Abstract
Neural progenitor cell (NPC) fate is influenced by a variety of biological cues elicited from the surrounding microenvironment and recent studies suggest their possible role in pediatric glioblastoma multiforme (GBM) development. Since a few GBM cells also display NPC characteristics, it is not clear whether NPCs transform to tumor cell phenotype leading to the onset of GBM formation, or NPCs migrate to developing tumor sites in response to paracrine signaling from GBM cells. Elucidating the paracrine interactions between GBM cells and NPCs in vivo is challenging due to the inherent complexity of the CNS. Here, we investigated the interactions between human NPCs (ReNcell) and human pediatric GBM-derived cells (SJ-GBM2) using a Transwell® coculture setup to assess the effects of GBM cells on ReNcells (cytokine and chemokine release, viability, phenotype, differentiation, migration). Standalone ReNcell or GBM cultures served as controls. Qualitative and quantitative results from ELISA®, Live/Dead® and BrdU assays, immunofluorescence labeling, western blot analysis, and scratch test suggests that although ReNcell viability remained unaffected in the presence of pediatric GBM cells, their morphology, phenotype, differentiation patterns, neurite outgrowth, migration patterns (average speed, distance, number of cells) and GSK-3β expression were significantly influenced. The cumulative distance migrated by the cells in each condition was fit to Furth’s formula, derived formally from Ornstein-Uhlenbeck process. ReNcell differentiation into neural lineage was compromised and astrogenesis promoted within cocultures. Such coculture platform could be extended to identify the specific molecules contributing to the observed phenomena, to investigate whether NPCs could be transplanted to replace lesions of excised tumor sites, and to elucidate the underlying molecular pathways involved in GBM-NPC interactions within the tumor microenvironment.
Keywords: pediatric glioblastoma cells, neural progenitor cells, differentiation, migration, GSK-3β
Introduction
Glioblastoma multiforme (GBM) is a highly malignant form of cancer found within the central nervous system (CNS) which, when diagnosed, has a median patient survival time of less than a year [1]. GBM affects populations of all ages, although the pediatric form is understudied due to the histopathological diversity of the ailment. Pediatric GBM is also more challenging to treat due to intrinsic drug resistance [2]. Although the cellular origins of pediatric GBM are unclear, one theory suggests that they could arise from the transformation of proliferating NPCs during embryogenesis [3–6]. In addition to cancer stem cell markers (e.g., CD133) [7, 8], GBM cells also express a variety of lineage markers including pre-neural and astrocyte, and a variety of mature neuronal markers including GABA and GalC [4].
NPCs respond to various spatio-temporal cues to determine their progeny, and their maturation is driven by a combination of intrinsic temporal factors as well as extracellular signals from the developing brain microenvironment. For example, at mid-gestation, young neurons migrate above the germinal ventricular zone (VZ) and eventually to the subventricular zone (SVZ). By postnatal stage, radial glia transform into astrocytes and the VZ disappears, but the SVZ remains into adulthood where NPCs continue to proliferate [9], and respond to a variety of growth factors mimicking responses seen during embryogenesis. When cultured in vitro, NPCs have been shown to expand and maintain an undifferentiated phenotype in the presence of epidermal growth factor (EGF) and/or basic fibroblast growth factor (bFGF), and could differentiate into both neuronal and glial lineages upon removal of these mitogens [10]. NPCs have been shown to migrate and integrate into the surrounding tumor microenvironment, and therefore offer potential to impede GBM progression by delivering relevant suppression molecules to the tumor site [11]. For example, studies have shown that GBM cells have a BMP cell-cycle exit pathway similar to that of NPCs, which could impede tumor progression if manipulated [12].
The therapeutic potential of NPCs in a cancerous microenvironment is currently limited by the potential transformation of NPCs to tumorigenic phenotype, which is thought to be triggered by a variety of molecular mechanisms [6]. Holland et al. reported that NPCs can produce similar histological characteristics as GBM in a rodent model, via transduction proteins such as Akt and KRas [13]. Genetic analysis suggested that Olig2 regulates both NPC and GBM lineages and is critical for cell proliferation in both populations [14]. Dai et al. found that mature mouse astrocytes transfected with the platelet-derived growth factor appeared to be more susceptible to GBM transformation in vivo [15]. Other studies suggest that the mechanisms driving GBM transformation are based on increased production of the glycoprotein CD133, which is also a NPC marker. Thus, the origin of GBM is likely either a derivation from CD133 expressing cells which are normally not present in the adult brain, or from CD133-positive ependymal cells in the adult brain [16]. In general, NPCs are at risk for malignant transformation based on activated pro-mitotic genes, telomerase activity, and anti-apoptotic genes, which can be triggered by a combination of less than seven mutations [17, 18].
Currently, limited information exists pertaining to the influence of GBMs on NPCs and vice versa [6]. NPCs injected in vivo to mouse tumor sites have been shown to aggressively migrate into tumor sites and overexpress several cytokines and chemokines, which has the potential to trigger antitumor NPC-mediated immunity [11, 19]. GBMs also appear to transmit signals to the surrounding environment that cause the co-expression of several unique neuronal markers in both themselves (autocrine) as well as surrounding stem cells (paracrine) [3, 20]. Investigating the paracrine signaling between GBM cells and NPCs in a controlled microenvironment (e.g., cocultures) might help in direct quantification of the influence of each cell type on the other, in the identification of molecules which arrest GBM growth, metastasis and tumor formation suppression, and in evaluating the potential of NPC transplantation in restoring lost cell populations at the lesions site after surgical removal of CNS tumor [6]. Using a biomimetic coculture system, limitations associated with in vivo studies could be overcome and direct cell-cell contact could be eliminated, while effectively exposing GBM cells and NPCs to the signaling molecules released by the other.
This study is based on the hypothesis that the biochemical signals released by pediatric GBM-derived cells influence the normal NPC phenotype by altering their morphology, survival, migration, differentiation patterns, and release of various cytokines and chemokines. Given the similarities in the cellular pathways which regulate both NPC and GBM cell differentiation and proliferation, such coculture studies would also provide key insights into the effect NPCs have on GBM cells. The outcomes from such studies might help elucidate the conditions leading to the onset and progression of pediatric GBMs, identify the target molecules and pathways which might help impede GBM progression, and unlock the interactions of NPCs in tumor microenvironment [21].
Materials and Methods
NPC and GBM cell expansion
Human NPCs (ReNcell VM Human Neural Progenitor Cell Line; SCC008) and all media components were purchased from EMD Millipore. ReNcells were maintained in an undifferentiated state by culturing on laminin-coated T-75 flasks in the presence of non-differentiating maintenance media (Millipore Cat. No. SCM005) containing 20 ng/mL of freshly-thawed bFGF and EGF. Media was changed every 24 h, and after 7 days, ReNcells were gently detached using Accutase (Life Technologies, Carlsbad, CA) and frozen in ReNcell freezing medium (Millipore Cat. No. SCM007). Differentiation media was ReNcell maintenance media without any bFGF or EGF. All cells used in this study were before passage 10.
Human pediatric glioblastoma multiforme cells (GBM) were obtained from the Children’s Oncology Group (COG) Cell Culture and Xenograft Repository at Texas Tech University Health Sciences Center School of Medicine. Cells were derived from a five-year old female and labeled as the “SJ-GBM2” cell line [22]. The SJ-GBM2 cell line was expanded in uncoated T-75 flasks. All media products were purchased from Life Technologies unless otherwise noted. GBM media was prepared using Iscove’s modified Dulbecco’s medium containing 20% fetal bovine serum, 4 mM L-glutamine, and ITS supplement (5 μg/mL insulin, 5 μg/mL transferrin, 5 ng/mL selenous acid). Media was replaced every 3 days, cells detached using Pucks EDTA (140 nM NaCl, 5 mM KCl, 5.5 mM glucose, 4 mM NaHCO3, 13 μM phenol red, 0.8 mM EDTA, and 9 mM HEPES) after 7 days, and frozen in a solution containing 50% fetal bovine serum and 7.5% DMSO in Iscove’s modified Dulbecco’s medium. Cells were used prior to passage 20. Henceforth, SJ-GBM2 cells will be referred to as GBM, and undifferentiated ReNcells as ReNcell-bFGF. All cultures in this study were performed on 2D tissue culture grade plastic, and ReNcells were always plated on laminin-coated dishes.
Transwell® coculture conditions
After initial expansion, test cultures were run for 5 or 10 days under these conditions, in parallel: ReNcells cultured alone in non-differentiating media (ReNcell bFGF), ReNcells cultured alone in differentiation media (ReNcells alone), GBMs cultured alone in GBM media (GBM alone), ReNcells cocultured with GBMs seeded in transwell insert (ReNcell cocultures), and GBMs cocultured with ReNcells seeded in transwell insert (GBM cocultures). For these cocultures, ReNcell differentiation media and GBM media were mixed in 1:1 ratio and supplemented. To ensure no direct cell-cell interactions, 1-μm PET membrane Transwell® cell culture inserts (Flacon/Corning, Durham, NC) were used. NPCs were first seeded in laminin-coated 24-well plates at a density of 4 × 104 cells/well and cultured for 3 h with 200 μL of differentiation media or non-differentiating media. For cocultures, after 3 h, Transwell® inserts were placed and 4 × 104 GBMs were seeded within the uncoated inserts. Respective media for both cell types was replaced daily. In all the cases, cells were cultured on 2D substrates and not within 3D scaffolds.
