Abstract
Ubiquitination of TNFR1-signaling-complex (TNF-RSC) controls the activation of RIPK1, a kinase critically involved in mediating multiple TNFα activated deleterious events. However, the molecular mechanism that coordinates different types of ubiquitination modifications to regulate the activation of RIPK1 kinase remains unclear. Here, we show that ABIN-1/NAF-1, a ubiquitin-binding protein, is recruited rapidly into TNF-RSC in a manner dependent upon M1 ubiquitinating complex LUBAC to regulate the recruitment of A20 to control K63 deubiquitination of RIPK1. ABIN-1 deficiency reduces the recruitment of A20 and licenses cells to die through necroptosis by promoting K63 ubiquitination and activation of RIPK1 with TNFα stimulation under conditions that would otherwise exclusively activate apoptosis in wild-type cells. Inhibition of RIPK1 kinase and RIPK3 deficiency block the embryonic lethality of Abin-1−/− mice. We propose that ABIN-1 provides a critical link between M1 ubiquitination mediated by LUBAC complex and K63 deubiquitination by phospho-A20 to modulate the activation of RIPK1.
RIPK1 is a critical regulator of necroptosis and apoptosis, two distinct regulated cell death mechanisms which may be activated under alternative conditions when cells are stimulated by TNFα, an important proinflammatory cytokine involved in mediating multitude of human diseases 1, 2. Under apoptotic deficient conditions, however, the kinase activity of RIPK1 is activated to interact with RIPK3 to induce the formation of a RIPK1/RIPK3 complex, known as complex IIb or necrosome, which promotes the execution of necroptosis 3–5. A20 is an important ubiquitin editing enzyme involved in regulating RIPK1 ubiquitination 6. Activation of RIPK1 has been shown to be critical for major human inflammatory degenerative diseases such multiple sclerosis, Crohn’s disease and amyotrophic lateral sclerosis (ALS) 7–10. Human clinical trials on small molecule inhibitors of RIPK1 are underway in pharmaceutical and biotech companies for developing drugs targeting ALS, Alzheimer’s disease, rheumatoid arthritis and Crohn’s disease 11. On the other hand, inactivation of the TNFAIP3 gene, encoding the A20 protein, is associated with inflammatory diseases including multiple sclerosis, rheumatoid arthritis and Crohn’s disease 12, 13. Understanding the interactive relationship between RIPK1 and A20 may be important for developing effective drugs to treat these conditions. However, the molecular mechanism that regulates the interaction of A20 in TNF-RSC to control the activation of RIPK1 kinase remains unclear.
TNFα stimulation induces the trimerization of TNFR1 to promote the formation of a transient intracellular multi-protein complex, the TNFR1 signaling complex - TNF-RSC (also called complex I), which recruits RIPK1 via homotypic interactions of their death domains 14. TNF-RSC is modified by a complex pattern of ubiquitination chains with different linkages, including M1, K48 and K63 15. In the TNF-RSC, cIAP1/2 mediate K63 ubiquitination of RIPK1 16, which in turn promotes the recruitment of linear ubiquitin chain assembly complex (LUBAC), composed of HOIP, HOIL-1 and SHARPIN, and subsequent conjugation of M1 ubiquitin chains 17. Deficiency in LUBAC complex leads to impaired activation of NF-κB and increased sensitivity to cell death in response to TNFα stimulation, which can be inhibited by RIPK1 inhibitor Nec-1 17–19. Knockdown of HOIL-1 and HOIP leads to significant reduction in the recruitment of IKK complex, the kinase critical for the activation of NF-κB. On the other hand, it is not clear how deficiency in LUBAC sensitizes RIPK1-mediated cell death.
ABIN-1 is a ubiquitin binding protein that can bind to both K63 and M1 ubiquitin chains but not to K48 chains 20. Specific risk haploid types in the gene Abin-1 (TNFAIP3-interacting protein 1, or Tnip1) are strongly associated with a predisposition for autoimmune inflammatory diseases such as systemic lupus erythematosus (SLE) and psoriasis 21, 22. These risk haplotypes produce lower levels of Abin-1 mRNA and ABIN-1 protein, suggesting that the susceptibility to autoimmunity is due to their hypomorphic function. Abin-1−/− mice die during late embryogenesis due to fetal liver cell death, anaemia and hypoplasia, which can be blocked by TNFα deficiency 23.
In this study, we show that ABIN-1 functions as a critical link between M1/K63 ubiquitination and deubiquitination of TNF-RSC to control the activation of RIPK1 kinase activity. ABIN-1 deficiency sensitizes cells to necroptosis by promoting the K63 ubiquitination levels and activation of RIPK1 upon TNFα stimulation. The recruitment of ABIN-1 into TNF-RSC requires the LUBAC complex whereas ABIN-1 deficiency inhibits the association of phospho-A20 with TNF-RSC. Importantly, genetic inactivation of RIPK1 or RIPK3 deficiency prolongs the survival of Abin-1−/− MEFs under various TNFα-mediated paradigms of necroptosis and apoptosis, as well as rescues the embryonic lethality of Abin-1−/− mice. Thus, ABIN-1 deficiency provides a unique paradigm where necroptosis may be activated by normally pro-apoptotic conditions and blocking necroptosis is sufficient to rescue cell death and embryonic lethality.
RESULTS
ABIN-1 deficiency promotes necroptosis
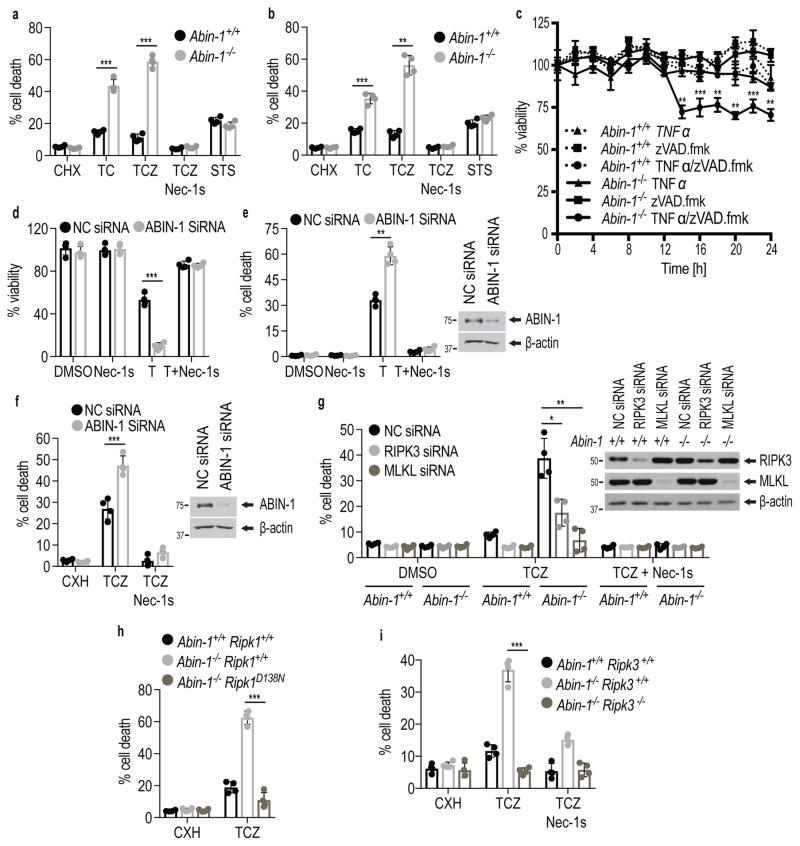
ABIN-1 deficiency has been proposed to promote caspase-8 mediated apoptosis in TNFα stimulated cells 23. However, zVAD.fmk did not inhibit, but rather further sensitized Abin-1−/− mouse embryonic fibroblasts (MEFs) to cell death induced by TNFα and CHX significantly more than that of WT MEFs (Fig. 1a, b). Furthermore, Abin-1−/− MEFs showed increased sensitivity to cell death induced by TNFα/zVAD compared to WT controls (Fig. 1c). Similar data were obtained using both immortalized and primary Abin-1−/− MEFs. In contrast, Abin-1−/− MEFs showed no difference from that of WT MEFs in their sensitivity to apoptosis induced by staurosporine (STS), suggesting no change in sensitivity to apoptosis in general. In addition, we tested the effect of ABIN-1 knockdown in murine fibrosarcoma L929 cells, which undergo necroptosis by stimulation with TNFα alone 19. We found that knockdown of ABIN-1 in L929 cells significantly sensitized them to cell death induced by TNFα (Fig. 1d,e). Similarly, knockdown of ABIN-1 also sensitized the death of RGC-5 to TNFα/CHX/zVAD induced cell death (Fig. 1f). Finally, treatment with TNFα/zVAD/SM-164, an IAP antagonist 24, also made Abin-1−/− MEFs died earlier than that of WT (Supplementary Fig. 1a).
