Summary
Microbial natural products are genetically encoded by dedicated biosynthetic gene clusters (BGCs). A given BGC usually produces a family of related compounds that share a core but contain variable substituents. Though common, the reasons underlying this divergent biosynthesis are in general unknown. Herein, we have addressed this issue using the hydroxyalkylquinoline family of natural products synthesized by Burkholderia thailandensis. Investigations into the detailed functions of two analogs show that they act synergistically in inhibiting bacterial growth. One analog is a nM inhibitor of pyrimidine biosynthesis and at the same time disrupts the proton motive force. A second analog inhibits the cytochrome bc1 complex as well as pyrimidine biogenesis. These results provide a functional rationale for the divergent nature of HAQs. They imply that synergy and target promiscuity are driving forces for the evolution of tailoring enzymes that diversify the products of the HAQ biosynthetic pathway.
Keywords: natural products, Burkholderia thailandensis, mode of action, antibiotics, synergy
Graphical abstract
Introduction
Bacteria are a dominant source of secondary metabolites with exquisite, sometimes clinically-useful, biological activities. These compounds are encoded by cognate biosynthetic gene clusters (BGCs), and the number of BGCs in a genome varies greatly depending on the bacterium (Liu and Cheng, 2014; Nett et al., 2009). Some bacteria harbor as many as fifty BGCs, although not all are actively-expressed at any given time. Why one organism has the capacity to synthesize up to fifty distinct families of secondary metabolites has remained a mystery, in part because the complete secondary metabolome has not been identified for any given prolific bacterium. Moreover, the natural functions of even well-known secondary metabolites are often unknown, making it difficult to find answers to this question (Davies, 2006; Williams et al., 1989; Yim et al., 2007). A related, but different question pertains to the diversity-oriented biosynthesis of a specific secondary metabolite. A BGC rarely gives rise to a single product and the number of analogs from one pathway can range from zero to >100 (Fischbach and Clardy, 2007). Why have bacteria evolved to synthesize so many variations on the same theme within a given biosynthetic pathway? What benefits are derived from molecules that share the same scaffold but diverge at a few substituent sites?
Burkholderia thailandensis provides a setting in which these questions can be addressed. It contains >20 BGCs, most of which are silent and do not give rise to appreciable levels of product during normal growth (Liu and Cheng, 2014). One of the few constitutively active BGCs is the quorum sensing-regulated hmq cluster, which gives rise to a diverse set of hydroxyalkylquinolines (HAQs, Fig. 1A) (Okada et al., 2016; Vial et al., 2008; Majerczyk et al., 2014). HAQs, which exist predominantly in the 4(1H)-quinolone form at neutral pH, are a well-known family of both naturally-occurring and synthetically-prepared bioactive metabolites (Heeb et al., 2011). The synthetic fluoroquinolone antibiotics have been used for several decades in the fight against infectious disease (Fàbrega et al., 2009). Likewise, the naturally-occurring HAQs have been investigated for over a century and their functions are perhaps best-characterized in Pseudomonas aeruginosa, where they exhibit Fe-binding and antibacterial activities, in addition to playing a critical role in quorum sensing (Cao et al., 2001; Heeb et al., 2011; Pesci et al., 1999; Williams et al., 1989). Specifically, the pseudomonas quinolone signal (PQS, Fig. 1A), along with its response regulator PqsR, regulates a variety of cellular behaviors, such as virulence factor production, as a function of cell density. In contrast, the roles of HAQs are poorly understood in other bacterial producers. Aside from B. thailandensis, Burkholderia ambifaria, and Burkholderia pseudomallei also synthesize diverse quinolones, but functional studies have remained scarce for the variants from these microbes.
Figure 1.
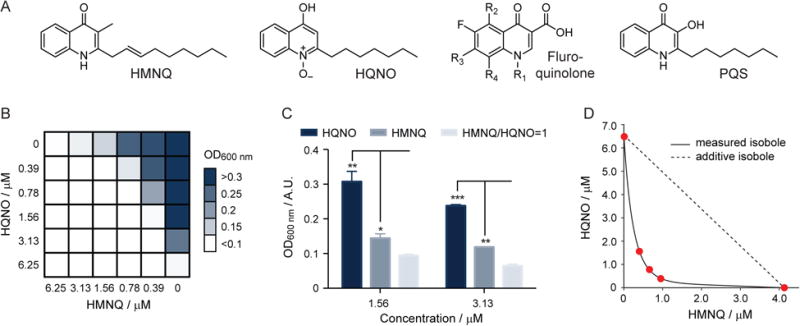
HMNQ and HQNO act synergistically. (A) Structures of HMNQ, HQNO, a generic fluoroquinolone antibiotic, and P. aeruginosa-derived PQS. (B) Checkerboard analysis measuring the OD600 nm of B. subtilis in the presence of various concentrations of HMNQ or HQNO. A similar result was obtained with E. coli (see also Fig. S2). The data represent averages of two independent replicates. (C) Bar chart representation of the data in panel (B) to emphasize the synergy observed. Bacterial growth, as determined by OD600 nm, is shown as a function of treatment with HQNO alone, HMNQ alone, or HMNQ and HQNO. The average of three biological replicates is shown. Error bars represent standard deviation (SD). One, two, and three stars represent p-values of <0.05, <0.01, and <0.001, respectively. (D) Isobologram analysis of the data in panel (B). The additive isobole is shown. The measured isobole clearly falls in the positive synergy region of the plot (below the dotted line).
In an insightful report, Challis and Hopwood provided an answer to the first question posed in the opening paragraph, that is, the reasons underlying the accumulation of numerous BGCs within a given bacterial genome (Challis and Hopwood, 2003). They posed synergy and contingency as evolutionary driving forces for the emergence of multiple BGCs in a microbe. For example, production of a β-lactam antibiotic and a β-lactamase inhibitor provides two synergistically-acting small molecules. Likewise, production of contingently-acting siderophores is common in bacteria. Herein we provide an answer to the second question posed above, that is, the reasons underlying the diversity-oriented nature of secondary metabolite biogenesis. By examining the detailed functions of two different HAQ variants from the same BGC in B. thailandensis, we identify synergy and target promiscuity as the underlying functional rationale for the divergent biosynthesis of quinolone secondary metabolites.
