. Links between acetyl-CoA metabolism, histone acetylation, and gene regulation have been documented, although how specificity in gene regulation is achieved by a metabolite has been challenging to answer. Recent studies suggest that acetyl-CoA metabolism is tightly regulated both spatially and temporally, to elicit responses to nutrient availability and signaling cues. Here we discuss evidence that acetyl-CoA production is distinctly regulated in the nucleus and cytosol in mammalian cells. Recent findings indicate that acetyl-CoA availability for site-specific histone acetylation is influenced through post-translational modification of acetyl-CoA producing enzymes, as well as through dynamic regulation of the nuclear localization and chromatin recruitment of these enzymes.
Keywords: Acetyl-CoA, compartmentalization, metabolism, epigenetics, acetylation
Metabolic regulation of the epigenome
Control of nutrient uptake and utilization is fundamental for cell viability and function. In addition to maintaining bioenergetics and supplying the building blocks and reducing equivalents needed for macromolecular biosynthesis, it is increasingly recognized that metabolites also play diverse signaling roles [1,2]. For example, the chemical modifications on DNA and histones are highly sensitive to changes in cellular metabolism, since many chromatin-modifying enzymes use metabolites as substrates. Global levels of histone acetylation are responsive to acetyl-CoA abundance, while levels of the methyl donor S-adenosylmethionine (SAM) can influence histone and DNA methylation. Additionally, JmjC domain-containing histone demethylases and TET 5-methylcytosine hydroxylases employ α-ketoglutarate (α-KG) as a co-substrate and are competitively inhibited by the structurally similar metabolites fumarate, succinate, (R)-2-hydroxyglutarate, and (S)-2-hydroxyglutarate [3–8].
Numerous studies have described roles for metabolic regulation of the epigenome in both normal physiology and in disease over the past several years [3–8]. Notably, many of these studies have reported that metabolic alterations regulate both global chromatin modification levels and expression of specific sets of genes. Yet, mechanisms that account for specificity in metabolic regulation of gene expression have largely remained a mystery. Emerging evidence, however, suggests that tight spatial and temporal control of metabolite production may be key to mediating specific molecular and functional outcomes, and this evidence will be discussed in this article.
Several recent reviews have covered the current knowledge of metabolism and its influence on chromatin modification in depth [3–8]. In this review, we discuss the evidence that spatiotemporal control of acetyl-CoA production and utilization contributes to precise regulation of gene expression and DNA repair, focusing on mammalian systems. We review the evidence that changes in acetyl-CoA production can impact transcriptional programs in a cell type- and context (i.e. nutrition, disease, microenvironment)-dependent manner. We then discuss data that point to a necessity for acetyl-CoA generation within the nucleus for efficient regulation of histone acetylation. Finally, we highlight recent studies that describe mechanisms of regulation of acetyl-CoA production within the nucleus to facilitate site-specific histone acetylation.
Acetyl-CoA compartmentalization and metabolism in mammalian cells
In mammalian cells, acetyl-CoA is produced within the mitochondria, cytosol, and nucleus. Mitochondrial acetyl-CoA can be generated through several pathways, depending on cell type and conditions [9,10], including: 1) pyruvate entry into mitochondria and its enzymatic conversion to acetyl-CoA by the pyruvate dehydrogenase complex (PDC), 2) fatty acid β-oxidation, 3) amino acid metabolism [10], 4) ketone body metabolism, and 5) ligation of acetate to CoA by mitochondrial acyl-CoA synthetase short chain family member 1 (ACSS1). Acetyl-CoA generated in mitochondria condenses with oxaloacetate to produce citrate, which is oxidized in the TCA cycle, enabling ATP production through oxidative phosphorylation.
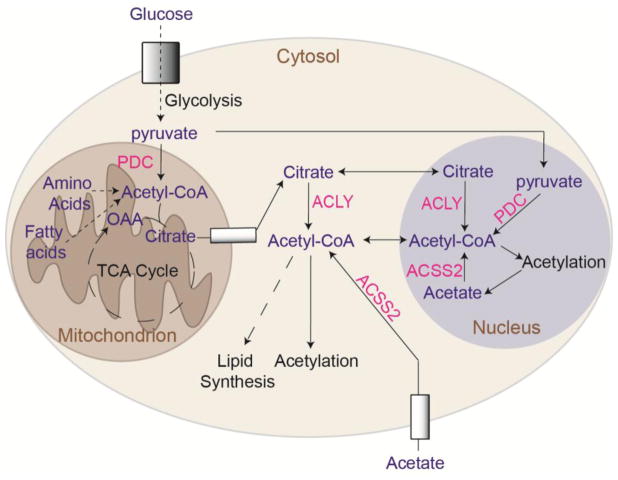
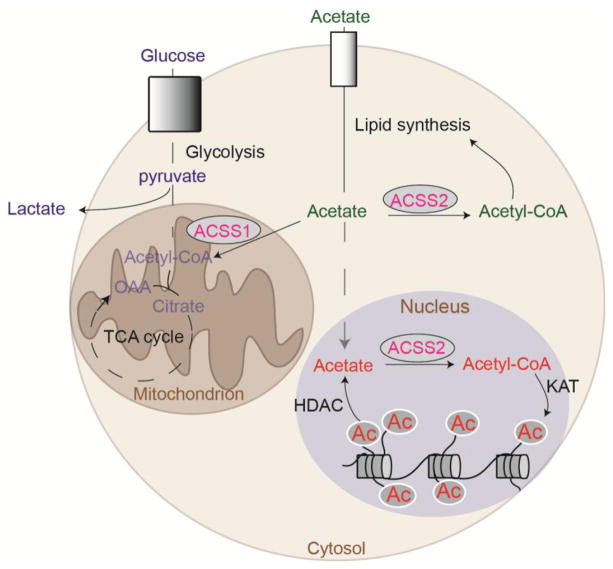
Citrate can be exported from mitochondria via the mitochondrial tricarboxylate transporter (SLC25A1), and acetyl-CoA and oxaloacetate are then regenerated in the cytosol and nucleus by ATP-citrate lyase (ACLY). Since acetyl-CoA cannot be directly transported across mitochondrial membranes, citrate export and cleavage by ACLY is a major mechanism by which acetyl-CoA is generated outside of mitochondria [9,11]. Within the cytosol, acetyl-CoA is used in biosynthetic processes, including synthesis of fatty acids and cholesterol. Acetyl-CoA is also the required acetyl donor for lysine acetylation, a post-translational modification that regulates the activity, stability, or localization of numerous cellular proteins [9,12]. Global histone acetylation levels are sensitive to the availability of acetyl-CoA in the cell, which fluctuates in response to nutrient availability or metabolic reprogramming [13–16]. Both lipid synthesis and histone acetylation depend on ACLY activity in many cell types [4,17] (Figure 1).
Figure 1. Acetyl-CoA production and utilization in mitochondria, cytosol, and nucleus.
Glucose, fatty acids, and amino acids can all contribute to mitochondrial acetyl-CoA production. Transfer of acetyl-CoA produced in mitochondria to the cytosol and nucleus relies on citrate export and cleavage by ACLY. ACSS2 generates acetyl-CoA from acetate in the nucleus and cytosol. The mitochondrial PDC has also been shown to translocate to the nucleus and to produce nuclear acetyl-CoA directly from pyruvate [9,11].