Cytokine/chemokine analysis
Cytokine and chemokine analyses were performed using Discovery Assays® (Eve Technologies, Alberta, Canada). Cell culture supernatants (150 μL) were collected from each well after 24 h culture, spun down at 3000 g for 5 min, and stored at −20 °C. The supernatants were then processed using multiplexing LASER bead technology and processed on a dual-laser flow-cytometry system (Bio-Plex 200). The technology works by utilizing different combinations of red and infrared fluorophore beads conjugated to specific antibodies targeted to the cytokine or chemokine of choice and were read using a flow-cytometry based system. The quantity of the specific analyte generated was based off of a series of standards set forth by the company. The following cases were tested: ReNcells alone, GBM alone, ReNcells bFGF, ReNcell cocultures and GBM cocultures. For ReNcell cocultures, ReNcells were plated in the bottom dish of the 24-well plates and GBM cells were seeded in the Transwell inserts. Similarly, for GBM cocultures, GBM cells were plated in the bottom dish of the 24-well plates and ReNcells were seeded in the Transwell inserts. The presence of following 42 cytokines and chemokines (EGF, Eotaxin-1, FGF-2, Flt-3L, Fractalkine, G-CSF, GM-CSF, GRO, IFNα2, IFN-γ, IL-1α, IL-1β, IL-1ra, IL-2, IL-3, IL-4, IL-5, IL-6, IL-7, IL-8, IL-9, IL-10, IL-12 (p40), IL-12 (p70), IL-13, IL-15, IL-17A, IL-18, IP-10, MCP-1, MCP-3, MDC, MIP-1α, MIP-1β, PDGF-AA, PDGF-AB/BB, RANTES, sCD40L, TGF-α, TNF-α, TNF-β, and VEGF) and 3 cancer markers (Galectin-3, MPO, and FAP) were tested. Cytokine and chemokine expression levels from these bead assays were plotted as a heat map using R software.
Cell viability
NSC and GBM viability was measured using the LIVE/DEAD® viability and cytotoxicity kit (Life Technologies) on days 5 and 10, for all the culture conditions. Cells were incubated with calcein AM and ethidium homodimer-1 for 30 min and immediately underwent fluorescent imaging using an Axio Vert.A1 fluorescence microscope (Carl Zeiss Microimaging; NY). For coculture conditions, the Transwell® inserts containing the GBMs were removed and placed in a separate well, stained using the aforementioned procedure and imaged. Viability of ReNcells and GBMs was expressed as a percentage of the number of live cells counted over the total number of cells, using a custom-written macro in ImageJ (> 1500 cells counted per condition).
Cellular morphology
Cell shape index (CSI) was assessed from the staining of live cells (FITC-channel) using the images acquired from all of the aforementioned LIVE/DEAD® staining cases. Images were processed using the particle analyzer function feature in ImageJ (> 1000 cells counted per condition). The projected area (A) and perimeter (P) for each cell was measured and the average cell morphology was quantified using the formula: CSI = 4πA/P2. CSI values range from zero (infinitely elongated polygon) to one (perfect circle).
Immunofluorescence imaging
On days 5 and 10, the following cultures were processed to identify and quantify the distinct neural and glial lineages: ReNcells alone, GBM alone, ReNcell cocultures, and GBM cocultures. Cells were washed once in sterile 1× PBS, fixed with 4% PFA (4 °C, 2 h), washed twice with 1× PBS (5 min), and incubated with blocking buffer (0.5% Triton-X, 5% serum, 1× PBS) for 2 h at room temperature. Serum selection was based on primary and secondary antibody host species. After removing the blocking buffer, cells were incubated with respective primary antibodies (4 °C, 24 h): rabbit monoclonal anti-TUJ1 (Abcam), mouse monoclonal anti-Nestin (Millipore), mouse monoclonal anti-CD133 (Prominin-1; glioblastoma and stem cell glycoprotein; Millipore), goat polyclonal anti-GFAP (Abcam), mouse monoclonal anti-SOX2 (ThermoFisher Scientific), rabbit monoclonal anti-EGFR (ThermoFisher Scientific), chicken monoclonal anti-MAP2 (ThermoFisher Scientific) and actin-staining Alexa Flour™ 488 Phalloidin (ThermoFisher Scientific). Cells were washed three times in PBS, incubated with appropriate secondary antibodies (Santa Cruz Biotechnology, Dallas, TX) at room temperature for 2 h, washed again three times with PBS, and then counterstained with 4,6-diamidino-2-phenylindole (DAPI, Sigma). In addition, double-immunostaining was performed in select cases – TUJ1/GFAP, Nestin/GFAP, Nestin/TUJ1 – using appropriate protocols as per vendor’s specifications. For coculture conditions, Transwell® inserts containing GBMs were removed, bottom mesh cutout and placed in separate wells, and stained using the above procedure. Negative controls were also processed in parallel, which were not stained with primary antibodies in each case. All cells were imaged using the Axio Vert.A1 fluorescence microscope under both phase contrast and fluorescence channels using a digital camera (Axiocam C1, Carl Zeiss) and Axiovision data acquisition software. Total number of cells per well was quantified using batch processing in ImageJ which quantified all the cells in the well emitting DAPI signal, and comparing that number to the total number of cells in that same well positively stained for the aforementioned anti-body markers (> 1000 cells counted per condition).
Axonal outgrowth
Axonal outgrowth was quantified using the fluorescence images acquired from the TUJ1 antibody staining of ReNcell cultures: ReNcells alone and ReNcell cocultures. Images from each respective condition were then imported into ImageJ and analyzed via the NeuronJ plugin [23]. This program traces the length of neurite branching from the soma to the end of axon using the bright intensity of the stained pixels (n = 3 wells/condition; >100 axons per condition counted).
Western blot analysis
Semi-quantitative western blot analysis was performed to identify GSK-3β, β-catenin, and Notch-1 expression levels in ReNcells and GBM cells, and assess if any of these pathways were involved in mediating the interactions between the two cell types. Briefly, at the end of standalone or cocultures, cells were washed with PBS, exposed to 1× passive lysis buffer (Promega), and the pooled cell lysate was centrifuged at 12,000 rpm for 20 min at 4 °C to extract protein content in the supernatant. Protein bands for GSK-3β (primary antibody from BD Biosciences), β-catenin (primary antibody from BD Biosciences) and notch-1 (primary antibody from Abcam) were detected using SDS/PAGE western blot procedures and appropriate secondary antibodies, using protocols we reported [24].
Cell proliferation assay
Cell proliferation was assessed using a BrdU cell proliferation assay (EMD Millipore). Assays for this experiment were run in 96-well plates as recommended by the manufacturer. The assay was first run on cells cultured for only 2 h to validate that each cell type was able to uptake the BrdU reagent and the assay would be able to provide reliable readings. ReNcells and GBMs were serially-diluted and seeded independently at the following densities per well: 1, 2, 5, 10, 20, 40, 80, 100, 150, and 300 thousand cells. After 2 h culture, the assay was run and the plates were read at 450/550 nm using a spectrophotometer (Biotek Synergy Winooski, VT) and analyzed using Gen 5 software. The data acquired followed a linear trend of cell population size vs. optical density, which was in agreement with the manufacturer’s recommendations, and thus the test was deemed valid to use for each specific case. The data collected from the serial dilution of the cell populations for 2 h was called “initial seeding” and was only performed on standalone GBMs and NSCs in differentiating media. In lieu of commercial Transwell® inserts for 96-well plates, ReNcell coculture conditions with GBM were achieved by taking 100 μL of spent media from the GBM cultures every 24 h and adding it to the ReNcell culture. Similarly, 100 μL of spent ReNcell media was added to GBM cultures. At this time, 100 μL of fresh media was replaced in each respective culture condition, and a full media change (200 μL) was performed on stand-alone culture conditions for consistency. All cultures were seeded at an initial density of 2 × 104 cells per well, which was measured using a Moxi-z mini-automated cell counter (Orflo, Ketchum, ID). The proliferation outcomes were tested at 3 different time points – days 1, 5 and 10.
Migration assay and cell tracking
To understand migration of GBM and ReNcells, a scratch test assay was performed to quantify cell migration in their respective microenvironments. Cells were seeded in 24-well plates, allowed to expand to confluence, and a 500 μm wide channel was scratched in the center of the confluent monolayer within each well. Time-lapse imaging in bright-field mode was performed using a 10× objective to ensure that the scratch was in the field of view. The scratch was performed on cell layers within the 24-well plate and not in the transwell. Thus, to quantify GBM migration within cocultures, they were seeded in the lower well and ReNcells were seeded in the Transwell® insert, and vice versa for quantifying ReNcell migration within cocultures.
For migration analysis, cells were individually tracked by selecting images taken from the similar field of view at various time points over 56 h. Images were analyzed in NIH ImageJ software by identifying the location of cells at each time point with reference to the original line of the scratch in each condition, thus allowing for all the images to be uniformly compared. Migration distances were excluded if a cell could not be easily identified from the previous time point or out of focus in the image. At least 30 cells were manually tracked per time point/condition. In addition to quantifying the average cumulative distance of cell migration at each time point under each culture condition, cell migration was also quantified by categorizing cells into two zones: those within the first 100 μm of the interface of scratch (termed as frontline), and those directly behind the frontline cells (termed as midline) at the start of the assay. Data was plotted as the individual migration distance vs. time with respect to the cell’s starting position and fitted with a mean and standard deviation using Graphpad prism software (San Diego, CA).
Statistical analysis
Except the cytokine/chemokine analysis, all other tests were performed in triplicate, with at least three wells per condition per replicate. Data acquired was expressed as mean ± standard error. Unless indicated otherwise. Statistical analysis was performed with Graphpad Prism 6 (La Jolla, CA) and plotted in Sigmaplot (San Jose, CA). Statistical significance between varying experimental conditions was tested using a student’s t-test (two-tailed, unequal distribution) or a two-way ANOVA with multiple comparisons (Fishers least significant difference). Statistically significant data was marked by p < 0.05.