Results and Discussion
Recently a large diversity of HAQs has been found in B. thailandensis E264. The diversity is provided by the presence of a proton or methyl group at the 3-position, a variety of saturated and unsaturated alkyl groups at the 2-position, as well as N-oxides in some derivatives at the N1 (Okada et al., 2016; Vial et al., 2008). Given that HAQ production is both abundant and divergent in this and other organisms, we thought it would present a suitable system for addressing the functional reasons underlying their diversity-oriented biogenesis. We began by selecting two analogs, HMNQ (4-hydroxy-3-methyl-2-(2-nonenyl)-quinoline) and HQNO (2-heptyl-4(1H)-quinolone N-oxide) (Fig. 1A), both of which are produced by B. thailandensis, especially when challenged by antibiotics (Fig. S1). Note that, as pointed out by Cámara and colleagues, the nomenclature of quinolones reflects preference and precedent, rather than the correct tautomeric form of each variant (Heeb et al., 2011). At neutral pH, HMNQ exists predominantly as the ketone tautomer, whereas HQNO is mostly in the enol form. Functional studies have not been conducted with HMNQ. HQNO is also produced by P. aeruginosa and previous reports have shown it to inhibit the cytochrome bc1 complex (Hacker et al., 1993; Hazan et al., 2016; Reil et al., 1997; Vallieres et al., 2012; Van Ark and Berden, 1977). To delve into the functions of HAQs, we obtained HQNO commercially and purified HMNQ to homogeneity from B. thailandensis cultures grown in the presence of trimethoprim, which increased production and purification yields (Seyedsayamdost, 2014).
We first assessed the bioactivities of the two derivatives using antimicrobial assays. Both HMNQ and HQNO showed potent antibiosis against Gram positive and some Gram negative bacteria. Specifically, they exhibited half maximal inhibitory concentration (IC50) values of 1.1 μM (HMNQ) and 4.1 μM (HQNO) against Bacillus subtilis 168. While they showed detectable but weak bioactivity against Escherichia coli K12, IC50s of 0.3 μM and 9.1 μM were determined against a cell wall-weakened E. coli strain, containing a mutation in the LPS assembly protein lptD. To examine whether the bioactivities of the two HAQs are synergistic or additive, a checkerboard assay was conducted, in which the minimum inhibitory concentrations (MICs) of varying ratios of HMNQ and HQNO were tested against B. subtilis and E. coli (Figs. 1B & S2) (Jayaraman et al., 2010; Orhan et al., 2005). If HMNQ and HQNO impinge on the same target, one would expect additive results, that is, a linear relationship between the sum of the concentrations of HMNQ and HQNO, relative to the concentration of each drug alone, on growth-inhibition. If however, they have different targets, synergistic bioactivity may be observed. In checkerboard analysis, a synergistic relationship between two drugs is expressed in the fractional inhibitory concentration index (ΣFIC), which is given by the sum of the fractional inhibitory concentration for HMNQ and HQNO (see SI). The drug combination is synergistic if the ΣFIC is <0.5, indifferent if ΣFIC is 0.5-2, and antagonistic when ΣFIC >2. The results of this analysis are shown (Fig. 1B); they yield a ΣFIC <0.25, consistent with a synergistic interaction between the two HAQ derivatives. For example, at 3.13 μM of HMNQ or HQNO, 73% and 27% growth inhibition was observed, respectively. However, at a 1:1 ratio of the two drugs (1.56 μM each), 96% growth inhibition occurred, rather than 27-73% (Fig. 1C). Similar results were obtained with E. coli: at 12.5 μM HMNQ or HQNO, 62% and 8% growth inhibition was observed, respectively. However, at a 1:1 ratio of the two drugs (6.25 μM each), 92% growth inhibition occurred (Fig. S2).
The checkerboard synergy data were also analyzed via the isobologram method, with the same outcome (Tallarida, 2006). As shown, the observed isobole significantly deviated from the additive isobole and fell in the positive synergy region of the plot below the dotted line (Fig. 1D). Together, these results show that HMNQ and HQNO act synergistically to inhibit bacterial growth. They further suggest that the two derivatives, generated by the same BGC, may have distinct biological targets, and therefore different modes of action.
Despite the history of bacterial quinolones, surprisingly little is known about their modes of antibiotic action. As alluded to above, biochemical and structural studies have shown that HQNO inhibits the cytochrome bc1 complex, one of the oxidoreductases involved in the electron transport chain of oxidative phosphorylation (Hacker et al., 1993; Hazan et al., 2016; Van Ark and Berden, 1977). However, other variants have not been examined in any detail, perhaps because the prevailing hypothesis has been that they inhibit the same target. Given our results above, we set out to determine the mode of action (MoA) of HMNQ. To do so, we first carried out bacterial cytological profiling (BCP) on both derivatives. This correlative method was recently pioneered by the Pogliano groups, and its main tenet is that bacterial cells that look the same – as determined from the treatment of drug-exposed cells with three distinct dyes – likely died by the same mechanism (Nonejuie et al., 2013). This approach has emerged as an effective MoA-determining tool and has recently been applied to secondary metabolites of diverse functions (Nonejuie et al., 2016; Wilson et al., 2016).
To apply BCP, we treated E. coli with HMNQ, HQNO, and a suite of eleven other antimicrobials representing various known modes of action. The cells were then exposed to FM4-64, which discretely stains the cell membrane; Sytox green, which reports on the permeability of the cell membrane; and DAPI, a DNA stain. Subsequent quantitative imaging of the dye-exposed cells by fluorescence microscopy gave a total of 14 parameters for each drug (Fig. 2A). Multivariate analysis of these parameters allowed us to group the antibiotics by their cytological profiles, a proxy for MoA. Consistent with the studies above, treatment of E. coli with HQNO and HMNQ gave different phenotypes and the two drugs grouped with other antibiotics rather than with each other (Fig. 2A, B). HQNO exhibited a unique cytological profile within the set of antimicrobials examined, in agreement with its unique target, the cytochrome bc1 complex. By contrast, the cytological profile obtained for HMNQ was very similar to that of monensin, and to a lesser degree, to the CCCP profile.
Figure 2.
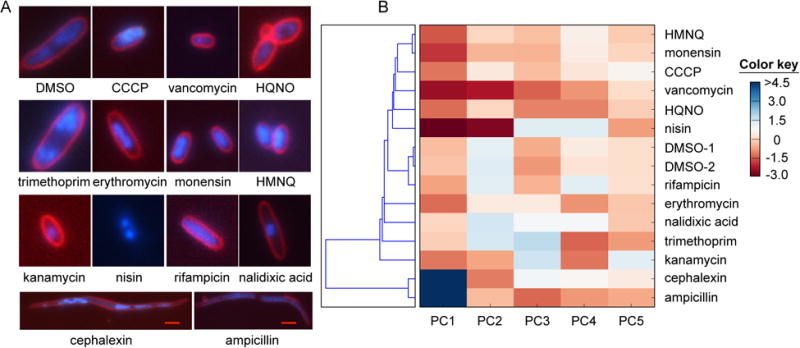
Modes of action of HMNQ and HQNO determined by bacterial cytological profiling. (A) Typical cell cytology of E. coli visualized with three fluorophores 2 h after treatment with the indicated drug. The chosen antibiotics represent a number of known modes of antibiotic action. Note that HMNQ- and HQNO-treated cells reveal different cytological profiles. White scale bar, 2 m; red scale bar, 4 m. (B) Principal component analysis of the features in panel (A) clusters the antibiotics by mode of action. The relative contribution of each principal component is indicated by the color key. HMNQ clusters most closely with monensin and CCCP.
