Abstract
Lack of prominent developmental defects arising from loss of many individual miRNAs is consistent with the observations of collaborative networks between miRNAs and roles for miRNAs in regulating stress responses. However, these characteristics may only partially explain the seemingly non-essential nature of many miRNAs. Non-miRNA gene expression regulatory mechanisms also collaborate with miRISC to support robust gene expression dynamics. Genetic enhancer screens have revealed roles of miRNAs and other gene repressive mechanisms in development or other cellular processes that were masked by genetic redundancy. Besides discussing the breadth of the non-miRNA genes, we will use LIN-28 as an example to illustrate how distinct regulatory systems, including miRNAs and multiple protein stability mechanisms, work at different levels to target expression of a given gene and provide tissue-specific and stage-specific regulation of gene expression.
Keywords: protease, caspase, Arg/N-end rule, developmental timing, heterochrony
The enigma of miRNAs
Outside of developmental timing (heterochrony), roles for many miRNAs (microRNAs) are still unclear. Using C. elegans, many individual miRNAs were shown to be non-essential [1] and even entire families of miRNAs were shown to be non-essential [2]. In mice, deletions of highly abundant miRNAs in certain muscle tissues were sometimes found to yield no discernable phenotype [3]. Though obtaining such results leaves one very unsettled, these findings strongly imply that functional redundancy is commonly associated with miRNAs, assuming that every expressed miRNA has some function. The notion of redundancy is consistent with the fact that loss of specific miRNAs typically results in less than two-fold changes in protein expression for relevant targets [3]. However, despite their apparent redundancy, genetic studies have implicated miRNAs in diverse diseases including multiple types of cancer, numerous cardiovascular disease phenotypes, and even a form of hearing loss due to a defect in hair cell sensory neurons [4]. Thus, understanding and overcoming the difficulty presented by genetic redundancy associated with miRNAs is very critical to understanding their roles in specific physiological events.
Lack of prominent developmental phenotypes associated with mutating individual miRNA or miRNA families may be partly due to the fact that many miRNAs and global miRISC have been shown to function in response to stressors or environmental changes [5]. Examples from more recent studies in C. elegans include roles in recovery from starvation [6], longevity regulation [7,8], pathogen resistance [9,10], and neuronal regulation of the decision to enter dauer (an alternative larval phase for enduring harsh conditions) [11]. Additionally, miRNAs are important in ensuring fecundity at elevated temperatures [12] and keeping the cell death pathway in check [13]. In these examples, specific physiological functions of miRNAs are also often executed through multi-miRNA-target interaction networks [9,11–13].
For a specific physiological function, it has been well documented that a group of miRNAs often functions to repress the expression of a group of targets or even a single target [3]. As an example of the latter, the mRNA of the lin-28 pluripotency factor is targeted by multiple miRNAs in C. elegans, including LIN-4 and LET-7 families that function in the conserved developmental timing pathway [14–17]. The molecular and genetic mechanisms related to developmental timing and the consequent cellular events provide an exciting case study of gene regulatory networks. The C. elegans model has arguably provided the most insight into the genetics of the deeply conserved developmental timing circuitry.
The balance between LIN-28 and LET-7 is important for the timing of the early and late larval programs in C. elegans. This balance is necessary for seam cell divisions and vulval development. The L1 stage in C. elegans is specified by the LIN-14 transcription factor that, among other things, promotes the LIN-28-dependent L2 stage-specific symmetric division of lateral seam cells (a stem-like cell type of the epidermis). A rapid down-regulation of LIN-28 protein by the mid-L3 stage leads to the subsequent upregulation of LET-7, and results in a final transition from proliferation to differentiation in seam and vulval tissues [18]. Prolonged LIN-28 levels cause additional symmetric divisions of seam cells leading to supernumerary seam cells by the L4 stage and gapped adult alae [14]. Prolonged expression of LIN-41 (TRIM71), a target of LET-7 in the mid-L3 stage, levels lead to inappropriate vulval morphogenesis that ultimately causes rupturing due to a loss of vulval-uterine integrity [19].
Although mutations in the developmental timing pathway may cause different phenotypes across metazoans, the critical genetic wiring has been highly conserved. Thus, understanding additional factors in this network is important. Moreover, understanding how compensatory regulators collaborate with miRNAs to ensure robust outcomes is fundamentally important to understand the architecture of post-transcriptional gene expression dynamics. Several recent genetic experiments have utilized strong tools to tackle the problem of miRNA-related redundancy. Very recent molecular analyses of non-miRNA collaborators have begun to unravel the mechanistic basis of the associated dynamic post-transcriptional gene regulation. This review will consider some of these new findings.