Figure 1. ABIN-1 deficiency sensitizes cells to necroptosis.
(a–c) Immortalized (a, c) and primary (b) WT (Abin-1+/+) or Abin-1−/− MEFs were treated for 7 h (a) and 8 h (b) with TNFα/CHX(TC), TNFα/CHX/zVAD.fmk (TCZ), Staurosporine (STS) (a,b), in the presence or absence of Nec-1s or (c) TNFα (T), zVAD.fmk (Z) or TNFα/zVAD.fmk (TZ) as indicated. L929 cells (d,e) or RGC5 cells (f) were transfected with siRNAs targeting ABIN-1 to knockdown ABIN-1, or control (NC), for 48 hrs. The cells were then treated with TNFα (d,e) or TNFα/CHX/zVAD.fmk (f) with or without Nec-1s for 8 h. Knockdown efficiency is shown on the right of e,f. (g) WT (Abin-1+/+) and Abin-1−/− MEFs were transfected with control non-targeting (NC), RIPK3 or MLKL siRNAs to knockdown of RIPK3 and MLKL. The death of MEFs treated with or without Nec-1s was measured. The knockdown efficiency is shown by western blotting with indicated abs on the right. (h,i) The death of MEFs with indicated genotypes induced by TNFα/CHX/zVAD.fmk with or without Nec-1s for 8 h was measured. TNFα (T): 10 ng/ml; Cycloheximide (CHX or C): 1 μg/ml (a, c, g, h, i) or 10 μg/ml (b, f); zVAD.fmk (Z): 20 μM; Nec-1s: 10 μM; Staurosporine (STS): 1 μM. The graphs (a–i) depict mean (± s.e.m.) of n=4 independent biological experiments (a–i). P values were calculated by two-tailed Student’s t-test (*P < 0.05, **P < 0.01, ***P < 0.001). Cell death was measured by ToxiLight™ (a,b,e,f,g,h,i) or CellTiter-Glo® (c,d) assays. Source data together with precise P values can be found in Supplementary Table 2. Unprocessed original scans of blots are shown in Supplementary Fig. 7.
Treatment of TNFα/CHX/zVAD or TNFα/zVAD can induce necroptosis independent of caspases which can be effectively inhibited by RIPK1 inhibitor Nec-1s 5, 19. We found that Nec-1s strongly blocked the increased cell death of Abin-1−/− MEFs induced by TNFα/CHX/zVAD, TNFα/zVAD or TNFα/zVAD/SM-164 (Fig. 1a–c; Supplementary Fig. 1a) and also TNFα induced cell death of L929 cells (Fig. 1d,e), suggesting that ABIN-1 deficiency sensitized cells to necroptosis mediated by RIPK1 activity.
Consistently, the increased death of Abin-1−/− MEFs or RGC5 cells with ABIN-1 knockdown induced by TNFα/CHX/zVAD.fmk was blocked by knockdown of either RIPK3 or MLKL (Fig. 1g and Supplementary Fig. 1b,c). Finally, the increased sensitivity of Abin-1−/− MEFs to death induced by TNFα/CHX/zVAD.fmk was blocked by genetic inactivation of RIPK1 kinase activity 25 and RIPK3 deficiency 26 in Abin-1−/−; Ripk1D138N/D138N MEFs and Abin-1−/−; Ripk3−/− MEFs (Fig. 1h,i). Thus, the sensitization of ABIN-1 deficient cells to TNFα-induced necroptosis requires RIPK1 kinase activity, RIPK3 and MLKL.
ABIN-1 deficiency promotes the activation of RIPK1
We next characterized the involvement of the necroptotic machinery in TNFα-induced cell death of ABIN-1 deficient cells. We first examined if ABIN-1 may regulate RIPK1 activation using anti-phospho-S166-RIPK1, a marker for its activation 27, 28. We found that the levels of p-S166 RIPK1 were elevated at earlier time points and higher in Abin-1−/− MEFs than that in WT MEFs and in RGC5 cells with ABIN-1-knockdown treated with TNFα/CHX/zVAD.fmk (Fig. 2a–c). Furthermore, increased p-S166 RIPK1 was inhibited in the presence of Nec-1s and by genetic inactivation mutant D138N RIPK1.
Figure 2. Elevated biochemical hallmarks of necroptosis in Abin-1−/− cells stimulated by TNFα/CHX/zVAD.fmk.
(a,b) WT (Abin-1+/+; Ripk1+/+), Abin-1−/−; Ripk1+/+, Abin-1+/+ ; Ripk1D138N and Abin-1−/−; Ripk1D138N MEFs were treated with TNFα/CHX/zVAD.fmk (TCZ) in the presence or absence of Nec-1s for indicated periods of time. The cell lysates were probed with anti-RIPK1(p-S166) or total RIPK1. (c) RGC-5 cells were transfected with indicated siRNA or non-targeting control (NC) siRNA for 48 hrs and treated with TNFα (T)/CHX(C)/zVAD.fmk(Z) for indicated time periods in the presence or absence of Nec-1s as shown. The cell lysates were analyzed by anti-pRIPK1(p-S166), anti-RIPK1, anti-p-MLKL and anti-MLKL. (d,e) Complex IIb in WT (Abin-1+/+) or Abin-1−/− MEFs treated with TNFα/CHX/zVAD.fmk(TCZ) was immunoprecipitated using anti-RIPK3 (d), or anti-FADD (e). The immunocomplexes were analyzed by western blotting using antibodies against RIPK1 and RIPK3 (d) or anti-phospho-S166 RIPK1, total RIPK1, p-MLKL and MLKL (e). (f) WT (Abin-1+/+) and Abin-1−/− MEFs were treated with TNFα/CHX/zVAD.fmk (TCZ) in the presence or absence of Nec-1s for indicated periods of time. The cell lysates were probed with anti-p-MLKL and MLKL. (g, i) WT (Abin-1+/+; Ripk1+/+), Abin-1−/−; Ripk1+/+, Abin-1+/+ ; Ripk1D138N and Abin-1−/−; Ripk1D138N MEFs as indicated were treated with TNFα/CHX/zVAD.fmk (TCZ). The cell lysates were analyzed by western blotting using anti-p-MLKL and MLKL (g). Anti-RIPK3 was used to immunoprecipitate complex IIb and immunocomplexes were analyzed by western blotting using indicated abs (i). (h) WT (Abin-1+/+) and Abin-1−/− MEFs were treated with TNFα/zVAD in the presence or absence of Nec-1s as indicated The cell lysates were analyzed by western blotting using anti-p-MLKL, MLKL and pRIPK1(p-S166). β-actin was a loading control. TNFα(T): 10 ng/ml; Cycloheximide (CHX or C): 1 μg/ml; zVAD.fmk (Z): 20 μM ; Nec-1s: 10 μM. Experiments were repeated independently with similar results twice (e) and at least three times (a–d,f–i). Unprocessed original scans of blots are shown in Supplementary Fig. 7.
Since RIPK1 is an upstream activator of RIPK3 and MLKL, we next tested if the formation of complex IIb, the complex consisting of RIPK1, RIPK3 and FADD necroptosis 3, 4, 29, was elevated in Abin-1−/− MEFs. We found that the RIPK1/RIPK3 interaction appeared earlier and at higher levels in Abin-1−/− MEFs stimulated by TNFα/CHX/zVAD than that of WT cells (Fig. 2d). Furthermore, we detected increased interaction of p-S166 RIPK1 with FADD in Abin-1−/− MEFs stimulated by TNFα/CHX/zVAD compared to that of WT control (Fig. 2e).
Higher levels of phospho-MLKL, a marker for its activation 30, were detected in Abin-1−/− MEFs than that of WT stimulated by TNFα/CHX/zVAD.fmk (Fig. 2f,g). Furthermore, we found that Abin-1−/− MEFs treated with TNFα/zVAD.fmk also showed increased p-MLKL and p-S166 RIPK1 compared to that of WT MEFs (Fig. 2h). Importantly, the formation of complex IIb and p-MLKL in both Abin-1−/− and WT MEFs were effectively blocked by Nec-1s, as well as in Abin-1−/−; Ripk1D138N/D138N MEFs (Fig. 2c–i). Based on these results, we conclude that ABIN-1 deficiency promotes the activation of RIPK1 to mediate necroptosis.
Inhibition of necroptosis prolongs the survival of Abin-1−/− cells with activated caspases
Nec-1s can inhibit necroptosis of WT MEFs induced by TNFα/CHX/zVAD but not that of apoptosis induced by TNFα/CHX 19. Surprisingly, we found that treatment with Nec-1s also reduced the death of Abin-1−/− MEFs induced by TNFα/CHX in the absence of zVAD.fmk, a condition which normally induces apoptosis independent of RIPK1. Similar data were obtained using both immortalized and primary Abin-1−/− MEFs (Fig. 3a–c). TNFα/CHX induced death of RGC5 cells was also sensitized by knockdown of ABIN-1 and inhibited by Nec-1s (Fig. 3d).