Results
Cytokine and chemokine production
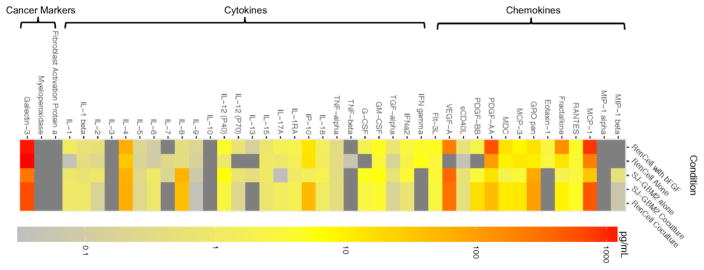
The analytes released by ReNcells and GBMs within standalone and cocultures was clustered based on their type (Fig. 1) or abundance levels (Fig SI-4: A: < 2.5 pg/mL; B: < 15 pg/mL; C: < 1500 pg/mL). When cultured alone, analytes except IL-7 and IL-8 were expressed by ReNcells in non-differentiating media (presence of bFGF). However, post-removal of bFGF to induce differentiation, ReNcells released lower amounts of IL-17a, PDGF-AA, MCP-1 and Fractalkine, and higher amounts of MCP-3, but no detectable levels of other analytes. Compared to ReNcells within standalone cultures, GBMs released higher amounts of IL-2, IL-6, IL-18, and G-CSF, and lower amounts of IL-17a, MCP-3, Galectin-3, PDGF-AA, and MCP-1. GBMs did not release any Eotaxin-1, IFN-γ, IP-10, and Fractalkine. Eotaxin-1, TGF-α and IFN-γ were not detectable in cocultures although they were quantified in GBM standalone cultures. In general, the analyte amounts in cocultures were within the ranges measured in standalone cultures of ReNcells or GBMs (except IL-1B), suggesting that the levels of these analytes in cocultures were not simply a combination of their individual levels in standalone cultures. The analyte amounts in GBM cocultures was similar to that in ReNcell cocultures, mostly due to similarities in culture conditions. Galectin-3 was the most abundantly released analyte in the pooled media for all the cases tested, and the only cancer-specific marker found; MPO and FAP were undetectable in all cases. Surprisingly, ReNcells released higher amounts of Galectin-3 than GBMs.
Figure 1.
Heat map representing cytokine and chemokine amounts released by ReNcells alone (presence or absence of bFGF), SJ-GBM2 cells alone, ReNcell cocultures, and SJ-GBM2 cocultures, as measured via laser bead immunoassay. Each row corresponds to one culture condition, and each column corresponds to one analyte. Analytes were clustered based on their type – cytokines, chemokines and cancer markers. Cocultures were performed using a Transwell® setup. The actual analyte concentration levels were shown in Supplementary Figure 4. No detectable levels of IL-10, IL-3, MIP1α, Myeloperoxidase, and Fibroblast Activation Protein were found.
Cell viability and proliferation
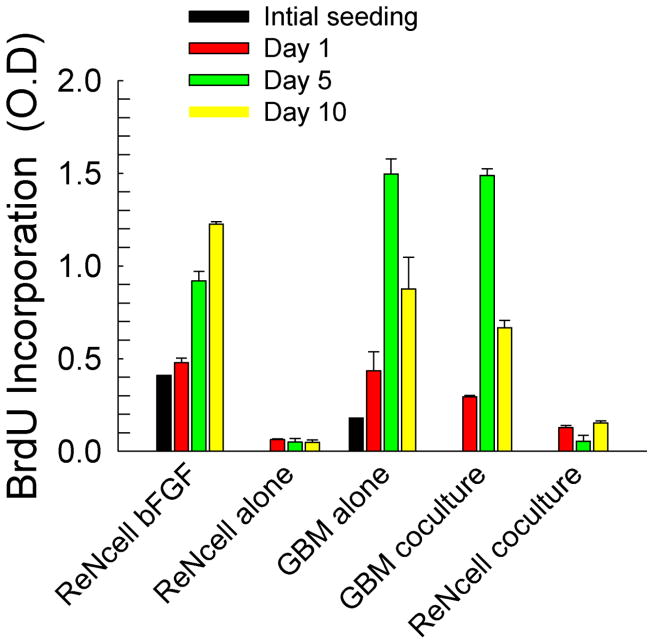
Representative images from the Live/Dead® assay were shown in Fig. SI-1A. Although cell viability in both the standalone and cocultures was higher than 80% (Fig. SI-1B), viability in cocultures appeared higher at both time points. However, no differences in cell survival between day 5 and day 10 were evident. Cell proliferation, on the other hand, appears to be significantly influenced both by the duration of culture as well as cocultures (Fig. 2). When ReNcells were cultured alone in non-differentiation media (containing bFGF), the number of actively dividing cells increased in an exponential fashion from days 1 to 10 (R2 = 0.96). Conversely, when ReNcells were cultured in differentiation media (absence of bFGF), no active cell proliferation was noted over the ten day cultures. In both standalone and cocultures, GBM cells aggressively multiplied as demonstrated by the almost 3-fold change in optical density between day 1 and 5 in both culture conditions. The significant decrease in O.D. within these GBM cultures by day 10 could be attributed to cell confluence within culture dishes, as evident from microscopy images. Finally, the presence of GBM cells (cocultures) appears to slightly stimulate ReNcell proliferation by day 10 compared to their standalone culture in differentiation media (p < 0.05).
Figure 2.
Cellular proliferation was measured via BrdU uptake and expressed as optical density (O.D) in various culture conditions. Measurements were taken immediately after seeding (4 h), and at days 1, 5 and 10. Error bars represent mean ± SEM for all cases.
Cell phenotype and differentiation within standalone and cocultures
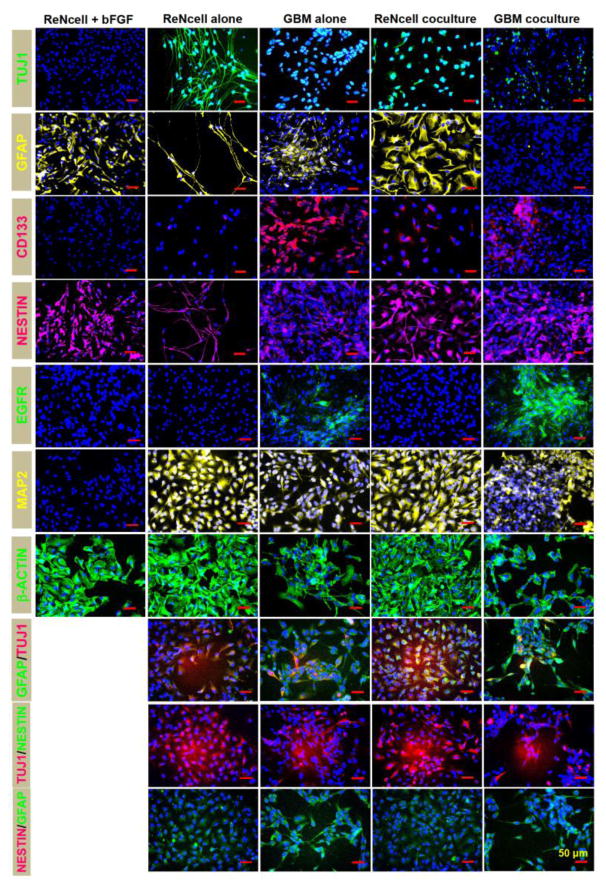
NPCs have been shown to differentiate and commit to distinct neural and glial lineages upon removal of EGF and bFGF from the culture media. Immunofluorescence imaging (Fig. 3) qualitatively revealed the lineage commitment of ReNcells and GBM cells within standalone and coculture conditions after a 10-day culture. The native phenotype of undifferentiated ReNcells (cultured with bFGF) at 24 h time point was also shown in select cases for comparison. It could be seen that the as-received ReNcells stained positive for Nestin, GFAP and β-actin, but negative for TUJ1, CD133, EGFR and MAP2, after 24 hours in culture. Double-immunostaining was not performed for as-received ReNcell cultures as they didn’t express TUJ1. ReNcells exhibited strong β-actin staining with well-spread cell morphology, distinct from GBM cells. However, after differentiation was induced by bFGF removal, significant morphological changes in ReNcells were noted by the end of ten day culture. A strong TUJ1 and MAP2 staining was evident, with axonal outgrowth in both TUJ1 and Nestin positive images. Astrocyte phenotype (GFAP marker) was also remarkably different in differentiated cultures compared to their undifferentiated counterparts. However, no CD133 or EGFR staining was noted in undifferentiated ReNcell standalone cultures. Double labeling revealed quite a few cells co-expressing GFAP and TUJ1 markers, but almost none co-expressing TUJ1/Nestin or GFAP/Nestin. On the other hand, within ten-day standalone cultures, GBM cells positively stained for CD133 and EGFR (cancer cell markers), GFAP (mature astrocyte), and Nestin, TUJ1 and MAP2 (evolution of neuron maturation and phenotype). GBM cells co-expressing TUJ1 and GFAP were also evident from double-immunolabeling images.
Figure 3.
Representative immunofluorescence images of ReNcells and GBMs, in standalone and cocultures, on day 10. For comparison, in select cases, ReNcells cultures in the presence of bFGF were also stained and imaged after 24 h in culture. Cultures were counterstained with DAPI for cell identification. Primary antibodies for TUJ1, GFAP, Nestin, CD133, EGFR, MAP2, and β-actin were used, with appropriate secondary antibodies. Double-immunolabeling was performed under select conditions. Scale bar: 50 μm.
Within cocultures, similar to their standalone counterparts, both ReNcells and GBMs stained positive for Nestin, although the morphology in ReNcells was more spread. Remarkably, compared to their standalone counterparts: (a) axonal outgrowth in ReNcell cocultures was significantly suppressed although TUJ1 expression could still be seen, (b) ReNcell cocultures exhibited strong and mature GFAP-staining indicative of astrocyte lineage morphology, (c) GBM cocultures had a reduced GFAP staining, (d) significantly higher ReNcells stained CD133-positive but not EGFR-positive in cocultures, (e) EGFR-staining was enhanced in GBM cocultures, (f) CD133 expression in GBM cocultures was slightly suppressed, (g) MAP2 and Nestin expressions remained largely unaffected in cocultures, and (h) the number of ReNcells co-expressing TUJ1 and GFAP increased. These results are important and relevant from numerous standpoints, as elaborated in the discussion section.