Complexity within the miRNA pathway
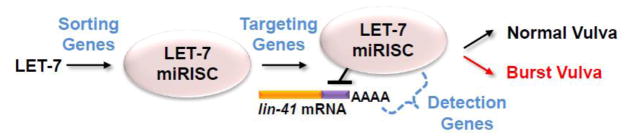
In addition to redundancy between miRNAs, redundancy also appears to be associated with proteins involved in miRNA-related functions. Such redundancy was demonstrated in C. elegans using a genomic-scale RNAi screen that identified enhancers of a weak let-7 mutation that alone did not produce developmental timing defects [20]. This enhancer screen focused on the vulva bursting phenotype associated with strong loss-of-function let-7 alleles. It was found that mutations in many candidate genes, showed a very strong and reliable synthetic vulval bursting phenotype when combined with the weak let-7 mutation. Since the let-7 mutant had a modest reduction in mature LET-7 expression and the enhanced vulval bursting phenotype was suppressed by loss of the key LET-7 target, LIN-41, it was concluded that many of the enhancer genes likely function in steps downstream of LET-7. Subsequent work has since shown that lin-41 overexpression is solely responsible for vulval bursting as a result of let-7(−) mutation [19], making lin-41(−) suppression of the synthetic phenotype a strong demonstration of let-7-pathway function. It was hypothesized that these genes may be important for sorting (loading) LET-7 into miRISC, for targeting of LET-7-miRISC to relevant mRNAs, or for detecting LET-7-miRISC target mRNA interactions (Figure 1).
Figure 1. Robustness within a miRNA pathway.
Genes involved in functions downstream of LET-7 including sorting, targeting, and detection of miRISCs were identified in an enhancer screen using a weak reduction-of-function allele (see text). Reduction in each of these genes alone displayed no severe phenotype. When the heterochronic function of LET-7-miRISC insufficiently limits LIN-41 levels, a burst vulva phenotype is observed.
In a similar experiment using C. elegans, mutation of several non-essential miRNAs produced severe viability phenotypes, including defective gonad migration or adult lethality, when in the context of a sensitized genetic background such as mutations in alg-1, an argonaute gene [21]. Argonaute proteins are required for miRISC formation and function. Since ALG-1 is one of the two argonaute proteins specifically involved in miRNA function, such enhanced phenotypes also likely reflect genetic redundancy within miRNA systems. Intriguingly, loss of multiple other non-essential miRNAs actually suppressed alg-1(−) mutant defects including gapped adult alae and/or adult lethality [21]. The observation of enhancement or suppression with distinct miRNAs suggests the possibility of epistatic functions within the miRNA pathway. Moreover, a few miRNAs exhibited both suppression of an alg-1(−) phenotype, such as gapped adult alae, with concurrent enhancement of another alg-1(−) phenotype, such as embryonic lethality [21]. The percentages involved between the phenotypes rule out a possibility of a directly inverse relationship between the two phenotypes. Altogether, these findings imply a complex relationship of factors within the miRNA pathway.
Early signs of pathways converging with miRNA regulation
A possibility to consider is that miRNAs may collaborate with non-miRNA pathways to convergently regulate targets. In this model, a miRNA and a non-miRNA pathway act on the same target, but at different levels of gene regulation. This convergent regulation can be considered as two genetic pathways acting in parallel.
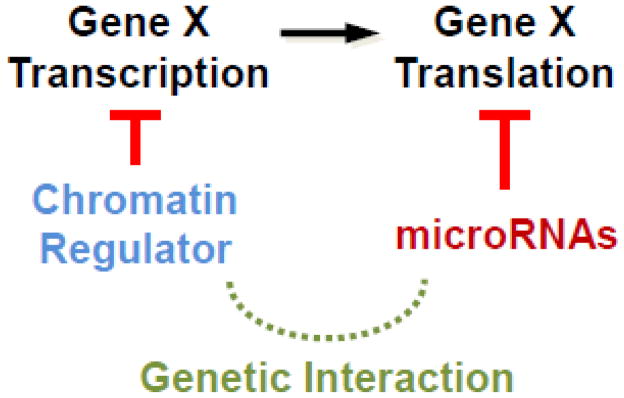
Another interaction screen was done in C. elegans by treating multiple viable miRNA mutants with RNAi against several chromatin regulatory hub genes to reveal synthetic sterile phenotypes [21]. Such convergent gene regulation is depicted in Figure 2. An important distinction in genetic interactions needs to be made here. Many genes identified using the weak reduced-function allele of let-7, which results in slightly reduced processing of let-7 miRNA, likely further impaired processing of the mature miRNA, including loading into miRISC, for example [20]. However, the study that utilized miRNA null mutations (large deletions) to identify synthetic interactions, was more likely to identify targets convergently regulated by miRNAs and different chromatin regulators, as the given miRNAs were abolished in the null mutants [21]. However, there still remains the potential caveat that the observed interactions between miRNAs and chromatin regulators may still be due to redundancy between different miRNAs, as the chromatin regulators could potentially be involved in the expression of other specific miRNAs.
Figure 2. Genetic redundancy due to potential parallel regulation by miRNAs and chromatin factors on the same target genes.
Synergistic phenotypes were observed for viable miRNA mutants when treated with RNAi for various chromatin hub regulatory genes. It is also possible that the regulation by chromatin regulators is indirect, mediated by other miRNAs.
miRNAs get a little help from a lot of friends
To identify non-miRNA genes working in parallel to the general miRNA/miRISC pathway to regulate the same target gene, a genomic-scale RNAi screen was used to systematically reveal genes that are essential when global miRISC function is compromised in C. elegans [22]. In order to identify a potentially broad range of functional collaborators, this miRISC enhancer screen examined multiple synthetic phenotypes including developmental stall, sterility, embryonic lethality, or other catastrophic outcomes. The miRISC screen identified 126 genes that become essential with loss of one of the GW182 proteins, AIN-1 or AIN-2. Importantly, only 8 of these interactors were also identified by the screen for miRNA pathway factors using the weak reduction-of-function let-7 allele [20]. Thus, more than 90% of the factors identified in the miRISC enhancer screen were not known to function with miRNA-related pathways, indicating a strong enrichment for genes working convergently with miRNA/miRISC pathways. The identified non-miRNA genes distribute over broad functional categories including cell migration, protein stability, and apoptosis. Interestingly, the largest category “unknown function”, includes almost a third of the identified genes. These findings clearly underscore the difficulty in understanding genomic function masked by genetic redundancy and pleiotropism.