Figure 3. Inhibition of RIPK1, RIPK3 and MLKL prolongs the survival of apoptotic ABIN-1 deficient cells induced by TNFα/CHX.
(a–c) Immortalized (a,c) and primary (b) WT (Abin-1+/+) or Abin-1−/− MEFs were treated for 7 h (a) and 8 h (b) with TNFα/CHX(TC) in the presence or absence of Nec-1s as indicated. (d) RGC5 cells with ABIN-1 or control non-targeting siRNA knockdown were treated with TNFα/CHX in the presence or absence of Nec-1s for 8 h. The knockdown control is shown in Supplementary Fig. 1c. (e) WT and Abin-1−/− MEFs were transfected with siRNA for RIPK3, MLKL, or control non-targeting (NC), to knockdown of RIPK3 and MLKL. The knockdown control is shown in Fig. 1d. (f,g) MEFs with indicated genotypes were treated with TNFα alone, CHX alone and TNFα/CHX as indicated. Depicted P values were calculated for Abin-1−/− and Abin-1−/−Ripk1D138N (f) or Abin-1−/− and Abin-1−/−Ripk3−/− MEFs (g) treated with TC. Cell death was measured by ToxiLight™ (a–e) or CellTiter-Glo® (f,g) assays. TNFα(T): 10 ng/ml; Cycloheximide (CHX or C): 1 μg/ml (a, c, e, f, g) or 10 μg/ml (b, d,); Nec-1s: 10 μM. . The graphs depict mean (± s.e.m.) of n=4 independent biological experiments. P values were calculated by two-tailed Student’s t-test (*P < 0.05, **P < 0.01, ***P < 0.001) Source data together with precise P values can be found in Supplementary Table 2.
TNFα stimulation under certain conditions may induce the activation of RIPK1 to mediate apoptosis, termed RIPK1-dependent apoptosis (RDA). However, RDA induced by TNFα stimulation combined with cIAP1/2 deficiency, TAK1 inhibition or IKKα/IKKβ deficiency, cannot be blocked by knockdown of RIPK3 or MLKL 31–33 (Supplementary Fig. 2a). Unexpectedly, we found that the survival of Abin-1−/− MEFs stimulated by TNFα/CHX was prolonged by knockdown of RIPK3 or MLKL (Fig. 3e) and by genetic mutations in Abin-1−/−; Ripk1D138N/D138N mutant MEFs or in Abin-1−/−; Ripk3−/− MEFs (Fig. 3f,g). Thus, blocking key mediators of necroptosis can prolong the survival of TNFα/CHX treated ABIN-1−/− cells, a condition that normally induces RIPK1-independent apoptosis.
Since TNFα/CHX-induced apoptosis involves the activation of caspases, we next characterized the biochemical evidence of caspase activation in Abin-1−/− cells. Although the binding of RIPK1 with caspase-8 was not different in Abin-1−/− MEFs stimulated by TNFα/CHX compared that of WT MEFs (Supplementary Fig. 2b), the stimulation of Abin-1−/− MEFs treated with TNFα/CHX led to not only earlier and stronger cleavage of PARP-1, caspase-3 and RIPK1, mediated by caspase-8 34, but also the simultaneous appearance of phosphorylated MLKL, which, as expected, was not detected in Abin-1+/+ MEFs (Fig. 4a). In addition, we also detected the formation of complex IIb, defined by the interaction between RIPK1/RIPK3 that is normally only found under necroptotic conditions after inhibition of caspases 35 in Abin-1−/− MEFs stimulated by TNFα/CHX (Fig. 4b). The formation of complex IIb and phosphorylation of MLKL induced by TNFα/CHX in Abin-1−/− MEFs were blocked by Nec-1s and in Abin-1−/−; Ripk1D138N/D138N MEFs (Fig. 4c,d).
Figure 4. Inhibition of necroptosis prolongs cell survival but has no effect on caspase activation in Abin-1−/− cells treated with TNFα/CHX.
(a,c,e) MEFs of indicated genotypes were treated for indicated periods of times with TNFα/CHX (TC) in the presence or absence of Nec-1s. The cell lysates were analyzed by western blotting using antibodies against PARP-1, cleaved caspase-3, p-MLKL, MLKL, RIPK1, ABIN-1 and β-actin as indicated. (b,d) MEFs of indicated genotypes were treated with TNFα/CHX (TC) in the presence or absence of Nec-1s for indicated periods of time. RIPK3 was immunoprecipitated with anti-RIPK3 antibody. The immunocomplexes were analyzed by western blotting using antibodies against RIPK1, RIPK3, p-RIPK1 (p-S166), ABIN-1 and β-actin as indicated. (f) WT (Ripk1+/+ Abin-1+/+), Ripk1+/+ Abin-1−/−, Ripk1D138N Abin-1−/− and Ripk1D138N Abin-1+/+ or (g) WT (Ripk3+/+ Abin-1+/+), Ripk3+/ Abin-1−/−, Ripk3−/− Abin-1−/− and Ripk3−/− Abin-1+/+ MEFs were treated with TNFα/CHX (TC) in presence or absence of Nec-1s. The cell lysates were analyzed by western blotting using anti-p-MLKL and MLKL, PARP-1, RIPK1, RIPK3, ABIN-1, activated caspase-3 and β-actin as indicated. TNFα(T): 10 ng/ml; Cycloheximide (C): 1 μg/ml; Nec-1s: 10 μM. Experiments were repeated independently with similar results twice (d) and at least three times (a–c,e–g). Unprocessed original scans of blots are shown in Supplementary Fig. 7.
We next investigated the effect of inhibiting RIPK1 kinase activity on the hallmarks of caspase activation in Abin-1−/− MEFs. Interestingly, although the treatment of Nec-1s, genetically inactivating RIPK1 kinase or RIPK3 deficiency prolonged the cell survival of Abin-1−/− MEFs treated with TNFα/CHX (Fig. 3) and blocked the phosphorylation of MLKL, the activation of caspase-3 and PARP cleavage was unaffected by blocking RIPK1 activity (Fig. 4e–g), suggesting that pro-apoptotic stimulus TNFα/CHX can activate both apoptosis and necroptosis in ABIN-1 deficient cells. Thus, inhibition of RIPK1 kinase activity and RIPK3 can promote the survival of Abin-1−/− MEFs stimulated by TNFα/CHX without blocking caspase activation.
Recruitment of ABIN-1 into TNF-RSC is dependent upon LUBAC and RIPK1
We next investigated if ABIN-1 was recruited into TNF-RSC (complex I) of TNFα stimulated cells. ABIN-1 was rapidly recruited into TNF-RSC within 5 min of TNFα stimulation (Fig. 5a). Although ABIN-1 deficiency had no effect on the recruitment of RIPK1 into TNF-RSC, the high molecular weight ubiquitination of RIPK1 was increased in Abin-1−/− MEFs. Furthermore, we found that significantly increased phosphorylation of S166 RIPK1 was associated with TNF-RSC in Abin-1−/− MEFs (Fig. 5b). Knockdown of Abin-1 in RGC5 cells had a similar effect (Supplementary Fig. 3a). Inhibition of RIPK1 by Nec-1s blocked the appearance of phospho-S166 RIPK1, but had no effect on the high molecular weight ubiquitinated RIPK1 (Fig. 5c).
Figure 5. Increased K63 ubiquitination of RIPK1 in Abin-1−/− MEFs stimulated by TNFα.
(a–c) WT (Abin-1+/+) and Abin-1−/− MEFs were stimulated by Flag-TNFα (100 ng/ml) for indicated periods of time and (c) Nec-1s was added in selected samples as indicated. TNF-RSC (Complex I) was immunoprecipitated using anti-FLAG resin and the recruitment of ABIN-1 and RIPK1 was analyzed by western blotting using indicated antibodies (a). The activation of RIPK1 was analyzed by western blotting using anti-p-RIPK1(p-S166) ab (b–c). TNFR1 was a control for TNF-RSC (c). β-actin is a loading control. (d) WT (Abin-1+/+) and Abin-1−/− MEFs were stimulated by Flag-TNFα (100 ng/ml) for indicated periods of time and the cell lysates were immunoprecipitated using anti-FLAG resin. The immunocomplexes were then denatured in 6M urea. K63-chain specific ubiquitin antibody was used in the second immunoprecipitation in 3M urea to analyze RIPK1 ubiquitination in TNF-RSC by western blotting using anti-RIPK1. (e) Hoip+/+ and Hoip−/− MEFs were stimulated by TNFα and anti-TNFR1 was used to immunoprecipitate TNF-RSC. The immunocomplexes were analyzed by western blotting using anti-ABIN-1, A20, RIPK1, phospho-S166 RIPK1, SHARPIN and TNFR1. (f) WT (Abin-1+/+) MEFs were pre-treated with SM-164 (S) for 4 h, followed by TNFα for indicated periods of time. Anti-TNFR1 was used to immunoprecipitate TNF-RSC. The immunocomplexes were analyzed by western blotting using anti-ABIN-1, HOIP, SHARPIN and TNFR1. TNFα(T): 100 ng/ml; Nec-1s: 10 μM; SM-164: 200 nM. Experiments were repeated independently with similar results twice (d) and at least three times (a–c,e,f). Unprocessed original scans of blots are shown in Supplementary Fig. 7.
