Abstract
Epigenetic mechanisms can stably maintain gene expression states even after the initiating conditions have changed. Often epigenetic information is transmitted only to daughter cells, but evidence is emerging, in both vertebrate and invertebrate systems, for transgenerational epigenetic inheritance (TEI), the transmission of epigenetic gene regulatory information across generations. Each new description of TEI helps uncover the properties, molecular mechanisms and biological roles for TEI. The nematode Caenorhabditis elegans has been particularly instrumental in the effort to understand TEI, as multiple environmental and genetic triggers can initiate an epigenetic signal that can alter the expression of both transgenes and endogenous loci. Here, we review recent studies of TEI in C. elegans.
Keywords: C. elegans, transgenerational, epigenetic, nuclear RNAi, HRDE-1
C. elegans as a model for transgenerational inheritance
Gene expression can be altered within just a single generation in response to an organism’s changing environment via epigenetic mechanisms that alter gene expression without altering DNA sequence. These mechanisms include modifications to chromatin structure and short interfering RNA (siRNA, see Glossary Box)-mediated regulation of transcription and mRNA stability. Remarkably, several examples in Caenorhabditis elegans and other animals [1] suggest that epigenetic modifications can be inherited across generations in the absence of the environmental or genetic signal that initiated the epigenetic modification. One way to propagate a transgenerational signal is via maternal effect, in which the progeny receives a maternal signal in utero that, if present in the germline of the progeny, can potentially instruct epigenetic changes in the next two generations. However, in C. elegans and in other species [2–7], epigenetic memories of altered gene expression can be propagated across three or more generations, suggesting that active mechanisms exist to maintain an epigenetic ancestral memory. These studies have been particularly productive in C. elegans due to its short generation time, ease of genetic analysis, and powerful molecular genetic tools. In fact, various environmental and genetic manipulations can heritably alter the expression of both transgenes and endogenous genes for several generations. In this review, we summarize epigenetically regulated transgenes and endogenous loci in C. elegans and discuss the molecular identity of the transmitted information and the mechanisms by which this transmitted information alters gene expression. We conclude with open questions in the field.
Heritable silencing of germline transgenes
When RNA interference (RNAi) was first described in C. elegans, two remarkable properties were noted regarding its transmission. Not only could the silencing signal be transmitted between cells, but it could also be transmitted to the progeny [8]. In RNAi, double stranded RNA (dsRNA) is processed into primary siRNAs stabilized by Argonaute proteins [9–13]. These siRNA/Argonaute complexes direct the RNA-directed RNA polymerase-dependent production of secondary siRNAs antisense to a homologous RNA template. Argonaute-stabilized secondary siRNA can then promote post-transcriptional silencing of the targeted mRNA or inhibit transcription [13–18]. In addition to dsRNA-derived exogenous primary siRNAs, endogenous primary small RNAs, including piRNAs, can direct the production of endogenous secondary siRNAs [13,19–22] (Figure 1). The transmission of dsRNA-initiated silencing between cells requires a highly conserved transmembrane protein SID-1, and SID-1 function in both the parent and progeny is required for the efficient transmission of dsRNA-initiated silencing from parent to progeny [23,24].
Figure 1.
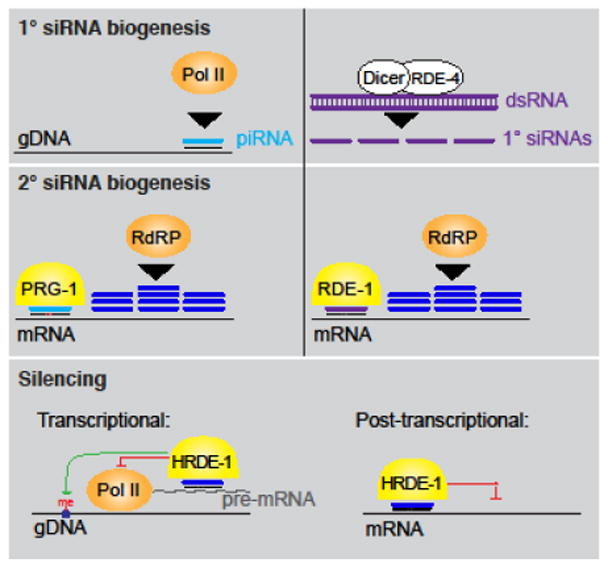
In C. elegans, piRNAs are transcribed by RNA Polymerase II (Pol II) and primary siRNAs can be produced from exogenous dsRNA. piRNAs and primary siRNAs are stabilized by unique Argonaute proteins (yellow) and can direct the production of highly amplified secondary siRNAs by RNA-directed RNA polymerases (RdRPs). Secondary siRNAs bound to the Argonaute HRDE-1 can direct transcriptional and post-transcriptional silencing of homologous loci. HRDE-1 is required for the heritable transmission of gene regulatory information in C. elegans.
Remarkably, dsRNA-initiated gene silencing can result in a heritable signal that persists over several generations (Figure 2A, C). In C. elegans, a germline gfp transgene that is present in multiple copies and silenced by gfp dsRNA can remain silenced for several generations in the absence of the initiating dsRNA signal. Strong silencing is transmitted to all progeny of dsRNA-exposed worms and to 30% of the next generation. Although most worms in subsequent generations re-express the transgene, silencing can persist indefinitely in a small fraction of worms [25]. Similarly, a germline-expressed transgene integrated in the genome as a single copy can be silenced with a 60–80% transmission rate for at least four generations after exposure to gfp dsRNA [26,27], after which the transgene becomes re-expressed in the absence of selection for the silenced state [28]. Both the partial transmission of the silencing signal and the observed reversion to the expressed state, suggest that an epigenetic signal is heritably transmitted, as the initiating signal would be quickly diluted among several hundred progeny at each generation.