In addition to ACLY, another major source of acetyl-CoA outside of mitochondria is acyl-CoA synthetase short chain family member 2 (ACSS2), which is localized to the cytosol and nucleus. ACSS2 is involved in the capture and use of exogenous acetate, as well as in the recycling of acetate produced inside the cell by histone deacetylases (HDAC) reactions [18]. In addition to ACLY and ACSS2, which are present in the cytosol and nucleus, the PDC can translocate to the nucleus in certain conditions, such as mitochondrial stress, where it contributes acetyl-CoA for histone acetylation [19–21] (Figure 1).
Acetyl-CoA abundance impacts global histone acetylation levels while also modulating expression of specific genes
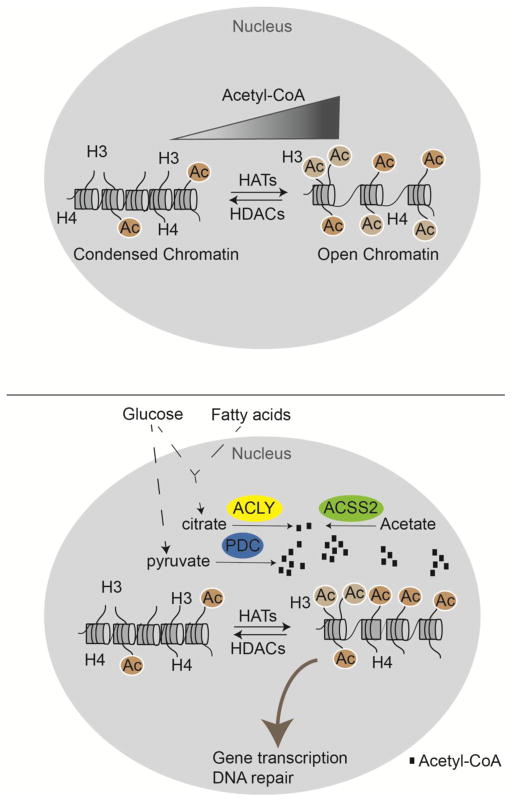
A close relationship between global levels of histone acetylation and acetyl-CoA production is observed in various cancer cell types, immortalized fibroblasts, and in normal cells of the mammalian body including adipocytes, endothelial cells, mesangial cells, macrophages, and T cells [14,16,22–29]. A link between acetyl-CoA and histone acetylation is also observed in lower organisms such as yeast and Drosophila [13,15,30]. As noted above, conditions that induce high global histone acetylation frequently also promote expression of specific sets of genes that facilitate an appropriate cellular response to availability of key nutrients, discussed below (Figure 2).
Figure 2. Acetyl-CoA availability is nutrient-dependent and dynamically regulates histone acetylation levels.
(A) Acetyl-CoA abundance correlates with global histone acetylation [13,14,27]. (B) ACLY, ACSS2, and PDC within the nucleus contribute to production of acetyl-CoA for histone acetylation, influencing gene transcription and DNA repair[15,16,19]. Substrates of these enzymes presumably can enter the nucleus through nuclear pores. The close proximity of mitochondria to the nucleus may facilitate citrate nuclear entry, possibly underlying the dominant role of ACLY in supplying acetyl-CoA for histone acetylation in many cell types. Acetate is produced within the nucleus by deacetylation of histones.
Glucose
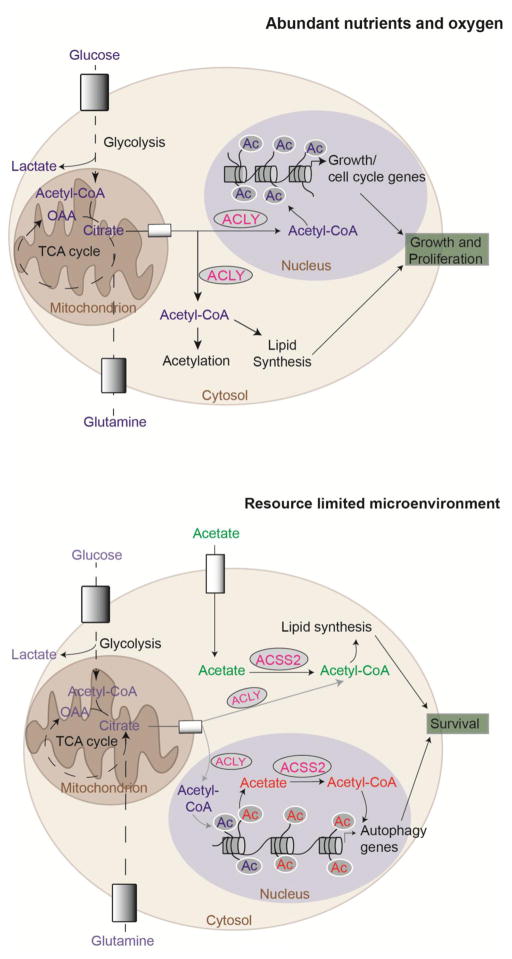
In proliferating cells in culture, glucose fuels the majority of acetyl-CoA production used for acetylating histones [14,31,32]. Glucose limitation or glycolytic inhibition suppresses both acetyl-CoA and histone acetylation levels [14,27], while AKT-dependent phosphorylation of ACLY at serine 455 (S455) enhances the ability to sustain acetyl-CoA production and histone acetylation under glucose limitation [14]. In addition to altering global levels of histone acetylation, several studies (discussed below) have reported that glucose-dependent acetyl-CoA production impacts expression of specific sets of genes to influence biological outcomes. For example, in cancer cells, histone acetylation levels decline with glucose limitation, and supplementation of acetate rescues both acetyl-CoA availability and global histone acetylation [14]. RNA-sequencing in glioblastoma cells revealed that about 10% of glucose- regulated genes were also responsive to acetate, suggesting that their expression is modulated by acetyl-CoA availability [14]. These acetyl-CoA-sensitive genes included many involved in cell cycle, DNA replication, and cell adhesion and migration [14], suggesting that acetyl-CoA levels serve as a signal to the cell that metabolic resources are sufficient for growth and proliferation (Figure 3).
Figure 3. Acetyl-CoA production supports proliferation when nutrients are abundant and survival when resources are limited.
(A) Under abundant nutrient and oxygen conditions, ACLY is the predominant source of acetyl-CoA in the cytosol and nucleus. ACLY may support proliferation by coordinating acetyl-CoA production for lipid synthesis and histone acetylation in concert. High acetyl-CoA abundance correlates with expression of growth and/or cell cycle genes [13,14,22,41]. (B) Under glucose and/or oxygen limited conditions, acetate becomes an important contributor to acetyl-CoA in the cytoplasm and nucleus, via ACSS2. ACSS2 promotes survival under metabolic stress conditions through leveraging extracellular acetate to supply acetyl-CoA for lipid synthesis and by utilizing acetate salvaged from deacetylase reactions to regulate histone acetylation at and expression of autophagy and lysosomal biogenesis genes [60,66,57]. ACLY also contributes to lipid synthesis in hypoxia using citrate generated by glutamine reductive carboxylation [87,88]. When phosphorylated at S455 (an AKT site), ACLY also promotes sustained histone acetylation under glucose limitation [14].