Since RIPK1 is known to be predominantly modified by K63 ubiquitination in TNF-RSC 36, 37, we next examined the impact of ABIN-1 deficiency on RIPK1 K63 ubiquitination using tandem immunoprecipitation. TNF-RSC was first immunoprecipitated in native condition. The Flag-TNFα-immunocomplexes were then dissociated in 6M urea and second immunoprecipitation was conducted in denatured condition using K63 linkage-specific polyubiquitin antibody 37. The K63 ubiquitination levels of RIPK1 were analyzed by western blotting for RIPK1. This experiment revealed that K63 ubiquitination of TNFR1-associated RIPK1 was increased in Abin-1−/− MEFs compared to that of WT (Fig. 5d). The increased K63 ubiquitination levels of RIPK1 in Abin-1−/− MEFs were further confirmed by directly immunoprecipitating K63 ubiquitinated proteins in cells lysed in 6M urea and followed by western blotting for RIPK1 (Supplementary ig. 3b). In addition, we also analyzed the K63 ubiquitination pattern of RIPK1 in complex IIb; consistently, increased K63 ubiquitination of RIPK1 in Abin-1−/− MEFs stimulated by TCZ was detected before the activation of RIPK1, detected by anti-p-S166 RIPK1 (Supplementary Fig. 3c).
Since ABIN-1 deficiency had no effect on the recruitment of HOIP and SHARPIN (Supplementary Fig. 3d), the key components of LUBAC complex that mediates M1 ubiquitination of TNF-RSC, we next investigated if the recruitment of ABIN-1 into TNF-RSC was affected by LUBAC. We found that the recruitment of ABIN-1 was blocked in Hoip−/− MEFs (Fig. 5e), as reported for A20 38 (Fig. 5e). Furthermore, SM-164 treatment, which eliminates cIAP1/2, also reduced the recruitment of ABIN-1 into TNF-RSC (Fig. 5f). Thus, the recruitment of ABIN-1 depends on the presence and/or output of the LUBAC complex and cIAP1/2 in the TNF-RSC.
ABIN-1 stabilizes the recruitment of phospho-A20 in TNF-RSC
Since HOIP deficient cells are unable to recruit neither ABIN-1 nor A20 to TNF-RSC, we next investigated if there was a relationship between the recruitment of these two proteins. Like that of ABIN-1 deficient cells, the p-S166 RIPK1 in A20−/− MEFs stimulated by TNFα/CHX/zVAD was enhanced compared to that of WT cells (Supplementary Fig. 3e). Thus, we considered the possibility that ABIN-1 might control the activation of RIPK1 by regulating the recruitment of A20 to the TNF-RSC. While the expression of A20 was largely normal in Abin-1−/− MEFs, the levels of A20 in TNF-RSC were significantly reduced by ABIN-1 deficiency (Fig. 6a).
Figure 6. ABIN-1 is important for the recruitment of p-A20 into TNF-RSC.
(a) WT (Abin-1+/+) and Abin-1−/− MEFs were stimulated by Flag-TNFα (100ng/ml) for indicated periods of time and TNF-RSC (Complex I) was immunoprecipitated using anti-FLAG resin and the recruitment of A20, p-A20(p-S381) and IKKβ was analyzed using indicated antibodies by western blotting analysis. (b) WT (Abin-1+/+) and Abin-1−/− MEFs were stimulated by Flag-TNFα (100 ng/ml) for indicated periods of time in the presence or absence of Nec-1s as indicated and TNF-RSC (Complex I) was immunoprecipitated using anti-FLAG resin and analyzed using indicated antibodies. β-actin is a loading control. (c) A20−/− MEFs complemented with FLAG-A20 WT (FLAG-A20) or FLAG-A20-S381A mutant (S381A) were stimulated by TNFα (100 ng/ml) for indicated periods of time. Complex I was immunoprecipitated by anti-TNFR1 antibody from the lysates and analyzed by anti-A20 and anti-TNFR1. (d) The TNF-RSC in WT (Abin-1+/+) and Abin-1−/− MEFs expressing FLAG-A20-S381D mutant was analyzed by anti-TNFR1. (e) WT (Abin-1+/+) MEFs were stimulated by TNFα (100 ng/ml) for indicated periods of time. ABIN-1 was immunoprecitated from cell lysates using anti-ABIN-1 and the immunocomplex was analyzed by anti-p-A20(p-S381) and anti-A20. (f) A20−/− MEFs complemented with FLAG-A20 WT (FLAG-A20) or FLAG-A20-S381A mutant (S381A) as in (c) were treated with TNFα for indicated periods of time. The cell lysates were analyzed by anti-ABIN-1 immunoprecipitation followed by western blotting using anti-A20 and anti-ABIN-1. β-actin is a loading control. Experiments were repeated independently with similar results twice (c,d,f) and at least three times (a,b,e). Unprocessed original scans of blots are shown in Supplementary Fig. 7.
TNFα stimulation has also been shown to lead to phosphorylation of A20 and CYLD by IKK complex as well as activation of p38 39–42. While the phosphorylation of A20, CYLD and p38 induced by TNFα was largely unaffected by ABIN-1 deficiency (Supplementary Fig. 4a), using an anti-phospho-S381 A20 antibody 40, the recruitment of phospho-A20 to TNF-RSC was significantly reduced in Abin-1−/− MEFs compared to that of WT (Fig. 6a). On the other hand, the recruitment of IKKβ and NEMO to TNF-RSC in Abin-1−/− MEFs was not altered (Fig. 6a–b). Importantly, the increased RIPK1 ubiquitination in the TNF-RSC of Abin-1−/− MEFs was not affected by the treatment of Nec-1s (Fig. 6b). Thus, regulating the recruitment of phosphorylated A20 into TNF-RSC by ABIN-1 affects the ubiquitination of RIPK1.
We next asked if phosphorylation of A20 played a role in its recruitment into TNF-RSC. Using A20−/− MEFs complemented with WT or S381A A20 mutant, we found that the stimulation of TNFα induced the recruitment of WT, but not S381A A20 mutant, into TNF-RSC (Fig. 6c). Furthermore, the recruitment of S381D mutant in the WT cells was highly effective but remained deficient in Abin-1−/− MEFs (Fig. 6d). To directly test if ABIN-1 might be able to interact with phosphorylated A20, we immunoprecipitated ABIN-1 from TNFα stimulated cells and found that A20 interacting with ABIN-1 was effectively phosphorylated and furthermore, the binding of ABIN-1 with S381A mutant was significantly reduced (Fig. 6e–f), suggesting that a critical function of S381 phosphorylation of A20 is to promote its recruitment into TNF-RSC by interacting with ABIN-1.
Truncated ABIN-1 (1-420a.a.) was unable to bind to A20 (Supplementary Fig. 4b) 43. Consistently, unlike that of full length ABIN-1, truncated ABIN-1 (1-420a.a) was unable to suppress the enhanced sensitivity of Abin-1−/− MEFs to necroptosis induced by TNFα/CHX/zVAD (Supplementary Fig. 4c). Furthermore, the truncated ABIN-1 (1-420a.a.) could not be recruited into complex I; nor could the expression of truncated ABIN-1 rescue the ability of Abin-1−/− MEFs to recruit A20 (Supplementary Fig. 4d). Thus, the domain of ABIN-1 that is required for binding with A20 is also required for the recruitment of A20 and ABIN-1 into complex I.
Taken together, we conclude that the recruitment of phospho-A20 by ABIN-1 into TNRF-RSC may play a critical role in regulating K63 ubiquitination and activation of RIPK1.