Interestingly, monensin and CCCP are both disrupters of the proton motive force (PMF), which is universally used to generate energy during oxidative phosphorylation. The PMF is composed of the transmembrane potential (ΔΨ) and the transmembrane proton gradient (ΔpH). Monensin is a so-called electroneutral proton anti-porter; it dissipates the proton gradient without affecting the membrane potential via an exchange of protons for intracellular monovalent cations (Dutton et al., 1995; Mollenhauer et al., 1990). CCCP, on the other hand, is an electrogenic proton antiporter; it collapses both the ΔpH and the ΔΨ by importing protons without exporting cations (Terada, 1981). Our data show that HMNQ dissipates the PMF via a monensin-like electroneutral proton antiport mechanism. This is markedly distinct from the MoA of HQNO, which acts as a ubiquinone mimic and interferes with the function of the cytochrome bc1 complex. Both HAQs affect energy generation, but interfere with different targets. HQNO shuts down electron transfer required for setting up the PMF, while HMNQ dissipates the PMF. This ‘combination therapy’ dispensed by B. thailandensis is an effective two-pronged antibiotic strategy. It suggests that the HAQ BGC in B. thailandensis generates two structurally similar but functionally distinct molecules with disparate and synergistic biological activities.
We previously showed that PMF-disrupting antibiotics could also be identified by primary metabolite analysis (Wilson et al., 2016). Because these drugs inhibit nucleoside triphosphate (NTP) synthesis, primary metabolite analysis revealed a decrease in NTP levels and a corresponding increase in nucleoside monophosphate (NMP) levels. We sought to obtain further evidence for the PMF-disrupting role of HMNQ by performing primary metabolite analysis. The observed changes in the primary metabolomes of E. coli after treatment with HMNQ and HQNO relative to a control sample are shown in the heatmap in Fig. 3A. Consistent with the ideas above, we found a clear reduction in NTP levels and a mild increase in intracellular NMP levels. Specifically, ATP, UTP, and CTP levels were lowered 5-fold, 10-fold, and 2.5-fold, respectively, upon exposure to HMNQ. A corresponding increase in the levels of AMP (2.8-fold), UMP (1.6-fold), CMP (1.6-fold), and GMP (3.3-fold) was also observed. Likewise, HQNO also lowered ATP, UTP, and CTP levels (4-14-fold) by interfering with a different process in oxidative phosphorylation. These data provide further support for the interference of HMNQ and HQNO with energy production pathways in E. coli.
Figure 3.
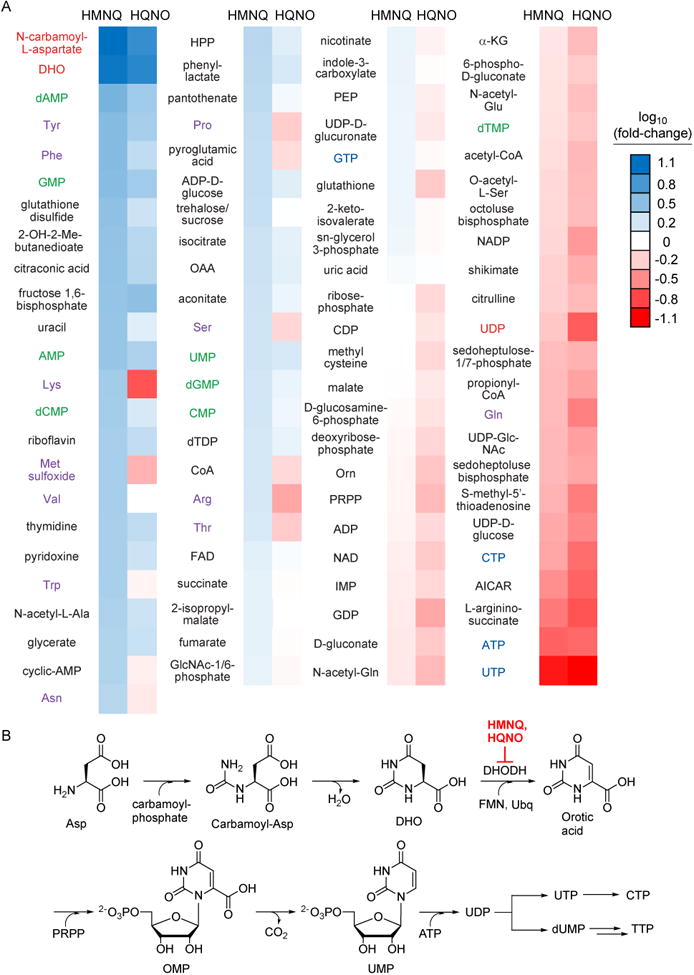
Effect of HMNQ and HQNO on the primary metabolome of E. coli. (A) Shown is the fold-change in the levels of 93 primary metabolites as a function of HMNQ or HQNO treatment, with respect to a no-treatment control, determined by HR-HPLC-MS. Three independent biological replicates for both drug-treated and untreated cells were used. The metabolites are sorted by fold-change. The color-coding is as follows: pyrimidine biosynthetic intermediates, red; NMPs, green; canonical amino acids, purple; NTPs, blue. (B) Biosynthetic pathway for pyrimidine nucleotides. HMNQ and HQNO inhibit DHODH, which results in accumulation of DHO and N-carbamoyl-L-aspartate (carbamoyl-Asp).