Figure 2.
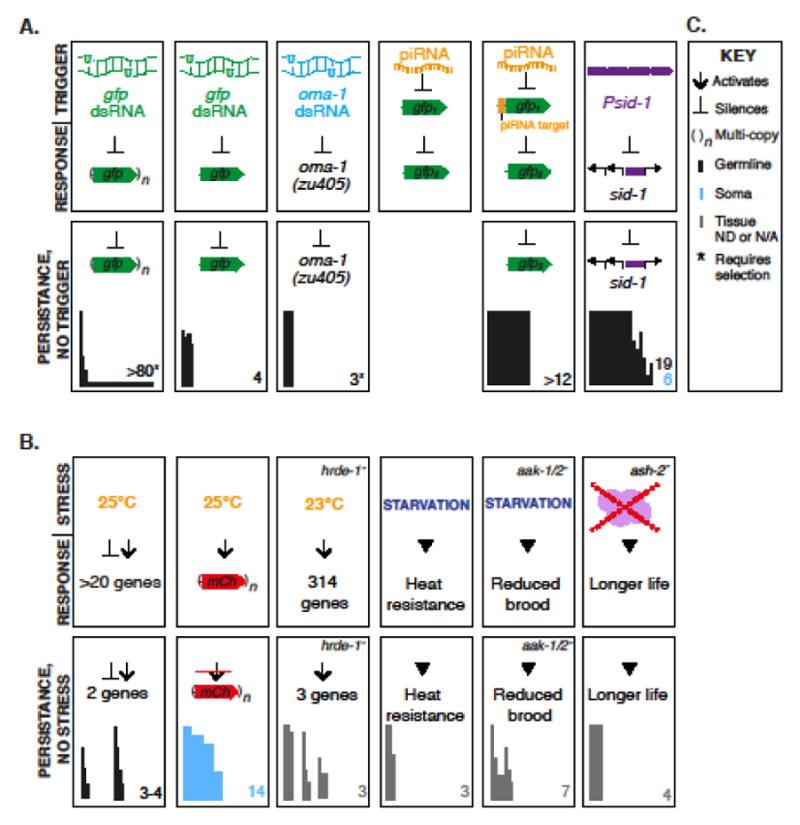
A summary of transgenerational epigenetic inheritance in C. elegans. A. Epigenetic silencing of transgenes and endogenous loci can be initiated by exogenous RNA, endogenous piRNAs, and a multi-copy intergenic region. Silencing persists after loss of the initiating trigger in the germline (black bars) or soma (blue bars). Bars represent transmission efficiency at each generation. B. Gene expression and physiological changes initiated by an environmental stress or methyltransferase inactivity can be persist transgenerationally. C. Key for parts A and B. ND: Not determined. N/A: Not applicable.
Although most transgenes present in single copy in the genome remain expressed in the germline [29], some single copy transgenes encoding gfp can become spontaneously epigenetically silenced in the germline in a process called “RNAe” for “epigenetic” [30] (Figure 2A, C). RNAe silencing is initiated by endogenous piRNAs, which are stabilized by the Argonaute protein PRG-1 [31]. Single copy transgenes introduced into prg-1 mutants never become silenced. However, inactivation of prg-1 after RNAe silencing does not result in re-expression of the transgene. Thus, piRNAs participate only in the initiation of transgene silencing in RNAe [30].
Downstream of both dsRNA-initiated silencing and piRNA-initiated silencing, is the nuclear RNAi Argonaute HRDE-1/WAGO-9 (hereafter called HRDE-1), which is required to maintain germline transgene silencing across generations. Progeny of worms fed gfp dsRNA can only silence a germline-expressed gfp transgene if HRDE-1 is functional [26,27]. HRDE-1 binds both endogenous secondary siRNAs and secondary siRNAs antisense to the targeted RNA [27], leading to the model that dsRNA-derived primary siRNAs direct the production of HRDE-1-stablized secondary siRNAs. A germline transgene engineered to be targeted by an endogenous piRNA (“piRNA sensor”) reveals that endogenous piRNAs can also direct the production of HRDE-1-stabilized siRNAs, which are required to silence the piRNA sensor [19] and HRDE-1 also contributes to piRNA-initiated RNAe [30]. One RNAe-silenced transgene becomes fully de-silenced when hrde-1 is mutated. Another transgene is partially de-silenced in a hrde-1 mutant, and even more strongly expressed when a cytoplasmic Argonaute protein is also compromised, suggesting that both cytoplasmic and nuclear Argonaute proteins can contribute to RNAe silencing [30].
The requirement for the siRNA-stabilizing Argonaute HRDE-1 suggests that small RNAs likely participate in the heritable, epigenetic silencing of germline transgenes. HRDE-1 was initially described as a germline-expressed nuclear Argonaute that is functionally similar to the somatic Argonaute NRDE-3 [32]. Together with additional nuclear RNAi factors, nuclear Argonaute proteins silence targeted genes transcriptionally by inhibiting RNA polymerase II and by promoting the deposition of the silencing histone H3K9me3 mark [27,32,33]. Indeed, in HRDE-1 dependent RNAe, the silencing is largely transcriptional, as pre-mRNA levels of a silenced single copy transgene decrease dramatically [30]. Later studies suggested that nuclear Argonautes can also contribute to post-transcriptional gene silencing [34,35]. Post-transcriptional silencing likely contributes to the dsRNA-initiated heritable silencing of a germline gfp transgene because mRNA levels decrease more severely than do pre-mRNA levels in silenced animals [26].