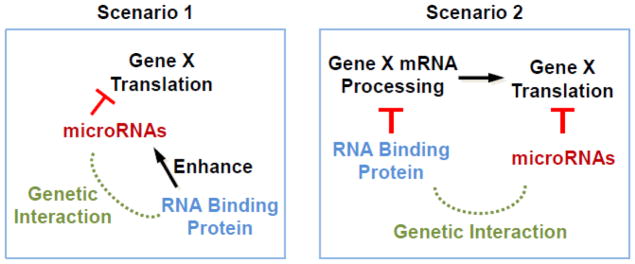
Several studies also observed strong enhancement of developmental defects between mutations in miRNAs and RNA binding proteins (RBPs) [23–27]. There are two basic models to explain the interactions between these RBPs and miRNAs. In the first model, specific RBPs work physically with miRISCs to enhance miRISC function (Figure 3). As an example of this scenario, it was shown that NHL-2, a TRIM-NHL protein, functions as a cofactor for the miRISC [23]. NHL-2 is required for efficient down-regulation of miRISC target mRNAs such as hbl-1(hunchback) and let-60(Ras) by LET-7 [23]. NHL-2 interacts physically with CGH-1, a key P-body component, and miRISC components including ALG-1 and AIN-1 [23]. In the second model, some RBPs work in parallel to (and independent of) miRISC to regulate common targets through direct or indirect interactions with the targets. The latter scenario has the potential to result in severe synergistic phenotypes when both the RBP and miRISC function are compromised (Figure 3). As an example of this situation, one study showed that vgln-1(vigilin) likely acted in parallel to miRISC function [27]. The authors found that loss of both vgln-1 and ain-2 resulted in 100 percent L1 arrest, while the individual mutants showed little or no arrest [27].
Figure 3. Potential redundancy between miRNAs and RNA binding proteins.
RNA binding proteins (RBPs) may positively enhance the effects of miRNA regulation (first scenario). This enhancement may involve physical interaction of the RBP with the miRISC to upregulate its activity. Alternatively, RBPs may act in parallel to miRNAs by acting at a different level of gene expression such as processing of mRNA (second scenario).
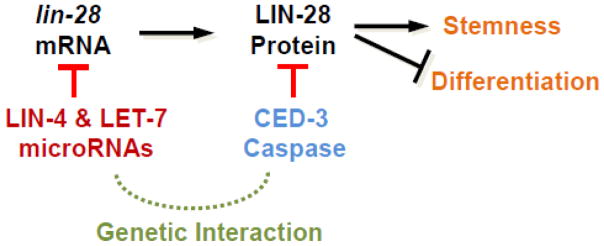
Importantly, the miRISC enhancer studies revealed previously unknown functions for many non-essential miRNAs, and the robustness of gene expression dynamics often requires collaborative efforts from miRNAs and other non-miRNA-related repressive mechanisms. In many cases, miRNAs regulate one level of gene regulation (production of new protein) while another factor regulates another level of gene expression (protein stability, for example) (Figure 4) [22].
Figure 4. Convergent gene regulation by non-homologous genes at distinct levels of gene expression.
Two pathways acting at different levels of gene expression ensure robust biological responses. In this example, miRNAs working in the miRISC complex negatively regulate lin-28 translation while CED-3 caspase (or LEP-2) function to proteolytically inactivate LIN-28 protein already present.
Unexpected support from a caspase
In the miRISC enhancer study, one intriguing interacting gene was the ced-3 caspase [22]. CED-3 was originally identified as the classic programmed cell death caspase [28–30]. As a caspase, CED-3 functions as a cysteine aspartase to cleave proteins C-terminal to a caspase cleavage sequence, DxxD [31]. Intriquingly, it was found that ain-1(−) mutation did not alter programmed cell death, thus suggesting that CED-3 had a non-apoptotic function in parallel to miRISC [22].
Non-apoptotic functions for ced-3 have also been implicated in neuronal regeneration [32], stress responses [33], and ageing [34]. Although those studies did not identify specific non-apoptotic targets, genetic experiments suggested that CED-3 may work in these processes as a cell signaling regulator. Specific targets for a non-apoptotic function of CED-3 were first reported in the developmental timing pathway where CED-3 functioned in parallel to AIN-1/miRISC to repress several developmental factors, including LIN-28 [22]. This finding enhanced the argument for the presence of mechanistic non-apoptotic functions associated with CED-3. By screening many miRNA mutants for developmental defects when treated with ced-3(RNAi), mutants of the let-7 family showed synergistic developmental timing defects including supernumerary seam cells and gapped adult alae [22]. Genetic and biochemical data suggested that CED-3 caspase likely cleaved LIN-28 to inactivate the pluripotency factor. In addition, purified CED-3 cleaved the DISL-2 RNAse and partially cleaved the LIN-14 transcription factor in vitro [22]. Interestingly, human Dis3l2 (DISL-2 ortholog) has been shown to function with (but downstream of) LIN-28 to degrade poly-uridylated let-7 [35]. LIN-14 is a well-established upstream positive regulator of LIN-28. Importantly, these findings established a non-apoptotic role for CED-3 caspase in the developmental timing pathway—likely working on multiple targets. This non-apoptotic role is also non-canonical in that it functions as a direct, negative gene expression regulator and not in cell signaling.