Aside from changes in NTP and NMP levels, the primary metabolite analysis upon HMNQ or HQNO treatment also showed drastic accumulation of N-carbamoyl-L-aspartate (12.6-fold) and dihydroorotate (11.3-fold), both products of the first two dedicated steps in the de novo biosynthesis of pyrimidine nucleotides (Fig. 3A, B). Their accumulation suggested to us that dihydroorotate dehydrogenase (DHODH) may be inhibited by the HAQs. To test this hypothesis, enzymatic activity assays were carried out with E. coli DHODH, which catalyzes the coenzyme Q-dependent reduction of dihydroorotate to orotate (Fagan and Palfey, 2009; Fagan et al., 2006; Palfey et al., 2001). In the absence of inhibitors, and using the soluble quinone dichlorophenolindophenol (DCIP), a common coenzyme Q surrogate, we obtained a kcat of 5 s-1, similar to previously reported results. Assays in the presence of HMNQ and HQNO showed potent inhibition of DHODH activity. Detailed enzymatic inhibition analysis showed that the drugs were competitive with DCIP (Fig. 4A, B), but not dihydroorotate (Fig. S3). With the mode of inhibition established, we determined Ki values of 51 and 239 nM for HMNQ and HQNO, respectively. Thus, aside from disrupting and preventing formation of the PMF, these drugs simultaneously inhibit pyrimidine biosynthesis by competing for the coenzyme Q binding site in DHODH. As such, HMNQ and HQNO are synergistic in interfering with energy production and additive in inhibiting DHODH function, and overall synergistic in antibiotic activity.
Figure 4.
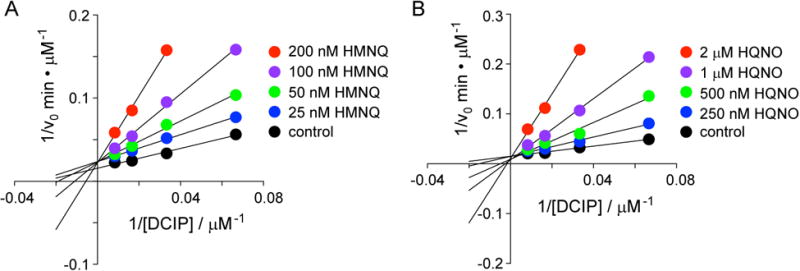
HMNQ and HQNO efficiently inhibit E. coli DHODH by competing for the quinone binding site. Lineweaver-Burke analysis for the inhibition of E. coli DHODH by HMNQ (A) or HQNO (B). Both drugs are competitive with the ubiquinone surrogate DCIP, but not with substrate DHO (see also Fig. S3). Data represent the average of two replicates; error bars are within the size of the circles.
Finally, we sought to find additional evidence for the in vivo inhibition of pyrimidine biosynthesis by HMNQ and HQNO. We surmised that both drugs would exhibit synergy with the drug monensin, which inhibits cell growth by dissipating the PMF. Indeed, bioactivity assays using B. subtilis showed synergistic interactions in both cases, with ΣFIC of 0.37 and 0.19 for the monensin/HMNQ and monensin/HQNO combinations, respectively (Fig. S4). Both values are significantly below the synergy threshold of ∼0.5. Similarly, synergistic interactions between the HAQs and monensin were also observed against E. coli (Fig. S5). These results are in agreement with the disparate modes of action proposed for the two HAQs. They further highlight the importance of using complementary profiling approaches in order to understand the multiple targets of a given antibiotic and its detailed effects on bacterial cells.
Conclusions
Our results have important implications for HAQ biosynthesis and function. They support a model in which structurally divergent HAQs represent an antibiotic cocktail with components that act synergistically to inhibit microbial growth. Both HAQs examined have dual targets. One of these, pyrimidine biosynthesis is shared, while the other, energy generation, is inhibited at different target sites in a synergistic manner. This is accomplished by a simple modification to the HAQ scaffold, namely hydroxylation of the quinoline-nitrogen, which results in a compound (HQNO) that exhibits a different bioactivity profile compared with HMNQ. These insights suggest that the diversity-oriented synthesis of HAQs is evolutionarily driven by synergy and target promiscuity. Together with results from other BGCs that generate synergistically-acting antibiotics, such as the virginiamycins or thuricins (Pulsawat et al., 2007; Rea et al., 2010), these results provide a functional rationale for the evolution of divergence in some secondary metabolite production pathways.
With the functions of two HAQ analogs established, it remains to be seen what bioactivities the remaining ∼20 or so HAQs in B. thailandensis exhibit. It is tempting to speculate that the diverse array of HAQs could mimic a variety of ubiquinones and menaquinones that are used in prokaryotic and eukaryotic cells, thereby acting as broad-spectrum antibiotics that provide the producing bacterium with a competitive advantage in colonizing a niche. Consistent with this hypothesis are the growth-inhibitory effects observed against both prokaryotes and some eukaryotes, such as the fungus Aspergillus niger and the microalga Emiliania huxleyi (Dekker et al., 1998; Harvey et al., 2016; Kilani-Feki et al., 2012). To verify this effect, we examined the effect of HMNQ and HQNO on Saccharomyces cerevisiae and found IC50s of 0.36 and 0.63 μM, respectively. Examination of additional HAQ analogs in the future will provide further insights into this aspect of HAQ biology.
Several lines of evidence suggest that the synergistic activities of HMNQ and HQNO are physiologically relevant: Production of a variety of HAQ derivatives, including HQNO, has been detected in the sputum of cystic fibrosis patients or burn victims infected by P. aeruginosa, suggesting that HAQ divergence occurs naturally (Collier et al., 2002; Machan et al., 1992; Que et al., 2011). While HAQ production has not been examined from Burkholderia spp. in a natural setting, the concentrations that we determine for HMNQ and HQNO in the lab (∼35 μM and ∼2 μM, respectively) are sufficient to exhibit antibiotic and DHODH-inhibitory effects. The HAQ BGC in P. aeruginosa (pqs) is highly homologous to hmq in B. thailandensis. The enzymes encoded by pqs are reasonably well-studied and a biochemical basis for the observed variants has been proposed (Dulcey et al., 2013). Both gene clusters are QS-activated in B. thailandensis and P. aeruginosa, indicating that as long as these bacteria establish a quorum, production of HAQs will ensue.
Lastly, our results expand the functions of quinolone metabolites. The fluoroquinolones, which always bear a 3-carboxylic acid moiety, are potent inhibitors of DNA gyrases. PQS, which contains a 3-hydroxyl group, acts as an antibiotic and a signaling molecule in P. aeruginosa, while HMNQ and HQNO, bearing either a 3-methyl group or hydrogen, halt pyrimidine biosynthesis, dissipate the PMF, and inhibit the cytochrome bc1 complex. Together, these data showcase quinolones as privileged scaffolds, which Nature has leveraged in order to carry out a myriad of functions (Fig. 1A). Given the availability of many naturally-occurring HAQ structures and that small modifications on this scaffold can have significant functional consequences, it is safe to assume that additional roles will be discovered for quinolone natural products in the future.