Several lines of evidence suggest that changes to chromatin structure also contribute to the epigenetic silencing of transgenes. Silencing of the piRNA sensor requires the Heterochromatin Protein 1 (HP1) homolog hpl-2, the methyltransferase set-25 and the predicted methyltransferase set-32 [26]. RNAe silencing is associated with an increase in histone H3K9me3 levels along the transgene DNA and methyltransferases (mes-3, mes-4) and HP1 homologs (hpl-1, hpl-2) are required to maintain transgene silencing in RNAe [30]. dsRNA-mediated silencing of a single copy germline gfp transgene also requires set-25 [26] while silencing of a multi-copy gfp transgene requires a predicted histone deacetylase (hda-4), a histone acetyltransferase (K03D10.3), a chromodomain-containing protein (mrg-1) and a homolog of a chromatin remodeling protein in yeast (isw-1) [25]. The mechanisms for heritable silencing of multi-copy and single copy gfp transgenes are at least partially distinct, as mrg-1 and hda-4 are not required for the heritable silencing of a single copy gfp transgene [26].
The epigenetic signal that heritably silences germline transgenes is paramutagenic; this signal can dominantly convert an active locus to the silenced state. When homologous expressed and RNAe-silenced single copy transgenes are introduced into a single animal by mating, both transgenes become stably silenced [30]. Although a maternally contributed silenced transgene results in more robust paramutation in the first generation, a paternally contributed silenced transgene can also silence the homologous transgene [30]. Like RNAe, a silenced piRNA sensor dominantly silences a homologous transgene [26], and the transgene remains stably silenced for many generations after the sensor is crossed away [35]. Although chromatin modifications contribute to maintenance of heritable silencing, the paramutagenic signal that silences the piRNA sensor is not encoded in the chromatin structure of the silenced locus. Even if progeny of silenced worms do not inherit the silenced locus, the dominant silencing signal is still successfully transmitted [35].
Heritable silencing of endogenous genes
In addition to heritably silenced transgenes, several heritably silenced endogenous loci have been identified in C. elegans (Figure 2A, C). A comparison between TEI of transgene and endogenous gene silencing is crucial, as the initiating events, mechanisms and biological roles of silencing a foreign DNA element and an endogenous locus may be distinct. While transmission of dsRNA-initiated silencing is often restricted to a single generation [8], a mutant allele of the germline-expressed oocyte maturation factor oma-1 can be silenced for several generations [36,37]. oma-1 dsRNA targeting a dominant negative oma-1 allele (zu405) [36] suppresses the embryonic lethality associated with this allele. Viable progeny of oma-1(zu405) worms injected with oma-1 dsRNA are recovered for three generations after dsRNA injection, indicating that oma-1(zu405) remains silenced by a signal propagated across generations [37]. Silencing is transmitted to only a fraction of progeny and the locus becomes re-expressed by the fourth generation [37]. The silencing signal is dominant, can be transmitted through the maternal and paternal germlines, and requires the nuclear RNAi Argonaute HRDE-1 [27,37,38]. In contrast to heritable silencing of a gfp transgene, heritable silencing of oma-1 does not require the methyltransferase set-25 and the predicted methyltransferase set-32 [38].
sid-1 encodes the protein required to transport dsRNA between generations in C. elegans [23] and can itself be silenced by an epigenetic mechanism [39] (Figure 2A, C, Figure 3). When sid-1 is silenced, animals can no longer respond effectively to RNAi [23]. sid-1 is the first described endogenous locus that can be stably epigenetically silenced. Epigenetic silencing of sid-1 is initiated by the introduction of a transgenic multi-copy array that contains only the sid-1 promoter and 5′ UTR sequence (“Psid-1”) [39]. Such initiation of silencing is unprecedented; multi-copy arrays of transcriptional fusions lacking a coding sequence have never been reported to silence the endogenous locus [40,41]. Once silenced, the sid-1 locus remains silenced in progeny that do not inherit the initiating array for up to 13 generations in the germline in all progeny and for up to 19 generations in some progeny. Transmission of silencing is robust and does not require selection for the silenced state at each generation. A silenced sid-1 locus is paramutagenic and the silencing signal can be transmitted even when the silenced locus is not transmitted. In contrast to RNAe [30] and silencing of the endogenous oma-1 locus [37], the silencing signal is transmitted only through the maternal germline [39].
Figure 3.
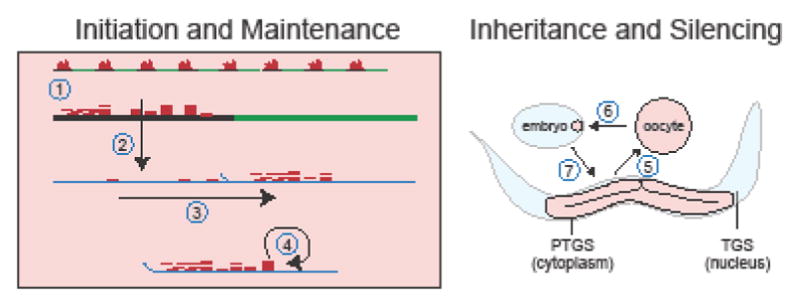
sid-1 TEI as a model of RNA mediated inheritance.
(1) Amplified siRNAs transcribed from a multicopy sid-1-promoter transgene array (2) target rare sid-1 pre-mRNA and (3) prime the RdRP dependent spread and amplification of siRNA targeting the sid-1 coding region prior to mRNA trans-splicing. (4) siRNA targeting the coding region are maintained by an RdRP-dependent autocatalytic amplification cycle. Mother to progeny transfer of silencing is accomplished by transfer of germline siRNA via ooctyes. (5) PTGS-produced germline siRNA are transmitted to oocytes.
(6) siRNA persist in embryonic germline precursor (pink) and initiate nuclear TGS to silence sid-1 in somatic cells (blue).
