Abstract
Abundant evidence has accumulated showing that fetal alcohol exposure broadly modifies DNA methylation profiles in the brain. DNA methyltransferases (DNMTs), the enzymes responsible for DNA methylation, are likely implicated in this process. However, their regulation by ethanol exposure has been poorly addressed. Here, we show that alcohol exposure modulates DNMT protein levels through multiple mechanisms. Using a neural precursor cell line and primary mouse embryonic fibroblasts (MEFs), we found that ethanol exposure augments the levels of Dnmt3a, Dnmt3b, and Dnmt3l transcripts. We also unveil similar elevation of mRNA levels for other epigenetic actors upon ethanol exposure, among which the induction of lysine demethylase Kdm6a shows heat shock factor dependency. Furthermore, we show that ethanol exposure leads to specific increase in DNMT3A protein levels. This elevation not only relies on the upregulation of Dnmt3a mRNA but also depends on posttranscriptional mechanisms that are mediated by NADPH oxidase-dependent production of reactive oxygen species (ROS). Altogether, our work underlines complex regulation of epigenetic actors in response to alcohol exposure at both transcriptional and posttranscriptional levels. Notably, the upregulation of DNMT3A emerges as a prominent molecular event triggered by ethanol, driven by the generation of ROS.
Electronic supplementary material
The online version of this article (doi:10.1007/s12192-017-0829-2) contains supplementary material, which is available to authorized users.
Keywords: DNA methyltransferases, Ethanol exposure, Transcription, Heat shock factors, Reactive oxygen species (ROS), NADPH oxidase, Epigenetic regulators
Introduction
Exposure to alcohol during pregnancy is a severe challenge for neurodevelopment and can result in neurobehavioral to structural brain defects in the offspring, a continuum of conditions referred to as fetal alcohol spectrum disorder (FASD). Ethanol (EtOH), as a small molecule with both hydrophilic and lipophilic properties, is able to cross the placental barrier at any point during pregnancy and produces structural or neurochemical defects in multiple areas during cerebral development (reviewed in Lemoine et al. 1968; Jones and Smith 1973; Streissguth and O’Malley 2000). Extensive work on mouse models unraveled a variety of cellular processes affected by EtOH in the developing brain, such as cell survival, neural precursors proliferation and differentiation, neuronal migration, synaptogenesis, redox balance, and gene expression (reviewed in Guerri et al. 2009; Kleiber et al. 2014). Recently, we and others have demonstrated that heat shock factors (HSFs), a family of transcription factors discovered as stress sensors, but also involved in normal development, are activated in the murine embryonic cortex exposed to alcohol in vivo and mediate both protective and detrimental effects in the developing cerebral cortex or in neural cell systems exposed to alcohol (Pignataro et al. 2007; El Fatimy et al. 2014; Hashimoto-Torii et al. 2014).
Both in the presence or in the absence of overt structural or neurological brain abnormalities, functional disabilities of the central nervous system (CNS), including behavioral and cognitive deficits as well as psychiatric vulnerability, are consistently observed in FASD individuals (Clarke and Gibbard 2003; Mattson et al. 2011). Especially when not attributable to specific brain defects, these permanent consequences of prenatal alcohol exposure (PAE) are thought to be mediated by stable epigenetic signatures, established in utero upon stress and propagated throughout development, in which DNA methylation plays a major role (Kleiber et al. 2014).
Methylation of the DNA plays a quintessential role in the control of gene expression (Jones 2012). Four DNA methyltransferases (DNMTs) have been identified in mammals: DNMT1, DNMT3A, DNMT3B, and DNMT3-like (DNMT3L). DNMT1 is traditionally regarded as the maintenance DNMT, which perpetuates DNA methylation upon cell division, while DNMT3A and DNMT3B are responsible for de novo methylation (Guibert and Weber 2013). DNMT3L lacks a functional catalytic domain on its own, but can associate with DNMT3A/3B and stimulate their activity in the germ line and in embryonic stem cells (ESCs) (Bourc’his et al. 2001; Hata et al. 2002). DNA methylation is critical for long-term silencing of gene expression during various developmental processes in mammals, including neurogenesis and brain formation (Okano et al. 1999; Fan et al. 2001; reviewed in Guibert and Weber 2013). As a consequence, altered DNA methylation profiles or defects in the methylation machinery have been associated to neurodevelopmental syndromes, cognitive and behavioral impairment, as well as to psychiatric disorders (Rudenko and Tsai 2014; Peña et al. 2014).
Robust evidence demonstrates that EtOH can genome-widely modify DNA methylation signatures in the fetal brain, with enduring effects in adulthood. This includes in vivo studies in rodent models of PAE and ex vivo EtOH exposure of mouse embryos, human or murine embryonic stem cells and neural stem cells (Kaminen-Ahola et al. 2010; Hicks et al. 2010; reviewed in Kleiber et al. 2014; Khalid et al. 2014), and analyses of FASD patients (Laufer et al. 2015). DNMTs are likely implicated in this process. The increase or reduction in the levels of Dnmt transcripts or proteins has been reported in studies of fetal alcohol exposure paradigms (Bielawski et al. 2002; Mukhopadhyay et al. 2013; reviewed in Mead and Sarkar 2014) or alcohol abuse behaviors (Bönsch et al. 2006; Ponomarev et al. 2012; Warnault et al. 2013; Barbier et al. 2016). However, the regulation of all DNMT members, both at the mRNA and protein levels, and, moreover, its kinetics have not been systematically explored, and the mode of regulation of DNMT levels by EtOH is still obscure.
Here, we showed that EtOH exposure of neural precursors increased the levels of Dnmt3a, Dnmt3b, and Dnmt3l transcripts, but not Dnmt1. Because the same doses and timing of EtOH exposure triggered HSF activation, we investigated the role of the EtOH-responsive HSFs in the accumulation of Dnmt3a/b/l mRNAs. We found that the EtOH-induced elevation in Dnmt mRNA levels occurred independently of HSF. Such HSF independency was also observed for other epigenetic actors, which also exhibited alteration of their transcript levels in response to EtOH exposure. One marked exception concerned the lysine demethylase KDM6A (also called UTX, ubiquitously transcribed tetratricopeptide repeat, X chromosome), whose elevation of mRNA levels was dependent on the HSF pathway. Finally, at the protein level, we found that the most prominent output of EtOH exposure was the upregulation of DNMT3A—but not of other DNMTs—which did not only result from augmented transcript levels but was also mediated by posttranscriptional mechanisms dependent on reactive oxygen species (ROS) production by NADPH oxidase.
Materials and methods
Cell culture and experimental treatments
1C11 cells are immortalized teratocarcinoma-derived murine cells that have the properties of committed progenitors of serotoninergic or catecholaminergic neuronal lineages (Buc-Caron et al. 1990). Murine neural progenitors 1C11 were cultured at 37 °C, in a 10% CO2 atmosphere, in DMEM high glucose pyruvate glutamax, supplemented with 10% FBS, 100 U/ml penicillin and 100 μg/ml streptomycin (all from Invitrogen).
Mouse Hsf1 tm1Ijb and Hsf2 tm1Mmr knockout models have been previously described by McMillan et al. (1998) and Kallio et al. (2002), respectively. Hsf1 tm1Ijb/tm1Ijb will be named Hsf1 −/− and Hsf2 tm1Mmr/tm1Mmr will be named Hsf2 −/−. Hsf1 and Hsf2 knockout mice were used to prepare primary mouse embryonic fibroblasts (MEFs). Primary MEFs deleted for both Hsf1 and Hsf2 (Hsf1 −/− Hsf2 −/−) were obtained by intercrossing single heterozygous knockout mice. Primary MEFs isolated from E13.5 Hsf1 WT, Hsf1 −/−, Hsf1 WT Hsf2 WT, or Hsf1 −/− Hsf2 −/− embryos were cultured at 37 °C, 5% CO2, in DMEM high glucose pyruvate glutamax, supplemented with 10% FBS, 100 U/ml penicillin and 100 μg/ml streptomycin (all from Invitrogen). Primary MEFs were used for experiments at passages 1–3. As the genetic background of the strain in which the mutation is maintained can sensibly influence the phenotypes of the Hsf knockout mice (Abane and Mezger 2010), Hsf1 −/−, and Hsf1 −/− Hsf2 −/− MEFs were compared with the corresponding WT MEFs derived from the same genetic background (Balb/c x C57Bl/6N).