Blocking necroptosis rescues the embryonic lethality of Abin-1−/− mice
Since Abin-1−/− deficiency in mice leads to TNFα–mediated embryonic lethality after post-coital day E18.5 23, we considered the possibility that necroptosis might contribute to the embryonic lethality of Abin-1-deficient mice. We generated Abin-1−/−; Ripk3−/− double knockout (DKO) mice and Abin-1−/−; Ripk1D138N/D138N mice. As expected, we were unable to obtain viable Abin-1−/−; Ripk3+/+ or Abin-1−/−; Ripk1+/+ mice. Consistent with the activation of both apoptosis and necroptosis upon loss of ABIN-1, Abin-1−/− E16.5 embryonic tissues exhibited hallmarks of apoptosis, including the cleavage of caspase-3 and PARP-1, and necroptosis, including the increased levels of RIPK1, RIPK3 and MLKL in the insoluble fraction, which is known to occur when cells undergo necroptosis 27, 35 – as well as the phosphorylation of MLKL (Fig. 7a). Interestingly, these biochemical hallmarks of both apoptosis and necroptosis were completely absent in E16.5 Abin-1−/−; Ripk3−/− double knockout (DKO) and Abin-1−/−; Ripk1D138N/D138N embryos (Fig. 7b). Furthermore, Abin-1−/−; Ripk3−/− DKO mice and Abin-1−/−; Ripk1D138N/D138N mice were born at expected Mendelian ratios (Supplementary Table 1) and were indistinguishable from their WT littermates when examined at ~8 weeks of age (Supplementary Fig. 5). Furthermore, histolopathological analysis and TUNEL staining of embryonic E18 livers revealed massive necrosis in Abin-1−/− mice which was suppressed by the RIPK1D138N mutation (Fig. 7c–e). Thus, inhibiting necroptosis and apoptosis effectively rescues the embryonic lethality and allows the normal development of Abin-1−/− mice.
Figure 7. The presence of hallmarks of both apoptosis and necroptosis in Abin-1−/− embryos (E16.5).
(a,b) The embryos of indicated genotypes at ~E18 were isolated and dissected. Tissue lysates from head (a) and the rest of the body (a,b) were subject to analysis by western blotting using indicated abs. Unprocessed original scans of blots are shown in Supplementary Fig. 7. (c) Gross appearance of E17.5 Abin-1+/+, Abin-1−/− and Abin-1−/− Ripk1D138N embryos. (d,e) H&E (d) and TUNEL (e) staining of liver sections from (c). Abin-1+/+, Abin-1−/− and Abin-1−/− Ripk1D138N as indicated. Scale bars: 200 μm (d,e). Magnification: 20X (d, e). Experiments were repeated independently with similar results twice (a,b) and at least three times (c–e).
DISCUSSION
Understanding the molecular mechanism that regulates the dynamics of ubiquitination-deubiquitination of TNF-RSC has been a critical goal of the TNFα research. We show here that the dominant function of ABIN-1 in the TNF–RSC is to connect M1 ubiquitinating complex LUBAC in TNF-RSC with the recruitment of A20, a deubiquitinating enzyme that modulates K63 ubiquitination of RIPK1 (Supplementary Fig. 6). Phosphorylation of A20 has been shown to promote its deubiquitination activity 36, 40; however, the mechanism by which TNF-RSC recruits this important modulator was unclear. The recruitment of ABIN-1 into TNF-RSC requires LUBAC complex whose recruitment depends on cIAP1/2, which provides K63, as well as K11, ubiquitination of TNF-RSC 17, 44. The ability of ABIN-1 to bind to both K63 and M1 ubiquitin chains 20 fits perfectly for its role as a modulator of TNF-RSC ubiquitination to connect TNFR1 complex decorated with both K63 and M1 ubiquitinations with phospho-A20 deubiquitinating enzyme to modulate the activation of RIPK1. Thus, ABIN-1 is a key missing piece in connecting M1/K63 ubiquitinated TNFR1 with A20 mediated deubiquitination.
A20 was identified as a regulator of NF-κB 45, 46. Recently, it was reported that A20 deficiency sensitized cells to necroptosis 47. Thus, the inability to effectively recruit A20 provides an important mechanism for the increased sensitivity of ABIN-1 deficient cells and Abin-1−/− mice to both apoptosis and necroptosis. These results also suggest that a critical mechanism mediated by the NF-κB pathway in suppressing the activation of RIPK1 and cell death is through up-regulation of A20 expression and phosphorylation of A20 mediated by activated IKK complex to promote its recruitment into TNF-RSC by interacting with ABIN-1 which in turn modulates the ubiquitination pattern of RIPK1 to prevent its activation. Importantly, since ABIN-1 deficiency has no effect on the activation of NF-κB mediated by TNFα, these results suggest that the dominant functions of ABIN-1 and A20 in the TNF-RSC are to control the activation of RIPK1.
LUBAC mediated M1 ubiquitination is important for efficient recruitment of NEMO and activation of the NEMO/IKKα/IKKβ (NEMO/IKK) complex to promote downstream NF-κB pathway 17. Our results suggest that mediating the recruitment of ABIN-1 might be an additional role of LUBAC-mediated M1 ubiquitination in TNF-RSC, independent of its role in regulating NF-κB activation. On the other hand, inactivation of LUBAC not only blocks the recruitment of ABIN-1, but also inhibits the phosphorylation of A20 mediated by IKK complex. Thus, a common consequence of deficiency in either LUBAC or ABIN-1 is to lead to imbalanced ubiquitination pattern that includes dysregulated K63 ubiquitination of RIPK1. Since deficiencies in HOIP, ABIN-1 and A20 all promote the activation of RIPK1, we propose that an imbalanced ubiquitination of RIPK1 that includes increased K63 ubiquitination and/or decreased M1 ubiquitination of RIPK1 may be critical in mediating its activation to promote cell death. Future studies are needed to identify the sites of RIPK1 modification by different E3 ubiquitin ligases, characterize the types of ubiquitin linkages and elucidate their functional significances in regulating the activation of RIPK1.
Our study described here demonstrates a unique paradigm that ABIN-1 deficiency sensitizes cells to both apoptosis and necroptosis. Furthermore, blocking RIPK1 or RIPK3 is sufficient to offer complete protection against embryonic lethality of Abin-1−/− mice. Necroptosis may be activated in ABIN-1 deficient cells stimulated by TNFα/CHX, which is a classical pro-apoptotic stimulus in WT cells. Furthermore, blocking necroptosis might allow cell survival with activated caspases. The ability of cells to tolerate certain levels of activated caspases is consistent with the increasing acknowledgement of caspases in mediating non-apoptotic functions, such as in mediating cytokine production, cell migration and neuronal function 48.
Our results suggest that a key function of ABIN-1 during embryonic development is to suppress cell death by regulating TNFR1 signaling. Since suppression of necroptosis by RIPK1D138N mutation or RIPK3 deficiency is sufficient to protect embryonic survival of Abin-1−/− mice, our results also suggest that the activation of necroptosis is highly detrimental to embryonic development. Since the activation of necroptosis mediates the lethality of mice with mutations in multiple genes such as caspase-8, FADD or the non-catalytically active caspase-8 homologue cFLIP and ABIN-1 during embryonic development 49–52, we propose that necroptosis may be evolved as a key quality control mechanism during embryogenesis to ensure the development of normal vertebrate animals.
Methods
Animals
Ripk3−/− mice were kindly provided by Dr. Vishva Dixit of Genentech. Ripk1D138N mice were kindly provided by Drs. Manolis Pasparakis of U. Cologne, Germany, and Michelle Kelliher of U. Mass. 8–10 weeks old Abin-1+/− and Ripk1D138N (C57BL/6), Ripk3−/− (C57BL/6N), male and female mice were used in mating. All animals were maintained in a pathogen-free environment, and experiments on mice were conducted according to the protocols approved by the Harvard Medical School Animal Care Committee.
Mouse genotyping
1–3 mm mouse tails were cut and digest overnight at 55° C in tail lysis buffer (10 mM Tris-HCl pH 8.0, 10 mM EDTA pH 8.0, 50 mM NaCl, 0.5% SDS and 20 μg/ml Proteinase K). Cell debris was removed by centrifugation in a microfuge for 30min in 4° C. DNA was precipitated by isopropanol (mixed 1:1), washed with 500 μl 70% ethanol and dried in SpeedVac. Pellets were incubated in TE buffer at 55° C for 2–4 h. Abin-1 genotyping used following primers (5′>3′): ATGGGTGGGTAGGCATAGGGATAG (common), CCTCAAACAGCAGAAGAGGAAAGC (WT) and TTGATTCCCCTTCGCCCATTCCAGC (KO). For Ripk3 (5′>3′): CGCTTTAGAAGCCTTCAGGTTGAC (common), GCAGGCTCTGGTGACAAGATTCATGG (WT) and CCAGAGGCCACTTGTGTAGCG (KO). For Ripk1D138N (5′>3′): TACCTTCTAACAAAGCTTTCC (common), AATGGAACCACAGCATTGGC (WT) and CCCTCGAAGAGGTTCACTAG (KD).
Cell lines, plasmids, siRNA and cell culture
L929, immortalized MEFs, HEK293T and RGC5 were cultured under 5% CO2 at 37°C in Dulbecco’s modified Eagle medium (DMEM) (Corning Cellgro, Cat# 10-017-CV) containing 10% (v/v) fetal bovine serum (FBS) (Gibco, Cat# 10437028), 1% (v/v) penicillin (100 IU/ml)/streptomycin (100 μg/ml), 4.5 g/l D-glucose and 2 mM L-glutamine. Cells were routinely tested for mycoplasma contamination using MycoAlertTM Mycoplasma Detection Kit (Lonza, Cat# LT07). L929, HEK23(T and RGC-5 cell lines were purchased from ATCC and were not authenticated. Generated MEFs were validated by genotyping and western blot.