(7) PTGS silencing is maintaned and silences sid-1 in the cytoplasm of the developing germline.
Like all other cases of heritable gene and transgene silencing, silencing of sid-1 in the germline requires the nuclear Argonaute HRDE-1 and downstream nuclear RNAi pathway components [39]. However, the chromatin modifying enzymes previously implicated in epigenetic transgene silencing (set-25, set-32), are not required for sid-1 silencing in the germline. All silenced animals, whether they carry the Psid-1 array or not, show a three-fold increase in siRNAs targeting the sid-1 coding region. HRDE-1-mediated silencing of sid-1 can be initiated without dsRNA and piRNAs, likely because HRDE-1-dependent siRNAs can be transcribed antisense to the multi-copy transgene array (Figure 3). These siRNAs can then target the sid-1 5′ UTR and direct the production of secondary siRNAs targeting the sid-1 coding sequence. siRNAs targeting the sid-1 coding sequence are maintained even in the absence of the array for many generations [39]. Although piRNAs are not absolutely required for the initiation of sid-1 silencing, an endogenous piRNA antisense to the Psid-1 transcript contributes to the efficient initiation of silencing [39].
Epigenetic regulation of somatically expressed genes is rare. In fact, the paramutagenic signal that silences a germline gfp transgene is unable to silence a somatically expressed gfp transgene [26]. Furthermore, while RNAi silencing of the somatically expressed gene ceh-13 can be transmitted with low penetrance, it is unknown whether silencing occurs in the soma or whether germline silencing of ceh-13 contributes to the phenotype [25]. In addition to sid-1 silencing in the germline, Psid-1 worms also silence sid-1 in the soma and somatic silencing persists for four generations in non-array progeny with complete transmission. Maternal germline silencing directs somatic sid-1 silencing in the progeny, but the somatic nuclear RNAi Argonaute NRDE-3 [32] is not required for somatic sid-1 silencing. Instead, somatic silencing of sid-1 specifically requires the chromatin modifying enzymes that have previously been implicated in germline transgene silencing (set-25, set-32), indicating that the mechanisms of transgenerational silencing in the soma and germline are distinct [39].
Stress-induced transgenerational gene expression changes
In addition to initiation by nucleic acids, epigenetic alteration of gene expression can also be initiated by physiological stress (Figure 2B, C). C. elegans is most comfortable at a temperature of 20°C, while growth at 25°C is considered mildly stressful and results in gene expression changes [42]. After resumed growth at 20°C, expression levels of 20 genes remain significantly altered in young embryos. Notably, two genes investigated in detail remain altered for three to four generations after return to 20°C, and these heritable gene expression changes are more dramatic when ancestors are heat stressed for two consecutive generations [42]. Heat-initiated gene expression changes require endogenous RNAi components and the levels of siRNAs antisense to the two heritably altered genes also change in response to the stress and persist for multiple generations, correlating with the silenced or elevated levels of the gene. Interestingly, unlike dsRNA induced oma-1 silencing and RNAe, but similar to epigenetic silencing at the sid-1 locus, inheritance is restricted to the maternal germline. Thus, heat stress results in a maternally heritable, siRNA-dependent signal that alters gene expression in subsequent generations [42]. In related experiments, some genes can remain up-regulated for up to three generations following an ancestral heat stress specifically in hrde-1 mutants but not in wild-type worms, leading to the proposal that germline nuclear RNAi can prevent large fluctuations in gene expression in response to heat stress [43].
Heat stress at 25°C can also epigenetically increase the expression of several somatically expressed multi-copy transgenes [44] (Figure 2B, C). This increase in expression can persist for up to seven or fourteen generations after return 20°C if ancestors are exposed to heat stress for 1 or 5 generations, respectively. The activated locus is not paramutagenic, and the epigenetic signal responsible for the elevated expression is likely present in the chromatin structure. Consistent with this model, the repressive SET-25-dependent histone H3K9me3 mark is reduced along arrays with increased expression [44].
Like heat stress, starvation can also result in persistent gene expression and siRNA level changes (Figure 2B, C). When starved, C. elegans L1 larvae enter a reversible larval arrest phase that can last for weeks in the absence of food [45,46]. Small RNA sequencing in starved larvae and subsequent fed progeny reveals that starvation alters the expression levels of many small RNAs (predicted to target >1000 genes) and expression of some small RNAs remains altered in fed worms for three generations [47]. Transgenerational persistence of starvation-modulated siRNA expression changes depends on the nuclear RNAi Argonaute HRDE-1 and the endogenous small RNA pathway protein RDE-4. Some mRNAs that can be targeted by siRNAs decreased in starved worms show the expected transgenerational increase in expression. These mRNAs include genes involved in nutrient regulation, providing a possible yet unverified physiological role for heritable siRNA changes in response to starvation conditions [47].
Transgenerational modification of physiological traits
Despite an impressive ability to survive starvation, L1 larvae that experience starvation exhibit several phenotypes once they are returned to food, including reduced brood size [48] (Figure 2B, C). Reduced brood size does not persist in subsequent fed generations in wild-type worms. However, aak-1/2 worms, which lack the metabolic kinase AMPK, exhibit post-starvation reduced brood size for up to seven generations [49]. Worms lacking AMPK function have elevated levels of the histone H3K4me3 modification in the germline and soma and these elevated levels persist in non-starved conditions for at least 6 generations [49]. Genetic reduction of H3K4me3 levels by RNAi targeting methyltransferase complex components can partially restore post-starvation brood size in aak-1/2 worms, indicating that increased H3K4me3 levels contribute to brood size reduction across generations [49]. The transgenerational signal responsible for this increase in H3K4me3 and the genetic consequences of increased H3K4me3 in starved aak-1/2 worms and their progeny remain unknown [49], but the unregulated accumulation of histone methylation marks in worms lacking a demethylase enzyme similarly results in a progressive reduction in fertility [50]. Starvation in wild-type worms results in increased heat resistance in the great grand-progeny of select starved worms (those most physiologically affected) suggesting that this trait is also subject to transgenerational epigenetic control by unknown mechanisms [48].