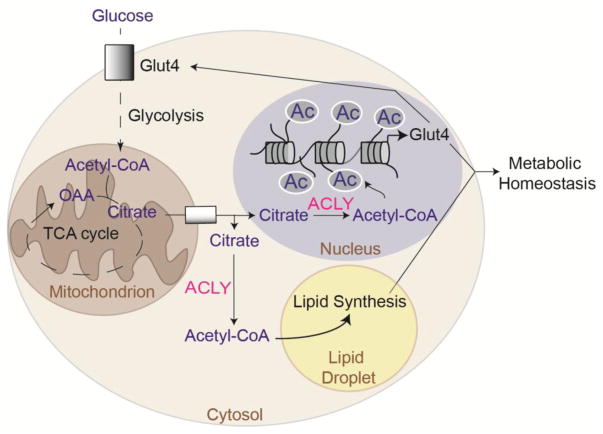
During adipocyte differentiation in vitro, global histone acetylation increases in a glucose- and ACLY-dependent manner [16]. Although at least a portion of the adipogenesis transcriptional program can be implemented in the absence of ACLY, expression of Slc2a4, encoding the insulin responsive glucose transporter GLUT4, is upregulated during differentiation in a glucose- and ACLY-dependent manner, correlating with global histone acetylation levels and histone acetylation at the Slc2a4 locus [16]. Slc2a4 is also selectively suppressed upon genetic deletion of Acly from adipocytes in vivo [22]. ACLY levels in adipocytes increase upon carbohydrate feeding and are potently suppressed in response to high fat feeding [25,33–35], suggesting that ACLY participates in a positive feedback loop to fine-tune the transcriptional response to carbohydrates in adipocytes (Figure 4). Notably, both GLUT4 expression and glucose-dependent de novo lipid synthesis (DNL) in adipocytes are associated with the maintenance of systemic insulin sensitivity [35–38]; hence, it is plausible that the suppression of ACLY in adipocytes in obesity contributes to the development of insulin resistance, although this remains to be tested. Interestingly, in high glucose conditions, ACLY was also found to promote histone acetylation globally and at profibrogenic genes in kidney mesangial cells, potentially contributing to renal complications associated with diabetes and hyperglycemia [28].
Figure 4. ACLY mediates a feed-forward mechanism to regulate GLUT4 levels in response to glucose uptake in adipocytes.
Adipocytes take up glucose in response to insulin via the GLUT4 glucose transporter. Glucose-dependent lipid synthesis and histone acetylation are ACLY-dependent, and GLUT4 expression is regulated in an ACLY-dependent manner in adipocytes[16,22,25]. Both adipocyte de novo lipogenesis and adipocyte GLUT4 expression are associated with improved systemic insulin sensitivity [35,36,38]. Since ACLY levels in adipocytes are suppressed by high fat feeding and obesity [25], it is plausible that low ACLY levels contribute to development of insulin resistance via ACLY’s dual roles in gene regulation and lipid synthesis.
Activated T cells also rely heavily on aerobic glycolysis. Deletion of Ldha, encoding lactate dehydrogenase A (LDHA), reduces rates of glucose consumption and suppresses expression of the cytokine IFN-γ in murine T cells differentiated under T helper 1 (TH1) conditions. LDHA knockout TH1 cells exhibited lower levels of acetyl-CoA and reduced histone acetylation at the Ifng locus. Supplementation with acetate rescued H3K9ac at the Ifng promoter, as well as expression of Ifng. Mice lacking LDHA in T cells were protected from IFNγ-related autoinflammatory disease. These data indicate that metabolic signaling through acetyl-CoA plays a key role in modulating T cell differentiation [39].
Links between glucose availability, acetyl-CoA abundance, histone acetylation, and gene regulation, appear to be conserved from yeast to mammals. When grown under glucose-limited conditions, yeast enter into metabolic cycles in which oxygen consumption oscillates [40]. Acetyl-CoA abundance rises and falls over these metabolic cycles, peaking during periods of growth [13]. Histone acetylation levels also oscillate, both globally and at specific genomic loci corresponding to genes involved in growth and cell cycle progression[13,41]. This oscillation is in close alignment with acetyl-CoA abundance and is dependent on the acetyltransferase Gcn5 [13]. Notably, supplementation of acetate raises acetyl-CoA and histone acetylation levels and is sufficient to drive yeast into growth [13].
Fatty Acids
Fatty acid oxidation is an important source of mitochondrial and extra-mitochondrial acetyl-CoA in certain contexts, such as during M2 macrophage polarization or lymphangiogenesis, discussed below. Isotope-tracer analysis using [U-13C]-octanoate demonstrated that when fatty acids are the predominant source of acetyl-CoA for the TCA cycle, they also supply acetyl-CoA for histone acetylation [42].
One cell type that relies heavily on fatty acid oxidation is the M2 macrophage. Macrophage polarization is associated with distinct metabolic features. The pro-inflammatory M1 phenotype is associated with reliance on glycolysis, while the M2 phenotype, which is involved in resolving inflammation, utilizes fatty acid oxidation as a major source of ATP [43]. In M2 macrophages, interleukin 4 (IL-4) signaling was found to modulate expression of a subset of M2 genes through activation of AKT-ACLY signaling, which enhanced acetyl-CoA production and promoted histone acetylation at these genes [23]. Interestingly, the AKT- and ACLY-responsive genes in M2 macrophages included many genes involved in chemokine production, as well as cell cycle and DNA replication, suggesting that macrophage function is fine-tuned in response to metabolic resources [23,44].
Lymphangiogenesis was also recently shown to depend on fatty acid oxidation [26]. PROX1, a transcription factor that regulates development of the lymphatic system, activates expression of CPT1A, which is required for fatty acid import into mitochondria for β-oxidation. Fatty acid oxidation leads to increased acetyl-CoA production in an ACLY-dependent manner, promoting histone acetylation at PROX1 target genes to further reinforce lymphatic development [26].
The gastrointestinal tract is colonized by bacteria that ferment dietary fiber to produce short chain fatty acids such as butyrate [45]. Bacterially produced butyrate is transported into colonocytes (specialized epithelial cells of the colon that aid in absorption of nutrients and metabolites), where it is oxidized and serves as an energy source. Notably, 14C-butyrate tracer experiments revealed that butyrate is used to supply acetyl-CoA for histone acetylation in colonocytes in an ACLY-dependent manner [46]. In addition to supplying acetyl-CoA for use by KATs, butyrate can also boost histone acetylation through HDAC inhibition. HDAC inhibition by butyrate is pro-apoptotic, whereas butyrate oxidation to generate acetyl-CoA promotes expression of genes involved in cell proliferation, even though both processes favor increased global histone acetylation levels [46]. Cancerous colonocytes do not catabolize butyrate to a significant extent, allowing its levels to build up in the cell and inhibit HDAC enzymes. As a consequence of butyrate functioning predominantly as an HDAC inhibitor in cancer cells, it exerts tumor-suppressive effects; high fiber feeding of mice with colorectal cancer suppressed tumor growth in a manner dependent on microbial butyrate production [47].
Acetate
Acetate is normally present at ~50–200 μM in circulation[18,48–52], and can rise further upon ethanol consumption (to more than 500 μM] [53,54]), high fat feeding (to 300–400 μM [48]), intermittent fasting (~2–4 fold increase [55]), or during acute bacterial infection (to as high as 5 mM [56]). Acetate in circulation is primarily generated through the breakdown of dietary fiber by the gut microbiota [18], and changes in serum acetate following high fat feeding or intermittent fasting have been attributed to these microbes [48,55]. The liver also produces acetate during ethanol metabolism [53,54]. Moreover, within mammalian cells, the dynamic deacetylation of histones and other proteins results in acetate production [18,32,58]. Acetate is also generated through the hydrolysis of acetyl-CoA or other acetylated metabolites, and in certain contexts (i.e. virus infection), it can be generated directly from pyruvate [18,59]. For a more in depth discussion of sources of acetate, the reader is referred to reference [18].