1C11 and MEFs were grown to 70–80% confluence and exposed to heat shock (HS) or EtOH. For thermal stress, cells were subjected to HS at 42 °C in a water bath. For EtOH treatment, a tenth of the medium was replaced by fresh medium containing 10× EtOH of the desired final concentration. Cells were maintained in presence of EtOH for 2, 4, 6, 8, 16, or 24 h. The permanence of EtOH in the medium during the time course was monitored with the EnzyChromTM EtOH Assay Kit (BioAssay Systems ECET-100) and never descended below 70% of the initial concentration upon 24 h. 1C11 treated with 50 mM (0.29% v/v) to 300 mM (1.76%v/v) EtOH did not display anomalies in cell morphology or obvious cell mortality based on the XTT Cell Viability Assay (Roche, according to the manufacturer’s instructions) and continued to proliferate (Online Resource 1, Suppl. Fig. S1). At the end of the HS or EtOH treatment, cells were immediately placed on ice, rinsed twice in cold PBS, collected as a dry pellet, frozen in dry ice, and stored at −80 °C, for successive RNA or protein preparation.
As indicated, cells were treated with 5 mM 4-methyl pyrazole (4-MP; Sigma M1387), 250 μM cyanamide (CYA; Sigma 187,364), 5 mM diallyl sulfite (DAS; Sigma A35501), or 4 μM diphenyleneiodonium (DPI; Sigma D2926). Cells were preincubated with drugs 45 min before the beginning of the EtOH treatment, and drugs were left in the medium all along the treatment.
RNA purification and quantitative RT-PCR (RT-qPCR)
The entire RT-qPCR workflow was designed to fit to MIQE guidelines (minimum information for publication of quantitative real-time PCR experiments; Bustin et al. 2009; Derveaux et al. 2010). RNAs from MEFs and 1C11 cells were purified with Trizol Reagent (Ambion 15596–018) according to manufacturer’s instructions and treated one or two times with Turbo DNase for 45 min at 37 °C (Turbo DNA-free kit, Applied Biosystem AM1907). RNA concentrations were quantified using the NanoDrop 1000 Overview (Thermo Scientific), and reverse transcription was performed from 500 to 1000 ng RNA using the Maxima Reverse Transcriptase (Thermo Scientific #EP0741) and oligodT primers, according to manufacturer’s instructions. RNA integrity was assessed by qPCR-based 3′:5′gene assay on cDNA (Nolan et al. 2006), and samples showing a 3′:5′ ratio higher than 7 were discarded.
Primers were designed with Primer3 software (http://bioinfo.ut.ee/primer3/) and in silico validated for specificity by electronic PCR (e-PCR; http://www.ncbi.nlm.nih.gov/tools/epcr/) and by performing qPCR and inspecting the amplicon length via gel electrophoresis. Primers efficiency was calculated with LinRegPCR software (http://www.hartfaalcentrum.nl/index.php?main=files&sub=LinRegPCR). Primers with efficiency lower to 1.8 were discarded and redesigned. If not differently specified, primers were designed in regions common to all the splicing variants described for that gene. For primers sequences, see Online Resource 2, Suppl. Table S1.
Quantitative PCR (qPCR) was performed on the Light Cycler LC480 (Roche) using the ABsolute Blue qPCR SYBR Green Mixes (Thermo Scientific AB-4167/B). Each reaction was performed in triplicate, in a total reaction volume of 10 μl (primers 200 μM). The cycling program (45 cycles) consisted in 10 s at 95 °C (denaturation), 20 s at 64 °C (annealing), and 20 s at 72 °C (elongation).
Relative quantification and normalization were performed with efficiency-corrected multiple reference genes model enabled by qBasePLUS software (http://www.qbaseplus.com). For each experience, the optimal number and choice of reference genes was determined with qBasePLUS within a panel of 7–9 putative reference genes. The selected reference genes were hprt1 and rpl13a for 6 h, 50–300 mM EtOH treatments on 1C11 cells, hprt1 and pgk1 for 0 h–24 h EtOH treatment on 1C11 cells, hmbs and pgk1 for H2O2 treatment on 1C11 cells, and B2m and cyclophilin for EtOH treatments on MEFs.
Western blot analysis
Nuclear and cytosolic extracts from 1C11 cells were generated as previously described (Méndez and Stillman 2000). Briefly, cell pellets were resuspended in buffer A (10 mM Hepes pH 8, 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 0.1% triton) supplemented with protease inhibitors cocktail (Roche), 1 mM DTT (Sigma), 1 mM PMSF (Sigma), 10 mM NaF (Sigma), and 1 mM Na3VO4 (NEB). Nuclei were pelleted by centrifugation at 1300 g for 5 min at 4 °C. Supernatants corresponding to cytosolic fractions were transferred into fresh tubes and boiled in Laemmli buffer 2× (100 mM Tris pH 7, 20% glycerol, 4% SDS, 0.004% bromophenol blue, 10% 2-mercaptoethanol) for 5 min. Nuclei were resuspended in buffer A complemented with 1 mM CaCl2, treated with 1 U of micrococcal nuclease (Sigma M0247S) for 10 min at 37 °C, and then pelleted by centrifugation at 1300 g for 5 min at 4 °C. Nuclear and cytosolic proteins were boiled in Laemmli buffer 2× (100 mM Tris pH 7, 20% glycerol, 4% SDS, 0.004% bromophenol blue, 10% 2-mercaptoethanol) for 5 min. Nuclear extracts were sonicated (Bioruptor, Diagenode, 5× 30 s ON/30 s OFF, High). Thirty to forty micrograms of proteins per sample was loaded in 7 or 8% Tris-tricine SDS/PAGE gels. Western blotting was performed with pan-anti-AKT (Cell Signaling Technology # 4691; 1:1000) and phosphoSerine473Akt (Cell Signaling Technology #4060; 1:1000), anti-β-Actin (Abcam ab20272; 1:1000), anti-DNMT1 (Imgenex IMG-261A; 1:1000), anti-DNMT3A (Imgenex IMG-268A; 1:250), anti-DNMT3B (Abcam ab13604; 1:250), anti-DNMT3L (non-commercial rabbit antibody, kind gift from Prof. S. Tajima), anti-HSC70 (Enzo ADI-SPA-815; 1:1000), anti-HSF1 (Cell Signaling 4356; 1:1000), anti-HSF2 (non-commercial rabbit antibody #57, kind gift from L. Sistonen), anti-HSP70 (Enzo ADI-SPA-810; 1:1000), anti-TET1 (Millipore 09-872, 1:1000), anti-TET2 (Abcam ab12497, 1:1000), and anti-TET3 (Abcam 135033, 1:500). The intensity of the signals was quantified with ImageJ software (http://imagej.nih.gov/ij/index.html).
Immunofluorescence
1C11 cultured on coverslips were fixed with 3% paraformaldehyde in phosphate-buffered saline (PBS) for 15 min at room temperature. After two washes with cold PBS, cells were incubated in blocking solution (5% bovine serum albumin in PBS-0.2% Triton) for 1 h at room temperature. Antibodies were diluted in blocking solution and added overnight (monoclonal anti-HSF1 (NeoMarkers ab4; 1:200) and rabbit anti-HSF2 (non-commercial polyclonal antibody #57, kind gift from L. Sistonen)). After three washes with PBS-0.05% Tween20, cover slips were incubated with secondary antibodies anti-rabbit Alexa 488 and anti-rat Alexa 568 (Invitrogen) for 1 h. The cover slips were mounted and DNA was visualized using Vectashield mounting medium with 4,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories, Burlingame, CA). Relative fluorescence intensity was quantified in nuclei using ImageJ (NIH), after defining regions of interest with DAPI staining. Pictures were taken with a Leica SP5 confocal microscope (magnification ×40) equipped with Metamorph.
Measure of ROS levels
ROS levels in 1C11 cells were estimated using the cell-permeant 2′,7′-dichlorodihydrofluorescein diacetate (H2DCFDA) molecular probe according to manufacturer’s instructions (Life technologies, D-399). Upon cleavage of the acetate groups by intracellular oxidation, the non-fluorescent H2DCFDA is converted to the highly fluorescent 2′,7′-dichlorofluorescein (DCF). Fluorescence was monitored using a microplate reader (Glomax, Promega).
Statistical analysis
For each experiment, the number of independent experiments and the statistical methods are described in the corresponding legends of figures.