Perturbations in histone modifying complexes can also lead to a heritable increase in longevity (Figure 2B, C). The ASH-2 complex methylates histone H3K4 in C. elegans and other animals. Disruption of this complex results in a decrease in histone H3K4me3 marks and also extends lifespan in C. elegans [51]. Notably, increased longevity persists for four generations even after the function of this histone modifying complex is restored [51]. This persistent increase in longevity requires an intact germline and depends on the presence of a histone H3K4me3 demethylase. Despite a global restoration of H3K4me3 levels to wild-type levels in wild-type progeny from ancestors that lacked ASH-2 complex activity, these progeny still have longer life spans, suggesting that regulation at a few genes may contribute to longevity. Consistent with this hypothesis, many genes whose proper expression depends on the ASH-2 complex function in longevity and growth and a significant portion of these genes remains misregulated three generations after restoration of the ASH-2 complex [51].
Concluding Remarks and Future Perspectives
Studies in C. elegans reveal that RNAi-based mechanisms can allow the organism to maintain a gene expression memory for many generations. Identification of additional heritably silenced loci will help to answer remaining questions regarding how and why such stable heritable epigenetic states are established and maintained (See Outstanding Questions Box). To date, a limited number of endogenous epigenetically silenced loci have been identified in C. elegans [25,37,39]. In one report, of 171 genes targeted by RNAi, silencing of 13 loci was inherited [25]. It is not yet clear why dsRNA targeting of some genes is more amenable to producing a heritable signal than others, although current models suggest that a gene must be expressed in the germline for the heritable signal to be sufficiently amplified and transmitted by a germline-restricted HRDE-1-dependent process in each generation. In addition to sid-1, the two genes located upstream of sid-1 are also heritably silenced by the same intergenic fragment that silences sid-1 [39]. No other intergenic locus has been reported to stably silence an endogenous gene. It is possible that other heritably silenced germline genes have not yet been identified because silencing of many genes is difficult to detect. Alternatively, it is possible that unidentified features of heritably silenced loci or the initiating silencing signals contribute to an enhanced ability to silence the locus. It may be easier to identify additional endogenous heritably silenced loci in a recently described sensitized mutant background in which germline transgene silencing can be extended by several generations [28].
Outstanding Questions Box.
Why are some endogenous loci more amenable to heritable silencing than others?
What are the physical signals that are transmitted across generations?
What mechanisms control the duration of epigenetic changes to gene expression?
What are the physiological consequences of stress-induced heritable gene expression changes?
Are the mechanisms that support TEI in C. elegans and in mammals conserved?
Heritably silenced endogenous genes and transgenes often rely on small RNA-based pathways and the silenced state is associated with an increase in small RNAs that can target the silenced gene. Despite this strong correlation, it is difficult to determine whether these small RNAs are the physical molecule transmitted across generations or whether these small RNAs are the downstream product re-generated in response to some other physical signal. A genetic analysis of the role of HRDE-1 in transgenerational silencing at the sid-1 locus suggests that RNA molecules, likely small RNAs themselves, embody the heritably transmitted entity. The transmitted sid-1 silencing signal can persist in the absence of zygotic HRDE-1 for a single generation presumably due to the presence of maternal HRDE-1, but requires HRDE-1 for stability and for the execution of silencing in subsequent generations [39]. Although all cases of transgenerational epigenetic gene silencing require HRDE-1, different transmission properties, including the persistence of transmission and the differential ability of males to transmit the signal, suggest that the identity or strength of the signal may vary among cases of heritable silencing.
Because the environmental signal that epigenetically alters gene expression is likely to be transient, a mechanism that ensures a transient increase in phenotypic diversity and the eventual re-expression of an epigenetically silenced gene is preferable to a permanent alteration. In fact, transgenerational epigenetic regulation of most transgenes and all endogenous genes is not permanent. One possibility is that an epigenetic signal is not transmitted across generations with 100% efficiency and thus diluted over time. However, a more precise mechanism may also limit transgenerational transmission. For example, endogenous RNAi pathways can limit the stability of heritable silencing by competing for shared pathway components [28,52,53]. Upon initiation of exogenous RNAi, endogenous siRNAs initially decrease but become re-expressed in the next generation [28]. Many of these endogenous siRNAs can target RNAi pathway components, potentially directly limiting the inheritance of silencing in response to exogenous dsRNA. This model is further supported by the observation that silencing and the persistence of antisense small RNAs targeting a germline gene can be extended by initiation of a second exogenous RNAi response targeting an unrelated gene [28]. It is not yet known what ensures the specific regulation of anti-RNAi factor small RNAs in response to initiation of exogenous RNAi nor whether such a competition model can be extended to other cases of transgenerational information transfer.