Exogenous acetate is normally dispensable to most cells, but can become a key source of acetyl-CoA in tumors or under metabolic stress conditions, particularly hypoxia or fasting [60–64]. Several recent studies have implicated acetate conversion to acetyl-CoA by ACSS2 in gene regulation in low oxygen and glucose conditions (Figure 3). ACSS2 was shown to regulate erythropoiesis under conditions of hypoxia and anemia by promoting the expression of EPO, encoding erythropoietin, in a hypoxia-inducible factor-2 (HIF-2) and CREB binding protein (CBP)-dependent manner [65]. In hepatocellular carcinoma cell lines, acetate enhanced histone acetylation at promoters of key lipogenic genes in hypoxia, promoting cell survival [66]. Moreover, under glucose deprivation, AMPK was found to phosphorylate ACSS2, promoting its nuclear translocation and facilitating expression of genes associated with autophagy and lysosomal biosynthesis [57].
Cellular acetyl-CoA levels and regulation of KATs
The large body of evidence that histone acetylation levels are sensitive to acetyl-CoA availability raises questions on the cellular concentrations of acetyl-CoA and whether they are within range to regulate KAT activity. Over the yeast metabolic cycle, acetyl-CoA levels oscillate in the range of 3–30 μM [13]. Similarly, whole cell acetyl-CoA concentrations in human cancer cells have been measured at ~3–20 μM, depending on nutrient availability, with acetyl-CoA levels declining upon nutrient limitation [14,67]. There is evidence that much of this acetyl-CoA is extra-mitochondrial in proliferating cells in culture [22,67], although measurement of the absolute concentrations of acetyl-CoA specifically within nuclei has not yet been achieved. Many KATs have a Kd for acetyl-CoA in the low μM or high nM range [68,69]. KATs are also inhibited by their product CoA, with varying sensitivity, and thus some KATs are likely sensitive to fluctuations in the acetyl-CoA: CoA ratio [14,69,70]. A positive relationship between histone acetylation levels and the acetyl-CoA: CoA ratio has been documented in several studies [14,25,26], although analysis of metabolite levels upon glycolysis inhibition in colon cancer cells found that histone acetylation correlated positively with acetyl-CoA levels, but negatively with the acetyl-CoA: CoA ratio [27]. Although these results may appear discrepant, whole cell metabolite measurements may reflect regulation in different subcellular compartments, underscoring the importance of elucidating how acetyl-CoA and other metabolite substrates of chromatin-modifying enzymes are regulated within the nucleus.
Are nuclear and cytosolic pools of acetyl-CoA distinct?
Nuclear and cytosolic pools of acetyl-CoA are unlikely to be physically separated, since small molecules can passively diffuse through nuclear pores. Consistently, in budding yeast, targeting of acetyl-CoA synthetase to either the cytosol or the nucleus is sufficient to maintain histone acetylation [15]. Moreover, reducing acetyl-CoA use in fatty acid synthesis by inhibition of acetyl-CoA carboxylase increases histone and transcription factor acetylation in yeast and mammalian cells, further indicating that nuclear and cytosolic pools interact [71–74]. Nevertheless, metabolite diffusion can be constrained in the cell by macromolecular crowding, and the nucleus in particular contains very high concentrations of proteins and DNA. Thus, the complex intracellular milieu could render spatial coordination of metabolism critical for specific functions [75,76].
Indeed, recent evidence suggests that lipid synthesis in the cytosol and histone acetylation in the nucleus may not access a uniform nuclear-cytosolic pool of acetyl-CoA. A kinetic flux analysis of 13C-glucose, 13C-glutamine, and 13C-acetate use for fatty acid synthesis under hypoxic conditions found that the demand for acetyl-CoA for lipogenesis was 2–3 fold higher than the rate of acetate uptake into the cell, suggesting that upon entering the cell, much of exogenous acetate is promptly captured for lipid synthesis in the cytosol by ACSS2, as well as for use in the TCA cycle by ACSS1 [32]. Although 13C-acetate use for histone acetylation increased in hypoxia, it remained a minor contributor to new histone acetylation, relative to glucose and glutamine [32]. Notably, hypoxia or low glucose conditions promote ACSS2 nuclear translocation. Under these conditions, nuclear ACSS2 facilitates the recycling of the acetate produced by HDAC reactions, instead of incorporating exogenous acetate [32,57]. These data indicate that ACSS2 in the nucleus may primarily rely on a locally generated acetate pool for histone acetylation, while ACSS2 in the cytosol accesses acetate taken up from the environment for lipid synthesis (Figure 5). Accordingly, silencing of ACSS2 in hypoxia suppresses global histone acetylation levels and promotes loss of acetate from the cell, in a manner reversible by HDAC inhibition [32].
Figure 5. Acetate is recycled within the nucleus for regulation of histone acetylation while exogenous acetate is taken up and used for lipid synthesis.
In hypoxia or in the absence of ACLY, ACSS2 is upregulated. Exogenous acetate becomes an important source of acetyl-CoA for fatty acid synthesis, and recent evidence indicates that it is inefficiently used for histone acetylation. Nuclear ACSS2 may instead primarily utilize acetate produced locally by HDAC reactions, allowing the recycling of acetate for the turnover of histone acetylation. Thus, acetyl-CoA may be produced very near to the site of its utilization [22,32].
Consistent with these findings, evidence from Acly−/− mouse embryonic fibroblasts (MEFs) also points to an inefficient use of exogenous acetate for histone acetylation [22]. Genetic deletion of Acly in MEFs potently inhibited 13C-glucose incorporation into fatty acids, HMG-CoA (an intermediate in the mevalonate pathway), and histone acetylation. At a physiologically relevant concentration of exogenous acetate (i.e. 100 μM), ACLY-deficient cells switched from using predominantly glucose-derived acetyl-CoA to mainly using acetate-derived acetyl-CoA for fatty acid and HMG-CoA synthesis. Despite the fact that extra-mitochondrial acetyl-CoA was maintained by acetate in Acly−/− MEFs, histone acetylation levels remained low and exogenous acetate was inefficiently used for histone acetylation; Acly−/− cells required substantially higher concentrations of acetate (i.e. 1 mM) to restore histone acetylation levels to that in wild type cells [22]. Together, these data support a model in which nuclear and cytosolic acetyl-CoA pools may be to a large extent functionally distinct and suggest that acetyl-CoA might need to be produced in close proximity to where it is used (Figure 5).
Spatiotemporal regulation of acetyl-CoA producing enzymes
If local production of acetyl-CoA is needed for histone acetylation, then specificity in gene regulation and other chromatin-dependent processes could be achieved through spatial and temporal control of acetyl-CoA producing enzymes. Indeed, recent studies have revealed that the nuclear accumulation of these enzymes, as well as their specific recruitment to target genes, is subject to precise regulation.