Results
Alcohol exposure increases Dnmt3a at transcript and protein levels in neural precursor cells and MEFs
First, to choose doses of EtOH that will be used along this study, we conducted pilot experiments with the 1C11 cells, a murine model of committed neuronal progenitors (Buc-Caron et al. 1990; Mouillet-Richard et al. 2000). Cells were exposed to a range of EtOH concentrations to determine the EtOH doses effective in triggering the activation of the HSF pathway, which has been observed in diverse neural PAE paradigms, in or ex vivo (Pignataro et al. 2007; El Fatimy et al. 2014; Hashimoto-Torii et al. 2011, 2014). The doses of 150 mM (0.88% v/v) and 300 mM EtOH were chosen for our following investigations, as exposure of 1C11 cells to both 150 or 300 mM EtOH led to increased HSF1 and HSF2 DNA-binding activity (HSR, Online Resource 1, Suppl. Fig. S2a) and Hsp70 mRNA and Hsp70 protein induction (Online Resource 1, Suppl. Fig. S2b, c). Note that blood alcohol concentrations (BAC) produced by in vivo EtOH treatments in pregnant mice are generally lower, comprised in a range between 60 and 100 mM (Green et al. 2007; El Fatimy et al. 2014; Hashimoto-Torii et al. 2011; Boehm et al. 1997). However, 150–300 mM EtOH concentrations are within the range attained by human alcoholics (Lindblad and Olsson 1976; Hofmann and Hofmann 1975). In addition, ex vivo cell systems often require higher doses of EtOH to mimic EtOH effects that are observed in vivo at lower concentrations (Cheema et al. 2000; Hicks et al. 2010). Of note, 1C11 cell treatment with 150 to 300 mM elicited an approximately twofold induction in hsp70 transcripts, which was comparable to observations in the fetal brain in vivo upon chronic alcohol intoxication (El Fatimy et al. 2014).
Analogously, EtOH doses for experiments on primary MEFs were chosen for their capacity to trigger a HSR, as they do in immortalized iMEFs (El Fatimy et al. 2014, see below).
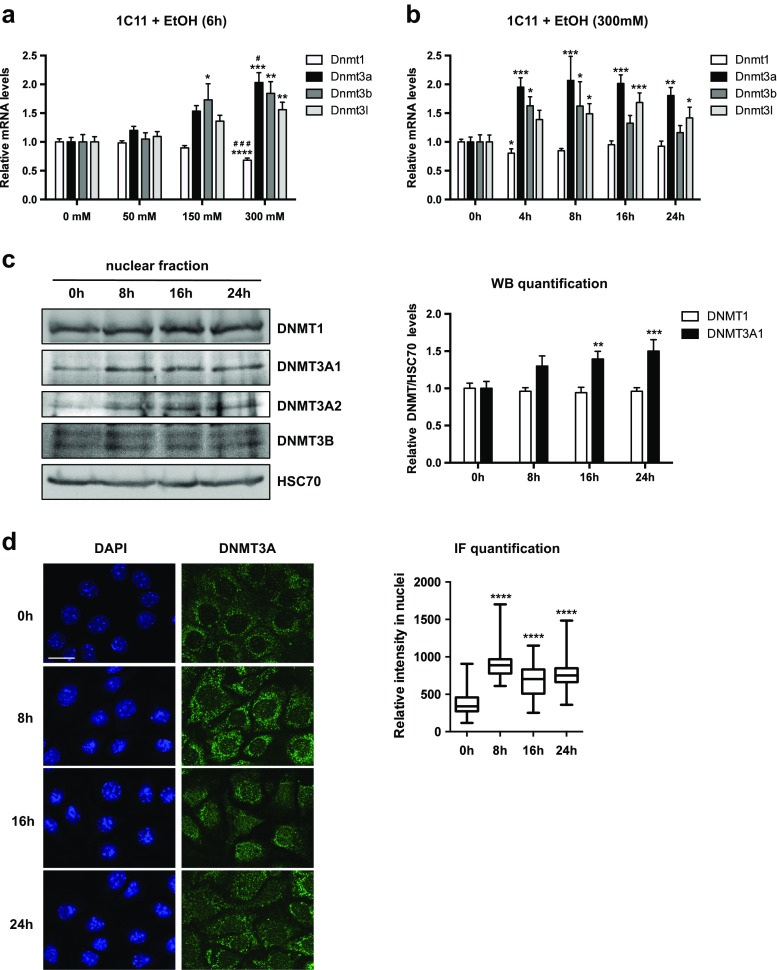
We next probed the impact of EtOH on DNMTs expression. Exposure of 1C11 progenitors for 6 h to 50, 150, and 300 mM of EtOH led to a significant dose-dependent increase, up to twofold, in the transcript levels of the DNA methyltransferases Dnmt3a, Dnmt3b, and Dnmt3l (Fig. 1a). In contrast, Dnmt1 mRNA levels were not significantly affected, except for a transient moderate reduction at 300 mM (Fig. 1a, b at 4 h).
Fig. 1.
Alcohol exposure increases Dnmt3a at transcript and protein levels in neural precursors. a Dnmt mRNA levels in 1C11 cells exposed from 50 to 300 mM EtOH for 6 hours (6 h). RNA preparations were assayed by RT-qPCR for Dnmt1, Dnmt3a, Dnmt3b, and Dnmt3l mRNAs. Results are shown as the average Dnmt mRNA levels ± SEM over unstimulated cells in three independent experiments, normalized to hprt1 and rpl13a levels. One-way ANOVA followed by adjustment for multiple comparisons using Dunnett’s method was performed: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 vs. 0 mM. #p < 0.05, ###p < 0.001 vs. 150 mM. b Dnmt mRNA levels in 1C11 cells exposed to 300 mM EtOH for 4, 8, 16, or 24 h. RNA preparations were assayed by RT-qPCR for Dnmt1, Dnmt3a, Dnmt3b, and Dnmt3l mRNA. Results are shown as the average Dnmt mRNA levels ± SEM over unstimulated cells in five independent experiments, normalized to hprt1 and pgk1 levels. One-way ANOVA followed by adjustment for multiple comparisons using Dunnett’s method was performed: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 vs. 0 h. c Immunoblot analysis of DNMT protein levels in nuclear extracts of 1C11 cells treated with EtOH (300 mM) for 8, 16, or 24 h. A representative immunoblot of four independent experiments (left panel) and quantification of the average DNMT/HSC70 ratios ± SEM over unstimulated cells (right panel) are shown. DNMT3L was not detected. The low signal/background ratio for DNMT3A2 and DNMT3B proteins prevented reliable quantification. HSC70 is a cognate heat shock protein that partially localizes in the nucleus; reviewed in Liu et al. 2012). As alcohol exposure does not affect its nucleocytoplasmic localization, nuclear HSC70 was therefore used as an internal loading control. One-way ANOVA followed by adjustment for multiple comparisons using Dunnett’s method was performed: **p < 0.01, ***p < 0.001 vs. 0 h. d DNMT3A was detected by immunofluorescence in 1C11 cells treated with 300 mM EtOH for 8, 16, or 24 h. Representative confocal microscopy images (left; scale bar 20 μm) and quantification of the average DNMT3A nuclear signal (right) are shown. Number of cells counted in each conditions: 320 in control conditions, 162 at 8 h, 142 at 16 h, and 176 at 24 h. One-way ANOVA followed by adjustment for multiple comparisons using Tukey’s method was performed: ****p < 0.0001 vs. 0 h
In time course experiments at 300 mM EtOH, Dnmt3a, Dnmt3b, and Dnmt3l mRNA levels were increased from 4 h of exposure (Fig. 1b). Analogous but less pronounced variations were also observed upon 150 mM EtOH time course (Online Resource 1, Suppl. Fig. S3a; see also Suppl. Fig. S3b for the evaluation of the respective abundance of each transcript population). Remarkably, Dnmt3a and Dnmt3l transcript levels were still elevated upon 16 and 24 h of EtOH exposure, in contrast to Dnmt3b transcripts, whose levels returned to basal values before 16 h, after a transient augmentation (Fig. 1b).
The prolonged increase in Dnmt3a transcripts induced by EtOH exposure was paralleled by an augmentation of protein levels in the nuclear compartment, for both DNMT3A1 and DNMT3A2 (Fig. 1c), two isoforms generated from alternative transcription start sites in the Dnmt3a gene (Chen et al. 2002). On the contrary, DNMT1 and DNMT3B signals were not affected by EtOH treatment in 1C11 cells (Fig. 1c). A global increase in DNMT3A protein levels was also observed in immunofluorescence experiments, in the nucleus of 1C11 cells upon EtOH exposure (Fig. 1d; note that a DNMT3A signal was also observed in the cytoplasm of 1C11 cells, a localization previously reported in the literature, (Wong et al. 2013; Jia et al. 2016).