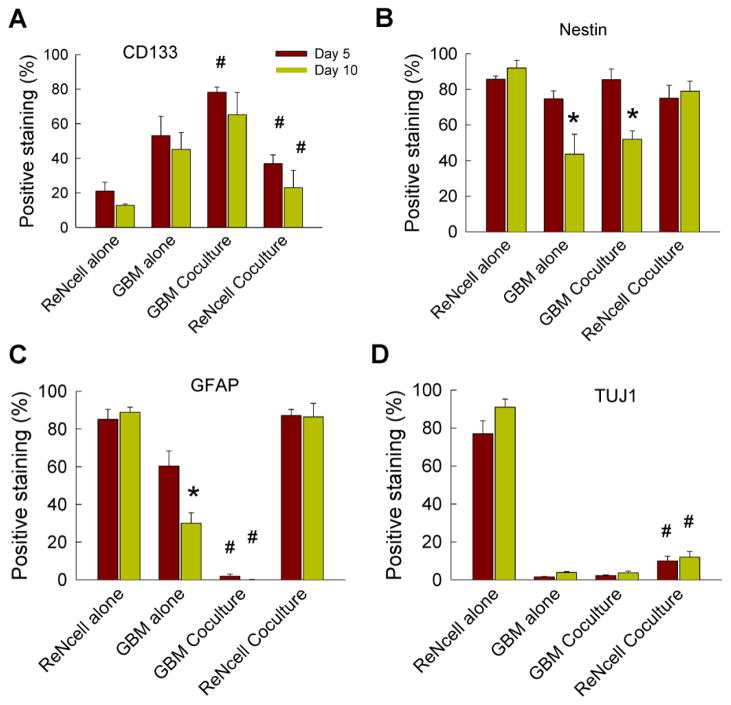
In general, trends in quantitative data closely matched the observations from qualitative imaging. Quantification of images indicated no time-dependent differences in CD133 expressing cells except for ReNcell alone cultures (Fig. 4A). GBM cultures, standalone or cocultures, had the highest number of cells positively-staining for CD133. CD133 expression in ReNcells was higher within cocultures compared to standalone counterparts. Of all the markers stained, Nestin was most abundantly found across all cases (Fig. 4B). Significant differences were not observed when comparing standalone cases to coculture cases on the same day, however a significant decrease in Nestin marker was observed from day 5 to day 10 within GBM cultures (p < 0.05). ReNcells expressed significant Nestin marker staining even after a 10-day differentiation induction. A significant reduction in GFAP expression was seen from days 5 to 10 in GBM cultures (Fig. 4C, p < 0.05) but not in ReNcell cultures. ReNcells had significant GFAP expression in both standalone and cocultures. GFAP expression was significantly suppressed within GBM cocultures compared to their standalone counterparts, on both day 5 and day 10 (p < 0.05). While TUJ1 expressing cells were the highest in standalone ReNcell cultures (Fig. 4D), coculturing with GBMs significantly reduced TUJ1 expression in these cultures. In both cocultures and standalone cultures, several cells appeared to be expressing “stemness” markers (CD-133 or Nestin) in conjunction with specific lineage markers (neural or glial).
Figure 4.
Quantification of ReNcell and GBM phenotype in standalone and cocultures on days 5 and 10, staining for (A) CD-133, (B) Nestin, (C) GFAP and (D) TUJ1. Values are expressed as a percentage of the number of cells stained positively for the respective marker (normalized to total number cells staining positive for DAPI). * indicates significant differences (p < 0.05) between day 5 vs. day 10, while # indicates significance in differences (p < 0.05) between standalone and cocultures of ReNcells or GBMs, at a given time point. Error bars represent mean ± SEM for all cases (n = 3/case).
Cell morphology and axonal outgrowth
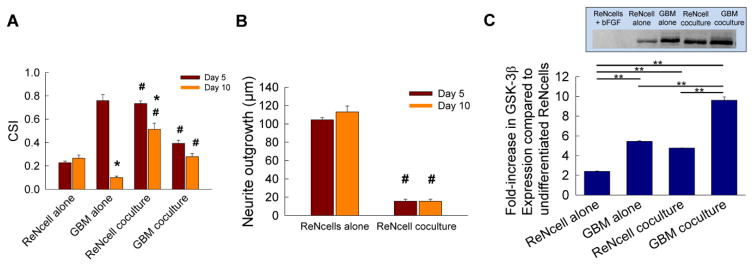
Significant time-dependent and culture condition dependent effects on cell shape index (CSI) were noted (Fig. 5A). ReNcells in standalone cultures had a more elongated shape highlighted by robust axonal extensions on both days 5 and 10. When ReNcells were cocultured with GBMs they became significantly more rounded at both the time points tested (p < 0.05). Standalone GBMs were more rounded on day 5, but became significantly more elongated by day 10, a pattern that was also noted in ReNcell cocultures (p < 0.05 for both cases). Although no significant changes in CSI were observed in GBM cocultures for day 5 vs. day 10, GBMs in cocultures on day 5 were significantly more elongated than their standalone counterparts. GBM cells in cocultures were significantly more rounded than their standalone counterparts by day 10.
Figure 5.
(A) Cell shape index (CSI) was measured using a particle analyzer plugin in ImageJ software, from images captured on days 5 and 10. CSI equal to one indicates infinitely elongated cell shape and a value of zero indicates perfectly rounded shape. (B) Average neurite outgrowth (μm) on days 5 and 10 was measured from TUJ1-stained cells within ReNcell cultures, in both standalone and coculture conditions. A NeuronJ plugin in ImageJ software was used to measure average outgrowth. * indicates significant differences (p < 0.05) between day 5 vs. day 10, while # indicates significant differences (p < 0.05) between coculture vs. standalone cultures at a given time point. Error bars represent mean ± SEM for all cases (n = 3/case). (C) Semi-quantitative western blot analysis of GSK-3β protein expression in ReNcells and GBM cells, cultured alone or in cocultures, at the end of 10 day culture period. Undifferentiated ReNcells (i.e., the presence of bFGF) were used as controls. Multi-fold increases in GSK-3β expression were noted in all the cases, with significantly higher levels in cocultures, specifically within GBM cells. Inset shows representative protein bands under respective culture conditions. ** indicates p < 0.05 vs. controls.
From the immunostaining images, neurite outgrowth within TUJ1-stained ReNcell cultures was quantified at days 5 and 10 (Fig. 5B). In general, at both the time points, neurites within standalone cultures were 7-fold longer than in cocultures (p < 0.001).
Of the three proteins tested using western blot analysis, β-catenin and notch-1 expressions were not detectable in both the cell types. However, GSK-3β expression was evident in both ReNcells and GBM cells (Fig. 5C), and representative bands under respective culture conditions were shown in inset. Undifferentiated ReNcells (i.e., cultured in bFGF presence) served as controls, and they did not express GSK-3β. Within standalone cultures, GBM cells expressed significantly higher levels (2.3-fold; p < 0.05) of GSK-3β compared to ReNcells. Similarly, GSK-3β expression in cocultures was significantly higher compared to standalone cultures (p < 0.05), with GBM cocultures having the highest expression (p < 0.05 vs. all other cases). This suggests that GSK-3β pathway might be involved in mediating the interactions between these two cell types, although further studies are required to validate this hypothesis.
Cell migration, speed and persistence
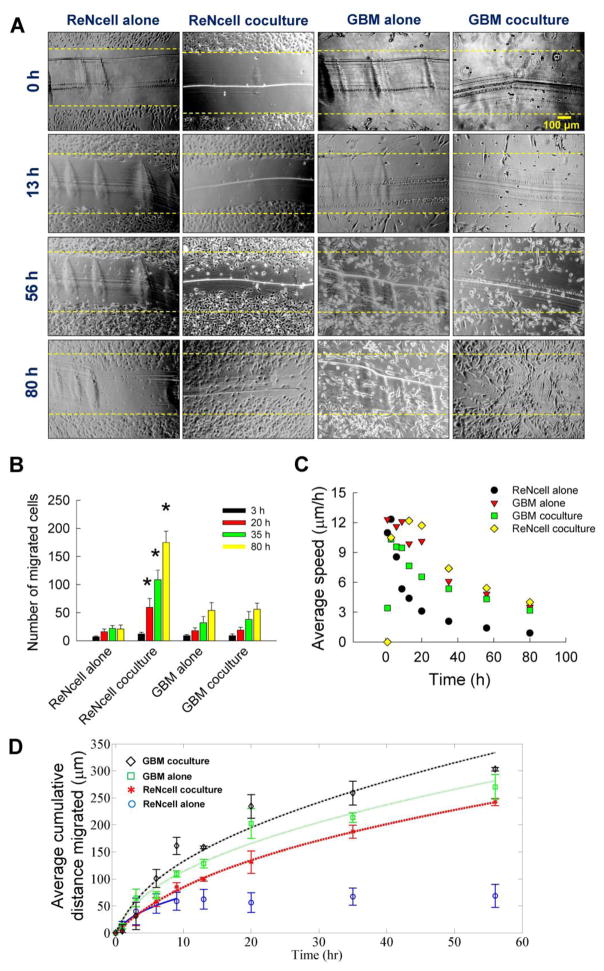
From the scratch test assay, the number of migrating cells in standalone and cocultures were assessed over a 80 h duration using time-lapse imaging, along with the cumulative distance migrated and their average speed. Representative images of ReNcells and GBMs at selected time points were shown in Fig. 6A. The dotted line in each image indicates the demarcation of original scratch from cell monolayers. Compared to that in standalone cultures, ReNcells migrated faster and in more numbers within cocultures, although such clear distinction could not be made in GBM cell cultures.
Figure 6.
(A) Representative bright-field images at selected time points, obtained from a scratch test assay. ReNcells or GBMs were cultured in 24-well plates, either standalone or in coculture (Transwell® setup) with the other cell type. Images were taken at regular intervals over an 80 h period to visualize cell migration. Yellow dotted lines in each image represent the starting position of the cells (at t = 0) when the scratch was initially made. Scale bar is 100 μm. (B) The number of ReNcells or GBMs, in standalone or cocultures, which migrated from their initial starting point into the cell-free zone created by scratch were quantified at various time points. * indicates significant differences in coculture vs. standalone cultures. Error bars represent mean ± SEM for all cases (n = 3/case). (C) The average cell speed (μm/h) was quantified in these cultures at various time points (n = 3 wells/time point). (D) The average cumulative distance (μm) covered by the cells was quantified at various time points (n = 3 wells/time point). Lines indicate the Ornstein-Uhlenbeck process model fit to the data shown in symbols. Error bars represent mean ± SEM for all cases.