ACLY
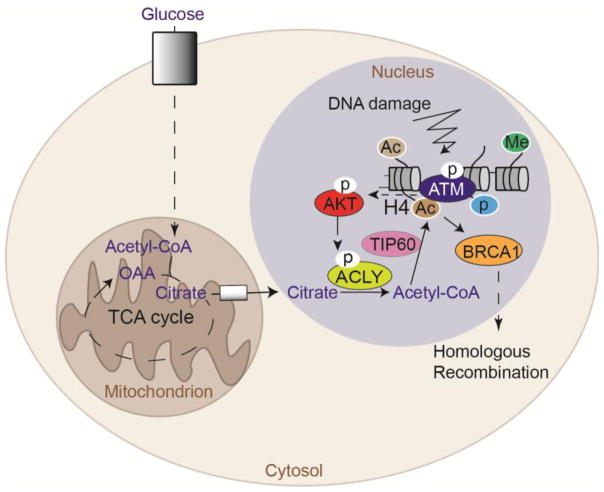
ACLY is present in both the nucleus and cytosol, although it lacks a canonical nuclear localization signal (NLS) and the conditions governing its nuclear localization remain poorly understood. Initial insight into the spatial and temporal regulation of ACLY within the nucleus has come from recent examination of its role in the DNA damage response [77]. Upon exposure to ionizing radiation, nuclear ACLY is dynamically phosphorylated at S455, a modification that enhances its activity [78], in an ataxia telangiectasia mutated (ATM)- and AKT-dependent manner [77]. Acetylation of histone H4 lysine 16 and H2A lysine 15 at nucleosomes flanking sites of DNA double strand breaks (DSBs) promotes repair by homologous recombination (HR) by interfering with the binding of non-homologous end joining (NHEJ) factor 53BP1, allowing recruitment of the pro-HR factor BRCA1 [79,80]. ACLY was found to be required for damage- induced histone H4 acetylation near DSBs, and in the absence of ACLY, BRCA1 recruitment and repair by HR were impaired. Moreover, ACLY failed to promote BRCA1 recruitment if it was excluded from the nucleus or if S455 was mutated to prevent phosphorylation, pointing to a tight spatial and temporal control of acetyl-CoA production during the DNA damage response. Intriguingly, nuclear ACLY levels were found to oscillate over the cell cycle, accumulating in S- and G2- cell cycle phases, thus potentially allowing cells to provision acetyl-CoA near DSB sites to promote histone acetylation and HR in S and G2, when HR is favored [77] (Figure 6).
Figure 6. Nuclear ACLY provides acetyl-CoA for histone acetylation near DNA double strand break sites to promote DNA repair by homologous recombination.
Nuclear ACLY S455 phosphorylation, which occurs downstream of ATM and AKT following exposure of cells to ionizing radiation, promotes histone acetylation near double strand break sites to enable BRCA1 recruitment and double strand break repair by homologous recombination [77].
ACSS2
ACSS2 is also present in both the nucleus and cytosol, and increased nuclear localization is observed under conditions of oxygen and/or glucose limitation [32,57,81]. As noted above, AMPK was recently shown to interact with and promote phosphorylation of ACSS2 at a conserved serine residue (S659), leading to ACSS2 nuclear translocation. Within the nucleus, ACSS2 interacts with transcription factor EB (TFEB), which binds to a consensus motif in promoters of autophagy and lysosomal biogenesis genes. This interaction with TFEB enables ACSS2 to recycle acetate produced in the nucleus by HDACs in order to facilitate local histone acetylation at TFEB target genes [57] (Figure 3).
Nuclear ACSS2 is also observed in neurons, where it is implicated in memory formation. ACSS2 was found to promote neuronal gene expression in a differentiation model, and ChIP-Seq analysis of its binding to chromatin revealed that 80% of ACSS2 peaks were overlapping with or were near H3 and H4 acetylation peaks, pointing to a role for ACSS2 in providing a localized acetyl-CoA pool for site-specific histone acetylation [82]. ACSS2 silencing in the mouse hippocampus suppressed expression of memory-related genes and impaired the formation of spatial memories [82].
Finally, there is evidence in yeast that Acs2, the orthologue of mammalian ACSS2, exists as a larger complex of metabolic enzymes called SESAME that associates with the Set1 histone methyltransferase complex to mediate dynamic chromatin modification in a manner responsive to glycolysis and serine synthesis. SESAME-- which includes the yeast homolog of PKM2 (Pyk1), Acs2, and various metabolic enzymes involved in serine and SAM synthesis-- is recruited to target genes such as PYK1, facilitating a positive feedback loop [83].
PDC
Pyruvate dehydrogenase complex (PDC) is a multi-enzyme complex crucial for conversion of pyruvate to acetyl-CoA in mitochondria. The presence of a functional PDC in the nucleus was recently identified [19], and the PDC has been found to enter the nucleus during conditions of mitochondrial stress, in response to growth factor or oncogenic signaling, and at distinct developmental stages [19–21]. Nuclear PDC enables direct production of acetyl-CoA from pyruvate and can thus bypass the canonical ACLY-dependent production of acetyl-CoA [19] (Figure 2).
Nuclear PDC has been found to form a complex with the arylhydrocarbon receptor (AhR), the acetyltransferase p300, and PKM2 to regulate histone acetylation at AhR target genes and promote their expression. The data suggest that PKM2 produces pyruvate in proximity to the PDC to enable local acetyl-CoA production and histone acetylation [84], providing additional evidence that multi-step metabolite channeling within nuclei may be engaged for chromatin modification and gene regulation.
Conclusions
The dynamic regulation of the post-translational modification and nuclear localization of acetyl-CoA-producing enzymes suggest a need for spatiotemporal control of acetyl-CoA generation for different functions. While only a few studies to date have described the recruitment of acetyl-CoA producing enzymes to specific genomic loci using chromatin immunoprecipitation experiments, the available data suggest that the provisioning of acetyl-CoA locally may be a key mechanism of gene regulation. Considering constraints on metabolite diffusion within the nucleus, actively regulating acetyl-CoA production at the right time and place may be crucial to ensuring that substrate is available to KATs when needed.
A key goal for future investigation will be to delineate the advantages gained by cells through engaging ACLY, ACSS2, or PDC for chromatin regulation in different conditions. For example, it is notable that when nutrient availability is high and cells are proliferating, ACLY regulates histone acetylation by controlling net acetyl-CoA input, while acetate recycling by ACSS2 becomes important when metabolic resources (glucose, oxygen) are limited (Fig 3). The proximity of mitochondria to the nucleus may facilitate the nuclear provisioning of citrate to allow the ACLY-dependent regulation of histone acetylation to occur in concert with ACLY-dependent lipid synthesis in the proliferating cell (Fig 3). On the other hand, when nutrients are limited, cells may rely on acetate recycling in order to devote available nutrients to ATP production rather than to chromatin regulation. Dependence on ACSS2 for histone acetylation in cells that are not proliferating (such as post-mitotic neurons) could also simply reflect a lesser need for net carbon input into histone acetylation. Nuclear PDC has been found to be important during a distinct developmental stage (2-cell embryo in mice) in which mitochondria are thought to be minimally active, as well as in cancer cells in response to stimuli such as mitochondrial stressors [19,20]. It is thus tempting to speculate that cancer cells may coopt a mechanism normally employed at discrete times in embryonic development to bring the PDC to the nucleus, potentially enabling new acetyl-CoA input into histone acetylation to sustain survival, growth, and proliferation when faced with mitochondrial stress. Further work will be needed to better understand the interplay between these enzymes and their capacity to compensate for one another during stress responses and in disease.
Additional investigation is needed to elucidate the mechanisms through which acetyl-CoA producing enzymes are trafficked in and out of the nucleus, to define their compartment-specific interacting partners, and to understand the distinct mechanisms of regulation (i.e. activity, stability, etc.) of these enzymes within each compartment- see outstanding questions. In addition, it will be important to consider how systemic changes in metabolism can impact the activity of acetyl-CoA producing enzymes. Food intake, dietary composition, alcohol consumption, and changes in the composition of the gut microbiome can all alter acetyl-CoA abundance and chromatin modification within tissues [25,48,54,85]. Thus, while ACSS2 and ACLY levels are regulated by nutrition, how changes in their levels impact the transcriptome in different tissues is an open question. From a therapeutic perspective, there is growing interest in targeting metabolic enzymes, including ACLY and ACSS2, in cancer and metabolic diseases [61,86]. Delineating how these enzymes co-operate and compensate for one another within distinct cellular compartments will be crucial for effective targeting of these enzymes in oncology. Our understanding of the spatiotemporal control of acetyl-CoA metabolism is still in its infancy; yet, recent advances in this area provide an impetus for further research into the roles of local metabolite production in epigenetic regulation.