Significance
Primary metabolism is in general target-oriented: the histidine biosynthetic pathway, for example, only gives rise to histidine, and not any analogs. Secondary metabolism, on the other hand, is inherently diversity-oriented. A single pathway can give rise to a large number of derivatives, but the reasons for this divergence are in almost all cases unknown. The HAQs are a family of secondary metabolites with a well-established promiscuous biosynthetic pathway that operates in a number of Proteobacteria. But the biological activity of only one of these variants from Burkholderia spp. has previously been examined and so the reasons for their divergent production have remained unknown. Here we have addressed this question by investigating the detailed functions of two HAQ analogs from B. thailandensis. We find that the two derivatives, HMNQ and HQNO, act synergistically. One variant (HQNO) inhibits cytochrome bc1 and pyrimidine biosynthesis. A second analog (HMNQ) dissipates the PMF and also inhibits pyrimidine biosynthesis. By interfering with energy production pathways via two distinct mechanisms and simultaneously inhibiting pyrimidine, and thereby, DNA biosynthesis, the HAQs synergistically inhibit bacterial growth. Given their multiple targets, the development of HAQ resistance is likely to be difficult as several simultaneous mutations in various proteins would be necessary to overcome HAQ-mediated growth inhibition. Our results provide a functional rationale for the emergence of non-specific biosynthetic enzymes that give rise to numerous HAQ analogs and suggest that synergy and target promiscuity evolutionarily drive the diversity-oriented biogenesis of HAQs, and by extension, perhaps other groups of secondary metabolites.
Star Methods
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Mohammad R. Seyedsayamdost (mrseyed@princeton.edu).
Experimental Model and Subject Details
Burkholderia thailandensis E264 culture
B. thailandensis E264 was routinely cultured in LB-MOPS liquid medium (consisting of LB + 50 mM, pH 7.0) at 30°C and 250 rpm.
Escherichia coli MC4100 lptd4213 culture
E. coli MC4100 lptd4213 strain was kindly provided by the Silhavy lab, and was routinely cultured in LB liquid medium at 37°C and 250 rpm.
Bacillus subtilis 168 culture
B. subtilis 168 was routinely cultured in LB liquid medium at 30°C and 250 rpm.
Saccharomyces cerevisiae ZSR3385 culture
S. cerevisiae ZSR3385 was kindly provided by the Kolter lab, and was cultured in YPD liquid medium containing 2% (w/v) glucose at 25°C and 250 rpm.
Method Details
Purification of HMNQ
Wild-type B. thailandensis E264 colonies on an LB agar plate were used to inoculate a 5 mL LB-MOPS overnight culture in a 14 mL sterile bacterial culture tube. After 12-13 h, the culture was diluted to an OD600 nm of 0.05 in a 250 mL Erlenmeyer flask containing 50 mL of LB-MOPS. After overnight growth, the culture was diluted to an OD600 nm of 0.05 into each of 8 × 4 L Erlenmeyer flasks containing 700 mL LB-Mops and 30 μM of trimethoprim. The cultures were grown for 28 h at 30°C and 200 rpm, then extracted with one volume of EtOAc, dried over Na2SO4, and evaporated completely in vacuo. The dried EtOAc fraction was re-dissolved in a small amount of MeOH, mixed 100 mg of Seppak-C18 resin, dried in vacuo, and applied to a pre-packed Seppak-C18 column (Sigma, 5 g), which had been washed with MeOH, then equilibrated with 20% MeOH in H2O. Step-wise elution was carried out with 50 mL of 20%, 35%, 50%, 75%, and 100% MeOH (in H2O). The 100% MeOH fraction, which contained HMNQ, was dried in vacuo, resuspended in MeOH, and further purified on an Agilent preparative HPLC using a Luna C18 column (250 × 21.2mm, 5 μm) operating at a flow rate of 12mL/min. Upon injection, the sample was resolved using a gradient of 40% MeCN in H2O to 100% MeCN over 32 min. Peaks containing HMNQ, as judged by HPLC-MS analysis, were pooled, dried in vacuo and purified to homogeneity on an Agilent semi-preparative HPLC using an Eclipse XDB-C8 column (250 × 9.4mm, 5 μm) operating at 2.5 mL/min and an isocratic elution step of 60% MeCN (in H2O). Pure HMNQ was verified by NMR and HR-MS giving similar results as previously published (Vial et al., 2008; Seyedsayamdost, 2014).
Bioactivity assay
All bioactivity assays were carried out in flat-bottom Costar 96-well polystyrene plates using a Biotek Synergy HT plate reader. Wells were filled with 100 μL or 200 μL of the appropriate liquid media, and inoculated from overnight cultures with a starting OD600 nm of 0.005-0.02 depending on the strain. A serial dilution of the compound of interest in DMSO in a range of 0–100 μM was added to the wells. The plates were sealed with Breathe-Easy sealing membrane (Sigma) and incubated at the appropriate temperature. Optical density at 600 nm was then recorded when the DMSO-only treated wells reached an OD600 nm of ∼0.3.
Checkerboard assay
All checkerboard assays were carried out in flat-bottom Costar 96-well polystyrene plates using a Biotek Synergy HT plate reader according to previously published methods (Orhan et al., 2005). Briefly, the wells were filled with 200 μL of the appropriate liquid media and inoculated from an overnight culture to a starting OD600 nm of 0.01. A serial dilution of compound 1 in DMSO in a range of 0–50 μM was added into each well in a column-wise fashion. Subsequently, a serial dilution of compound 2 in DMSO in a range of 0–50 μM was added into each well row-wise manner. Throughout the whole plate, the concentrations of compound 1 and compound 2 varied from no compounds to 50 μM of compound 1 mixed with 50 μM of compound 2. The plates were sealed with Breathe-Easy membranes (Sigma) and incubated at the appropriate temperature. Optical density at 600nm was then recorded after 10 hr.
Bacterial Cytological Profiling
Cytological profiling was preformed as previously described with minor modifications (Nonejuie et al., 2013). Briefly, saturated overnight cultures of E. coli MC4100 lptd4213 were subcultured (1:100 dilution) in LB and allowed to grow for 1.5 hr at 30°C/200 rpm. The cultures were then treated with antibiotics at 5x the MIC, as determined by the microdilution method, and allowed to grow at 30°C and 60 rpm for 2 hrs. The E. coli cells were then stained with final concentrations of 1 μg/mL FM4-64 (Life Technologies), 2 μg/mL DAPI (Sigma), and 0.5 μM Sytox Green (Life Technologies) and allowed to incubate for 20 minutes at room temperature. The cells were concentrated 3-fold by centrifuging at 3300g for 1 min and resuspended in 20% LB. Individual antibiotic treatment groups were then spotted onto 1.2% agarose pads prepared in 20% LB. Exposure times for each detection channel were kept constant for all samples. To prevent variations, all samples were imaged in a single sitting. The E. coli cells were segmented from their phase images and features were extracted from segmented cells using a custom Matlab code. Principal component analysis was performed in Matlab using the pca function and clustering was carried out using the single-linkage method.