Persistent transgenerational gene expression changes in response to an environmental stress in C. elegans suggest that a transgenerational memory of an ancestor’s environment may be beneficial to future progeny. Although multiple genes alter expression in response to these environmental stresses [42,43,47], it is not yet known whether the heritable gene expression changes have a physiological impact. Are these genes a byproduct of experienced stress or do they contribute to the ability to survive an environmental stress? Furthermore, it is still unknown how the physiological stress specifically targets a restricted set of small RNAs for transgenerational regulation. Nonetheless, the ability of both C. elegans and of other species to transmit a gene expression memory across many generations in the absence of the initiating signal presents the fascinating possibility that the experiences of an ancestor can affect the gene expression, physiology, and potentially the survival of many future generations. Analogous to findings in C. elegans, in mice expression changes in small RNAs transmitted by sperm in response to environmental stresses can result in gene expression and behavioral changes in progeny [5–7]. Mendel’s studies of transmission of simple traits in peas allowed him to formulate and test the fundamental rules of genetic inheritance. TEI in C. elegans similarly presents the opportunity to study the strategies and mechanisms of intergenerational transmission of epigenetic information that are likely preserved in more complex organisms.
Box 1.
We use the term epigenetics, literally “beyond” genetics, to refer to the transfer of non-DNA sequence information between cells and organisms. Multiple strategies enable epigenetics and numerous mechanisms have been described. These often operate in parallel, ensuring robust inheritance and obfuscating analysis and understanding. Here we list four known epigenetic strategies and provide examples of specific mechanisms.
Autocatalytic feed forward loop
Examples include transcription factors that drive their own expression [54], metabolites that induce the expression of their own transporter [55], and proteases required for their own activation [56]. In each case, daughter cells that inherit the protein product (regulator) maintain the positive feed forward loop.
Structured templating
Examples include prions [57] and cortical templates [58]. Prions are extremely stable protein aggregates that induce new subunits to adopt an alternate, often non-functional or toxic, structure. The cortical structure of ciliates is composed of regular patterns of ciliary units. Alterations in these patterns produce new morphologies that are stably propagated to daughter cells.
Chromatin marks
Examples include DNA methylation (5-methylcytosine, [59]), often associated with gene silencing, and chemical modifications of histones (e.g. methylation and acetylation) [60], which can promote or silence transcription. De novo DNA methyltransferases establish (by largely mysterious mechanisms) DNA methylation patterns in early mammalian embryos, while maintenance DNA methyltransferases methylate post-replicated hemi-methylated DNA. Unlike mammals, C. elegans and Drosophila lack significant 5-methylcytosine. Histone modification patterns are initially responsive to and directed by gene expression states. Once established, histone modification patterns are maintained autocatalytically, whereby the modifications are binding sites for the modifying enzyme complexes.
Sequence specific RNA guides
Examples include genome rearrangements [61] and RNAi [62]. Ciliates contain an unassembled micronuclear “germline” genome and a transcriptionally active assembled macronuclear “somatic” genome. Following conjugation, a duplicated micronuclei becomes a new macronucleus, using RNA guides from the previous macronucleus to direct its assembly. Thus, changes in the old macronucleus can produce “new” RNA guides that direct genome rearrangements to copy the change, enabling the inheritance of an epigenetically controlled genome structure. In RNAi, Argonaute-bound small interfering RNAs interact with additional proteins, including methyltransferases, to regulate gene expression. siRNAs are most often produced from and target homologous genes. Thus, RNAi can target any gene that can be induced to produce siRNAs. Transgenerational RNAi occurs in organisms that can replicate and amplify siRNAs, establishing an autocatalytic feed forward loop.
Trends Box.
In C. elegans, epigenetic information that alters the expression of transgenes and endogenous loci can sometimes be propagated for many generations even when the initiating environment or genetic event is no longer present.
The transgenerational signal is often, but not always, paramutagenic and extra-genomic.
Transgenerational epigenetic inheritance at endogenous loci and transgenes can be initiated by small RNA pathways, multi-copy transgenes and by physiological stress. Maintenance of the altered expression state requires small RNA and chromatin modifying pathways.
Changes to physiological traits can also persist transgenerationally in the absence of the initiating genetic alteration or physiological stress. The causative genetic alterations that result in the altered physiological state are not yet known.
Acknowledgments
We thank J. Malin for comments on this manuscript.
Glossary
- epigenetic inheritance
Transfer of gene regulatory information across generations without a change to DNA sequence.
- paramutation
A trans interaction between two homologous loci that leads to a heritable change in expression at one of the loci
- short interfering RNA (siRNA)
A short RNA molecule that participates in transcriptional and post-transcriptional gene silencing. In C. elegans, siRNAs are single stranded molecules
- RNA interference
Gene silencing by double stranded RNA
- small RNA
see siRNA
- piRNA
An endogenous 21-nucleotide small RNA stabilized by PRG-1
- HRDE-1
A nuclear RNAi Argonaute proteins that binds secondary siRNAs
- PRG-1
An Argonaute protein that binds piRNAs
- oma-1
A germline-expressed gene encoding an oocyte maturation factor
- sid-1
A gene encoding the transmembrane dsRNA-transporting protein SID-1.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Olga Minkina, Department of Biology, New York University, New York, New York 10003, USA.