Outstanding questions box.
What are acetyl-CoA concentrations within the nucleus and how is nuclear production of acetyl-CoA impacted by nutrient availability and signaling cues?
Do niches exist within the nucleus where the production of acetyl-CoA is concentrated?
What are the mechanisms regulating the nuclear localization and chromatin recruitment of acetyl-CoA producing enzymes?
What other proteins physically interact with acetyl-CoA producers within the nucleus?
How rapid is acetyl-CoA turnover in the cell? Are the kinetics of acetyl-CoA production and utilization different in the nucleus and cytosol?
Can acetate deposited on chromatin be mobilized for use in metabolic processes depending on the needs of the cell?
Trend Box.
Global levels of histone acetylation are responsive to acetyl-CoA abundance. Acetyl-CoA production has been shown to modulate transcriptional responses in various conditions and cell types, although the mechanisms underlying specificity in gene regulation by acetyl-CoA have been unclear.
Despite the permeability of nuclear pores to acetyl-CoA, recent metabolic evidence suggests that nuclear and cytosolic pools of acetyl-CoA may be, in large part, functionally distinct.
The acetyl-CoA producing enzymes ACLY, ACSS2, and PDC are present in the nucleus, and the nuclear accumulation of each of these enzymes is regulated by specific stimuli.
Recent studies have found that ACSS2 is recruited to chromatin, where it facilitates local histone acetylation and gene regulation.
Acknowledgments
The authors acknowledge NCI grants R01 CA174761 and R21CA 194973. We thank members of the Wellen lab for helpful discussion and feedback on the manuscript.
Glossary
- PDC
Pyruvate dehydrogenase complex. Multi-subunit complex comprised of three enzymes (E1, pyruvate dehydrogenase; E2, dihydrolipoyl transacetylase; E3, dihydrolipoyl dehydrogenase) that converts pyruvate into acetyl-CoA in the mitochondria. PDC can also be localized and functional in the nucleus.
- ACSS1
Acyl-CoA synthetase short-chain family member 1, catalyzes ATP-dependent ligation of acetate to CoA to generate mitochondrial acetyl-CoA.
- ACLY
ATP-citrate lyase. Enzyme that catalyzes the production of acetyl-CoA and oxaloacetate from citrate and CoA, in an ATP-dependent manner, in the cytosol and nucleus.
- ACSS2
Acyl-CoA synthetase short chain family member 2, catalyzes ATP-dependent ligation of acetate to CoA to generate acetyl-CoA in the cytosol and nucleus.
- HDAC
Histone deacetylase, catalyzes the removal of acetyl groups from acetylated lysine residues of histone and non-histone proteins.
- AKT
Serine/threonine protein kinase, also called protein kinase B (PKB). The name AKT is derived from the mouse strain used (Ak) and for the transformative potential of the kinase to generate thymic lymphomas (t). AKT is a key activator of pro-proliferative and pro-survival pathways.
- KAT
Lysine (K) acetyltransferase. KATs use acetyl-CoA as a substrate to catalyze the transfer of acetyl groups to lysine residues on histones and non-histone proteins.
- CBP
CREB binding protein. CBP is a lysine acetyltransferase and functions as a transcriptional coactivator, scaffolding protein, and chromatin remodeler. CBP acetylates both histone and non-histone proteins and has diverse biological roles.
- Kd
Dissociation constant for formation of the enzyme-substrate complex. The Kd is a measure of affinity, with a high Kd indicating lower affinity. Thus, the Kd of an enzyme for its substrate indicates the concentration of substrate at which half of the molecules of enzyme are associated with substrate.
- ATM
Serine/threonine protein kinase encoded by the gene ataxia telangiectasia mutated (ATM). ATM belongs to the PI3/PI4-kinase family and phosphorylates numerous substrates in response to activation of the DNA damage response pathway. ATM also regulates cell cycle progression in response to DNA damage.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pavlova NN, Thompson CB. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016;23:27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wellen KE, Thompson CB. A two-way street: reciprocal regulation of metabolism and signalling. Nat Rev Mol Cell Biol. 2012;13:270–276. doi: 10.1038/nrm3305. [DOI] [PubMed] [Google Scholar]
- 3.Berger SL, Sassone-Corsi P. Metabolic signaling to chromatin. Cold Spring Harb Perspect Biol. 2016;8:1–23. doi: 10.1101/cshperspect.a019463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kinnaird A, et al. Metabolic control of epigenetics in cancer. Nat Rev Cancer. 2016;16:694–707. doi: 10.1038/nrc.2016.82. [DOI] [PubMed] [Google Scholar]
- 5.Etchegaray JP, Mostoslavsky R. Interplay between Metabolism and Epigenetics: A Nuclear Adaptation to Environmental Changes. Mol Cell. 2016;62:695–711. doi: 10.1016/j.molcel.2016.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao X, et al. Metabolic interactions with cancer epigenetics. Mol Aspects Med. 2017;54:50–57. doi: 10.1016/j.mam.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van der Knaap JA, Verrijzer CP. Undercover: Gene control by metabolites and metabolic enzymes. Genes Dev. 2016;30:2345–2369. doi: 10.1101/gad.289140.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Su X, et al. Metabolic control of methylation and acetylation. Curr Opin Chem Biol. 2016;30:52–60. doi: 10.1016/j.cbpa.2015.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pietrocola F, et al. Acetyl coenzyme A: A central metabolite and second messenger. Cell Metab. 2015;21:805–821. doi: 10.1016/j.cmet.2015.05.014. [DOI] [PubMed] [Google Scholar]
- 10.Corbet C, Feron O. Cancer cell metabolism and mitochondria: Nutrient plasticity for TCA cycle fueling. Biochim Biophys Acta - Rev Cancer. 2017;1868:7–15. doi: 10.1016/j.bbcan.2017.01.002. [DOI] [PubMed] [Google Scholar]
- 11.Carrer A, Wellen KE. Metabolism and epigenetics: A link cancer cells exploit. Curr Opin Biotechnol. 2015;34:23–29. doi: 10.1016/j.copbio.2014.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choudhary C, et al. The growing landscape of lysine acetylation links metabolism and cell signalling. Nat Rev Mol Cell Biol. 2014;15:536–50. doi: 10.1038/nrm3841. [DOI] [PubMed] [Google Scholar]
- 13.Cai L, et al. Acetyl-CoA Induces Cell Growth and Proliferation by Promoting the Acetylation of Histones at Growth Genes. Mol Cell. 2011;42:426–437. doi: 10.1016/j.molcel.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee JV, et al. Akt-dependent metabolic reprogramming regulates tumor cell Histone acetylation. Cell Metab. 2014;20:306–319. doi: 10.1016/j.cmet.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takahashi H, et al. Nucleocytosolic Acetyl-Coenzyme A Synthetase Is Required for Histone Acetylation and Global Transcription. Mol Cell. 2006;23:207–217. doi: 10.1016/j.molcel.2006.05.040. [DOI] [PubMed] [Google Scholar]
- 16.Wellen KE, et al. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324:1076–1080. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lemus NH, Mendivil CO. Adenosine triphosphate citrate lyase: Emerging target in the treatment of dyslipidemia. J Clin Lipidol. 2015;9:384–389. doi: 10.1016/j.jacl.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 18.Schug ZT, et al. The metabolic fate of acetate in cancer. Nat Rev Cancer. 2016;16:708–717. doi: 10.1038/nrc.2016.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sutendra G, et al. A Nuclear Pyruvate Dehydrogenase Complex Is Important for the Generation of Acetyl-CoA and Histone Acetylation. Cell. 2014;158:84–97. doi: 10.1016/j.cell.2014.04.046. [DOI] [PubMed] [Google Scholar]