A20−/− pBABE, A20−/− FLAG-A20 and A20−/− A20-FLAG(S381A) cell lines expressing plasmids pBABE, pBABE FLAG-A20 (S381D) were established as described previously 53. HA-A20 (Cat# DU8411) and ABIN1 (Cat# DU12906) were obtained from MRC Protein Phosphorylation and Ubiquitylation Unit (University of Dundee, UK). ABIN-1 (0–420) was generated by PCR using following primers:
AAAGGATCCATGGAAGGGAGAGGACCCTACCGG and
TTTGCGGCCGCTCACTTCGATTTGGCCAGGAGGAGTTT.
FLAG-ABIN1 and FLAG-ABIN1 (0–420) expression vectors were generated by cloning into pcDNATM5/FRT/TO (Thermo Fisher Scientific, Cat# V652020) plasmid containing 3xFLAG cassette (5′ AGCTTATGGACTACAAAGACCATGACGGTGATTATAAAGATCATGACATCGATTACAAGGATGACGATGACAAGA, 3′ ATCCTGATGTTTCTGGTACTGCCACTAATATTTCTAGTACTGTAGCTAATGTTCCTACTGCTACTGTTCTTCGA), which was previously cloned into HindIII site. Abin-1−/− FLAG-ABIN-1, Abin-1−/− FLAG-ABIN-1 (0–420), Abin-1+/+ A20 (S381D) and Abin-1−/− A20 (S381D) MEFs stable cell lines were obtained by the use of Lipofectamine® 2000 (Invitrogen, Cat# 11668019) transfection following manufactures protocol. All plasmids were validated by DNA sequencing.
Knockdowns were generated using Lipofectamine® RNAiMAX (Invitrogen, Cat# 13778-150) and following siRNA sequences: Abin-1: GAACUGCGCCAGAAGGUCATT, Ripk3: CGACGAUGUCUUCUGUCA, MLKL: GAGAUCCAGUUCAACGAUA. Knockdowns efficiency was authenticated by western blotting.
Human kidney HEK293T cells were transfected using PEI (Polysciences, Cat# 24765-2g, PEI MAX (MW 40,000)). In brief: 20 μg of indicated plasmid was mixed with 40 μl of 1 mg/ml PEI in 1 ml of Opti-MEM (gipco, Cat# 31985-700). After incubation at 20 min RT, transfection mixtures were applied to cultured cells at approximately 50% confluency. Cells were collected for analysis 72 h post transfection.
Generation and immortalization of MEFs
Abin-1+/− mice were mated with Abin-1+/− Ripk1D138N or Abin-1+/− Ripk3−/− mice and pregnancy was terminated at E11–13 stage. Embryos were homogenized individually and treated with trypsin/EDTA, sieved through 70-micron filter and primary MEFs were cultured in high glucose DMEM supplemented with 15% FBS, non-essential amino acids, sodium pyruvate, penicillin, streptomycin and amphotericin B. At passages 4–6, primary MEFs were immortalized by transfection with SV40 small + large T antigen-expressing plasmid (Addgene, Cat# 22298), using Lipofectamine 2000 (Invitrogen, Cat# 11668019) and following manufacturer’s instruction.
Reagents
Recombinant mouse mTNFα (T) (Cell sciences Cat# CRT192C) was resuspended in PBS at a concentration of 100 μg/ml. zVAD.fmk (Z) (Sigma, Cat# V116) was dissolved in DMSO at a concentration of 20 mM and used at a final concentration of 20 μM. Cycloheximide (Sigma, Cat# C-6255) was resuspended in DMSO at a concentration of 10 mg/ml and used as indicated. 7-Cl-O-Nec-1 (Nec-1s) was made by custom synthesis, dissolved in DMSO at a concentration of 10 mM and used at a final concentration of 10 μM. Custom-synthesized (Selleckchem) Smac mimetic SM-164 54 was resuspended in DMSO at a concentration of 10 mM and used at a final concentration of 500 nM. 5Z-7-Oxozeaenol (Sigma, Cat# 09890) was dissolved in DMSO at a concentration of 25 mM and used at a final concentration of 500 nM.
Cell death assays
Cell viability was determined by using CellTiter-Glo® Luminescent Cell Viability Assay (Promega, Cat# G7571) and cell death by using ToxiLight™ Non-destructive Cytotoxicity BioAssay Kit (Lonza, Cat# LT07). All experiments were conducted on 96-well plates using at least four biological replicates. Data was collected using Synergy 2: multimode plate reader from BioTek.
Antibodies, immuoprecipitiation and immunoblots
The following primary antibodies were used: ABIN-1 (1:1,000 Ubiquigent, Cat# 68-0001-100), p-RIPK1 (1:1,000 Cell Signaling, Cat#65746), p-MLKL (1:1,000 Abcam, Cat# ab196436), MLKL (1:1,000 Abcam, Cat# ab67942), PARP-1 (1:1,000 Cell Signaling, Cat# 9542), cleaved Caspase-3 (1:1,000 Cell Signaling, Cat# 9661), Caspase-8 (1:2,000 Enzo, Cat# ALX-804-447-C100), SHARPIN (1:1,000 Proteintech, Cat# 14626-1-AP), HOIP (1:1,000 Abcam, Cat# ab46322), NEMO (1:1,000 Cell Signaling, Cat# 2685S), p-CYLD (1:1,000 Cell Signaling, Cat# 4500S), CYLD (1:1,000 Cell Signaling, Cat# 8462), p-p38 (1:1,000 Cell Signaling, Cat# 9211), p38 (1:1,000 Cell Signaling, Cat# 9212), K63 ubiquitin antibody (3 μg/ml, kindly provided by Genentech described in 37), RIPK1 (1:1,000 Cell Signaling, Cat# 3493 and 1:1,000 BD Biosciences, Cat# 610459), RIPK3 (1:1,000 Serotec, Cat# AHP1797 and 1:300 Santa Cruz, Cat# sc-374639), TNFR1 (1:1,000 Cell Signaling, Cat# 13377S and R&D Systems, Cat# AF-425-PB), A20 (1:1,000 Cell Signaling, Cat# 5630), IKKβ (1:1,000 BD Technologies, Cat# 611254), cIAP1 (1:5 custom made antibody described in 53), FADD (1:300 Santa Cruz, Cat# sc-6036), FLAG-M2 (1:5,000 Sigma, Cat# F3165), HA-probe (1:1,000 Santa Cruz, Cat# sc-805), β-actin (1:5,000 Santa Cruz, Cat# sc-81178). The following secondary antibodies were used: anti-rabbit (1:5,000 SouthernBiotech, Cat# 4050-05), anti-mouse (1:5,000 SouthernBiotech, Cat# 1031-05), anti-rat (1:5,000 SouthernBiotech, Cat# 3010-05) and anti-sheep (1:5,000 SouthernBiotech, Cat# 2018-02).
For experiments not involving immunoprecipitation (IP) cell pellets were lysed in the RIPA lysis buffer containing 10mM Tris-HCl (pH 8.0), 1 mM EDTA, 0.5 mM EGTA, 1% Triton X-100, 150mM NaCl, 0.1% SDS, 0.2% sodium deoxycholate, 50mM NaF, 10mM β-glycerophosphate, 5mM sodium pyrophosphate, 1mM Na3VO4, cOmpleteTM Protease Inhibitor Cocktail (Sigma-Aldrich, Cat# 11697498001), 1 mM PMSF and 5mM N-ethylmaleimide (NEM)(Sigma-Aldrich, Cat# E3876). Protein concentration in collected supernatants was measured using PierceTM BCA Protein Assay Kit (Thermo Scientific, Cat# 23225) and normalized to the lowest concentration. Samples were diluted with 4X SDS-PAGE sample loading buffer (240 mM Tris-HCl pH 6.8, 40% (v/v) glycerol, 8% (v/v) SDS, 0.04% bromophenol blue and 5% (v/v) β-mercaptoethanol).