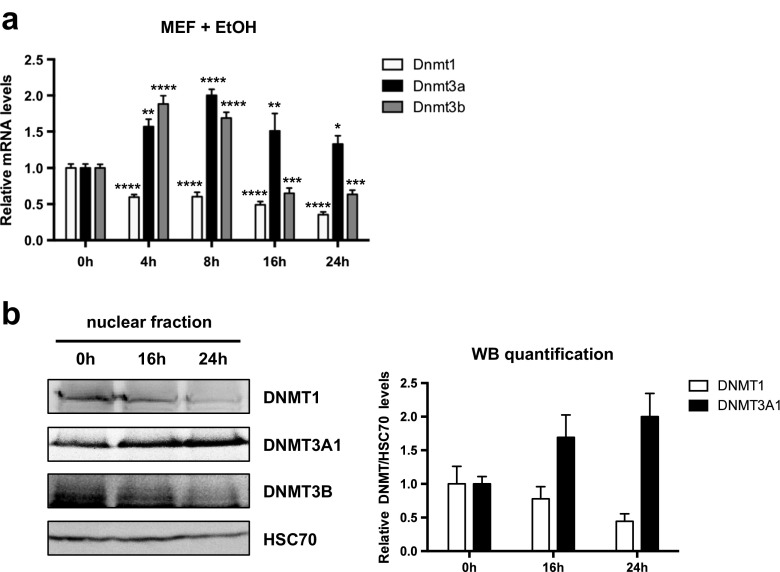
To enlarge our observations in the 1C11 cell line, we treated wild-type (WT) primary MEFs with EtOH and analyzed Dnmt levels. MEFs exposed to EtOH displayed statistically significant, prolonged increase in Dnmt3a transcript levels, similarly to 1C11 cells. In addition, MEFs displayed a transient increase in Dnmt3b mRNA, followed by marked downregulation, as well as a reduction in Dnmt1 mRNA levels (Fig. 2a). As expected, Dnmt3l mRNA was not detected in these cells, since it is quickly downregulated during the differentiation of somatic cells (Aapola et al. 2000). Importantly, the increase in DNMT3A1 protein levels was reproducibly observed in WT MEFs exposed to EtOH for 16 and 24 h (Fig. 2b), as they were in 1C11 cells (Fig. 1c). In WT MEFs, and in contrast to 1C11 cells, DNMT1 and DNMT3B levels decreased (Fig. 2b), underlying some cell-type specificity in DNMT induction profiles, at EtOH concentrations that trigger DNMT3A upregulation.
Fig. 2.
EtOH exposure increases Dnmt3a at transcript and protein levels in primary MEFs. a Dnmt mRNA levels in primary MEFs exposed to 430 mM (2.5%) EtOH for 4, 8, 16, or 24 h. RNA preparations were assayed by RT-qPCR for Dnmt1, Dnmt3a, Dnmt3b, and Dnmt3l mRNA. Dnmt3l transcripts could not be detected in MEFs (Aapola et al. 2000). Results are shown as the average Dnmt mRNA levels ± SEM over unstimulated cells in four independent experiments, normalized to B2m and cyclophilin levels. One-way ANOVA followed by adjustment for multiple comparisons using Dunnett’s method was performed: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 vs. 0 h. b. Immunoblot analysis of DNMT protein levels in nuclear extracts of primary MEFs treated with EtOH (430 mM) for 16 or 24 h (right). DNMT3L and DNMT3A2 were not detected. The low DNMT3B signal/background ratio prevented reliable quantification. HSC70 was used as an internal loading control. A representative immunoblot of three independent experiments (left panel) and quantification of the average DNMT/HSC70 ratios ± SEM over unstimulated cells (right panel) are shown
Steady-state methylation of DNA reflects a balance between methylation and demethylation activities. Cytosine hydroxymethylation (5hmC) can serve as an intermediate in DNA demethylation process (Guibert and Weber 2013; Lister et al. 2013) and is catalyzed by TET enzymes (ten–eleven translocation TET1, TET2, and TET3), which convert 5mC into 5hmC. 5hmC is a very abundant epigenetic mark in the brain and hydroxymethylation is potentially disturbed by alcohol, at least in adult tissues, or by other prenatal stress (Tammen et al. 2014; Massart et al. 2014). For these reasons, we investigated the dynamics of TET levels in response to EtOH exposure. Although we detected a tendency to increased levels of TET transcripts in response to exposure to 150 or 300 mM EtOH in 1C11 cells (Online Resource 1, Suppl. Fig. S4a), we could not detect any alteration in TET enzyme levels (Online Resource 1, Suppl. Fig. S4b). This suggests that it is unlikely that EtOH would disturb hydroxymethylation through changes in TET levels, in 1C11 cells.
Altogether, these results indicate that EtOH exposure upregulates both DNMT3A transcript and protein levels in 1C11 neural precursor cells and MEFs. Remarkably, despite cell-type differences in the EtOH-induced modulation of Dnmt transcript and protein levels, the increase in DNMT3A emerged as a shared robust feature triggered by EtOH.
The elevation of Dnmt transcript levels in response to EtOH is HSF-independent
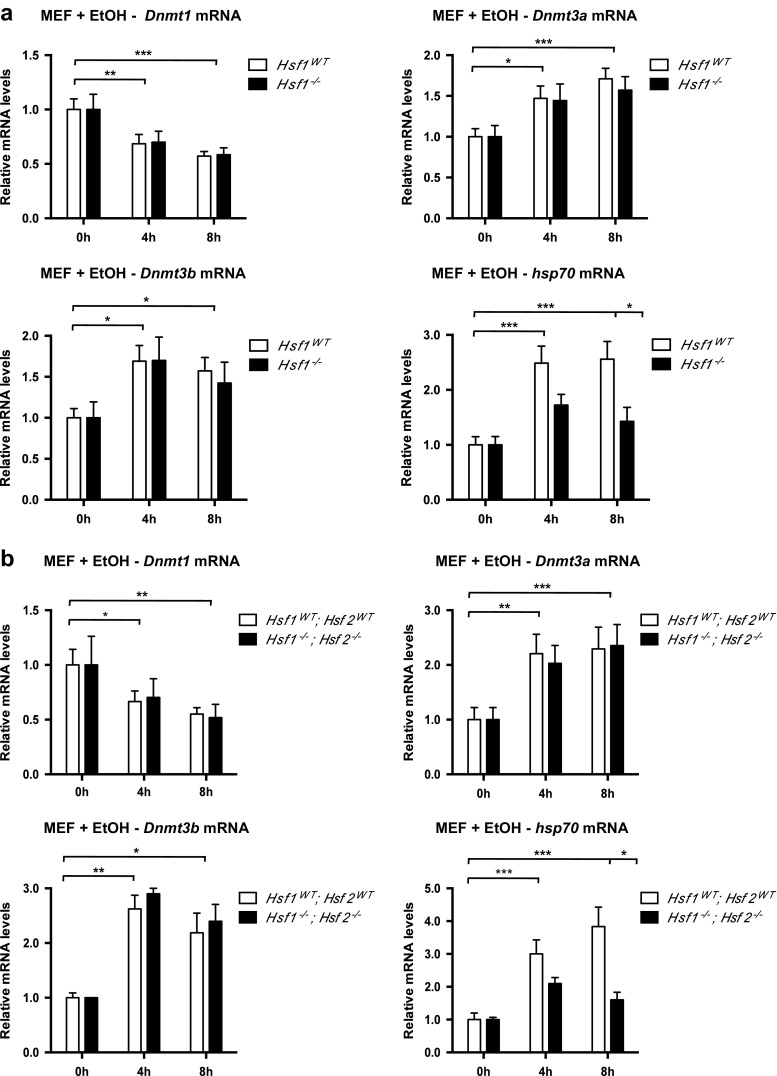
We and others previously showed that HSFs are activated by alcohol (El Fatimy et al. 2014; Pignataro et al. 2007; Hashimoto-Torii et al. 2014). Because Dnmt genes have been identified as potential HSF targets in various HSF1 or HSF2 ChIP-seq analyses (Online Resource 2, Suppl. Table S2), we hypothesized that the modulation of Dnmt mRNA levels could be HSF-dependent. Taking advantage of the mouse Hsf1 and Hsf2 knockout models (McMillan et al. 1998; Kallio et al. 2002), we generated and analyzed early passage Hsf1 −/− and Hsf1 −/− Hsf2 −/− (double knockout) MEFs. Early passages were chosen, since the epigenome is quickly remodeled in culture, with alteration in Dnmt gene expression (Nestor et al. 2015), which could be a bias in our studies. Although a clear HSF1-dependent induction of Hsp70 mRNA levels was observed upon EtOH exposure (Fig. 3a, b), showing the correct activation of the HSF pathway in WT MEFs, the changes in Dnmt1, Dnmt3a, or Dnmt3b levels displayed no HSF dependency (Fig. 3a, b).