Craig P. Hunter, Department of Molecular and Cellular Biology, Harvard University, Cambridge, MA 02138, USA
References
- 1.Jablonka E, Raz G. Transgenerational epigenetic inheritance: prevalence, mechanisms, and implications for the study of heredity and evolution. Q Rev Biol. 2009;84:131–176. doi: 10.1086/598822. [DOI] [PubMed] [Google Scholar]
- 2.Hermant C, et al. Paramutation in Drosophila Requires Both Nuclear and Cytoplasmic Actors of the piRNA Pathway and Induces Cis-spreading of piRNA Production. Genetics. 2015;201:1381–1396. doi: 10.1534/genetics.115.180307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Vanssay A, et al. Paramutation in Drosophila linked to emergence of a piRNA-producing locus. Nature. 2012;490:112–115. doi: 10.1038/nature11416. [DOI] [PubMed] [Google Scholar]
- 4.Ciabrelli F, et al. Stable Polycomb-dependent transgenerational inheritance of chromatin states in Drosophila. Nat Genet. 2017 doi: 10.1038/ng.3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Q, et al. Epigenetic inheritance of acquired traits through sperm RNAs and sperm RNA modifications. Nature Reviews Genetics. 17:733–743. doi: 10.1038/nrg.2016.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gapp K, et al. Implication of sperm RNAs in transgenerational inheritance of the effects of early trauma in mice. Nature Publishing Group. 2014;17:667–669. doi: 10.1038/nn.3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharma U, et al. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science. 2016;351:391–396. doi: 10.1126/science.aad6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fire A, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 9.Bernstein E, et al. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 10.Knight SW, Bass BL. A Role for the RNase III Enzyme DCR-1 in RNA Interference and Germ Line Development in Caenorhabditis elegans. Science. 2001;293:2269–2271. doi: 10.1126/science.1062039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yigit E, et al. Analysis of the C. elegans Argonaute Family Reveals that Distinct Argonautes Act Sequentially during RNAi. Cell. 2006;127:747–757. doi: 10.1016/j.cell.2006.09.033. [DOI] [PubMed] [Google Scholar]
- 12.Tabara H, et al. The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-box helicase to direct RNAi in C. elegans. Cell. 2002;109:861–871. doi: 10.1016/s0092-8674(02)00793-6. [DOI] [PubMed] [Google Scholar]
- 13.Grishok A. Biology and Mechanisms of Short RNAs in Caenorhabditis elegans. 1. Elsevier Inc; 2013. p. 83. [DOI] [PubMed] [Google Scholar]
- 14.Parrish S, Fire A. Distinct roles for RDE-1 and RDE-4 during RNA interference in Caenorhabditis elegans. RNA. 2001;7:1397–1402. [PMC free article] [PubMed] [Google Scholar]
- 15.Sijen T, et al. Secondary siRNAs Result from Unprimed RNA Synthesis and Form a Distinct Class. Science. 2007;315:244–247. doi: 10.1126/science.1136699. [DOI] [PubMed] [Google Scholar]
- 16.Sijen T, et al. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell. 2001;107:465–476. doi: 10.1016/s0092-8674(01)00576-1. [DOI] [PubMed] [Google Scholar]
- 17.Smardon A, et al. EGO-1 is related to RNA-directed RNA polymerase and functions in germ-line development and RNA interference in C. elegans. Curr Biol. 2000;10:169–178. doi: 10.1016/s0960-9822(00)00323-7. [DOI] [PubMed] [Google Scholar]
- 18.Pak J, Fire A. Distinct Populations of Primary and Secondary Effectors During RNAi in C. elegans. Science. 2007;315:241–244. doi: 10.1126/science.1132839. [DOI] [PubMed] [Google Scholar]
- 19.Bagijn MP, et al. Function, Targets, and Evolution of Caenorhabditis elegans piRNAs. Science. 2012;337:574–578. doi: 10.1126/science.1220952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee HC, et al. C. elegans piRNAs Mediate the Genome-wide Surveillance of Germline Transcripts. Cell. 2012;150:78–87. doi: 10.1016/j.cell.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gent JI, et al. Distinct Phases of siRNA Synthesis in an Endogenous RNAi Pathway in C. elegans Soma. Molecular Cell. 2010;37:679–689. doi: 10.1016/j.molcel.2010.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vasale JJ, et al. Sequential rounds of RNA-dependent RNA transcription drive endogenous small-RNA biogenesis in the ERGO-1/Argonaute pathway. Proc Natl Acad Sci USA. 2010;107:3582–3587. doi: 10.1073/pnas.0911908107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Winston WM, et al. Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science. 2002;295:2456–2459. doi: 10.1126/science.1068836. [DOI] [PubMed] [Google Scholar]
- 24.Feinberg EH, Hunter CP. Transport of dsRNA into Cells by the Transmembrane Protein SID-1. Science. 2003;301:1545–1547. doi: 10.1126/science.1087117. [DOI] [PubMed] [Google Scholar]
- 25.Vastenhouw NL, et al. Gene expression: long-term gene silencing by RNAi. Nature. 2006;442:882. doi: 10.1038/442882a. [DOI] [PubMed] [Google Scholar]
- 26.Ashe A, et al. piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell. 2012;150:88–99. doi: 10.1016/j.cell.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buckley BA, et al. A nuclear Argonaute promotes multigenerational epigenetic inheritance and germline immortality. Nature. 2012;489:447–451. doi: 10.1038/nature11352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Houri-Ze’evi L, et al. A Tunable Mechanism Determines the Duration of the Transgenerational Small RNA Inheritance in C. elegans. Cell. 2016;165:88–99. doi: 10.1016/j.cell.2016.02.057. [DOI] [PubMed] [Google Scholar]