Microscopy
Images were taken using a Nikon Eclipse Ti-E microscope with a 100x objective and an Andor Zyla sCMOS camera. The microscope was installed with an external phase contrast unit to enable phase contrast imaging. To prevent evaporation, coverslips were sealed to slides with valap (1:1:1 lanolin/paraffin/petroleum jelly).
Primary Metabolomics Experiments
Saturated overnight cultures of E. coli strain MC4100 lptd4213 were subcultured (1:100 dilution) into Gutnick Minimal Medium and grown to an OD600 nm ∼0.1 at 37°C and 200 rpm. At t=0, three out of nine replicate cultures were treated with 100 μM HMNQ in DMSO, another three were treated with 100 μM HQNO in DMSO, and the rest three replicate control cultures were treated with an equivalent volume of only DMSO. Primary metabolites were isolated after treatment for 1 hr by filtering 5 mL of each culture onto a 50 mm nylon membrane filter and immediately quenching the membrane in extraction solvent (2:2:1 acetonitrile/methanol/water) that had been equilibrated at -20°C. The extraction process conti nued for 15 min. Each extract was centrifuged at 10,000g for 10 min and the supernatants were analyzed by reversed-phase ion-pairing liquid chromatography (LC) coupled to a high resolution, high-accuracy electrospray ionization (ESI) mass spectrometer (Thermo Q Exactive mass spectrometer), which was operated in negative ion detection in full scan mode, as previously described in detail (Lu et al., 2010). Primary metabolite identities were verified by high-resolution mass and retention time matches to authenticated standards as previously described. Metabolites were quantified in MAVEN software using integrated peak areas from extracted ion chromatograms. Variations in the absolute mass spectral counts from run-to-run were avoided by direct comparison of samples from different sequences without spiking in isotopically-labeled metabolites. Metabolite levels were determined as relative fold-changes to the DMSO treated samples. Three replicates were averaged before fold-changes were calculated.
DHODH enzymatic assay
Continuous DHODH enzymatic assays were carried as previously reported (Palfey et al., 2001). Briefly, the assays contained 300 μL reaction buffer (50 mM Tris-HCl, 150 mM KCl, 0.1% Triton X-100, 5% glycerol, pH=8.0) and final concentrations of 50 nM E. coli DHODH, kindly provided by Prof. B. Palfey, 125 μM DCIP, 125 μM dihydroorotate, and the inhibitors HMNQ or HQNO dissolved in DMSO. The inhibitors were varied from 25–200 nM (HMNQ) or 250–2000 nM (HQNO). For competitive inhibition assays, DCIP and dihydroorotate were varied from 15–125 μM and 12.5–125 μM, respectively. For the control reactions, an equivalent volume of DMSO only was added. Upon initiating the reaction with DHO, UV-vis absorbance at 600 nm were monitored on an Agilent Cary 60 UV-Vis for 1 min. The decrease at 600 nm, corresponding to reduction of DCIP, was used to calculate initial velocities. Lineweaver-Burke analysis was analyzed in Graphpad Prism software.
Quantification and Statistical Analysis
Bioactivity assays were performed in biological triplicates, with error bars reporting SD. Checkerboard assays were performed in biological duplicates, with the colors in the checkerboard plots reporting averages from duplicates. For bacterial cytological profiling, about 100 cells were analyzed for each treatment, and averaged parameters were used for principle component analysis. For primary metabolomics measurements, three independent biological samples were analyzed for each treatment, and the means of triplicates were calculated for comparison. Enzymatic assays were performed in duplicate with two independent samples, and error bars represent the SD. P-values were calculated for triplicates in Graphpad Prism software. In all figures: *, p-value < 0.05; **, p-value < 0.01; ***, p-value < 0.001.
Key Resources Table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Bacterial and Virus Strains | ||
| Escherichia coli MC4100 lptd4213 | Silhavy Lab | N/A |
| Bacillus subtilis 168 | ATCC | Cat#23857 |
| Saccharomyces cerevisiae ZSR3385 | Kolter Lab | N/A |
| Burkholderia thailandensis E264 | ATCC | Cat#700388 |
| Biological Samples | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| octadecyl-functionalized silica gel | Sigma Aldrich | Cat#377635 |
| HQNO | Santa Cruz Biotech | Cat#202654;CAS:34 1-88-8 |
| FM4-64 | Thermo Fisher | Cat#T13320;CAS:16 2112-35-8 |
| DAPI | Sigma Aldrich | Cat#D9542;CAS:287 18-90-3 |
| Sytox green | Thermo Fisher | Cat#S7020 |
| trimethoprim | Sigma Aldrich | Cat#T7883;CAS:738 -70-5 |
| 2,6-Dichloroindophenol sodium salt | Sigma Aldrich | Cat#D1878;CAS:126 6615-56-8 |
| L-dihydroorotic acid | Sigma Aldrich | Cat#D7128;CAS:598 8-19-2 |
| Escherichia coli dihydroorotate dehydrogenase | Palfey Lab | N/A |
| Critical Commercial Assays | ||
| Deposited Data | ||
| Experimental Models: Cell Lines | ||
| Experimental Models: Organisms/Strains | ||
| Oligonucleotides | ||
| Recombinant DNA | ||
| Software and Algorithms | ||
| Matlab | https://www.mathworks.com/ | N/A |
| Graphpad Prism | https://www.graphpad.com/ | N/A |
| MAVEN | http://genomics-pubs.princeton.edu/mzroll/index.php | N/A |
| Other | ||
Supplementary Material
Highlights.
HMNQ and HQNO, two natural quinolones, act synergistically to inhibit bacterial growth
Bacterial cytological profiles show they inhibit distinct steps in energy production
Primary metabolite analysis reveals that both also inhibit pyrimidine biosynthesis
Our results provide a functional rational for the structural divergence in quinolones
In Brief.
Secondary metabolism has long been known to be diversity-oriented. Here, Wu & Seyedsayamdost provide an explanation for this phenomenon by demonstrating that two structurally-related quinolone natural products from the same biosynthetic pathway have different targets and synergistically inhibit bacterial growth.