Fig. 3.
HSF1 and HSF2 do not regulate Dnmt transcription in response to EtOH in primary MEFs. Dnmt mRNA levels in primary Hsf1 WT and Hsf1 −/− MEFs (a) or Hsf1 WT Hsf2 WT and Hsf1 −/− Hsf2 −/− MEFs (b), exposed to 430 mM EtOH for 4 or 8 h. Knocked-out cells were compared with WT MEFs arisen from the corresponding genetic background (see the “Methods” section). RNA preparations were assayed by RT-qPCR for Dnmt and hsp70 mRNA. Results are shown as the average mRNA level ± SEM over unstimulated cells in three independent experiments, normalized to B2m and cyclophilin levels. One-way ANOVA followed by adjustment for multiple comparisons using Sidak’s method was performed: *p < 0.05, **p < 0.01, ***p < 0.001
We also explored the EtOH-induced changes in the transcript levels of other epigenetic actors, which have been identified as potential HSF target genes in the above-cited ChIP-seq analyses: enzymes involved in histone acetylation, deacetylation, methylation, or demethylation (Online Resource 2, Suppl. Table S2). Although many of them exhibited increased mRNA levels in response to EtOH, this elevation occurred in an HSF-independent manner (Online Resource 1, Suppl. Fig. S5). The sole exception was the lysine demethylase Kdm6a gene (UTX; reviewed in Black et al. 2012), whose increase in mRNA levels was dependent on HSF1 (Online Resource 1, Suppl. Fig. S5).
Altogether, these data indicate that the upregulation of Dnmt mRNA levels, as well as most of other epigenetic actors upon EtOH treatment, is not connected to the HSF pathway.
The increase in DNMT3A levels is dependent on EtOH-induced ROS production in 1C11 neural precursor cells
EtOH metabolism is known to generate ROS in the liver and brain in adult mice, and cytochrome P4502E1 (CYP2E1) has been largely suspected to entail ROS production, along with alcohol dehydrogenase (ADH), mitochondrial aldehyde-dehydrogenase (ALDH), and NADPH oxidase (for review see Caro and Cederbaum (2004); Goodlett et al. 2005; Das and Vasudevan 2007). This prompted us to investigate whether the robust and specific induction of DNMT3A1 at the protein level observed upon EtOH exposure in 1C11 neural progenitors and WT MEFs (Figs. 1 and 2) would be mediated by ROS.
We verified that EtOH exposure, at the concentrations used, was able to induce ROS accumulation in 1C11 cells (300 mM, Fig. 4a or 150 mM, Fig. 4b). Specific inhibition of ALDH by CYA (Dawson 1983), of CYP2E1 by DAS, or combined inhibition of ADH and CYP2E1 by 4-MP (Swaminathan et al. 2013), did not prevent ROS production in 1C11 cells exposed to 150 mM EtOH (Fig. 4b). In contrast, two inhibitors of NADPH oxidase (Ragan 1980), DPI (Fig. 4a) or APO (Fig. 4b), abolished the EtOH-induced production of ROS, suggesting that NADPH oxidase is one major enzyme driving the production of ROS in 1C11 neural progenitors upon EtOH exposure.
Fig. 4.
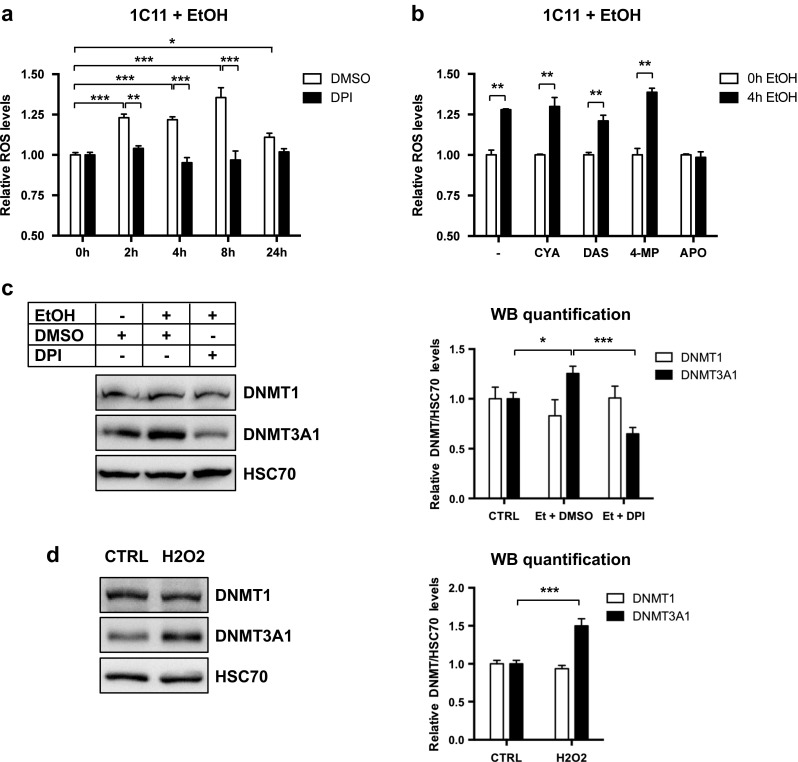
DNMT3A1 protein levels are controlled by NADPH oxidase-mediated ROS production. a ROS levels in 1C11 cells treated with EtOH (300 mM) for 2, 4, 8, or 24 h, in the presence of 4 μM DPI or the same volume of DMSO. ROS levels were measured using the CM-H2DCFDA fluorogenic probe. Results are shown as the average ROS levels ± SEM over unstimulated cells treated with DMSO of three independent experiments. One-way ANOVA followed by adjustment for multiple comparisons using Sidak’s method was performed: *p < 0.05, **p < 0.01, ***p < 0.001. b ROS levels in 1C11 cells treated with EtOH (150 mM) for 4 h, in presence of 250 μM cyanimide (CYA, 5 mM DAS, 5 mM 4-MP, or 500 μM APO. ROS levels were measured using the CM-H2DCFDA fluorogenic probe. Results are shown as the average ROS levels ± SEM over unstimulated cells treated with DMSO of three to five independent experiments. Two-way ANOVA followed by Bonferroni post hoc test was performed: **p < 0.01. c Immunoblot analysis of DNMT1 and DNMT3A protein levels in nuclear extracts of 1C11 cells treated with EtOH (300 mM) for 16 h, in presence of 4 μM DPI or the same volume of DMSO. HSC70 was used as an internal loading control. A representative immunoblot of four independent experiments (left) and quantification of the average DNMT/HSC70 ratios ± SEM over unstimulated cells (right) are shown. One-way ANOVA followed by adjustment for multiple comparisons using Sidak’s method was performed: *p < 0.05, ***p < 0.001. d Immunoblot analysis of DNMT1 and DNMT3A protein levels in nuclear extracts of 1C11 cells treated with H2O2 (100 μM) for 16 h. HSC70 was used as an internal loading control. A representative immunoblot of four independent experiments (left) and quantification of the average DNMT/HSC70 ratios ± SEM over unstimulated cells (right) are shown. Unpaired t test was performed: ***p < 0.001
We next asked whether EtOH-induced ROS production could mediate the increase in DNMT3A protein. Remarkably, DPI counteracted EtOH-induced DNMT3A1 protein upregulation in 1C11 progenitors (Fig. 4c). The protein level of DNMT1 remained unchanged whatever the treatment. To confirm that ROS were involved in the accumulation of DNMT3A protein, we exposed 1C11 cells to H2O2 (100 μM, 16 h). We showed that H2O2 treatment had no impact on DNMT1 expression but specifically increased DNMT3A protein level, as did EtOH (Fig. 4d).