- 29.Single-copy insertion of transgenes in Caenorhabditis elegans. 2008;40:1375–1383. doi: 10.1038/ng.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shirayama M, et al. piRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell. 2012;150:65–77. doi: 10.1016/j.cell.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Batista PJ, et al. PRG-1 and 21U-RNAs Interact to Form the piRNA Complex Required for Fertility in C. elegans. Molecular Cell. 2008;31:67–78. doi: 10.1016/j.molcel.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guang S, et al. An Argonaute Transports siRNAs from the Cytoplasm to the Nucleus. Science. 2008;321:537–541. doi: 10.1126/science.1157647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guang S, et al. Small regulatory RNAs inhibit RNA polymerase II during the elongation phase of transcription. Nature. 2010;465:1097–1101. doi: 10.1038/nature09095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhuang JJ, et al. The nuclear argonaute NRDE-3 contributes to transitive RNAi in Caenorhabditis elegans. Genetics. 2013;194:117–131. doi: 10.1534/genetics.113.149765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sapetschnig A, et al. Tertiary siRNAs Mediate Paramutation in C. elegans. PLoS Genet. 2015;11:e1005078. doi: 10.1371/journal.pgen.1005078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin R. A gain-of-function mutation in oma-1, a C. elegans gene required for oocyte maturation, results in delayed degradation of maternal proteins and embryonic lethality. Developmental Biology. 2003;258:226–239. doi: 10.1016/s0012-1606(03)00119-2. [DOI] [PubMed] [Google Scholar]
- 37.Alcazar RM, et al. Transmission Dynamics of Heritable Silencing Induced by Double-Stranded RNA in Caenorhabditis elegans. Genetics. 2008;180:1275–1288. doi: 10.1534/genetics.108.089433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kalinava N, et al. Decoupling the downstream effects of germline nuclear RNAi reveals that H3K9me3 is dispensable for heritable RNAi and the maintenance of endogenous siRNA-mediated transcriptional silencing in Caenorhabditis elegans. Epigenetics Chromatin. 2017;10:6. doi: 10.1186/s13072-017-0114-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Minkina O, Hunter CP. Stable Heritable Germline Silencing Directs Somatic Silencing at an Endogenous Locus. Molecular Cell. 2017;65:659–670. e5. doi: 10.1016/j.molcel.2017.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ketting RF, Plasterk RH. A genetic link between co-suppression and RNA interference in C. elegans. Nature. 2000;404:296–298. doi: 10.1038/35005113. [DOI] [PubMed] [Google Scholar]
- 41.Dernburg AF, et al. Transgene-mediated cosuppression in the C. elegans germ line. Genes & Development. 2000;14:1578–1583. [PMC free article] [PubMed] [Google Scholar]
- 42.Schott D, et al. Natural RNA interference directs a heritable response to the environment. Sci Rep. 2014;4:7387. doi: 10.1038/srep07387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ni JZ, et al. A transgenerational role of the germline nuclear RNAi pathway in repressing heat stress-induced transcriptional activation in C. elegans. Epigenetics Chromatin. 2016 doi: 10.1186/s13072-016-0052-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klosin A, et al. Transgenerational transmission of environmental information in C. elegans. Science. 2017;356:320–323. doi: 10.1126/science.aah6412. [DOI] [PubMed] [Google Scholar]
- 45.Johnson TE, et al. Arresting development arrests aging in the nematode Caenorhabditis elegans. Mech Ageing Dev. 1984;28:23–40. doi: 10.1016/0047-6374(84)90150-7. [DOI] [PubMed] [Google Scholar]
- 46.Baugh LR. To Grow or Not to Grow: Nutritional Control of Development During Caenorhabditis elegans L1 Arrest. Genetics. 2013;194:539–555. doi: 10.1534/genetics.113.150847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rechavi O, et al. Starvation-induced transgenerational inheritance of small RNAs in C. elegans. Cell. 2014;158:277–287. doi: 10.1016/j.cell.2014.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jobson MA, et al. Transgenerational Effects of Early Life Starvation on Growth, Reproduction, and Stress Resistance in Caenorhabditis elegans. Genetics. 2015;201:201–212. doi: 10.1534/genetics.115.178699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Demoinet E, et al. AMPK blocks starvation-inducible transgenerational defects in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2017 doi: 10.1073/pnas.1616171114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Katz DJ, et al. A C. elegans LSD1 Demethylase Contributes to Germline Immortalityby Reprogramming Epigenetic Memory. Cell. 2009;137:308–320. doi: 10.1016/j.cell.2009.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Greer EL, et al. Transgenerational epigenetic inheritance of longevity in Caenorhabditis elegans. Nature. 2011;479:365–371. doi: 10.1038/nature10572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee RC, et al. Interacting endogenous and exogenous RNAi pathways in Caenorhabditis elegans. RNA. 2006;12:589–597. doi: 10.1261/rna.2231506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhuang JJ, Hunter CP. The Influence of Competition Among C. elegans Small RNA Pathways on Development. Genes. 2012;3:671–685. doi: 10.3390/genes3040671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weintraub H. The MyoD family and myogenesis: redundancy, networks, and thresholds. Cell. 1993;75:1241–1244. doi: 10.1016/0092-8674(93)90610-3. [DOI] [PubMed] [Google Scholar]
- 55.Novick A, Weiner M. Enzyme Induction as an All-Or-None Phenomenon. Proc Natl Acad Sci USA. 1957;43:553–566. doi: 10.1073/pnas.43.7.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roberts BT, Wickner RB. Heritable activity: a prion that propagates by covalent autoactivation. Genes & Development. 2003;17:2083–2087. doi: 10.1101/gad.1115803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Masison DC, et al. The prion model for [URE3] of yeast: spontaneous generation and requirements for propagation. Proc Natl Acad Sci USA. 1997;94:12503–12508. doi: 10.1073/pnas.94.23.12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Beisson J, Sonneborn TM. Cytoplasmic Inheritance of the Organization of the Cell Cortex In Paramecium Aurelia. Proc Natl Acad Sci USA. 1965;53:275–282. doi: 10.1073/pnas.53.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nature Reviews Genetics. 2013;14:204–220. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- 60.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modi cations. Cell Research. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chalker DL, et al. Epigenetics of Ciliates. Cold Spring Harbor Perspectives in Biology. 2013;5:a017764–a017764. doi: 10.1101/cshperspect.a017764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wilson RC, Doudna JA. Molecular Mechanisms of RNA Interference. Annu Rev Biophys. 2013;42:217–239. doi: 10.1146/annurev-biophys-083012-130404. [DOI] [PMC free article] [PubMed] [Google Scholar]