The number of migrating cells in standalone and cocultures were quantified from these images (Fig. 6B). Cell migration within standalone ReNcell and GBM cultures was low in general, but increased with time over the duration of the study. However, among all the four test cases, significantly higher number of ReNcells migrated within cocultures at any time point, suggesting the influence of paracrine signaling from GBM cells. Cell speed was also found to be time-dependent in each culture condition. For the first 8 hours of study, cells in all conditions moved rapidly at an average speed of 8 μm/h or higher (Fig. 6C). After that, GBMs in standalone and cocultures, and ReNcells in cocultures, migrated at an average speed of 4 μm/h.
The cell population movement was also quantified from these images (Fig. 6D). The cumulative distance migrated by GBM cells in cocultures was the highest among the four cases over the first 52 hours (symbols in Fig. 6D). Cell migration was aggressive over the first 20 h covering almost 50 – 250 μm, after which it significantly slowed down. Expectedly, ReNcells remained dormant in standalone cultures for the remainder of the culture period, which is strikingly different from the additional 170 μm they traveled in cocultures. Although there were no significant differences in the total distance traveled by GBMs in standalone vs. cocultures (except at the 36 h time point), ReNcells in cocultures traveled significantly more distance than their standalone counterparts (244 μm vs. 69 μm, respectively, p < 0.05).
The cumulative distance migrated by cell populations in these cultures were fitted to a model based on Furth’s formula [25], which was more formally derived in the Ornstein-Uhlenbeck process [26], and describes the expected value of the mean squared displacement of the cell, 〈d⃗2(t)〉, as
Here, μ is the random motility coefficient that describes the rate at which cell population randomly migrates (analogous to diffusion coefficient), and P is the persistence time of the motion (i.e., average time between significant changes in direction). At t ≫ P, 〈d⃗2(t)〉 ≈ 4μt. Although this equation is typically used to model cell population motility in the absence of signaling molecules, this model could also be used when the biomolecular presence is global, as is the case in our experimental setup. This enables direct comparison of the parameters μ and P. The fitting of the data to the model to obtain the parameters μ and P was done using fminsearch – a method for finding the minimum of unconstrained multivariable functions using a derivative-free approach [27] in MATLAB (version 8.6). The fitting of the model is shown in Fig. 6D (lines), and the model parameters obtained are shown in Table 1. The data for only the first 13 hours of ReNcell movement in the absence of GBM cells was used in this model fit, as these cells are no longer motile after this time.
Table 1.
Model parameters derived for ReNcells and GBM cell population migration within standalone and cocultures.
| Random motility coefficient (μm2/h) | Persistence time (h) | |
|---|---|---|
| ReNcells alone (first 13 h) | 122.1 | 0.76 |
| ReNcell coculture | 280.5 | 3.94 |
| GBM cells alone | 360.3 | 1.02 |
| GBM coculture | 509.9 | 1.41 |
The model offered an excellent fit with the data, and the parameter values obtained are comparable in magnitude to other mammalian cell movements [28]. For example, the persistence time is 4 min for rabbit neutrophils, 30 min for rat alveolar macrophages, 1 h for mouse fibroblasts, 4–5 h for human microvessel endothelial cells, and 250 min for smooth muscle cells [29]. The random motility coefficient is >10,000 μm2/h for neutrophils, 3600 for μm2/h macrophages, 400–900 μm2/h for fibroblasts, 1250–2250 μm2/h for human microvessel endothelial cells, and 2200 μm2/h for smooth muscle cells [29].
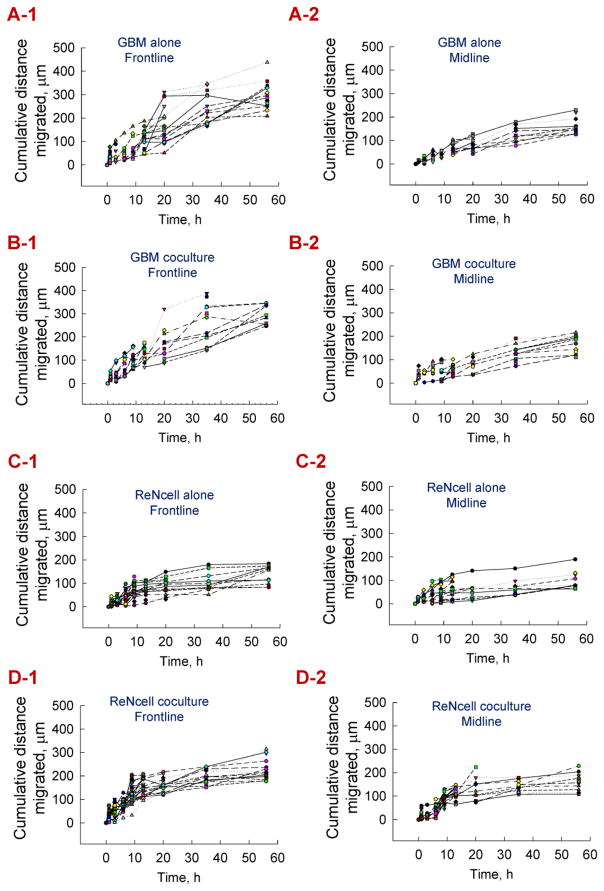
The individual cell migration over this period was analyzed by grouping the cells within these cultures into two separate but adjacent zones: those within the first 100 μm of the scratch interface (frontline), and those directly behind the frontline cells (midline) at the start of the assay. Individual cells in each of these zones were tracked (at least n = 20 per condition) over the first 56 hours, and their migration patterns were shown in Fig. 7. Each symbol represents the migration of a single cell under those respective culture conditions over 56 hours. The average distance migrated by the frontline and midline cells over 56 h, in each case, were shown in Fig. SI-2. The key findings from this individual cell migration analysis were that (i) frontline cells in each condition migrated more distance compared to their midline counterparts (A-1 vs. A2, etc.); (ii) distance migrated by both frontline and midline GBM cells were higher than their ReNcell counterparts (A-1 vs. C-1, B-1 vs. D-1, etc.), in standalone and coculture conditions; (iii) both frontline and midline ReNcells in standalone and cocultures exhibited a dormant phenotype with little migration after the first 20 hours, in contrast to their GBM counterparts, and (iv) the migration patterns of frontline cells but not midline cells broadly mimics that of overall cell population dynamics shown in Fig. 6D.
Figure 7.
From scratch test assay, cell migration within standalone and cocultures of ReNcells and GBMs was divided into two zones – frontline (first 100 μm from scratch interface) and midline (next 100 μm behind frontline). Cumulative distance migrated by individual frontline cells over 56 h was tracked in GBMs cultured alone (A-1), GBM cocultures (B-1), ReNcells cultured alone (C-1), and ReNcell cocultures (D-1). Similarly, distance migrated by individual midline cells was tracked within GBMs cultured alone (A-2), GBM cocultures (B-2), ReNcells cultured alone (C-2), and ReNcell cocultures (D-2). Each symbol corresponds to a cell tracked over the test duration, and at least 15 individual cells were randomly selected and plotted for each culture condition.
ReNcells cultured alone, in undifferentiated medium in a similar experimental setup, were tracked for their frontline and midline cell movement patterns and the results were shown in Fig. SI-3. It was noted that most of the cell migration ceased beyond 13 h of culture in both the cases, and a majority of the frontline cells traveled 110–190 μm on average, while the midline cells migrated 45–110 μm on average. These migration distances were comparable to those by ReNcells cultured alone in the presence of differentiating medium (Fig. 7, C-1 and C-2).
Discussion
Brain tumors are the second most common malignancy in younger populations and treated routinely by surgery in conjunction with chemo-radiation therapies. Such removal of malignant brain tissue could result in loss of cognitive function as well as the unintentional death of surrounding healthy neurons [30]. In lieu of pharmacological options, surgical or radiological intervention is necessary for a majority of these patients, therefore researchers have begun exploring the utility of stem cells to restore the lost neurons and their functionality and improve the patient quality of life [31]. On the other hand, NPCs have been shown to be capable of tracking tumors which has led to the notion that these cells could also be engineered as delivery vehicles for targeting therapeutic agents to tumor sites [32]. Therefore, towards developing an expedient in vitro model that mimics tumor microenvironment, the current study describes a coculture model of human NPCs and pediatric GBM cells to ascertain the effects of paracrine signaling on their phenotype and functionality.
Previous studies have reported on the tumor development of SJ-GBM2 cells in mice models [33]. When tumor pieces of SJ-GBM-2, derived at autopsy from a 5-year old female patient, were engrafted in the dorsal lateral flanks of CBA/CaJ mice (4 weeks of age) to initiate tumor growth, each tumor grew in over 90% of the recipient mice and exhibited human-origin characteristics as validated by karyotype and isoenzyme patterns [34].
Cytokines and chemokines influence the development of primary tumors in malignant diseases, which has prompted further research in identifying the types, amounts and effects of such markers in a tumor microenvironment [35, 36]. Unfortunately, limited information exists in literature on the cytokine and chemokine production levels by NPCs, as it is assumed that microglial cells are the primary contributors of these analytes in the CNS. The concentrations of several cytokines and chemokines produced by GBMs in this study (e.g., MCP-1, interleukins, Galactin-3) and the absence of some markers (e.g., IFN-γ) is in broad agreement with prior studies in this regard [37–40]. We also noted that ReNcells expressed high levels of several analytes (e.g., MCP-1, PDGF-AA) and the cancer marker Galectin-3, even when cultured in non-differentiating media. In agreement with our results, Sheng et al. reported that NPCs exposed to inflammatory stimuli such as TNF-α or IL-1β produced significant concentrations of MCP-1 and IP-10 [41]. Altering the types and amounts of these analytes in a microenvironment could regulate tumor cell phenotype (invasion, proliferation, migration, neoangiogenesis, immune cell infiltration) as well as influence NPC development and behavior, presumably via activation of similar biochemical pathways (i.e. JAK/STAT or NF-κB) [42, 43]. Our results indicate that coculturing with GBMs and the presence of various cytokines and chemokines significantly influenced the patterns of NPC differentiation, morphology, and migration.