Interestingly, NADPH oxidase inhibition by DPI did not prevent the increase in Dnmt3a transcript levels upon EtOH exposure in 1C11 cells (Online Resource 1, Suppl. Fig. 6). This suggests that the reduction of DNMT3A protein level observed after NADPH oxidase inhibition in EtOH exposed cells is not associated with decrease of Dnmt3a mRNA. EtOH-induced accumulation of DNMT3A1 protein rather reflects ROS-dependent action at the level of Dnmt3a mRNA translation or DNMT3A protein stability.
These data thus suggest that EtOH exposure, beyond modulating Dnmt3a transcript levels by yet-to-be identified mechanisms, favors the accumulation of DNMT3A protein in 1C11 cells, by a process that depends on the production of ROS by NADPH oxidase. Moreover, because NADPH oxidase has been shown to modulate Akt activity (Chen et al. 2007), we checked whether exposure of MEFs and 1C11 cells to EtOH, in conditions that lead to elevated DNMT3A levels, provoked Akt activation. Indeed, we observed an increase in Akt phosphorylation on residue Ser473 in both cell systems (Online Resource 1, Suppl. Fig. 6B and C), suggesting that at least part of the effect of NADPH oxidase on DNMT3A levels might be mediated by Akt activation.
Discussion
Dysregulation of DNA methylation in response to alcohol exposure has been documented in the literature, but the impact of EtOH on the expression of all members of the DNMT family has been poorly explored at the mRNA and protein levels.
Because neural development is extremely sensitive to stress, including ethanol (Gräff et al. 2011), here we analyzed the variations in Dnmt transcript and DNMT protein levels, induced by EtOH in the 1C11 cell line, a murine model of committed neuronal progenitors. We also took advantage of primary MEFs derived from Hsf1 and Hsf2 knockout mice, in order to investigate the role of HSFs on DNMT expression.
First, we bring evidence that the different DNMTs are not affected in the same manner upon EtOH exposure. Fundamentally, de novo Dnmt3a, Dnmt3b, and Dnmt3l mRNA levels tend to increase both in 1C11 cells and in primary MEFs, while the amount of Dnmt1 transcripts remains stable or is downregulated. Second, prolonged upregulation of DNMT3A by ethanol is observed in both systems, despite 1C11 cells and MEFs display some cell-type differences for the other DNMTs. The augmentation of DNMT3A in the nucleus could potentially increase DNMT3A availability in the cell for de novo DNA methylation.
In the case of MEFs, similar observations have been reported for the downregulation of DNMT1 levels, while inverse variations concerning DNMT3A and DNMT3B have been shown by Mukhopadhyay et al. (2013). Such discrepancies might root in the different culture conditions that have been applied, that is the use of primary MEFs of early passages (in our case) vs. immortalized MEFs, as well as the different kinetics that have been investigated, that is a series of time points within the first 24 h in study vs. a single time point at 48 h in Mukhopadhyay et al. (2013).
We also show that EtOH exposure disturbs the mRNA levels of many epigenetic actors, involved in (de)methylation and/or acetylation. Interestingly, most of these alterations result in the upregulation of transcript levels (except for Kdm8; Online Resource 1, Suppl. Fig. 5). One can thus speculate that these transcription events might be under the control of specific transcription factors or posttranscriptional events affecting mRNA stability. However, for most of these epigenetic actors, these transcript perturbations occur independently of the HSF pathway, although this pathway is activated upon EtOH exposure in this study, as well as in others (El Fatimy et al. 2014; Pignataro et al. 2007; Hashimoto-Torii et al. 2014). One notable exception is the histone lysine demethylase Kdm6a, whose elevation in mRNA levels in response to alcohol exposure is dependent on HSF1 (Online Resource 1, Suppl. Fig. 5). Of note, mutations in Kdm6a are linked to intellectual disabilities (Miyake et al. 2013). Another explanation to account for the accumulation of mRNAs encoding epigenetic actors might be the alterations in the abundance of microRNAs, induced by alcohol exposure (Sathyan et al. 2007).
Although Dnmt and TET mRNA levels are increased upon EtOH exposure, we find that many of their protein products remain unaffected by alcohol exposure, which suggests that some compensatory mechanisms at the steps of translation or protein stability could prevent the accumulation of epigenetic actors (Gutala et al. 2004; Green et al. 2007). One marked exception concerns DNMT3A, whose levels are robustly upregulated in response to alcohol exposure both in 1C11 neural progenitors and in MEFs.
To gain further insight into DNMT3A regulation, we investigated whether the EtOH-induced ROS levels might take part in this process. In agreement with the well-described capacity of ethanol to promote the synthesis of ROS at the root of some deficits observed in FASD (Brocardo et al. 2011), we show that alcohol stimulates the production of ROS by NADPH oxidase, which in turn contributes to the increase in DNMT3A protein expression in 1C11 cells. Of note, because inhibition of NADPH oxidase does not prevent the EtOH-induced accumulation of Dnmt3a transcripts, it appears that the rise of Dnmt3a transcript depends on yet-to-be identified redox-insensitive mechanisms. In contrast, the EtOH-induced increase in DNMT3A protein levels mobilizes redox-sensitive posttranscriptional mechanisms. Because NADPH oxidase has been shown to modulate Akt activity (Chen et al. 2007), one attractive possibility is that NADPH oxidase impact on DNMT3A levels would be mediated by Akt. We indeed observed Akt activation by phosphorylation on Ser473 in MEFs and 1C11 cells exposed to EtOH, which is in line with previous reports in other cell systems (Ron and Messing 2013).
Overall, we provide a thorough picture of the impact of ethanol on the expression levels of Dnmts, in cell systems. Our finding that DNMT3A accumulates upon alcohol exposure paves the way towards a mechanism that could mediate either protective or detrimental effects of ethanol, through modifications of the epigenome. Indeed, on the one hand, it remains to be understood whether DNMT3A accumulation represents a global mechanism that aims at minimizing the perturbations induced by alcohol on DNA methylation, since alcohol exposure results in reduced availability in S-adenosyl methionine (SAM), the principal methyl-donor, for DNA or histone methylation. On the other hand, alcohol exposure results in hypomethylation or hypermethylation of DNA, depending on the genomic regions considered, with potential impact on gene expression. This suggests that DNMT3A accumulation could be hijacked for DNMT3A redistribution and leads to the deposition of DNA methylation “scars” in a genome-wide manner, with potential deleterious effects in gene expression, as suggested by the persistence of altered DNA methylation regions at temporal distance from alcohol exposure (Kleiber et al. 2014).
Electronic supplementary material
(PDF 4325 kb)
(PDF 267 kb)
(PDF 105 kb)
(PDF 200 kb)
(PDF 93 kb)
(PDF 340 kb)
(PDF 148 kb)
Acknowledgements
We thank Dr. Slimane Ait-Si-Ali and Dr. Véronique Dubreuil for helpful comments and discussion. We are grateful to Professor Lea Sistonen (Turku University, Finland) for kindly providing us with rabbit anti-HSF2 antibody #57, to Professor Elisabeth Christians (UPMC Univ Paris 06, Villefranche-sur-mer, France) for the Hsf1 tm1Ijb mouse strain, and to Professor Shoji Tajima (Osaka University, Japan) for anti-DNMT3L antibody. We thank Anne Le Mouël (IR CNRS, UMR7216) for her advices in the RT-PCR experiments.
We acknowledge the ImagoSeine core facility of the Institut Jacques Monod, member of the infrastructure France BioImaging (http://imagoseine.ijm.fr/en/), for the use of the Leica SP5 confocal microscope. We acknowledge the Functional Epigenomics Platform of the UMR7216 Epigenetics and Cell Fate. VM laboratory has been supported by the Agence Nationale pour la Recherche (Program SAMENTA ANR-13-SAMA-0008-01), Fondation Jérôme Lejeune, IREB/FRA (2014/18 and 2012/19). BS laboratory has been supported by the Agence Nationale pour la Recherche (Program SAMENTA ANR-13-SAMA-0001-01). FM has been supported by a PhD Fellowship from the Centre National de Recherche Scientifique (CNRS) and the Fondation pour la Recherche Médicale; FM and ALS, by a postdoctoral fellowship by SAMENTA ANR-13-SAMA-0008-01.
Compliance with ethical standards
Competing interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1007/s12192-017-0829-2) contains supplementary material, which is available to authorized users.
Contributor Information
Benoît Schneider, Phone: 33 (0)1 42 86 22 09, Email: benoit.schneider@parisdescartes.fr.