Acknowledgments
We thank Dr. M. Z. Wilson for assistance with bacterial cytological profiling, Dr. G. Laevsky in the Princeton Microscopy Core Facility for assistance with collecting microscopic images, Prof. B. A. Palfey for the kind gift of E. coli DHODH, Prof. J. D. Rabinowitz and L. Chen for assistance with primary metabolite analysis, as well as the Searle Scholars Program and the National Institutes of Health (DP2-AI-124786) for support of this work.
Footnotes
Author Contributions: Y.W. and M.R.S. designed, performed, and analyzed experiments, and wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Cao H, Krishnan G, Goumnerov B, Tsongalis J, Tompkins R, Rahme LG. A quorum sensing-associated virulence gene of Pseudomonas aeruginosa encodes a LysR-like transcription regulator with a unique self-regulatory mechanism. Proc Natl Acad Sci U S A. 2001;98:14613–14618. doi: 10.1073/pnas.251465298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challis GL, Hopwood DA. Synergy and contingency as driving forces for the evolution of multiple secondary metabolite production by Streptomyces species. Proc Natl Acad Sci U S A. 2003;100:14555–14561. doi: 10.1073/pnas.1934677100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier DN, Anderson L, McKnight SL, Noah TL, Knowles M, Boucher R, Schwab U, Gilligan P, Pesci EC. A bacterial cell to cell signal in the lungs of cystic fibrosis patients. FEMS Microbiol Lett. 2002;215:41–46. doi: 10.1111/j.1574-6968.2002.tb11367.x. [DOI] [PubMed] [Google Scholar]
- Davies J. Are antibiotics naturally antibiotics? J Ind Microbiol Biotechnol. 2006;33:496–499. doi: 10.1007/s10295-006-0112-5. [DOI] [PubMed] [Google Scholar]
- Dekker KA, Inagaki T, Gootz TD, Huang LH, Kojima Y, Kohlbrenner WE, Matsunaga Y, McGuirk PR, Nomura E, Sakakibara T, et al. New quinolone compounds from Pseudonocardia sp with selective and potent anti-Helicobacter pylori activity: taxonomy of producing strain, fermentation, isolation, structural elucidation and biological activities. J Antibiot (Tokyo) 1998;51:145–152. doi: 10.7164/antibiotics.51.145. [DOI] [PubMed] [Google Scholar]
- Dulcey CE, Dekimpe V, Fauvelle DA, Milot S, Groleau MC, Doucet N, Rahme LG, Lépine F, Déziel E. The End of an Old Hypothesis: The Pseudomonas Signaling Molecules 4-Hydroxy-2-Alkylquinolines Derive from Fatty Acids, Not 3-Ketofatty Acids. Chem Biol. 2013;20:1481–1491. doi: 10.1016/j.chembiol.2013.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutton CJ, Banks BJ, Cooper CB. Polyether ionophores. Nat Prod Rep. 1995;12:165–181. doi: 10.1039/np9951200165. [DOI] [PubMed] [Google Scholar]
- Fàbrega A, Madurga S, Giralt E, Vila J. Mechanism of action of and resistance to quinolones. Microbiol Biotechnol. 2009;2:40–61. doi: 10.1111/j.1751-7915.2008.00063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagan RL, Palfey BA. Roles in Binding and Chemistry for Conserved Active Site Residues in the Class 2 Dihydroorotate Dehydrogenase from Escherichia coli. Biochemistry. 2009;48:7169–7178. doi: 10.1021/bi900370s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagan RL, Nelson MN, Pagano PM, Palfey BA. Mechanism of Flavin Reduction in Class 2 Dihydroorotate Dehydrogenases. Biochemistry. 2006;45:14926–14932. doi: 10.1021/bi060919g. [DOI] [PubMed] [Google Scholar]
- Fischbach MA, Clardy J. One pathway, many products. Nat Chem Biol. 2007;3:353–355. doi: 10.1038/nchembio0707-353. [DOI] [PubMed] [Google Scholar]
- Hacker B, Barquera B, Crofts AR, Gennis RB. Characterization of mutations in the cytochrome b subunit of the bc1 complex of Rhodobacter sphaeroides that affect the quinone reductase site (Qc) Biochemistry. 1993;32:4403–4410. doi: 10.1021/bi00067a033. [DOI] [PubMed] [Google Scholar]
- Harvey EL, Deering RW, Rowley DC, El Gamal A, Schorn M, Moore BS, Johnson MD, Mincer TJ, Whalen KE. A Bacterial Quorum-Sensing Precursor Induces Mortality in the Marine Coccolithophore, Emiliania huxleyi. Front Microbiol. 2016;7:59. doi: 10.3389/fmicb.2016.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazan R, Que YA, Maura D, Strobel B, Majcherczyk PA, Hopper LR, Wilbur DJ, Hreha TN, Barquera B, Rahme LG. Auto Poisoning of the Respiratory Chain by a Quorum-Sensing-Regulated Molecule Favors Biofilm Formation and Antibiotic Tolerance. Curr Biol. 2016;26:195–206. doi: 10.1016/j.cub.2015.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heeb S, Fletcher MP, Chhabra SR, Diggle SP, Williams P, Cámara M. Quinolones: from antibiotics to autoinducers. FEMS Microbiol Rev. 2011;35:247–274. doi: 10.1111/j.1574-6976.2010.00247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaraman P, Sakharkar MK, Lim CS, Tang TH, Sakharkar KR. Activity and interactions of antibiotic and phytochemical combinations against Pseudomonas aeruginosa in vitro. Int J Biol Sci. 2010;6:556–568. doi: 10.7150/ijbs.6.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilani-Feki O, Zouari I, Culioli G, Ortalo-Magné A, Zouari N, Blache Y, Jaoua S. Correlation between synthesis variation of 2-alkylquinolones and the antifungal activity of a Burkholderia cepacia strain collection. World J Microbiol Biotechnol. 2012;28:275–281. doi: 10.1007/s11274-011-0817-0. [DOI] [PubMed] [Google Scholar]
- Liu X, Cheng YQ. Genome-guided discovery of diverse natural products from Burkholderia sp J Ind Microbiol Biotechnol. 2014;41:275–284. doi: 10.1007/s10295-013-1376-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Clasquin MF, Melamud E, Amador-Noguez D, Caudy AA, Rabinowitz JD. Metabolomic analysis via reversed-phase ion-pairing liquid chromatography coupled to a stand alone orbitrap mass spectrometer. Anal Chem. 2010;82:3212–3221. doi: 10.1021/ac902837x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machan ZA, Taylor GW, Pitt TL, Cole PJ, Wilson R. 2-Heptyl-4-hydroxyquinoline N -oxide, an antistaphylococcal agent produced by Pseudomonas aeruginosa. J Antimicrob Chemother. 1992;30:615–623. doi: 10.1093/jac/30.5.615. [DOI] [PubMed] [Google Scholar]