Previous reports have suggested that the release of cytokines (e.g., TNF-α) and interleukins by activated microglial cells after injury (both in vivo and in vitro) could be detrimental to NPC survival [44]. In agreement with the results from our study, other studies involving in vitro cocultures or in vivo glioblastoma injection have reported high NSC survival rates in a cancerous microenvironment [19, 45]. On the contrary, Glass et al. observed a significant suppression of adult GBM cell proliferation and apoptosis induction by NPCs within cocultures on coverslips [20]. Our data suggests that the panel of cytokines and chemokines produced by the GBM/NPC cocultures did not suppress their viability; in fact, cocultures marginally promoted cell survival. SJ-GBM2 cells in this study might have released a panel of analytes unique to the cancer milieu, which helped maintain ReNcell viability. In agreement with literature [46], we noted significant viable and rapidly proliferating GBM cells in standalone and cocultures during the initial five days of culture. A probable contributing factor for the decrease in OD readings from day 5 to day 10 in GBM cultures might be the lack of available space for cells to expand in the culture wells, which was confirmed by examination under the light microscope. NPCs continued to proliferate in non-differentiation media although the number of actively-dividing cells (BrdU-incorporated) was significantly reduced in cultures with differentiation media, as reported in literature [47, 48]. A few ReNcells in cocultures were actively dividing even on day 10, possibly due to dysregulation of the STAT3 or Wnt pathway from signals elicited from the surrounding GBMs, suggesting these NPCs could be exhibiting a cancerous phenotype [1, 49].
NPCs respond to a variety of biochemical and spatiotemporal cues to induce differentiation into specific neural and glial lineages [48]. In vivo studies demonstrated the feasibility of NPC transplantation to repair diseased tissues in the brain, and coupled with the evidence that NPCs could survive in a cancerous microenvironment, it could be inferred that similar strategies could be used for regenerative purposes during or after cancer treatment [50, 51]. However, research within the cancer milieu is limited to studies focusing on the potential integration and delivery of anti-cancer drugs to the lesion site [19, 52]. Glass et al. observed in NSC-GBM explant cocultures that approximately 30% of NSCs stained positive for Nestin (“stemness” marker), and within that Nestin-positive subpopulation, 60% co-expressed GFAP and another 30% co-expressed the pre-neural markers (PSA-NCAM and DCx) [20]. Here we see similar patterns in co-expression of specific neuronal or glial lineages in conjunction with stemness marker. It has also been documented that pediatric GBMs express GFAP marker as well as the stemness markers Nestin and CD133 [2]. CD133 is a cell surface glycoprotein and has been utilized as a surface marker to identify cancer cells from various solid tumors including breast, pancreas, liver and colorectal cancers [53–56]. Studies have shown that glioma-initiating cells and NPCs isolated from GBMs express CD133, and both CD133+ and CD133− cells represent various differentiation stages for tumor cells [57]. In fact, glioblastoma cells positive for both EGFR (variant III) and CD133 were shown to exhibit cancer stemness, i.e., tumor-initiating ability and self-renewal [58].
The co-expression of CD133 and Nestin markers by rhabdomyosarcoma tissue and derived cell lines point to the existence of cancer stem cells within these tumors [59]. The expression of CD133, Nestin, EGFR and GFAP by standalone pediatric GBM cells (in the absence of cocultures) in our study suggest to the potential presence of such cancer stem cells. While coculturing with ReNcells appeared to still retain the co-expression of CD133 and Nestin in these cells, GFAP expression was almost suppressed. Hemmati et al. isolated cells from pediatric GBMs which displayed qualities more commonly observed in healthy NSC cultures, including the ability to form neurospheres and express neural markers [3]. Given that CD133-expression is not normally present in the adult brain, rather only in GBMs and NSCs, cells expressing these markers may indicate a potential transformation of NSCs to a cancer phenotype [16]. Furthermore, the cytokine/chemokine production in coculture conditions appears to influence cell fate [60, 61], i.e., downregulation of TUJ1 commitment in NPCs, reduced expression of GFAP in GBMs, as well as increased expression of CD133. However, more studies are required to understand the precise mechanisms driving these changes, so that it could lead to integration of NSCs into this microenvironment without transformation to undesired lineages.
In standalone cultures, NPCs displayed an elongated shape highlighted by axonal and dendrite extensions [62], whereas confluent GBMs were generally small and rounded with star like filopodia [63]. However, in cocultures, their morphologies switched, as GBMs appeared elongated and NSCs more rounded. This suggests that NPCs were forced to differentiate into glial lineage, initiated by the presence of cytokines via a notch signaling pathway [64]. Given the similarities in the GBM proliferation within standalone and cocultures, and the lack of elevated staining for specific neuronal markers, the morphological data gathered from GBM cocultures is not enough evidence to suggest these cells are losing their cancer phenotype.
Alterations in axonal outgrowth have been attributed to the Rho signaling pathway [65], and could be differentially regulated by the presence of several cytokines and chemokines [66]. Since ReNcells were found to have a more rounded morphology in cocultures, axons from TUJ1-positive cells were measured and compared, which revealed reduced neurite outgrowth in coculture conditions. This suggests the role of cancer cell microenvironment in inhibiting the axonal elongation, which is a crucial obstacle that must be overcome to regenerate any lost neuronal network in vivo. We hypothesize that a strategic combination of growth factors [67] and biological scaffolds [68] might help in shielding and promoting robust axonal outgrowth to overcome this obstacle.
Numerous studies attest to the complex and controversial role of GSK-3β in regulating cellular functions (e.g., apoptosis, migration), specifically in tumor environments [69]. Only recently, the role of GSK-3β in regulating NPCs [70] and adult glioma cells [71] has been reported. To our knowledge, GSK-3β expression in pediatric GBM cells and NPCs in a coculture environment has not been reported earlier. Although we noted that GSK-3β was expressed in both NPCs and GBM cells, with significantly higher expressions in cocultures, the mechanistic role underlying GSK-3β signaling in their survival, proliferation and migration yet remains to be elucidated.
Research quantifying NPC migration in a cancerous microenvironment has recently gained momentum as NPCs could act as a delivery vehicle for chemotherapeutics [11, 19, 32]. NPCs have been shown to chemotaxis towards the tumor site and envelop the tumors both in vivo and in vitro, depending on the severity of tumor pathology [20]. Aboody et al. observed the ability of NPCs to surround, track and target glioblastoma cells, both in vivo and in vitro, likely due to a complex mixture of biochemical signals acting as attractants, adhesion and substrate molecules, and chemokines [11]. Although they did not perform further analysis, our cytokine and chemokine data supports their notion that a complex cocktail of signaling molecules were released in this coculture microenvironment. Using a scratch test, Natarajan et al. observed comparable number of migrating GBM cells, which they attributed to changes in focal adhesion kinases (FAK) modulated by several key analytes in the microenvironment [72]. Using Boyden chambers, Heese et al. observed an increase in NSC migration when exposed to several GBM cell lines, or under exposure to analytes such as TGF-α and PDGF-AA [73]. Debray et al. noted that GBM cell motility on laminin-coated surfaces was directly affected by the presence of Galectin-3, which has the ability to modulate α6 and β1 integrin expression and cause cellular locomotion [74]. Our results on GBM speed were strikingly similar to that of Chicoine et al. [75]. In summary, the data presented here demonstrated an increased NPC migration in the presence of GBMs, in terms of cell speed and number of cells migrating, which could have practical applications in targeted cell therapy and enhanced regeneration of lost tissue.
Conclusions
Alternate strategies to disrupt the progression of pediatric glioblastomas could lead to increased patient survival rates. Given the similarities in the molecular pathways driving NPC and GBM differentiation, survival, and proliferation, studies like this provide insight into the ideal microenvironment needed for NPCs to perform their intended functions. Our results suggest that within cocultures, a unique cocktail of analytes is produced which was regulating NPC migration, lineage commitment (less neuronal and more astrocyte), and axonal outgrowth. Given that NPC survival and migration was not compromised in this coculture microenvironment, they still offer a suitable platform to deliver anti-tumor drugs. Importantly, GBM cocultures appeared to promote CD133 expression in ReNcells, suggesting their potential transformation to cancer phenotype. Given the unpredictability of NPC differentiation in this microenvironment, coupled with the lack of axonal outgrowth, further analysis of the biochemical markers produced by both cell types in cocultures, as well as an increased understanding of the molecular pathways involved, could provide the requisite details needed to fully utilize NPCs for regenerative medicine applications in brain cancer.
Supplementary Material
Live/Dead® analysis of ReNcells and GBMs, in standalone and cocultures, at 5 and 10 day time points. Representative images of the stained cells for all conditions on day 10 were shown (A). Green indicates live cells and red indicates dead cells. The images were quantified and the cell survival in respective cases plotted (B). Error bars represent mean ± SEM for all cases.
The average distance migrated by frontline (A) and midline (B) cells within standalone and cocultures were tracked over 56 h and plotted for comparison. These averages were obtained from the data shown in Fig. 7, where individual cells in frontline and midline zones within standalone and cocultures were tracked over 56 hours. Frontline and midline GBM cells traveled significantly higher distance on average, compared to ReNcells in standalone cultures (p < 0.05). Frontline cells also migrated significantly more distance than their midline counterparts (p < 0.05), in all culture conditions. Error bars represent mean ± SEM for all cases.
ReNcells cultures in the presence of non-differentiation media, i.e., containing bGFG, were subjected to scratch test assay and the frontline (A) and midline (B) cells were traced and their migration distances quantified (similar to Fig. SI-2). At least 15 cells in each respective zone were traced. Significantly higher migration distance was noted in frontline cells compared to midline counterparts (p < 0.01). The average distance migrated by ReNcells (both frontline and midline) in the presence of non-differentiating media is similar to that by ReNcells alone exposed to differentiating media (C-1 and C-2, respectively, in Fig. 7), but significantly lower than that ReNcells in cocultures with GBM cells (D-1 and D-2, respectively, in Fig. 7).
The cytokines, chemokines and cancer cell markers released by ReNcells and GBM cells, either in standalone or in cocultures, were clustered and plotted depending on their abundance levels: A – low; B – medium; C – high amounts.