Valérie Mezger, Phone: 33 (0)1 57 27 89 14, Phone: 33 (0)6 75 77 11 98, Email: valerie.mezger@univ-paris-diderot.fr.
References
- Aapola U, Kawasaki K, Scott HS, et al. Isolation and initial characterization of a novel zinc finger gene, DNMT3L, on 21q22.3, related to the cytosine-5-methyltransferase 3 gene family. Genomics. 2000;65:293–298. doi: 10.1006/geno.2000.6168. [DOI] [PubMed] [Google Scholar]
- Abane R, Mezger V. Roles of heat shock factors in gametogenesis and development. FEBS J. 2010;277:4150–4172. doi: 10.1111/j.1742-4658.2010.07830.x. [DOI] [PubMed] [Google Scholar]
- Barbier E, Johnstone AL, Khomtchouk BB et al (2016) Dependence-induced increase of alcohol self-administration and compulsive drinking mediated by the histone methyltransferase PRDM2. Mol Psychiatry. doi:10.1038/mp.2016.131 [DOI] [PMC free article] [PubMed]
- Bielawski DM, Zaher FM, Svinarich DM, Abel EL. Paternal alcohol exposure affects sperm cytosine methyltransferase messenger RNA levels. Alcohol Clin Exp Res. 2002;26:347–351. doi: 10.1111/j.1530-0277.2002.tb02544.x. [DOI] [PubMed] [Google Scholar]
- Black JC, Van Rechem C, Whetstine JR. Histone lysine methylation dynamics: establishment, regulation, and biological impact. Mol Cell. 2012;48:491–507. doi: 10.1016/j.molcel.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm SL, Lundahl KR, Caldwell J, Gilliam DM. Ethanol teratogenesis in the C57BL/6J, DBA/2J, and A/J inbred mouse strains. Alcohol. 1997;14:389–395. doi: 10.1016/S0741-8329(97)87950-5. [DOI] [PubMed] [Google Scholar]
- Bönsch D, Lenz B, Fiszer R, et al. Lowered DNA methyltransferase (DNMT-3b) mRNA expression is associated with genomic DNA hypermethylation in patients with chronic alcoholism. J Neural Transm Vienna Austria. 2006;113:1299–1304. doi: 10.1007/s00702-005-0413-2. [DOI] [PubMed] [Google Scholar]
- Bourc’his D, Xu GL, Lin CS, et al. Dnmt3L and the establishment of maternal genomic imprints. Science. 2001;294:2536–2539. doi: 10.1126/science.1065848. [DOI] [PubMed] [Google Scholar]
- Brocardo PS, Gil-Mohapel J, Christie BR. The role of oxidative stress in fetal alcohol spectrum disorders. Brain Res Rev. 2011;67:209–225. doi: 10.1016/j.brainresrev.2011.02.001. [DOI] [PubMed] [Google Scholar]
- Buc-Caron MH, Launay JM, Lamblin D, Kellermann O. Serotonin uptake, storage, and synthesis in an immortalized committed cell line derived from mouse teratocarcinoma. Proc Natl Acad Sci U S A. 1990;87:1922–1926. doi: 10.1073/pnas.87.5.1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Caro AA, Cederbaum AI. Oxidative stress, toxicology, and pharmacology of CYP2E1. Annu Rev Pharmacol Toxicol. 2004;44:27–42. doi: 10.1146/annurev.pharmtox.44.101802.121704. [DOI] [PubMed] [Google Scholar]
- Cheema ZF, West JR, Miranda RC. Ethanol induces Fas/Apo [apoptosis]-1 mRNA and cell suicide in the developing cerebral cortex. Alcohol Clin Exp Res. 2000;24:535–543. doi: 10.1111/j.1530-0277.2000.tb02022.x. [DOI] [PubMed] [Google Scholar]
- Chen T, Ueda Y, Xie S, Li E. A novel Dnmt3a isoform produced from an alternative promoter localizes to euchromatin and its expression correlates with active de novo methylation. J Biol Chem. 2002;277:38746–38754. doi: 10.1074/jbc.M205312200. [DOI] [PubMed] [Google Scholar]
- Chen JX, Zeng H, Tuo QH, Yu H, Meyrick B, Aschner JL. NADPH oxidase modulates myocardial Akt, ERK1/2 activation, and angiogenesis after hypoxia-reoxygenation. Am J Physiol Heart Circ Physiol. 2007;292:H1664–H1674. doi: 10.1152/ajpheart.01138.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke ME, Gibbard WB. Overview of fetal alcohol spectrum disorders for mental health professionals. Can Child Adolesc Psychiatry Rev. 2003;12:57–63. [PMC free article] [PubMed] [Google Scholar]
- Das SK, Vasudevan DM. Alcohol-induced oxidative stress. Life Sci. 2007;81:177–187. doi: 10.1016/j.lfs.2007.05.005. [DOI] [PubMed] [Google Scholar]
- Dawson AG. Ethanol oxidation in systems containing soluble and mitochondrial fractions of rat liver. Regulation by acetaldehyde. Biochem Pharmacol. 1983;32:2157–2165. doi: 10.1016/0006-2952(83)90221-6. [DOI] [PubMed] [Google Scholar]
- Derveaux S, Vandesompele J, Hellemans J. How to do successful gene expression analysis using real-time PCR. Methods San Diego Calif. 2010;50:227–230. doi: 10.1016/j.ymeth.2009.11.001. [DOI] [PubMed] [Google Scholar]
- El Fatimy R, Miozzo F, Le Mouël A, et al. Heat shock factor 2 is a stress-responsive mediator of neuronal migration defects in models of fetal alcohol syndrome. EMBO Mol Med. 2014;6:1043–1061. doi: 10.15252/emmm.201303311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan G, Beard C, Chen RZ, et al. DNA hypomethylation perturbs the function and survival of CNS neurons in postnatal animals. J Neurosci. 2001;21:788–797. doi: 10.1523/JNEUROSCI.21-03-00788.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodlett CR, Horn KH, Zhou FC. Alcohol teratogenesis: mechanisms of damage and strategies for intervention. Exp Biol Med Maywood NJ. 2005;230:394–406. doi: 10.1177/15353702-0323006-07. [DOI] [PubMed] [Google Scholar]
- Gräff J, Kim D, Dobbin MM, Tsai L-H. Epigenetic regulation of gene expression in physiological and pathological brain processes. Physiol Rev. 2011;91:603–649. doi: 10.1152/physrev.00012.2010. [DOI] [PubMed] [Google Scholar]
- Green ML, Singh AV, Zhang Y, et al. Reprogramming of genetic networks during initiation of the fetal alcohol syndrome. Dev Dyn. 2007;236:613–631. doi: 10.1002/dvdy.21048. [DOI] [PubMed] [Google Scholar]
- Guerri C, Bazinet A, Riley EP. Foetal alcohol spectrum disorders and alterations in brain and behaviour. Alcohol Alcohol. 2009;44:108–114. doi: 10.1093/alcalc/agn105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guibert S, Weber M. Functions of DNA methylation and hydroxymethylation in mammalian development. Curr Top Dev Biol. 2013;104:47–83. doi: 10.1016/B978-0-12-416027-9.00002-4. [DOI] [PubMed] [Google Scholar]
- Gutala R, Wang J, Kadapakkam S, et al. Microarray analysis of ethanol-treated cortical neurons reveals disruption of genes related to the ubiquitin-proteasome pathway and protein synthesis. Alcohol Clin Exp Res. 2004;28:1779–1788. doi: 10.1097/01.ALC.0000148117.17707.B4. [DOI] [PubMed] [Google Scholar]
- Hashimoto-Torii K, Kawasawa YI, Kuhn A, Rakic P. Combined transcriptome analysis of fetal human and mouse cerebral cortex exposed to alcohol. Proc Natl Acad Sci U S A. 2011;108:4212–4217. doi: 10.1073/pnas.1100903108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto-Torii K, Torii M, Fujimoto M, et al. Roles of heat shock factor 1 in neuronal response to fetal environmental risks and its relevance to brain disorders. Neuron. 2014;82:560–572. doi: 10.1016/j.neuron.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata K, Okano M, Lei H, Li E. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development. 2002;129:1983–1993. doi: 10.1242/dev.129.8.1983. [DOI] [PubMed] [Google Scholar]
- Hicks SD, Middleton FA, Miller MW. Ethanol-induced methylation of cell cycle genes in neural stem cells. J Neurochem. 2010;114:1767–1780. doi: 10.1111/j.1471-4159.2010.06886.x. [DOI] [PubMed] [Google Scholar]
- Hofmann FG, Hofmann AD. A handbook on drug and alcohol abuse: the biomedical aspects. New York: Oxford University Press; 1975. [Google Scholar]
- Jia Y, Li P, Fang L, et al. Negative regulation of DNMT3A de novo DNA methylation by frequently overexpressed UHRF family proteins as a mechanism for widespread DNA hypomethylation in cancer. Cell Discov. 2016;2:16007. doi: 10.1038/celldisc.2016.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- Jones KL, Smith DW. Recognition of the fetal alcohol syndrome in early infancy. Lancet Lond Engl. 1973;302:999–1001. doi: 10.1016/S0140-6736(73)91092-1. [DOI] [PubMed] [Google Scholar]