Eliminating LIN-28: The task of unmasking the new proteolytic players
LIN-28 (LIN28) is critical to development across diverse phyla including roles in developmental timing (heterochrony) and promoting pluripotency [18,36], as well as important roles in various cancers [37]. The lin-28 mRNA is targeted by miRNAs in both C. elegans and humans [15]. However, recent findings show several examples of proteolytic pathways acting to regulate LIN-28 levels in worms and humans.
How CED-3 recognizes LIN-28 was an interesting question, as this caspase has well-established functions in programmed cell death. Therefore, how a caspase with both apoptotic and non-apoptotic functions is able to differentially recognize targets is fundamentally important to understand. Although CED-3 was shown to be necessary for the robust inactivation of LIN-28 [22], alone it was found to be insufficient for this inactivation role [38], suggesting the involvement of an additional proteolytic system.
The Arg/N-end rule pathway controls protein stability largely as a function of certain N-terminal amino acids [39]. Specifically, this proteolytic pathway has three important determinants for substrates, including a destabilizing N-terminal residue, an internal Lysine for poly-ubiquitination, and an unstructured N-terminus [39]. Amongst the required factors within the Arg/N-end rule pathway, the N-terminal arginine transferase (ATE-1) adds an N-terminal Arg to substrates that are then recognized by a UBR-type E3 ligase (UBR-1) for poly-ubiquitination [39]. Genetically, both ATE-1 and UBR-1 were found to function in parallel to miRISC to negatively regulate symmetric seam cell divisions [38]. Biochemically, both ATE-1 and UBR-1 were found to physically and specifically interact with CED-3 caspase [38]. Surprisingly, the Arg/N-end rule E3 ubiquitin ligase, UBR-1, was found to be required for efficient CED-3-mediated cleavage of LIN-28 substrate that triggers further degradation of LIN-28 by the proteasome [38]. Moreover, human caspase 8 and human UBR2 were also found to physically interact [38], suggesting that caspase-UBR interactions may be conserved. Human caspase 8 has been implicated in non-apoptotic functions including monocyte differentiation and trophoblast fusion [40,41], adding even more interest to these findings. In fact, non-apoptotic caspase functions represent a rapidly emerging field with critical functions throughout eukaryotes [42].
In addition to regulating seam cell development, LIN-28 is important in other aspects of developmental timing including the juvenile-to-adult transition [43]. The LEP-2 gene in C. elegans is the sole Makorin (Mkrn) ortholog and encodes an E3 ubiquitin ligase with RING and C3H1-type zinc fingers. The functions of Makorins are not well established. Interestingly, lep-2 mutations from several forward screens were found to affect the timing of male tail morphogenesis [43]. In fact, LEP-2 was found to function as an important developmental timing regulator with a critical role in normal molting processes in both sexes [43]. Expression analyses indicated a role for the LEP-2 Mkrn protein in regulating LIN-28 protein stability in certain non-seam cell tissues [43]. While the mechanism for LEP-2 regulation of LIN-28 stability remains to be understood, comparing the CED-3-UBR-1 findings with the LEP-2 findings demonstrates an exciting feature of non-miRNA genes in collaborating with the miRISC by imparting tissue-specific functions.
An important demonstration of signal transduction in regulating LIN28 stability in human cells was recently reported [44]. Using a targeted screen of kinase inhibitors in human embryonal carcinoma cells, the ERK signaling pathway was found to control LIN28 phosphorylation [44]. Intriguingly, this phosphorylation increased stability of LIN28 [44]. Moreover, the ERK-mediated phospho-regulation separates LET-7-dependent from LET-7-independent LIN28-mediated effects [44]. A phospho-mimetic mutation in LIN28 also enhanced reprogramming efficiency in induced pluripotent stem cells [44]. However, the proteolytic pathway acting on LIN28 in a phospho-dependent manner is not yet known.
Altogether, the findings of multiple protein stability mechanisms acting on LIN-28 (LIN28) provide a strong case study for convergent regulation with the miRISC pathway. Clearly, miRNAs require substantial collaboration to ensure robust and dynamic gene regulation. This conclusion is not meant to undermine or trivialize the importance of miRNAs, rather the opposite. The miRNA pathway seems particularly adept at integrating with other levels of gene regulation. The extensive network converging on an individual target gene, such as lin-28, also clearly demonstrates the difficulty of identifying genetic functions of specific regulatory inputs for a specific physiological function.
Concluding remarks
Although robustness is an innate property of all life-ensuring outcomes, its mechanistic underpinnings remain an important topic of investigation. Importantly, the role of non-homologous genes in convergent gene regulation provides an extra layer of complexity for gene expression dynamics. Additionally, the ability of one gene to assume many roles, such as the CED-3 caspase working in programmed cell death and non-apoptotic regulation of pluripotency, imparts enhanced versatility but also imposes a regulatory quandary. That is to say, how does a protease with such dichotomous functions—promoting death of some cells while supporting the vigor of other cells—achieve these distinct outcomes? In other words, what differences in the cellular milieu or upstream signaling account for the different functions? Recent findings that demonstrate coupling mechanisms, such as the CED-3 complex formation with UBR-1, begin to address this regulatory dilemma.