For Complex I IP, cells were either harvested in NP-40 buffer (25 mM HEPES-KOH pH 7.5, 0.2% NP-40, 120 mM NaCl, 0.27 M sucrose, 1 mM EDTA, 1 mM EGTA, 50 mM NaF, 10 mM β-glycerophosphate, 5 mM sodium pyrophosphate, 2mM Na3VO4, 2X cOmpleteTM Protease Inhibitor Cocktail, 2 mM PMSF and 10 mM NEM) and incubated with FLAG-tagged beads (Sigma, Cat# A2220) for 4 h, or overnight with TNFR1 antibody followed by 4 h incubation with Protein A/G ultra link resin (Thermo Scientific, Cat# 53133). Complex IIb was immunoprecipitated by overnight incubation with RIPK3 or FADD abs in 4 C with rotation followed by 4 h incubation with Protein A/G ultra link resin For K63 ubiquitin IP, cells were harvested in 6 M urea lysis buffer (6 M urea, 20 mM Tris-HCl, pH 7.5, 135 mM NaCl, 1.5 mM MgCl2, 1 mM EGTA, 1% Triton X-100, 50 mM NaF, 10 mM β-glycerophosphate, 5 mM sodium pyrophosphate, 2mM Na3VO4, 2X cOmpleteTM Protease Inhibitor Cocktail, 2 mM PMSF and 20 mM NEM) and flash frozen in liquid nitrogen. Thawed lysates were centrifuged at 16,000xg for 30 min in 4C and supernatants were diluted with lysis buffer (without urea) to bring urea concentration to 3 M. For K63 IP, 3 μg of the chain-specific antibody was incubated at 4 C overnight followed by 4 h incubation with Proterase A agarose resin (Pierce, Cat# 20333). Beads were washed and proteins were eluted in 70° with 2X SDS-PAGE sample buffer using Thermomixer (Eppendorf).
The primary antibodies were incubated in 2% BSA overnight at 4° C. Membranes were washed three times (8 min each time) with TBST. Appropriate HRP-conjugated secondary antibodies were incubated for 1 h at room temperature. After subsequent TBST washes (three times, 8min each time) proteins were visualized with the enhanced chemiluminescence substrate ECL and imaged using the Kodak M35A X-OMAT X-Ray Film Processor
H&E and TUNEL tissue staining
Collected mouse tissue was fixed in 3% PFA and processed for paraffin embedding. The 6-micron thick histological sections were stained with hematoxyin and eosin by Rodent Histopathology Core at Harvard Medical School (Boston, MA). TUNEL assay was used to detect dead cells with DNA fragmentation using In Situ Cell Death Detection Kit, POD (Roche, Cat# 11684795910) by following manufacture’s protocol. Images were taken on an inverted Nikon Ti fluorescence microscope at Nikon Imaging Center at Harvard Medical School (Boston, MA) and analyzed using MetaMorph image acquisition software. Camera: Hamamatsu ORC-ER cooled CCD; light source: Lumencor SOLA fluorescence light source; objective: 20X
Statistics and Reproducibility
Each individual experiment was repeated independently with similar results at least three times, except experiments shown in Figs 2e, 4d, 5d, 6c, d, f, 7a, b and Supplementary Figs 2b, and 3c were performed twice. Statistical analysis was performed using Microsoft Excel 2011. Prism 7 software was used to generate graphs. Statistical significance was evaluated by using two-tailed Student’s t-test. Differences were considered statistically significant if P < 0.05(*); P < 0.01 (**) or P < 0.001(***). Data are expressed as the mean ± standard error of the mean (SEM). Exact P values for each experiment (where applicable) can be found in Supplementary Table 2.
Data availability
All data that support the conclusions are available from the authors on reasonable request. Source data for Figs 1a–i, 3a–g and Supplementary Figs 1a–c, 2a, 4c and 5a,b have been provided as Supplementary Table 2.
Supplementary Material
Acknowledgments
This work was supported in part by grants (to JY) from the NINDS (1R01NS082257) and the NIA (1R01AG047231) and from the Chinese Academy of Sciences, the China Ministry of Science and Technology Program (2014ZX09102001-002), the China National Natural Science Foundation (31530041), the National Key R&D Program of China, and the National Key Research and Development Program (2016YFA0501900). We thank Dr. Vishva Dixit of Genentech for Ripk3−/− mice and for ab against K63 ubiquitin chains, Drs. Pasparakis M of U. Cologne, Germany, and Kelliher MA of U. Mass for providing Ripk1-D138N mice and Dr. Henning Walczak of Imperial College, UK, for providing Hoip−/− MEFs. We thank Drs. Gary Kasof of Cell Signaling for generating p-S166 antibody and Dr. Roderick Bronson for mouse histopathology analysis. We thank Drs. Dimitry Ofengeim and Albert D. Yu for comments on the manuscript and the members of Yuan laboratory for stimulating discussions.
Footnotes
Author contributions
J. Y. conceived the concept, designed the experiments and wrote the manuscript. S. A. D. designed the experiments, executed majority of the experiments and prepared the figures. Z. S., V. J. B., A. N., A.K.M., P.A., H. P., and L. S. conducted specific experiments. D.W.A. provided p-S381 A20 ab, A20 plasmids and A20 mutant complemented cells. A.M. provided Abin-1−/+ mice. H.Z. and V. J. B. did genotyping.
References
- 1.Ofengeim D, Yuan J. Regulation of RIP1 kinase signalling at the crossroads of inflammation and cell death. Nat Rev Mol Cell Biol. 2013;14:727–736. doi: 10.1038/nrm3683. [DOI] [PubMed] [Google Scholar]
- 2.Wallach D, Kang TB, Dillon CP, Green DR. Programmed necrosis in inflammation: Toward identification of the effector molecules. Science. 2016;352:aaf2154. doi: 10.1126/science.aaf2154. [DOI] [PubMed] [Google Scholar]
- 3.Cho YS, et al. Phosphorylation-driven assembly of the RIP1–RIP3 complex regulates programmed necrosis and virus-induced inflammation. Cell. 2009;137:1112–1123. doi: 10.1016/j.cell.2009.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He S, et al. Receptor interacting protein kinase-3 determines cellular necrotic response to TNF-alpha. Cell. 2009;137:1100–1111. doi: 10.1016/j.cell.2009.05.021. [DOI] [PubMed] [Google Scholar]
- 5.Degterev A, et al. Identification of RIP1 kinase as a specific cellular target of necrostatins. Nat Chem Biol. 2008;4:313–321. doi: 10.1038/nchembio.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vucic D, Dixit VM, Wertz IE. Ubiquitylation in apoptosis: a post-translational modification at the edge of life and death. Nat Rev Mol Cell Biol. 2011;12:439–452. doi: 10.1038/nrm3143. [DOI] [PubMed] [Google Scholar]
- 7.Ito Y, et al. RIPK1 mediates axonal degeneration by promoting inflammation and necroptosis in ALS. Science. 2016;353:603–608. doi: 10.1126/science.aaf6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pasparakis M, Vandenabeele P. Necroptosis and its role in inflammation. Nature. 2015;517:311–320. doi: 10.1038/nature14191. [DOI] [PubMed] [Google Scholar]
- 9.Welz PS, et al. FADD prevents RIP3-mediated epithelial cell necrosis and chronic intestinal inflammation. Nature. 2011;477:330–334. doi: 10.1038/nature10273. [DOI] [PubMed] [Google Scholar]
- 10.Vlantis K, et al. NEMO Prevents RIP Kinase 1-Mediated Epithelial Cell Death and Chronic Intestinal Inflammation by NF-kappaB-Dependent and -Independent Functions. Immunity. 2016;44:553–567. doi: 10.1016/j.immuni.2016.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harris PA, et al. DNA-Encoded Library Screening Identifies Benzo[b][1,4]oxazepin-4-ones as Highly Potent and Monoselective Receptor Interacting Protein 1 Kinase Inhibitors. J Med Chem. 2016;59:2163–2178. doi: 10.1021/acs.jmedchem.5b01898. [DOI] [PubMed] [Google Scholar]
- 12.Matmati M, et al. A20 (TNFAIP3) deficiency in myeloid cells triggers erosive polyarthritis resembling rheumatoid arthritis. Nat Genet. 2011;43:908–912. doi: 10.1038/ng.874. [DOI] [PubMed] [Google Scholar]
- 13.Catrysse L, Vereecke L, Beyaert R, van Loo G. A20 in inflammation and autoimmunity. Trends Immunol. 2014;35:22–31. doi: 10.1016/j.it.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 14.Hsu H, Huang J, Shu HB, Baichwal V, Goeddel DV. TNF-dependent recruitment of the protein kinase RIP to the TNF receptor-1 signaling complex. Immunity. 1996;4:387–396. doi: 10.1016/s1074-7613(00)80252-6. [DOI] [PubMed] [Google Scholar]
- 15.Dondelinger Y, Darding M, Bertrand MJ, Walczak H. Poly-ubiquitination in TNFR1-mediated necroptosis. Cell Mol Life Sci. 2016 doi: 10.1007/s00018-016-2191-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bertrand MJ, et al. cIAP1 and cIAP2 facilitate cancer cell survival by functioning as E3 ligases that promote RIP1 ubiquitination. Mol Cell. 2008;30:689–700. doi: 10.1016/j.molcel.2008.05.014. [DOI] [PubMed] [Google Scholar]