- 20.Nagaraj R, et al. Nuclear Localization of Mitochondrial TCA Cycle Enzymes as a Critical Step in Mammalian Zygotic Genome Activation. Cell. 2017;168:210–223. doi: 10.1016/j.cell.2016.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi WY, et al. NOK mediates glycolysis and nuclear PDC associated histone acetylation. Front Biosci (Landmark Ed. 2017;22:1792–1804. doi: 10.2741/4572. [DOI] [PubMed] [Google Scholar]
- 22.Zhao S, et al. ATP-Citrate Lyase Controls a Glucose-to-Acetate Metabolic Switch. Cell Rep. 2016;17:1037–1052. doi: 10.1016/j.celrep.2016.09.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Covarrubias AJ, et al. Akt-mTORC1 signaling regulates Acly to integrate metabolic input to control of macrophage activation. Elife. 2016;5:1–19. doi: 10.7554/eLife.11612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Osinalde N, et al. Nuclear Phosphoproteomic Screen Uncovers ACLY as Mediator of IL-2-induced Proliferation of CD4+ T lymphocytes. Mol Cell Proteomics. 2016;15:2076–2092. doi: 10.1074/mcp.M115.057158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carrer A, et al. Impact of a high-fat diet on tissue Acyl-CoA and histone acetylation levels. J Biol Chem. 2017;292:3312–3322. doi: 10.1074/jbc.M116.750620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wong BW, et al. The role of fatty acid β-oxidation in lymphangiogenesis. Nature. 2016;542:49–54. doi: 10.1038/nature21028. [DOI] [PubMed] [Google Scholar]
- 27.Cluntun AA, et al. The rate of glycolysis quantitatively mediates specific histone acetylation sites. Cancer Metab. 2015;3:1–12. doi: 10.1186/s40170-015-0135-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deb DK, et al. ATP-citrate lyase is essential for high glucose-induced histone hyperacetylation and fibrogenic gene upregulation in mesangial cells. Am J Physiol Renal Physiol. 2017;313:423–429. doi: 10.1152/ajprenal.00029.2017. [DOI] [PubMed] [Google Scholar]
- 29.Gao X, et al. Acetate functions as an epigenetic metabolite to promote lipid synthesis under hypoxia. Nat Commun. 2016;7:1–14. doi: 10.1038/ncomms11960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peleg S, et al. Life span extension by targeting a link between metabolism and histone acetylation in Drosophila. EMBO Rep. 2016;17:455–469. doi: 10.15252/embr.201541132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Evertts AG, et al. Quantitative dynamics of the link between cellular metabolism and histone acetylation. J Biol Chem. 2013;288:12142–12151. doi: 10.1074/jbc.M112.428318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bulusu V, et al. Acetate Recapturing by Nuclear Acetyl-CoA Synthetase 2 Prevents Loss of Histone Acetylation during Oxygen and Serum Limitation. Cell Rep. 2017;18:647–658. doi: 10.1016/j.celrep.2016.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang L, et al. Leptin contributes to the adaptive responses of mice to high-fat diet intake through suppressing the lipogenic pathway. PLoS One. 2009;4:1–9. doi: 10.1371/journal.pone.0006884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fukuda H, et al. Effects of nutrients and hormones on gene expression of ATP citrate-lyase in rat liver. Eur J Biochem. 1992;209:217–222. doi: 10.1111/j.1432-1033.1992.tb17279.x. [DOI] [PubMed] [Google Scholar]
- 35.Herman MA, et al. A novel ChREBP isoform in adipose tissue regulates systemic glucose metabolism. Nature. 2012;484:333–338. doi: 10.1038/nature10986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abel ED, et al. Adipose-selective targeting of the GLUT4 gene impairs insulin action in muscle and liver. Nature. 2001;409:729–733. doi: 10.1038/35055575. [DOI] [PubMed] [Google Scholar]
- 37.Shepherd PR, et al. Adipose cell hyperplasia and enhanced glucose disposal in transgenic mice overexpressing GLUT4 selectively in adipose tissue. J Biol Chem. 1993;268:22243–22246. [PubMed] [Google Scholar]
- 38.Cao H, et al. Identification of a Lipokine, a Lipid Hormone Linking Adipose Tissue to Systemic Metabolism. Cell. 2008;134:933–944. doi: 10.1016/j.cell.2008.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peng M, et al. Aerobic glycolysis promotes T helper 1 cell differentiation through an epigenetic mechanism. Science. 2016;354:481–484. doi: 10.1126/science.aaf6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tu BP. Logic of the Yeast Metabolic Cycle: Temporal Compartmentalization of Cellular Processes. Science. 2005;310:1152–1158. doi: 10.1126/science.1120499. [DOI] [PubMed] [Google Scholar]
- 41.Shi L, Tu BP. Acetyl-CoA induces transcription of the key G1 cyclin CLN3 to promote entry into the cell division cycle in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2013;110:7318–23. doi: 10.1073/pnas.1302490110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McDonnell E, et al. Lipids Reprogram Metabolism to Become a Major Carbon Source for Histone Acetylation. Cell Rep. 2016;17:1463–1472. doi: 10.1016/j.celrep.2016.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O’Neill LAJ, Pearce EJ. Immunometabolism governs dendritic cell and macrophage function. J Exp Med. 2016;213:15–23. doi: 10.1084/jem.20151570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Torres A, et al. Metabolism fine-tunes macrophage activation. Elife. 2016;5:2015–2017. doi: 10.7554/eLife.14354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Postler TS, Ghosh S. Understanding the Holobiont: How Microbial Metabolites Affect Human Health and Shape the Immune System. Cell Metab. 2017;26:110–130. doi: 10.1016/j.cmet.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Donohoe DR, et al. The Warburg Effect Dictates the Mechanism of Butyrate-Mediated Histone Acetylation and Cell Proliferation. Mol Cell. 2012;48:612–626. doi: 10.1016/j.molcel.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Donohoe DR, et al. A gnotobiotic mouse model demonstrates that dietary fiber protects against colorectal tumorigenesis in a microbiota- and butyrate-dependent manner. Cancer Discov. 2014;4:1387–1397. doi: 10.1158/2159-8290.CD-14-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Perry RJ, et al. Acetate mediates a microbiome-brain-β cell axis promoting metabolic syndrome. Nature. 2016;534:213–217. doi: 10.1038/nature18309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scheppach W, et al. The contribution of the large intestine to blood acetate in man. Clin Sci. 1991;80:177–182. doi: 10.1042/cs0800177. [DOI] [PubMed] [Google Scholar]
- 50.Skutches CL, et al. Plasma acetate turnover and oxidation. J Clin Invest. 1979;64:708–713. doi: 10.1172/JCI109513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tollinger CD, et al. Measurement of acetate in human blood by gas chromatography: Effects of sample preparation, feeding, and various diseases. Clin Chem. 1979;25:1787–1790. [PubMed] [Google Scholar]
- 52.Herrmann DBJ, et al. Role of gastrointestinal tract and liver in acetate metabolism in rat and man. Eur J Clin Invest. 1985;15:221–226. doi: 10.1111/j.1365-2362.1985.tb00172.x. [DOI] [PubMed] [Google Scholar]
- 53.Nuutinen H, et al. Elevated blood acetate as indicator of fast ethanol elimination in chronic alcoholics. Alcohol. 1985;2:623–626. doi: 10.1016/0741-8329(85)90090-4. [DOI] [PubMed] [Google Scholar]