Highlights.
Interactions between human NPCs and human pediatric GBM-derived cells investigated using a coculture setup.
NPC morphology, phenotype, differentiation patterns, neurite outgrowth, and migration patterns significantly influenced by GBMs.
Cumulative migration distance fit to Furth’s formula derived formally from Ornstein-Uhlenbeck process.
Significant applications in NPC transplantation efforts to replace lesions of excised tumor sites.
Such cocultures could help elucidate the underlying molecular pathways involved in GBM-NPC interactions in a tumor microenvironment.
Acknowledgments
Cell Lines were obtained from the Children’s Oncology Group Cell Culture and Xenograft Repository (cogcell.org), which is supported by Alex’s Lemonade Stand Foundation. Kurt Farrell was partially supported by the Cellular and Molecular Medicine Fellowship (CMMA) and Dissertation Research Awards (DRA) from CSU. Research reported in this study was partially supported by National Institute of Environmental Health Sciences of the National Institutes of Health under award number R01ES025779 to Drs. Kothapalli and Lee.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sandberg CJ, Altschuler G, Jeong J, et al. Comparison of glioma stem cells to neural stem cells from the adult human brain identifies dysregulated Wnt- signaling and a fingerprint associated with clinical outcome. Exp Cell Res. 2013;319:2230–2243. doi: 10.1016/j.yexcr.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 2.Bax DA, Little SE, Gaspar N, et al. Molecular and phenotypic characterisation of paediatric glioma cell lines as models for preclinical drug development. PLoS One. 2009;4:e5209. doi: 10.1371/journal.pone.0005209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hemmati HD, Nakano I, Lazareff JA, et al. Cancerous stem cells can arise from pediatric brain tumors. Proc Natl Acad Sci U S A. 2003;100:15178–15183. doi: 10.1073/pnas.2036535100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Galli R, Binda E, Orfanelli U, et al. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 2004;64:7011–7021. doi: 10.1158/0008-5472.CAN-04-1364. [DOI] [PubMed] [Google Scholar]
- 5.Singh SK, Hawkins C, Clarke ID, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 6.Goffart N, Kroonen J, Rogister B. Glioblastoma-initiating cells: relationship with neural stem cells and the micro-environment. Cancers (Basel) 2013;5:1049–1071. doi: 10.3390/cancers5031049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu Y, Wu PY. CD133 as a marker for cancer stem cells: progresses and concerns. Stem Cells Dev. 2009;18:1127–1134. doi: 10.1089/scd.2008.0338. [DOI] [PubMed] [Google Scholar]
- 8.Mizrak D, Brittan M, Alison M. CD133: molecule of the moment. J Pathol. 2008;214:3–9. doi: 10.1002/path.2283. [DOI] [PubMed] [Google Scholar]
- 9.Temple S. The development of neural stem cells. Nature. 2001;414:112–117. doi: 10.1038/35102174. [DOI] [PubMed] [Google Scholar]
- 10.Clarke DL, Johansson CB, Wilbertz J, et al. Generalized potential of adult neural stem cells. Science. 2000;288:1660–1663. doi: 10.1126/science.288.5471.1660. [DOI] [PubMed] [Google Scholar]
- 11.Aboody KS, Brown A, Rainov NG, et al. Neural stem cells display extensive tropism for pathology in adult brain: evidence from intracranial gliomas. Proc Natl Acad Sci U S A. 2000;97:12846–12851. doi: 10.1073/pnas.97.23.12846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caren H, Stricker SH, Bulstrode H, et al. Glioblastoma Stem Cells Respond to Differentiation Cues but Fail to Undergo Commitment and Terminal Cell-Cycle Arrest. Stem Cell Reports. 2015 doi: 10.1016/j.stemcr.2015.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holland EC, Celestino J, Dai C, et al. Combined activation of Ras and Akt in neural progenitors induces glioblastoma formation in mice. Nat Genet. 2000;25:55–57. doi: 10.1038/75596. [DOI] [PubMed] [Google Scholar]
- 14.Ligon KL, Huillard E, Mehta S, et al. Olig2-regulated lineage-restricted pathway controls replication competence in neural stem cells and malignant glioma. Neuron. 2007;53:503–517. doi: 10.1016/j.neuron.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dai C, Celestino JC, Okada Y, et al. PDGF autocrine stimulation dedifferentiates cultured astrocytes and induces oligodendrogliomas and oligoastrocytomas from neural progenitors and astrocytes in vivo. Genes Dev. 2001;15:1913–1925. doi: 10.1101/gad.903001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pfenninger CV, Roschupkina T, Hertwig F, et al. CD133 is not present on neurogenic astrocytes in the adult subventricular zone, but on embryonic neural stem cells, ependymal cells, and glioblastoma cells. Cancer Res. 2007;67:5727–5736. doi: 10.1158/0008-5472.CAN-07-0183. [DOI] [PubMed] [Google Scholar]
- 17.Sanai N, Alvarez-Buylla A, Berger MS. Neural stem cells and the origin of gliomas. N Engl J Med. 2005;353:811–822. doi: 10.1056/NEJMra043666. [DOI] [PubMed] [Google Scholar]
- 18.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 19.Ehtesham M, Kabos P, Kabosova A, et al. The use of interleukin 12-secreting neural stem cells for the treatment of intracranial glioma. Cancer Res. 2002;62:5657–5663. [PubMed] [Google Scholar]
- 20.Glass R, Synowitz M, Kronenberg G, et al. Glioblastoma-induced attraction of endogenous neural precursor cells is associated with improved survival. J Neurosci. 2005;25:2637–2646. doi: 10.1523/JNEUROSCI.5118-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reya T, Morrison SJ, Clarke MF, et al. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 22.Kang MH, Smith MA, Morton CL, et al. National Cancer Institute pediatric preclinical testing program: model description for in vitro cytotoxicity testing. Pediatr Blood Cancer. 2011;56:239–249. doi: 10.1002/pbc.22801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meijering E, Jacob M, Sarria JC, et al. Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytometry A. 2004;58:167–176. doi: 10.1002/cyto.a.20022. [DOI] [PubMed] [Google Scholar]
- 24.Simmers P, Gishto A, Vyavahare N, et al. Nitric oxide stimulates matrix synthesis and deposition by adult human aortic smooth muscle cells within three-dimensional cocultures. Tissue Eng Part A. 2015;21:1455–1470. doi: 10.1089/ten.tea.2014.0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fürth R. Die Brownsche Bewegung bei Berücksichtigung einer Persistenz der Bewegungsrichtung. Mit Anvendungen auf die. Z Physik. 1920;2:244–256. [Google Scholar]
- 26.Uhlenbeck GE, Ornstein LS. On the theory of Brownian Motion. Phys Rev. 1930;21:823–841. [Google Scholar]
- 27.Lagarias J, Reeds JA, Wright MH, et al. Convergence Properties of the Nelder-Mead Simplex Method in Low Dimensions. SIAM Journal of Optimization. 1998;9:112–147. [Google Scholar]
- 28.Rosello C, Ballet P, Planus E, et al. Model driven quantification of individual and collective cell migration. Acta biotheoretica. 2004;52:343–363. doi: 10.1023/B:ACBI.0000046602.58202.5e. [DOI] [PubMed] [Google Scholar]
- 29.Lauffenburger DA, Linderman JJ. Receptors: Models for binding, trafficking and signaling. New York: Oxford University Press; 1993. [Google Scholar]
- 30.Ellenberg L, McComb JG, Siegel SE, et al. Factors affecting intellectual outcome in pediatric brain tumor patients. Neurosurgery. 1987;21:638–644. doi: 10.1227/00006123-198711000-00006. [DOI] [PubMed] [Google Scholar]
- 31.Gardner SL. Application of stem cell transplant for brain tumors. Pediatr Transplant. 2004;8(Suppl 5):28–32. doi: 10.1111/j.1398-2265.2004.00202.x. [DOI] [PubMed] [Google Scholar]
- 32.Li SC, Kabeer MH, Vu LT, et al. Training stem cells for treatment of malignant brain tumors. World J Stem Cells. 2014;6:432–440. doi: 10.4252/wjsc.v6.i4.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Middlemas DS, Stewart CF, Kirstein MN, et al. Biochemical correlates of temozolomide sensitivity in pediatric solid tumor xenograft models. Clin Cancer Res. 2000;6:998–1007. [PubMed] [Google Scholar]
- 34.Houghton PJ, Cheshire PJ, Hallman JD, et al. Efficacy of topoisomerase I inhibitors, topotecan and irinotecan, administered at low dose levels in protracted schedules to mice bearing xenografts of human tumors. Cancer Chemother Pharmacol. 1995;36:393–403. doi: 10.1007/BF00686188. [DOI] [PubMed] [Google Scholar]
- 35.Balkwill F. Cancer and the chemokine network. Nat Rev Cancer. 2004;4:540–550. doi: 10.1038/nrc1388. [DOI] [PubMed] [Google Scholar]
- 36.Lippitz BE. Cytokine patterns in patients with cancer: a systematic review. Lancet Oncol. 2013;14:e218–228. doi: 10.1016/S1470-2045(12)70582-X. [DOI] [PubMed] [Google Scholar]
- 37.Bresalier RS, Yan PS, Byrd JC, et al. Expression of the endogenous galactose-binding protein galectin-3 correlates with the malignant potential of tumors in the central nervous system. Cancer. 1997;80:776–787. [PubMed] [Google Scholar]
- 38.Gustafson MP, Lin Y, New KC, et al. Systemic immune suppression in glioblastoma: the interplay between CD14+HLA-DRlo/neg monocytes, tumor factors, and dexamethasone. Neuro Oncol. 2010;12:631–644. doi: 10.1093/neuonc/noq001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yeung YT, McDonald KL, Grewal T, et al. Interleukins in glioblastoma pathophysiology: implications for therapy. Br J Pharmacol. 2013;168:591–606. doi: 10.1111/bph.12008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamanaka R, Tanaka R, Saitoh T, et al. Cytokine gene expression on glioma cell lines and specimens. J Neurooncol. 1994;21:243–247. doi: 10.1007/BF01063773. [DOI] [PubMed] [Google Scholar]
- 41.Sheng WS, Hu S, Ni HT, et al. TNF-alpha-induced chemokine production and apoptosis in human neural precursor cells. J Leukoc Biol. 2005;78:1233–1241. doi: 10.1189/jlb.0405221. [DOI] [PubMed] [Google Scholar]