- 17.Haas TL, et al. Recruitment of the linear ubiquitin chain assembly complex stabilizes the TNF-R1 signaling complex and is required for TNF-mediated gene induction. Mol Cell. 2009;36:831–844. doi: 10.1016/j.molcel.2009.10.013. [DOI] [PubMed] [Google Scholar]
- 18.Gerlach B, et al. Linear ubiquitination prevents inflammation and regulates immune signalling. Nature. 2011;471:591–596. doi: 10.1038/nature09816. [DOI] [PubMed] [Google Scholar]
- 19.Degterev A, et al. Chemical inhibitor of nonapoptotic cell death with therapeutic potential for ischemic brain injury. Nat Chem Biol. 2005;1:112–119. doi: 10.1038/nchembio711. [DOI] [PubMed] [Google Scholar]
- 20.Nanda SK, et al. Polyubiquitin binding to ABIN1 is required to prevent autoimmunity. J Exp Med. 2011;208:1215–1228. doi: 10.1084/jem.20102177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Callahan JA, et al. Cutting edge: ABIN-1 protects against psoriasis by restricting MyD88 signals in dendritic cells. J Immunol. 2013;191:535–539. doi: 10.4049/jimmunol.1203335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caster DJ, et al. ABIN1 dysfunction as a genetic basis for lupus nephritis. Journal of the American Society of Nephrology : JASN. 2013;24:1743–1754. doi: 10.1681/ASN.2013020148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oshima S, et al. ABIN-1 is a ubiquitin sensor that restricts cell death and sustains embryonic development. Nature. 2009;457:906–909. doi: 10.1038/nature07575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu J, et al. SM-164: a novel, bivalent Smac mimetic that induces apoptosis and tumor regression by concurrent removal of the blockade of cIAP-1/2 and XIAP. Cancer Res. 2008;68:9384–9393. doi: 10.1158/0008-5472.CAN-08-2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Polykratis A, et al. Cutting edge: RIPK1 Kinase inactive mice are viable and protected from TNF-induced necroptosis in vivo. J Immunol. 2014;193:1539–1543. doi: 10.4049/jimmunol.1400590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Newton K, Sun X, Dixit VM. Kinase RIP3 is dispensable for normal NF-kappa Bs, signaling by the B-cell and T-cell receptors, tumor necrosis factor receptor 1, and Toll-like receptors 2 and 4. Mol Cell Biol. 2004;24:1464–1469. doi: 10.1128/MCB.24.4.1464-1469.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ofengeim D, et al. Activation of necroptosis in multiple sclerosis. Cell reports. 2015;10:1836–1849. doi: 10.1016/j.celrep.2015.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berger SB, et al. Cutting Edge: RIP1 kinase activity is dispensable for normal development but is a key regulator of inflammation in SHARPIN-deficient mice. J Immunol. 2014;192:5476–5480. doi: 10.4049/jimmunol.1400499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang DW, et al. RIP3, an energy metabolism regulator that switches TNF-induced cell death from apoptosis to necrosis. Science. 2009;325:332–336. doi: 10.1126/science.1172308. [DOI] [PubMed] [Google Scholar]
- 30.Sun L, et al. Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell. 2012;148:213–227. doi: 10.1016/j.cell.2011.11.031. [DOI] [PubMed] [Google Scholar]
- 31.Dondelinger Y, et al. NF-kappaB-Independent Role of IKKalpha/IKKbeta in Preventing RIPK1 Kinase-Dependent Apoptotic and Necroptotic Cell Death during TNF Signaling. Mol Cell. 2015;60:63–76. doi: 10.1016/j.molcel.2015.07.032. [DOI] [PubMed] [Google Scholar]
- 32.Mihaly SR, Ninomiya-Tsuji J, Morioka S. TAK1 control of cell death. Cell Death Differ. 2014;21:1667–1676. doi: 10.1038/cdd.2014.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang L, Du F, Wang X. TNF-alpha induces two distinct caspase-8 activation pathways. Cell. 2008;133:693–703. doi: 10.1016/j.cell.2008.03.036. [DOI] [PubMed] [Google Scholar]
- 34.Lin Y, Devin A, Rodriguez Y, Liu ZG. Cleavage of the death domain kinase RIP by caspase-8 prompts TNF-induced apoptosis. Genes Dev. 1999;13:2514–2526. doi: 10.1101/gad.13.19.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li J, et al. The RIP1/RIP3 Necrosome Forms a Functional Amyloid Signaling Complex Required for Programmed Necrosis. Cell. 2012;150:339–350. doi: 10.1016/j.cell.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wertz IE, et al. Phosphorylation and linear ubiquitin direct A20 inhibition of inflammation. Nature. 2015;528:370–375. doi: 10.1038/nature16165. [DOI] [PubMed] [Google Scholar]
- 37.Newton K, et al. Ubiquitin chain editing revealed by polyubiquitin linkage-specific antibodies. Cell. 2008;134:668–678. doi: 10.1016/j.cell.2008.07.039. [DOI] [PubMed] [Google Scholar]
- 38.Draber P, et al. LUBAC-Recruited CYLD and A20 Regulate Gene Activation and Cell Death by Exerting Opposing Effects on Linear Ubiquitin in Signaling Complexes. Cell reports. 2015;13:2258–2272. doi: 10.1016/j.celrep.2015.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reiley W, Zhang M, Wu X, Granger E, Sun SC. Regulation of the deubiquitinating enzyme CYLD by IkappaB kinase gamma-dependent phosphorylation. Mol Cell Biol. 2005;25:3886–3895. doi: 10.1128/MCB.25.10.3886-3895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hutti JE, et al. IkappaB kinase beta phosphorylates the K63 deubiquitinase A20 to cause feedback inhibition of the NF-kappaB pathway. Mol Cell Biol. 2007;27:7451–7461. doi: 10.1128/MCB.01101-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raingeaud J, et al. Pro-inflammatory cytokines and environmental stress cause p38 mitogen-activated protein kinase activation by dual phosphorylation on tyrosine and threonine. J Biol Chem. 1995;270:7420–7426. doi: 10.1074/jbc.270.13.7420. [DOI] [PubMed] [Google Scholar]
- 42.Hutti JE, et al. Phosphorylation of the tumor suppressor CYLD by the breast cancer oncogene IKKepsilon promotes cell transformation. Mol Cell. 2009;34:461–472. doi: 10.1016/j.molcel.2009.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heyninck K, Kreike MM, Beyaert R. Structure-function analysis of the A20-binding inhibitor of NF-kappa B activation, ABIN-1. FEBS Lett. 2003;536:135–140. doi: 10.1016/s0014-5793(03)00041-3. [DOI] [PubMed] [Google Scholar]
- 44.Dynek JN, et al. c-IAP1 and UbcH5 promote K11-linked polyubiquitination of RIP1 in TNF signalling. EMBO J. 2010;29:4198–4209. doi: 10.1038/emboj.2010.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bosanac I, et al. Ubiquitin binding to A20 ZnF4 is required for modulation of NF-kappaB signaling. Mol Cell. 2010;40:548–557. doi: 10.1016/j.molcel.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 46.Lee EG, et al. Failure to regulate TNF-induced NF-kappaB and cell death responses in A20-deficient mice. Science. 2000;289:2350–2354. doi: 10.1126/science.289.5488.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Onizawa M, et al. The ubiquitin-modifying enzyme A20 restricts ubiquitination of the kinase RIPK3 and protects cells from necroptosis. Nat Immunol. 2015;16:618–627. doi: 10.1038/ni.3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yi CH, Yuan J. The Jekyll and Hyde functions of caspases. Dev Cell. 2009;16:21–34. doi: 10.1016/j.devcel.2008.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dillon CP, et al. Survival function of the FADD-CASPASE-8-cFLIP(L) complex. Cell reports. 2012;1:401–407. doi: 10.1016/j.celrep.2012.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang J, Cado D, Chen A, Kabra NH, Winoto A. Fas-mediated apoptosis and activation-induced T-cell proliferation are defective in mice lacking FADD/Mort1. Nature. 1998;392:296–300. doi: 10.1038/32681. [DOI] [PubMed] [Google Scholar]
- 51.Yeh WC, et al. Requirement for Casper (c-FLIP) in regulation of death receptor-induced apoptosis and embryonic development. Immunity. 2000;12:633–642. doi: 10.1016/s1074-7613(00)80214-9. [DOI] [PubMed] [Google Scholar]
- 52.Varfolomeev EE, et al. Targeted disruption of the mouse Caspase 8 gene ablates cell death induction by the TNF receptors, Fas/Apo1, and DR3 and is lethal prenatally. Immunity. 1998;9:267–276. doi: 10.1016/s1074-7613(00)80609-3. [DOI] [PubMed] [Google Scholar]
- 53.Xu L, et al. c-IAP1 cooperates with Myc by acting as a ubiquitin ligase for Mad1. Mol Cell. 2007;28:914–922. doi: 10.1016/j.molcel.2007.10.027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data that support the conclusions are available from the authors on reasonable request. Source data for Figs 1a–i, 3a–g and Supplementary Figs 1a–c, 2a, 4c and 5a,b have been provided as Supplementary Table 2.