- 54.Lundquist F, et al. ETHANOL METABOLISM AND PRODUCTION OF FREE ACETATE IN THE HUMAN LIVER. J Clin Invest. 1962;41:955–961. doi: 10.1172/JCI104574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li G, et al. Intermittent Fasting Promotes White Adipose Browning and Decreases Obesity by Shaping the Gut Microbiota. Cell Metab. 2017;26:672–685. doi: 10.1016/j.cmet.2017.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Balmer ML, et al. Memory CD8+ T Cells Require Increased Concentrations of Acetate Induced by Stress for Optimal Function. Immunity. 2016;44:1312–1324. doi: 10.1016/j.immuni.2016.03.016. [DOI] [PubMed] [Google Scholar]
- 57.Li X, et al. Nucleus-Translocated ACSS2 Promotes Gene Transcription for Lysosomal Biogenesis and Autophagy. Mol Cell. 2017;66:684–697. doi: 10.1016/j.molcel.2017.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McBrian MA, et al. Histone Acetylation Regulates Intracellular pH. Mol Cell. 2013;49:310–321. doi: 10.1016/j.molcel.2012.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vysochan A, et al. ACSS2-mediated acetyl-CoA synthesis from acetate is necessary for human cytomegalovirus infection. Proc Natl Acad Sci U S A. 2017;114:1528–1535. doi: 10.1073/pnas.1614268114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schug ZT, et al. Acetyl-CoA synthetase 2 promotes acetate utilization and maintains cancer cell growth under metabolic stress. Cancer Cell. 2015;27:57–71. doi: 10.1016/j.ccell.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Comerford SA, et al. Acetate dependence of tumors. Cell. 2014;159:1591–1602. doi: 10.1016/j.cell.2014.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mashimo T, et al. Acetate is a bioenergetic substrate for human glioblastoma and brain metastases. Cell. 2014;159:1603–1614. doi: 10.1016/j.cell.2014.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kamphorst JJ, et al. Quantitative analysis of acetyl-CoA production in hypoxic cancer cells reveals substantial contribution from acetate. Cancer Metab. 2014;2:1–8. doi: 10.1186/2049-3002-2-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sakakibara I, et al. Fasting-Induced Hypothermia and Reduced Energy Production in Mice Lacking Acetyl-CoA Synthetase 2. Cell Metab. 2009;9:191–202. doi: 10.1016/j.cmet.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 65.Xu M, et al. An acetate switch regulates stress erythropoiesis. Nat Med. 2014;20:1018–1026. doi: 10.1038/nm.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gao X, et al. Acetate functions as an epigenetic metabolite to promote lipid synthesis under hypoxia. Nat Commun. 2016;7:1–14. doi: 10.1038/ncomms11960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen WW, et al. Absolute Quantification of Matrix Metabolites Reveals the Dynamics of Mitochondrial Metabolism. Cell. 2016;166:1324–1337. doi: 10.1016/j.cell.2016.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Langer MR, et al. Modulating acetyl-CoA binding in the GCN5 family of histone acetyltransferases. J Biol Chem. 2002;277:27337–27344. doi: 10.1074/jbc.M203251200. [DOI] [PubMed] [Google Scholar]
- 69.Albaugh BN, et al. KAT(ching) metabolism by the tail: insight into the links between lysine acetyltransferases and metabolism. Chembiochem. 2011;12:290–298. doi: 10.1002/cbic.201000438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Montgomery DC, et al. Global Profiling of Acetyltransferase Feedback Regulation. J Am Chem Soc. 2016;138:6388–6391. doi: 10.1021/jacs.6b03036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Galdieri L, Vancura A. Acetyl-CoA carboxylase regulates global histone acetylation. J Biol Chem. 2012;287:23865–23876. doi: 10.1074/jbc.M112.380519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Galdieri L, et al. Activation of AMP-activated protein kinase by metformin induces protein acetylation in prostate and ovarian cancer cells. J Biol Chem. 2016;291:25154–25166. doi: 10.1074/jbc.M116.742247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang M, et al. The yeast AMPK homolog SNF1 regulates acetyl coenzyme A homeostasis and histone acetylation. Mol Cell Biol. 2013;33:4701–17. doi: 10.1128/MCB.00198-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rios Garcia M, et al. Acetyl-CoA Carboxylase 1-Dependent Protein Acetylation Controls Breast Cancer Metastasis and Recurrence. Cell Metab. 2017 doi: 10.1016/j.cmet.2017.09.018. [DOI] [PubMed] [Google Scholar]
- 75.Katada S, et al. Connecting threads: Epigenetics and metabolism. Cell. 2012;148:24–28. doi: 10.1016/j.cell.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 76.Theillet FX, et al. Physicochemical Properties of Cells and Their Effects on Intrinsically Disordered Proteins (IDPs) Chem Rev. 2014;114:6661–6714. doi: 10.1021/cr400695p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sivanand S, et al. Nuclear Acetyl-CoA Production by ACLY Promotes Homologous Recombination. Mol Cell. 2017;67:252–265. doi: 10.1016/j.molcel.2017.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Potapova IA, et al. Phosphorylation of recombinant human ATP:citrate lyase by cAMP-dependent protein kinase abolishes homotropic allosteric regulation of the enzyme by citrate and increases the enzyme activity. Allosteric activation of atp:citrate lyase by phosphorylated sug. Biochemistry. 2000;39:1169–1179. doi: 10.1021/bi992159y. [DOI] [PubMed] [Google Scholar]
- 79.Jacquet K, et al. The TIP60 Complex Regulates Bivalent Chromatin Recognition by 53BP1 through Direct H4K20me Binding and H2AK15 Acetylation. Mol Cell. 2016;62:409–421. doi: 10.1016/j.molcel.2016.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tang J, et al. Acetylation limits 53BP1 association with damaged chromatin to promote homologous recombination. Nat Struct Mol Biol. 2013;20:317–25. doi: 10.1038/nsmb.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chen R, et al. The acetate/ACSS2 switch regulates HIF-2 stress signaling in the tumor cell microenvironment. PLoS One. 2015;10:1–24. doi: 10.1371/journal.pone.0116515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mews P, et al. Acetyl-CoA synthetase regulates histone acetylation and hippocampal memory. Nature. 2017;546:381–386. doi: 10.1038/nature22405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li S, et al. Serine and SAM Responsive Complex SESAME Regulates Histone Modification Crosstalk by Sensing Cellular Metabolism. Mol Cell. 2015;60:408–421. doi: 10.1016/j.molcel.2015.09.024. [DOI] [PubMed] [Google Scholar]
- 84.Matsuda S, et al. Nuclear pyruvate kinase M2 complex serves as a transcriptional coactivator of arylhydrocarbon receptor. Nucleic Acids Res. 2016;44:636–647. doi: 10.1093/nar/gkv967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Krautkramer KA, et al. Diet-Microbiota Interactions Mediate Global Epigenetic Programming in Multiple Host Tissues. Mol Cell. 2016;64:982–992. doi: 10.1016/j.molcel.2016.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pinkosky SL, et al. Liver-specific ATP-citrate lyase inhibition by bempedoic acid decreases LDL-C and attenuates atherosclerosis. Nat Commun. 2016;7:1–13. doi: 10.1038/ncomms13457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Metallo CM, et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2012;481:380–4. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wise DR, et al. Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of α-ketoglutarate to citrate to support cell growth and viability. Proc Natl Acad Sci U S A. 2011;108:19611–6. doi: 10.1073/pnas.1117773108. [DOI] [PMC free article] [PubMed] [Google Scholar]