The collaboration of non-miRNA genes with the miRISC provides several important features, including tissue-specific regulation, synergy, and dynamic control of gene expression. As an example of tissue-specific functions, the CED-3-UBR-1 function supports the transition of stem-like cells from proliferative to differentiated fates during mid-larval development; whereas, LEP-2 supports the juvenile-to-adult molting that promotes tail tip morphogenesis. Additionally, the miRISC acts on lin-28 mRNA while CED-3-UBR-1 irreversibly eliminates LIN-28 protein, supporting a fully committed mid-larval transition of the stem-like seam cells. Thus, by acting at different levels of gene expression, the two systems confer collaborative and dynamic temporal control of gene expression (Figure 5). Importantly, as seen with CED-3-UBR-1 function, proteolytic regulators have the distinct advantage to rapidly commit gene expression programs by executing an irreversible form of gene regulation.
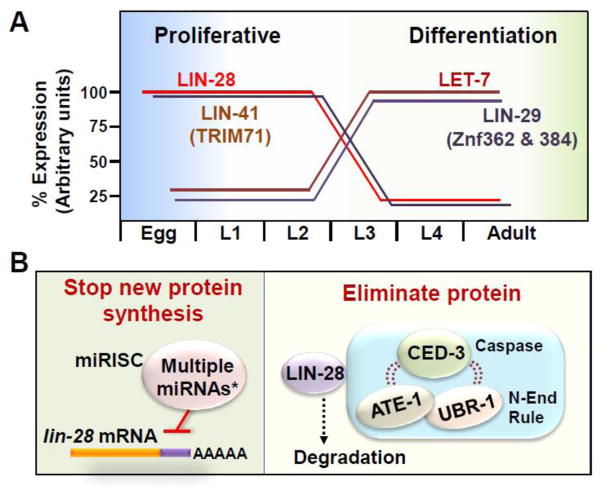
Key Figure: Figure 5. Robust gene expression dynamics ensured by convergent pathways.
Transitioning from proliferative to differentiated states requires dynamic changes in gene expression. (A) As an example from C. elegans, in the stem-like cell type, seam cells, LIN-28 levels are rapidly downregulated toward the end of the second larval stage and nearly absent by the middle of the third larval stage. Temporal expression patterns for several other factors are also shown. When names differ, mammalian orthologs are indicated in parentheses. (B) The miRNA pathway prevents production of additional LIN-28 protein. *Multiple miRNAs, including LET-7 family members and LIN-4, collaborate within the miRNA pathway. CED-3 caspase, coupled in a complex with Arg/N-end rule components (ATE-1 and UBR-1), is able to recognize LIN-28 to downregulate the protein in a non-canonical function that negatively regulates pluripotency. The collaboration between the miRNA and caspase pathways in seam cells provides a tissue-specific function and dynamic gene regulation. Other tissues utilize additional factors such as LEP-2.
As an interesting possibility, collaborating gene regulators provide ample material for evolutionary processes to act upon. Since gene regulation is often the result of complex networks of factors, minor alterations within a regulatory network or additional interactions across networks could allow new functions to emerge without substantial detriment to the organism.
Trends Box.
miRNA pathways converge with diverse non-miRNA regulatory mechanisms to regulate common targets.
Convergent regulation at different levels of gene expression imparts tissue-specific functions, synergy, and precise temporal gene regulation.
Proteolytic pathways provide a strong commitment in gene regulation since proteolysis is irreversible without new protein synthesis.
Genetic redundancy remains a critical barrier in understanding genomic architecture and regulatory networks.
Genes with known functions may have additional important non-canonical functions.
Outstanding questions.
How do miRNA pathways converge with non-miRNA pathways in evolution?
Based on findings from multiple genetic studies, collaboration of regulatory systems is not unique to miRNAs. However, miRNAs may be particularly well-suited for this adaptation due to the small seed sequence needed to target an mRNA and the abundance of different miRNAs providing many possible seed sequences.
Are apoptotic caspases widely used to regulate gene expression?
Studies in various organisms suggest that usage of caspases to limit gene expression in non-apoptotic process is unlikely to represent a rare phenomenon, but the broadness of the usage may need to be addressed by systematic methods with both genetic and biochemical approaches.
What other mechanisms underlie the differential recognition of non-apoptotic targets from apoptotic targets by caspases?
The coupling between CED-3 and the Arg/N-end rule pathway shown by Weaver and colleagues (2017) provided one likely mechanism. However, other mechanisms may also exist because not all targets may be good Arg/N-end rule substrates.
How are tissue-specific functions differentially regulated?
Based on emerging findings, tissue-specific regulation is a wide-spread phenomenon. Identifying upstream factors regulating the tissue-specific effectors has proven to be particularly difficult.
How do genes acquire additional non-canonical functions?
This is difficult to understand fully since acquisition of a novel function cannot compromise the original function. As a speculation, a non-miRNA gene, such as a caspase, may drift in substrate recognition while a miRNA-based mechanism acts as a buffer to ensure gene regulation of the relevant targets. The concept of miRNAs acting as buffers has been previously considered as an analog to chaperones buffering mis-folded proteins as discussed by Ebert and Sharp (2012). Conversely, it is also possible that the non-miRNA-based mechanism may buffer regulation while a miRNA mechanism is altered.