- Kallio M, Chang Y, Manuel M, et al. Brain abnormalities, defective meiotic chromosome synapsis and female subfertility in HSF2 null mice. EMBO J. 2002;21:2591–2601. doi: 10.1093/emboj/21.11.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminen-Ahola N, Ahola A, Maga M, et al. Maternal ethanol consumption alters the epigenotype and the phenotype of offspring in a mouse model. PLoS Genet. 2010;6:e1000811. doi: 10.1371/journal.pgen.1000811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalid O, Kim JJ, Kim H-S, et al. Gene expression signatures affected by alcohol-induced DNA methylomic deregulation in human embryonic stem cells. Stem Cell Res. 2014;12:791–806. doi: 10.1016/j.scr.2014.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiber ML, Diehl EJ, Laufer BI, et al. Long-term genomic and epigenomic dysregulation as a consequence of prenatal alcohol exposure: a model for fetal alcohol spectrum disorders. Front Genet. 2014;5:161. doi: 10.3389/fgene.2014.00161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufer BI, Kapalanga J, Castellani CA, et al. Associative DNA methylation changes in children with prenatal alcohol exposure. Epigenomics. 2015;7:1259–1274. doi: 10.2217/epi.15.60. [DOI] [PubMed] [Google Scholar]
- Lemoine P, Harousseau H, Borteyru J (1968) Les enfants de parents alcooliques: anomalies observées. 476–482
- Lindblad B, Olsson R. Unusually high levels of blood alcohol? JAMA. 1976;236:1600–1602. doi: 10.1001/jama.1976.03270150034027. [DOI] [PubMed] [Google Scholar]
- Lister R, Mukamel EA, Nery JR, et al. Global epigenomic reconfiguration during mammalian brain development. Science. 2013;341:1237905. doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Daniels CK, Cao S. Comprehensive review on the HSC70 functions, interactions with related molecules and involvement in clinical diseases and therapeutic potential. Pharmacol Ther. 2012;136:354–374. doi: 10.1016/j.pharmthera.2012.08.014. [DOI] [PubMed] [Google Scholar]
- Massart R, Suderman M, Provencal N, et al. Hydroxymethylation and DNA methylation profiles in the prefrontal cortex of the non-human primate rhesus macaque and the impact of maternal deprivation on hydroxymethylation. Neuroscience. 2014;268:139–148. doi: 10.1016/j.neuroscience.2014.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattson SN, Crocker N, Nguyen TT. Fetal alcohol spectrum disorders: neuropsychological and behavioral features. Neuropsychol Rev. 2011;21:81–101. doi: 10.1007/s11065-011-9167-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan DR, Xiao X, Shao L, et al. Targeted disruption of heat shock transcription factor 1 abolishes thermotolerance and protection against heat-inducible apoptosis. J Biol Chem. 1998;273:7523–7528. doi: 10.1074/jbc.273.13.7523. [DOI] [PubMed] [Google Scholar]
- Mead EA, Sarkar DK. Fetal alcohol spectrum disorders and their transmission through genetic and epigenetic mechanisms. Front Genet. 2014;5:154. doi: 10.3389/fgene.2014.00154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méndez J, Stillman B. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol Cell Biol. 2000;20:8602–8612. doi: 10.1128/MCB.20.22.8602-8612.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake N, Koshimizu E, Okamoto N, et al. MLL2 and KDM6A mutations in patients with Kabuki syndrome. Am J Med Genet A. 2013;161A:2234–2243. doi: 10.1002/ajmg.a.36072. [DOI] [PubMed] [Google Scholar]
- Mouillet-Richard S, Mutel V, Loric S, et al. Regulation by neurotransmitter receptors of serotonergic or catecholaminergic neuronal cell differentiation. J Biol Chem. 2000;275:9186–9192. doi: 10.1074/jbc.275.13.9186. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay P, Rezzoug F, Kaikaus J, et al. Alcohol modulates expression of DNA methyltranferases and methyl CpG-/CpG domain-binding proteins in murine embryonic fibroblasts. Reprod Toxicol. 2013;37:40–48. doi: 10.1016/j.reprotox.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestor CE, Ottaviano R, Reinhardt D, et al. Rapid reprogramming of epigenetic and transcriptional profiles in mammalian culture systems. Genome Biol. 2015;16:11. doi: 10.1186/s13059-014-0576-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan T, Hands RE, Bustin SA. Quantification of mRNA using real-time RT-PCR. Nat Protoc. 2006;1:1559–1582. doi: 10.1038/nprot.2006.236. [DOI] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/S0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Peña CJ, Bagot RC, Labonté B, Nestler EJ. Epigenetic signaling in psychiatric disorders. J Mol Biol. 2014;426:3389–3412. doi: 10.1016/j.jmb.2014.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pignataro L, Miller AN, Ma L, et al. Alcohol regulates gene expression in neurons via activation of heat shock factor 1. J Neurosci. 2007;27:12957–12966. doi: 10.1523/JNEUROSCI.4142-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponomarev I, Wang S, Zhang L, et al. Gene coexpression networks in human brain identify epigenetic modifications in alcohol dependence. J Neurosci. 2012;32:1884–1897. doi: 10.1523/JNEUROSCI.3136-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragan CI. The molecular organization of NADH dehydrogenase. Subcell Biochem. 1980;7:267–307. doi: 10.1007/978-1-4615-7948-9_6. [DOI] [PubMed] [Google Scholar]
- Ron D, Messing RO. Signaling pathways mediating alcohol effects. Curr Top Behav Neurosci. 2013;13:87–126. doi: 10.1007/978-3-642-28720-6_161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudenko A, Tsai L-H. Epigenetic regulation in memory and cognitive disorders. Neuroscience. 2014;264:51–63. doi: 10.1016/j.neuroscience.2012.12.034. [DOI] [PubMed] [Google Scholar]
- Sathyan P, Golden HB, Miranda RC. Competing interactions between micro-RNAs determine neural progenitor survival and proliferation after ethanol exposure: evidence from an ex vivo model of the fetal cerebral cortical neuroepithelium. J Neurosci. 2007;27:8546–8557. doi: 10.1523/JNEUROSCI.1269-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streissguth AP, O’Malley K. Neuropsychiatric implications and long-term consequences of fetal alcohol spectrum disorders. Semin Clin Neuropsychiatry. 2000;5:177–190. doi: 10.1053/scnp.2000.6729. [DOI] [PubMed] [Google Scholar]
- Swaminathan K, Kumar SM, Clemens DL, Dey A. Inhibition of CYP2E1 leads to decreased advanced glycated end product formation in high glucose treated ADH and CYP2E1 over-expressing VL-17A cells. Biochim Biophys Acta. 2013;1830:4407–4416. doi: 10.1016/j.bbagen.2013.05.022. [DOI] [PubMed] [Google Scholar]
- Tammen SA, Dolnikowski GG, Ausman LM, et al. Aging and alcohol interact to alter hepatic DNA hydroxymethylation. Alcohol Clin Exp Res. 2014;38:2178–2185. doi: 10.1111/acer.12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnault V, Darcq E, Levine A, et al. Chromatin remodeling—a novel strategy to control excessive alcohol drinking. Transl Psychiatry. 2013;3:e231. doi: 10.1038/tp.2013.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong M, Gertz B, Chestnut BA, Martin LJ. Mitochondrial DNMT3A and DNA methylation in skeletal muscle and CNS of transgenic mouse models of ALS. Front Cell Neurosci. 2013;7:279. doi: 10.3389/fncel.2013.00279. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 4325 kb)
(PDF 267 kb)
(PDF 105 kb)
(PDF 200 kb)
(PDF 93 kb)
(PDF 340 kb)
(PDF 148 kb)