- Majerczyk C, Brittnacher M, Jacobs M, Armour CD, Radey M, Schneider E, Phattarasokul S, Bunt R, Greenberg EP. Global Analysis of the Burkholderia thailandensis Quorum Sensing-Controlled Regulon. J Bacteriol. 2014;196:1412–1424. doi: 10.1128/JB.01405-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollenhauer HH, Morré DJ, Rowe LD. Alteration of intracellular traffic by monensin; mechanism, specificity and relationship to toxicity. Biochim Biophys Acta. 1990;1031:225–246. doi: 10.1016/0304-4157(90)90008-Z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nett M, Ikeda H, Moore BS. Genomic basis for natural product biosynthetic diversity in the actinomycetes. Nat Prod Rep. 2009;26:1362–1384. doi: 10.1039/b817069j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonejuie P, Burkart M, Pogliano K, Pogliano J. Bacterial cytological profiling rapidly identifies the cellular pathways targeted by antibacterial molecules. Proc Natl Acad Sci U S A. 2013;110:16169–16174. doi: 10.1073/pnas.1311066110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonejuie P, Trial RM, Newton GL, Lamsa A, Ranmali Perera V, Aguilar J, Liu WT, Dorrestein PC, Pogliano J, Pogliano K. Application of bacterial cytological profiling to crude natural product extracts reveals the antibacterial arsenal of Bacillus subtilis. J Antibiot (Tokyo) 2016;69:353–361. doi: 10.1038/ja.2015.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada BK, Wu Y, Mao D, Bushin LB, Seyedsayamdost MR. Mapping the Trimethoprim-Induced Secondary Metabolome of Burkholderia thailandensis. ACS Chem Biol. 2016;11:2124–2130. doi: 10.1021/acschembio.6b00447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orhan G, Bayram A, Zer Y, Balci I. Synergy Tests by E Test and Checkerboard Methods of Antimicrobial Combinations against Brucella melitensis. J Clin Microbiol. 2005;43:140–143. doi: 10.1128/JCM.43.1.140-143.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palfey BA, Björnberg O, Jensen KF. Insight into the chemistry of flavin reduction and oxidation in Escherichia coli dihydroorotate dehydrogenase obtained by rapid reaction studies. Biochemistry. 2001;40:4381–4390. doi: 10.1021/bi0025666. [DOI] [PubMed] [Google Scholar]
- Pesci EC, Milbank JB, Pearson JP, McKnight S, Kende AS, Greenberg EP, Iglewski BH. Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 1999;96:11229–11234. doi: 10.1073/pnas.96.20.11229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulsawat N, Kitani S, Nihira T. Characterization of biosynthetic gene cluster for the production of virginiamycin M, a streptogramin type A antibiotic, in Streptomyces virginiae. Gene. 2007;393:31–42. doi: 10.1016/j.gene.2006.12.035. [DOI] [PubMed] [Google Scholar]
- Que YA, Hazan R, Ryan CM, Milot S, Lépine F, Lydon M, Rahme LG. Production of Pseudomonas aeruginosa Intercellular Small Signaling Molecules in Human Burn Wounds. J Pathog. 2011;2011:549302. doi: 10.4061/2011/549302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea MC, Sit CS, Clayton E, O'Connor PM, Whittal RM, Zheng J, Vederas JC, Ross RP, Hill C. Thuricin CD, a posttranslationally modified bacteriocin with a narrow spectrum of activity against Clostridium difficile. Proc Natl Acad Sci U S A. 2010;107:9352–9357. doi: 10.1073/pnas.0913554107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reil E, Höfle G, Draber W, Oettmeier W. Quinolones and their N-oxides as inhibitors of mitochondrial complexes I and III. Biochim Biophys Acta. 1997;1318:291–298. doi: 10.1016/s0005-2728(96)00150-8. [DOI] [PubMed] [Google Scholar]
- Seyedsayamdost MR. High-throughput platform for the discovery of elicitors of silent bacterial gene clusters. Proc Natl Acad Sci U S A. 2014;111:7266–7271. doi: 10.1073/pnas.1400019111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallarida RJ. An Overview of Drug Combination Analysis with Isobolograms. J Pharmacol Exp Ther. 2006;319:1–7. doi: 10.1124/jpet.106.104117. [DOI] [PubMed] [Google Scholar]
- Terada H. The interaction of highly active uncouplers with mitochondria. Biochim Biophys Acta. 1981;639:225–242. doi: 10.1016/0304-4173(81)90011-2. [DOI] [PubMed] [Google Scholar]
- Vallieres C, Fisher N, Antoine T, Al-Helal M, Stocks P, Berry NG, Lawrenson AS, Ward SA, O'Neill PM, Biagini GA, et al. HDQ, a Potent Inhibitor of Plasmodium falciparum Proliferation, Binds to the Quinone Reduction Site of the Cytochrome bc1 Complex. Antimicrob Agents Chemother. 2012;56:3739–3747. doi: 10.1128/AAC.00486-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ark G, Berden JA. Binding of HQNO to beef-heart sub-mitochondrial particles. Biochim Biophys Acta. 1977;459:119–127. doi: 10.1016/0005-2728(77)90014-7. [DOI] [PubMed] [Google Scholar]
- Vial L, Lepine F, Milot S, Groleau MC, Dekimpe V, Woods DE, Deziel E. Burkholderia pseudomallei, B. thailandensis, and B. ambifaria Produce 4-Hydroxy-2-Alkylquinoline Analogues with a Methyl Group at the 3 Position That Is Required for Quorum-Sensing Regulation. J Bacteriol. 2008;190:5339–5352. doi: 10.1128/JB.00400-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams DH, Stone MJ, Hauck PR, Rahman SK. Why Are Secondary Metabolites (Natural Products) Biosynthesized? J Nat Prod. 1989;52:1189–1208. doi: 10.1021/np50066a001. [DOI] [PubMed] [Google Scholar]
- Wilson MZ, Wang R, Gitai Z, Seyedsayamdost MR. Mode of action and resistance studies unveil new roles for tropodithietic acid as an anticancer agent and the γ-glutamyl cycle as a proton sink. Proc Natl Acad Sci U S A. 2016;113:1630–1635. doi: 10.1073/pnas.1518034113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yim G, Wang HH, Davies J. Antibiotics as signalling molecules. Philos Trans R Soc Lond B Biol Sci. 2007;362:1195–1200. doi: 10.1098/rstb.2007.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