Acknowledgments
We are eminently thankful for the thought-provoking suggestions of previous and current anonymous peer-reviewers. We apologize to our colleagues whose excellent work was not cited due to space limitations. We also thank Y. Weaver and A. Sewell for helpful discussions and critical reading of the manuscript. This work was supported in part by a postdoctoral fellowship 121631-PF-12-088-01-RMC from the American Cancer Society (BPW), National Institutes of Health grant 5R01GM047869 (MH), and the Howard Hughes Medical Institute (MH).
Glossary
- AIN Genes
Encode the C. elegans GW182 orthologs, AIN-1 and AIN-2. ALG-1 INteracting proteins.
- ALG Genes
Encode several of the C. elegans argonaute orthologs. Argonaute (plant)-Like Gene.
- Argonaute
First identified in the Arabidopsis plant, a family of proteins conserved throughout eukaryotes and prokaryotes. Required for miRNA or siRNA-mediated gene silencing.
- Arg/N-End rule
A multi-enzyme pathway that recognizes N-terminal destabilizing residues in the context of an unstructured N-terminus and available internal lysine residue for poly-ubiquitination. For type 1 N-degrons, tertiary destabilizing residues (Asn and Gln) can be oxidized (by an NTAN or NTAQ homologs) to secondary destabilizing residues (Asp and Glu), and then arginylated by an ATE homolog (Arginyl-TransferasE) to generate a primary destabilizing residue. Positively charged primary destabilizing residues (Arg, Lys, His) are type 1 N-degrons; whereas bulky aliphatic primary destabilizing residues (Phe, Trp, Tyr, Leu, Ile) are type 2 N-degrons. The N-degrons are recognized by an N-recognin of the UBR-type E3 ubiquitin ligase family.
- Caspase
Class of proteases that are cysteine aspartases required for programmed cell death and certain non-apoptotic or non-canonical functions. The sole caspase in C. elegans, CED-3, was first identified as Cell Death defective mutant 3.
- Developmental timing
Developmental processes resulting from heterochrony. See heterochronic pathway.
- Genetic Interaction
When two (or more) genetic mutations in distinct genes are combined and result in a phenotype that is not seen (or only marginally seen) with the individual mutants. The interaction can suppress or enhance a phenotype. In addition, reduction of gene expression for a given gene by RNAi in a mutant for another gene can also reveal a genetic interaction.
- Genetic Redundancy
A situation where one gene can substitute or partially substitute for another gene. Also known as genetic masking.
- GW182 Genes
Mammalian TNR6A-C proteins (TriNucleotide Repeat-containing gene 6 proteins), Drosophila Gawky, C. elegans AIN-1 and AIN-2 all contain GW-type repeats.
- Heterochronic pathway
Also referred to as developmental timing pathway. Elaborate genetic network regulating rates of developmental processes including cell/tissue specification. LIN-4 and LET-7 family of miRNAs are critical in this pathway. As an example, LIN-28 is a critical miRNA target. LIN-28 is also targeted at the protein level by the CED-3 caspase-UBR-1 E3 ligase complex in some tissues and LEP-2 Makorin in other tissues.
- Heterochrony
Developmental processes owing to changes in the timing or rate of certain events relative to other events, resulting in alterations in size, shape, or cell/tissue specification. Regulated by factors in the heterochronic pathway.
- miRNA or microRNA
Class of small non-coding RNA molecules 21–24 nucleotides in length. Mature miRNAs are functional in the miRISC.
- miRISC
miRNA-Induced Silencing Complex. A ribonucleoprotein complex consisting of a mature miRNA in specific association with an argonaute protein, a GW182 protein, mRNA deadenylases, a poly-A nuclease (PAN), and other factors.
- Pleiotropism
A situation where one gene has more than one physiological function within an organism giving rise to numerous phenotypic traits. Often the traits may appear unrelated to each other. Some traits are often masked by the manifestation of other traits.
- RNAi
RNA-interference. Used to reduce gene expression and is often achieved by introducing an exogenous RNA fragment through various means into an organism and results in reduction or elimination of expression of an endogenous RNA or mRNA.
- Robustness
An innate property of all living systems that ensures outcomes. Genetic redundancy is a major contributor to robustness. This form of robustness is also known as canalization. Robustness can also refer to the strength of a phenotypic outcome.