- 42.Zhu VF, Yang J, Lebrun DG, et al. Understanding the role of cytokines in Glioblastoma Multiforme pathogenesis. Cancer Lett. 2012;316:139–150. doi: 10.1016/j.canlet.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 43.Iwami K, Natsume A, Wakabayashi T. Cytokine networks in glioma. Neurosurg Rev. 2011;34:253–263. doi: 10.1007/s10143-011-0320-y. discussion 263–254. [DOI] [PubMed] [Google Scholar]
- 44.Breton JM-DY. Impact of Cytokines on Neural Stem/Progenitor Cell Fate. Neurol Neurophysiol. 2011:S4. [Google Scholar]
- 45.Ehtesham M, Kabos P, Gutierrez MA, et al. Induction of glioblastoma apoptosis using neural stem cell-mediated delivery of tumor necrosis factor-related apoptosis-inducing ligand. Cancer Res. 2002;62:7170–7174. [PubMed] [Google Scholar]
- 46.Colin C, Baeza N, Tong S, et al. In vitro identification and functional characterization of glial precursor cells in human gliomas. Neuropathol Appl Neurobiol. 2006;32:189–202. doi: 10.1111/j.1365-2990.2006.00740.x. [DOI] [PubMed] [Google Scholar]
- 47.Breier JM, Radio NM, Mundy WR, et al. Development of a high-throughput screening assay for chemical effects on proliferation and viability of immortalized human neural progenitor cells. Toxicol Sci. 2008;105:119–133. doi: 10.1093/toxsci/kfn115. [DOI] [PubMed] [Google Scholar]
- 48.Donato R, Miljan EA, Hines SJ, et al. Differential development of neuronal physiological responsiveness in two human neural stem cell lines. BMC Neurosci. 2007;8:36. doi: 10.1186/1471-2202-8-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sherry MM, Reeves A, Wu JK, et al. STAT3 is required for proliferation and maintenance of multipotency in glioblastoma stem cells. Stem Cells. 2009;27:2383–2392. doi: 10.1002/stem.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hovakimyan M, Muller J, Wree A, et al. Survival of transplanted human neural stem cell line (ReNcell VM) into the rat brain with and without immunosuppression. Ann Anat. 2012;194:429–435. doi: 10.1016/j.aanat.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 51.Liang Y, Agren L, Lyczek A, et al. Neural progenitor cell survival in mouse brain can be improved by co-transplantation of helper cells expressing bFGF under doxycycline control. Exp Neurol. 2013;247:73–79. doi: 10.1016/j.expneurol.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Frank RT, Edmiston M, Kendall SE, et al. Neural stem cells as a novel platform for tumor-specific delivery of therapeutic antibodies. PLoS One. 2009;4:e8314. doi: 10.1371/journal.pone.0008314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wright M, Calcagno A, Salcido C, et al. Brca1 breast tumors contain distinct CD44+/CD24− and CD133+ cells with cancer stem cell characteristics. Breast Cancer Res. 2008;10:R10. doi: 10.1186/bcr1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Olempska M, Eisenach PA, Ammerpohl O, et al. Detection of tumor stem cell markers in pancreatic carcinoma cell lines. Hepatobiliary Pancreat Dis Int. 2007;6:92–97. [PubMed] [Google Scholar]
- 55.Suetsugu A, Nagaki M, Aoki H, et al. Characterization of CD133+ hepatocellular carcinoma cells as cancer stem/progenitor cells. Biochem Biophys Res Comm. 2006;351:820–824. doi: 10.1016/j.bbrc.2006.10.128. [DOI] [PubMed] [Google Scholar]
- 56.Ricci-Vitiani L, Lombardi DG, Pilozzi E, et al. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111–115. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- 57.Singh SK, Clarke ID, Terasaki M, et al. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821–5828. [PubMed] [Google Scholar]
- 58.Emlet DR, Gupta P, Holgado-Madruga M, et al. Targeting a glioblastoma cancer stem-cell population defined by EGF receptor variant III. Cancer Res. 2014;74:1238–1249. doi: 10.1158/0008-5472.CAN-13-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sana J, Zambo I, Skoda J, et al. CD133 expression and identification of CD133/nestin positive cells in rhabdomyosarcomas and rhabdomyosarcoma cell lines. Anal Cell Pathol (Amst) 2011;34:303–318. doi: 10.3233/ACP-2011-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Levina V, Marrangoni AM, DeMarco R, et al. Drug-selected human lung cancer stem cells: cytokine network, tumorigenic and metastatic properties. PLoS One. 2008;3:e3077. doi: 10.1371/journal.pone.0003077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Deverman BE, Patterson PH. Cytokines and CNS development. Neuron. 2009;64:61–78. doi: 10.1016/j.neuron.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 62.Christopherson GT, Song H, Mao HQ. The influence of fiber diameter of electrospun substrates on neural stem cell differentiation and proliferation. Biomaterials. 2009;30:556–564. doi: 10.1016/j.biomaterials.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 63.Machado CM, Schenka A, Vassallo J, et al. Morphological characterization of a human glioma cell line. Cancer Cell Int. 2005;5:13. doi: 10.1186/1475-2867-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nagao M, Sugimori M, Nakafuku M. Cross talk between notch and growth factor/cytokine signaling pathways in neural stem cells. Mol Cell Biol. 2007;27:3982–3994. doi: 10.1128/MCB.00170-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gu H, Yu SP, Gutekunst CA, et al. Inhibition of the Rho signaling pathway improves neurite outgrowth and neuronal differentiation of mouse neural stem cells. Int J Physiol Pathophysiol Pharmacol. 2013;5:11–20. [PMC free article] [PubMed] [Google Scholar]
- 66.Gonzalez-Perez O, Quinones-Hinojosa A, Garcia-Verdugo JM. Immunological control of adult neural stem cells. J Stem Cells. 2010;5:23–31. [PMC free article] [PubMed] [Google Scholar]
- 67.Srinivasan P, Zervantonakis IK, Kothapalli CR. Synergistic effects of 3D ECM and chemogradients on neurite outgrowth and guidance: a simple modeling and microfluidic framework. PLoS One. 2014;9:e99640. doi: 10.1371/journal.pone.0099640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Horn EM, Beaumont M, Shu XZ, et al. Influence of cross-linked hyaluronic acid hydrogels on neurite outgrowth and recovery from spinal cord injury. J Neurosurg Spine. 2007;6:133–140. doi: 10.3171/spi.2007.6.2.133. [DOI] [PubMed] [Google Scholar]
- 69.Doble BW, Woodgett JR. GSK-3: tricks of the trade for a multi-tasking kinase. J Cell Sci. 2003;116:1175–1186. doi: 10.1242/jcs.00384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Holowacz T, Alexson TO, Coles BL, et al. The responses of neural stem cells to the level of GSK-3 depend on the tissue of origin. Biol Open. 2013;2:812–821. doi: 10.1242/bio.20131941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kotliarova S, Pastorino S, Kovell LC, et al. Glycogen synthase kinase-3 inhibition induces glioma cell death through c-MYC, nuclear factor-kappaB, and glucose regulation. Cancer Res. 2008;68:6643–6651. doi: 10.1158/0008-5472.CAN-08-0850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Natarajan M, Stewart JE, Golemis EA, et al. HEF1 is a necessary and specific downstream effector of FAK that promotes the migration of glioblastoma cells. Oncogene. 2006;25:1721–1732. doi: 10.1038/sj.onc.1209199. [DOI] [PubMed] [Google Scholar]
- 73.Heese O, Disko A, Zirkel D, et al. Neural stem cell migration toward gliomas in vitro. Neuro Oncol. 2005;7:476–484. doi: 10.1215/S1152851704000754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Debray C, Vereecken P, Belot N, et al. Multifaceted role of galectin-3 on human glioblastoma cell motility. Biochem Biophys Res Commun. 2004;325:1393–1398. doi: 10.1016/j.bbrc.2004.10.181. [DOI] [PubMed] [Google Scholar]
- 75.Chicoine MR, Silbergeld DL. Assessment of brain tumor cell motility in vivo and in vitro. J Neurosurg. 1995;82:615–622. doi: 10.3171/jns.1995.82.4.0615. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Live/Dead® analysis of ReNcells and GBMs, in standalone and cocultures, at 5 and 10 day time points. Representative images of the stained cells for all conditions on day 10 were shown (A). Green indicates live cells and red indicates dead cells. The images were quantified and the cell survival in respective cases plotted (B). Error bars represent mean ± SEM for all cases.
The average distance migrated by frontline (A) and midline (B) cells within standalone and cocultures were tracked over 56 h and plotted for comparison. These averages were obtained from the data shown in Fig. 7, where individual cells in frontline and midline zones within standalone and cocultures were tracked over 56 hours. Frontline and midline GBM cells traveled significantly higher distance on average, compared to ReNcells in standalone cultures (p < 0.05). Frontline cells also migrated significantly more distance than their midline counterparts (p < 0.05), in all culture conditions. Error bars represent mean ± SEM for all cases.
ReNcells cultures in the presence of non-differentiation media, i.e., containing bGFG, were subjected to scratch test assay and the frontline (A) and midline (B) cells were traced and their migration distances quantified (similar to Fig. SI-2). At least 15 cells in each respective zone were traced. Significantly higher migration distance was noted in frontline cells compared to midline counterparts (p < 0.01). The average distance migrated by ReNcells (both frontline and midline) in the presence of non-differentiating media is similar to that by ReNcells alone exposed to differentiating media (C-1 and C-2, respectively, in Fig. 7), but significantly lower than that ReNcells in cocultures with GBM cells (D-1 and D-2, respectively, in Fig. 7).
The cytokines, chemokines and cancer cell markers released by ReNcells and GBM cells, either in standalone or in cocultures, were clustered and plotted depending on their abundance levels: A – low; B – medium; C – high amounts.