- UBR-type E3 ligase
The E3 ubiquitin ligases that function as N-recognins and act in one or more branches of the N-End rule (Arg, Ac, or Pro). Canonical UBR-type ligases contain several structural features including the atypical zinc finger UBR-box motif, a ClpS core adaptor motif, and a RING/FYVE/PHD-type Zinc finger motif. Human UBR1, human UBR2, yeast UBR1, and C. elegans UBR-1 all share significant homology.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Miska EA, Alvarez-Saavedra E, Abbott AL, Lau NC, Hellman AB, McGonagle SM, Bartel DP, Ambros VR, Horvitz HR. Most Caenorhabditis elegans microRNAs are individually not essential for development or viability. PLoS Genet. 2007;3:e215. doi: 10.1371/journal.pgen.0030215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alvarez-Saavedra E, Horvitz HR. Many families of C. elegans microRNAs are not essential for development or viability. Curr Biol. 2010;20:367–373. doi: 10.1016/j.cub.2009.12.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ebert MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012;149:515–524. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mendell JT, Olson EN. MicroRNAs in stress signaling and human disease. Cell. 2012;148:1172–1187. doi: 10.1016/j.cell.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leung AK, Sharp PA. MicroRNA functions in stress responses. Mol Cell. 2010;40:205–215. doi: 10.1016/j.molcel.2010.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang X, Zabinsky R, Teng Y, Cui M, Han M. microRNAs play critical roles in the survival and recovery of Caenorhabditis elegans from starvation-induced L1 diapause. Proc Natl Acad Sci U S A. 2011;108:17997–18002. doi: 10.1073/pnas.1105982108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pincus Z, Smith-Vikos T, Slack FJ. MicroRNA predictors of longevity in Caenorhabditis elegans. PLoS Genet. 2011;7:e1002306. doi: 10.1371/journal.pgen.1002306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boulias K, Horvitz HR. The C. elegans microRNA mir-71 acts in neurons to promote germline-mediated longevity through regulation of DAF-16/FOXO. Cell Metab. 2012;15:439–450. doi: 10.1016/j.cmet.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kudlow BA, Zhang L, Han M. Systematic analysis of tissue-restricted miRISCs reveals a broad role for microRNAs in suppressing basal activity of the C. elegans pathogen response. Mol Cell. 2012;46:530–541. doi: 10.1016/j.molcel.2012.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ren Z, Ambros VR. Caenorhabditis elegans microRNAs of the let-7 family act in innate immune response circuits and confer robust developmental timing against pathogen stress. Proc Natl Acad Sci U S A. 2015;112:E2366–E2375. doi: 10.1073/pnas.1422858112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Than MT, Kudlow BA, Han M. Functional analysis of neuronal microRNAs in Caenorhabditis elegans dauer formation by combinational genetics and Neuronal miRISC immunoprecipitation. PLoS Genet. 2013;9:e1003592. doi: 10.1371/journal.pgen.1003592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McJunkin K, Ambros V. The embryonic mir-35 family of microRNAs promotes multiple aspects of fecundity in Caenorhabditis elegans. G3 (Bethesda) 2014;4:1747–1754. doi: 10.1534/g3.114.011973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sherrard R, Luehr S, Holzkamp H, McJunkin K, Memar N, Conradt B. miRNAs cooperate in apoptosis regulation during C. elegans development. Genes Dev. 2017;31:209–222. doi: 10.1101/gad.288555.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moss EG, Lee RC, Ambros V. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell. 1997;88:637–646. doi: 10.1016/s0092-8674(00)81906-6. [DOI] [PubMed] [Google Scholar]
- 15.Moss EG, Tang L. Conservation of the heterochronic regulator Lin-28, its developmental expression and microRNA complementary sites. Dev Biol. 2003;258:432–442. doi: 10.1016/s0012-1606(03)00126-x. [DOI] [PubMed] [Google Scholar]
- 16.Abbott AL, Alvarez-Saavedra E, Miska EA, Lau NC, Bartel DP, Horvitz HR, Ambros V. The let-7 MicroRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditis elegans. Dev Cell. 2005;9:403–414. doi: 10.1016/j.devcel.2005.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morita K, Han M. Multiple mechanisms are involved in regulating the expression of the developmental timing regulator lin-28 in Caenorhabditis elegans. EMBO J. 2006;25:5794–5804. doi: 10.1038/sj.emboj.7601451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rougvie AE, Moss EG. Developmental Transitions in C. elegans Larval Stages. Curr Top Dev Biol. 2013;105:153–180. doi: 10.1016/B978-0-12-396968-2.00006-3. [DOI] [PubMed] [Google Scholar]
- 19.Ecsedi M, Rausch M, Grosshans H. The let-7 microRNA directs vulval development through a single target. Dev Cell. 2015;32:335–344. doi: 10.1016/j.devcel.2014.12.018. [DOI] [PubMed] [Google Scholar]
- 20.Parry DH, Xu J, Ruvkun G. A whole-genome RNAi Screen for C. elegans miRNA pathway genes. Curr Biol. 2007;17:2013–2022. doi: 10.1016/j.cub.2007.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brenner JL, Jasiewicz KL, Fahley AF, Kemp BJ, Abbott AL. Loss of individual microRNAs causes mutant phenotypes in sensitized genetic backgrounds in C. elegans. Curr Biol. 2010;20:1321–1325. doi: 10.1016/j.cub.2010.05.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weaver BP, Zabinsky R, Weaver YM, Lee ES, Xue D, Han M. CED-3 caspase acts with miRNAs to regulate non-apoptotic gene expression dynamics for robust development in C. elegans. Elife. 2014;3:e04265. doi: 10.7554/eLife.04265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hammell CM, Lubin I, Boag PR, Blackwell TK, Ambros V. nhl-2 Modulates microRNA activity in Caenorhabditis elegans. Cell. 2009;136:926–938. doi: 10.1016/j.cell.2009.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Kouwenhove M, Kedde M, Agami R. MicroRNA regulation by RNA-binding proteins and its implications for cancer. Nat Rev Cancer. 2011;11:644–656. doi: 10.1038/nrc3107. [DOI] [PubMed] [Google Scholar]
- 25.Akay A, Craig A, Lehrbach N, Larance M, Pourkarimi E, Wright JE, Lamond A, Miska E, Gartner A. RNA-binding protein GLD-1/quaking genetically interacts with the mir-35 and the let-7 miRNA pathways in Caenorhabditis elegans. Open Biol. 2013;3:130151. doi: 10.1098/rsob.130151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ren Z, Veksler-Lublinsky I, Morrissey D, Ambros V. Staufen Negatively Modulates MicroRNA Activity in Caenorhabditis elegans. G3 (Bethesda) 2016;6:1227–1237. doi: 10.1534/g3.116.027300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zabinsky RA, Weum B, Cui M, Han M. RNA Binding Protein Vigilin Collaborates with miRNAs To Regulate Gene Expression for Caenorhabditis elegans Larval Development. G3 (Bethesda) 2017 doi: 10.1534/g3.117.043414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ellis HM, Horvitz HR. Genetic control of programmed cell death in the nematode C. elegans. Cell. 1986;44:817–829. doi: 10.1016/0092-8674(86)90004-8. [DOI] [PubMed] [Google Scholar]
- 29.Yuan JY, Horvitz HR. The Caenorhabditis elegans genes ced-3 and ced-4 act cell autonomously to cause programmed cell death. Dev Biol. 1990;138:33–41. doi: 10.1016/0012-1606(90)90174-h. [DOI] [PubMed] [Google Scholar]
- 30.Shaham S, Reddien PW, Davies B, Horvitz HR. Mutational analysis of the Caenorhabditis elegans cell-death gene ced-3. Genetics. 1999;153:1655–1671. doi: 10.1093/genetics/153.4.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xue D, Shaham S, Horvitz HR. The Caenorhabditis elegans cell-death protein CED-3 is a cysteine protease with substrate specificities similar to those of the human CPP32 protease. Genes Dev. 1996;10:1073–1083. doi: 10.1101/gad.10.9.1073. [DOI] [PubMed] [Google Scholar]
- 32.Pinan-Lucarre B, Gabel CV, Reina CP, Hulme SE, Shevkoplyas SS, Slone RD, Xue J, Qiao Y, Weisberg S, Roodhouse K, et al. The core apoptotic executioner proteins CED-3 and CED-4 promote initiation of neuronal regeneration in Caenorhabditis elegans. PLoS Biol. 2012;10:e1001331. doi: 10.1371/journal.pbio.1001331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Judy ME, Nakamura A, Huang A, Grant H, McCurdy H, Weiberth KF, Gao F, Coppola G, Kenyon C, Kao AW. A shift to organismal stress resistance in programmed cell death mutants. PLoS Genet. 2013;9:e1003714. doi: 10.1371/journal.pgen.1003714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yee C, Yang W, Hekimi S. The intrinsic apoptosis pathway mediates the pro-longevity response to mitochondrial ROS in C. elegans. Cell. 2014;157:897–909. doi: 10.1016/j.cell.2014.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang HM, Triboulet R, Thornton JE, Gregory RI. A role for the Perlman syndrome exonuclease Dis3l2 in the Lin28-let-7 pathway. Nature. 2013;497:244–248. doi: 10.1038/nature12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ambros V. MicroRNAs and developmental timing. Curr Opin Genet Dev. 2011;21:511–517. doi: 10.1016/j.gde.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Balzeau J, Menezes MR, Cao S, Hagan JP. The LIN28/let-7 Pathway in Cancer. Front Genet. 2017;8:31. doi: 10.3389/fgene.2017.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weaver BP, Weaver YM, Mitani S, Han M. Coupled Caspase and N-End Rule Ligase Activities Allow Recognition and Degradation of Pluripotency Factor LIN-28 during Non-Apoptotic Development. Dev Cell. 2017;41:665–673. doi: 10.1016/j.devcel.2017.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Varshavsky A. The N-end rule pathway and regulation by proteolysis. Protein Sci. 2011;20:1298–1345. doi: 10.1002/pro.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Black S, Kadyrov M, Kaufmann P, Ugele B, Emans N, Huppertz B. Syncytial fusion of human trophoblast depends on caspase 8. Cell Death Differ. 2004;11:90–98. doi: 10.1038/sj.cdd.4401307. [DOI] [PubMed] [Google Scholar]
- 41.Sordet O, Rebe C, Plenchette S, Zermati Y, Hermine O, Vainchenker W, Garrido C, Solary E, Dubrez-Daloz L. Specific involvement of caspases in the differentiation of monocytes into macrophages. Blood. 2002;100:4446–4453. doi: 10.1182/blood-2002-06-1778. [DOI] [PubMed] [Google Scholar]
- 42.Bell RA, Megeney LA. Evolution of caspase-mediated cell death and differentiation: twins separated at birth. Cell Death Differ. 2017 doi: 10.1038/cdd.2017.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Herrera RA, Kiontke K, Fitch DH. Makorin ortholog LEP-2 regulates LIN-28 stability to promote the juvenile-to-adult transition in Caenorhabditis elegans. Development. 2016;143:799–809. doi: 10.1242/dev.132738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsanov KM, Pearson DS, Wu Z, Han A, Triboulet R, Seligson MT, Powers JT, Osborne JK, Kane S, Gygi SP, et al. LIN28 phosphorylation by MAPK/ERK couples signalling to the post-transcriptional control of pluripotency. Nat Cell Biol. 2017;19:60–67. doi: 10.1038/ncb3453. [DOI] [PMC free article] [PubMed] [Google Scholar]